Abstract
Deep brain stimulation is a treatment for Parkinson's disease and other related disorders, involving the surgical placement of electrodes in the deeply situated basal ganglia or thalamic structures. Good clinical outcome requires accurate targeting. However, due to limited visibility of the target structures on routine clinical MR images, direct targeting of structures can be challenging. Non-clinical MR scanners with ultra-high magnetic field (7T or higher) have the potential to improve the quality of these images. This technology report provides an overview of the current possibilities of visualizing deep brain stimulation targets and their related structures with the aid of ultra-high field MRI. Reviewed studies showed improved resolution, contrast- and signal-to-noise ratios at ultra-high field. Sequences sensitive to magnetic susceptibility such as T2* and susceptibility weighted imaging and their maps in general showed the best visualization of target structures, including a separation between the subthalamic nucleus and the substantia nigra, the lamina pallidi medialis and lamina pallidi incompleta within the globus pallidus and substructures of the thalamus, including the ventral intermediate nucleus (Vim). This shows that the visibility, identification, and even subdivision of the small deep brain stimulation targets benefit from increased field strength. Although ultra-high field MR imaging is associated with increased risk of geometrical distortions, it has been shown that these distortions can be avoided or corrected to the extent where the effects are limited. The availability of ultra-high field MR scanners for humans seems to provide opportunities for a more accurate targeting for deep brain stimulation in patients with Parkinson's disease and related disorders.
Keywords: ultra-high field, magnetic resonance imaging, basal ganglia, thalamus, deep brain stimulation
Introduction
The basal ganglia are a group of nuclei deep in the brain, which play an important role in specific motor, limbic, and associative processes (Temel et al., 2005). Anatomically, they consist of the caudate nucleus-putamen (also referred to as striatum), external and internal globus pallidus (GPe and GPi, respectively), substantia nigra (SN), and the subthalamic nucleus (STN). Structural or functional impairments of basal ganglia structures can lead to neurological and psychiatric disorders, e.g., Parkinson's disease (PD) (Obeso et al., 2008), dystonia (Wichmann and Dostrovsky, 2011), Tourette's syndrome (Mink, 2006), and obsessive-compulsive disorder (Maia et al., 2008). Although most of the patients with basal ganglia diseases can be managed by drug and/or behavioral therapy, an increasing number of patients are referred to specialized teams for deep brain stimulation (DBS) (Lee et al., 2007; Ackermans et al., 2008; Limousin and Martinez-Torres, 2008; Denys et al., 2010). The main reasons for DBS referral include the proven benefit of DBS over best medical treatment (Deuschl et al., 2006; Schuepbach et al., 2013) or insufficient response to non-surgical therapies. DBS is a minimally invasive surgical procedure and involves the implantation of stimulating electrodes with millimeter precision into a specific brain target. The brain regions targeted most often are located in the basal ganglia, and include the ventral parts of the striatum (Malone et al., 2009; Denys et al., 2010), post-eroventral part of the GPi (Damier et al., 2007; Lee et al., 2007; Ackermans et al., 2008), ventral and anterior parts of the pallidum (Ackermans et al., 2008), the STN (Follett and Torres-Russotto, 2012), and surrounding structures such as the ventrolateral and anterior parts of the thalamus (Fisher et al., 2010).
Currently, there are three methods to locate the target for DBS: (a) using intraoperative neurophysiological mapping tools, (b) using stereotactic coordinates derived from post-mortem or magnetic resonance imaging (MRI) based atlases (indirect targeting), and (c) via direct visualization on individual magnetic resonance (MR) images (direct targeting). Combinations of these methods are generally used. Direct targeting has the advantage over indirect targeting in that it accounts for differences in individual anatomy, which is especially critical when small structures such as those in DBS are targeted. However, at standard clinical magnetic field strengths (1.5T and 3T) direct visualization often lacks contrast for very high precision DBS targeting. The increasing availability of ultra-high magnetic field (7T or higher) MR scanners promises direct, accurate visualization of target regions with a very high specificity. A better understanding of the structural and functional components of the basal ganglia and related structures at ultra-high resolution approaching the microscopic level, is not only expected to increase the accuracy of DBS, shorten surgery, and potentially improve the clinical outcomes (Yokoyama et al., 2006; Wodarg et al., 2012), but also to enhance our understanding of brain function and disease states. In this technology report, we present the current options for detailed visualization of deep-brain structures using multiple MRI contrasts at ultra-high magnetic field, based on a literature review.
English-language studies were searched on PubMed using combinations of title and abstract key words related to basal ganglia, thalamus, and ultra-high field MRI. Publications were selected by screening of titles and abstracts. Additional studies were found through the references cited in the selected articles.
In this technology report, anatomical structures are denoted in English, unless their Latin names are commonly used. In the first sections, we provide background information on the basic concepts of MRI, which we consider important to understand the different image types that can be obtained, and on the conventional methods of MR imaging of the basal ganglia. Subsequently, we review the current literature on in vivo and ex vivo (i.e., post-mortem) ultra-high field imaging of the basal ganglia and related structures.
Summary of the principles of magnetic resonance imaging
Whether and how well a certain brain structure is visible on an MR image depends on biophysical tissue parameters and MRI acquisition protocols (see Table 1).
Table 1.
Important concepts in MR imaging.
| Variable | Definition | Specific for | Relevance |
|---|---|---|---|
| T1 | Spin-lattice relaxation time | Tissue | Influences MR signal in tissue |
| T2 | Spin-spin relaxation time | ||
| T2* | T2* relaxation time | ||
| R1 | 1/T1 | ||
| R2 | 1/T2 | ||
| R2* | 1/T2* | ||
| TE | Echo time | Sequence | Determines the generated contrast |
| TR | Repetition time | ||
| Flip angle | Flip angle | ||
| χ | Magnetic susceptibility | Tissue | Gives extra contrast to certain substances |
| SNR | Signal-to-noise ratio | Image | Quantifies the quality of the image |
| CNR | Contrast-to-noise ratio |
The relaxation times, T1, T2, and T2*, are time constants that describe magnetic spin interaction properties of nuclei, which depend, among other things, on the molecular composition and organization of the tissue and the strength of the main magnetic field. Often, the relaxation rates R1, R2, and R2* are used, defined as 1/T1, 1/T2, and 1/T2* respectively. MRI uses the dependencies of these relaxation times on tissue properties to generate contrast within an image.
The actual type of contrast is determined by the MRI pulse sequence that is used, such as spin-echo (SE) and gradient echo (GE) sequences. These MRI contrasts are sensitive to different biophysical properties of the tissue and it is a matter of intense research to quantitatively relate tissue composition and MRI contrasts. Thus, individual and combinations of MRI contrasts provide a window to examine microstructural properties of brain tissue. The different sequences are described by the combination of the properties of the gradient, radio-frequency pulses and timing parameters. Properties that are often varied are the echo time (TE), repetition time (TR), and flip angle. This can result in T1-, T2- or T2*-weighted images in which the contrast is mainly caused by differences in T1, T2, or T2* values of the tissue. The variability in sequences therefore facilitates optimization of the protocol for each structure of interest individually.
Susceptibility weighted imaging
Susceptibility-weighted (SW) images can also be acquired (Haacke and Reichenbach, 2011). These images are based on the principle that MR images are generally complex-valued, i.e., effectively two images are always acquired: a commonly used magnitude image that often directly displays the anatomical structures and a phase image that is usually disregarded. The phase image however is sensitive to the so-called magnetic susceptibility (χ). This property of tissues and substances alters the local magnetic field values. Paramagnetic materials have a positive χ and strengthen the magnetic field, and diamagnetic materials have a negative χ and weaken the magnetic field. Tissues with a susceptibility that differs from their surrounding structures, such as tissues with myelin and iron-containing substances, cause local deviations in the magnetic field inside and outside of the structures. This leads to local phase differences, which can then be extracted from the original phase images. In susceptibility-weighted imaging (SWI), these phase images are combined with the magnitude images, which can result in additional contrast, which particularly enhances the brain's (micro)vessels and the small deep brain structures.
Quantitative maps
Furthermore, post-processing techniques can be employed, to produce so-called T1, T2, T2*, or (quantitative) susceptibility maps, which display the quantitative T1, T2, T2*, or susceptibility values of each voxel in an image respectively. Sometimes R1-, R2- or R2*-values are computed instead, which are defined as 1/T1, 1/T2, and 1/T2* respectively.
Other techniques
In addition to structural imaging, diffusion-weighted imaging (DWI), which is directionally sensitive to water diffusion, gives complementary information (Le Bihan, 2003). It can provide information on the location and orientation of neuronal fibers, aiding in visualization of these pathways (tractography) (Mori et al., 1999) or super-resolution track-density imaging (TDI) (Calamante et al., 2010). Furthermore, functional MRI (fMRI) can provide information on localized brain activity (Buxton, 2013). Finally, DWI and fMRI can be used to compute the connectivity between two areas by computing the fiber paths between them (structural connectivity) or the correlation of functional activity (functional connectivity) respectively.
Field strength
In most DBS centers, the MR images are obtained from 1.5T or 3T MR scanners. However, in specialized neuroimaging centers, the possibilities of scanning at ultra-high field are increasingly being explored (Duyn, 2012). Although the number keeps growing, at present an estimate of 61 human ultra-high field MR scanners has been installed or will be installed in the near future (see Table 2). At ultra-high field alterations of physical properties can influence measurements both positively and negatively. Several issues including field strength dependent changes in relaxation times T1, T2, and T2*; increased B0 and B1 magnetic field inhomogeneities; and increased risks of tissue heating (Duyn, 2012) make ultra-high field scanning more sensitive to inhomogeneous signal-to-noise ratios (SNR) and contrast-to-noise ratios (CNR), geometric distortions, and movement artifacts. This limits the use of T1-weighted, T2-weighted, and proton density weighted turbo spin echo (TSE) scan protocols that are commonly used in clinics (Hennig et al., 1986). However, the alterations in relaxation times and the increased sensitivity to magnetic susceptibility have stimulated the focus of ultra-high field imaging to shift to susceptibility and T2*-dependent gradient echo sequences (Haase et al., 2011). Furthermore, the SNR increases close to linearly with field strength, which offers the option to scan with higher spatial resolution (Vaughan et al., 2001) and/or CNR in a shorter time (Duyn, 2012). This makes ultra-high field MRI especially beneficial for detailed imaging of structures with altered magnetic susceptibility, such as the basal ganglia, myelin, and blood, which is also important for ultra-high field high-resolution fMRI imaging. Finally, side effects might occur during movement through the gradients of the strong field. The majority of the subjects have been reported to feel sensations when moving into or out of the bore, which was rated as unpleasant vertigo in 5–20% of the subjects (Glover et al., 2007; Theysohn et al., 2008) and a small number of subjects (approximately 3%) experienced a medium or strong metallic taste (Theysohn et al., 2008).
Table 2.
Overview of ultra-high magnetic field (7T or higher) human MR scanners that have been installed or will be installed in the future according to the institutions' websites.
| Nr | Country | City | Institution, department | Manufacturer | Field strength (T) | Publication |
|---|---|---|---|---|---|---|
| 1 | Australia | Melbourne | Melbourne Brain Centre, Melbourne Brain Centre Imaging Unit | Siemens | 7 | |
| 2 | Australia | Brisbane | University of Queensland, Centre for Advanced Imaging | Siemens | 7 | |
| 3 | Austria | Vienna | Medical University of Vienna, MR Center of Excellence | Siemens | 7 | Hahn et al., 2013 |
| 4 | Brazil | Sao Paulo | University of Sao Paulo | Siemens | 7 | |
| 5 | Canada | London | Western University, Robarts Research Institute, Centre for Functional and Metabolic Mapping | Siemens | 7 | Goubran et al., 2014 |
| 6 | Canada | Toronto | Toronto Western Hospital, Krembil Neuroscience Centre | Siemens | 7 | |
| 7 | China | Beijing | Chinese Academy of Sciences, State Key Laboratory of Brain and Cognitive Science | Siemens | 7 | He et al., 2014 |
| 8 | Denmark | Copenhagen | Hvidovre Hospital, Danish Research Centre for Magnetic Resonance | Philips | 7 | |
| 9 | France | Marseille | Center for Magnetic Resonance in Biology and Medicine | Siemens | 7 | |
| 10 | France | Saclay | Alternative Energies and Atomic Energy Commission, Life Sciences Division, Neurospin | Siemens | 7 | Boulant et al., 2011 |
| 11 | France | Saclay | Alternative Energies and Atomic Energy Commission, Life Sciences Division, Neurospin | Custom built | 11.7 | Vedrine et al., 2014 |
| 12 | Germany | Berlin | Max-Delbrueck-Center for Molecular Medicine, Berlin Ultrahigh Field Facility | Siemens | 7 | Dieringer et al., 2011 |
| 13 | Germany | Bonn | German Center for Neurodegenerative Diseases | Siemens | 7 | |
| 14 | Germany | Essen | Erwin L. Hahn Institute for Magnetic Resonance Imaging | Siemens | 7 | Dammann et al., 2011 |
| 15 | Germany | Heidelberg | German Cancer Research Center | Siemens | 7 | Hoffmann et al., 2011 |
| 16 | Germany | Jülich | Research Centre Jülich, Institute of Neuroscience and Medicine | Siemens | 9.4 | Arrubla et al., 2013 |
| 17 | Germany | Leipzig | Max Planck Institute for Human Cognitive and Brain Sciences, | Siemens | 7 | Deistung et al., 2013a |
| 18 | Germany | Magdeburg | Leibniz Institute for Neurobiology, Center for Advanced Imaging | Siemens | 7 | Hoffmann et al., 2009 |
| 19 | Germany | Tübingen | Max Planck Institute for Biological Cybernetics | Siemens | 9.4 | Budde et al., 2014 |
| 20 | Italy | Pisa | Imago7 Foundation | GE | 7 | Costagli et al., 2014 |
| 21 | Japan | Niigata | University of Niigata, Center for Integrated Human Brain Science | GE | 7 | Kabasawa et al., 2006 |
| 22 | Japan | Morioka | Iwate Medical University, Institute for Biomedical Sciences | GE | 7 | Sato and Kawagishi, 2014 |
| 23 | Japan | Suita City | Center for Information and Neural Networks | 7 | ||
| 24 | Netherlands | Leiden | Leiden University Medical Center, C.J. Gorter Center for High Field Magnetic Resonance in the LUMC | Philips | 7 | Dzyubachyk et al., 2013 |
| 25 | Netherlands | Utrecht | UMC Utrecht | Philips | 7 | de Bresser et al., 2013 |
| 26 | Netherlands | Amsterdam | Spinoza Centre for Neuroimaging | Philips | 7 | |
| 27 | Netherlands | Maastricht | Maastricht University, Maastricht Brain Imaging Centre | Siemens | 7 | Ivanov et al., 2014 |
| 28 | Netherlands | Maastricht | Maastricht University, Maastricht Brain Imaging Centre | Siemens | 9.4 | Cloos et al., 2014 |
| 29 | Republic of Korea | Icheon | Gachon University of Medicine and Science, Neuroscience Research Institute | Siemens | 7 | Cho et al., 2008 |
| 30 | Sweden | Lund | Lund University, Lund University Bioimaging Center | Philips | 7 | |
| 31 | Switzerland | Lausanne | Centre d'Imagerie BioMédicale | Siemens | 7 | Kickler et al., 2010 |
| 32 | Switzerland | Zürich | Swiss Federal Institute of Technology and University of Zurich, Institute for Biomedical Engineering | Philips | 7 | Wyss et al., 2014 |
| 33 | UK | Nottingham | University of Nottingham, Sir Peter Mansfield Magnetic Resonance Centre | Philips | 7 | Lotfipour et al., 2012 |
| 34 | UK | Oxford | University of Oxford, Oxford Centre for Functional Magnetic Resonance Imaging of the Brain | Siemens | 7 | Berrington et al., 2014 |
| 35 | USA | Auburn | Auburn University, Magnetic Resonance Imaging Research Center | Siemens | 7 | Denney et al., 2014 |
| 36 | USA | Baltimore | Kennedy Krieger Institute, FM Kirby Center for Functional Brain Imaging | Philips | 7 | Intrapiromkul et al., 2013 |
| 37 | USA | Bethesda | National Institute of Health, Functional MRI Facility | Siemens | 7 | Gaitan et al., 2013 |
| 38 | USA | Bethesda | National Institutes of Health, National Institute of Neurological Disorders and Stroke | Siemens | 11.7 | |
| 39 | USA | Boston | Massachusetts General Hospital, Martinos Center for Biomedical Imaging | Siemens | 7 | Augustinack et al., 2005 |
| 40 | USA | Chapel Hill | University of North Carolina | 7 | ||
| 41 | USA | Chicago | University of Illinois, Center for MR Research | Custom built | 9.4 | Lu et al., 2013 |
| 42 | USA | Cleveland | Cleveland Clinic | Siemens | 7 | |
| 43 | USA | Columbus | Ohio State University, Department of Radiology | Bruker | 8 | Bourekas et al., 1999; Robitaille et al., 1999 |
| 44 | USA | Columbus | Ohio State University, Department of Radiology | Philips | 7 | |
| 45 | USA | Minneapolis | University of Minnesota, Center for Magnetic Resonance Research | Siemens | 7 | Abosch et al., 2010 |
| 46 | USA | Minneapolis | University of Minnesota, Center for Magnetic Resonance Research | Siemens | 7 | |
| 47 | USA | Minneapolis | University of Minnesota, Center for Magnetic Resonance Research | Siemens | 10.5 | |
| 48 | USA | Minneapolis | University of Minnesota, Center for Magnetic Resonance Research | Varian | 9.4 | Deelchand et al., 2010 |
| 49 | USA | Nashville | Vanderbilt University, Institute of Imaging Science | Philips | 7 | Eapen et al., 2011 |
| 50 | USA | New Haven | Yale University, Magnetic Resonance Research Center | Varian | 7 | Pan et al., 2010 |
| 51 | USA | New York | New York University School of Medicine, Center for Biomedical Imaging | Siemens | 7 | Pakin et al., 2006 |
| 52 | USA | New York | Icahn School of Medicine at Mount Sinai, Translational and Molecular Imaging Institute | Siemens | 7 | |
| 53 | USA | Philadelphia | University of Pennsylvania, Center For Magnetic Resonance And Optical Imaging | Siemens | 7 | Bhagat et al., 2011 |
| 54 | USA | Pittsburgh | University of Pittsburgh, Magnetic Resonance Research Center | Siemens | 7 | Moon et al., 2013 |
| 55 | USA | Portland | Oregon Health & Science University, Advanced Imaging Research Center | Siemens | 7 | |
| 56 | USA | San Francisco | San Francisco Veterans Affairs Medical Center, Center for Imaging of Neurodegenerative Diseases | Siemens | 7 | |
| 57 | USA | San Francisco | University of California, Department of Radiology and Biomedical Imaging | GE | 7 | Metcalf et al., 2010 |
| 58 | USA | Dallas | University of Texas Southwestern Medical Center, Advanced Imaging Research Center | Philips | 7 | Ren et al., 2013 |
| 59 | USA | Iowa City | University of Iowa, Iowa Institute for Biomedical Imaging | GE | 7 | |
| 60 | USA | Milwaukee | Medical College of Wisconsin, Center for Imaging Research | GE | 7 | |
| 61 | USA | Stanford | Stanford University, Richard M. Lucas Center for Imaging | GE | 7 | Kerchner et al., 2012 |
Because not all scanners are operational yet, the last column refers to publications in which the mentioned scanner is used.
MRI of DBS targets at clinical field strengths of 1.5T and 3T
Direct visualization and targeting of DBS structures based on 1.5T or 3T MR images obtained in clinical practice can be challenging. Several studies compared different scanning sequences for the visibility of the STN (Kerl et al., 2012a; Liu et al., 2013), GPi (Nolte et al., 2012; Liu et al., 2013), GPe (Nolte et al., 2012), and zona incerta (ZI) (Kerl et al., 2012b) and showed that T2* (Kerl et al., 2012a,c; Nolte et al., 2012) and quantitative susceptibility maps (Liu et al., 2013) outperformed T1- and T2-weighted images. Furthermore, 3T functional and structural connectivity maps have been measured in healthy volunteers to visualize the functional subdivision of the STN, although higher spatial resolution is expected to reveal a more detailed anatomy (Brunenberg et al., 2012). Also, a literature review concluded that there is no consensus whether 1.5T and 3T MRI are reliable and accurate enough to be employed for direct targeting of the STN, due to serious shortcomings in the contrast between the STN and surrounding structures (Brunenberg et al., 2011). Visualization of the small substructures in the thalamus at lower field strengths is even less straightforward, primarily due to lack of contrast. One study identified four large thalamic nuclei groups on 3T magnetization-prepared rapid acquisition of gradient echo (MPRAGE) images (Bender et al., 2011) and another study identified the centromedian nucleus directly on 3T proton density weighted MR images (Kanowski et al., 2010). The thalamus was also segmented at 1.5T and 3T using DWI (Wiegell et al., 2003; Unrath et al., 2008; Pouratian et al., 2011; Mang et al., 2012) or a combination of ten different sequences (Yovel and Assaf, 2007).
Although several sequences have been investigated for the visualization of basal ganglia structures at clinical field strengths, DBS structures such as the motor part of the STN, and certain regions within the thalamus, such as the ventrolateral nuclei, need to be displayed more distinctively in order to rely on these images solely for targeting.
Ultra-high field imaging of the deep-brain structures
Several studies identified deep-brain (sub)structures at ultra-high field using different MRI contrasts. These studies, reviewed below, show the high potential of ultra-high field MRI to accurately identify and delineate thalamic, parathalamic and subthalamic nuclei. Table 3 shows detailed scanning parameters of the described studies, referred to by line numbers.
Table 3.
Overview of acquisition parameters used in the described studies.
Ax, axial; Cor, coronal; EPI, echo planar imaging; FLAIR, fluid attenuation inversion recovery; FLASH, fast low angle shot; FLASH-HB, FLASH with high bandwidth; FOV, field of view; GE, gradient echo; GRASE, gradient and spin echo; M-GE, multi-echo GE; MPRAGE, magnetization prepared rapid gradient echo; MP2RAGE, magnetization prepared 2 rapid acquisition gradient echoes; PD, Parkinson's disease; PIAF, parallel imaging acceleration factor; Sag, sagittal; SD, spin density; SE, spin-echo; S-GE, single-echo GE; SPACE, sampling perfection with application of optimized contrasts using different flip angle evolutions; SWI, susceptibility weighted imaging; T1w, T1-weighted; T2w, T2-weighted; T2*w, T2*-weighted; TDI, track density imaging; TI, inversion time; TR, repetition time; TSE, turbo spin-echo; χ-map, susceptibility map. Bandwidths that were originally reported in kHz have been converted to Hz/pixel and are denoted with an asterisk (*).
Visualization of deep-brain structures at ultra-high field in vivo
Since the installation of the first ultra-high field MR scanner, several studies investigated the visualization of deep-brain structures at ultra-high field in vivo (Table 4).
Table 4.
Overview of the basal ganglia and related (sub)structures that have been identified using different protocols at ultra-high field MRI.
| Study | Image type | Findings | Line |
|---|---|---|---|
| Bourekas et al., 1999 | T2*w | GP, SN, and RN appear hypointense | 1 |
| Novak et al., 2001 | GP, SN, and RN appear hypointense | 2 | |
| Abduljalil et al., 2003 | GE Magnitude | SN and RN appear hypointense | 3 |
| GE Phase | Substructures within SN and RN | 3 | |
| Cho et al., 2008 | GE | SN and RN in coronal plane hypointense | 4 |
| Cho et al., 2010 | Coronal GE | Discrimination of STN and SN | 38 |
| Abosch et al., 2010 | SWI | Clear delineation of STN | 7–8 |
| Boundary between STN and SN | |||
| Lamina pallidi medialis and lamina pallidi incompleta | |||
| Vim, anterior and medial boundaries of pulvinar, boundary of the nucleus ventralis caudalis | |||
| Eapen et al., 2011 | T2w and T2*w | Subregions within RN | 9 |
| T2*w | Subregions within RN and SN | 10 | |
| Schafer et al., 2012 | χ-map | Boundary between STN and SN | 11 |
| Deistung et al., 2013b | χ-map | Subnuclei within the SN | 12 |
| Discrimination of the STN from the SN and surrounding gray and white matter | |||
| Lamina pallidi medialis and lamina pallidi incompleta | |||
| Medullary lamina in RN | |||
| Vim, pulvinar, lateral and medial geniculate nucleus, dorsomedial nucleus and dorsal nuclei group | |||
| R2*-map | Substructures in RN | 13 | |
| Lenglet et al., 2012 | Tractography | Projection based subdivisions of the SN, STN, GP and thalamus | 14 |
| Calamante et al., 2012 | TDI | Signal intensity differences within thalamus | 15 |
| POST-MORTEM STUDIES | |||
| Rijkers et al., 2007 | T2w | Visualization of the pulvinar, the lateral and medial geniculate bodies, cerebral peduncle, habenulointerpeduncular tract, periaquaductal gray, the medial lemniscus, the spinothalamic tract, the mammillothalamic tract, and the superior colliculus. | 16:18 |
| Soria et al., 2011 | T1w | Visibility of SN and RN | 19 |
| Massey et al., 2012 | T2w | Hypointense band between SN and STN | 21 |
| High detailed visibility of STN and surrounding | |||
| Intensity differences between anteromedial and posterolateral part of STN | |||
| T2w | Fibers of the subthalamic fasciculus | 20 | |
| Foroutan et al., 2013 | FLASH GE | High-detail images of SN, RN, putamen, and a clear separation of the GP into its external and internal part. | 22 |
The last column refers to the line of Table 3 that gives more details about the scan protocols used. FLASH, fast low angle shot; GE, gradient echo; GP, globus pallidus; RN, red nucleus; SN, substantia nigra; STN, subthalamic nucleus; SWI, susceptibility-weighted imaging; T1w, T1-weighted; T2w, T2-weighted; T2*w, T2*-weighted; TDI, track density imaging; Vim, ventral intermediate nucleus; χ-map, susceptibility map.
In 1999, the basal ganglia were visualized at ultra-high field (8T) using a two-dimensional (2D) multi-slice GE sequence, where high-resolution (195 × 195 μm in-plane) T2*-weighted axial images of one volunteer were obtained in 13 min (Table 3-1) (Bourekas et al., 1999). On these images the globus pallidus (GP), SN and red nucleus (RN) appeared as hypointense regions. These findings were later confirmed in sagittally recorded slices with similar acquisition parameters (Table 3-2) (Novak et al., 2001). In 2003, the same group showed that on GE phase images (Table 3-3), within the SN, the SN pars dorsalis and SN pars lateralis had a higher signal intensity than the matrix of the SN, and within the RN, the medullary lamella showed a higher signal intensity than the RN pars oralis (Abduljalil et al., 2003). A few years later, again the SN and RN appeared hypointense on 7T axial, sagittal, and coronal GE images (Table 3-4) (Cho et al., 2008) and in 2010, 7T coronal GE images (Table 3-38) were obtained on which the STN and SN could be well distinguished (Cho et al., 2010).
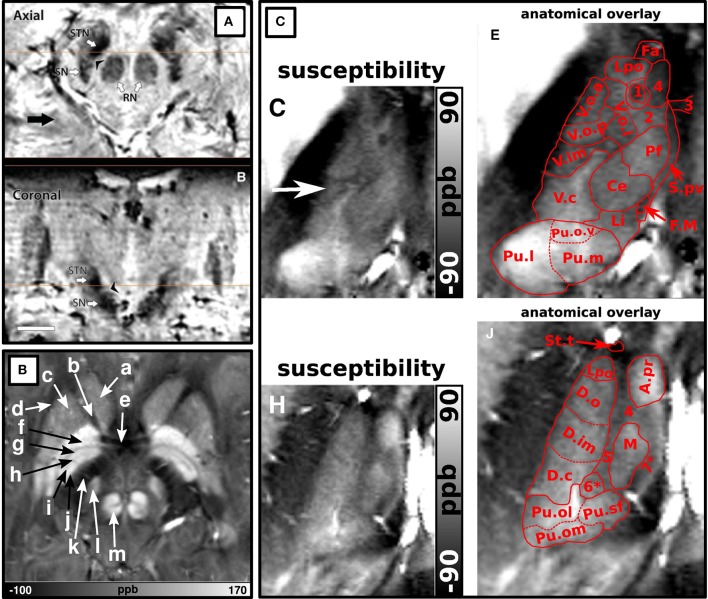
A more detailed description of the visualization of the basal ganglia at 7T with three different scanning sequences, exploiting T1-weighted, T2-weighted and susceptibility-weighted imaging, was published in 2010 (Table 3-5:8) (Abosch et al., 2010). Using SWI, a clear delineation of the STN and the boundary dividing it from the SN were visualized in both axial and coronal planes (Figure 1A). Also, SWI allowed visualization of varying levels of contrast within the RN and two of the laminae within the GP (lamina pallidi medialis and incompleta), thus also distinguishing between the GPi and the GPe. Within the thalamus, it showed intensity variations corresponding to the locations of the ventral intermediate nucleus (Vim), the anterior and medial boundaries of the pulvinar, and the boundary of the nucleus ventralis caudalis as identified with the Schaltenbrand and Wahren atlas (Schaltenbrand et al., 1977).
Figure 1.
Examples of structures identified at ultra-high field. (A) Adopted with permission from Abosch et al. (2010). Ultra-high field (7T) susceptibility-weighted axial and coronal images show a clearly delineated subthalamic nucleus (STN), a boundary between the STN and substantia nigra, and heterogeneous signal intensity in the red nucleus. (B) Adopted with permission from Deistung et al. (2013b). Axial 7T susceptibility map displaying (a) the head of the caudate nucleus, (b) anterior limb of the internal capsule, (c) putamen, (d) external capsule, (e) anterior commissure, (f) external globus pallidus, (g) lamina pallidi medialis, (h) pallidum mediale externum, (i) lamina pallidi incompleta, (j) pallidum mediale internum, (k) posterior limb of internal capsule, (l) subthalamic nucleus, and (m) red nucleus. (C) Adopted with permission from Deistung et al. (2013b). Ultra-high field (7T) susceptibility maps of inferior (C,E) and superior (H,J) sections of the thalamus. (E,J) show overlays of substructures of the thalamus according to the Schaltenbrand et al. (1977) on the images shown in (C,H) respectively. The pulvinar (Pu.l) can be distinguished from (C,E) and the dorsomedial nucleus (M) and dorsal nuclei group (D.o and D.im) can be seen in (H,J).
In 2011, Eapen et al., imaged several deep-brain structures with two different sequences at 7T: T2- and T2*-weighted gradient and spin-echo (GRASE) and T2*-weighted GE (Table 3-9:10) (Eapen et al., 2011). Both GRASE and GE scans showed a clear distinction between the densely and the poorly vascularized regions of the RN, but only the GE scan also showed signal intensity differences within the SN, possibly representing the SN pars compacta and SN pars reticulata. In two later studies, susceptibility maps were investigated. Using a multi-echo GE sequence (Table 3-11), a boundary between the STN and the SN was shown (Schafer et al., 2012). The use of susceptibility maps generated from three single-echo GE phase data sets with different head positions (Table 3-12) also facilitated detailed visualization of structures (Deistung et al., 2013b). It provided discrimination between the subnuclei within the SN, and allowed for accurate discrimination of the STN from the SN and surrounding gray matter and white matter. Furthermore, within the GP, these maps showed the lamina pallidi medialis and lamina pallidi incompleta (Figure 1B). The RN displayed substructures in the susceptibility maps, facilitating identification of the medullary lamella, and the RN pars oralis and RN pars dorsomedialis showed a significantly increased susceptibility, compared to the RN pars caudalis. Finally, within the thalamus clear intensity variations were observed on these susceptibility maps corresponding to the Vim, pulvinar, lateral and medial geniculate nucleus, dorsomedial nucleus, and dorsal nuclei group as identified with the Schaltenbrand and Wahren atlas (Schaltenbrand et al., 1977) (Figure 1C).
In two other studies by Kerl et al., investigating the STN and ZI with different sequences at 7T (Kerl et al., 2012c, 2013), a distinction between the STN and the SN and ZI and a clear boundary dividing the rostral ZI from the internal capsule, STN and the pallidofugal fibers could be seen on T2*-weighted images and the latter also on coronal SW images.
Finally, two studies employed DWI properties to identify substructures within the DBS related structures. In one study, DWI (Table 3-14) was used to estimate the pathways between seven regions of interest: caudate nucleus, putamen, GPe, GPi, SN, STN, and thalamus (Lenglet et al., 2012). Seven pathways could be successfully identified: the nigrostriatal, nigropallidal, nigrothalamic, subthalamopallidal, pallidothalamic, striatopallidal, and thalamostriatal pathway. These projections were also used to create subparcellations of the SN, possibly corresponding to the SN pars reticulata and SN pars compacta; subdivisions of the STN into a dorsolateral and ventromedial part; subdivisions of the GPe into medial, lateral and rostro-ventral parts; subdivisions of the GPi into laterocaudal, rostral, and mid portions; and many subdivisions within the thalamus. In another study, 7T DWI (Table 3-15) was used to construct track-density images of the thalamus (Calamante et al., 2012). These showed high-resolution (200 μm isotropic) substructures within the thalamus with clear intensity differences, not only related to track-density, but also to the directionality of the fibers.
Visualization of deep-brain structures at ultra-high field ex vivo
When scanning ex vivo, even higher resolution and higher SNR can be obtained due to the possibility of longer scan times and less movement artifacts. Although fixed tissue may suffer from altered tissue properties, such as decreases in T1 and T2 (Tovi and Ericsson, 1992) and a decreased diffusion coefficient (D'Arceuil et al., 2007), which is especially challenging for DWI, it also has great advantages over in vivo MRI. Several studies employed ex-vivo imaging for investigating the deep-brain structures at ultra-high field (Table 4).
In 2007, the STN and its surroundings were explored at 9.4T with a T2-weighted sequence (Table 3-16:18) in a post-mortem brain sample (Rijkers et al., 2007). Acquiring a high in-plane resolution of 100 × 100 μm, not only the most prominent structures of the basal ganglia were visualized, but also the pulvinar, the lateral and medial geniculate bodies, cerebral peduncle, habenulointerpeduncular tract (fasciculus retroflexus), periaquaductal gray, the medial lemniscus, the spinothalamic tract, the mammillothalamic tract, and the superior colliculus.
Three post-mortem brain stems have also been imaged at 7T for 119 min, acquiring 150 × 150 μm images. On these T1-weighted images (Table 3-19), the RN and SN, which displayed heterogeneous signal intensity, could be visualized (Soria et al., 2011). Even higher in-plane resolutions of 44 × 44 and 88 × 88 μm (Table 3-20:21) were achieved in a different study after scanning post-mortem brain samples for 72 and 10 h respectively (Massey et al., 2012). The obtained T2-weighted images facilitated visualization of the STN, SN, RN, ZI, and thalamus but also allowed a highly detailed identification of many smaller structures surrounding the STN. Furthermore, a hypointense signal band was seen between the SN and STN facilitating easy separation of the two structures. Also the anteromedial part of the STN was relatively hypointense compared to the posterolateral portion, which might be related to the subdivision of the STN in a limbic, associative and sensorimotor part. On the 44 × 44 μm resolution images even the fibers of the subthalamic fasciculus were visualized accurately.
Finally, one study that focused on differences in T2 and T2* values and iron content between post-mortem brains of progressive supranuclear palsy patients and controls, showed high-resolution (50 μm isotropic) fast low-angle shot (FLASH) GE images (Table 3-22), displaying with much detail the SN, RN, putamen, and the GP with a clear separation into the GPe and GPi (Foroutan et al., 2013).
These studies show that ultra-high field MRI can aid substantially in the identification of small (sub)structures including the separation between the STN and SN and the laminae within the GP both ex vivo and in vivo.
Comparison between sequences for ultra-high field imaging
In addition to the qualitative description of the visibility of deep-brain structures with ultra-high field MRI, comparisons between different sequences and image reconstruction methods have been made (see Table 5).
Table 5.
Overview of comparative studies at ultra-high field.
| Study | Sequences | Line | Measure | Findings |
|---|---|---|---|---|
| Abduljalil et al., 2003 | GE magnitude | 3 | Qualitative | Phase images show additional structures to magnitude images |
| GE SWI | 3 | |||
| GE phase | 3 | Magnitude + Phase ≥ SWI | ||
| Wharton and Bowtell, 2010 | MO χ-map | 23 | Artifacts and Δχ | Least noise related artifact and most accurate Δχ in MO |
| RSO χ-map | 23 | |||
| TSO χ-map | 23 | MO≈RSO≈TSO | ||
| Abosch et al., 2010 | T1w | 5 | Qualitative | SWI > T2w > T1w |
| T2w | 6 | |||
| SWI | 7:8 | |||
| Eapen et al., 2011 | T2w + T2*w | 9 | CNR of RN/VTA | T2w + T2*w > T2*w |
| T2*w | 10 | |||
| Schafer et al., 2012 | T2*w | 11 | CNR | χ-map > T2*w > T2*-map |
| T2*-maps | 11 | |||
| χ-map | 11 | |||
| Kerl et al., 2012c, 2013 | T1w | 24 | SNR STN | T2*w‡ > T1w‡ > SWI-MIP‡ > SWI cor‡ > T2w |
| T2w | 25 | CNR STN | T2*w‡ > SWI-MIP‡ > T2 > SWI > T1w | |
| T2*w | 26:28 | SNR rZI | T2*w‡ > SWI-MIP‡>T1‡>SWI>T2w | |
| SWI | 29 | CNR rZI | T2*w‡ > SWI-MIP‡ >T2>SWI>T1w | |
| SWI-MIP | 29 | |||
| Deistung et al., 2013b | GE magnitude | 12 | Qualitative | χ-map showed most detail |
| GE phase | 12 | |||
| χ-map | 12 | |||
| R2*-map | 13 | |||
| Deistung et al., 2013a | T2w | 30 | CNR SN | χ-map > R2*-map > T2w > R1-map |
| R1-map | 31 | CNR RN | χ-map > R2*-map > T2w > R1-map | |
| R2*-map | 32 | |||
| χ-map | 32 |
The third column refers to the line of Table 3 that gives more details about the scan protocols used. Sequences that give significantly better results than T2-weighted images are denoted with a double dagger (‡). CNR, contrast-to-noise ratio; cor, coronal; GE, gradient echo; MIP, minimum intensity projection; MO, multi-orientation; RN, red nucleus; RSO, regularized single-orientation; rZI, rostral part of zona incerta; SNR, signal-to-noise ratio; STN, subthalamic nucleus; SWI, susceptibility-weighted imaging; T1w, T1-weighted; T2w, T2-weighted; T2*w, T2*-weighted; TSO, threshold based single orientation; VTA, ventral tegmental area; χ-map, susceptibility map.
In a previously mentioned study from 2003, magnitude, phase-weighted magnitude (SWI), and phase images of a GE dataset (Table 3-3), were compared for their capability to visualize (sub)structures (Abduljalil et al., 2003). On magnitude images the SN and RN showed up hypointense and on phase images, substructures within the SN could be distinguished as well. The combined magnitude and phase images added little extra to the magnitude and phase images separately.
Later, in 2010, the mean susceptibility difference (Δχ) between compartments in an agar phantom, and between white matter and deep-brain structures of healthy subjects were compared among three different susceptibility mapping methods applied to GE FLASH images acquired at 7T (Table 3-23) (Wharton and Bowtell, 2010). The mapping methods consisted of (a) a multi-orientation method using images acquired with differing head positions, (b) a regularized single-orientation method, and (c) a threshold-based single-orientation method. Although all three methods showed large Δχ in the GP, SN, RN, internal capsule, putamen and caudate nucleus, the multi-orientation method resulted in the least noise related artifacts and good estimation of Δχ values in the phantom.
In another 2010 study, T1-weighted, T2-weighted, and SW imaging (Table 3-5:8) were compared (Abosch et al., 2010). Most structures were identified in the SW images (see Table 4), followed by the T2-weighted images (Figure 2). The T1-weighted images showed no obvious structures. Eapen et al., also quantitatively compared their T2 + T2*- and T2*-weighted images (Table 3-9:10) (Eapen et al., 2011). No difference between both sequences could be found in the CNR between the SN and ventral tegmental area (VTA) and between the SN and RN, but in the T2 + T2*-weighted images, the CNR between RN and VTA was significantly better than in the T2*-weighted images.
Figure 2.
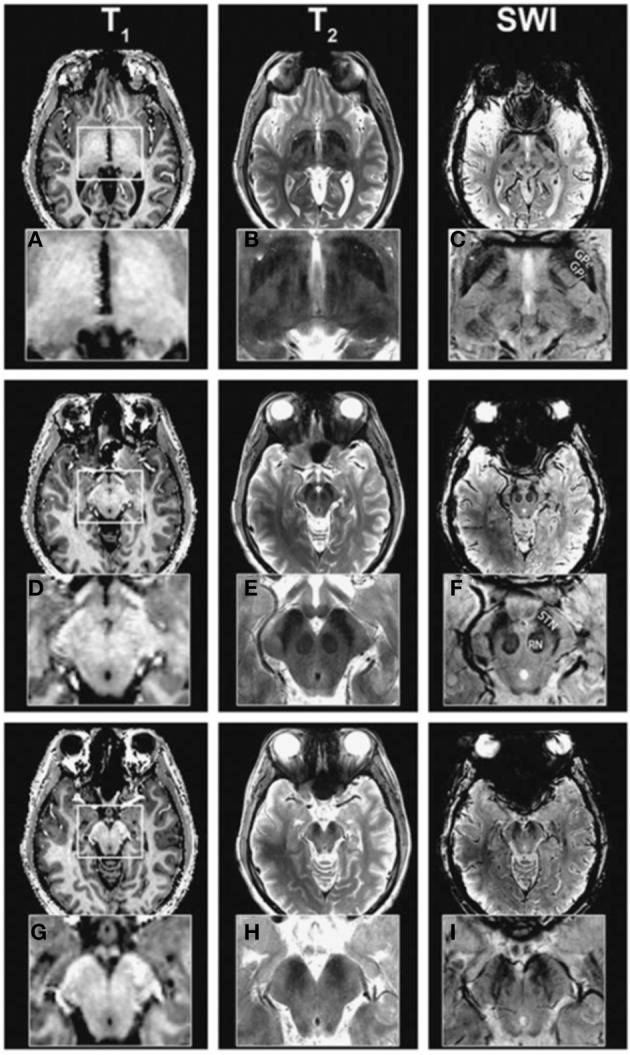
Ultra-high field (7T) T1-weighted (A,D,G), T2-weighted (B,E,H), and susceptibility-weighted (C,F,I) images at different levels. Adopted with permission from Abosch et al. (2010). The susceptibility-weighted images show the highest detail followed by the T2-weighted images.
In 2012, again differently reconstructed images derived from a multi-echo GE sequence (Table 3-11) were compared, consisting of T2*-weighted magnitude images, T2*-maps, and susceptibility maps (Schafer et al., 2012). In most subjects, the CNR between the SN and STN was highest in the susceptibility maps, suggesting that these are most suitable for differentiating the STN from the SN. The SNR of the STN and the rostral part of the ZI (rZI) and the CNR between these structures and white matter, imaged with different sequences, were investigated in two recent studies that compared T1-weighted GE, T2-weighted TSE, T2*-weighted FLASH and SW images (Table 3-24:29) (Kerl et al., 2012c, 2013). Furthermore, minimum intensity projections (MIPs) of the SW images were computed. After adjusting the SNR and CNR for differences in voxel size, they were highest on the T2*-weighted images for both structures. Furthermore, the SNRs of both structures on the T2*-weighted, T1-weighted, SWI-MIP (and for the STN also on the coronal SW images) were significantly higher than those of the T2-weighted images. The CNRs of both structures on the T2*-weighted and for the rZI also on the SWI-MIP images were also significantly higher than on the T2-weighted images. Also, a 2013 study compared image reconstruction techniques at 7T consisting of (a) magnitude, (b) frequency, and (c) susceptibility maps derived from GE scans (Table 3-12), and (d) R*2 maps derived from multi-echo GE scans (Table 3-13) (Deistung et al., 2013b). Qualitative analysis by a neuroanatomist revealed that susceptibility maps in general facilitated the most detailed visualization of structures. Finally, in a recent study by the same group, the CNR between several brain stem structures and their surroundings were compared between sequences (Table 3-30:32) (Deistung et al., 2013a). For the RN and the SN, the CNR of the R2*-map and the susceptibility map outperformed those of the R1-map and the T2-weighted image.
Although comparison between studies is difficult due to the differences in scanning conditions, the majority of these studies show that sequences that are sensitive to magnetic susceptibility such as SWI and T2* related images are most suitable for targeting basal ganglia structures and their subdivisions in DBS at ultra-high field.
Comparison between field strengths
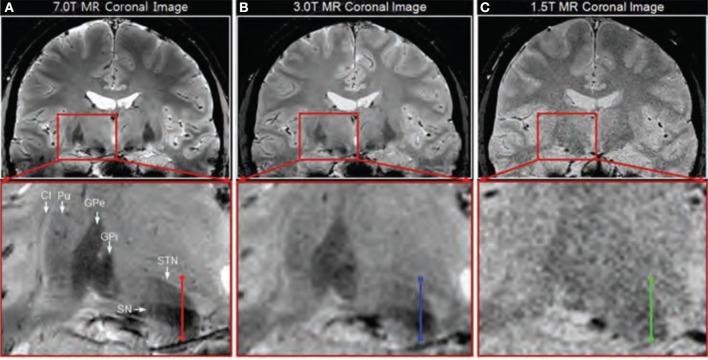
In addition to comparisons between different sequences, some studies compared similar sequences between different field strengths (see Table 6). In a 2008 study, the difference between a 7T GE image (Table 3-4) and a 1.5 T image was briefly treated (Cho et al., 2008). Visual inspection showed that the 7T image displayed better contrast, SNR and resolution. However, comparison is difficult because the acquisition parameters of the 1.5T image were unfortunately not provided. In the same year, T2*-weighted GE images were investigated, acquired at several echo times at three different field strengths: 1.5T, 3T, and 7T (Table 3-33:35) (Yao et al., 2009). This showed that increasing field strength resulted in a higher influence of iron on the value of R2*, making this contrast useful for iron-rich deep-brain structures, such as the GP, RN, SN, and putamen (Hallgren and Sourander, 1958). A thorough quantitative investigation of the visibility of the STN related to field strength was performed in 2010 (Cho et al., 2010), comparing the contrast between the STN and a baseline (containing the ZI and thalamus), the contrast between the STN and SN, the SNR in gray matter areas, and the slope of signal increase between STN and baseline among 1.5T, 3T, and 7T T2*-weighted GE images (Table 3-36:38). At higher field strengths, the STN, SN, putamen, GPi, and GPe could be visualized while the boundaries of these structures were unclear on the 1.5T images (Figure 3). Furthermore, all quantitative measures increased with field strength, and the SNR and contrast were significantly improved at 7T compared to 1.5 and 3T. Finally, the two studies by Kerl et al., investigating the STN and rZI at 7T (Kerl et al., 2012c, 2013) were additionally performed at 3T. Again, they compared the SNR and CNR of these structures between different sequences: T1-weighted MPRAGE, T2-weighted fluid attenuated inversion recovery (FLAIR), T2-weighted sampling perfection with application of optimized contrasts using different flip angle evolutions (SPACE), two T2*-weighted 2D FLASH (FLASH2D) sequences, and SW images and their MIPs (Table 3-39:46) (Kerl et al., 2012a,b). This makes it possible to compare the SNRs and CNRs of the different studies between field strengths, when adjusted for voxel size, although it should be noted that for the T1- and T2-weighted images different sequences were used between field strengths. For both structures, the SNRs of the T2*-weighted, SWI-MIP and SW images of the 7T images were higher than those of the 3T images, but the SNRs of the 3T T2-weighted SPACE image and T1-weighted images were higher at 3T than at 7T. However, the CNRs of both structures were substantially higher on all the 7T sequences than on the corresponding 3T sequences.
Table 6.
Overview of studies that compare scan protocols between field strengths.
| Study | Sequence | Line | Measure | Findings |
|---|---|---|---|---|
| Cho et al., 2008 | 1. 1.5T | 4 | Qualitative | 7T has better contrast, SNR and resolution than 1.5T |
| 2. 7T T2*w | ||||
| Yao et al., 2009 | 1. 1.5T T2*w | 33 | R2* | R2* becomes more sensitive to iron with increasing field strength |
| 2. 3T T2*w | 34 | |||
| 3. 7T T2*w | 35 | |||
| Cho et al., 2010 | 1. 1.5T T2*w | 36 | Contrast | 7T‡>3T>1.5T |
| 2. 3T T2*w | 37 | Slope of signal increase | 7T>3T>1.5T | |
| 3. 7T T2*w | 38 | SNR | 7T‡>3T>1.5T | |
| Kerl et al., 2012a,b,c, 2013 | 1. 3T T1w | 39 | SNR | 3T T1w > 7T T1w |
| 2. 3T T2w FLAIR | 40 | 7T T2*w > 3T T2*w | ||
| 3. 3T T2w SPACE | 41 | 7T SWI-MIP > 3T SWI-MIP axial | ||
| 4. 3T T2*w | 41:45 | 3T T2w SPACE > 7T T2w > 3T T2w FLAIR | ||
| 5. 3T SWI | 46 | 7T SWI > 3T SWI | ||
| 6. 3T SWI-MIP | 46 | |||
| 7. 7T T1w | 24 | CNR | 7T T2*w > 3T T2*w | |
| 8. 7T T2w TSE | 25 | 7T SWI-MIP > 3T SWI-MIP | ||
| 9. 7T T2*w | 26:28 | 7T T2w > 3T T2w SPACE > 3T T2w FLAIR | ||
| 10. 7T SWI | 29 | 7T SWI > 3T SWI | ||
| 11. 7T SWI-MIP | 29 | 7T T1 > 3T T1 |
The third column refers to the line of Table 3 that gives more details about the scan protocols used. Sequences that significantly improve imaging at 7T compared to 1.5T and 3T are denoted with a double dagger (‡). CNR, contrast-to-noise ratio; FLAIR, fluid attenuated inversion recovery; MIP, minimum intensity projection; rZI, rostral part of zona incerta; SNR, signal-to-noise ratio; SPACE, sampling perfection with application of optimized contrasts using different flip angle evolutions; STN, subthalamic nucleus; SWI, susceptibility-weighted imaging; T1w, T1-weighted; T2w, T2-weighted; T2*w, T2*-weighted; TSE, turbo spin-echo.
Figure 3.
Coronal T2*-weighted images obtained at 7.0T (A), 3.0T (B), and 1.5T (C). Adapted with permission from Cho et al. (2010). Visual inspection shows clearer identification of the substantia nigra (SN), subthalamic nucleus (STN), internal globus pallidus (GPi), external globus pallidus (GPe), and putamen (Pu) at 7T compared to 3T and 1.5T.
These studies suggest that 7T MRI can better facilitate accurate targeting of deep brain structures than 1.5T or 3T MRI.
Discussion
Accurate visualization of deep-brain structures is important to improve our understanding of their anatomy, connectivity and function, and for improved surgical targeting for DBS in movement and psychiatric disorders. To date, targeting based on direct visualization of DBS targets with T2-weighted 1.5T or 3T MRI can be difficult. However, studies at ultra-high field showed good visibility of these structures on SW images based on T2* and phase contrast. Structures that have been identified at ultra-high field include: a separation between the STN and SN (Abosch et al., 2010; Cho et al., 2010; Massey et al., 2012; Schafer et al., 2012; Deistung et al., 2013b); the lamina pallidi medialis and lamina pallidi incompleta within the GP (Abosch et al., 2010; Deistung et al., 2013b); a subdivision of the STN in two halves (Lenglet et al., 2012; Massey et al., 2012); subdivisions of the SN possibly representing the SN pars reticulata and SN pars compacta (Eapen et al., 2011; Lenglet et al., 2012; Deistung et al., 2013b); substructures in the RN (Abosch et al., 2010; Eapen et al., 2011) including the medullary lamella (Abduljalil et al., 2003; Deistung et al., 2013b), RN pars oralis (Abduljalil et al., 2003), and RN pars caudalis (Deistung et al., 2013b); and several regions in the thalamus (Lenglet et al., 2012) including the Vim (Abosch et al., 2010; Deistung et al., 2013b), the pulvinar (Deistung et al., 2013b) and its anterior and medial boundaries (Abosch et al., 2010), the boundary of the nucleus ventralis caudalis (Abosch et al., 2010), the lateral and medial geniculate nucleus (Deistung et al., 2013b), the dorsomedial nucleus (Deistung et al., 2013b) and the dorsal nuclei group (Deistung et al., 2013b). Furthermore, 7T T2*-weighted and SW images have displayed improved CNR, SNR and resolution in the deep-brain regions, compared to 1.5T and 3T images (Cho et al., 2010; Kerl et al., 2012a,b,c, 2013).
Based on a descriptive evaluation of different MR images, more and smaller structures can be identified on T2*-weighted, GE phase, SW images, and susceptibility and R2* maps than on T1- and T2-weighted images (Abduljalil et al., 2003; Abosch et al., 2010; Kerl et al., 2012c, 2013; Deistung et al., 2013b). Although quantitative comparison between studies is difficult due to variations in scan protocols, the CNRs of deep-brain structures on T2* and SW images and corresponding maps are generally higher than those of T2- and T1-weighted images (Kerl et al., 2012c, 2013). For the SNR, the same trend can be seen, although T1-weighted images seem to have a higher SNR than SW images (Kerl et al., 2012c, 2013).
Perspectives
The improved visualization of the basal ganglia with ultra-high field MRI discussed here provides good perspectives for clinical practice. The clear delineation of DBS target structures and their possible subdivisions may aid in more accurate targeting, which may reduce negative side effects and shorten surgery duration, or it may even allow surgery under general anesthesia. Furthermore, ultra-high field MRI also shows potential for more accurate diagnosis and monitoring of basal ganglia diseases due to, for example, improved identification of the SN pars compacta and SN pars reticulata, which may in its turn facilitate improved patient specific treatments.
In addition, ultra-high field MRI promises to be a versatile tool in clinically oriented research of the deep brain nuclei. It might help us to improve our current understanding of the functionality of the healthy basal ganglia and its disease processes with high resolution functional MRI and connectivity analyses.
Recommendations
When in the end considering the optimal scan protocol for visualizing the DBS targets for clinical purposes at ultra-high field, both image quality and practical requirements need to be taken into account. In terms of hardware, it is recommended to use a head coil with a high number of receive channels (i.e., 16 or higher). This has been shown to improve the SNR (de Zwart et al., 2004; Wiggins et al., 2006) which is also reflected from the studies described in Table 3. In terms of scan protocol, based on the described literature, we recommend to use a 3D multi-echo GE sequence with an isotropic resolution of 0.5 mm3 and partial brain coverage. The 3D sequence facilitates small and isotropic voxel sizes, which ensures good resolution in every plane which is important for distinguishing the STN from the SN. From the multi-echo GE scan, both T2*-weighted and susceptibility weighted images as well as T2*-maps, R2*-maps, and susceptibility maps can be computed, which were shown in the reviewed literature to display best basal ganglia visibility. Since the basal ganglia are located within the same axial oblique slab of approximately 4–5 cm thickness, we advise to shorten scan time by covering only this part of the brain. If more time reduction is required, partial Fourier imaging, elliptical k-space coverage, or parallel imaging can be considered as well.
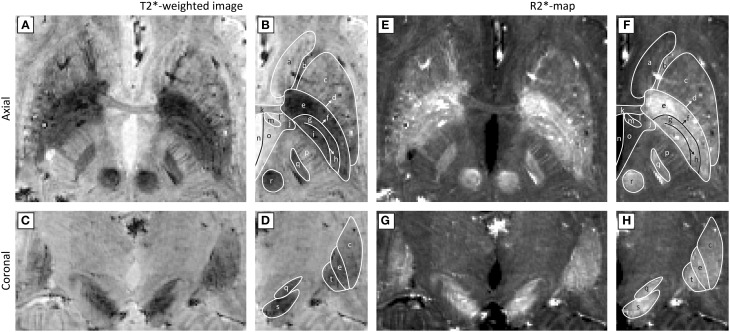
To support these guidelines, Figure 4 shows an example of a T2*-weighted image and an R2*-map created with these recommendations. The images were obtained by scanning a healthy volunteer on a 7T MR scanner (Magnetom 7T, Siemens, Erlangen, Germany) at Scannexus (Maastricht, The Netherlands) using a 32-channel phased-array coil (Nova Medical, Wilmington, United States) with a multi-echo 3D GE sequence. Scan time was reduced to 12 min and 23 s by partial brain coverage and 75% partial Fourier imaging (other scanning parameters can be found in Table 3–line 47:48). On these 0.5 mm3 isotropic resolution images, the STN can be distinguished from the SN in the coronal plane, and the three laminae of the GP can be identified.
Figure 4.
Ultra-high field (7T) axial (A,B,E,F) and coronal (C,D,G,H) T2*-weighted images (A–D) and R2*-maps (E–H). Panels (B,D,F,H) show the anatomical structures that can be identified with the Schaltenbrand and Wahren atlas (Schaltenbrand and Wahren, 2005): (a) caudate nucleus, (b) anterior limb of internal capsule, (c) putamen, (d) lamina pallidi lateralis, (e) external globus pallidus, (f) lamina pallidi medialis, (g) pallidum mediale externum, (h) lamina pallidi incompleta, (i) pallidum mediale internum, (j) inferior thalamic peduncle, (k) anterior commissure, (l) prothalamus, (m) fornix, (n) third ventricle, (o) hypothalamus, (p) posterior limb of internal capsule, (q) subthalamic nucleus, (r) red nucleus, (s) substantia nigra, (t) internal globus pallidus. (Courtesy D. Ivanov).
When planning a DBS surgery, the MR images are often registered to CT images, resulting in images that both display the stereotactic frame from the CT image as well as contrast within the brain. This registration may be more reliable, however if a whole brain MR image is available as an intermediate step. Abosch et al. (2010) showed that it is already possible to perform 1 mm3 whole brain T1-weighted imaging in 3.5 min, which may be a good candidate for coregistration.
Limitations
Despite these promising results concerning accurate and high-resolution visualization of the small deep brain (sub)structures, several issues still need to be addressed before they can routinely be employed in direct targeting for DBS.
Firstly, ultra-high field images have an increased risk of geometrical distortions compared to 1.5T images. The severity of these distortions at 7T in deep-brain regions has been investigated in several studies. One study compared the coordinates of marker points in a phantom imaged with 1.5T and 7T MRI to their locations on computed tomography (CT) images (Dammann et al., 2011). The maximum distortion in either x-, y-, or z-direction at 7T was 1.6 mm, which was slightly larger than at 1.5T (0.9 mm). Furthermore, the fewest distortions were observed in the center of the phantom. In another study the distortions in an anthropomorphic phantom between T2*-weighted 7T MR and CT images were investigated, revealing a maximum deviation of 0.78 mm (Cho et al., 2010). Finally, registration of 7T T1- and T2-weighted images of the midbrain of PD patients to 1.5T T1- and T2-weighted images showed that mainly rigid body transformations were required and that scaling and skew deformations were small (Duchin et al., 2012). Furthermore, the midbrain region, containing many DBS targets, required the least correction. Quantitative comparison showed that the distances of the T2-weighted images were significantly less than 1 mm suggesting that affine registration of T1- and T2-weighted 7T images to CT images can already provide MR images with midbrain distortions comparable to those of 1.5T images. These few studies suggest that at 7T images can be acquired with distortions smaller than 1 mm in the deep-brain areas.
Secondly, some of the mentioned imaging techniques pose additional challenges in the clinical context. Most studies were performed on young and healthy volunteers. In patients, movement during image acquisition can be less controlled, counteracting the gain in SNR and spatial specificity obtained with ultra-high field. However, newer techniques, such as prospective motion correction might remedy this problem (Maclaren et al., 2013). This approach monitors movement in the scanner with high accuracy and corrects the new image acquisition adaptively according to the new head position. That is, even with large head movements—as observed in many patients—the resulting images are already coregistered and movement artifact free.
In addition, the availability of ultra-high field MR scanners is currently limited. Firstly, the number of scanners that have been installed in the world is limited itself (see Table 2), which is inherent to its high cost in purchase and in operation. Secondly, due to the novel status of ultra-high field MRI, safety precautions regarding metallic objects are often more strict than on 3T systems and the use of ultra-high field MRI is currently only allowed for research purposes.
Finally, direct targeting in DBS suffers from brain shift, intra-operative deformation of the brain compared to preoperative MR images due to difference in head position and cerebrospinal fluid loss. Without compensation for this, it will eventually still limit targeting accuracy. However, this effect is independent of the magnetic field strength and even the pre-operative imaging modality.
Conclusion
Ultra-high field MRI can reliably and accurately display subdivisions within the basal ganglia and related structures, which especially benefits from T2*- and phase-related contrasts. If the limitations concerning image distortions and the availability of the scanners are solved, these technical advances have the potential to improve accuracy of targeting in DBS surgery and the clinical outcome.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the Joint Scientific Thematic Research Programme (JSTP) of the Netherlands Organization for Scientific Research (NWO) and by the Limburg University Fund/Foundation for Higher Education in Limburg (SWOL). The authors would like to thank Björn Falkenburger for his contribution to the manuscript.
Glossary
Abbreviations
- CNR
contrast-to-noise ratio
- DBS
deep brain stimulation
- DWI
diffusion-weighted imaging
- FLAIR
fluid attenuated inversion recovery
- FLASH
fast low-angle shot
- GE
gradient echo
- GP
globus pallidus
- GPe
external globus pallidus
- GPi
internal globus pallidus
- GRASE
gradient and spin-echo
- MIP
minimum intensity projection
- MPRAGE
magnetization-prepared rapid acquisition of gradient echo
- PD
Parkinson's disease
- PIAF
parallel imaging acceleration factor
- RN
red nucleus
- rZI
rostral part of ZI
- SE
spin-echo
- SN
substantia nigra
- SNR
signal-to-noise ratio
- SPACE
sampling perfection with application of optimized contrasts using different flip angle evolutions
- SW
susceptibility-weighted
- SWI
susceptibility-weighted imaging
- TDI
track-density imaging
- TE
echo time
- TR
repetition time
- TSE
turbo spin-echo
- ZI
zona incerta.
References
- Abduljalil A. M., Schmalbrock P., Novak V., Chakeres D. W. (2003). Enhanced gray and white matter contrast of phase susceptibility-weighted images in ultra-high-field magnetic resonance imaging. J. Magn. Reson. Imaging 18, 284–290. 10.1002/jmri.10362 [DOI] [PubMed] [Google Scholar]
- Abosch A., Yacoub E., Ugurbil K., Harel N. (2010). An assessment of current brain targets for deep brain stimulation surgery with susceptibility-weighted imaging at 7 tesla. Neurosurgery 67, 1745–1756. discussion: 56. 10.1227/NEU.0b013e3181f74105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermans L., Temel Y., Visser-Vandewalle V. (2008). Deep brain stimulation in Tourette's syndrome. Neurotherapeutics 5, 339–344. 10.1016/j.nurt.2008.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrubla J., Neuner I., Hahn D., Boers F., Shah N. J. (2013). Recording visual evoked potentials and auditory evoked P300 at 9.4T static magnetic field. PLoS ONE 8:e62915. 10.1371/journal.pone.0062915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustinack J. C., van der Kouwe A. J., Blackwell M. L., Salat D. H., Wiggins C. J., Frosch M. P., et al. (2005). Detection of entorhinal layer II using 7Tesla [corrected] magnetic resonance imaging. Ann. Neurol. 57, 489–494. 10.1002/ana.20426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender B., Manz C., Korn A., Nagele T., Klose U. (2011). Optimized 3D magnetization-prepared rapid acquisition of gradient echo: identification of thalamus substructures at 3T. AJNR Am. J. Neuroradiol. 32, 2110–2115. 10.3174/ajnr.A2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrington A., Jezzard P., Clare S., Emir U. (2014). Two-voxel Hadamard encoded semi-LASER spectroscopy for in vivo MRS at ultra-high field, in Joint Annual Meeting ISMRM-ESMRMB (Milan, Italy: ). [Google Scholar]
- Bhagat Y. A., Rajapakse C. S., Magland J. F., Love J. H., Wright A. C., Wald M. J., et al. (2011). Performance of muMRI-Based virtual bone biopsy for structural and mechanical analysis at the distal tibia at 7T field strength. J. Magn. Reson. Imaging 33, 372–381. 10.1002/jmri.22439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant N., Cloos M. A., Amadon A. (2011). B1 and B0 inhomogeneity mitigation in the human brain at 7 T with selective pulses by using average Hamiltonian theory. Magn. Reson. Med. 65, 680–691. 10.1002/mrm.22658 [DOI] [PubMed] [Google Scholar]
- Bourekas E. C., Christoforidis G. A., Abduljalil A. M., Kangarlu A., Chakeres D. W., Spigos D. G., et al. (1999). High resolution MRI of the deep gray nuclei at 8 Tesla. J. Comput. Assist. Tomogr. 23, 867–874. 10.1097/00004728-199911000-00009 [DOI] [PubMed] [Google Scholar]
- Brunenberg E. J., Platel B., Hofman P. A., Ter Haar Romeny B. M., Visser-Vandewalle V. (2011). Magnetic resonance imaging techniques for visualization of the subthalamic nucleus. J. Neurosurg. 115, 971–984. 10.3171/2011.6.JNS101571 [DOI] [PubMed] [Google Scholar]
- Brunenberg E. J. L., Moeskops P., Backes W. H., Pollo C., Cammoun L., Vilanova A., et al. (2012). Structural and resting state functional connectivity of the subthalamic nucleus: identification of motor STN parts and the hyperdirect pathway. PLoS ONE 7:e39061. 10.1371/journal.pone.0039061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde J., Shajan G., Scheffler K., Pohmann R. (2014). Ultra-high resolution imaging of the human brain using acquisition-weighted imaging at 9.4T. Neuroimage 86, 592–598. 10.1016/j.neuroimage.2013.08.013 [DOI] [PubMed] [Google Scholar]
- Buxton R. B. (2013). The physics of functional magnetic resonance imaging (fMRI). Rep. Prog. Phys. 76:096601. 10.1088/0034-4885/76/9/096601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamante F., Oh S. H., Tournier J. D., Park S. Y., Son Y. D., Chung J. Y., et al. (2012). Super-resolution track-density imaging of thalamic substructures: comparison with high-resolution anatomical magnetic resonance imaging at 7.0T. Hum. Brain Mapp. 34, 2538–2548. 10.1002/hbm.22083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamante F., Tournier J. D., Jackson G. D., Connelly A. (2010). Track-density imaging (TDI): super-resolution white matter imaging using whole-brain track-density mapping. Neuroimage 53, 1233–1243. 10.1016/j.neuroimage.2010.07.024 [DOI] [PubMed] [Google Scholar]
- Cho Z.-H., Kim Y.-B., Han J.-Y., Min H.-K., Kim K.-N., Choi S.-H., et al. (2008). New brain atlas—Mapping the human brain in vivo with 7.0 T MRI and comparison with postmortem histology: will these images change modern medicine? Int. J. Imaging Syst. Technol. 18, 2–8 10.1002/ima.20143 [DOI] [Google Scholar]
- Cho Z. H., Min H. K., Oh S. H., Han J. Y., Park C. W., Chi J. G., et al. (2010). Direct visualization of deep brain stimulation targets in Parkinson disease with the use of 7-tesla magnetic resonance imaging. J. Neurosurg. 113, 639–647. 10.3171/2010.3.JNS091385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos M., Wiggins C., Wiggins G., Sodickson D. (2014). Plug and play parallel transmission at 7 and 9.4 Tesla based on principles from MR fingerprinting, in Joint Annual Meeting ISMRM-ESMRMB (Milan, Italy: ). [Google Scholar]
- Costagli M., Kelley D. A., Symms M. R., Biagi L., Stara R., Maggioni E., et al. (2014). Tissue border enhancement by inversion recovery MRI at 7.0 Tesla. Neuroradiology 56, 517–523. 10.1007/s00234-014-1365-8 [DOI] [PubMed] [Google Scholar]
- Damier P., Thobois S., Witjas T., Cuny E., Derost P., Raoul S., et al. (2007). Bilateral deep brain stimulation of the globus pallidus to treat tardive dyskinesia. Arch. Gen. Psychiatry 64, 170–176. 10.1001/archpsyc.64.2.170 [DOI] [PubMed] [Google Scholar]
- Dammann P., Kraff O., Wrede K. H., Ozkan N., Orzada S., Mueller O. M., et al. (2011). Evaluation of hardware-related geometrical distortion in structural MRI at 7 Tesla for image-guided applications in neurosurgery. Acad. Radiol. 18, 910–916. 10.1016/j.acra.2011.02.011 [DOI] [PubMed] [Google Scholar]
- D'Arceuil H. E., Westmoreland S., de Crespigny A. J. (2007). An approach to high resolution diffusion tensor imaging in fixed primate brain. Neuroimage 35, 553–565. 10.1016/j.neuroimage.2006.12.028 [DOI] [PubMed] [Google Scholar]
- de Bresser J., Brundel M., Conijn M. M., van Dillen J. J., Geerlings M. I., Viergever M. A., et al. (2013). Visual cerebral microbleed detection on 7T MR imaging: reliability and effects of image processing. AJNR Am. J. Neuroradiol. 34, E61–E64. 10.3174/ajnr.A2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelchand D. K., van de Moortele P. F., Adriany G., Iltis I., Andersen P., Strupp J. P., et al. (2010). In vivo 1H NMR spectroscopy of the human brain at 9.4 T: initial results. J. Magn. Reson. 206, 74–80. 10.1016/j.jmr.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deistung A., Schafer A., Schweser F., Biedermann U., Gullmar D., Trampel R., et al. (2013a). High-resolution MR imaging of the human brainstem in vivo at 7 Tesla. Front. Hum. Neurosci. 7:710. 10.3389/fnhum.2013.00710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deistung A., Schafer A., Schweser F., Biedermann U., Turner R., Reichenbach J. R. (2013b). Toward in vivo histology: a comparison of quantitative susceptibility mapping (QSM) with magnitude-, phase-, and R2*-imaging at ultra-high magnetic field strength. Neuroimage 65, 299–314. 10.1016/j.neuroimage.2012.09.055 [DOI] [PubMed] [Google Scholar]
- Denney T. S., Bolding M., Beyers R., Salibi N., Li M., Zhang X., et al. (2014). Differential motion in orbital and global layers of extraocular muscles measured by tagged MRI At 7T, in Joint Annual Meeting ISMRM-ESMRMB (Milan, Italy: ). [Google Scholar]
- Denys D., Mantione M., Figee M., van den Munckhof P., Koerselman F., Westenberg H., et al. (2010). Deep brain stimulation of the nucleus accumbens for treatment-refractory obsessive-compulsive disorder. Arch. Gen. Psychiatry 67, 1061–1068. 10.1001/archgenpsychiatry.2010.122 [DOI] [PubMed] [Google Scholar]
- Deuschl G., Schade-Brittinger C., Krack P., Volkmann J., Schafer H., Botzel K., et al. (2006). A randomized trial of deep-brain stimulation for Parkinson's disease. N. Engl. J. Med. 355, 896–908. 10.1056/NEJMoa060281 [DOI] [PubMed] [Google Scholar]
- de Zwart J. A., Ledden P. J., van Gelderen P., Bodurka J., Chu R., Duyn J. H. (2004). Signal-to-noise ratio and parallel imaging performance of a 16-channel receive-only brain coil array at 3.0 Tesla. Magn. Reson. Med. 51, 22–26. 10.1002/mrm.10678 [DOI] [PubMed] [Google Scholar]
- Dieringer M. A., Renz W., Lindel T., Seifert F., Frauenrath T., von Knobelsdorff-Brenkenhoff F., et al. (2011). Design and application of a four-channel transmit/receive surface coil for functional cardiac imaging at 7T. J. Magn. Reson. Imaging 33, 736–741. 10.1002/jmri.22451 [DOI] [PubMed] [Google Scholar]
- Duchin Y., Abosch A., Yacoub E., Sapiro G., Harel N. (2012). Feasibility of using ultra-high field (7 T) MRI for clinical surgical targeting. PLoS ONE 7:e37328. 10.1371/journal.pone.0037328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyn J. H. (2012). The future of ultra-high field MRI and fMRI for study of the human brain. Neuroimage 62, 1241–1248. 10.1016/j.neuroimage.2011.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzyubachyk O., Lelieveldt B. P., Blaas J., Reijnierse M., Webb A., van der Geest R. J. (2013). Automated algorithm for reconstruction of the complete spine from multistation 7T MR data. Magn. Reson. Med. 69, 1777–1786. 10.1002/mrm.24404 [DOI] [PubMed] [Google Scholar]
- Eapen M., Zald D. H., Gatenby J. C., Ding Z., Gore J. C. (2011). Using high-resolution MR imaging at 7T to evaluate the anatomy of the midbrain dopaminergic system. AJNR Am. J. Neuroradiol. 32, 688–694. 10.3174/ajnr.A2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R., Salanova V., Witt T., Worth R., Henry T., Gross R., et al. (2010). Electrical stimulation of the anterior nucleus of thalamus for treatment of refractory epilepsy. Epilepsia 51, 899–908. 10.1111/j.1528-1167.2010.02536.x [DOI] [PubMed] [Google Scholar]
- Follett K. A., Torres-Russotto D. (2012). Deep brain stimulation of globus pallidus interna, subthalamic nucleus, and pedunculopontine nucleus for Parkinson's disease: which target? Parkinsonism Relat. Disord. 18(Suppl. 1), S165–S167 10.1016/S1353-8020(11)70051-7 [DOI] [PubMed] [Google Scholar]
- Foroutan P., Murray M. E., Fujioka S., Schweitzer K. J., Dickson D. W., Wszolek Z. K., et al. (2013). Progressive supranuclear palsy: high-field-strength MR microscopy in the human substantia nigra and globus pallidus. Radiology 266, 280–288. 10.1148/radiol.12102273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitan M. I., Sati P., Inati S. J., Reich D. S. (2013). Initial investigation of the blood-brain barrier in MS lesions at 7 tesla. Mult. Scler. 19, 1068–1073. 10.1177/1352458512471093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover P. M., Cavin I., Qian W., Bowtell R., Gowland P. A. (2007). Magnetic-field-induced vertigo: a theoretical and experimental investigation. Bioelectromagnetics 28, 349–361. 10.1002/bem.20316 [DOI] [PubMed] [Google Scholar]
- Goubran M., Rudko D. A., Santyr B., Gati J., Szekeres T., Peters T. M., et al. (2014). In vivo normative atlas of the hippocampal subfields using multi-echo susceptibility imaging at 7 Tesla. Hum. Brain Mapp. 35, 3588–3601. 10.1002/hbm.22423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haacke E. M., Reichenbach J. R. (2011). Susceptibility Weighted Imaging in MRI: Basic Concepts and Clinical Applications. Hoboken, NJ: Wiley. [Google Scholar]
- Haase A., Frahm J., Matthaei D., Hanicke W., Merboldt K. D. (2011). FLASH imaging: rapid NMR imaging using low flip-angle pulses. 1986. J. Magn. Reson. 213, 533–541. 10.1016/j.jmr.2011.09.021 [DOI] [PubMed] [Google Scholar]
- Hahn A., Kranz G. S., Seidel E. M., Sladky R., Kraus C., Kublbock M., et al. (2013). Comparing neural response to painful electrical stimulation with functional MRI at 3 and 7 T. Neuroimage 82, 336–343. 10.1016/j.neuroimage.2013.06.010 [DOI] [PubMed] [Google Scholar]
- Hallgren B., Sourander P. (1958). The effect of age on the non-haemin iron in the human brain. J. Neurochem. 3, 41–51. 10.1111/j.1471-4159.1958.tb12607.x [DOI] [PubMed] [Google Scholar]
- He L., Zuo Z., Chen L., Humphreys G. (2014). Effects of number magnitude and notation at 7T: separating the neural response to small and large, symbolic and nonsymbolic number. Cereb. Cortex 24, 2199–2209. 10.1093/cercor/bht074 [DOI] [PubMed] [Google Scholar]
- Hennig J., Nauerth A., Friedburg H. (1986). RARE imaging: a fast imaging method for clinical MR. Magn. Reson. Med. 3, 823–833. 10.1002/mrm.1910030602 [DOI] [PubMed] [Google Scholar]
- Hoffmann M. B., Stadler J., Kanowski M., Speck O. (2009). Retinotopic mapping of the human visual cortex at a magnetic field strength of 7T. Clin. Neurophysiol. 120, 108–116. 10.1016/j.clinph.2008.10.153 [DOI] [PubMed] [Google Scholar]
- Hoffmann S. H., Begovatz P., Nagel A. M., Umathum R., Schommer K., Bachert P., et al. (2011). A measurement setup for direct 17O MRI at 7 T. Magn. Reson. Med. 66, 1109–1115. 10.1002/mrm.22871 [DOI] [PubMed] [Google Scholar]
- Intrapiromkul J., Zhu H., Cheng Y., Barker P. B., Edden R. A. (2013). Determining the in vivo transverse relaxation time of GABA in the human brain at 7T. J. Magn. Reson. Imaging 38, 1224–1229. 10.1002/jmri.23979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D., Poser B., Huber L., Pfeuffer J., Uludağ K. (2014). Whole-brain perfusion measurements at 7T using pulsed arterial spin labelling and simultaneous multi-slice multi-echo echo planar imaging, in Joint Annual Meeting ISMRM-ESMRMB (Milan, Italy: ). [Google Scholar]
- Kabasawa H., Nabetani A., Matsuzawa H., Nakada T. (2006). Imaging optimization for in-vivo human micro imaging at 7T, in Joint Annual Meeting ISMRM-ESMRMB (Seattle, Washington: ). [Google Scholar]
- Kanowski M., Voges J., Tempelmann C. (2010). Delineation of the nucleus centre median by proton density weighted magnetic resonance imaging at 3 T. Neurosurgery 66(3 Suppl. Operative), E121–E123. discussion: E3. 10.1227/01.NEU.0000348560.85056.63 [DOI] [PubMed] [Google Scholar]
- Kerchner G. A., Deutsch G. K., Zeineh M., Dougherty R. F., Saranathan M., Rutt B. K. (2012). Hippocampal CA1 apical neuropil atrophy and memory performance in Alzheimer's disease. Neuroimage 63, 194–202. 10.1016/j.neuroimage.2012.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerl H. U., Gerigk L., Brockmann M. A., Huck S., Al-Zghloul M., Groden C., et al. (2013). Imaging for deep brain stimulation: the zona incerta at 7 Tesla. World J. Radiol. 5, 5–16. 10.4329/wjr.v5.i1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerl H. U., Gerigk L., Huck S., Al-Zghloul M., Groden C., Nolte I. S. (2012b). Visualisation of the zona incerta for deep brain stimulation at 3.0 Tesla. Clin. Neuroradiol. 22, 55–68. 10.1007/s00062-012-0136-3 [DOI] [PubMed] [Google Scholar]
- Kerl H. U., Gerigk L., Pechlivanis I., Al-Zghloul M., Groden C., Nolte I. (2012a). The subthalamic nucleus at 3.0 Tesla: choice of optimal sequence and orientation for deep brain stimulation using a standard installation protocol: clinical article. J. Neurosurg. 117, 1155–1165. 10.3171/2012.8.JNS111930 [DOI] [PubMed] [Google Scholar]
- Kerl H. U., Gerigk L., Pechlivanis I., Al-Zghloul M., Groden C., Nölte I. S. (2012c). The subthalamic nucleus at 7.0 Tesla: evaluation of sequence and orientation for deep-brain stimulation. Acta Neurochir. (Wien). 154, 2051–2062. 10.1007/s00701-012-1476-0 [DOI] [PubMed] [Google Scholar]
- Kickler N., van der Zwaag W., Mekle R., Kober T., Marques J. P., Krueger G., et al. (2010). Eddy current effects on a clinical 7T-68 cm bore scanner. MAGMA 23, 39–43. 10.1007/s10334-009-0192-0 [DOI] [PubMed] [Google Scholar]
- Le Bihan D. (2003). Looking into the functional architecture of the brain with diffusion MRI. Nat. Rev. Neurosci. 4, 469–480. 10.1038/nrn1119 [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Deogaonkar M., Rezai A. (2007). Deep brain stimulation of globus pallidus internus for dystonia. Parkinsonism Relat. Disord. 13, 261–265. 10.1016/j.parkreldis.2006.07.020 [DOI] [PubMed] [Google Scholar]
- Lenglet C., Abosch A., Yacoub E., De Martino F., Sapiro G., Harel N. (2012). Comprehensive in vivo mapping of the human basal ganglia and thalamic connectome in individuals using 7T MRI. PLoS ONE 7:e29153. 10.1371/journal.pone.0029153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limousin P., Martinez-Torres I. (2008). Deep brain stimulation for Parkinson's disease. Neurotherapeutics 5, 309–319. 10.1016/j.nurt.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Eskreis-Winkler S., Schweitzer A. D., Chen W., Kaplitt M. G., Tsiouris A. J., et al. (2013). Improved subthalamic nucleus depiction with quantitative susceptibility mapping. Radiology 269, 216–223. 10.1148/radiol.13121991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfipour A. K., Wharton S., Schwarz S. T., Gontu V., Schafer A., Peters A. M., et al. (2012). High resolution magnetic susceptibility mapping of the substantia nigra in Parkinson's disease. J. Magn. Reson. Imaging 35, 48–55. 10.1002/jmri.22752 [DOI] [PubMed] [Google Scholar]
- Lu A., Atkinson I. C., Zhou X. J., Thulborn K. R. (2013). PCr/ATP ratio mapping of the human head by simultaneously imaging of multiple spectral peaks with interleaved excitations and flexible twisted projection imaging readout trajectories at 9.4 T. Magn. Reson. Med. 69, 538–544. 10.1002/mrm.24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclaren J., Herbst M., Speck O., Zaitsev M. (2013). Prospective motion correction in brain imaging: a review. Magn. Reson. Med. 69, 621–636. 10.1002/mrm.24314 [DOI] [PubMed] [Google Scholar]
- Maia T. V., Cooney R. E., Peterson B. S. (2008). The neural bases of obsessive-compulsive disorder in children and adults. Dev. Psychopathol. 20, 1251–1283. 10.1017/S0954579408000606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone D. A., Jr., Dougherty D. D., Rezai A. R., Carpenter L. L., Friehs G. M., Eskandar E. N., et al. (2009). Deep brain stimulation of the ventral capsule/ventral striatum for treatment-resistant depression. Biol. Psychiatry 65, 267–275. 10.1016/j.biopsych.2008.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mang S. C., Busza A., Reiterer S., Grodd W., Klose A. U. (2012). Thalamus segmentation based on the local diffusion direction: a group study. Magn. Reson. Med. 67, 118–126. 10.1002/mrm.22996 [DOI] [PubMed] [Google Scholar]
- Massey L. A., Miranda M. A., Zrinzo L., Al-Helli O., Parkes H. G., Thornton J. S., et al. (2012). High resolution MR anatomy of the subthalamic nucleus: imaging at 9.4 T with histological validation. Neuroimage 59, 2035–2044. 10.1016/j.neuroimage.2011.10.016 [DOI] [PubMed] [Google Scholar]
- Metcalf M., Xu D., Okuda D. T., Carvajal L., Srinivasan R., Kelley D. A., et al. (2010). High-resolution phased-array MRI of the human brain at 7 tesla: initial experience in multiple sclerosis patients. J. Neuroimaging 20, 141–147. 10.1111/j.1552-6569.2008.00338.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink J. W. (2006). Neurobiology of basal ganglia and Tourette syndrome: basal ganglia circuits and thalamocortical outputs. Adv. Neurol. 99, 89–98. [PubMed] [Google Scholar]
- Moon C. H., Kim J. H., Zhao T., Bae K. T. (2013). Quantitative (23) Na MRI of human knee cartilage using dual-tuned (1) H/(23) Na transceiver array radiofrequency coil at 7 tesla. J. Magn. Reson. Imaging 38, 1063–1072. 10.1002/jmri.24030 [DOI] [PubMed] [Google Scholar]
- Mori S., Crain B. J., Chacko V. P., van Zijl P. C. (1999). Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann. Neurol. 45, 265–269. [DOI] [PubMed] [Google Scholar]
- Nolte I. S., Gerigk L., Al-Zghloul M., Groden C., Kerl H. U. (2012). Visualization of the internal globus pallidus: sequence and orientation for deep brain stimulation using a standard installation protocol at 3.0 Tesla. Acta Neurochir. (Wien) 154, 481–494. 10.1007/s00701-011-1242-8 [DOI] [PubMed] [Google Scholar]
- Novak P., Novak V., Kangarlu A., Abduljalil A. M., Chakeres D. W., Robitaille P. M. (2001). High resolution MRI of the brainstem at 8 T. J. Comput. Assist. Tomogr. 25, 242–246. 10.1097/00004728-200103000-00016 [DOI] [PubMed] [Google Scholar]
- Obeso J. A., Marin C., Rodriguez-Oroz C., Blesa J., Benitez-Temino B., Mena-Segovia J., et al. (2008). The basal ganglia in Parkinson's disease: current concepts and unexplained observations. Ann. Neurol. 64(Suppl. 2), S30–S46. 10.1002/ana.21481 [DOI] [PubMed] [Google Scholar]
- Pakin S. K., Cavalcanti C., La Rocca R., Schweitzer M. E., Regatte R. R. (2006). Ultra-high-field MRI of knee joint at 7.0T: preliminary experience. Acad. Radiol. 13, 1135–1142. 10.1016/j.acra.2006.06.007 [DOI] [PubMed] [Google Scholar]
- Pan J. W., Avdievich N., Hetherington H. P. (2010). J-refocused coherence transfer spectroscopic imaging at 7 T in human brain. Magn. Reson. Med. 64, 1237–1246. 10.1002/mrm.22534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouratian N., Zheng Z., Bari A. A., Behnke E., Elias W. J., Desalles A. A. (2011). Multi-institutional evaluation of deep brain stimulation targeting using probabilistic connectivity-based thalamic segmentation. J. Neurosurg. 115, 995–1004. 10.3171/2011.7.JNS11250 [DOI] [PubMed] [Google Scholar]
- Ren J., Lakoski S., Haller R. G., Sherry A. D., Malloy C. R. (2013). Dynamic monitoring of carnitine and acetylcarnitine in the trimethylamine signal after exercise in human skeletal muscle by 7T 1H-MRS. Magn. Reson. Med. 69, 7–17. 10.1002/mrm.24249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers K., Temel Y., Visser-Vandewalle V., Vanormelingen L., Vandersteen M., Adriaensens P., et al. (2007). The microanatomical environment of the subthalamic nucleus. Technical note. J. Neurosurg. 107, 198–201. 10.3171/JNS-07/07/0198 [DOI] [PubMed] [Google Scholar]
- Robitaille P. M., Warner R., Jagadeesh J., Abduljalil A. M., Kangarlu A., Burgess R. E., et al. (1999). Design and assembly of an 8 tesla whole-body MR scanner. J. Comput. Assist. Tomogr. 23, 808–820. 10.1097/00004728-199911000-00002 [DOI] [PubMed] [Google Scholar]
- Sato H., Kawagishi K. (2014). Labyrinthine artery detection in patients with idiopathic sudden sensorineural hearing loss by 7-T MRI. Otolaryngol. Head Neck Surg. 150, 455–459. 10.1177/0194599813517990 [DOI] [PubMed] [Google Scholar]
- Schafer A., Forstmann B. U., Neumann J., Wharton S., Mietke A., Bowtell R., et al. (2012). Direct visualization of the subthalamic nucleus and its iron distribution using high-resolution susceptibility mapping. Hum. Brain Mapp. 33, 2831–2842. 10.1002/hbm.21404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaltenbrand G., Wahren W. (2005). Atlas for Stereotaxy of the Human Brain: with an Accompanying Guide. Stuttgart: Thieme. [Google Scholar]
- Schaltenbrand G., Wahren W., Hassler R. (1977). Atlas for Stereotaxy of the Human Brain, 2nd Edn. Stuttgart: Thieme. [Google Scholar]
- Schuepbach W. M., Rau J., Knudsen K., Volkmann J., Krack P., Timmermann L., et al. (2013). Neurostimulation for Parkinson's disease with early motor complications. N. Engl. J. Med. 368, 610–622. 10.1056/NEJMoa1205158 [DOI] [PubMed] [Google Scholar]
- Soria G., De Notaris M., Tudela R., Blasco G., Puig J., Planas A. M., et al. (2011). Improved assessment of ex vivo brainstem neuroanatomy with high-resolution MRI and DTI at 7 Tesla. Anat. Rec. (Hoboken) 294, 1035–1044. 10.1002/ar.21383 [DOI] [PubMed] [Google Scholar]
- Temel Y., Blokland A., Steinbusch H. W. M., Visser-Vandewalle V. (2005). The functional role of the subthalamic nucleus in cognitive and limbic circuits. Prog. Neurobiol. 76, 393–413. 10.1016/j.pneurobio.2005.09.005 [DOI] [PubMed] [Google Scholar]
- Theysohn J. M., Maderwald S., Kraff O., Moenninghoff C., Ladd M. E., Ladd S. C. (2008). Subjective acceptance of 7 Tesla MRI for human imaging. MAGMA 21, 63–72. 10.1007/s10334-007-0095-x [DOI] [PubMed] [Google Scholar]
- Tovi M., Ericsson A. (1992). Measurements of T1 and T2 over time in formalin-fixed human whole-brain specimens. Acta Radiol. 33, 400–404. 10.3109/02841859209172021 [DOI] [PubMed] [Google Scholar]
- Unrath A., Klose U., Grodd W., Ludolph A. C., Kassubek J. (2008). Directional colour encoding of the human thalamus by diffusion tensor imaging. Neurosci. Lett. 434, 322–327. 10.1016/j.neulet.2008.02.013 [DOI] [PubMed] [Google Scholar]
- Vaughan J. T., Garwood M., Collins C. M., Liu W., DelaBarre L., Adriany G., et al. (2001). 7T vs. 4T: RF power, homogeneity, and signal-to-noise comparison in head images. Magn. Reson. Med. 46, 24–30. 10.1002/mrm.1156 [DOI] [PubMed] [Google Scholar]
- Vedrine P., Aubert G., Belorgey J., Berriaud C., Bourquard A., Bredy P., et al. (2014). Manufacturing of the Iseult/INUMAC Whole Body 11.7 T MRI Magnet. IEEE Trans. Appl. Supercond. 24, 1–6 10.1109/TASC.2013.2286256 [DOI] [Google Scholar]
- Wharton S., Bowtell R. (2010). Whole-brain susceptibility mapping at high field: a comparison of multiple- and single-orientation methods. Neuroimage 53, 515–525. 10.1016/j.neuroimage.2010.06.070 [DOI] [PubMed] [Google Scholar]
- Wichmann T., Dostrovsky J. O. (2011). Pathological basal ganglia activity in movement disorders. Neuroscience 198, 232–244. 10.1016/j.neuroscience.2011.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegell M. R., Tuch D. S., Larsson H. B., Wedeen V. J. (2003). Automatic segmentation of thalamic nuclei from diffusion tensor magnetic resonance imaging. Neuroimage 19(2 Pt 1), 391–401. 10.1016/S1053-8119(03)00044-2 [DOI] [PubMed] [Google Scholar]
- Wiggins G. C., Triantafyllou C., Potthast A., Reykowski A., Nittka M., Wald L. L. (2006). 32-channel 3 Tesla receive-only phased-array head coil with soccer-ball element geometry. Magn. Reson. Med. 56, 216–223. 10.1002/mrm.20925 [DOI] [PubMed] [Google Scholar]
- Wodarg F., Herzog J., Reese R., Falk D., Pinsker M. O., Steigerwald F., et al. (2012). Stimulation site within the MRI-defined STN predicts postoperative motor outcome. Mov. Disord. 27, 874–879. 10.1002/mds.25006 [DOI] [PubMed] [Google Scholar]
- Wyss M., Bruegger M., Daeubler B., Vionnet L., Brunner D., Pruessmann K. (2014). Visualization of human brainstem substructures using gray matter nulling 3D-MPRAGE at 7Tesla, in Joint Annual Meeting ISMRM-ESMRMB (Milan, Italy: ). [Google Scholar]
- Yao B., Li T. Q., Gelderen P., Shmueli K., de Zwart J. A., Duyn J. H. (2009). Susceptibility contrast in high field MRI of human brain as a function of tissue iron content. Neuroimage 44, 1259–1266. 10.1016/j.neuroimage.2008.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama T., Ando N., Sugiyama K., Akamine S., Namba H. (2006). Relationship of stimulation site location within the subthalamic nucleus region to clinical effects on parkinsonian symptoms. Stereotact. Funct. Neurosurg. 84, 170–175. 10.1159/000094956 [DOI] [PubMed] [Google Scholar]
- Yovel Y., Assaf Y. (2007). Virtual definition of neuronal tissue by cluster analysis of multi-parametric imaging (virtual-dot-com imaging). Neuroimage 35, 58–69. 10.1016/j.neuroimage.2006.08.055 [DOI] [PubMed] [Google Scholar]