Abstract
Background and objectives
X-linked Alport syndrome is caused by mutations in the COL4A5 gene. Although many COL4A5 mutations have been detected, the mutation detection rate has been unsatisfactory. Some men with X-linked Alport syndrome show a relatively mild phenotype, but molecular basis investigations have rarely been conducted to clarify the underlying mechanism.
Design, setting, participants, & measurements
In total, 152 patients with X-linked Alport syndrome who were suspected of having Alport syndrome through clinical and pathologic investigations and referred to the hospital for mutational analysis between January of 2006 and January of 2013 were genetically diagnosed. Among those patients, 22 patients had suspected splice site mutations. Transcripts are routinely examined when suspected splice site mutations for abnormal transcripts are detected; 11 of them showed expected exon skipping, but others showed aberrant splicing patterns. The mutation detection strategy had two steps: (1) genomic DNA analysis using PCR and direct sequencing and (2) mRNA analysis using RT-PCR to detect RNA processing abnormalities.
Results
Six splicing consensus site mutations resulting in aberrant splicing patterns, one exonic mutation leading to exon skipping, and four deep intronic mutations producing cryptic splice site activation were identified. Interestingly, one case produced a cryptic splice site with a single nucleotide substitution in the deep intron that led to intronic exonization containing a stop codon; however, the patient showed a clearly milder phenotype for X-linked Alport syndrome in men with a truncating mutation. mRNA extracted from the kidney showed both normal and abnormal transcripts, with the normal transcript resulting in the milder phenotype. This novel mechanism leads to mild clinical characteristics.
Conclusions
This report highlights the importance of analyzing transcripts to enhance the mutation detection rate and provides insight into genotype-phenotype correlations. This approach can clarify the cause of atypically mild phenotypes in X-linked Alport syndrome.
Keywords: Alport syndrome, genetic renal disease, glomerular disease
Introduction
Alport syndrome is a hereditary disorder of type IV collagen that progresses to CKD and usually develops into ESRD. Symptoms include sensorineural hearing loss and ocular abnormalities. X-linked Alport syndrome (XLAS) accounts for approximately 85% of patients with Alport syndrome. These patients have mutations in the COL4A5 gene, which encodes the type IV collagen-α5 [α5(IV)] chain. COL4A5 mutations result in abnormal α5(IV) expression, with a typically complete absence of α5(IV) in the glomerular basement membrane (GBM) and Bowman’s capsule in men and a mosaic expression pattern in women (1).
To date, >500 different COL4A5 mutations have been identified in patients with XLAS, but mutation detection rates vary widely, ranging between approximately 40%–82% (2–6). The low detection rate is unsatisfactory, because it hampers a deeper understanding of the genotype-phenotype relationship in patients with XLAS. Previous studies have reported that missense and in-frame mutations have milder phenotypes compared with truncating mutations (7–9). However, these studies did not fully consider the effect of splice site mutations on COL4A5 expression. This study requires careful analysis of COL4A5 transcripts and assessment of intronic mutations, which have been increasingly identified as a cause of aberrant splicing for various disorders, including Alport syndrome (10–12).
Recently, we reported that 29% of patients with XLAS showed α5(IV) expression on kidney glomeruli, and these patients displayed milder phenotypes (13). All of these patients positive for α5(IV) had nontruncating mutations, including in-frame deletions or in-frame mutations resulting from splice site mutations and exon skipping. This finding suggests that patients with XLAS with in-frame mutations can show a milder phenotype, even when derived from a splice site mutation (13). Therefore, it is important to clarify splice site mutations as either in-frame or out-of-frame mutations after examining transcripts to estimate the phenotype.
It is well known among clinical nephrologists that some men with XLAS show a relatively mild phenotype, leading to the development of ESRD after the age of 60 years old. The reason for the mild phenotype has been attributed to missense or in-frame mutations or somatic mosaicism (8,9,13,14). One of our patients showed a novel mechanism leading to a mild phenotype revealed by transcript analysis extracted from the kidney. These patients showed both normal and abnormal spliced transcripts, and the normal transcript prevented progression to the typically severe phenotype of XLAS.
In this study, we report on 11 patients showing aberrant splicing of primary transcripts, providing additional insight into genotype-phenotype correlations in XLAS.
Materials and Methods
Ethical Considerations
All procedures were reviewed and approved by the Institutional Review Board of Kobe University School of Medicine. Informed consent was obtained from patients or their parents.
Inclusion Criteria
Clinical and laboratory findings of Japanese patients with XLAS were obtained from their medical records. Patients were referred to our hospital for clinical evaluation or genetic analysis. Most patients were followed in various local hospitals in Japan. DNA and data sheets were sent to our laboratory after acceptance of the request for mutational analysis. We routinely conduct genetic analysis of clinically diagnosed patients with XLAS. When we detect mutations that may possibly affect RNA processing, we routinely analyze transcripts to confirm the mutation-induced splicing abnormalities. When we fail to detect mutations in typical patients with XLAS using DNA-based mutation assays, we conduct transcript analyses as well. All patients with splice site mutations that led to an atypical splicing pattern were included in this study.
One hundred fifty-two patients were genetically diagnosed with XLAS at our laboratory; 22 patients had splice site mutations, and 11 of them showed atypical splicing patterns. These 11 patients were included in this study.
The degree of urinary protein excretion was evaluated using the urinary protein-to-creatinine ratio. eGFR was calculated using the Schwartz equation for patients ≤17 years old (15,16) and GFR-estimating equations for Japanese for patients ≥18 years old (17). Images of kidney α5(IV) staining were sent to us for evaluation of the staining patterns and assessed by the same person (K.N.). eGFR was measured on the basis of the data on data sheets.
Mutational Analyses
Mutational analyses of COL4A5 were carried out using the following methods. (1) PCR and direct sequencing of genomic DNA of all exons and exon-intron boundaries were performed. (2) When we detected a suspected splicing site mutation or failed to detect mutations with step 1 analysis, RT-PCR of mRNA and direct sequencing of abnormal mRNA products were carried out.
Genomic DNA was isolated from peripheral blood leukocytes from patients using the Quick Gene Mini 80 System (Fujifilm Corporation, Tokyo, Japan) according to the manufacturer’s instructions. For genomic DNA analysis, all 51 specific exons of COL4A5 were amplified by PCR as described previously (5). PCR-amplified products were then purified and subjected to direct sequencing using a Dye Terminator Cycle Sequencing Kit (Amersham Biosciences, Piscataway, NJ) with an automatic DNA sequencer (ABI Prism 3130; Perkin Elmer Applied Biosystems, Foster City, CA). Total RNA was extracted from blood leukocytes, kidneys, and/or hair roots. RNA from leukocytes was isolated using a Paxgene Blood RNA Kit (Qiagen Inc., Chatsworth, CA) and then reverse transcribed into cDNA using random hexamers and a Superscript III Kit (Invitrogen, Carlsbad, CA). RNA from kidneys and hair roots was isolated as described previously (14). cDNA was amplified by nested PCR using primer pairs for COL4A5 as described previously with slight modifications (sequences available on request) (18). PCR-amplified products were purified and subjected to direct sequencing.
Immunohistochemical Analyses
Immunohistochemical analyses were performed using frozen sections of kidney tissue. The immunohistochemical procedure has been described previously (19–21). A mixture of fluorescein isothiocyanate–conjugated rat mAb for the human α5(IV) chain (H53) and Texas red–conjugated rat mAb for the human α2(IV) chain (H25) was purchased from Shigei Medical Research Institute (Okayama, Japan). The epitopes were EAIQP at positions 675–679 of the α2(IV) chain and IDVEF at positions 251–255 of the α5(IV) chain.
Results
Patients’ Clinical Features and Pathologic Findings
In total, 11 patients were included in the study. Mutations creating or eliminating the 3′ splice site consensus sequences were detected in six patients (patient IDs 7, 17, 27, 28, 128, and 158), including two patients who had relatively deep intron mutations (patient IDs 128 and 158 with mutation of intervening sequence (IVS)27–18 and IVS29–8, respectively). One patient had a mutation at the last nucleotide of exon 41, altering the 5′ splice site consensus (patient ID 21). These patients proceeded to transcript analysis to determine the influence of the mutations on splicing. Detection of mutations in another four patients failed with genomic DNA–based assays, prompting transcript analyses that detected intronic exonization in patient IDs 19, 48, 126, and 217. All mutations together with patients’ clinical, laboratory, and pathologic data are shown in Table 1. All patients showed typical clinical features of XLAS.
Table 1.
Clinical and pathologic findings
| Patient ID | Sex | Age (yr) | ESRD Age (yr) | Hearing Loss (Detected Age) | sCr (mg/dl) | U-P/Cr (g/g) | eGFR | EM | α5 | Family History | Mutation Position | Nucleotide Change | Transcript | Truncating Mutation | Expected Change |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Splicing acceptor site mutations | |||||||||||||||
| 7 | M | 25 | 9 | Mild | ESRD | — | — | BWC | Negative | Mother: OB | Intron 10 | IVS11−2a>t | 19-bp deletion (frame shift) | Yes | Exon 11 skipping (36 bp) |
| 17 | W | 11 | — | — | 0.35 | 1 | 151 | BWC | Mosaic | None | Intron 38 | IVS39−1g>a | 1-bp deletion (frame shift) | Yes | Exon 39 skipping (99 bp) |
| 27 | M | 5 | — | — | 0.21 | 0.71 | 155 | BWC | Negative | None | Intron 18 | IVS18−1g>a | 1-bp deletion (frame shift) | Yes | Exon 18 skipping (42 bp) |
| 28 | W | 6 | — | — | 0.32 | 0.2 | 158 | TBM | Mosaic | Sister: OB | Intron 27 | IVS28−2a>g | 18-bp deletion | No | Exon 28 skipping (98 bp) |
| 128 | M | 46 | — | Mild | 1.53 | 1.07 | 41 | ND | Negative (skin) | Brother: 40 yr ESRD; daughters: OB | Intron 26 | IVS27−18a>g | 105-bp deletion (exon 27 skipping) | No | — |
| 158 | W | 14 | — | — | 0.53 | 0.35 | 114 | BWC | Mosaic | None | Intron 28 | IVS29−8t>a | 6-bp insertion | No | Exon 29 skipping (151 bp) |
| Exonic mutation | |||||||||||||||
| 21 | M | 41 | 24 | Mild | ESRD | — | — | BWC | Negative | Grandfather: 20 yr ESRD; mother: OB | Exon 41 | c.3790G>A | 186-bp deletion (exon 41 skipping) | No | Missense mutation |
| Deep intronic mutations | |||||||||||||||
| 19 | M | 22 | — | Mild | 0.56 | 0.55 | 98 | BWC | Negative | Mother: pro/OB | Intron 25 | IVS25+894c>g | 106-bp insertion with stop codona | Yes | — |
| 48 | M | 6 | — | — | 0.3 | 1.87 | 122 | ND | Negative (skin) | Mother: 28 yr ESRD | Intron 47 | IVS48−345a>g | 74-bp insertion with stop codon | Yes | — |
| 126 | M | 12 | — | Severe | 0.79 | 0.24 | 96 | BWC | Negative | Mother: OB; sister: OB | Intron 10 | IVS10+875g>t | 123-bp insertion with stop codon | Yes | — |
| 217 | M | 4 | — | — | 0.88 | 0.5 | 50 | BWC | Negative | Mother and sisters: pro/OB | Intron 47 | IVS47+1754t>g | 84-bp insertion with stop codon | Yes | — |
M, man; W, woman; sCr, serum creatinine; U-P/Cr, urinary protein-to-creatinine ratio; EM, electron microscopic findings; BWC, basket-weave change; ND, not determined; OB, occult blood; pro, protein; TBM, thin basement membrane.
RT-PCR results showed both normal and abnormal transcripts only in cDNA extracted from the kidney, and this case showed an extremely mild phenotype for a man with X-linked Alport syndrome with a truncating mutation.
Acceptor Splice Site Mutations
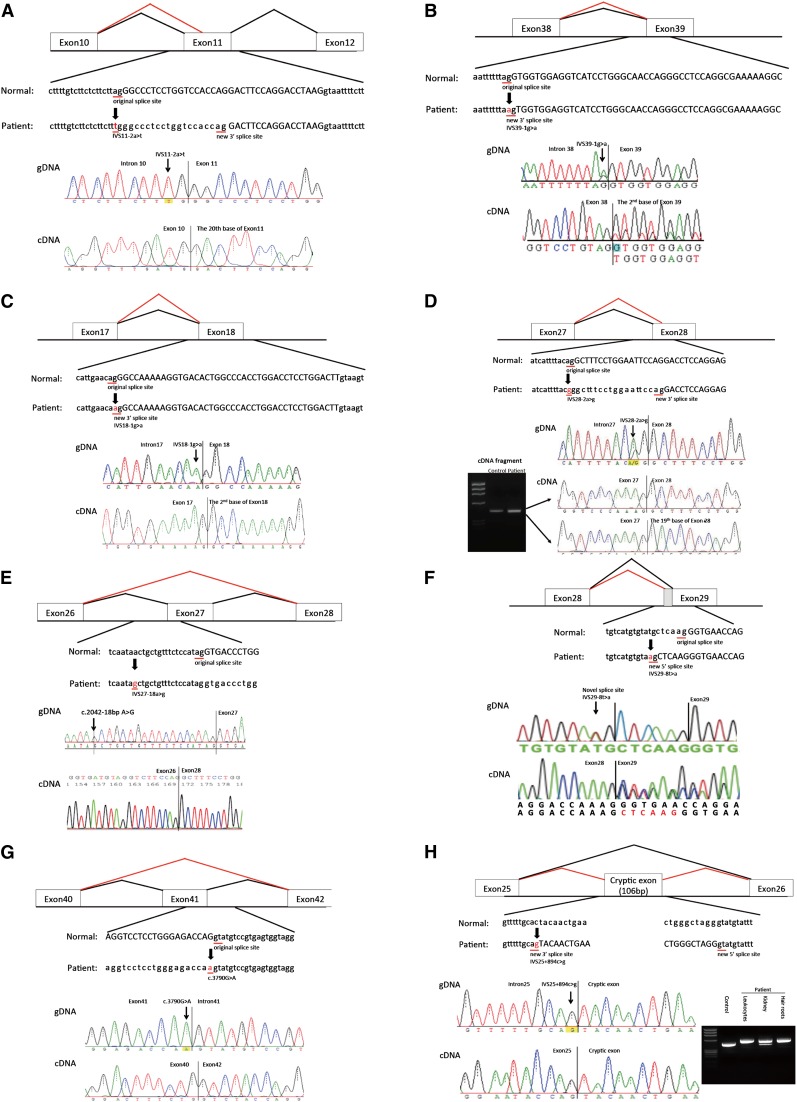
Six patients had mutations at 3′ splice sites (Figure 1, A–F). Four of them had mutations in the last intron dinucleotides (patient IDs 7, 17, 27, and 28) that did not result in skipping of complete exonic sequences but produced new 3′ splice sites downstream of authentic sites, creating small deletions in COL4A5 mRNA (19, 1, 1, and 18 bp, respectively) (Figure 1, A–D). One patient had an IVS27–18a>g mutation, which resulted in exon 27 skipping (patient ID 128) (Figure 1E). The IVS29–8t>a mutation (patient ID 158) produced a new splice acceptor site, resulting in a 6-bp insertion in the mature transcript (Figure 1F).
Figure 1.
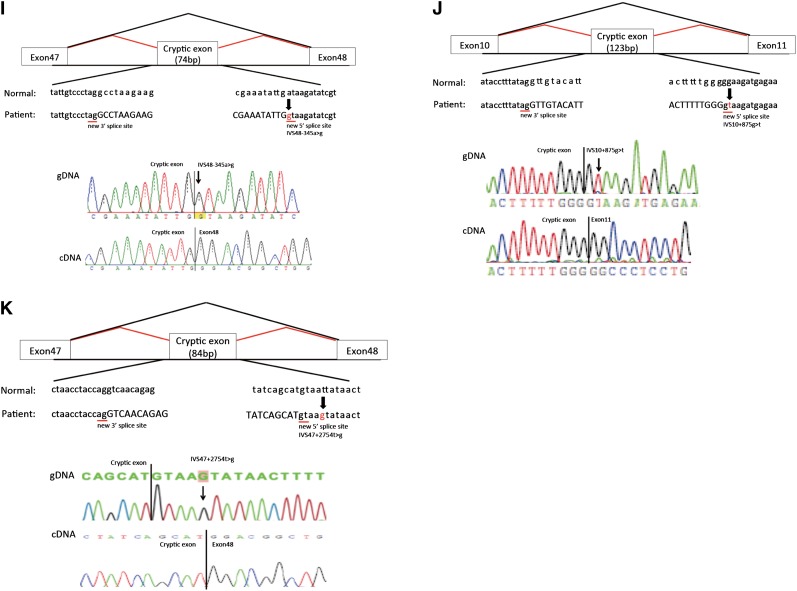
Mutations and their consequences. Upper panels show schematics of aberrant splicing (red lines). Normal splicing is indicated by black lines. The original and new splice sites and flanking sequences are shown below. A patient’s flanking genomic DNA and cDNA sequences are shown in lower panels. (A) Patient ID 7. IVS11–2A>T eliminated the splice acceptor site of intron 10 to activate a new splice site 19 nucleotides upstream. (B) Patient ID 17. IVS39–1G>A eliminated the splice acceptor site of intron 38, producing a transcript with a 1-bp deletion. (C) Patient ID 27. IVS18–1G>A removed the splice acceptor site of intron 17, generating a transcript with a 1-bp deletion. (D) Patient ID 28. IVS28–2A>G altered the splice acceptor site of intron 27, producing an 18-bp deletion in mature transcripts. (E) Patient ID 128. IVS27–18A>G potentially disrupted the splice acceptor site of intron 26, resulting in exon 27 skipping, which creates a transcript 105-bp deletion. (F) Patient ID 158. IVS29–8T>A changed the splice acceptor site of intron 28 to IVS29–7, which creates a transcript 6-bp insertion. (G) Patient ID 21. The last nucleotide of exon 41 mutation, C3790G>A, disrupted the splice donor site of intron 41, resulting in exon 41 skipping, which creates a transcript with a 186-bp deletion. (H) Patient ID 19. IVS25+894C>G produced a new splice acceptor site, resulting in a cryptic exon activation between exons 25 and 26 and creating a transcript with a 106-bp insertion that contains a stop codon. Lower right panel shows RT-PCR products from leukocytes, kidneys, and hair roots. Only the transcript from the kidney shows a normal-sized band, meaning that the kidney produced both abnormal and normal transcripts without a cryptic exon. (I) Patient ID 48. IVS48–345A>G made a new splice donor site, resulting in the production of a cryptic exon between exons 47 and 48, which creates a transcript with a 74-bp insertion that contains a stop codon. (J) Patient ID 126. IVS10+875G>T made a new splicing donor site, resulting in the production of a cryptic exon between exons 10 and 11 and creating a transcript with a 123-bp insertion that contains a stop codon. (K) Patient ID 217. IVS47+2754T>G made a new splicing donor site, resulting in the production of a cryptic exon between exons 47 and 48, which creates a transcript 84-bp insertion that contains a stop codon. IVS, intervening sequence.
Donor Splice Site Mutation
One patient had a single nucleotide substitution at the last position of exon 41 (Figure 1G). Although the mutation is a missense change (p.G1263S), cDNA analysis revealed skipping of exon 41 (186 bp). This finding is consistent with previous studies pointing to a substantial fraction of missense mutations that alter pre-mRNA processing (22).
Deep Intronic Mutations
A failure to detect mutations using genomic DNA analysis in four patients with typical features of XLAS (patient IDs 19, 48, 126, and 217) prompted us to conduct transcript analyses. RT-PCR showed insertions of cryptic exonic sequences in each case (Figure 1, H–K, Supplemental Figure 1). We then conducted genomic DNA analysis of intronic sequences and found point mutations that produced new splice sites (Figure 1, H–K). Interestingly, patient ID 19 showed a milder phenotype than a typical patient with XLAS with a truncating mutation. This patient had both normal and abnormal transcripts of the kidney that led to his phenotype being milder than that usually seen with a man with XLAS with a truncating mutation. Patient ID 217 had a mutation at position +5 of the new exon (Figure 1K). All cryptic exons had stop codons in them and thus, turned out to be truncating mutations (Supplemental Figure 1). From these results, we could group these four patients as having truncating mutations.
Discussion
This study provides the first case series report on aberrant splice site mutations. We identified six splice consensus sequence mutations, including two relatively deeper intronic nucleotide substitutions, one exonic mutation, and four deep intron mutations.
It is well established that genotype-phenotype shows a strong correlation in XLAS (7–9). Jais et al. (9) reported that large deletions and nonsense mutations confer a 90% probability of ESRD by the age of 30 years old compared with a 70% risk with splice site mutations and a 50% risk with missense mutations. Gross et al. (8) grouped men with XLAS into three groups as follows. (1) Large rearrangements, frame shift, nonsense, and splice donor site mutations had a mean ESRD age of 19.8±5.7 years. (2) Nonglycine or 3′ glycine missense mutations, in-frame deletions/insertions, and splice acceptor site mutations had a mean ESRD age of 25.7±7.2 years, and (3) 5′ glycine substitutions had an even later onset of ESRD at a mean of 30.1±7.2 years (8). Recently, Bekheirnia et al. (7) reported the average onset of ESRD as 37 years old for those with missense mutations, 28 years old for those with splice site mutations, and 25 years old for those with truncating mutations. Although these reports very clearly show genotype-phenotype correlations, all studies have grouped splice site mutations together without considering their diverse consequences for collagen transcripts.
Recently, we reported that 29% of men with XLAS were positive for α5(IV) on the GBM and showed significantly milder phenotypes, including milder proteinuria, later onset of ESRD, and less occurrence of hearing loss (13). All of the α5(IV)-positive group had nontruncating mutations, including three deletion mutations (9, 36, and 384 bp) and one splice site mutation, which led to exon 9 (81 bp) skipping. From these results, it was suggested that in-frame mutations could show a milder phenotype, even if derived from a splice site mutation (13). One patient (patient ID 128) in this study had a 105-bp deletion (full exon 27 skipping) and showed an extremely mild phenotype for a man with XLAS. He is now 46 years old and has not developed ESRD (eGFR=41 ml/min per 1.73 m2).
One man (patient ID 19) showed an atypically mild phenotype of slight proteinuria with normal renal function at the age of 22 years old. The intron mutation in this patient created a cryptic exon that included a stop codon, creating a truncating mutation. Patients with truncating mutations usually show a severe disease course. Bekhernia et al. (7) reported that patients with truncating mutations develop to ESRD at an average age of 25 years old, and Gross et al. (8) reported that patients with nonsense or other large mutations develop to ESRD at an average age of 19.8 years old. To clarify the reason why our patient showed a milder phenotype while possessing a truncating mutation, we conducted additional transcript analysis using cDNA extracted from the kidney and hair roots. Interestingly, cDNA from the kidney showed both a larger-sized band and a normal-sized band on electrophoresis and confirmed the presence of normal transcripts lacking the cryptic exon. This finding suggests that this patient’s intronic exonization does not occur completely in the kidney and that some transcript escaped from producing the cryptic exon. This result could explain why this patient shows a milder phenotype of XLAS.
Patient ID 217 had a mutation at IVS cryptic exon position +5T>G that produced a cryptic splice donor site. The IVS+5G position is susceptible to aberrant splice site activation, and this point mutation of G to another nucleotide always changes the splice site, usually showing exon skipping (23). However, this patient created a cryptic splice site with the IVS+5T>G mutation and produced the cryptic splice site. This kind of mutation has been rarely reported (23).
In a recently published study, we reported that some men with XLAS were positive for α5(IV) at the GBM and that all were diagnosed with XLAS using a genetic approach (13). However, four patients in this study who showed no mutations using standard genome DNA direct sequencing analysis (patients ID 19, 48, 126, and 217) had abnormal α5(IV) expression, and we proceeded to transcript analysis and detected aberrant splicing in a deep intron. From these results, it is suggested that the genetic approach and immunohistochemical analysis provide some clues as to diagnosis of XLAS for patients with atypical α5(IV) expression and patients with an absence of any mutation by standard direct sequencing, respectively.
A recent case study reported a high sensitivity for detecting somatic mosaic mutations in the COL4A5 gene using next-generation sequencing (24). High-throughput sequencing technologies may not only lower the cost of DNA sequencing but also help identify those low-percentage somatic mosaic mutations, which may help increase the mutation detection rates for XLAS as well as transcript analysis.
The results of this study may have implications in future therapeutic trials of XLAS. In muscular dystrophies, clinical trials using antisense oligonucleotides to induce exon skipping of specific mutations or drugs developed to allow read through of nonsense mutations are ongoing and have the aim of changing truncating mutations into nontruncating mutations (25). Similar therapeutic strategies may be available for patients with genetically diagnosed XLAS in the future, and accurate descriptions of the transcriptomic consequences of somatic or germ-line DNA changes in relevant tissues should be a prerequisite for inclusion in any clinical trials.
In conclusion, we report 10 intronic mutations and one exonic mutation that produce aberrant splicing, including four deep intronic mutations that produced cryptic exons. One patient showed a milder phenotype, although he had a truncating mutation because of different splicing patterns in different tissues. With transcript analysis, we can determine splice site mutations as either truncating or nontruncating mutations. This study provides valuable data, which will help with future analysis of genotype-phenotype correlations, because only transcript analysis makes it possible to divide splice site mutations into two groups of either truncating or nontruncating mutations. This report also illustrates that the low mutation detection rate in XLAS can be improved by thorough transcript analysis and highlights the importance of distinct tissue-specific exon inclusion levels that may explain the severity of genetic diseases in general.
Disclosures
K.I. received grants from Pfizer Japan Inc.; Kyowa Hakko Kirion, Co., Ltd.; Abbot Japan Co., Ltd.; Takeda Pharmaceutical Co., Ltd.; Asahi Kasei Pharma Corp.; Astellas Pharma Inc.; Terumo Medical Care K.K.; Chugai Pharmaceutical Co., Ltd.; Benesis (currently Japan Blood Product Organization); Dainippon Sumitomo Pharma Co., Ltd.; Genzyme Japan K.K.; Novartis Pharma K.K.; Mizutori Clinic; AbbVie LLC; and Janssen Pharmaceutical K.K. and lecture fees from Novartis Pharma K.K.; Asahi Kasei Pharma Corp.; Baxter Ltd.; Sanofi K.K.; Pfizer Japan Inc.; Meiji Seika Pharma Co., Ltd.; Taisho Toyama Pharmaceutical Co., Ltd.; Kyorin Pharmaceutical Co., Ltd.; Kyowa Hakko Kirion Co., Ltd.; Dainippon Sumitomo Pharma Co., Ltd.; Astellas Pharma Inc.; and Chugai Pharmaceutical Co., Ltd. K.I. has recently become an advisor for Zenyaku Kogyo Co., Ltd.
Supplementary Material
Acknowledgments
The authors acknowledge the cooperation of the attending physicians in this study.
This study was supported by Ministry of Education, Culture, Sports, Science and Technology of Japan Grant-in-Aid for Scientific Research (KAKENHI) 25893131 (to K.N.), Mother and Child Health Foundation Grant 25-7 (to K.N.), and Ministry of Health, Labour and Welfare, Japan, for Research on Rare Intractable Diseases in Kidney and Urinary Tract Grant H24-nanchitou (nan)-ippan-041 (to K.I.) in the “Research on Measures for Intractable Diseases” Project.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
This article contains supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.04140414/-/DCSupplemental.
References
- 1.Kashtan CE: Alport syndrome and thin glomerular basement membrane disease. J Am Soc Nephrol 9: 1736–1750, 1998 [DOI] [PubMed] [Google Scholar]
- 2.Barker DF, Denison JC, Atkin CL, Gregory MC: Efficient detection of Alport syndrome COL4A5 mutations with multiplex genomic PCR-SSCP. Am J Med Genet 98: 148–160, 2001 [DOI] [PubMed] [Google Scholar]
- 3.Cheong HI, Park HW, Ha IS, Choi Y: Mutational analysis of COL4A5 gene in Korean Alport syndrome. Pediatr Nephrol 14: 117–121, 2000 [DOI] [PubMed] [Google Scholar]
- 4.Hertz JM, Juncker I, Persson U, Matthijs G, Schmidtke J, Petersen MB, Kjeldsen M, Gregersen N: Detection of mutations in the COL4A5 gene by SSCP in X-linked Alport syndrome. Hum Mutat 18: 141–148, 2001 [DOI] [PubMed] [Google Scholar]
- 5.Martin P, Heiskari N, Zhou J, Leinonen A, Tumelius T, Hertz JM, Barker D, Gregory M, Atkin C, Styrkarsdottir U, Neumann H, Springate J, Shows T, Pettersson E, Tryggvason K: High mutation detection rate in the COL4A5 collagen gene in suspected Alport syndrome using PCR and direct DNA sequencing. J Am Soc Nephrol 9: 2291–2301, 1998 [DOI] [PubMed] [Google Scholar]
- 6.Nagel M, Nagorka S, Gross O: Novel COL4A5, COL4A4, and COL4A3 mutations in Alport syndrome. Hum Mutat 26: 60, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Bekheirnia MR, Reed B, Gregory MC, McFann K, Shamshirsaz AA, Masoumi A, Schrier RW: Genotype-phenotype correlation in X-linked Alport syndrome. J Am Soc Nephrol 21: 876–883, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gross O, Netzer KO, Lambrecht R, Seibold S, Weber M: Meta-analysis of genotype-phenotype correlation in X-linked Alport syndrome: Impact on clinical counselling. Nephrol Dial Transplant 17: 1218–1227, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Jais JP, Knebelmann B, Giatras I, De Marchi M, Rizzoni G, Renieri A, Weber M, Gross O, Netzer KO, Flinter F, Pirson Y, Verellen C, Wieslander J, Persson U, Tryggvason K, Martin P, Hertz JM, Schröder C, Sanak M, Krejcova S, Carvalho MF, Saus J, Antignac C, Smeets H, Gubler MC: X-linked Alport syndrome: Natural history in 195 families and genotype- phenotype correlations in males. J Am Soc Nephrol 11: 649–657, 2000 [DOI] [PubMed] [Google Scholar]
- 10.King K, Flinter FA, Nihalani V, Green PM: Unusual deep intronic mutations in the COL4A5 gene cause X linked Alport syndrome. Hum Genet 111: 548–554, 2002 [DOI] [PubMed] [Google Scholar]
- 11.Lo YF, Nozu K, Iijima K, Morishita T, Huang CC, Yang SS, Sytwu HK, Fang YW, Tseng MH, Lin SH: Recurrent deep intronic mutations in the SLC12A3 gene responsible for Gitelman’s syndrome. Clin J Am Soc Nephrol 6: 630–639, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nozu K, Iijima K, Nozu Y, Ikegami E, Imai T, Fu XJ, Kaito H, Nakanishi K, Yoshikawa N, Matsuo M: A deep intronic mutation in the SLC12A3 gene leads to Gitelman syndrome. Pediatr Res 66: 590–593, 2009 [DOI] [PubMed] [Google Scholar]
- 13.Hashimura Y, Nozu K, Kaito H, Nakanishi K, Fu XJ, Ohtsubo H, Hashimoto F, Oka M, Ninchoji T, Ishimori S, Morisada N, Matsunoshita N, Kamiyoshi N, Yoshikawa N, Iijima K: Milder clinical aspects of X-linked Alport syndrome in men positive for the collagen IV α5 chain. Kidney Int 85: 1208–1213, 2014 [DOI] [PubMed] [Google Scholar]
- 14.Krol RP, Nozu K, Nakanishi K, Iijima K, Takeshima Y, Fu XJ, Nozu Y, Kaito H, Kanda K, Matsuo M, Yoshikawa N: Somatic mosaicism for a mutation of the COL4A5 gene is a cause of mild phenotype male Alport syndrome. Nephrol Dial Transplant 23: 2525–2530, 2008 [DOI] [PubMed] [Google Scholar]
- 15.Schwartz GJ, Gauthier B: A simple estimate of glomerular filtration rate in adolescent boys. J Pediatr 106: 522–526, 1985 [DOI] [PubMed] [Google Scholar]
- 16.Schwartz GJ, Haycock GB, Edelmann CM, Jr., Spitzer A: A simple estimate of glomerular filtration rate in children derived from body length and plasma creatinine. Pediatrics 58: 259–263, 1976 [PubMed] [Google Scholar]
- 17.Matsuo S, Imai E, Horio M, Yasuda Y, Tomita K, Nitta K, Yamagata K, Tomino Y, Yokoyama H, Hishida A, Collaborators developing the Japanese equation for estimated GFR : Revised equations for estimated GFR from serum creatinine in Japan. Am J Kidney Dis 53: 982–992, 2009 [DOI] [PubMed] [Google Scholar]
- 18.Inoue Y, Nishio H, Shirakawa T, Nakanishi K, Nakamura H, Sumino K, Nishiyama K, Iijima K, Yoshikawa N: Detection of mutations in the COL4A5 gene in over 90% of male patients with X-linked Alport's syndrome by RT-PCR and direct sequencing. Am J Kidney Dis 34: 854–862, 1999 [DOI] [PubMed] [Google Scholar]
- 19.Naito I, Kawai S, Nomura S, Sado Y, Osawa G, Japanese Alport Network : Relationship between COL4A5 gene mutation and distribution of type IV collagen in male X-linked Alport syndrome. Kidney Int 50: 304–311, 1996 [DOI] [PubMed] [Google Scholar]
- 20.Nakanishi K, Iijima K, Kuroda N, Inoue Y, Sado Y, Nakamura H, Yoshikawa N: Comparison of alpha5(IV) collagen chain expression in skin with disease severity in women with X-linked Alport syndrome. J Am Soc Nephrol 9: 1433–1440, 1998 [DOI] [PubMed] [Google Scholar]
- 21.Sado Y, Kagawa M, Kishiro Y, Sugihara K, Naito I, Seyer JM, Sugimoto M, Oohashi T, Ninomiya Y: Establishment by the rat lymph node method of epitope-defined monoclonal antibodies recognizing the six different alpha chains of human type IV collagen. Histochem Cell Biol 104: 267–275, 1995 [DOI] [PubMed] [Google Scholar]
- 22.Cartegni L, Chew SL, Krainer AR: Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet 3: 285–298, 2002 [DOI] [PubMed] [Google Scholar]
- 23.Buratti E, Chivers M, Královicová J, Romano M, Baralle M, Krainer AR, Vorechovsky I: Aberrant 5′ splice sites in human disease genes: Mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res 35: 4250–4263, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beicht S, Strobl-Wildemann G, Rath S, Wachter O, Alberer M, Kaminsky E, Weber LT, Hinrichsen T, Klein HG, Hoefele J: Next generation sequencing as a useful tool in the diagnostics of mosaicism in Alport syndrome. Gene 526: 474–477, 2013 [DOI] [PubMed] [Google Scholar]
- 25.Mercuri E, Muntoni F: Muscular dystrophy: New challenges and review of the current clinical trials. Curr Opin Pediatr 25: 701–707, 2013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.