Abstract
Serotonin 2C receptors (5-HT2CR) are G-protein-coupled receptors with various actions, including involvement in drug addiction. 5-HT2CR undergoes mRNA editing, converting genomically encoded adenosine residues to inosines via adenosine deaminases acting on RNA (ADARs). Here we show that enhanced alcohol drinking behaviour in mice is associated with the degree of 5-HT2CR mRNA editing in the nucleus accumbens and dorsal raphe nuceus, brain regions important for reward and addiction. Following chronic alcohol vapour exposure, voluntary alcohol intake increased in C57BL/6J mice, but remained unchanged in C3H/HeJ and DBA/2J mice. 5-HT2CR mRNA editing frequency in both regions increased significantly in C57BL/6J mice, as did expressions of 5-HT2CR, ADAR1 and ADAR2, but not in other strains. Moreover, mice that exclusively express the unedited isoform (INI) of 5-HT2CR mRNA on a C57BL/6J background did not exhibit increased alcohol intake compared with wild-type mice. Our results indicate that alterations in 5-HT2CR mRNA editing underlie alcohol preference in mice.
Keywords: Alcohol drinking, dorsal raphe nucleus, nucleus accumbens, RNA editing, serotonin 2C receptor
Introduction
Serotonin 2C receptors (5-HT2CR) are broadly expressed in the central nervous system (Abramowski et al., 1995; Clemett et al., 2000) and modulate behavioural and physiological processes, including emotion, locomotion, appetite and metabolic rate (Giorgetti and Tecott, 2004). 5-HT2CR is also linked to addictive drug behaviour, such as sensitization, conditioned place preference and drug self-administration (Muller and Huston, 2006). We recently demonstrated that 5-HT2CR in the nucleus accumbens (ACC) is involved in increased alcohol intake after chronic alcohol exposure in C57BL/6J mice (Yoshimoto et al., 2012). This increase might be caused by abnormalities of the levels of monoamines, dopamine, 5-HT and their metabolites in the ACC and the dorsal raphe nucleus (DRN) during chronic alcohol exposure (Yoshimoto et al., 2012). Moreover, local injection of the 5-HT2CR antagonist into the ACC suppressed voluntary alcohol-drinking behaviour in the alcohol-exposed mice, supporting an alteration in 5-HT2CR activity in the ACC being involved in this behaviour (Yoshimoto et al, 2012). Dopamine and serotonin (5-HT) in the ACC of the brain reward pathway were implicated in the mechanisms underlying abnormal alcohol drinking behaviour and the development of alcohol dependence (Yoshimoto et al, 1992a). 5-HT2CR also regulates the mesolimbic dopamine circuit (Navailles et al, 2008), and the ACC is important for the reinforcing action of alcohol (Koob et al., 1998a, b). The major source of 5-HT in the ACC is the 5-HT neurons in the DRN (Van Bockstaele et al, 1993), which is also involved in reward and alcohol intake (Pistis et al, 1997; Miyazaki et al, 2012). Furthermore, DRN electrical stimulation on dopamine release in the ACC are regulated via 5-HT2CR, suggesting that 5-HT2CR has an important role in the DRN-ACC circuit (De Deurwaerdere and Spampinato, 1999). Therefore, an alteration in 5-HT2CR activity after chronic alcohol exposure might cause aberration in this circuit, resulting in the enhancement of alcohol drinking.
Recently, epigenetic mechanisms, such as alterations in chromatin structure induced by histone tail modification and DNA methylation, and the regulation of gene expression by microRNA, were reported to be involved in the development of addiction (Robison and Nestler, 2011). The 5-HT2CR is a G-protein coupled receptor containing seven transmembrane domains. It is subjected to RNA editing at five sites (A, B, C, D and E) of the second intracellular loop by adenosine deaminases acting on RNA (ADARs), converting adenosine to inosine. Combinational editing at these five sites changes three amino acids (I156, N158, and I160) of the unedited receptor (INI), resulting in expression of 24 different editing isoforms in specific brain regions (Wang et al., 2000b; Nishikura, 2006; Werry et al., 2008). In vitro studies showed that RNA editing modulates receptor functions, including 5-HT potency, agonist binding affinity, constitutive activities and G protein coupling activity (Burns et al., 1997; Fitzgerald et al., 1999; Niswender et al., 1999; Wang et al., 2000b; Gurevich et al., 2002a), suggesting that RNA editing of 5-HT2CR mRNA may modulate serotonergic systems in the brain that have causative relevance to neuropsychiatric disorders (Maas et al., 2006; Werry et al., 2008). Moreover, the extent of 5-HT2CR mRNA editing that occurs in response to stress or the 5-HT selective reuptake inhibitor fluoxetine depends on the genetic background of mice (Englander et al., 2005). Inbred strains of mice (C57L/6J, C3H/He, and DBA/2Cr) are known to vary in voluntary alcohol consumption (Yoshimoto and Komura, 1989). To date there have been no studies investigating alcohol preference with regard to 5-HT2CR mRNA editing.
In the present study, we examined the involvement of 5-HT2CR expression and mRNA editing in alcohol drinking behaviour of these three strains of mice. The editing and expression of 5-HT2CR mRNAs were significantly increased in the ACC and the DRN of C57BL/6J mice that showed enhanced alcohol drinking behaviour following chronic alcohol exposure. Increased RNA editing in these regions resulted in a prevalence of the 5-HT2CR VNV isoform with lower basal activity. These results were dependent on the genetic background of the mice examined, as C3H/HeJ and DBA/2J mice did not show increased alcohol intake or increased 5-HT2CR mRNA editing and expression in the ACC and the DRN. Moreover, by examining the INI mutant mice, which express solely the unedited INI isoform of 5-HT2CR (Kawahara et al., 2008), we confirmed our observation that 5-HT2CR mRNA editing is crucial in determining alcohol drinking behaviour.
Materials and methods
Animals
C57BL/6J, C3H/HeJ and DBA/2J inbred male mice (CLEA Japan, Japan) were maintained on a 12 h light/dark cycle at 25 °C. Heterozygous female 5T-HT2CR-INI mice bred on a C57BL/6J back ground, were mated with wild-type male mice. Male littermates of mixed genotypes were housed in groups of seven per cage under a 12 h light/dark cycle with ad libitum food and water. Genotyping of 5-HT2CR was carried out with a PCR-based assay using primers, 5′-ACTGATACTACCATCACTGG-3′ and 5′-AGCATATATAGGAAATTGCAGTAACCCT-3′ (Kawahara et al., 2008). PCR products were digested with HindIII and separated by 2% agarose gel electrophoresis. All procedures were conducted in strict adherence to the Care and Use Guidelines of the Committee of Laboratory Animals, Kyoto Prefectural University of Medicine, Japan.
Chronic alcohol exposure model and alcohol-drinking behaviour test
Chronically alcohol-exposed mice were produced as previously described (Yoshimoto et al., 2012). Briefly, mice were exposed to 22–27 mg/l of ethanol vapour for 20 d using an intermittent 3–6 h/d schedule that mimics cyclical patterns of alcohol consumption. After alcohol exposure, mice were withdrawn from alcohol for 5 h in normal experimental conditions. For the alcohol-drinking behaviour test, mice were provided with 10% (v/v) ethanol solution for 4 h, and their consumption was measured. The total amount of alcohol intake and alcohol intake rate were represented as ethanol (g)/body weight (kg) and as ethanol (g)/body weight (kg)/hour, respectively.
Measurement of RNA editing efficiency
Coronal mouse brain sections were obtained by 2 mm punch biopsy and quickly frozen in liquid nitrogen (N=6 in each group). cDNAs were synthesized as previously described (Watanabe et al., 2009). Total RNAs were prepared using an RNAqueous Kit (Applied Biosystems, USA) and cDNAs were then synthesized using a Transcriptor First-Strand cDNA Synthesis Kit (Roche, Switzerland). cDNAs from each group were mixed in an equal volume, and the segment spanning the edited sites of 5-HT2CR (374–846 bp) was amplified by KOD FX DNA polymerase (Toyobo, Japan), using primers 5′-GATATTTGTGCCCCGTCTGG-3′ and 5′-GGGGTTGG-GAGCGTTCTCTT-3′. For the recovery of normal splicing form, 470 bp of PCR products were separated on lowmelting agarose after adding 3′-A overhangs using the A-attachment Mix (Toyobo). PCR products were ligated with pGEM-T easy vector (Promega, USA) and transformed into E. coli. 5-HT2CR cDNA was amplified from an individual colony using the M13-20 primer (5′-CGACGTTGTAAAACGACGGCCAGT-3′) and the M13 reverse primer (5′-CAGGAAACAGCTATGAC-3′), and then sequenced using the Big Dye Terminator v1 sequencing kit (Applied Biosystems).
Measurement of 5-HT2CR receptor activity
Expression vectors of 5-HT2CR isoforms (INI, VNI, VSI, VNV, VDV, VSV and VGV) were constructed by site-directed mutagenesis. First, we cloned a cDNA encoding the VNV isoform into the BamHI-EcoRV site of pEF1-mycHisB using the primer set 5′-GGATCCGCCAC-CATGGTGAACCTGGGCACT-3′/5′-GATATCCACACTA-CTAATCCTCTCGC-3′. Subsequently, other isoforms were generated by site-directed mutagenesis using mutation primers (Supplementary Table S1). Amplification was performed using KOD-Plus polymerase (Toyobo, Japan), followed by blunting with T4 DNA polymerase (New England Biolabs, USA) and DpnI-digestion. Amplified DNA was self-ligated using the DNA Ligation Kit Ver.2 (Takara Biomedicals, Japan) and transformed into the E. coli Top10 strain. The mutations were verified by sequencing.
For measurement of receptor activity, these plasmids were introduced into HeLa cells by an electroporator (CUY21; NEPA Gene, Japan). After 30 h, cells were washed with inositol-free DMEM (MP Biomedical, USA) and placed in inositol-free DMEM containing 1 μCi myo-[3H]inositol for 18 h; followed by the addition of 5-HT. One hour after 5-HT stimulation, cells were harvested with 10 mM formic acid, and the cell extract was applied to a column of Dowex 1×8 (Cl-form). The column was washed with 60mM sodium formate containing 5mM borax and then eluted with 100 mM formic acid containing 1 M ammonium formate. [3H]-labeled inositol phosphates were quantitated by liquid scintillation counting (Packard Tri-Carb 2700). The dose–response curve was fitted by a four-parameter logistic model (MasterPlex-ReaderFit; Hitachi Solutions, Japan).
Real-time PCR
Real-time quantitative PCR was performed using the Thermal Cycler Dice Real Time System (TaKaRa Bio, Japan) with SYBR Premix Ex Taq II (TaKaRa Bio). PCR reactions were carried out using 40 cycles of two-step PCR: 5 s at 95 °C and 30 s at 60 °C. For quantification of 5-HT2CR, ADAR1 or ADAR2 expression, the following primer sets were used: 5′-ATGCCCCTGTCTCTGCTTGC-3′/5′-GCATAGCCGGTTCAATTCGC-3′, 5′-ATCCTGGAT-GGCACCAGAGG-3′/5′-GAAACGCGCCCCACTATCAG-3′, and 5′-GGTGACGCCACCATTGAGGT-3′/5′-AGTGAAC-AGACGGGCCTTGG-3′, respectively. Based on analyses and calculations integrating adequate standard curves, expression levels of these genes were normalized to glyceraldehyde-3-phosphate dehydrogenase (G3PDH) expression, determined using primers 5′-GGCATTGCTCT-CAATGACAA-3′/5′-TGTGAGGGAGATGCTCAGTG-3′.
Immunoblot analysis
Coronal mouse brain sections (2 mm punch biopsy) were homogenized in RIPA buffer (50 mM Tris-Cl (pH7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), and clear lysates separated on 10% polyacrylamide gels. Proteins were then transferred onto a PVDF membrane (Millipore, USA). After blocking, the membranes were incubated with anti-ADAR1 (1:500; sc-73408; Santa Cruz Biotechnology, USA) or anti-ADAR2 (1:500; sc-73409; Santa Cruz Biotechnology) monoclonal antibody for 12 h at 25 °C. The membranes were then incubated with alkaline phosphatase-conjugated anti-mouse IgG (1:5000 dilution; Jackson ImmunoResearch, USA) for 1 h. Immunopositive signals were detected with nitrobluetetrazolium chloride and 5-bromo-4-chloro-3′- indolylphosphatase p-toluidine salt reagents.
Statistics
Statistical analyses were conducted using GraphPad Prism5 or Statview5 software. Differences in editing frequencies, amino acid frequencies and protein isoforms obtained from sequence analyses of clones were analysed using Fisher’s exact test. Real-time PCR data were tested for significance using ANOVA (Tukey post-hoc). Statistical analysis of alcohol drinking behaviour was performed using Student’s t-test, Mann–Whitney U-test, two-way ANOVA or ANOVA with repeated measures. The significance of differences between two groups was calculated by Student’s t-test, except when there was a significant difference between the variances of the two groups (F-test), in which case the Mann–Whitney U-test was used. Statistical significance was set at p<0.05.
Results
Increased RNA editing of 5-HT2CR following chronic alcohol exposure
Genetic factors contribute to alcohol preference and aberrant alcohol drinking behaviour (Lumeng et al., 1995). Among inbred mouse strains, C57BL/6J mice exhibit the highest alcohol preference, while the C3H/HeJ and DBA/2J strains have low alcohol preferences (McClearn, 1968; Yoshimoto and Komura, 1989). We measured alcohol intake of these strains of mice after chronic alcohol exposure. The mice were intermittently exposed to alcohol by inhalation for a period of 20 d, and then the intake of 10% ethanol was measured after recovery in ambient air for 5 h. Although the rates of alcohol metabolism were moderately different among the inbred mouse strains due to different levels of alcohol dehydrogenase in the DBA/2J mice (Sheppard et al., 1968), the blood alcohol levels of all strains were estimated to be zero within 3 h after recovery (Supplementary Fig. S1). There was, however, a significant increase in alcohol intake following chronic alcohol exposure in C57BL/6J mice (Fig. 1a; B6, p=0.009) compared to controls without chronic exposure. Neither the C3H/HeJ nor the DBA/2J mice, showed increased ethanol consumption after alcohol exposure (Fig. 1a; C3 and D2). Chronic alcohol exposure had no effect on water intake (Fig. 1b) or body weight (Supplementary Fig. S2) in any of the mouse strains. These results suggest there is a difference in alcohol intake among the three inbred strains of mice examined.
Fig. 1.
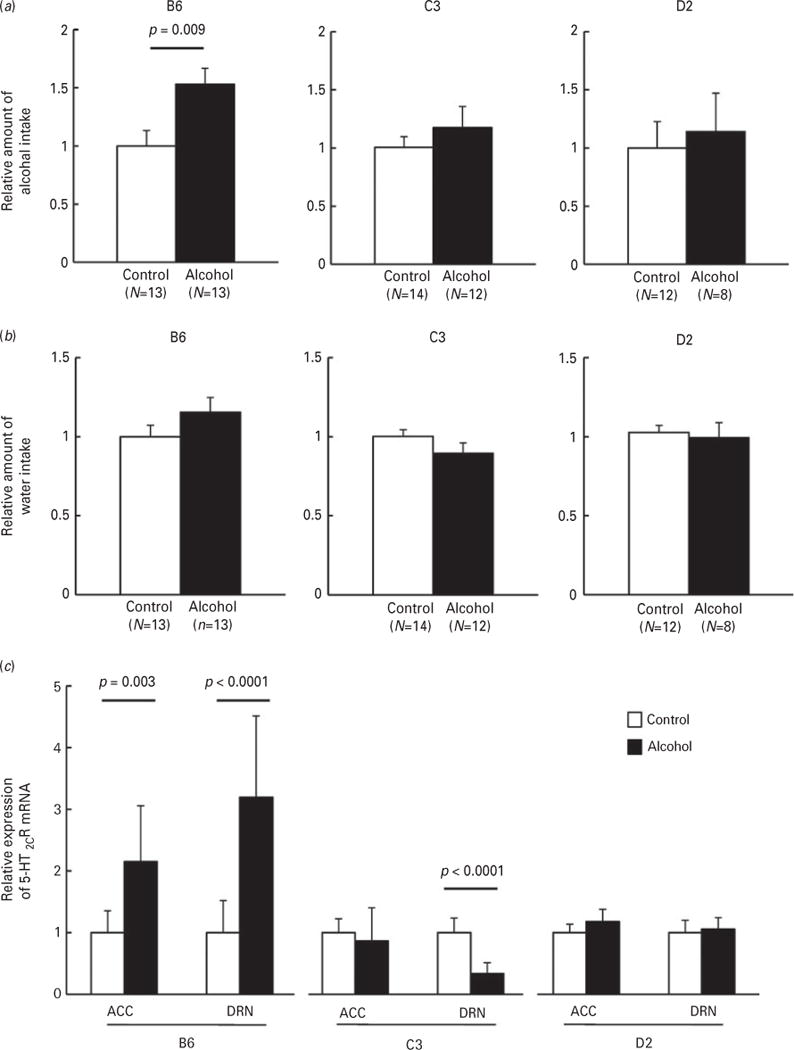
Comparison of alcohol intake among inbred mouse strains. Control (open bars) and chronic alcohol-exposed (solid bars) inbred mice of the C57BL/6J (B6), C3H/HeJ (C3) and DBA/2J (D2) strains were returned to ambient air for 5 h prior to the measurement of the intake of 10% ethanol (a) or water (b). Consumption of ethanol or water is expressed relative to control. Statistical analyses were performed using Student’s t-test or Mann–Whitney’s U-test. (c) Relative expressions of 5-HT2CR mRNA in the ACC and DRN of the three mouse strains were quantified by real-time PCR. Data are presented as mean±S.D. N=6–10 per group. Statistical analyses were performed using one-way ANOVA (Tukey post-hoc).
We previously reported that 5-HT2CR expression in the ACC and DRN is increased after chronic alcohol exposure (Yoshimoto et al., 2012); therefore, we examined whether chronic alcohol exposure resulted in increased 5-HT2CR expression in the ACC or DRN of mouse strains with different alcohol drinking behaviours. Although several splicing variant forms of 5-HT2CR have been reported (Flomen et al., 2004), the designed primer set allowed the amplification of all forms. Consistent with our previous data, the relative expression of 5-HT2CR mRNA was significantly increased in the ACC and the DRN of C57BL/6J mice after chronic alcohol exposure (Fig. 1c; B6-ACC, p=0.003; B6-DRN, p<0.0001). This increase was not observed in either the C3H/HeJ or DBA/2J mice with low alcohol consumptions (Fig. 1c; C3 and D2). However, there was a significant decrease in 5-HT2CR expression level in the DRN of C3H/HeJ mice (Fig. 1c; C3-DRN, p<0.0001). These results indicate the strain difference in the influence of chronic alcohol exposure on 5HT2CR expression.
It is well known that 5-HT2CR mRNA undergoes A-to-I RNA editing at five sites (sites A–E), resulting in the translation of multiple editing isoform receptors (Fig. 2a). Altered 5-HT2CR mRNA editing is observed in patients with mental disorders, including suicide victims, and also in animal models of depression, forced swimming, water maze and maternal separation (Iwamoto et al., 2009). Thus, we investigated whether 5-HT2CR mRNA editing was influenced by chronic alcohol exposure. Editing frequencies of 5-HT2CR mRNA in the ACC and the DRN were measured by cloning–sequencing analysis of RT-PCR products (Burns et al., 1997). In the ACC of alcohol-preferring C57BL/6J mice, the editing frequency at site D was significantly increased after chronic alcohol exposure (Fig. 2b; ACC, p=0.035, Fisher’s exact test). There also tended to be a slight increase in the editing frequencies at sites A and B in alcohol-exposed mice (Fig. 2b; ACC). Similarly in the DRN, the editing frequency at site B was significantly increased after chronic alcohol exposure (Fig. 2b; DRN, p=0.007, Fisher’s exact test). However, 5-HT2CR-editing did not increase throughout the entire brain; for example, there was no significant change in the hippocampus (Supplementary Fig. S3). By contrast, there were no significant increases in the editing frequencies at sites B and D in either the C3H/HeJ or DBA/2J mice after chronic alcohol exposure (Fig. 2c and d).
Fig. 2.
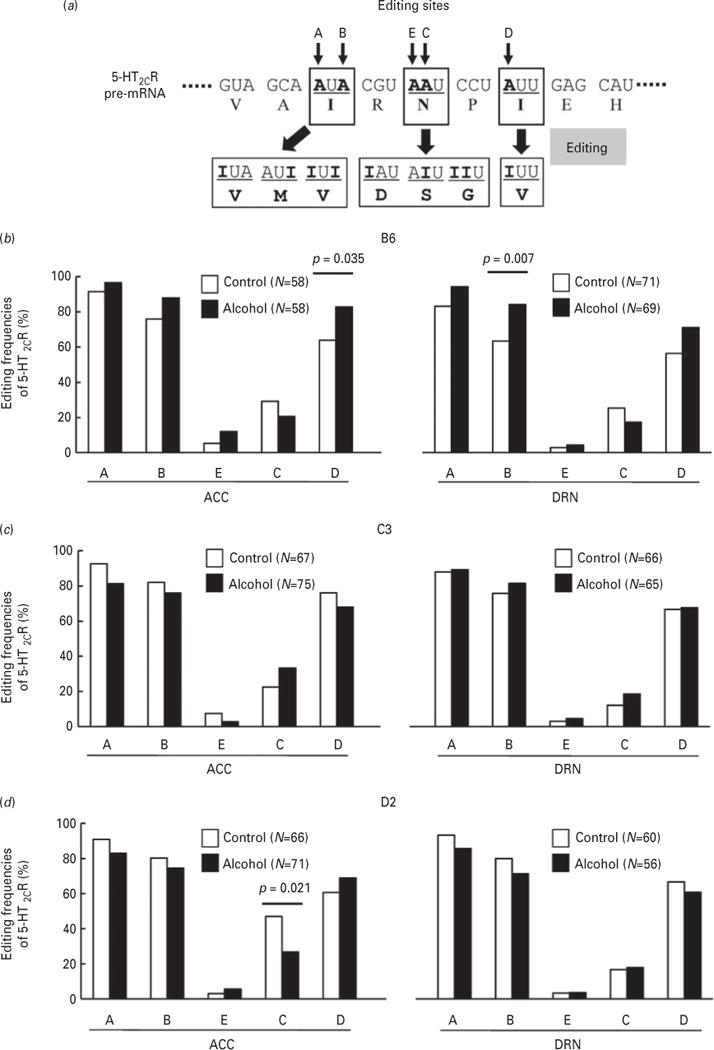
Alteration of 5-HT2CR mRNA editing frequency after chronic alcohol exposure. (a) RNA editing of 5-HT2CR occurs in five positions (A–E) and leads to amino acid substitutions at three sites, generating 24 possible protein isoforms. Frequencies of 5-HT2CR mRNA editing in control (open bars) and chronic alcohol exposed mice (solid bars) were measured by cloning–sequencing analysis of RT-PCR products. cDNAs were prepared from the ACC and the DRN of C57BL/6J (b), C3H/HeJ (c), and DBA/2J (d) mice. DNAs from individual clones (N=56–75) were sequenced, and editing frequencies at the five sites expressed as percentages. Statistical analyses were performed using Fisher’s exact test. Two-sided tests were used to calculate p-values.
ADAR1 edits 5-HT2CR mRNA specifically at sites A and B, whereas ADAR2 edits the D site (Hartner et al., 2004; Wang et al., 2004; Maas et al., 2006). Therefore, we also assessed ADAR1- and ADAR2-specific editing frequencies in the three mouse strains. A significant increase in the RNA editing frequency was detected at the ADAR2-specific site (site D) in the ACC but not at either of the ADAR1-specific sites (sites A or B) in both ACC and DRN of C57BL/6J mice (Supplementary Fig. S4).
Altered 5-HT2CR isoform patterns
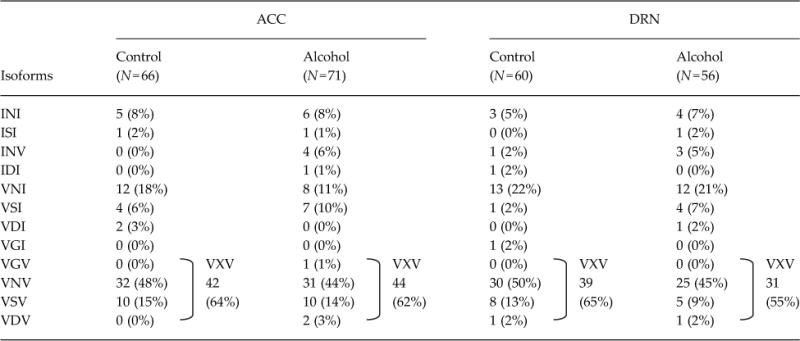
Based on individual sequence data of 5-HT2CR, we determined the amino acid sequences of the edited sites. In controls of all three mouse strains, the non-edited, INI, isoform was rare, while the VNV isoform was the major isoform detected in the ACC and DRN, as well as in the whole brain and amygdala (Du et al., 2006; Hackler et al., 2006) (Table 1). After chronic alcohol exposure, expression of the VNV isoform was significantly increased in the ACC and DRN of C57BL/6J mice (ACC, 40 to 62%; DRN, 34 to 55%), reflecting an increase in RNA editing at sites A and D (Table 1; ACC and DRN), while no such alteration in the isoform pattern was found in the hippocampus (Supplementary Table S2). By contrast, there was no significant change in expression of the VNV isoform in either the C3H/HeJ or DAB/2J mice, between control and alcohol groups, with expression frequencies of it being 35–55% (Tables 2 and 3). These results suggest that alterations in 5-HT2CR isoform patterns in the ACC and the DRN during alcohol exposure are involved in enhancement of alcohol intake.
Table 1.
Frequencies of 5-HT2C-isoforms in C57BL/6J
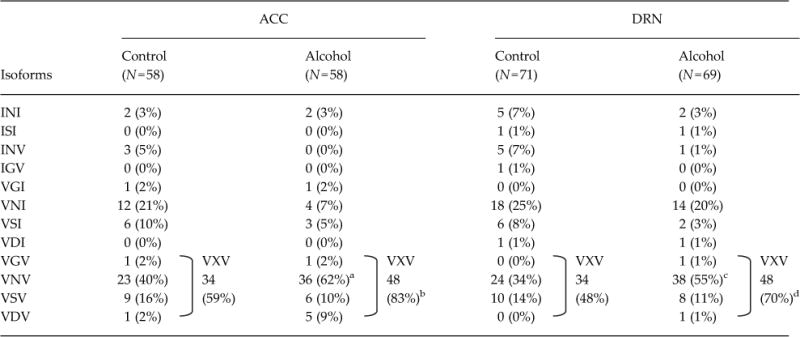
|
Number of samples is in parentheses. Amino acid sequences are represented by one-letter amino acid codes and wild-card character (X). Statistical analyses were performed with the use of Fisher’s exact test by GraphPad Prism 5 software. Two-sided tests were used to calculate p-values. Two-sided
p=0.025,
p=0.008,
p=0.017, and
p=0.011 by Fisher’s exact test.
Table 2.
Frequencies of 5-HT2C-isoforms in C3H/HeJ
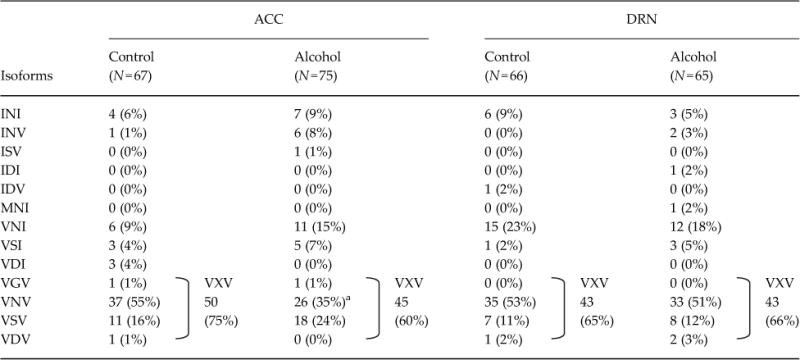
|
Number of samples is in parentheses. Amino acid sequences are represented by one-letter amino acid codes and wild-card character (X). Statistical analyses were performed with the use of Fisher’s exact test by GraphPad Prism 5 software. Two-sided tests were used to calculate p-values. Two-sided
p=0.018 by Fisher’s exact test.
Table 3.
Frequencies of 5-HT2C-isoforms in DBA/2J
|
Number of samples is in parentheses. Amino acid sequences are represented by one-letter amino acid codes and wild-card character (X). Statistical analyses were performed with the use of Fisher’s exact test by GraphPad Prism 5 software.
Activity of 5-HT2CR VNV isoform
The major isoforms of 5-HT2CR in the mouse ACC and DRN are VNI, VSI, VNV and VSV, as shown in Table 1. Although it is known that the VGV isoform is less potent than the non-edited INI isoform, there is no consensus about the activity of other isoforms, despite many studies (Burns et al., 1997; Fitzgerald et al., 1999; Herrick-Davis et al., 1999; Niswender et al., 1999; Wang et al., 2000a). We therefore reinvestigated the basal and 5-HT-stimulated activities of various isoforms. First, western blot analysis with anti-Myc antibody showed that there were no major differences in the expression levels of different isoforms of Myc/His-tagged 5-HT2CR isoforms (Fig. 3a).
Fig. 3.
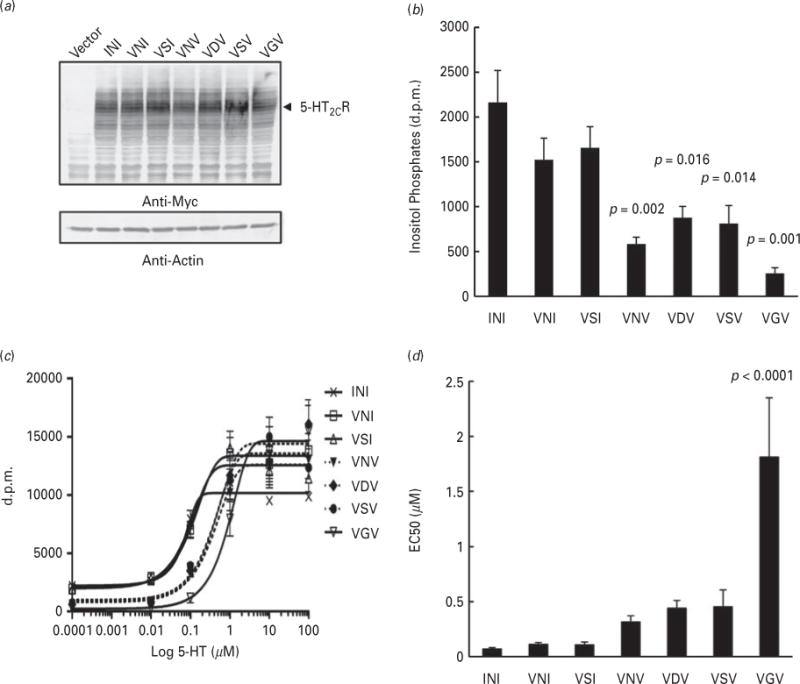
Receptor activity analysis of 5-HT2CR isoforms. (a) Myc/His-tagged 5-HT2CR isoforms were transiently expressed in HeLa cells prior to prelabelling with myo-[3H]inositol. Expression levels of isoforms were evaluated by western blot analysis with anti-Myc (upper panel) and anti-Actin (lower panel) antibodies. An arrowhead indicates 5-HT2CR isoforms bands. (b) For measurement of basal receptor activity, the production of 3H-labeled inositol phosphate was measured after lithium-treatment. Data are presented as mean±S.E.M. (N=7 in each group). Statistical analyses were performed using one-way ANOVA (Tukey post-hoc). p-values as compared to INI isoform are shown above the bars. (c) 5-HT-stimulated receptor activity was measured 1 h after treatment with 5-HT (1×10−4−102 μM) containing lithium chloride. Dose–response curves are described by the Four-Parameter Logistic (4PL) model. (d) The EC50 values are presented as mean±S.E.M. (N=8 in each group). Statistical analyses were performed using one-way ANOVA (Tukey post-hoc). p-value as compared to INI isoform is shown above the bar.
We then measured basal activities of the 5-HT2CR isoforms in HeLa cells (Fig. 3b). The basal activities of VXV isoforms containing two valine residues, such as VNV, VDV, VSV and VGV, were significantly decreased compared with that of the INI isoform (11.8 to 40.5%). By contrast, there were no significant changes in VNI or VSI isoforms. Interestingly, a significant reduction in 5-HT-stimulated receptor activity was detected only with the VGV isoform (Fig. 3c and d). The EC50 value for 5-HT-stimulation of the VGV isoform activity was approximately 26 times that of the INI isoform (Fig. 3d). Although not significant, other VXV isoforms, such as VNV, VDV, and VSV, tended to have EC50 values that were 4.5–6.5 times that of the INI isoform (Fig. 3d). Based on these findings, we grouped the 5-HT2CR isoforms according to activity levels. The proportion of VXV isoforms with lower basal activity was significantly increased in both ACC and DRN of alcohol-exposed C57BL/6J mice (Table 1, ACC, 59 to 83%; DRN, 48 to 70%). The result suggests that constitutive activity of 5-HT2CR is reduced in the brain of alcohol-exposed C57BL/6J mice, resulting in enhancement of alcohol drinking.
Upregulation of ADAR expression by chronic alcohol exposure
Increased 5-HT2CR mRNA editing in alcohol-exposed C57BL/6J mice might be accounted for by an increase in ADAR expression during chronic alcohol treatment. Therefore, we quantified the expression levels of ADARs using real-time PCR and immunoblot analysis (Fig. 4). Real-time PCR demonstrated that both ADAR1 and ADAR2 mRNA levels increased in the ACC and DRN of C57BL/6J mice following chronic alcohol exposure (Fig. 4a and b; B6). In contrast, no such increases were detected in either the C3H/HeJ or DBA/2J mice (Fig. 4a and b; C3 and D2). Consistent with these results, upregulations of ADAR1 and ADAR2 were confirmed in C57BL/6J mice by immunoblotting analyses (Fig. 4c and d; B6). In the ACC of alcohol-exposed C57BL/6J mice, ADAR1 and ADAR2 protein levels were 2.7 and 3.2 times those of the control mice, respectively. Similarly, chronic alcohol exposure increased levels of both proteins in the DRN of C57BL/6J mice. These results suggest that alterations in 5-HT2CR mRNA editing in C57BL/6J mice are associated with upregulation of ADARs during alcohol exposure.
Fig. 4.
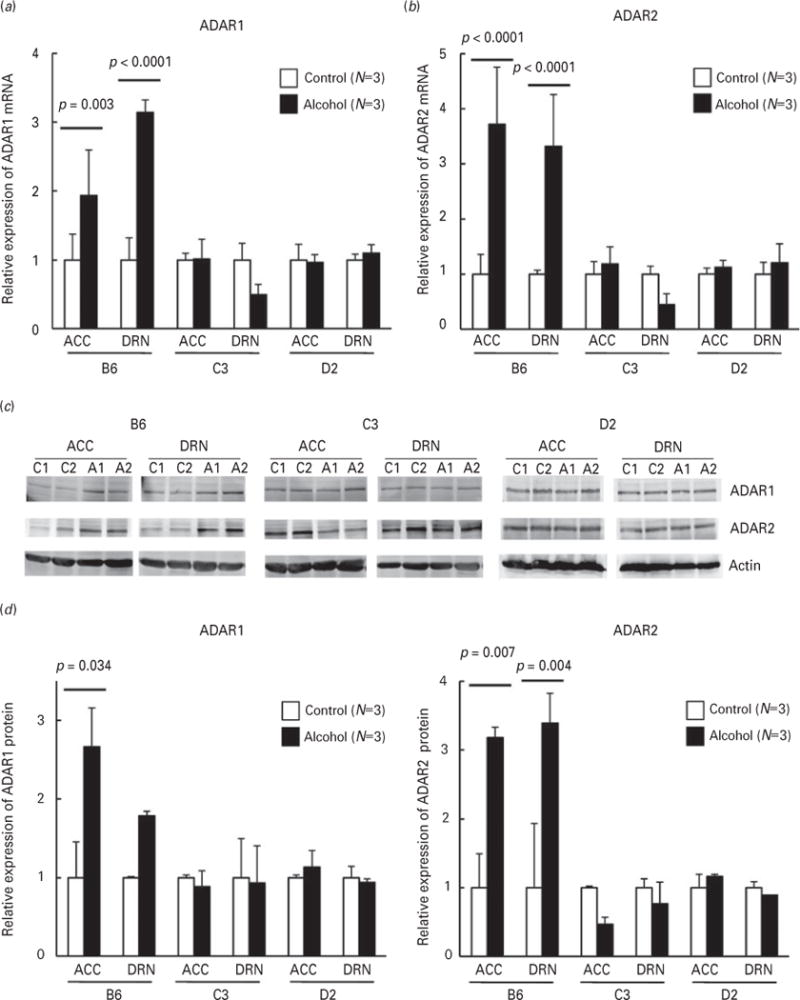
Expression analysis of adenosine deaminases acting on RNA (ADARs). Expression levels of ADAR1 (a) and ADAR2 (b) in control (open bars) and alcohol-exposed (solid bars) mice of the C57BL/6J (B6), C3H/HeJ (C3), and DBA/2J (D2) strains, were determined by real-time PCR. Expression levels of ADARs are expressed relative to control. Data are presented as mean±S.D. (N=3 per group). Statistical analyses were performed using one-way ANOVA (Tukey post-hoc). (c) Immunoblot analyses of ADAR1 (upper panels) and ADAR2 (middle panels). Samples were prepared from the ACC and DRN of control (C1 and C2) and alcohol-exposed (A1 and A2) mice. (d) Band intensity was measured with ImageJ software and normalized to the band intensity of actin (lower panels). Expression levels of ADAR1 (left) and ADAR2 (right) are expressed relative to control. Data are presented as mean±S.D. (N=3). Statistical analyses were performed using one-way ANOVA (Tukey post-hoc).
ADAR1 and ADAR2 also catalyse the RNA editing of other mRNAs, for example the glutamate receptor subunits (GluR2 and GluR5), BLCAP (bladder cancer associated protein) and CYFIP2 (cytoplasmic FMR1 interacting protein 2) (Sommer et al., 1991; Kwak et al., 2008). We next examined whether chronic alcohol exposure influences the RNA editing frequencies of these mRNAs. cDNAs from the ACC of control and chronic alcohol-exposed mice were prepared from mice of the three strains, and the editing sites of GluR2 Q/R, GluR5, BLCAP Y/C and CYFIP2 K/E were analysed using a direct sequencing method. The results suggest that there are no differences in RNA editing of GluR2 Q/R or CYFIP2 K/E, edited by ADAR2, between control and alcohol-exposed groups from all three mice strains (Supplementary Figs S5 and S6). Moreover, no differences in RNA editing frequency of GluR5 (ADAR1 and 2 target) or BLCAP (ADAR1 target) were detected between control and alcohol-exposed groups of all strains (Supplementary Figs S7 and S8).
Alcohol preference analysis of editing-blocked INI-mice
To verify our hypothesis that alterations in the 5-HT2CR isoform pattern contribute to alcohol intake, we compared alcohol drinking behaviour in INI-mice that express only the non-edited INI isoform of 5-HT2CR (Kawahara et al., 2008) with that in their wildtype littermates (WT). INI-mice and WT littermates, with a C57BL/6J genetic background, were chronically exposed to alcohol vapour for 20 d, and then alcohol consumption was measured. No significant alterations in alcohol metabolism or body weight were observed between WT and INI-mice (Supplementary Figs S1 and S2b). Interestingly, increased 5-HT2CR expression following alcohol exposure was not found in INI-mice (Fig. 5a). After chronic alcohol exposure, increased alcohol intake was observed in WT littermates but not in the INI-mice (Fig. 5b; WT, p=0.008, Tukey’s test after two-way ANOVA). Two-way ANOVA analysis of 5-HT2CR expression or alcohol intake showed that there was genotype-treatment interaction (F1,20 =6.817, p=0.017 or F1,36= 5.867, p=0.021, respectively). Similarly, the rate of alcohol intake was significantly different between control and alcohol-exposed WT mice (Fig. 5c; p=0.023, repeated measures ANOVA). There was no significant difference in the rate of alcohol consumption between control and alcohol-exposed INI-mice (Fig. 5b; INI, and 5d). These results suggest again that a change in the expression levels and RNA editing patterns of 5-HT2CR is involved in augmentation of alcohol drinking.
Fig. 5.
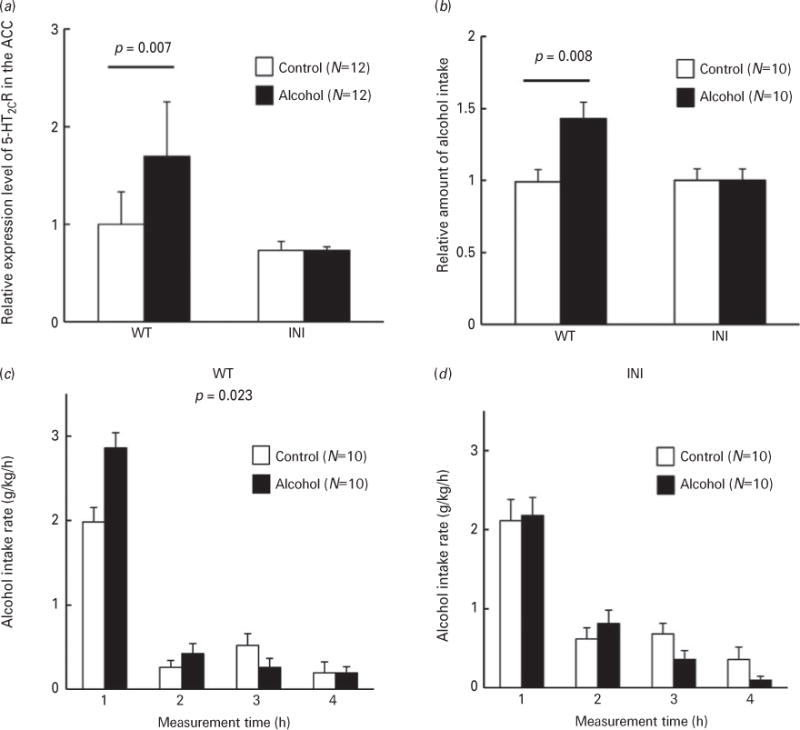
Drinking behaviour analysis of INI-mice. INI-mice (INI) and wildtype littermates (WT) were chronically exposed to alcohol vapour for 20 d. 5-HT2CR expression levels and alcohol intake of alcohol-exposed mice (solid bars) were compared with those of control mice (open bars). (a) Relative expression of 5-HT2CR mRNA in the ACC and DRN was quantified by real-time PCR. Data are presented as mean±S.D. (N=12). Statistical analyses were performed using two-way ANOVA, followed by Tukey’s post-hoc test. (b) Consumption of 10% ethanol is expressed relative to control. Data are presented as mean±S.E.M. Statistical analyses were performed using two-way ANOVA, followed by Tukey’s post-hoc test. (c) and INI-mice (d) were measured for 4 h. Data are presented as mean±S.E.M. Statistical analyses were performed with repeated measures ANOVA. There was a significant difference between WT control and alcohol groups (F(1,18)=6.216 p=0.023).
Discussion
Differences in alcohol preference, sensitivity and tolerance among inbred mouse strains (McClearn, 1968; Crabbe, 2002) suggest there is a genetic influence on these behaviours. Our present study shows that following chronic alcohol exposure, increased alcohol consumption and alterations in RNA editing of 5-HT2CR were detected only in C57BL/6J mice among the three inbred mouse strains examined. The RNA editing frequency at site D increased in the alcohol-preferring C57BL/6J mice following alcohol exposure, resulting in an increase in the proportion of VXV isoforms (41% increase) in the ACC and a similar increase (46% increase) in the DRN. Conversely, neither the C3H/HeJ nor the DBA/2J mice showed any changes in RNA editing. Although the reason for strain differences in RNA editing is unclear, our data suggest that upregulation of ADARs, observed in the C57BL/6J strain during chronic alcohol exposure, is likely to have causative relevance.
Previous studies demonstrated that mRNA editing of 5-HT2CR reduces receptor functions, including 5-HT potency, agonist binding affinity, constitutive activities, and G protein coupling activity (Burns et al., 1997; Fitzgerald et al., 1999; Niswender et al., 1999; Wang et al., 2000b; Berg et al., 2001; Gurevich et al., 2002b). The unedited INI isoform has the potency of the constitutive activity of the 5-HT2CR, while its activity is inhibited proportionally to the extent of editing, with the fully edited VGV isoform displaying the lowest degree of receptor activity (Herrick-Davis et al., 1999; Niswender et al., 1999; Wang et al., 2000b). Our study revealed that VXV isoforms, including VNV and VSV, had lower potencies of basal receptor activity. Considering these results, the basal activity of 5-HT2CR in the ACC and the DRN of C57BL/6J mice should be attenuated by chronic alcohol exposure, compared with that in C3H/HeJ or DBA/2J mice. The constitutive activity of 5-HT2CR in the ACC was reported to provide tonic suppression of dopamine release (De Deurwaerdere et al., 2004; Navailles et al., 2006). These reports speculate about the influence of 5-HT2CR mRNA editing on dopamine release. In particular, the ACC is a critical element of the mesocorticolimbic system, a brain circuit implicated in reward, suggesting that elevation of 5-HT2CR-RNA editing in the ACC is involved in determination of alcohol drinking behaviour. The DRN is also involved in the regulation of dopamine release in the ACC via 5-HT2CR (Yoshimoto and McBride, 1992; De Deurwaerdere and Spampinato, 1999). Taken together, our results suggest that increased RNA editing of 5-HT2CR during alcohol exposure alters dopamine release in the ACC, resulting in increased alcohol intake in wildtype C57BL/6J mice.
In addition to increased RNA editing, 5-HT2CR expression in the ACC of C57BL/6J mice was also significantly increased by chronic alcohol exposure, whereas no increase was observed in low alcohol preferring mice. There are strain differences in the release of 5-HT and dopamine by alcohol stimulation (Yoshimoto et al., 1992b; Kapasova and Szumlinski, 2008), suggesting that the increased 5-HT2CR expression may result in alterations in neurotransmission of 5-HT or dopamine. Indeed, 5-HT2CR expression shows an adaptive change following chronic administration of 5-HT2CR agonists or antagonists (Buckholtz et al., 1988; McKenna et al., 1989; Van Oekelen et al., 2003), or antagonists of the dopamine D2 receptor (Buckland et al., 1997). Alternatively, this enhancement might result from adaptation to the attenuation of 5-HT2CR activity brought about by RNA editing, as expression of 5-HT2CR in the ACC of INI-mice did not change with chronic alcohol exposure.
Alcohol intake did not increase in INI-mice bred on a C57BL/6J genetic background, even after chronic alcohol exposure. This suggests that alterations in RNA editing of 5-HT2CR in the ACC is crucial for determining alcohol drinking behaviour. INI-mice bred on a C57BL/6 background exhibit no significant differences from WT littermates in anxiety-like and feeding behaviours (Mombereau et al., 2010). Thus, our observation would not be attributed to another behavioural phenotype. In the ACC, the basal levels (i.e. before alcohol exposure) of the combined VXV isoforms of 5-HT2CR account for 60–75% of all isoforms in all inbred strains and 0% in the INI-mice; therefore, the increase in RNA editing in response to alcohol is potentially the most important factor in the acquisition of alcohol preference.
In conclusion, alterations in the 5-HT2CR isoform pattern and expression in the ACC and the DRN after chronic alcohol exposure is involved in increased alcohol-drinking behaviour. The precise mechanism of strain differences in the response of ADAR expression and RNA editing to alcohol will be clarified in future investigations.
Supplementary Material
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research to KY and MT and ‘Development of biomarker candidates for social behaviour’ carried out under the Strategic Research Program for Brain Sciences to MK from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The work was also supported by grants from the National Institutes of Health, the Ellison Medical Foundation and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health, to KN.
References
- Abramowski D, Rigo M, Duc D, Hoyer D, Staufenbiel M. Localization of the 5-hydroxytryptamine2C receptor protein in human and rat brain using specific antisera. Neuropharmacol. 1995;34:1635–1645. doi: 10.1016/0028-3908(95)00138-7. [DOI] [PubMed] [Google Scholar]
- Berg KA, Cropper JD, Niswender CM, Sanders-Bush E, Emeson RB, Clarke WP. RNA-editing of the 5-HT(2C) receptor alters agonist-receptor-effector coupling specificity. Br J Pharmacol. 2001;134:386–392. doi: 10.1038/sj.bjp.0704255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckholtz NS, Zhou DF, Freedman DX. Serotonin2 agonist administration down-regulates rat brain serotonin2 receptors. Life Sci. 1988;42:2439–2445. doi: 10.1016/0024-3205(88)90342-6. [DOI] [PubMed] [Google Scholar]
- Buckland PR, D’Souza U, Maher NA, McGuffin P. The effects of antipsychotic drugs on the mRNA levels of serotonin 5HT2A and 5HT2C receptors. Brain Res Mol Brain Res. 1997;48:45–52. doi: 10.1016/s0169-328x(97)00076-4. [DOI] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Clemett DA, Punhani T, Duxon MS, Blackburn TP, Fone KC. Immunohistochemical localisation of the 5-HT2C receptor protein in the rat CNS. Neuropharmacol. 2000;39:123–132. doi: 10.1016/s0028-3908(99)00086-6. [DOI] [PubMed] [Google Scholar]
- Crabbe JC. Alcohol and genetics: new models. Am J Med Genet. 2002;114:969–974. doi: 10.1002/ajmg.b.10984. [DOI] [PubMed] [Google Scholar]
- De Deurwaerdere P, Spampinato U. Role of serotonin(2A) and serotonin(2B/2C) receptor subtypes in the control of accumbal and striatal dopamine release elicited in vivo by dorsal raphe nucleus electrical stimulation. J Neurochem. 1999;73:1033–1042. doi: 10.1046/j.1471-4159.1999.0731033.x. [DOI] [PubMed] [Google Scholar]
- De Deurwaerdere P, Navailles S, Berg KA, Clarke WP, Spampinato U. Constitutive activity of the serotonin2C receptor inhibits in vivo dopamine release in the rat striatum and nucleus accumbens. J Neurosci. 2004;24:3235–3241. doi: 10.1523/JNEUROSCI.0112-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Davisson MT, Kafadar K, Gardiner K. A-to-I pre-mRNA editing of the serotonin 2C receptor: comparisons among inbred mouse strains. Gene. 2006;382:39–46. doi: 10.1016/j.gene.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Englander MT, Dulawa SC, Bhansali P, Schmauss C. How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. J Neurosci. 2005;25:648–651. doi: 10.1523/JNEUROSCI.3895-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald LW, Iyer G, Conklin DS, Krause CM, Marshall A, Patterson JP, Tran DP, Jonak GJ, Hartig PR. Messenger RNA editing of the human serotonin 5-HT2C receptor. Neuropsychopharmacol. 1999;21:82S–90S. doi: 10.1016/S0893-133X(99)00004-4. [DOI] [PubMed] [Google Scholar]
- Flomen R, Knight J, Sham P, Kerwin R, Makoff A. Evidence that RNA editing modulates splice site selection in the 5-HT2C receptor gene. Nucleic Acids Res. 2004;32:2113–2122. doi: 10.1093/nar/gkh536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti M, Tecott LH. Contributions of 5-HT(2C) receptors to multiple actions of central serotonin systems. Eur J Pharmacol. 2004;488:1–9. doi: 10.1016/j.ejphar.2004.01.036. [DOI] [PubMed] [Google Scholar]
- Gurevich I, Englander MT, Adlersberg M, Siegal NB, Schmauss C. Modulation of serotonin 2C receptor editing by sustained changes in serotonergic neurotransmission. J Neurosci. 2002a;22:10529–10532. doi: 10.1523/JNEUROSCI.22-24-10529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002b;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- Hackler EA, Airey DC, Shannon CC, Sodhi MS, Sanders-Bush E. 5-HT(2C) receptor RNA editing in the amygdala of C57BL/6J, DBA/2J, and BALB/cJ mice. Neurosci Res. 2006;55:96–104. doi: 10.1016/j.neures.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- Herrick-Davis K, Grinde E, Niswender CM. Serotonin 5-HT2C receptor RNA editing alters receptor basal activity: implications for serotonergic signal transduction. J Neurochem. 1999;73:1711–1717. doi: 10.1046/j.1471-4159.1999.731711.x. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Kato T. Serotonin receptor 2C and mental disorders: genetic, expression and RNA editing studies. RNA Biol. 2009;6:248–253. doi: 10.4161/rna.6.3.8370. [DOI] [PubMed] [Google Scholar]
- Kapasova Z, Szumlinski KK. Strain differences in alcohol-induced neurochemical plasticity: a role for accumbens glutamate in alcohol intake. Alcohol Clin Exp Res. 2008;32:617–631. doi: 10.1111/j.1530-0277.2008.00620.x. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Grimberg A, Teegarden S, Mombereau C, Liu S, Bale TL, Blendy JA, Nishikura K. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J Neurosci. 2008;28:12834–12844. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Sanna PP, Bloom FE. Neuroscience of addiction. Neuron. 1998a;21:467–476. doi: 10.1016/s0896-6273(00)80557-7. [DOI] [PubMed] [Google Scholar]
- Koob GF, Roberts AJ, Schulteis G, Parsons LH, Heyser CJ, Hyytia P, Merlo-Pich E, Weiss F. Neurocircuitry targets in ethanol reward and dependence. Alcohol Clin Exp Res. 1998b;22:3–9. [PubMed] [Google Scholar]
- Kwak S, Nishimoto Y, Yamashita T. Newly identified ADAR-mediated A-to-I editing positions as a tool for ALS research. RNA Biol. 2008;5:193–197. doi: 10.4161/rna.6925. [DOI] [PubMed] [Google Scholar]
- Lumeng L, Murphy J, McBride W, Li T-K. Genetic Influences on alcohol preference in animals. In: Begleiter H, Kissin B, editors. The genetics of alcoholism. New York: Oxford University Press; 1995. pp. 165–201. [Google Scholar]
- Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClearn GE. The use of strain rank orders in assessing equivalence of techniques. Behav Res Meth Instrum. 1968;1:49–51. [Google Scholar]
- McKenna DJ, Nazarali AJ, Himeno A, Saavedra JM. Chronic treatment with (+/−)DOI, a psychotomimetic 5-HT2 agonist, downregulates 5-HT2 receptors in rat brain. Neuropsychopharmacol. 1989;2:81–87. doi: 10.1016/0893-133x(89)90010-9. [DOI] [PubMed] [Google Scholar]
- Miyazaki KW, Miyazaki K, Doya K. Activation of dorsal raphe serotonin neurons is necessary for waiting for delayed rewards. J Neurosci. 2012;32:10451–10457. doi: 10.1523/JNEUROSCI.0915-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombereau C, Kawahara Y, Gundersen BB, Nishikura K, Blendy JA. Functional relevance of serotonin 2C receptor mRNA editing in antidepressant- and anxiety-like behaviours. Neuropharmacol. 2010;59:468–473. doi: 10.1016/j.neuropharm.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller CP, Huston JP. Determining the region-specific contributions of 5-HT receptors to the psychostimulant effects of cocaine. Trends Pharmacol Sci. 2006;27:105–112. doi: 10.1016/j.tips.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Navailles S, Moison D, Ryczko D, Spampinato U. Region-dependent regulation of mesoaccumbens dopamine neurons in vivo by the constitutive activity of central serotonin2C receptors. J Neurochem. 2006;99:1311–1319. doi: 10.1111/j.1471-4159.2006.04188.x. [DOI] [PubMed] [Google Scholar]
- Navailles S, Moison D, Cunningham KA, Spampinato U. Differential regulation of the mesoaccumbens dopamine circuit by serotonin2C receptors in the ventral tegmental area and the nucleus accumbens: an in vivo microdialysis study with cocaine. Neuropsychopharmacol. 2008;33:237–246. doi: 10.1038/sj.npp.1301414. [DOI] [PubMed] [Google Scholar]
- Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender CM, Copeland SC, Herrick-Davis K, Emeson RB, Sanders-Bush E. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J Biol Chem. 1999;274:9472–9478. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- Pistis M, Muntoni AL, Gessa G, Diana M. Effects of acute, chronic ethanol and withdrawal on dorsal raphe neurons: electrophysiological studies. Neuroscience. 1997;79:171–176. doi: 10.1016/s0306-4522(96)00643-4. [DOI] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nature Reviews Neuroscience. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard JR, Albersheim P, McClearn GE. Enzyme activities and ethanol preference in mice. Biochem Genet. 1968;2:205–212. doi: 10.1007/BF01474759. [DOI] [PubMed] [Google Scholar]
- Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Van Bockstaele EJ, Biswas A, Pickel VM. Topography of serotonin neurons in the dorsal raphe nucleus that send axon collaterals to the rat prefrontal cortex and nucleus accumbens. Brain Res. 1993;624:188–198. doi: 10.1016/0006-8993(93)90077-z. [DOI] [PubMed] [Google Scholar]
- Van Oekelen D, Luyten WH, Leysen JE. 5-HT2A and 5-HT2C receptors and their atypical regulation properties. Life Sci. 2003;72:2429–2449. doi: 10.1016/s0024-3205(03)00141-3. [DOI] [PubMed] [Google Scholar]
- Wang Q, O’Brien PJ, Chen CX, Cho DS, Murray JM, Nishikura K. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J Neurochem. 2000a;74:1290–1300. doi: 10.1046/j.1471-4159.2000.741290.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, O’Brien PJ, Chen CX, Cho DS, Murray JM, Nishikura K. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J Neurochem. 2000b;74:1290–1300. doi: 10.1046/j.1471-4159.2000.741290.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Ikegawa M, Naruse Y, Tanaka M. A novel splicing variant form suppresses the activity of full-length signal transducer and activator of transcription 5A. FEBS J. 2009;276:6312–6323. doi: 10.1111/j.1742-4658.2009.07339.x. [DOI] [PubMed] [Google Scholar]
- Werry TD, Loiacono R, Sexton PM, Christopoulos A. RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacol Ther. 2008;119:7–23. doi: 10.1016/j.pharmthera.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Komura S. Genetic differences in the effects of voluntary ethanol consumption on brain monoamine levels in inbred strains of mice, C57BL/6J, C3H/He and DBA/2Cr. Alcohol Alcohol. 1989;24:225–229. [PubMed] [Google Scholar]
- Yoshimoto K, McBride WJ. Regulation of nucleus accumbens dopamine release by the dorsal raphe nucleus in the rat. Neurochem Res. 1992;17:401–407. doi: 10.1007/BF00969884. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, McBride WJ, Lumeng L, Li TK. Ethanol enhances the release of dopamine and serotonin in the nucleus accumbens of HAD and LAD lines of rats. Alcohol Clin Exp Res. 1992a;16:781–785. doi: 10.1111/j.1530-0277.1992.tb00678.x. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, McBride WJ, Lumeng L, Li TK. Alcohol stimulates the release of dopamine and serotonin in the nucleus accumbens. Alcohol. 1992b;9:17–22. doi: 10.1016/0741-8329(92)90004-t. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Watanabe Y, Tanaka M, Kimura M. Serotonin2C receptors in the nucleus accumbens are involved in enhanced alcohol-drinking behaviour. Eur J Neurosci. 2012;35:1368–1380. doi: 10.1111/j.1460-9568.2012.08037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


