Abstract
Biological membranes organize and compartmentalize cell signaling into discrete microdomains, a process that often involves stable, cholesterol-rich platforms that facilitate protein-protein interactions. Polarized cells with distinct apical and basolateral cell processes rely on such compartmentalization to maintain proper function. In the cochlea, a variety of highly polarized sensory and non-sensory cells are responsible for the early stages of sound processing in the ear, yet little is known about the mechanisms that traffic and organize signaling complexes within these cells. We sought to determine the prevalence, localization, and protein composition of cholesterol-rich lipid microdomains in the cochlea. Lipid raft components, including the scaffolding protein caveolin and the ganglioside GM1, were found in sensory, neural, and glial cells. Mass spectrometry of detergent-resistant membrane (DRM) fractions revealed over 600 putative raft proteins associated with subcellular localization, trafficking, and metabolism. Among the DRM constituents were several proteins involved in human forms of deafness including those involved in ion homeostasis, such as the potassium channel KCNQ1, the co-transporter SLC12A2, and gap junction proteins GJA1 and GJB6. The presence of caveolin in the cochlea and the abundance of proteins in cholesterol-rich DRM suggest that lipid microdomains play a significant role in cochlear physiology.
Keywords: Hearing, cochlea, hair cell, microdomain, caveolin, lipid raft
INTRODUCTION
The membrane lipid bilayer serves as a gatekeeper in cell physiology, segregating biological reactions to specific extracellular and intracellular spaces and serving as a hub of cell signaling. Fittingly, over 30% of the genes in most genomes are estimated to code for integral membrane proteins [1], and the proportion of proteins associated with the membrane is even greater when including those anchored by glycosylphosphatidylinositol (GPI) [2], fatty acid modifications [3, 4], or other proteins. According to the fluid mosaic model, these membrane proteins diffuse freely within a fluid lipid bilayer unless compartmentalized by protein-protein interactions [5]. However, an accumulating amount of evidence suggests that lipid-protein and lipid-lipid interactions in the plasma membrane constrain the motion of membrane proteins and aid in their localization [6–8]. These observations and others gave rise to the concept of lipid rafts, discrete 10–200 nm lipid-ordered microdomains enriched with sphingomyelin, glycosphingolipids, GM1 ganglioside, and cholesterol [6, 7, 9–12]. The partitioning of specific proteins within these microdomains appears to be a primary means for compartmentalizing various processes in many, if not most, cell types. A large diversity in the structure and dynamics of lipid rafts [9, 10, 13] underlies their ability to organize a wide array of processes including protein and lipid trafficking, endocytosis, cellular recognition, and signaling [12].
The unique composition of lipid rafts renders them insoluble in micelles of non-ionic detergents. These so-called detergent-resistant membrane (DRM) fragments can be separated from soluble membrane by sucrose density gradient fractionation [9, 14]. Although DRM fragments and in situ lipid rafts are not to be equated, the analysis of DRM composition traditionally is the first step in identification of putative raft components. Within the DRM, there are at least two domain types, those that incorporate the scaffolding protein caveolin and those that do not. Moreover, caveolin-based domains can be further classified into two subtypes based on whether the scaffold assembles into planar rafts or morphologically distinct membrane invaginations termed caveolae [15].
In hair cells in the inner ear, observations of caveolae-like structures were reported decades ago [16, 17], but little is known about the role of cholesterol-enriched microdomains in the inner ear. Hair cells contain diverse machinery, with mechanotransduction of sound energy occurring at the apical end, synaptic transmission occurring at the basolateral end, and a complex network of ion channels functioning together to shape excitability. The polarity of these cells, their role in signal transduction, and the interplay of these ion channels necessitates the localization and co-localization of membrane proteins to specific regions of the cell. Multiple observations including the presence of caveolae, inhomogeneous distribution of membrane cholesterol [18, 19], and detection of extensive segregation of lipids in the membrane of the hair bundle [20] suggest that the lipid bilayer of these cells serves to modulate membrane protein distribution and functionality. While only caveolin and BK-type potassium channels have been biochemically identified in DRMs of cochlea [18], there is growing evidence that cholesterol-enriched microdomains may be involved in a wide range of processes in the inner ear, including sensory transduction[20] and cochlear mechanics [21–24]. Moreover, disruption of these microdomains with cholesterol-chelating cyclodextrins causes aberrant electrophysiological behavior [18, 22, 23, 25, 26], while systemic delivery of cyclodextrins causes profound hearing loss and outer hair cell death with no apparent effect on other systems [27].
To explore what processes may be affected by changes in membrane cholesterol distribution in the inner ear, and the mechanisms behind the inner ear’s unique sensitivity, the localization and composition of cholesterol-enriched membrane microdomains was analyzed. The presence of raft-like domains is supported by localization of the raft-associated ganglioside GM1 and the raft-scaffolding protein caveolin to sensory and non-sensory cells in the ear. Additionally, over 600 proteins were found in triplicate preparations of cochlear DRM, and gene ontology analysis suggested DRM involvement in cell signaling, protein localization, and metabolism. Three gene products associated with syndromic hearing loss and eight gene products associated with non-syndromic hearing loss were also found in all three samples.
METHODS
Tissue Preparations
All animal procedures were conducted with the express approval of the University Animal Care and Use Committee at the University of Michigan. White Leghorn chickens (Gallus gallus), 18–28 days old, were anesthetized with a ketamine/xylazine solution and euthanized by decapitation. Whole auditory organs and microdissected subcompartments were collected essentially as described previously [28]. Pectoral skeletal muscle, proventricular smooth muscle, heart, lung, and brain (cerebellum) were dissected rapidly on ice.
Gene expression assays
Total RNA was extracted from basilar papilla (10–12 per sample) and auditory nerve (8–12 per sample) using an RNEasy Micro Kit (Qiagen), and from 50–200 mg samples of all other organ tissues using Trizol reagent (Invitrogen) after mechanical homogenization. RNA integrity was assessed using an Agilent 2100 Bioanalyzer. Only samples with a 28S:18S ratios greater than 1.0 were included in the analyses. First-strand cDNA was synthesized using Superscript III Reverse Transcriptase (Invitrogen) according to manufacturer’s instructions.
Polymerase chain reaction (PCR) was used to verify the presence of caveolin expression in the cochlea. Specific primers designed to amplify G. gallus caveolins included: Cav1 (GenBank accession NM_0011056641) forward 5′-CAACATCTACAAGCCCAATAACAA-3′, reverse 5′-CTGAACACCTTGCCCATAGC-3′, amplicon 438 bp; Cav2 (GenBank accession NM_001007086) forward 5′-AAGAGCCTGACAGATGTTTTCGTT-3′, reverse 5′-AGCACAGTGAGGGCCAAAAATGAT-3′, amplicon 528 bp; Cav3(GenBank accession NM_204370) forward 5′-GTGGGGACGTACAGCTTTGATGGT-3′, reverse 5′-GAGGGAGTAGATGCGGCTGACACA-3′, amplicon 225 bp. PCR reactions were performed using Platinum Taq DNA polymerase (Invitrogen) and 200 ng cDNA. PCR products were electrophoresed on a 1% agarose gel with 0.005% ethidium bromide. Bands were visualized with an Alpha Innotech FluorChem SP Imaging System (ProteinSimple). The identity of the amplicons was confirmed by DNA sequencing.
Real-time, quantitative polymerase chain reaction (qPCR) was conducted to determine differential expression of the caveolin genes between tissues. These reactions were performed on an ABI Prism 7900HT Sequence Detection System using custom-designed TaqMan probes and commercial PCR master mix (TaqMan 2X PCR Master Mix, Applied Biosystems). Each sample was run in triplicate and was normalized to the housekeeping gene S16 (Rps16). Primer combinations amplified a single product of expected size, and amplification efficiency for each gene product was within 90–100%. Primer and probe sequences for target genes included: Cav1 forward 5′-ATGTCCGGCACCAAATACGT-3′, reverse 5′-GCTTGTAGATGTTGCCCTGTTC-3′, probe 5′-TCGGAGGGCTTTCTG-3′; Cav2 forward 5′-CCCCGCGGGCTGAA-3′, reverse 5′-CGGGCTCGGCGATCA-3′, probe 5′-CTCCAGCTGGGCTTC-3′; Cav3 forward 5′-CCTGGTGAACAGAGATCCAAAGAG-3′, reverse 5′-CCACGGGCTCAGCTATCAC-3′, probe 5′-ATCCTCGAAATCCACCTTTAC-3′. Genes of interest were normalized to the reference gene Rps16 (S16, Genbank accession XM_416113), which encodes a component of the small 40S ribosomal subunit [28]. Differences in expression level were calculated as fold-change relative to a control condition using the Δ ΔCT method [28, 29].
Immunohistochemistry
Temporal bones were dissected and dura, oval window, and middle ear structures were removed. Bones were fixed in 2% paraformaldehyde (PFA) solution for 3 hours at room temperature then decalcified for 2–3 days with 5% EDTA in 0.12M phosphate buffer (PB) with 0.2% PFA (pH 7.4 – 8.0) at room temperature. After rinsing with 0.5% sucrose in 0.12M PB, tissues were cryoprotected via incubation with increasing concentrations of sucrose (5%, 13%, 18%, 22%, and 30% in PB) for 30 min at room temperature and finally overnight at 4°C in 30% sucrose. Tissues were frozen in O.C.T. compound using liquid nitrogen-cooled isopentane and stored at −80°C. Thin cryosections (10–12 μm) were collected alternately on approximately 20 slides so that any one slide contained representative sections from 8 to 10 equally spaced regions along the length of the cochlear duct. To label sections, slides were warmed for 30 minutes at room temperature and washed in phosphate buffered saline (PBS; Sigma D5652). Sections were blocked in 5% normal goat serum and 1% bovine serum albumin in PBS with 0.1% Triton X-100 (TX-100) for 1 hour at room temperature. After removing the blocking buffer, tissue sections were exposed to primary antibody diluted in PBS plus 0.1% TX-100 overnight at 4°C. Primary antibody binding was identified with AlexaFluor-conjugated goat anti-rabbit or anti-mouse secondary antibodies (Invitrogen) diluted 1:500 in PBS plus 0.1% TX-100, applied for 2 hours at room temperature. Controls without primary antibody were prepared and analyzed in parallel.
Dissociated single hair cells were prepared for immunocytochemistry to investigate subcellular compartmentalization of microdomain structures. To dissociate single cells, basilar papillae were treated for less than 1 min with 0.01% protease type XXIV (Sigma, P8038) to facilitate removal of the tegmentum vasculosum and tectorial membrane. A borosilicate capillary tube was used to aspirate the sensory epithelium and mechanically dissociate the cells onto a slide containing (in mM) 150 NaCl, 6 KCl, 5 CaCl2, 2 MgCl2, and 5 HEPES, buffered to pH 7.4 with NaOH. Cells were allowed to settle on SuperfrostPlus slides for 10–15 minutes to promote adherence, then fixed with 2% PFA for 30 min at room temperature. Slides of dissociated cells were blocked and labeled as above, with the exception of using 0.01% saponin in place of 0.1% TX-100 when labeling fixed hair cells with anti-Cav1.
Primary antibodies included rabbit anti-caveolin (BD Transduction Laboratories, 610059), mouse anti-Cav1 (BD Transduction Laboratories, 610406), mouse anti-Cav2 (BD Transduction Laboratories, 610684), mouse anti-Cav3 (BD Transduction Laboratories, 610420), and rabbit anti-neurofilament M 145 kD (Chemicon, AB1987). In some cases, hair cell preparations were treated with 1:200 OG-phalloidin (Invitrogen, O7466) to identify actin-rich hair bundles. When required, nuclei were counterstained with 1:50 Hoechst 33342 (Invitrogen, H3570). Sections and cells were imaged on a Leica DM LB fluorescence microscope (Leica Microsystems). Images were acquired using single- and triple-band filter sets and a cooled-CCD color digital camera (MicroPublisher, QImaging).
Isolation of detergent resistant membrane fractions
Twelve whole cochlear ducts per sample were extracted, pooled into a single tube, and flash frozen on dry ice. Samples were thawed on ice in 120 μl of MES buffered saline (MBS; 25 mM MES, 150 mM NaCl, pH 6.5) containing 0.25%, 0.5%, 1%, or 2% (v/v) TX-100 and protease inhibitor cocktail (Thermo Fisher, PI-78410). After homogenization with a motorized pestle, lysates were incubated on ice for 30 minutes. The homogenate was placed on the bottom of a 2.2 ml ultracentrifuge tube and mixed 1:3 with 53.3% (w/v) sucrose/MBS to a final concentration of 40% (w/v) sucrose/MBS. For a discontinuous density gradient, this solution was overlaid with 900 μl of 30% sucrose/MBS, then overlaid with 1mL of 5% sucrose/MBS and balanced. Membrane proteins were separated using an Optima Max-E Ultracentrifuge with a swinging bucket rotor (TLS-55) (Beckman-Coulter, Fullerton, CA). The samples were ultracentrifuged at 54,000 rpm (~200,000g) for 24 hours at 4 °C. Twelve equal volume fractions (183 μl each) and a pellet fraction were collected from the top.
Immunoblot
Aliquots (25 μl) were taken from each fraction, mixed with Laemmli sample buffer (Bio-Rad 161-0737) containing 5% 2-mercaptoethanol, separated by SDS-PAGE on a 4–15% polyacrylamide gel, and transferred to nitrocellulose membranes (Pierce Biotechnology, Rockford, IL) for Western blotting. Membranes were probed with a primary antibody to caveolin (1:1000 anti-caveolin, BD Transduction Laboratories, 610059) or transferrin receptor (1:1000 anti-TfR, Invitrogen, 13-6800). Secondary antibodies included goat anti-mouse or anti-rabbit conjugated to horseradish peroxidase (1:10000, Thermo Scientific Pierce, 31432 or 31460). Reactions were visualized with SuperSignal West Femto chemiluminescent substrate (Thermo Fisher Pierce, 34095) and imaged on an Alpha Innotech FluorChem SP Imaging System (ProteinSimple).
Labeling of GM1-enriched domains
The localization of GM1-enriched domains was visualized with fluorescently conjugated cholera toxin subunit B (CTX) (FITC-CTX, Sigma C1655; Alexa Fluor 488-CTX, Invitrogen C34775). Dissociated cochlear hair cells were pre-fixed with 2% PFA for 30 minutes, treated with 0.01% saponin for 15 min, and exposed to 8–20 μg/ml CTX for 30 minutes at room temperature. In some cases, CTX was directly applied to freshly dissociated hair cells in the absence of PFA and saponin. Punctate aggregates of CTX label were counted on pre-fixed, 488-CTX-labeled cells. Clearly isolated puncta greater than 0.3 μm in diameter and located within the outline of the basolateral cell (i.e. excluding the hair bundle and cuticular plate) were included in these counts.
Dot blots of subcellular membrane fractions were labeled with HRP-conjugated CTX (Invitrogen, C34780). One microliter of each fraction was applied to strips of dry nitrocellulose membrane. The strips were blocked for 1 hour at room temperature in 5% (w/v) non-fat milk in TBS containing 0.1%(v/v) Tween (TBS-T) and incubated at 4°C overnight in 200 ng/mL HRP-CTX in 5% non-fat milk in TBS-T. Reactions were visualized with SuperSignal West Femto chemiluminescent substrate and imaged on an Alpha Innotech FluorChem SP Imaging System.
Protein quantification
The micro bicinchoninic acid (BCA) protein assay (Pierce Biotechnology, 23235) was used for protein quantification. Fractions were diluted with distilled water to reach protein concentrations within the linear range of the assay. Absorbances were measured at 590 nm using an automated microplate reader (EL311, Bio-Tek Instruments).
Cholesterol quantification
The Amplex Red cholesterol assay (Invitrogen, A12216) was used for cholesterol quantification according to manufacturer instructions, and fluorescence was measured using a Fluoroskan plate reader (Thermo Fischer Scientific Inc.). Fractions were diluted with the provided reaction buffer to reach cholesterol concentrations within the linear range of the assay.
Mass spectrometry
Detergent-resistant membrane fractions from three independent biological samples were submitted to MS Bioworks for protein mass spectrometry analysis. Following sucrose-gradient separation, fractions 5–7 for a given sample were pooled and dissolved in MBS with protease inhibitor cocktail (Thermo Fisher, PI-78410) for a final volume of 2.3 mL. The diluted samples were centrifuged for 1 hour at 54,000 rpm (~200,000g) at 4°C. The pellet was resolubilized in 100 μL of 2x Laemmli Sample Buffer (Bio-Rad, 161-0737) diluted 1:1 in MBS without 2-mercaptoethanol. Protein concentrations were measured and 20 μg of protein from each sample was separated ~1.5 cm on a 10% Bis-Tris Novex mini-gel (Invitrogen) using the MES buffer system. The gel was stained with Coomassie blue and each lane was excised into ten equally sized segments. Gel pieces were processed using a robot (ProGest, DigiLab), which washed with 20 mM ammonium bicarbonate followed by acetonitrile, then reduced with 10 mM dithiothreitol at 60°C, followed by alkylation with 50 mM iodoacetamide at room temperature, digested with trypsin (Promega) at 37°C for 4 hours, and quenched with formic acid. The supernatant was analyzed directly without further processing. Each fraction was analyzed by nano LC/MS/MS with a Waters NanoAcquity HPLC system interfaced to a ThermoFisher LTQ Orbitrap Velos Pro. Peptides were loaded on a trapping column and eluted over a 75 μm analytical column at 300 nL/min; both columns were packed with Jupiter Proteo resin (Phenomenex). The mass spectrometer was operated in data-dependent mode, with MS performed in the Orbitrap at 60,000 FWHM resolution and MS/MS performed in the LTQ. The fifteen most abundant ions were selected for MS/MS. Spectral data were searched using Mascot (Matrix Science) with the following parameters: Enzyme, Trypsin; Database, Uniprot Chicen (concatenated forward and reverse plus common contaminants); Fixed modification, Carbamidomethyl (C); Variable modifications, Oxidation (M), Acetyl (N-term), Pyro-Glu (N-term Q), Deamidation (N,Q); Mass values, Monoisotopic; Peptide mass tolerance, 10 ppm; Fragment mass tolerance, 0.8 Da; Max missed cleavages, 2.
Mascot DAT files were parsed into Scaffold4 (Proteome Software) for validation, filtering, and to create a non-redundant list per sample. Data were filtered using a minimum protein value of 99.0%, a minimum peptide value of 50.0% (Prophet scores), and a minimum of two unique peptides per protein. Protein and peptide false discovery rates were both set to 1%. The spectral abundance factor for each protein was calculated by dividing the number of spectral counts of assigned peptides by the protein molecular weight in kDa. The spectral abundance factor for each protein was divided by the sum of all of the spectral abundance factors in each biological sample, disregarding contaminants and decoy proteins, resulting in the normalized spectral abundance factor (NSAF) [30, 31]. The NSAF for each protein was averaged over all three biological samples.
Gene ontology and Prediction tools
Gene ontology (GO) analysis was conducted using blast2GO [32–35] and the ClueGO extension [36] for Cytoscape 3.0.1 [37]. Protein FASTA sequences for each UniProt entry were entered into blast2GO and all default settings for blast2GO were used, except when creating the combined graph for biological process GO terms in which a minimum of 3 sequences was required to create a node. All annotation tools in blast2GO were used, except GO-slim. Gene symbols inferred from homology were submitted into ClueGO and all default ClueGO settings were used, except for the following: GO terms were limited to one level at a time, and the minimum number of sequences per node was set to 1. GO term enrichment tests were conducted according to hypergeometric tests and corrected using Bonferroni step down analysis [36, 38].
Transmembrane helix prediction was conducted using the online TMHMM 2.0 prediction server(http://www.cbs.dtu.dk/services/TMHMM/) [1, 39] and palmitoylation was assessed using CSS-Palm 3.0 [40, 41]. Glypiation was predicted using the online predGPI prediction server (http://gpcr.biocomp.unibo.it/predgpi/) [2], and myristoylation was predicted using the online Expasy Myristoylator prediction server (http://web.expasy.org/myristoylator/) [42].
RESULTS
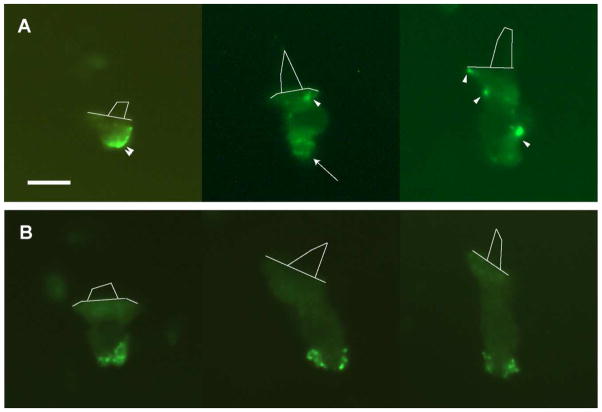
Dissociated hair cells were stained by fluorescently-conjugated cholera toxin β-subunit (CTX), a marker for GM1 gangliosides and lipid rafts. Live-cell preparations exhibited punctate CTX spots at subnuclear and supranuclear ends of the hair cell (Figure 1A). Exemplar cells classified as “short”, “intermediate”, and “tall” based on length/width ratios were similarly labeled. These morphologies reflect distinct functional roles in the chick cochlea [43]. The results, therefore, indicate that CTX shows no preference for hair cell subtype. More recently, CTX has been applied to fixed preparations in order to limit the possibility of CTX-induced endocytosis and translocation of GM1-rich membrane domains [44, 45]. Labeling in fixed hair cells resulted in more numerous, smaller, and more distinct puncta located primarily in subnuclear regions of the cell (Figure 1B). Approximately 60% of isolated, fixed hair cells demonstrated quantifiable punctate spots. On average, labeled hair cells exhibited 3–4 puncta (Figure 2). The remaining cells included those that were unlabeled, those masked by debris, and those with indistinct puncta (i.e. too small or without clear boundaries).
Figure 1. CTX labeling of isolated hair cells reveals punctate GM1-rich microdomains.
Unfixed (A) and pre-fixed (B), dissociated hair cells from throughout the abneural and neural regions of the cochlea exhibited bright, punctate spots following CTX application. Unlabeled preparations and those treated with unconjugated CTX were devoid of punctate staining (not shown). Live cells displayed punctate spots between 0.3 and 1 μm in diameter (arrow) as well as large, diffuse domains along the basolateral surface (arrowhead). Punctate spots were exclusively found on pre-fixed, CTX-labeled cells. Hair bundles are outlined with thin lines to show cell orientation. Scale bar = 10 μm.
Figure 2. Histogram of CTX puncta shows an average of 3–4 puncta per hair cell.
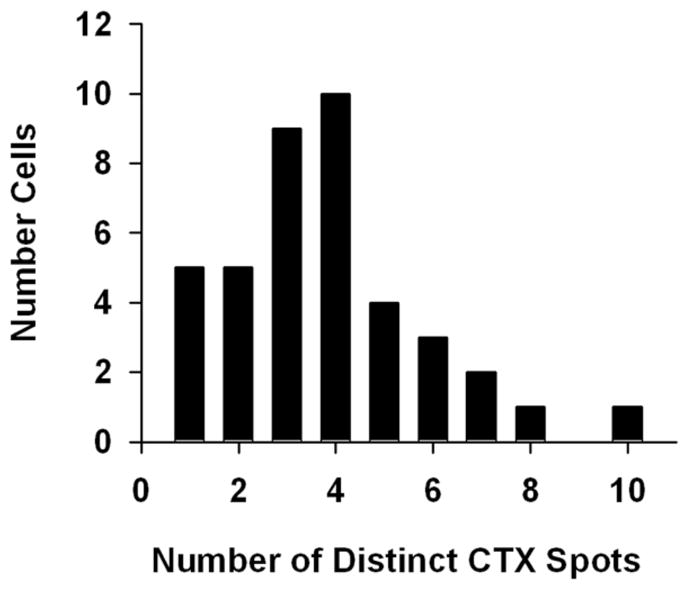
Pre-fixed, isolated hair cells displaying one or more CTX-puncta were analyzed, revealing a mean of 3.8 ± 0.3 (standard error of the mean) puncta per cell. N = 40.
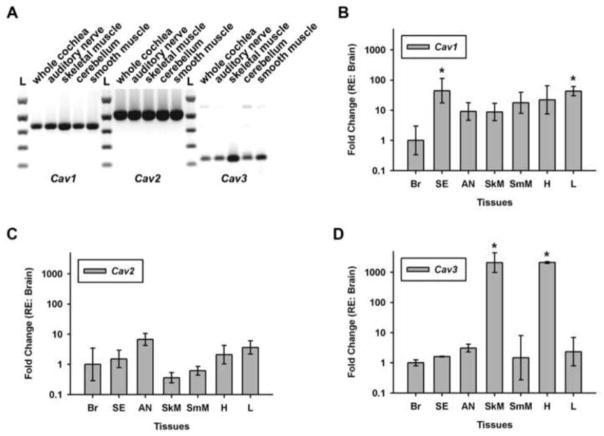
While GM1 is often found co-localized with the cholesterol-binding caveolin, there is evidence in chick brain that GM1 and caveolin are members of separate populations of DRM [46]. To probe caveolin expression, total RNA was collected from whole cochlea, auditory nerve, skeletal muscle, cerebellum, and smooth muscle and probed for the expression of caveolin 1–3 (Figure 3A). All three members of the caveolin gene family were detected in whole cochlea and auditory nerve. The presence of caveolin 3 in the cochlea was somewhat surprising since this product is primarily associated with muscle and glia. To probe further, qPCR was used to examine caveolin expression in the microdissected, hair cell-enriched sensory epithelium and compared to other tissues (Figure 3B–D). The auditory sensory epithelium and lung were significantly enriched in caveolin 1 compared to brain (P<0.05, one-way ANOVA), whereas caveolin 2 expression was more uniform between tissues. Skeletal muscle and heart were significantly enriched in caveolin 3 (P<0.05, one-way ANOVA), while the other tissues had relatively similar levels compared to brain.
Figure 3. All three members of the caveolin gene family are expressed in the chick cochlear sensory epithelium and auditory nerve.
(A) RT-PCR of whole cochlea, auditory nerve, and select control tissues reveal bands corresponding to Cav1, Cav2, and Cav3. Quantitative PCR shows the differential expression of (B) Cav1, (C) Cav2, and (D) Cav3 in sensory epithelium(SE), auditory nerve(AN), skeletal (SkM) and smooth (SmM) muscle, heart (H), and liver (L) tissue relative to cerebellum (Br). *, P<0.05, One-way ANOVA with post-hoc pairwise comparison to Br.
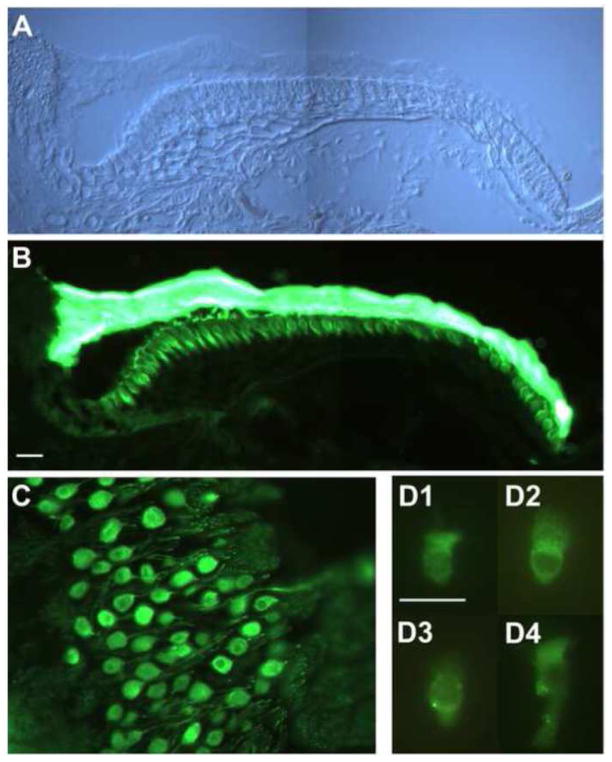
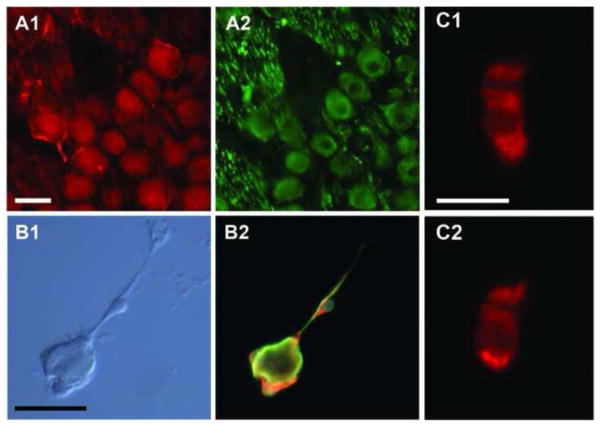
To localize the expression of caveolin in the inner ear, sections of chicken cochlea and dissociated hair cells were probed for the presence of caveolin 1 (Figure 4) and caveolin 3 (Figure 5); antibodies to caveolin 2 revealed no specific staining above background (data not shown). Radial frozen sections of the basilar papilla stained with anti-caveolin 1, showing bright label in hair cells throughout the neural (left) to abneural (right) cross-section (i.e. no preference for short or tall hair cells) (Figure 4A–B). Primary auditory neurons and their peripheral projections were strongly labeled by anti-caveolin 1 (Figure 4C). Similar to results with CTX, approximately 50% of isolated hair cells revealed several punctate caveolin-1-positive spots (Figure 4D). Anti-caveolin 3 labeled throughout the sensory epithelium making it difficult to distinguish between specific label in hair cells, supporting cells, and neuronal cell types (data not shown). The strongest caveolin 3 staining was found in regions surrounding the soma of primary auditory neurons (Figure 5A). To confirm that this label was associated with small diameter glial cells associated with these neurons, teased nerve preparations were stained for caveolin 3 and neurofilament. In some cases, large diameter neurofilament-positive neurons were found with tightly bound, small-diameter, neurofilament-negative/caveolin-3-positive cells indicative of caveolin 3 expression in glial cells (Figure 5B). Approximately 30% of isolated hair cells showed evidence of punctate caveolin-3-positive spots at the basolateral pole of the cell (Figure 5C).
Figure 4. Anti-caveolin 1 labels hair cells and auditory neurons in chick cochlea.
(A) A radial section from the distal (low-frequency) portion of the chick basilar papilla is shown to illustrate the position of hair cells (HC) and supporting cells (SC) of the sensory epithelium, neighboring homogene cells (HG), and the tectorial membrane (TM). (B) Anti-caveolin 1 brightly labeled the basolateral portion of the hair cells. As is often the case, the tectorial membrane was non-specifically reactive with the secondary antibody. Supporting cells were not immunoreactive. (C) Primary auditory neurons were specifically labeled by anti-caveolin 1. (D1-4) Isolated hair cells were diffusely labeled by anti-caveolin 1, with approximately 50% of the cells also displaying punctate basolateral spots (among 20 imaged hair cells). Hair bundles (all pointing upward) were not immunoreactive. Scale bars, A-C = 10 μm, D = 20 μm.
Figure 5. Anti-caveolin 3 labels hair cells and non-neuronal cells in the auditory ganglion.
A cryosection of the auditory ganglion is shown stained for caveolin 3 (A1) and neurofilament (A2), revealing caveolin staining in glia surrounding the neuron cell bodies. Teased nerve preparations of isolated neuron/glia complexes (B1) were stained with anti-caveolin 3 (red), neurofilament (green), and Hoechst (blue) and imaged with a triple-band filter (B2). Neurofilament-negative cells labeled strongly with anti-caveolin 3. Isolated hair cells were labeled throughout by anti-caveolin 3, except the cuticular plate at the base of the hair bundle (C1-2). Approximately 30% of the cells displayed punctate spots at the base of the cell (C2, among 20 imaged hair cells). Scale bars, A-B = 20 μM, C = 10 μM.
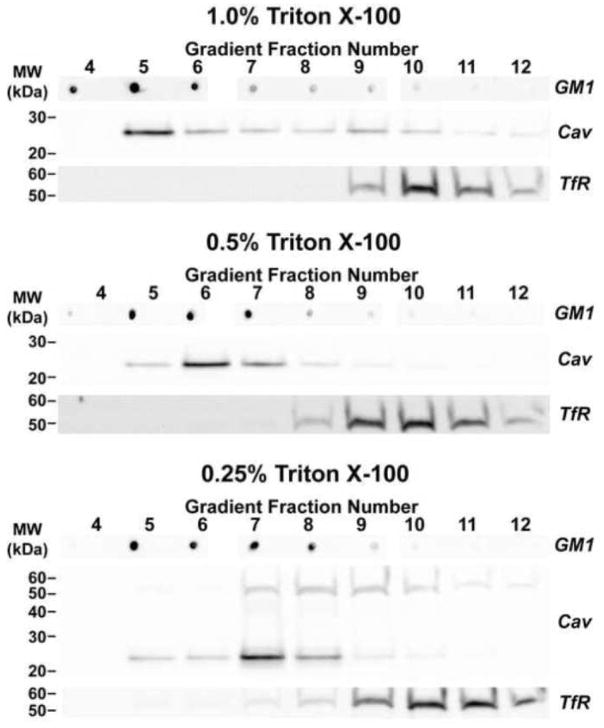
To begin unraveling the composition of caveolin-based and GM1-rich microdomains, whole cochlear lysates were treated with cold TX-100 and subjected to sucrose-gradient fractionation. Using 1% TX-100, the composition of proteins in DRM fractions differed from those in detergent-sensitive and pellet fractions (Figure 6). While this concentration of TX-100 is a useful starting point separating DRM and non-DRM, some optimization was required to (1) fully solubilize non-DRM while leaving DRMs intact, (2) identify those fractions that contain both caveolin and GM1, and (3) confirm the enrichment of cholesterol in those DRM/caveolin/GM1 fractions. High detergent concentrations (1% [Figure 7] and 2% TX-100 [not shown]) resulted in a broad distribution of caveolin and GM1 across low and high numbered fractions, overlapping with the non-DRM marker transferrin receptor. Similarly, at 0.25% TX-100, caveolin monomers and putative dimers were distributed across most fractions, overlapping with transferrin receptor. The separation was optimized with 0.5% TX-100 with faint overlap of caveolin/GM1 and transferrin receptor in fraction 8. Under these conditions, caveolin and GM1 were concentrated particularly in fractions 5, 6, and 7.
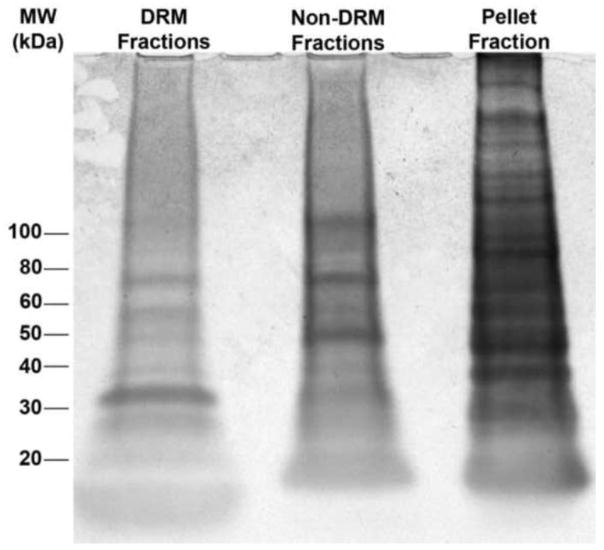
Figure 6. SDS-PAGE of cochlear fractions reveals heterogeneous protein distributions among DRM, non-DRM, and soluble protein pools.
Whole cochlea lysates treated with 1% TX-100 were separated by SDS-PAGE and stained using Coomassie Blue to reveal the diversity and abundance of proteins in pooled DRM fractions (4–6), pooled detergent sensitive fractions (9–11), and the pellet of soluble proteins.
Figure 7. The separation of DRM and non-DRM membrane fractions is dependent on TX-100 concentration.
Western blots and dot blots of fractionated whole cochlear lysates are shown following treatment with 1%, 0.5%, or 0.25% TX-100. Western blots were probed with pan-caveolin (Cav) and transferrin receptor (TfR) antibodies to identify DRM and non-DRM fractions, respectively. Dot blots were probed with HRP-conjugated CTX to identify GM1-enriched fractions.
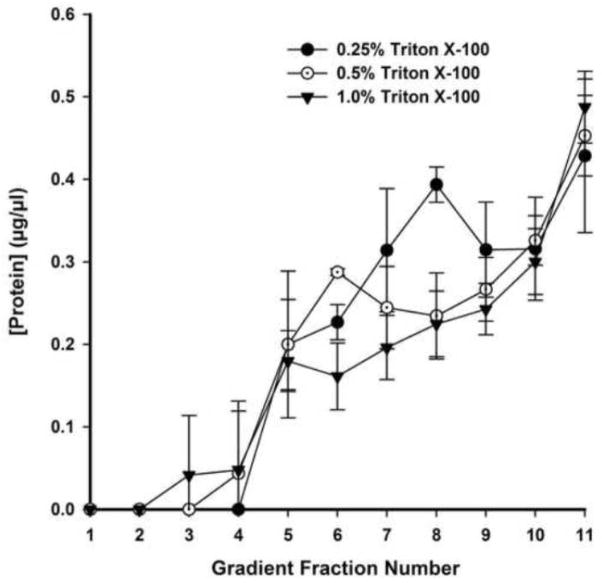
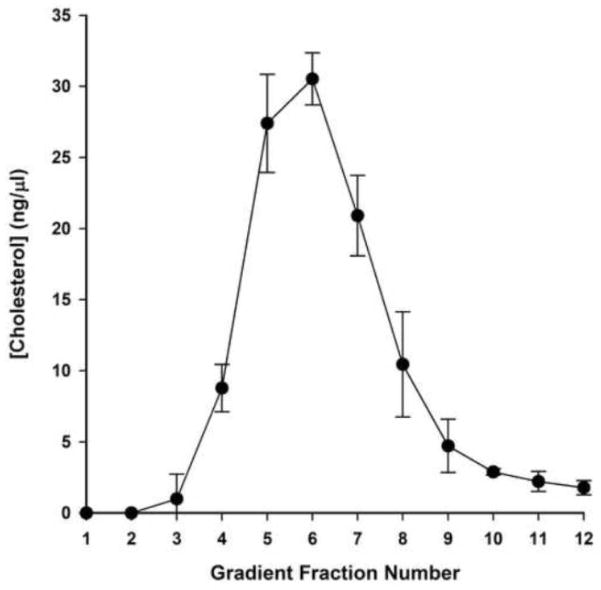
Likewise, bulk protein partitioning in DRM fractions was best separated from soluble fractions with 0.5% TX-100. The distribution of protein in fractions 4–11 was reliably dependent on TX-100 concentration (Figure 8) (P<0.05, two-way ANOVA). For 0.25% TX-100, total protein was undetectable in the lowest density fractions (1–4), peaking in fraction 8 and reducing only slightly before rising again in fractions 11–12. In higher concentrations of TX-100, protein was first detectable in fractions 3 and 4. Treatment with 1% TX-100 resulted in a gradually increasing distribution of protein from low- to-high-density fractions; there was no apparent bimodal separation of protein in DRM and non-DRM fractions. The 0.5% TX-100 condition, however, showed a distinct peak of protein concentration in fraction 6, consistent with the separation of caveolin and transferrin receptor seen in immunoblots. Likewise, cholesterol was concentrated in fractions 5–7 under these conditions, peaking in fraction 6, whereas higher numbered, detergent-soluble fractions were depleted of cholesterol (Figure 9). Together, these data depict an optimized separation between DRM and non-DRM pools using 0.5% TX-100.
Figure 8. Protein concentration among DRM and non-DRM fractions varies considerably with concentration of TX-100.
Protein concentration was quantified by BCA assay and averaged for fractions 1–11 of 0.25%, 0.5%, and 1% TX-100 treated whole cochlea lysates. N = 3 for each group. Errors represent one standard deviation from the mean.
Figure 9. Cholesterol is specifically associated with DRM fractions.
Cholesterol concentration was determined using Amplex Red enzyme assays for fractions 1–12 of 0.5% TX-100 treated whole cochlea lysates and averaged across 3 independent samples. Error bars represent one standard deviation from the mean.
LC-MS/MS of pooled DRM fractions 5, 6, and 7 identified 967 proteins out of three separate biological samples, with 14 contaminants and 9 decoy proteins using the criteria described (Supplementary Table 1). A total of 606 proteins were common to all three separate biological samples and were used for all further analyses. Using gene ontology (GO) analysis, twenty-six (4%) of these DRM proteins were well-characterized raft-related proteins, including caveolin-1, thy-1, and flotillin-2 (Table 1). Only 264 (44%) of the identified proteins had predicted transmembrane helices based on protein sequence (Supplementary Table 1). Protein sequences were examined for post-translational modifications that can target peripheral membrane proteins to DRM (glypiation, palmitoylation, and myristoylation) [12, 47] using prediction algorithms. The presence of sites specific to each of these three post-translational modifications and association with protein complexes is indicated in Supplementary Table 1 and summarized in Table 2. In total, 20 proteins were predicted to be myristoylated, and 26 were predicted to be glypiated with no overlap. In contrast, 462 proteins (76%) had predicted palmitoylation sites. Additionally, using GO analysis, 247 proteins (41%) were identified as well-characterized constituents of protein complexes, which could facilitate membrane association by incorporating integral membrane components into the complex. Importantly, there is significant overlap between proteins with predicted palmitoylation sites and all other categories. Only 41 proteins (7%) were not predicted to be associated with the membrane by any of these methods.
Table 1.
Identified DRM proteins associated with the GO term “membrane raft”
| Gene Description | Gene Name |
|---|---|
| ATPase, Na+/K+ transporting, alpha 2 polypeptide | ATP1A2 |
| ATPase, Ca2+ transporting, plasma membrane 1 | ATP2B1 |
| Caveolin 1 | CAV1 |
| Cadherin 13 | CDH13 |
| Dystrophin | DMD |
| Flotillin 2 | FLOT2 |
| Connexin 43 | GJA1 |
| G-protein subunits/polypeptides | GNA11, GNAI2, GNAI3, GNAS |
| Glypican 1 | GPC1 |
| Inositol 1,4,5-triphosphate receptors | ITPR1, ITPR3 |
| K-Ras P21 protein | KRAS |
| Late endosomal/ lysosomal adaptor and MAPK and MTOR Activator 3 | LAMTOR3 |
| Lck/Yes-related novel protein tyrosine kinase | LYN |
| Neimann-Pick C1 Protein | NPC1 |
| Protein Kinase C, alpha type | PRKCA |
| Major Prion Protein | PRNP |
| RAS-related Protein Rab-5A | RAB5A |
| HEPG2 Glucose transporter type 1, erythrocyte/brain | SLC2A1 |
| Syntaxins | STX2, STX12 |
| Thy-1 Membrane Glycoprotein/ Surface Antigen | THY1 |
Table 2.
Predicted transmembrane helices, post-translational modifications, and protein complex association of identified DRM proteins
| Transmembrane | Palmitoylation | Glypiation | Myristoylation | Protein Complex | Combined for Each Category | |
|---|---|---|---|---|---|---|
| Transmembrane | 4.6% | 34.7% | 1.7% | 0.2% | 15.2% | 43.6% |
| Palmitoylation | 19.6% | 4.0% | 2.1% | 17.5% | 76.2% | |
| Glypiation | 0.2% | 0% | 1.0% | 4.3% | ||
| Myristoylation | 0.8% | 0.3% | 3.3% | |||
| Protein Complex | 6.8% | 40.8% |
The 20 DRM proteins with highest NSAF are described in Table 3. Peptide spectra matched to these proteins accounted for 23.7% of the NSAF. The majority of these proteins are involved in mitochondrial metabolic processes including proteins from the respiratory chain, namely ATP synthase and cytochrome C oxidase components, and proteins involved in transporting organic molecules across the mitochondrial outer membrane. Well-known constituents of lipid microdomains in the plasma membrane and intracellular organelles, such as prohibitins, annexins and myelin protein zero were also abundant. GO analysis of all 606 DRM proteins by biological process revealed top level 2 categories (>5% of NSAF) of cellular processes, metabolic processes, localization and organization, biological regulation and response to stimulus, multicellular processes and signaling (Figure 10A). Top level 2 categories of cellular component (>1% of NSAF) GO analysis were cell, membrane, protein complexes, and organelles, specifically membrane-bounded organelles (Figure 10B).
Table 3.
Identified DRM proteins with highest NSAF
| Gene Name | Gene Description | NSAF |
|---|---|---|
| VDAC2 | Outer Mitochondrial Membrane Porin 2 (voltage dependent) | 2.59% |
| COX6C | Cytochrome C Oxidase, Subunit 6C | 2.02% |
| MPZ | Myelin Protein Zero | 1.91% |
| SLC25A4 | Mitrochondrial ADP/ATP Translocase 1 | 1.55% |
| ATP1A1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 1.52% |
| COX4I1 | Cytochrome C Oxidase Subunit 4, Isoform 1 | 1.20% |
| ANXA2 | Annexin A2 | 1.16% |
| ATP5A1 | ATP Synthase, H+ Transporting, F1 Complex, Alpha Subunit 1 | 1.12% |
| ATP5B | ATP Synthase, H+ Transporting, F1 Complex, Beta Subunit | 1.08% |
| CISD1 | CDGSH Iron Sulfur Domain 1 | 1.02% |
| ACTG1 | Cytoplasmic/Cytoskeletal Actin, Gamma 1 | 0.99% |
| COX6A1 | Cytochrome C Oxidase, Subunit 6A | 0.95% |
| SLC25A6 | Mitochondrial ADP/ATP Translocase 3 | 0.87% |
| VDAC1 | Outer Mitochondrial Membrane Porin 1 (voltage dependent) | 0.86% |
| CKAP4 | Cytoskeleton-associated protein 4 | 0.84% |
| SLC25A3 | Mitochondrial Phosphate Carrier Protein | 0.83% |
| PHB | Prohibitin | 0.82% |
| PHB2 | Prohibitin 2 | 0.80% |
| ATP5H | ATP Synthase, H+ Transporting, F0 Complex, Subunit D | 0.80% |
| ANXA6 | Annexin A6 | 0.79% |
Figure 10. GO associations are listed for identified DRM proteins.
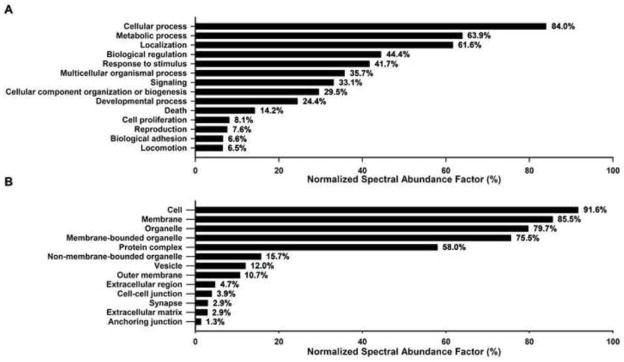
Annotated proteins were weighted by the normalized spectral abundance factor and analyzed using blast2GO to identify major GO associations. (A) Level 2 biological process GO terms are listed for terms with a total NSAF above 5%. (B) Level 3 cellular component GO terms are listed for terms with a total NSAF above 1%.
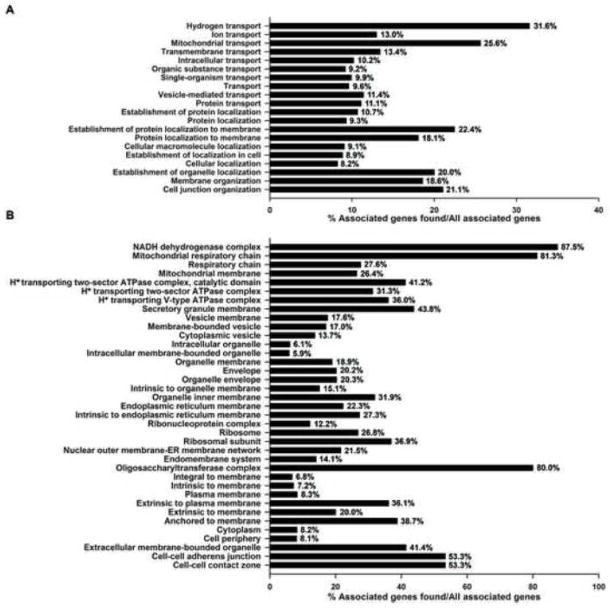
While the GO categories of DRM proteins illustrate the locations and processes in which cholesterol-enriched membrane microdomains may play a role, by focusing on the total spectral abundance of proteins associated with a specific GO category, this approach gives more importance to GO categories that have more database entries. Additionally, this method limits the utility of more specific, higher resolution GO categorization by providing an unmanageably large number of distinct, but related GO terms. Figure 11 depicts significantly enriched (P<0.01), level 3 GO categories by comparing the number of genes that are associated with each GO category with the total number of genes associated with that category in the GO database. Using biological processes GO terms, most of the top categories in Figure 10A are still represented, but now, other important GO categories are included such as those related to various transport processes and subcellular localization (Figure 11A). This approach to GO analysis highlights many smaller and more specific cellular component GO categories (Figure 11B), including those associated with cell-cell junctions and protein anchoring at the plasma membrane. The membrane raft component was also significantly enriched (P<0.05) (not shown).
Figure 11. Significantly enriched GO categories are listed for identified DRM proteins.
(A) Level 3 biological process GO terms and (B) Level 3 cellular component terms with significant enrichment (P<0.01). Terms are scored according to the number of genes identified in the DRM that are associated with a given term as a percent of the total number of genes associated with that term.
By far, processes associated with metabolism and bioenergetics involved the majority of identified DRM proteins (63.9%), which carries particular importance given the cochlea’s acute sensitivity to mitochondrial dysfunction. However, the analyses in Figure 11 highlight additional processes/components that play influential roles in the cochlea, including those involved in ion homeostasis, exocytosis and vesicular transport, and cell-cell interactions befitting an epithelial tissue separated by unique ionic environments. To further explore the DRM components involved in these processes, we examined those proteins lying at the key intersections of vesicular transport through which the trafficking of proteins and synaptic signaling in hair cells occurs (Supplementary Table 3), ion transport through which biological regulation, signaling and homeostasis occur (Supplementary Table 4), and cell junctions where signaling and protein localization are crucial (Supplementary Table 5). Over 100 proteins were associated with the GO terms “vesicle” and/or “vesicle-mediated transport” (Supplementary Table 3). Thirty-one were associated with the Golgi or endoplasmic reticulum membrane, where proteins may be embedded in nascent lipid microdomains. Interestingly, 19 of these vesicle-associated proteins were associated with the synapse, and 42 were associated with exocytosis/endocytosis, essential processes in normal cochlear physiology. Supplementary Table 4 depicts a complex set of sodium, potassium, calcium, chloride and magnesium transporters and channels. Over 100 proteins were associated with ion transport or ion flux, suggesting that DRM microdomains may be essential components of ion homeostasis and energy metabolism in the ear. Finally, the GO terms “cell junction” and “cell junction organization” were combined to examine DRM proteins that may be involved in the conservation of the endocochlear potential and potassium recycling in the inner ear [48] (Supplementary Table 5). Key cell junction proteins, including connexins, catenins, plakophilins, spectrins, and claudin-1, were present in inner ear DRM along with a number of cell adhesion related surface proteins, such as CD9 and thy-1. These results suggest that cholesterol-rich domains significantly contribute to the tight-junction and gap-junction networks involved in potassium recycling within the ear.
Among the proteins found in DRM microdomains, eight were associated with non-syndromic deafness and three with syndromic deafness (Table 4). These proteins are primarily involved in ion transport and homeostasis, cell-cell junctions, vesicular transport, and extracellular structures, broadly reflecting the range of enriched components/processes identified by the GO analysis.
Table 4.
Identified DRM proteins associated with deafness
| Gene Name | Gene Description | Deafness |
|---|---|---|
| ACTG1 | Cytoplasmic/Cytoskeletal Actin, Gamma 1 | Non-Syndromic |
| CISD2 | CDGSH Iron Sulfur Domain 2 | Non-Syndromic |
| CLCNKB | Chloride Channel, Voltage-Sensitive Kb | Syndromic |
| GJA1 | Connexin 43 | Non-Syndromic |
| GJB6 | Connexin 30 | Non-Syndromic |
| KCNQ1 | Voltage-gated Potassium Channel | Syndromic |
| MYH9 | Myosin, Heavy Chain 9, non-muscle | Non-Syndromic |
| MYO6 | Myosin 6 | Non-Syndromic |
| OTOA | Otoancorin | Non-Syndromic |
| SLC12A2 | Basolateral Na+-K+-Cl− Symporter | Syndromic |
| TECTA | Tectorin Alpha | Non-Syndromic |
DISCUSSION
Mounting evidence supports a role for cholesterol in the segregation of membrane domains in the cochlea and in modulating cochlear physiology [18, 20, 23, 25, 26]. Until now, such studies have only hinted at the involvement of raft-like, lipid-ordered microdomains that, in other systems, limit lateral diffusion of membrane-associated proteins and aid in the compartmentalization of cell signaling. Our study represents the first comprehensive analysis of the localization and composition of cholesterol-enriched membrane microdomains in a peripheral auditory organ. The raft markers caveolin and GM1 were found in both sensory and non-sensory cells in the inner ear, and a screen of cochlear DRM fractions revealed hundreds of proteins that co-segregated with cholesterol and GM1. Bioinformatics analysis of these microdomains identified proteins involved in metabolic, trafficking, and signaling processes that are essential to normal cochlear function.
Caveolin is a scaffolding protein that identifies a subclass of lipid raft, structurally supporting either planar domains or invaginating caveolae. Caveolin expression was relatively high in the cochlea and auditory ganglion, compared to other central and peripheral tissues. Immunohistochemistry detected Cav1 and Cav3 in sensory cells, Cav1 in auditory neurons, and Cav3 in glia. Caveolin-2 antibodies failed to reveal specific staining patterns, but this may be attributed to limited cross-reactivity with chick isoforms. Based on the expression of Cav2 mRNA in cochlea and similarities in the tissue distribution of Cav1 and Cav2 in other systems [49] it is quite possible that Cav2 also plays a role in inner ear wherever Cav1 is found. Caveolins are widely distributed in adipocytes, epithelial, and endothelial cells, as well as sensory, neural, and muscle cells [50–53]. Caveolin interacting proteins include Src family tyrosine kinases, nitric oxide synthase, receptor tyrosine kinases, phospholipase C, protein kinase C, Ras proteins, and G protein subunits [51]. Many of these proteins play important roles in cochlear signaling, development of hearing, and cochlear response to injury.
Caveolin and GM1-rich microdomains were identified in isolated hair cells as discrete puncta located along the basolateral membrane. Unfortunately, the size and subcellular localization of raft domains can be confounded by coalescence and redistribution during labeling. For example, when applied to live cells, CTX can induce the internalization of GM1-rich vesicles, whereas application of CTX to lightly fixed, cold preparations can limit redistribution of GM1 [45]. These observations may explain the appearance of large and more broadly distributed CTX-domains in live compared to fixed hair cells. Taken together, our data would suggest that GM1-rich domains are present at the synaptic pole of the cell, consistent with caveolin-1 and caveolin-3 staining patterns. However, it remains unclear whether these markers labeled the same structures [46, 54]. Nevertheless, it is tempting to associate these basolateral microdomains with synaptic active zones, calcium hotspots, and BK-channel puncta that all exist along the basal hair cell membrane and are similar to CTX-positive spots in both size and number [55–57]. This inference is supported by evidence that BK-channel subunits are incorporated into DRM and that both BK activity and calcium channel function are modulated by the DRM-disruption agent methyl-beta-cyclodextrin [18]. Further study is required to define the roles of GM1 and caveolin in hair cells and to test whether these domains organize channel clusters and synaptic machinery.
As a first step toward identifying the protein composition of lipid rafts in the cochlea, we optimized the separation of DRM from non-DRM fractions by titrating the concentration of TX-100. Although 1.0% TX-100 has been considered a gold-standard for defining DRM, protein recovery in DRM fractions is exquisitely sensitive to detergent:protein ratio [58, 59]. In chick cochlea under these homogenization conditions, a lower concentration of 0.5% TX-100 yielded optimal separation, reducing loss of DRM proteins from over- or under-solubilization. When subjected to mass spectrometry, over 960 proteins were identified in three separate cochlear DRM preparations. About 600 of these proteins were common to each biological repeat with the remainder largely representing low abundant proteins at the detection limits of our approach. We identified a relatively large set of DRM proteins compared to similar studies, which reported 70 DRM proteins in Jurkat T cells [60], 380 in kidney epithelial cells [61], and 216 from neonatal mouse brain [62]. The detection of Cav1 in our screen provides internal validation that DRM fractions contained sufficient quantities of DRM markers identified by qPCR, immunohistochemistry, and immunoblot. However, Cav3 was absent from the LC-MS/MS results as was the BK-channel α subunit previously reported in chick cochlear DRM [18]. These omissions suggest a detection limit in the proteomic analysis and the missed identification of some verifiable DRM proteins.
Our informatics predictions estimated that only about 40% of identified DRM proteins have transmembrane helices. However, post-translational modifications including palmitoylation, myristoylation, and glypiation anchor proteins to raft-like domains [12, 47]. Predictive algorithms suggested that these modifications could account for the targeting of the majority of non-integral membrane proteins to DRM fractions. Additionally, DRM are rich in protein complexes, which may also anchor non-integral membrane proteins to membrane microdomains. With this under consideration, only 7% of identified proteins were neither predicted to have transmembrane domains nor any of these post-translational modifications and were not well-characterized members of protein complexes. Interestingly, proteins in this minority group include prohibitin and apolipoproteins, which are purported lipid-raft constituents [63, 64], suggesting that the prediction and GO analyses may underestimate the number of proteins that may in fact be membrane, and specifically DRM, associated. It is especially interesting to note that these post-translational modifications are highly regulated and modulated processes, enabling the cell to exert another level of control over DRM incorporation. Palmitoylation is highly dynamic. Over its lifetime, a protein can be palmitoylated and depalmitoylated several times allowing for dynamic incorporation and exclusion from microdomains [47]. For example palmitoylation of inactive H-Ras targets the protein from Golgi to the plasma membrane, and depalmitoylation results in H-Ras internalization [4]. H-Ras is involved in gentamicin-related ototoxicity [65], and its presence in cochlear DRM could provide novel links between fatty-acid modifications, signaling compartmentalization, and hearing loss due to aminoglycosides. Similarly, myristoylation and glypiation can participate in domain targeting [12] and can operate in a regulated manner [66, 67]. Taken together, the prevalence of cochlear DRM proteins with putative fatty acid modifications highlights the possibility that lipid-organized platforms in the ear are highly dynamic in nature and contribute to a large array of regulatory processes.
The impact of cytoskeletal and organelle-associated proteins in DRM fractions is poorly understood. Indeed, their identification as raft constituents remains controversial. However, cytoskeletal proteins are known to interact with many well-known raft proteins, and such interactions may be vital for the stabilization and function of the microdomain [12, 68]. As an example, proteins involved in vesicle transport and actin-myosin motility were enriched in our DRM fractions. As a molecular motor, myosin is an essential component of membrane recycling, transport, and remodeling [69]. Some myosins have been implicated in the organization of cholesterol-rich membranes [70]. In hair cells specifically, myosin 1c, the hair cell’s adaptation motor, exhibits non-ionic detergent resistance [71, 72]. The presence of myosin 1c in our cholesterol-rich DRM supports the hypothesis that elements of the transduction apparatus rely on lipid-mediated compartmentalization [20, 73].
The enrichment of endoplasmic reticulum, Golgi, and mitochondrial proteins in DRM is even less well understood as these membranes contain little cholesterol [12]. In many cases, these proteins may be recruited to the cell surface [74–77]. On the other hand, several lines of evidence suggest that intracellular organelles contain raft-like domains. Raft-dependent endocytosis [78, 79] and Golgi membrane cycling [80] indicates that lipid microdomains are involved in protein sorting and vesicular trafficking between organelles and with the cell surface. Similarly, raft-associated lipids including glycosphingolipids and polysialogangliosides are present in mitochondria [12, 81] and raft-associated proteins like prohibitin and erlin have been identified in mitochondrial membrane and ER membrane, respectively [63]. These findings support the possibility that intracellular organelles contain raft-like microdomains that differ in composition from plasma membrane microdomains and require less cholesterol [63].
Two major themes emerge from the bioinformatics analysis, namely DRM involvement in metabolism and ion homeostasis. The large number of proteins associated with bioenergetics and metabolism included several respiratory chain and mitochondrial transport proteins, and the product of the non-syndromic deafness gene Cisd2. Oxidative stress, mediated by Rac/Rho pathways and dysfunctional energetics, is commonly associated with antibiotic-related ototoxicity, noise trauma, and age-related hearing loss [82–84]. The prevalence of metabolism-related proteins in inner ear DRM, as well as both Rac1 and RhoA, suggests a possible link between DRM organization and oxidative stress.
In addition to bioenergetics, proteins involved in ion homeostasis were major components of the cochlear DRM. The cochlea is comprised of three fluid filled chambers and differences in the ionic composition of these fluids as well as an associated electrical potential between the fluids is a critical feature of normal cochlear function. A hallmark of ion homeostasis in the ear is the recycling of potassium through these fluid compartments. Cochlear DRM included the major ion channel responsible for potassium secretion into endolymph (KCNQ1) [85], gap junction proteins essential for the flux of potassium through cellular compartments (GJA1 and GJB6) [86, 87], the co-transporter largely responsible for establishing the endocochlear potential (SLC12A2) [88], and tight-junction proteins that facilitate separation of the endolymphatic-perilymphatic compartments.
Our results provide new insights into the possible impact of cholesterol-rich microdomains in the ear and provide new hypotheses for uncovering the unique sensitivity of the ear to cholesterol-chelating cyclodextrins. In cats and mice, systemic application of 2-hydroxypropyl-beta-cyclodextrin causes substantial outer hair cell death and hearing loss with no obvious pathology in other systems [27, 89]. The unique sensitivity of the ear to cyclodextrins is perplexing given the ubiquitous distribution of cholesterol in cells throughout the body. However, the ear is particularly sensitive to oxidative stress and ion imbalance, two processes heavily linked to DRM composition. These data add to prior reports of cyclodextrin effects on cochlear mechanics [22, 23, 25] and the electrical activity of hair cells, specifically calcium influx and potassium efflux [18]. The ability of cyclodextrins to function as a cholesterol shuttle has been utilized as a treatment for Neiman-Pick C disease, a usually fatal disorder, where NPC1 or NPC2 deficiency prevents cholesterol efflux to the plasma membrane [90]. Hearing loss is part of the clinical diagnosis and spectrum of this disease and is associated with other genetic cholesterol synthesis disorders [91–94]. The profound sensitivity of the auditory organ to cyclodextrins implores further study into the source of cholesterol synthesis in the inner ear and the relationship between cholesterol homeostasis and hearing loss.
Conclusion
The presence of over 600 proteins in inner ear cholesterol-enriched membrane microdomains along with the prevalence of microdomains in a variety of inner ear cell types indicates the importance lipid-mediated compartmentalization in this organ. Increasing evidence of lipid and cholesterol segregation in the hair cell and the unique sensitivity of the auditory organ to changes in cholesterol localization motivates further study into the mechanism of cholesterol synthesis and regulation in this system.
Supplementary Material
SIGNIFICANCE.
Although mechanisms underlying cholesterol synthesis, homeostasis, and compartmentalization in the ear are poorly understood, there are several lines of evidence indicating that cholesterol is a key modulator of cochlear function. Depletion of cholesterol in mature sensory cells alters calcium signaling, changes excitability during development, and affects the biomechanical processes in outer hair cells that are responsible for hearing acuity. More recently, we have established that the cholesterol-modulator beta-cyclodextrin is capable of inducing significant and permanent hearing loss when delivered subcutaneously at high doses. We hypothesize that proteins involved in cochlear homeostasis and otopathology are partitioned into cholesterol-rich domains. The results of a large-scale proteomics analysis point to metabolic processes, scaffolding/trafficking, and ion homeostasis as particularly associated with cholesterol microdomains. These data offer insight into the proteins and protein families that may underlie cholesterol-mediated effects in sensory cell excitability and cyclodextrin ototoxicity.
HIGHLIGHTS.
We report for the first time evidence of raft-like microdomains in the cochlea.
Key raft markers are expressed as puncta in sensory and non-sensory cells.
Over 600 proteins were identified in cochlear detergent-resistant membrane factions.
Major gene ontology terms included energetics, transport, ion homeostasis, and cell contact.
Acknowledgments
This work was supported by grants from the National Institute of Deafness and Communication Disorders (R01 DC07432 to RKD and core grant P30 DC05188).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 2.Pierleoni A, Martelli PL, Casadio R. PredGPI: a GPI-anchor predictor. BMC bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. Journal of molecular biology. 2002;317:541–57. doi: 10.1006/jmbi.2002.5426. [DOI] [PubMed] [Google Scholar]
- 4.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–90. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 5.Singer SJ, Nicolson GL. The Fluid Mosaic Model of the Structure of Cell Membranes. Science. 1972;175:720–31. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 6.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–80. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 7.Kusumi A, Ike H, Nakada C, Murase K, Fujiwara T. Single-molecule tracking of membrane molecules: plasma membrane compartmentalization and dynamic assembly of raft-philic signaling molecules. Seminars in Immunology. 2005;17:3–21. doi: 10.1016/j.smim.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, et al. PARADIGM SHIFT OF THE PLASMA MEMBRANE CONCEPT FROM THE TWO-DIMENSIONAL CONTINUUM FLUID TO THE PARTITIONED FLUID: High-Speed Single-Molecule Tracking of Membrane Molecules. Annual Review of Biophysics and Biomolecular Structure. 2005;34:351–78. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 9.Pike LJ. Lipid rafts: heterogeneity on the high seas. The Biochemical journal. 2004;378:281–92. doi: 10.1042/BJ20031672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lingwood D, Simons K. Lipid Rafts As a Membrane-Organizing Principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 11.Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. Journal of lipid research. 2006;47:1597–8. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Pike LJ. The challenge of lipid rafts. Journal of lipid research. 2009;50:S323–S8. doi: 10.1194/jlr.R800040-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karnovsky MJ, Kleinfeld AM, Hoover RL, Klausner RD. The concept of lipid domains in membranes. The Journal of Cell Biology. 1982;94:1–6. doi: 10.1083/jcb.94.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–44. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 15.Head BP, Insel PA. Do caveolins regulate cells by actions outside of caveolae? Trends in Cell Biology. 2007;17:51–7. doi: 10.1016/j.tcb.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Saito K. Fine structure of the sensory epithelium of guinea-pig organ of Corti: subsurface cisternae and lamellar bodies in the outer hair cells. Cell and tissue research. 1983;229:467–81. doi: 10.1007/BF00207692. [DOI] [PubMed] [Google Scholar]
- 17.Saito K. Fine structure of the sensory epithelium of the guinea pig organ of Corti: afferent and efferent synapses of hair cells. Journal of ultrastructure research. 1980;71:222–32. doi: 10.1016/s0022-5320(80)90108-2. [DOI] [PubMed] [Google Scholar]
- 18.Purcell EK, Liu L, Thomas PV, Duncan RK. Cholesterol influences voltage-gated calcium channels and BK-type potassium channels in auditory hair cells. PloS one. 2011;6:e26289. doi: 10.1371/journal.pone.0026289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen T-VN, Brownell WE. Contribution of membrane cholesterol to outer hair cell lateral wall stiffness. Otolaryngology--Head and Neck Surgery. 1998;119:14–20. doi: 10.1016/S0194-5998(98)70167-6. [DOI] [PubMed] [Google Scholar]
- 20.Zhao H, Williams DE, Shin J-B, Brügger B, Gillespie PG. Large membrane domains in hair bundles specify spatially constricted radixin activation. The Journal of Neuroscience. 2012;32:4600–9. doi: 10.1523/JNEUROSCI.6184-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sturm AK, Rajagopalan L, Yoo D, Brownell WE, Pereira FA. Functional expression and microdomain localization of prestin in cultured cells. Otolaryngology -- Head and Neck Surgery. 2007;136:434–9. doi: 10.1016/j.otohns.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sfondouris J, Rajagopalan L, Pereira FA, Brownell WE. Membrane Composition Modulates Prestin-associated Charge Movement. Journal of Biological Chemistry. 2008;283:22473–81. doi: 10.1074/jbc.M803722200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajagopalan L, Greeson JN, Xia A, Liu H, Sturm A, Raphael RM, et al. Tuning of the Outer Hair Cell Motor by Membrane Cholesterol. Journal of Biological Chemistry. 2007;282:36659–70. doi: 10.1074/jbc.M705078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajagopalan L, Organ-Darling L, Liu H, Davidson A, Raphael R, Brownell W, et al. Glycosylation Regulates Prestin Cellular Activity. JARO. 2010;11:39–51. doi: 10.1007/s10162-009-0196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brownell WE, Jacob S, Hakizimana P, Ulfendahl M, Fridberger A. Membrane cholesterol modulates cochlear electromechanics. Pflügers Archiv-European Journal of Physiology. 2011;461:677–86. doi: 10.1007/s00424-011-0942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levic S, Yamoah EN. Plasticity in Membrane Cholesterol Contributes toward Electrical Maturation of Hearing. Journal of Biological Chemistry. 2011;286:5768–73. doi: 10.1074/jbc.M110.186486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crumling MA, Liu L, Thomas PV, Benson J, Kanicki A, Kabara L, et al. Hearing Loss and Hair Cell Death in Mice Given the Cholesterol-Chelating Agent Hydroxypropyl-β-Cyclodextrin. PloS one. 2012;7:e53280. doi: 10.1371/journal.pone.0053280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Atkin G, Morales M, Liu L, Tong M, Duncan RK. Developmental expression of BK channels in chick cochlear hair cells. BMC developmental biology. 2009;9:67. doi: 10.1186/1471-213X-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JM, Beyer R, Morales M, Chen S, Liu LQ, Duncan RK. Expression of BK-type calcium-activated potassium channel splice variants during chick cochlear development. Journal of Comparative Neurology. 2010;518:2554–69. doi: 10.1002/cne.22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paulo JA, Urrutia R, Banks PA, Conwell DL, Steen H. Proteomic analysis of an immortalized mouse pancreatic stellate cell line identifies differentially-expressed proteins in activated vs nonproliferating cell states. Journal of proteome research. 2011;10:4835–44. doi: 10.1021/pr2006318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical Analysis of Membrane Proteome Expression Changes in Saccharomyces c erevisiae. Journal of proteome research. 2006;5:2339–47. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 32.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic acids research. 2008;36:3420–35. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 34.Conesa A, Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. International journal of plant genomics 2008. 2008 doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Götz S, Arnold R, Sebastián-León P, Martín-Rodríguez S, Tischler P, Jehl M-A, et al. B2G-FAR, a species-centered GO annotation repository. Bioinformatics. 2011;27:919–24. doi: 10.1093/bioinformatics/btr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivals I, Personnaz L, Taing L, Potier M-C. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics. 2007;23:401–7. doi: 10.1093/bioinformatics/btl633. [DOI] [PubMed] [Google Scholar]
- 39.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings / International Conference on Intelligent Systems for Molecular Biology; ISMB International Conference on Intelligent Systems for Molecular Biology. 1998;6:175–82. [PubMed] [Google Scholar]
- 40.Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Engineering Design and Selection. 2008;21:639–44. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue Y, Liu Z, Cao J, Ren J. Computational Prediction of Post-Translational Modification Sites in Proteins. Ning-Sun Yang. 2011 [Google Scholar]
- 42.Bologna G, Yvon C, Duvaud S, Veuthey AL. N-Terminal myristoylation predictions by ensembles of neural networks. Proteomics. 2004;4:1626–32. doi: 10.1002/pmic.200300783. [DOI] [PubMed] [Google Scholar]
- 43.Fischer FP. Hair cell morphology and innervation in the basilar papilla of the emu (Dromaius novaehollandiae) Hear Res. 1998;121:112–24. doi: 10.1016/s0378-5955(98)00072-0. [DOI] [PubMed] [Google Scholar]
- 44.Selvaraj V, Asano A, Buttke DE, Sengupta P, Weiss RS, Travis AJ. Mechanisms underlying the micron-scale segregation of sterols and GM1 in live mammalian sperm. Journal of cellular physiology. 2009;218:522–36. doi: 10.1002/jcp.21624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blank N, Schiller M, Krienke S, Wabnitz G, Ho AD, Lorenz HM. Cholera toxin binds to lipid rafts but has a limited specificity for ganglioside GM1. Immunology and cell biology. 2007;85:378–82. doi: 10.1038/sj.icb.7100045. [DOI] [PubMed] [Google Scholar]
- 46.Henke RC, Hancox KA, Jeffrey PL. Characterization of two distinct populations of detergent resistant membrane complexes isolated from chick brain tissues. Journal of neuroscience research. 1996;45:617–30. doi: 10.1002/(SICI)1097-4547(19960901)45:5<617::AID-JNR11>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 47.Smotrys JE, Linder ME. PALMITOYLATION OF INTRACELLULAR SIGNALING PROTEINS: Regulation and Function. Annual Review of Biochemistry. 2004;73:559–87. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- 48.Zhao HB, Kikuchi T, Ngezahayo A, White TW. Gap junctions and cochlear homeostasis. The Journal of membrane biology. 2006;209:177–86. doi: 10.1007/s00232-005-0832-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scherer PE, Lewis RY, Volonte D, Engelman JA, Galbiati F, Couet J, et al. Cell-type and tissue-specific expression of caveolin-2. Caveolins 1 and 2 co-localize and form a stable hetero-oligomeric complex in vivo. The Journal of biological chemistry. 1997;272:29337–46. doi: 10.1074/jbc.272.46.29337. [DOI] [PubMed] [Google Scholar]
- 50.Head BP, Hu Y, Finley JC, Saldana MD, Bonds JA, Miyanohara A, et al. Neuron-targeted caveolin-1 protein enhances signaling and promotes arborization of primary neurons. The Journal of biological chemistry. 2011;286:33310–21. doi: 10.1074/jbc.M111.255976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okamoto T, Schlegel A, Scherer PE, Lisanti MP. Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. The Journal of biological chemistry. 1998;273:5419–22. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- 52.Li X, McClellan ME, Tanito M, Garteiser P, Towner R, Bissig D, et al. Loss of caveolin-1 impairs retinal function due to disturbance of subretinal microenvironment. The Journal of biological chemistry. 2012;287:16424–34. doi: 10.1074/jbc.M112.353763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikol DD, Hong HL, Cheng HL, Feldman EL. Caveolin-1 expression in Schwann cells. Glia. 1999;27:39–52. [PubMed] [Google Scholar]
- 54.Pang H, Le PU, Nabi IR. Ganglioside GM1 levels are a determinant of the extent of caveolae/raft-dependent endocytosis of cholera toxin to the Golgi apparatus. J Cell Sci. 2004;117:1421–30. doi: 10.1242/jcs.01009. [DOI] [PubMed] [Google Scholar]
- 55.Tucker T, Fettiplace R. Confocal imaging of calcium microdomains and calcium extrusion in turtle hair cells. Neuron. 1995;15:1323–35. doi: 10.1016/0896-6273(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 56.Martinez-Dunst C, Michaels RL, Fuchs PA. Release sites and calcium channels in hair cells of the chick’s cochlea. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17:9133–44. doi: 10.1523/JNEUROSCI.17-23-09133.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Atkin GM, Morales MM, Liu LQ, Tong M, Duncan RK. Developmental expression of BK channels in chick cochlear hair cells. BMC Dev Biol. 2009;9:67. doi: 10.1186/1471-213X-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ostermeyer AG, Beckrich BT, Ivarson KA, Grove KE, Brown DA. Glycosphingolipids are not essential for formation of detergent-resistant membrane rafts in melanoma cells. methyl-beta-cyclodextrin does not affect cell surface transport of a GPI-anchored protein. The Journal of biological chemistry. 1999;274:34459–66. doi: 10.1074/jbc.274.48.34459. [DOI] [PubMed] [Google Scholar]
- 59.Brady JD, Rich TC, Le X, Stafford K, Fowler CJ, Lynch L, et al. Functional role of lipid raft microdomains in cyclic nucleotide-gated channel activation. Molecular pharmacology. 2004;65:503–11. doi: 10.1124/mol.65.3.503. [DOI] [PubMed] [Google Scholar]
- 60.von Haller PD, Donohoe S, Goodlett DR, Aebersold R, Watts JD. Mass spectrometric characterization of proteins extracted from Jurkat T cell detergent-resistant membrane domains. Proteomics. 2001;1:1010–21. doi: 10.1002/1615-9861(200108)1:8<1010::AID-PROT1010>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 61.Blonder J, Hale ML, Lucas DA, Schaefer CF, Yu LR, Conrads TP, et al. Proteomic analysis of detergent-resistant membrane rafts. Electrophoresis. 2004;25:1307–18. doi: 10.1002/elps.200405891. [DOI] [PubMed] [Google Scholar]
- 62.Yu H, Wakim B, Li M, Halligan B, Tint GS, Patel SB. Quantifying raft proteins in neonatal mouse brain by ‘tube-gel’ protein digestion label-free shotgun proteomics. Proteome science. 2007;5:17. doi: 10.1186/1477-5956-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Browman DT, Hoegg MB, Robbins SM. The SPFH domain-containing proteins: more than lipid raft markers. Trends in Cell Biology. 2007;17:394–402. doi: 10.1016/j.tcb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 64.Browman DT, Resek ME, Zajchowski LD, Robbins SM. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. Journal of cell science. 2006;119:3149–60. doi: 10.1242/jcs.03060. [DOI] [PubMed] [Google Scholar]
- 65.Battaglia A, Pak K, Brors D, Bodmer D, Frangos JA, Ryan AF. Involvement of ras activation in toxic hair cell damage of the mammalian cochlea. Neuroscience. 2003;122:1025–35. doi: 10.1016/j.neuroscience.2003.08.041. [DOI] [PubMed] [Google Scholar]
- 66.Ames JB, Lim S, Ikura M. Molecular structure and target recognition of neuronal calcium sensor proteins. Frontiers in molecular neuroscience. 2012;5:10. doi: 10.3389/fnmol.2012.00010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Mayor S, Riezman H. Sorting GPI-anchored proteins. Nature reviews Molecular cell biology. 2004;5:110–20. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- 68.Oliferenko S, Paiha K, Harder T, Gerke V, Schwärzler C, Schwarz H, et al. Analysis of Cd44-Containing Lipid Rafts Recruitment of Annexin II and Stabilization by the Actin Cytoskeleton. The Journal of cell biology. 1999;146:843–54. doi: 10.1083/jcb.146.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diz-Munoz A, Fletcher DA, Weiner OD. Use the force: membrane tension as an organizer of cell shape and motility. Trends Cell Biol. 2013;23:47–53. doi: 10.1016/j.tcb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanzal-Bayer MF, Hancock JF. Lipid rafts and membrane traffic. FEBS Letters. 2007;581:2098–104. doi: 10.1016/j.febslet.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 71.Gillespie PG, Cyr JL. Myosin-1c, the Hair Cell’s Adaptation Motor. Annual Review of Physiology. 2004;66:521–45. doi: 10.1146/annurev.physiol.66.032102.112842. [DOI] [PubMed] [Google Scholar]
- 72.Gillespie PG, Wagner MC, Hudspeth A. Identification of a 120 kd hair-bundle myosin located near stereociliary tips. Neuron. 1993;11:581–94. doi: 10.1016/0896-6273(93)90071-x. [DOI] [PubMed] [Google Scholar]
- 73.Hirono M, Denis CS, Richardson GP, Gillespie PG. Hair Cells Require Phosphatidylinositol 4,5-Bisphosphate for Mechanical Transduction and Adaptation. Neuron. 2004;44:309–20. doi: 10.1016/j.neuron.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 74.Kim KB, Lee JW, Lee CS, Kim BW, Choo HJ, Jung SY, et al. Oxidation–reduction respiratory chains and ATP synthase complex are localized in detergent-resistant lipid rafts. Proteomics. 2006;6:2444–53. doi: 10.1002/pmic.200500574. [DOI] [PubMed] [Google Scholar]
- 75.Kim BW, Lee JW, Choo HJ, Lee CS, Jung SY, Yi JS, et al. Mitochondrial oxidative phosphorylation system is recruited to detergent-resistant lipid rafts during myogenesis. Proteomics. 2010;10:2498–515. doi: 10.1002/pmic.200900826. [DOI] [PubMed] [Google Scholar]
- 76.Bini L, Pacini S, Liberatori S, Valensin S, Pellegrini M, Raggiaschi R, et al. Extensive temporally regulated reorganization of the lipid raft proteome following T-cell antigen receptor triggering. Biochem J. 2003;369:301–9. doi: 10.1042/BJ20020503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bae T-J, Kim M-S, Kim J-W, Kim B-W, Choo H-J, Lee J-W, et al. Lipid raft proteome reveals ATP synthase complex in the cell surface. Proteomics. 2004;4:3536–48. doi: 10.1002/pmic.200400952. [DOI] [PubMed] [Google Scholar]
- 78.Helms JB, Zurzolo C. Lipids as targeting signals: lipid rafts and intracellular trafficking. Traffic. 2004;5:247–54. doi: 10.1111/j.1600-0854.2004.0181.x. [DOI] [PubMed] [Google Scholar]
- 79.Ikonen E. Roles of lipid rafts in membrane transport. Current Opinion in Cell Biology. 2001;13:470–7. doi: 10.1016/s0955-0674(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 80.Nichols BJ, Kenworthy AK, Polishchuk RS, Lodge R, Roberts TH, Hirschberg K, et al. Rapid cycling of lipid raft markers between the cell surface and Golgi complex. The Journal of cell biology. 2001;153:529–42. doi: 10.1083/jcb.153.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gillard BK, Thurmon LT, Marcus DM. Variable subcellular localization of glycosphingolipids. Glycobiology. 1993;3:57–67. doi: 10.1093/glycob/3.1.57. [DOI] [PubMed] [Google Scholar]
- 82.Jiang H, Sha SH, Schacht J. Rac/Rho pathway regulates actin depolymerization induced by aminoglycoside antibiotics. Journal of neuroscience research. 2006;83:1544–51. doi: 10.1002/jnr.20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen F-Q, Zheng H-W, Hill K, Sha S-H. Traumatic noise activates Rho-family GTPases through transient cellular energy depletion. The Journal of Neuroscience. 2012;32:12421–30. doi: 10.1523/JNEUROSCI.6381-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang H, Talaska AE, Schacht J, Sha SH. Oxidative imbalance in the aging inner ear. Neurobiology of aging. 2007;28:1605–12. doi: 10.1016/j.neurobiolaging.2006.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wangemann P. K+ cycling and the endocochlear potential. Hearing Research. 2002;165:1–9. doi: 10.1016/s0378-5955(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 86.Liu XZ, Xia XJ, Adams J, Chen ZY, Welch KO, Tekin M, et al. Mutations in GJA1 (connexin 43) are associated with non-syndromic autosomal recessive deafness. Human molecular genetics. 2001;10:2945–51. doi: 10.1093/hmg/10.25.2945. [DOI] [PubMed] [Google Scholar]
- 87.Teubner B, Michel V, Pesch J, Lautermann J, Cohen-Salmon M, Söhl G, et al. Connexin30 (Gjb6)-deficiency causes severe hearing impairment and lack of endocochlear potential. Human molecular genetics. 2003;12:13–21. doi: 10.1093/hmg/ddg001. [DOI] [PubMed] [Google Scholar]
- 88.Dixon MJ, Gazzard J, Chaudhry SS, Sampson N, Schulte BA, Steel KP. Mutation of the Na-K-Cl Co-Transporter Gene Slc12a2 Results in Deafness in Mice. Human Molecular Genetics. 1999;8:1579–84. doi: 10.1093/hmg/8.8.1579. [DOI] [PubMed] [Google Scholar]
- 89.Ward S, O’Donnell P, Fernandez S, Vite CH. 2-Hydroxypropyl-[bgr]-Cyclodextrin Raises Hearing Threshold in Normal Cats and in Cats With Niemann-Pick Type C Disease. Pediatr Res. 2010;68:52–6. doi: 10.1203/PDR.0b013e3181df4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rosenbaum AI, Maxfield FR. Niemann-Pick type C disease: molecular mechanisms and potential therapeutic approaches. Journal of neurochemistry. 2011;116:789–95. doi: 10.1111/j.1471-4159.2010.06976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Di Berardino F, Alpini D, Ambrosetti U, Amadeo C, Cesarani A. Sensorineural hearing-loss in the Smith–Lemli-Opitz syndrome. International Journal of Pediatric Otorhinolaryngology Extra. 2007;2:169–72. [Google Scholar]
- 92.McCall AA, Kirsch CF, Ishiyama G, Ishiyama A. Otologic findings in Antley-Bixler syndrome: a clinical and radiologic case report. International journal of pediatric otorhinolaryngology. 2007;71:1139–43. doi: 10.1016/j.ijporl.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 93.Yanjanin NM, Velez JI, Gropman A, King K, Bianconi SE, Conley SK, et al. Linear clinical progression, independent of age of onset, in Niemann-Pick disease, type C. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2010;153B:132–40. doi: 10.1002/ajmg.b.30969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brunetti-Pierri N, Corso G, Rossi M, Ferrari P, Balli F, Rivasi F, et al. Lathosterolosis, a novel multiple-malformation/mental retardation syndrome due to deficiency of 3beta-hydroxysteroid-delta5-desaturase. American journal of human genetics. 2002;71:952–8. doi: 10.1086/342668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.