Abstract
Most follicular lymphomas (FLs) are genetically defined by the t(14;18)(q32;q21) translocation that juxtaposes the BCL2 gene to the immunoglobulin heavy chain (IgH) 3' regulatory regions (IgH-3'RRs). Despite this recurrent translocation, FL cases are heterogeneous in terms of intratumoral clonal diversity for acquired mutations and variations in the tumor microenvironment. Here we describe an additional mechanism that contributes to inter- and intratumoral heterogeneity in FLs. By applying a novel single-molecule RNA fluorescence-based in situ hybridization (FISH) technique to detect mRNA molecules of BCL2 and IgH in single cells, we found marked heterogeneity in the number of BCL2 mRNA transcripts within individual lymphoma cells. Moreover, BCL2 mRNA molecules correlated with IgH mRNA molecules in individual cells both in t(14;18) lymphoma cell lines and in patient samples. Consistently, a strong correlation between BCL2 and IgH protein levels was found in a series of 205 primary FL cases by flow cytometry and immunohistochemistry. Inter- and intratumoral heterogeneity of BCL2 expression determined resistance to drugs commonly used in FL treatment and affected overall survival of FL patients. These data demonstrate that BCL2 and IgH expressions are heterogeneous and coregulated in t(14;18)-translocated cells, and determine the response to therapy in FL patients.
Introduction
Follicular lymphoma (FL) shows a remarkable diversity in phenotypic, genetic and microenvironment intratumoral heterogeneity. Phenotypically, it is well known that FL display striking inter- and intratumoral heterogeneity in terms of the expression of several FL markers including immunoglobulin heavy chain (IgH), CD10, CD20 and BCL2 proteins.1, 2, 3, 4 Genetically, copy number variation and exome sequencing studies have shown marked intratumoral clonal diversity within the FL.5,6 Analysis of intratumoral clonal diversity within FL cases has shown that the IgH-BCL2 t(14;18) translocation is a founder event at the top of the hierarchy of FL oncogenic events, whereas other mutations such as those in CREBBP, MLL2 and TNFRS14 genes are acquired only by a fraction of the cells during tumor evolution.6 Similarly, the FL microenvironment is highly heterogeneous being composed of stromal cells, macrophages and T/natural killer cell subsets that surround FL cells, and has roles in FL survival, growth, drug resistance and prognosis.7, 8, 9, 10
The hallmark of almost 90% of FL is the t(14;18)(q32;q21) translocation that juxtaposes the BCL2 gene to the IgH locus.11 Breakpoints at the BCL2 locus cluster in the major breakpoint region and in the minor breakpoint region, both regions being located downstream of the BCL2 gene.12 It is believed that regulatory elements in the IgH locus, such as enhancers in the 3′ regulatory regions (IgH-3'RRs), have a critical role in the deregulated expression of the translocated BCL2 allele.13 Indeed, the IgH-3'RRs increase BCL2 transcription by deregulating promoter usage. In normal cells, BCL2 transcription starts primarily from the P1 promoter, a TATA-less, GC-rich promoter located ~1400 bp upstream of the translational start site. In t(14;18)-translocated cells, BCL2 transcription instead originates primarily from the P2 promoter, a classical TATA plus CAAT box promoter located immediately before the translational start site in exon II.13 When the IgH-3'RRs are integrated in the BCL2 locus in mice, increased levels of BCL2 mRNA and protein are observed, and mice develop FL.14 Overexpression of BCL2 is pivotal for FL and diffuse large B-cell lymphoma (DLBCL) pathogenesis as it promotes survival of lymphoma cells.15,16
BCL2 expression shows significant intertumoral variability among t(14;18)-translocated FLs, ranging from cases with relatively low to very high expression.16 Furthermore, despite each FL case carries a clonal t(14;18) translocation that occurs as an early event in the development of the lymphoma and is constant among different subclonal populations within each FL tumor,6 FL shows a large degree of intratumoral heterogeneity of BCL2 expression, with cells displaying variable amounts of BCL2 within the same tumor.17,18 The molecular basis and the pathologic implications of such heterogeneity are poorly understood.
In this work, we demonstrate that such heterogeneity of BCL2 expression strongly correlates with heterogeneity of IgH expression, likely due to the activity of IgH-3'RRs that can simultaneously control BCL2 and IgH transcription in t(14;18) cells. We validated such correlations by a newly developed single-molecule RNA fluorescence-based in situ hybridization (smFISH) assay in individual lymphoma cells and by protein expression in a large series of FL cases. Importantly, we show that heterogeneity of BCL2 expression has implications in FL response to therapy and overall survival.
Materials and methods
Single-molecule RNA FISH
Human IgM+, t(14;18)-positive (SU-DHL-6, Ly8 and VAL) and -negative (RCK8, MAVER-1, TEKO-1) lymphoma cell lines were cultivated in RPMI-1640 with 10% fetal bovine serum. Cells were fixed in methanol–acetic acid (3:1 (vol/vol)), spotted on microscope slides by cytospin, let to dry and stored at room temperature. Frozen tissue sections (5 μm thick) were mounted onto a poly-L-lysine-coated coverglass, fixed in 4% formaldehyde prepared in 1 × phosphate-buffered saline and stored in 70% ethanol at 4 °C.
We designed probes targeting IGHM and BCL2 mRNAs using a customized algorithm we recently published.19 Probes consisted of the oligonucleotides listed in Supplementary Table 1. We purchased oligonucleotides with a 3′-TEG amino group from Biosearch Technologies (Novato, CA, USA), and coupled them to either Cy5 (GE Healthcare UK Ltd, Little Chalfont, Buckinghamshire, UK; cat. no. Q15108) or Alexa Fluor 594 (Molecular Probes, Eugene, OR, USA; cat. no. A20004). We covered cell spots with 22 × 22 mm2 SecureSeal hybridization chambers (Electron Microscopy Sciences, EMS, Hatfield, PA, USA; cat. no. 70333-10). Before hybridization, we briefly rehydrated cells by filling the chamber two times with 100 μl of 2 × saline-sodium citrate buffer (Ambion, Austin, TX, USA; cat. no. AM9765) supplemented with ribonucleoside–vanadyl complex (RVC) (NEB, New England Biolabs, Ipswich, MA, USA; cat. no. S1402S) diluted 1:20 (vol/vol) (saline-sodium citrate-RVC) at room temperature. Hybridization was performed as we recently described.19 We followed the same protocol for frozen tissue sections.
Microscopy and analysis
All images were acquired at x100 magnification (oil immersion, high numerical aperture objective) on an inverted epifluorescence microscope (Nikon Instruments, Melville, NY, USA) equipped with a high-resolution CCD camera (Pixis; Princeton Instruments, Trenton, NJ, USA), and controlled by MetaMorph software (Universal Imaging Corporation, Downingtown, PA, USA). Per region of interest, we typically acquired an image stack consisting of five image planes spaced 0.4 μm apart.
Image processing was carried out as previously described.20 Nuclei were manually segmented and uniformly dilated by 10 pixels (1.25 μm) to obtain an estimated cell boundary for single-cell mRNA counts assignment. As cells were not imaged throughout their entire thickness in the z direction, we computed mRNA densities by dividing single-cell counts by the volume of the prism with base corresponding to the polygonal segmentation of a given cell's nucleus and height equal to 0.4 μm multiplied by the number of image planes minus one. All data analyses were performed in MATLAB (The Mathworks, Inc., Natick, MA, USA) using custom-made scripts.
Case selection and immunohistochemical analysis
Two hundred and five consecutive cases of FL patients with available flow cytometry data, positivity for the t(14;18) translocation by PCR or FISH and for BCL2 by immunohistochemistry were selected from the database of the Surgical Pathology Unit, San Giovanni Battista Hospital of Turin (Turin, Italy) in the years between 2000 and 2011. The use of samples was approved by the Internal Ethical Committee. Patients were treated all in the same center of the San Giovanni Battista Hospital of Turin by multiple cycles of chemotherapy and rituximab. Cases totally negative for BCL2 by immunohistochemistry were excluded from the series as potential false negatives because of the presence of somatic mutations that impair the recognition of BCL2 by the monoclonal antibody used in the study, as previously described.21 The immunohistochemical panel included CD3 (1:50), CD5 (1:50), CD10 (1:50; all from Novocastra Lab, Newcastle, UK), CD20 (1:200), BCL2 (1:30), CD21 (1:30), BCL6 (1:20) and Ki-67 (1:100; all from Dako, Glostrup, Denmark). Sections of a tonsil with reactive lymphoid hyperplasia were used for positive and negative controls. Expression of the positive reaction was evaluated with a three-tie scoring system. FL cases were considered BCL2high when the intensity of BCL2 staining was higher than surrounding reactive T and B cells in more than 50% of the FL cells. BCL2low FL cases had BCL2 staining intensity equal or lower than surrounding reactive T and B cells in more than 50% of the FL cells. Immunohistochemistry was performed on all 205 cases. The expression of BCL2 before and after relapse was investigated in 33 cases.
Flow cytometry
Flow cytometry analysis was conducted on freshly isolated cells obtained from 156 lymph nodes of FL patients or from reactive lymph nodes as controls. Isolation, staining and analyses of lymphoid cells have been previously described.22 Briefly fresh tissues were allocated on saline solution and mechanically disaggregated with the BD Medimachine System (BD Biosystems, San Jose, CA, USA). Mononuclear cell suspension was obtained by the Ficoll-Hypaque density gradient method and resuspended in RPMI-1640 medium with 10% fetal calf serum. Staining was performed with directly conjugated monoclonal antibodies in 4-color (fluorescein isothiocyanate/phycoerythrin/peridinin chlorophyll protein or peridinin chlorophyll protein-cyanin5.5/allophycocyanin) or 6-color (fluorescein isothiocyanate/phycoerythrin/peridinin chlorophyll protein or peridinin chlorophyll protein–cyanine5.5/phycoerythrin–cyanine7/allophycocyanin/allophycocyanin–H7) combinations. The panels included antibodies for the screening and for the specific study of B-cell lymphoma and are indicated in Supplementary Table 2. Antigen expression was classified as negative, dimly positive, positive and strongly positive using arbitrary relative linear mean fluorescence intensity cutoff values of 1–5, 5–101, 101–102 and >102 (data obtained with the FACSCalibur analogical flow cytometer (BD Biosciences, San Jose, CA, USA): scale range, 100–104) and of 0–102, 102–103, 103–104 and >104 (data obtained with the FACSCantoII digital flow cytometer (BD Biosciences): scale range, 10–264,144), as previously reported.23
For IgH, we considered the expression as low (IgHlow) when it was negative or dimly positive, and high (IgHhigh) when it was positive or strongly positive. BCL2 evaluation was performed using a combined surface and intracellular staining on 156 out the 205 cases of the series studied by immunohistochemistry. After surface staining and washing, cells were resuspended, fixed, permeabilized and stained with BCL2-FITC for 15 min at room temperature. For BCL2 expression, the normal ratio of mean fluorescence intensity of normal B cells versus T cells in reactive lymph nodes was previously determined in the laboratory to be 1.21+0.18; an overexpression was considered when a ratio value greater than or equal to the mean±2 s.d. was detected.24 A minimum of 20 000 events was collected for each sample.
In vitro cell treatments and apoptosis detection
Histone deacetylase inhibitor trichostatin A (TSA) and lipopolysaccharide (LPS) extracted from Escherichia coli O111:B4 (both purchased from Sigma-Aldrich, St Louis, MO, USA) were resuspended in dimethyl sulfoxide or phosphate-buffered saline, respectively. For in vitro treatment with drugs, primary cells freshly obtained from FL patients were cultivated in RPMI-1640 containing 10% heat-inactivated fetal calf serum at 1 × 106 cells per ml concentration for 24 h with 2 μg/ml of doxorubicin or with 2 μg/ml of rituximab and then stained for intracellular BCL2 and cleaved caspase-3 (Asp 175, 1:200; Cell Signaling, Danvers, MA, USA). The staining for cleaved caspase-3 was performed using Cell permeabilization buffer (Caltag Laboratories, Caltag, Burlingame, CA, USA), a secondary biotinylated anti-rabbit antibody (1:400) and a phycoerythrin-conjugated streptavidin (1:400).
Quantitative RT-PCR and western blotting
For quantitative RT-PCR, total RNA was extracted from 12 cases from cells or tissue using Trizol according to the instructions of the manufacturers and 5 μg of RNA were purified with RNeasy Mini Kit (Qiagen, Hilden, Germany). Reverse transcription-polymerase chain reactions (RT-PCRs) were performed (1 μg RNA) according to the manufacturer's protocols (Invitrogen). Real-time RT-PCR was carried out on a Bio-Rad iCycler using IQ Sybr Green Supermix (Bio-Rad, Hercules, CA, USA). Bcl-2 was normalized to RPLP0 housekeeping gene mRNA as described previously. Briefly, to normalized Bcl-2 mRNA amount we used the formula 2−ΔΔCt, where ΔCt=Ct (threshold cycle) gene of interest−Ct internal control.25 Primers for BCL2 were: 5′-GTCATGTGTGTGGAGAGCGT-3′ and 5′-ACAGTTCCACAAAGGCATCC-3′, for RPLP0 were: 5′-GCTTCCTGGAGGGTGTCC-3′ and 5′-GGACTCGTTTGTACCCGTTG-3′. For western blotting, total cellular proteins were extracted from 11 cases and run on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, incubated with the specific antibody (BCL2 1:200 (Dako); α-tubulin 1 (Cell Signaling)) and then detected with peroxidase-conjugated secondary antibodies and chemiluminescent reagent (ECL; Amersham, GE Healthcare UK Ltd).
Statistical analyses
Correlation between BCL2 and IgH mRNA molecules was statistically evaluated by Spearman's rank correlation coefficient. The association between categorical variables, such as patient characteristics, immunoglobulin expression and BCL2 levels, was estimated by Fisher's exact test. Correlations between the mean value of proliferation index (PI), determined by Ki-67, were calculated using Student's t test. P<0.05 was considered statistically significant. The nonparametric Kolmogorov–Smirnov test was adopted to compare shift expression of BCL2 and IgM by flow cytometry. Survival distribution was estimated by the Kaplan–Meier method. For statistical analysis, overall survival was defined as the interval between the diagnosis to death or last follow-up visit.
Results and discussion
Heterogeneity of BCL2 mRNA correlates with IgH mRNA in t(14;18) lymphoma cells
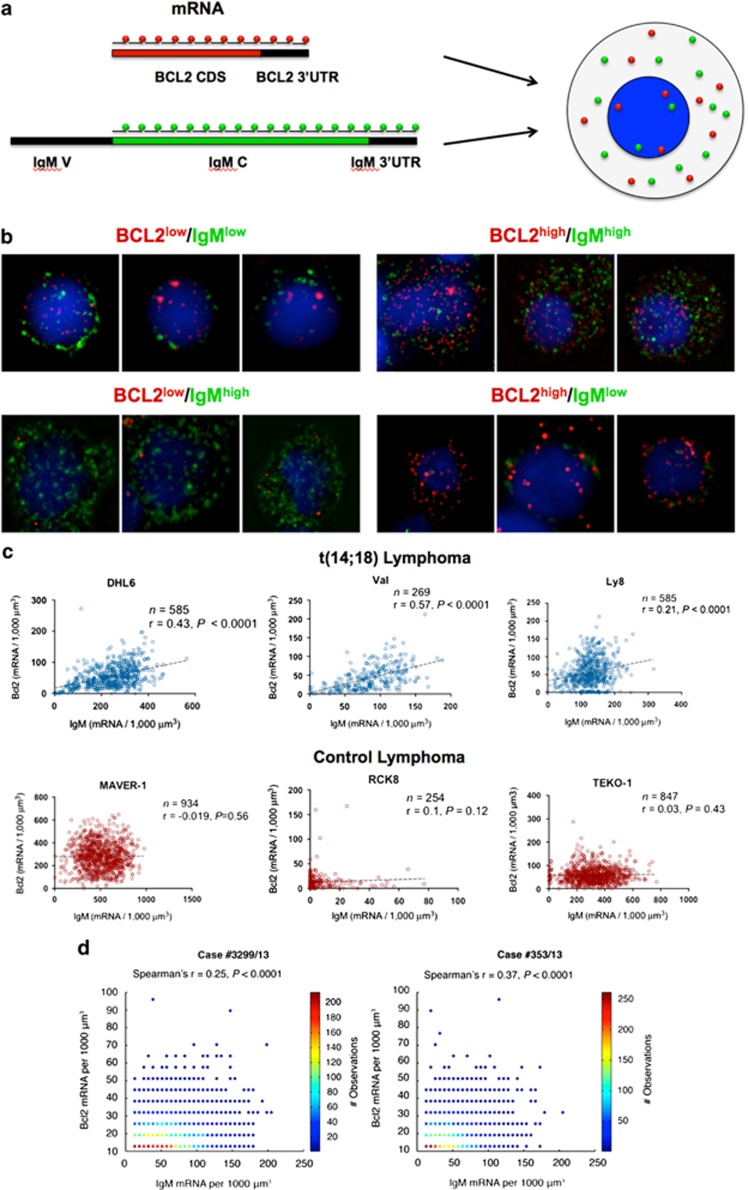
In t(14;18) lymphoma cells, the BCL2 wild-type allele is silent and transcription originates mostly from the BCL2-translocated allele, where the IgH-3'RRs are thought to regulate the expression of the BCL2 gene by providing long-range enhancer activity (>350 kb) and by inducing BCL2 promoter usage shift.13,26 However, IgH-3'RRs also regulates IgH expression, somatic hypermutation and class switch in the wild-type IgH allele.27, 28, 29, 30, 31 Therefore, we reasoned that intratumoral heterogeneity of BCL2 expression in lymphoma cells should at least in part originate from differential mRNA transcription from the t(14;18)-translocated allele, and correlate with IgH since both are controlled by IgH-3'RRs regions. To analyze the BCL2 and IgH mRNA transcripts in single lymphoma cells, we implemented an smFISH technique that allows absolute measurements of the number of mRNA molecules in individual cells.32 For BCL2, we designed arrays of labeled probes targeting the coding sequence and the 3'-untranslated region of BCL2 (Figure 1a). For IgH, we designed probes over the constant region of IgM because it is the most represented IgH type in FL patients and lymphoma cell lines1 (Figure 1a). As no true FL cell lines are currently available, we tested the probes on lymphoma cell lines derived from t(14;18)- translocated IgM+ DLBCL, and we used IgM+ t(14;18)-negative lymphoma cell lines as control. To account for differences in cell size, we computed transcript densities by dividing the number of transcripts per cell by the cell volume. In all cell lines examined, we observed a high degree of intratumoral heterogeneity, with densities ranging from few transcripts (BCL2low or IgMlow) to several hundred transcripts per 1000 μm3 (BCL2high or IgMhigh) (Figure 1b). The MAVER-1 cell line showed the highest expression of both BCL2 and IgM (max density=647 and 976 per 1000 μm3, for BCL2 and IgM, respectively) (Figure 1c). Remarkably, we found a strong correlation between BCL2 and IgM mRNA molecules only in t(14;18)-translocated cells. In t(14;18)-negative cells, we found some level of BCL2 and IgH heterogeneity because of intrinsic gene expression noise and extrinsic factors such as the cell cycle, but, as expected, we did not find no correlation between BCL2 and IgH mRNA transcripts (Figure 1c). These data indicate that translocated BCL2 and IgH might be under the same transcriptional control in standard growing condition. Next, we wanted to test whether BCL2 and IgM mRNA molecules correlated also in primary samples from FL patients by probing frozen sections obtained from freshly isolated FL cases. In all the cases analyzed (n=5), we found a significant correlation between BCL2 and IgH mRNA molecules, thus indicating that BCL2 and IgH are coregulated not only in lymphoma cell lines but also in FL patients (Figure 1d).
Figure 1.
Heterogeneity of BCL2 mRNA expression and correlation with IgM mRNA molecules in cells with t(14:18). (a) Schematic representation of smFISH for BCL2 and IgM. Single-fluorophore- (magenta, BCL2; green, IgM) labeled 20 nt oligonucleotides target the coding (CDS) and 3′-untranslated region (3′-UTR) BCL2 and IgM transcripts. For IgM, the oligonucleotides were designed in the constant region (IgM C); IgM V, IgM variable region; (b) BCL2 transcripts (red spots) and IgM transcripts (green spots) were detected within single cells. Images of single lymphoma cells are representative of different patterns of BCL2 and IgM expression. (c) BCL2 vs IgM expression in single cells of lymphoma cells lines carrying the translocation t(14;18)- (top) compared with t(14;18)-negative lymphoma cells (bottom); n, number of single cells counted; r, Spearman's rank correlation coefficient. (d) BCL2 vs IgM expression in single cells of FL patients carrying the t(14;18) translocation. Frozen sections from IgM+ FL patients were incubated with BCL2 and IgM probes. BCL2 and IgM mRNA molecules were counted as indicated in the Material and methods section and plotted in colorimetric scale.
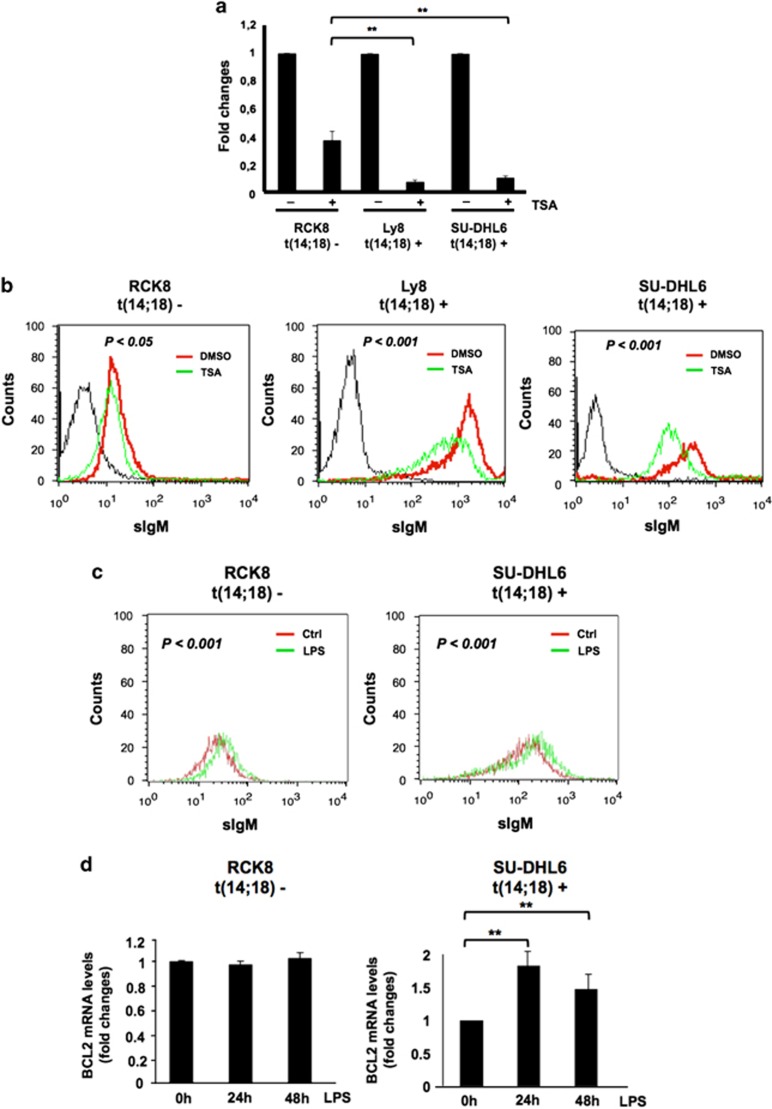
IgH-3'RRs have been shown to physically and functionally interact at long distance with the translocated BCL2 promoter region through transcription factors such as the POU2 family transcription factor Oct-2 and its co-factor Bob-1.33,34 Treatment with the histone deacetylase inhibitor TSA decreases BCL2 transcription from both the P1 and P2 promoters by decreasing Oct-2 protein levels and by reducing Oct-2 binding to BCL2 promoters.34 Also, TSA treatment impairs Oct-2 binding to IgH-3'RRs, thereby further repressing BCL2 transcription by blocking the interaction between BCL2 promoters and IgH-3'RRs.34 In this context, we reasoned that TSA treatment should simultaneously affect BCL2 and IgH mRNA levels with a more profound effect on BCL2 expression in t(14;18)-translocated lymphoma cells than in control cells, because of the disruption of the interaction between the BCL2 promoters and the IgH-3'RRs. Indeed, using quantitative PCR we found that treatment with TSA induced a significantly stronger reduction of BCL2 transcription in t(14;18) translocation positive than in translocation-negative lymphoma cells in which there is no interaction with the IgH-3'RRs, and the BCL2 transcription originates mainly from the P1 promoter (Figure 2a). As expected, the decrease of BCL2 transcription was accompanied by a decrease in IgM expression (Figure 2b). In contrast, when we treated lymphoma cells with LPS to increase IgH transcription by enhancing the effects of the IgH-3'RRs,35 we observed an increase of both IgH and BCL2 expressions in t(14:18)-translocated lymphoma cells, whereas translocation-negative lymphoma cells upregulated IgM levels but showed no changes in BCL2 transcription (Figures 2c and d). In conclusion, all these data indicate that IgH and BCL2 expression are coregulated in t(14;18) lymphoma cell lines likely because they are under the same transcriptional control if the IgH-3'RRs.
Figure 2.
BCL2 and sIgH show coordinated stimulation-induced changes. To modulate the activity of the IgH-3′RRs, t(14;18)-positive lymphoma cells SU-DHL-6 and Ly8 and t(14;18)-negative RCK8 were treated for 1 h with 500 ng/ml. TSA diluted from a stock solution in dimethyl sulfoxide (DMSO). (a) BCL2 mRNA levels were determined by quantitative real-time RT-PCR during TSA treatment. For each condition, BCL2 mRNA levels were normalized to the expression of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) housekeeping gene. Fold changes in BCL2 mRNA expression were calculated by comparing TSA-treated cells to control cells. Standard deviations and statistics were calculated from three independent experiments. **P<0.01. (b) Intensities of surface IgM (sIgM) immunoglobulin expression (sIgM) were measured by flow cytometry (black, unstained control; red, control DMSO-treated cells; green, TSA-treated cells). P-value was calculated using the nonparametric Kolmogorov–Smirnov test to compare shifted IgM expression. (c) t(14;18)-positive SU-DHL-6 and -negative RCK8 lymphoma cells were stimulated for 24 or 48 h with 1 μg/ml LPS and flow cytometry for sIgM was performed as described. P-value was calculated using the nonparametric Kolmogorov–Smirnov test to compare shifted IgM expression. (d) BCL2 mRNA levels were determined by quantitative real-time RT-PCR at the indicated time point of LPS treatment. For each condition, BCL2 mRNA levels were normalized to the expression of the GAPDH housekeeping gene. Fold changes in BCL2 mRNA expression were calculated by comparing LPS-treated to control cells. Standard deviations and statistics were calculated from three independent experiments. **P<0.01.
Heterogeneous BCL2 expression correlates with IgH expression in FL patients
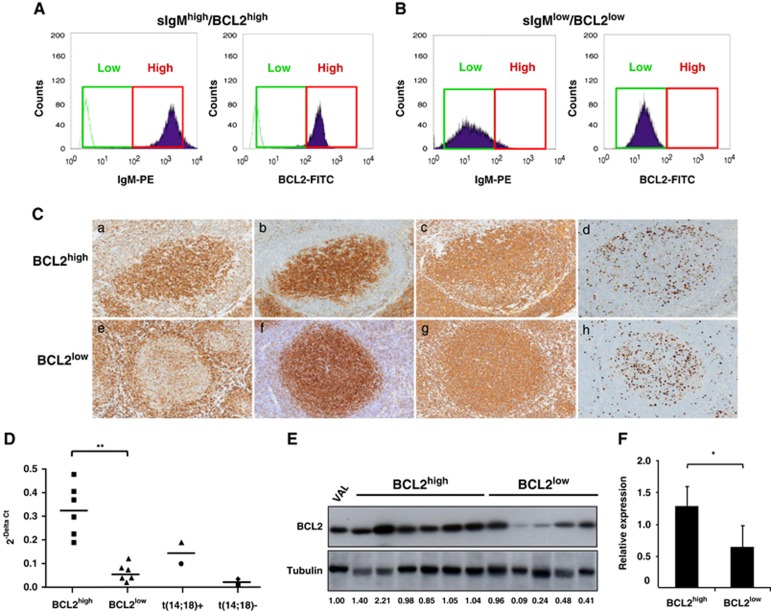
To extend these findings to primary FL patients, we characterized BCL2 and IgH protein expression levels in a series FL cases by flow cytometry and immunohistochemistry. To determine the intensity of BCL2 expression in FL by flow cytometry, we compared control reactive lymph nodes with lymph nodes obtained from FL patients. In reactive lymph nodes, flow cytometry showed that the intracytoplasmic expression of BCL2 was comparable in CD10− B cells and T cells according to the criteria indicated in Materials and methods. As expected, reactive CD10+ B cells from germinal centers showed lower BCL2 expression. In lymph nodes from FL patients, when the expression of BCL2 in the CD10+ B-cell clonal tumoral population was higher than in T cells or CD10− B cells, we considered the case as BCL2high. In contrast, when the tumoral population showed intensity comparable or lower than reactive T cells, we considered the case as BCL2low. We then validated this classification into BCL2high and BCL2low FL cases by immunohistochemistry on all the cases by staining with anti-BCL2 antibody on formalin-fixed, paraffin-embedded sections. We translated the criteria defined by flow cytometry, and considered the FL cases as BCL2high or BCL2low when the intensity of BCL2 expression in the neoplastic follicles was higher or lower/equal to the surrounding reactive T and B cells, respectively (Figures 3A–C). We analyzed 156 cases by flow cytometry and 205 cases by immunohistochemistry. By these criteria, the concordance between flow cytometry and immunohistochemistry was very high (Fisher's exact test: P<0.0001, Supplementary Table 3) allowing us to use immunohistochemistry as a reliable technique to quantitate BCL2 expression on the entire series. As a confirmation of the validity of such approach, FL cases classified as BCL2high by immunohistochemistry showed significantly higher levels of BCL2 mRNA (Figure 3D) and BCL2 protein (Figures 3E and F) as compared with BCL2low cases.
Figure 3.
Intertumoral heterogeneous BCL2 expression in FL cases by immunohistochemistry, flow cytometry, mRNA and western blot. (A and B) Representative cases of FL patients with either sIgHhigh/BCL2high (A) or sIgHlow/BCL2low (B) expression. Surface IgM (sIgM) and intracellular BCL2 expression were detected as described in Materials and methods. Intensity of expression was considered high (red square) for IgH and BCL2 when the mean intensity was higher than 102 FI. Green lines represent negative controls for the staining. (C) We stained formalin-fixed and paraffin-embedded sections from FL cases with the following primary antibodies: BCL2 (a and e), CD10 (b and f), CD20 (c and g) and Ki-67 (d and h). (a–d) Grade 2 BCL2high FL case in which the intensity of BCL2 expression in FL cells was higher than reactive T cells; (e–h) grade 2 BCL2low FL case with BCL2 expression lower than normal reactive cells. Microphotographs were taken with an Olympus BX51WI microscope with a x10 objective. (D) BCL2 mRNA levels in patients with high or low BCL2 expression as determined by immunohistochemistry. All cases were selected to have >70% neoplastic FL cells by flow cytometry stainings. BCL2 mRNA levels were analyzed by quantitative RT-PCR in purified cases classified as BCL2high or BCL2low based on immunohistochemistry. Expression of BCL2 mRNA was normalized to the expression of RPLP0 housekeeping gene. Control cell lines analyzed were either t(14;18)-positive (DHL-6, circle; Ly8, triangle) or t(14;18)-negative (Ly3, diamond; RCK8, square). (E and F) BCL2 protein levels in patients with BCL2 high or low expression. For western blot, we selected FL cases classified as BCL2high or BCL2low based on immunohistochemistry that had >70% neoplastic cells. Cells were lysed and western blot was performed with the indicated antibodies. The ratio of BCL2 and tubulin band intensities measured by densitometry in FL cases were calculated and expressed as a relative number to the t(14;18)-translocated VAL cell line that was used as a standard. (f) Histograms represent mean intensities (±s.d.) for BCL2high and BCL2low cases shown in (g) and calculated as relative expression to the VAL control cell line. *P<0.05
IgH expression was evaluated by flow cytometry in all 205 cases and considered low (IgHlow) or high (IgHhigh) as described in Materials and methods. When more than one IgH was expressed, we scored the IgH IgM, IgG, IgD or IgA that was expressed by the majority of lymphoma cells. By these criteria, 179 cases (87.3%) showed high levels of surface IgH expression (sIgHhigh), whereas 26 cases (12.7%) had low expression (sIgHlow) (Table 1). Among sIgHhigh cases, we found 129 (72.1%) BCL2high and only 50 (27.9%) BCL2low FL cases, whereas sIgHlow cases were mostly BCL2low (9 BCL2high vs 17 BCL2low; Fisher's exact test: P<0.0001) (Table 1). BCL2 expression was not skewed toward any IgH subclasses as a similar distribution of BCL2high and BCL2low cases was observed in the different IgH subtypes (Table 1). A strong correlation between IgH and BCL2 expression was also found when we scored both IgH and BCL2 expression by flow cytometry, although the series of cases was smaller (156 cases; Supplementary Table 4). Therefore, these data demonstrate that a strong correlation between IgH and BCL2 expression is found also at protein levels in a large series of primary FL patients.
Table 1. Patient characteristics, immunoglobulin expression and Bcl-2 levels.
| Category |
Total population |
BCL2high |
BCL2low |
P-value |
|||
|---|---|---|---|---|---|---|---|
| No. of patients | % | No. of patients | % | No. of patients | % | ||
| Sex | |||||||
| Male | 101 | 49.3 | 67 | 66.3 | 34 | 33.7 | χ2=0.021, P=0.882 |
| Female | 104 | 50.7 | 71 | 68.3 | 33 | 31.7 | P=0.882 |
| Age (years) | |||||||
| Median | 61 | 61 | 62 | ||||
| Range | 26–89 | 26–89 | 32–86 | ||||
| IgH surface levels | |||||||
| sIgHhigh | 179 | 87.3 | 129 | 72.1 | 50 | 27.9 | χ2=12.821, P<0.0001 |
| sIgHlow | 26 | 12.7 | 9 | 34.6 | 17 | 65.4 | P<0.0001 |
| IgH subtype | |||||||
| IgM | 106 | 51.7 | 68 | 64.2 | 38 | 35.8 | χ2=1.243, P=0.743 |
| IgG | 88 | 42.9 | 62 | 70.5 | 26 | 29.5 | P=0.743 |
| IgA | 6 | 2.9 | 4 | 66.7 | 2 | 33.3 | |
| IgD | 5 | 2.4 | 4 | 80 | 1 | 20 | |
| FL grading | |||||||
| Grades 1 and 2 | 139 | 67.8 | 103 | 74.1 | 36 | 25.9 | χ2=9.388, P=0.009 |
| Grade 3a | 51 | 24.9 | 28 | 54.9 | 23 | 45.1 | P=0.009 |
| Grade 3b | 15 | 7.3 | 7 | 46.7 | 8 | 53.3 | |
IgH expression levels and subtypes were determined by flow cytometry. BCL2 expression intensities were scored by immunohistochemistry. BCL2 expression was considered high when higher than the expression in surrounding normal and reactive cells T cells, and low when similar or lower than surrounding normal and reactive cells T cells.
Abbreviations:FL, follicular lymphoma; Ig, immunoglobulin; sIg, surface Ig.
FL cases are typically divided into three main histologic grades based on the relative proportion of centrocytes and centroblasts.1 Grade 1 and 2 FL cases are predominantly composed of centrocytes with low proliferation rates, whereas grade 3 FL cases (which are further subdivided into grades 3a and 3b) have increasing numbers of actively proliferating centroblasts with higher proliferation rates.1 As previous reports showed an inverse correlation between proliferation rates and BCL2 expression,2,36 we asked whether heterogeneity of BCL2 expression in our tumor cohort was independent of the proliferation rates of the tumor cells. To this end, we first measured the PI in all FL patient samples by staining with the proliferation marker Ki-67 (Figure 3C) and found, as expected, that grade 1 and 2 FL showed a lower mean PI as compared with grade 3a and 3b FL (20.08% for grades 1 and 2 vs 39.22% for grade 3a vs 56.8% for grade 3b). Overall, BCL2 expression inversely correlated with the PI, as suggested by previous studies on BCL2 expression and proliferation in FL cells 2,36 (Supplementary Table 5). When we correlated BCL2 expression and FL grading, we found that grade 1 and 2 FL more frequently showed BCL2high expression as compared with grade 3 FL (Table 1). Given the different rates of PI between grade 1 and 2 and grade 3 FL, we next wondered whether the correlation of BCL2 expression with IgH could be still detected regardless of the grading and proliferation rate. To this end, we repeated our analysis in low proliferating grade 1 and 2 FL cases, and found a distribution of BCL2high and BCL2low cases similar to the whole series, together with a conserved strong correlation between sIgH and BCL2 expression (Table 2). Therefore, these analyses support the conclusion that the correlation between IgH and BCL2 expression is not only independent of the proliferation rate, as suggested by the smFISH data, but also of the FL grade. Despite a similar trend, a statistical correlation was not observed between IgH and BCL2 expression in grade 3 FL, possibly because of the relatively low number of cases analyzed or to a different biology of grade 3 FL, which typically have an increased number and complex genetic aberrations as compared with grade 1 and 2 FL.37
Table 2. Correlation between sIgH and BCL2 expression in FL different grades.
|
Total population |
BCL2high |
BCL2low |
P-value (Fisher's exact test) |
||||
|---|---|---|---|---|---|---|---|
| No. of patients | % | No. of patients | % | No. of patients | % | ||
| Grades 1 and 2 | |||||||
| sIgHhigh | 123 | 88.5 | 97 | 78.8 | 26 | 21.2 | P=0.001 |
| sIgHlow | 16 | 11.5 | 6 | 37.5 | 10 | 62.5 | |
| Grade 3 | |||||||
| sIgHhigh | 56 | 84.8 | 32 | 57.1 | 24 | 42.9 | P=0.17 |
| sIgHlow | 10 | 15.2 | 3 | 30 | 7 | 70 | |
Abbreviations: FL, follicular lymphoma; Ig, immunoglobulin; sIg, surface Ig.
Heterogeneity of BCL2 expression in individual FL cases determines resistance to therapy
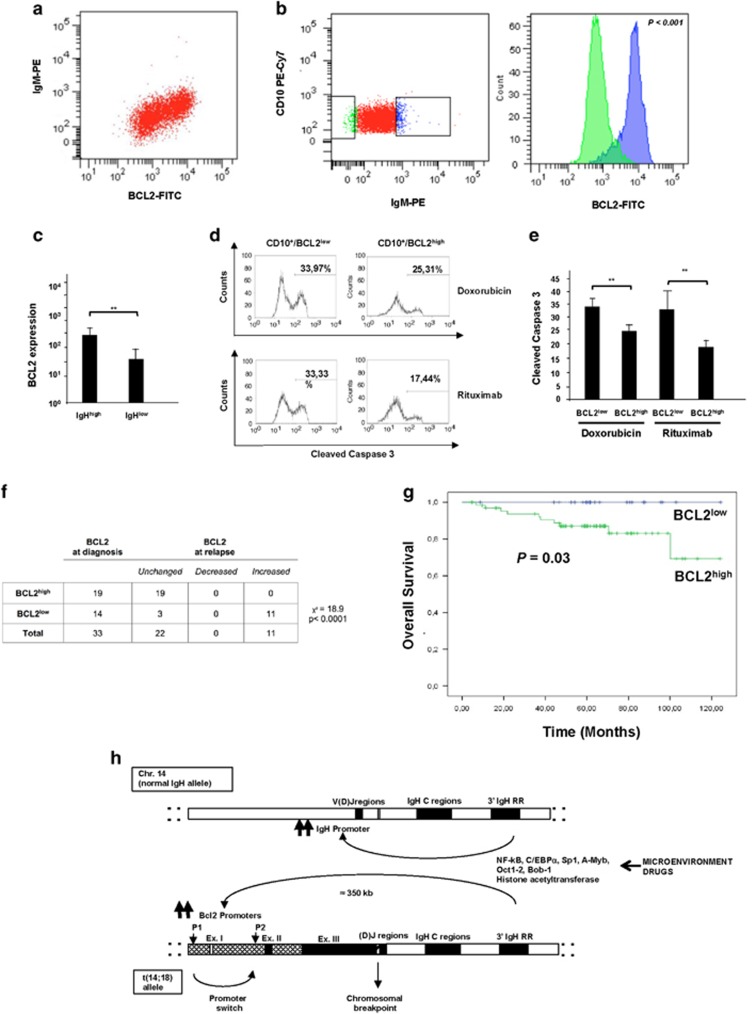
We showed that FL cases exhibit inter- and intratumoral heterogeneity of BCL2 expression that correlates with IgH both at the transcription and protein levels. As BCL2 levels in FL cells are critical to protect lymphoma cells from apoptosis, we then asked whether this heterogeneity would have therapeutic and prognostic impact. To this end, we first characterized resistance to therapy of FL cells from individual FL patients, to limit the confounding effect of variations in the genetic background among different FL cases. We studied by flow cytometry individual FL cases for BCL2 and IgH expression, and showed that, by gating on the CD19+CD10+ monoclonal lymphoma population, BCL2high cells were mostly sIgHhigh, whereas BCL2low cells were mostly sIgHlow (Figures 4a–c). Therefore, BCL2high and BCL2low cell populations could be detected in the same neoplastic clone in individual FL patients, and correlated with IgH expression.
Figure 4.
Intratumoral heterogeneity of BCL2 expression correlates with sIgH expression. (a and b) Levels of sIgH correlate with BCL2 protein expression within the neoplastic population derived from single FL patients. (a) Dot plot from one representative sIgM+ FL patient stained by flow cytometry with the indicated antibodies. Cells were freshly isolated from lymph nodes and the neoplastic clone was identified and gated by CD19, CD10 and Ig light-chain restriction. (b) Within the neoplastic population, cells with sIgHlow (green dots) or sIgHhigh (blue dots) expression were gated and studied for intracellular BCL2 expression. P-value was calculated using the nonparametric Kolmogorov–Smirnov test to compare the shift in BCL2 expression. (c) Histograms represent the mean±s.d. fluorescence intensities for BCL2 surface expression analyzed by flow cytometry in the sub-populations of sIgHhigh and sIgHlow cells in individual FL cases. Ten cases were analyzed; **P<0.01 (d and e) Differential sensitivity to therapeutic agents of BCL2high and BCL2low populations of FL cells within the same FL patient. Cells were freshly isolated from patients with >70% neoplastic population and cultivated with either 2 μg/ml of doxorubicin (top panels) or 2 μg/ml of rituximab (bottom panels). After 24 h, cells were stained with CD10 to identify the neoplastic cells, fixed and permeabilized to stain intracellular BCL2 and cleaved caspase-3 to detect apoptotic cells. Percentages of apoptotic cells (cleaved caspase-3 positive) in the CD10+/BCL2high or CD10+/BCL2low gate are indicated. (e) Mean percentages and standard deviations of cells showing cleaved caspase-3 induced by the indicated treatment in selective CD10+/BCL2high or CD10+/BCL2low tumor populations from eight cases of FL. **P<0.01. (f) The expression levels of BCL2 were characterized by immunohistochemistry in 33 FL cases with available biopsy before treatment and after relapse. All the BCL2high cases maintained a high expression at relapse, with no case showing a decreased BCL2 expression. In contrast, 11/14 cases with BCL2low expression at diagnosis showed a BCL2high expression at the relapsed biopsy. (g) Overall survival for patients (n=102) with BCL2low expression (blue) and for patients with BCL2high expression (green) (P=0.03). (h) Schematic representation of the model of coregulation of IgH and BCL2 expression in FL. In t(14;18)-translocated cells, the BCL2 gene is juxtaposed to the IgH locus. Typically, chromosomal breakpoint are located in the V(d)J region of the IgH locus and 3′ downstream of the BCL2 gene. As a result, the BCL2 gene is under the control of regulatory sequences located in the IgH locus, mainly the IgH-3'RRs. The IgH-3′RRs physically interact with the IgH promoters to induce IgH expression in the normal allele, and with the BCL2 promoter regions in the translocated allele. The IgH-3′RRs upregulate BCL2 transcription by a switch in the usage from the P1 to the P2 promoter. Stimuli from the microenvironment or modifications of endogenous transcription factors that act via the IgH-3′RRs could coregulate both the transcription levels of the IgH locus and the translocated BCL2 locus, thus inducing changes in protein expression levels.
Next, we compared the response to therapy of such BCL2high and BCL2low cell populations. We treated freshly isolated FL cells with doxorubicin or with rituximab, which are commonly used in FL therapy, and measured drug-induced Caspase 3 activation in BCL2high and BCL2low populations. In these conditions, BCL2high FL cells showed significantly higher resistance to treatment-induced apoptosis than BCL2low FL cells, thus indicating a differential sensitivity to therapy of distinct sub-populations of FL cells in individual patients (Figures 4d and e).
A prediction stemming from this observed differential sensitivity to therapy between BCL2high and BCL2low populations in FL cases is that BCL2high cells would be progressively enriched over time in treated FL patients. To test this hypothesis, we evaluated BCL2 expression at diagnosis and at relapse after chemotherapy and rituximab treatment in 33 FL patients for which sequential biopsies were available. Strikingly, all the 19 FL cases with BCL2high expression at diagnosis retained high expression after therapy, whereas 11/14 (78%) BCL2low cases showed a BCL2high expression at relapse (Figure 4f), suggesting that BCL2high FL cells are progressively selected during therapy.
To further study the therapeutic implications of BCL2 heterogeneity in terms of resistance to therapy, we sought to determine whether BCL2 levels could also have prognostic implications and determine survival in FL patients. To this end, we analyzed the overall survival of BCL2high or BCL2low FL patients in our series. Out of the 205 cases in the series, we included in the survival analyses all patients (n=103) for which a minimum follow-up of 5 years was available. All patients were treated with regimens of chemotherapy and rituximab, and clinical characteristics were evaluated in this series including involvement of bone marrow and extranodal sites, Ann Arbor stages, presence of B symptoms, International Prognostic Index), Follicular Lymphoma International Prognostic Index (FLIPI) and FLIPI-2 (ref. 38) (Supplementary Table 6). Remarkably, BCL2low cases had a significantly higher probability of survival as compared with BCL2high cases (Figure 4g). A recent work by Maeshima et al.39 analyzed BCL2 expression by immunohistochemistry in a series of FL and found 60% of FL cases as BCL2high, very similar to our rate of 67% (Table 1). They also found an inverse correlation between FL grade and BCL2 expression, very similar to our findings (Table 1). However, the authors did not report a significant association between Bcl-2 expression and disease outcome. In our series, together with BCL2 expression levels, other clinicopathologic features showing significance by univariate analysis were FLIPI, FLIPI-2 and the Ann Arbor stage. However, when we performed multivariate Cox proportional hazard analyses using the significant parameters emerging from the univariate analysis, the only independent prognostic factor for disease-free survival and overall survival was FLIPI-2 (disease-free survival: hazard ratio: 2.61; 95% confidence interval: 1.399–4.868; P=0.003; overall survival: hazard ratio: 9.381; 95% confidence interval: 1.975–44.553; P=0.005).
In conclusion, in this study we provide evidence for concurrent regulation of IgH and BCL2 that determines BCL2 heterogeneous expression in FL cells with implications for the biology and the therapy of FL. We suggest that the coregulation of IgH and BCL2 is mediated by the shared IgH-3'RRs (Figure 4h). The factors and mechanisms involved in the interaction of IgH-3'RRs with the BCL2 promoter are complex and still poorly characterized. Taken together with Oct1-2 and Bob-1 discussed above, other transcription factors such as c/EBPα, Sp1, A-Myb and the NF-kB pathway can regulate this physical interaction and, thereby, BCL2 transcription in the t(14;18)-translocated allele.33,40,41 In this context, it is conceivable that drugs or different stimuli from the microenvironment, such as growth factors and cytokines, could change the availability or the balance between these transcription factors and thus modify IgH and BCL2 expression in FL cells. It is possible that the important role of the tumor microenvironment in FL pathogenesis and prognosis7,8,42 could partially be explained though the modulation of IgH and BCL2 expression.
Finally, a similar coregulation could also explain heterogeneity of oncogene expression in other types of lymphoma where different oncogenes are involved in chromosomal translocations with the IgH locus.43 As examples, mantle cell lymphoma is genetically characterized by the t(11;14) translocation that juxtaposes the Cyclin D1/CCND1 gene to the IgH locus,1 whereas DLBCL carry BCL6/IgH translocations in about 30% of the cases.44 Cyclin D1 expression in mantle cell lymphoma and BCL6 expression in DLBCL are quite heterogeneous45 and might as well correlate with IgH expression as we showed for BCL2 and IgH in FL. Similarly, in Burkitt's lymphoma c-myc is translocated with IgH and its expression is under the control of IgH-3'RRs.46 Importantly, heterogeneous oncogene expression in sub-populations of mantle cell lymphoma, DLBCL and Burkitt's lymphoma could have an important role in the response to therapy and prognosis. Thus, the pharmacologic or genetic disruption of the binding of IgH-3'RRs to the BCL2 promoter, as to other oncogene promoters, could result in decreased oncogene transcription, and might therefore be exploited as a novel therapeutic tool in lymphoma.
Acknowledgments
We thank D Corino for his precious technical assistance. The work has been supported by Grants FP7 ERC-2009-StG (Proposal No. 242965—‘Lunely'), Associazione Italiana per la Ricerca sul Cancro (AIRC) Grant IG-12023 and Association for International Cancer Research (AICR) Grant 12-0216 (to RC), and by the US NIH/National Cancer Institute Physical Sciences Oncology Center at MIT (U54CA143874) (to AvO).
Author Contributions
AB, CM and LA contributed equally to this study. AB, CM and LR performed research and analyzed data; LA and NC conceived, designed, performed and analyzed smFISH; LC performed statistical analysis; AC and ML provided and revised clinical information on patients; AS and AD performed research and analyzed data; AvO provided technical and bioinformatics support for smFISH and acquisition of data; RC conceived the project, designed and performed research, analyzed data and wrote the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumors of the Haematopoietic and Lymphoid Tissues. Lyon, France; IARC Press; 2008. [Google Scholar]
- Masir N, Campbell LJ, Goff LK, Jones M, Marafioti T, Cordell J, et al. BCL2 protein expression in follicular lymphomas with t(14;18) chromosomal translocations. Br J Haematol. 2009;144:716–725. doi: 10.1111/j.1365-2141.2008.07528.x. [DOI] [PubMed] [Google Scholar]
- Camacho FI, Bellas C, Corbacho C, Caleo A, Arranz-Saez R, Cannata J, et al. Improved demonstration of immunohistochemical prognostic markers for survival in follicular lymphoma cells. Mod Pathol. 2011;24:698–707. doi: 10.1038/modpathol.2010.237. [DOI] [PubMed] [Google Scholar]
- Olejniczak SH, Stewart CC, Donohue K, Czuczman MS. A quantitative exploration of surface antigen expression in common B-cell malignancies using flow cytometry. Immunol Invest. 2006;35:93–114. doi: 10.1080/08820130500496878. [DOI] [PubMed] [Google Scholar]
- Eide MB, Liestol K, Lingjaerde OC, Hystad ME, Kresse SH, Meza-Zepeda L, et al. Genomic alterations reveal potential for higher grade transformation in follicular lymphoma and confirm parallel evolution of tumor cell clones. Blood. 2010;116:1489–1497. doi: 10.1182/blood-2010-03-272278. [DOI] [PubMed] [Google Scholar]
- Green MR, Gentles AJ, Nair RV, Irish JM, Kihira S, Liu CL, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121:1604–1611. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- Ame-Thomas P, Tarte K. The yin and the yang of follicular lymphoma cell niches: Role of microenvironment heterogeneity and plasticity. Semin Cancer Biol. 2013;24:23–32. doi: 10.1016/j.semcancer.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Gribben JG. Implications of the tumor microenvironment on survival and disease response in follicular lymphoma. Curr Opin Oncol. 2010;22:424–430. doi: 10.1097/CCO.0b013e32833d5938. [DOI] [PubMed] [Google Scholar]
- Gribben J, Rosenwald A, Gascoyne R, Lenz G. Targeting the microenvironment. Leuk Lymphoma. 2010;51 (Suppl 1:34–40. doi: 10.3109/10428194.2010.500072. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y, Cossman J, Jaffe E, Croce CM. Involvement of the bcl-2 gene in human follicular lymphoma. Science. 1985;228:1440–1443. doi: 10.1126/science.3874430. [DOI] [PubMed] [Google Scholar]
- Cleary ML, Galili N, Sklar J. Detection of a second t(14;18) breakpoint cluster region in human follicular lymphomas. J Exp Med. 1986;164:315–320. doi: 10.1084/jem.164.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Heckman CA, Boxer LM. The immunoglobulin heavy-chain gene 3' enhancers deregulate bcl-2 promoter usage in t(14;18) lymphoma cells. Oncogene. 2007;26:2635–2641. doi: 10.1038/sj.onc.1210061. [DOI] [PubMed] [Google Scholar]
- Xiang H, Noonan EJ, Wang J, Duan H, Ma L, Michie S, et al. The immunoglobulin heavy chain gene 3' enhancers induce Bcl2 deregulation and lymphomagenesis in murine B cells. Leukemia. 2011;25:1484–1493. doi: 10.1038/leu.2011.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JC, Kitada S, Takayama S, Miyashita T. Regulation of chemoresistance by the bcl-2 oncoprotein in non-Hodgkin's lymphoma and lymphocytic leukemia cell lines. Ann Oncol. 1994;5 (Suppl 1:61–65. doi: 10.1093/annonc/5.suppl_1.s61. [DOI] [PubMed] [Google Scholar]
- Kridel R, Sehn LH, Gascoyne RD. Pathogenesis of follicular lymphoma. J Clin Invest. 2012;122:3424–3431. doi: 10.1172/JCI63186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulard P, d'Agay MF, Peuchmaur M, Brousse N, Gisselbrecht C, Solal-Celigny P, et al. Expression of the bcl-2 gene product in follicular lymphoma. Am J Pathol. 1992;140:1089–1095. [PMC free article] [PubMed] [Google Scholar]
- Dogan A, Du MQ, Aiello A, Diss TC, Ye HT, Pan LX, et al. Follicular lymphomas contain a clonally linked but phenotypically distinct neoplastic B-cell population in the interfollicular zone. Blood. 1998;91:4708–4714. [PubMed] [Google Scholar]
- Semrau S, Crosetto N, Bienko M, Boni M, Bernasconi P, Chiarle R, et al. FuseFISH: robust detection of transcribed gene fusions in single cells. Cell Rep. 2014;6:18–23. doi: 10.1016/j.celrep.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol. 2012;14:106–114. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraders M, de Jong D, Kluin P, Groenen P, van Krieken H. Lack of Bcl-2 expression in follicular lymphoma may be caused by mutations in the BCL2 gene or by absence of the t(14;18) translocation. J Pathol. 2005;205:329–335. doi: 10.1002/path.1689. [DOI] [PubMed] [Google Scholar]
- Demurtas A, Aliberti S, Bonello L, Di Celle PF, Cavaliere C, Barreca A, et al. Usefulness of multiparametric flow cytometry in detecting composite lymphoma: study of 17 cases in a 12-year period. Am J Clin Pathol. 2011;135:541–555. doi: 10.1309/AJCPQKE25ADCFZWN. [DOI] [PubMed] [Google Scholar]
- Barrena S, Almeida J, Del Carmen Garcia-Macias M, Lopez A, Rasillo A, Sayagues JM, et al. Flow cytometry immunophenotyping of fine-needle aspiration specimens: utility in the diagnosis and classification of non-Hodgkin lymphomas. Histopathology. 2011;58:906–918. doi: 10.1111/j.1365-2559.2011.03804.x. [DOI] [PubMed] [Google Scholar]
- Demurtas A, Stacchini A, Aliberti S, Chiusa L, Chiarle R, Novero D. Tissue flow cytometry immunophenotyping in the diagnosis and classification of non-Hodgkin's lymphomas: a retrospective evaluation of 1,792 cases. Cytometry Part B. 2013;84:82–95. doi: 10.1002/cyto.b.21065. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Heckman CA, Cao T, Somsouk L, Duan H, Mehew JW, Zhang CY, et al. Critical elements of the immunoglobulin heavy chain gene enhancers for deregulated expression of bcl-2. Cancer Res. 2003;63:6666–6673. [PubMed] [Google Scholar]
- Pinaud E, Marquet M, Fiancette R, Peron S, Vincent-Fabert C, Denizot Y, et al. The IgH locus 3' regulatory region: pulling the strings from behind. Adv Immunol. 2011;110:27–70. doi: 10.1016/B978-0-12-387663-8.00002-8. [DOI] [PubMed] [Google Scholar]
- Vincent-Fabert C, Fiancette R, Cogne M, Pinaud E, Denizot Y. The IgH 3' regulatory region and its implication in lymphomagenesis. Eur J Immunol. 2010;40:3306–3311. doi: 10.1002/eji.201040778. [DOI] [PubMed] [Google Scholar]
- Cogne M, Lansford R, Bottaro A, Zhang J, Gorman J, Young F, et al. A class switch control region at the 3' end of the immunoglobulin heavy chain locus. Cell. 1994;77:737–747. doi: 10.1016/0092-8674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, Le Bert M, et al. Localization of the 3' IgH locus elements that effect long-distance regulation of class switch recombination. Immunity. 2001;15:187–199. doi: 10.1016/s1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- Rouaud P, Vincent-Fabert C, Saintamand A, Fiancette R, Marquet M, Robert I, et al. The IgH 3' regulatory region controls somatic hypermutation in germinal center B cells. J Exp Med. 2013;210:1501–1507. doi: 10.1084/jem.20130072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman CA, Duan H, Garcia PB, Boxer LM. Oct transcription factors mediate t(14;18) lymphoma cell survival by directly regulating bcl-2 expression. Oncogene. 2006;25:888–898. doi: 10.1038/sj.onc.1209127. [DOI] [PubMed] [Google Scholar]
- Duan H, Xiang H, Ma L, Boxer LM. Functional long-range interactions of the IgH 3' enhancers with the bcl-2 promoter region in t(14;18) lymphoma cells. Oncogene. 2008;27:6720–6728. doi: 10.1038/onc.2008.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SJ, Saleque S, Birshtein BK. Yin Yang 1 is a lipopolysaccharide-inducible activator of the murine 3' Igh enhancer, hs3. J Immunol. 2003;170:5549–5557. doi: 10.4049/jimmunol.170.11.5549. [DOI] [PubMed] [Google Scholar]
- Winter JN, Andersen J, Reed JC, Krajewski S, Variakojis D, Bauer KD, et al. BCL-2 expression correlates with lower proliferative activity in the intermediate- and high-grade non-Hodgkin's lymphomas: an Eastern Cooperative Oncology Group and Southwest Oncology Group cooperative laboratory study. Blood. 1998;91:1391–1398. [PubMed] [Google Scholar]
- Leich E, Ott G, Rosenwald A. Pathology, pathogenesis and molecular genetics of follicular NHL. Best Pract Res Clin Haematol. 2011;24:95–109. doi: 10.1016/j.beha.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Federico M, Bellei M, Marcheselli L, Luminari S, Lopez-Guillermo A, Vitolo U, et al. Follicular Lymphoma International Prognostic Index 2: a new prognostic index for follicular lymphoma developed by the International Follicular Lymphoma Prognostic Factor Project. J Clin Oncol. 2009;27:4555–4562. doi: 10.1200/JCO.2008.21.3991. [DOI] [PubMed] [Google Scholar]
- Maeshima AM, Taniguchi H, Nomoto J, Miyamoto K, Fukuhara S, Munakata W, et al. Prognostic implications of histologic grade and intensity of Bcl-2 expression in follicular lymphomas undergoing rituximab-containing therapy. Hum Pathol. 2013;44:2529–2535. doi: 10.1016/j.humpath.2013.06.013. [DOI] [PubMed] [Google Scholar]
- Heckman CA, Mehew JW, Boxer LM. NF-kappaB activates Bcl-2 expression in t(14;18) lymphoma cells. Oncogene. 2002;21:3898–3908. doi: 10.1038/sj.onc.1205483. [DOI] [PubMed] [Google Scholar]
- Heckman CA, Mehew JW, Ying GG, Introna M, Golay J, Boxer LM. A-Myb up-regulates Bcl-2 through a Cdx binding site in t(14;18) lymphoma cells. J Biol Chem. 2000;275:6499–6508. doi: 10.1074/jbc.275.9.6499. [DOI] [PubMed] [Google Scholar]
- de Jong D, Fest T. The microenvironment in follicular lymphoma. Best Pract Res Clin Haematol. 2011;24:135–146. doi: 10.1016/j.beha.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2011;29:319–350. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- Basso K, Dalla-Favera R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv Immunol. 2010;105:193–210. doi: 10.1016/S0065-2776(10)05007-8. [DOI] [PubMed] [Google Scholar]
- Klapper W. Histopathology of mantle cell lymphoma. Semin Hematol. 2011;48:148–154. doi: 10.1053/j.seminhematol.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Gostissa M, Yan CT, Bianco JM, Cogne M, Pinaud E, Alt FW. Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3' regulatory region. Nature. 2009;462:803–807. doi: 10.1038/nature08633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.