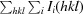SERK proteins play a central role in immune and developmental signaling pathways in plants. Structural studies have been performed in order to better understand the role of the OsSERK2 coreceptor in signaling with its partner receptors. Here, crystal structures of the LRR domains of OsSERK2 and a D128N OsSERK2 mutant, expressed as hagfish variable lymphocyte receptor (VLR) fusions, are reported.
Keywords: SERKs, leucine-rich repeats, OsSERK2
Abstract
Somatic embryogenesis receptor kinases (SERKs) are leucine-rich repeat (LRR)-containing integral membrane receptors that are involved in the regulation of development and immune responses in plants. It has recently been shown that rice SERK2 (OsSERK2) is essential for XA21-mediated resistance to the pathogen Xanthomonas oryzae pv. oryzae. OsSERK2 is also required for the BRI1-mediated, FLS2-mediated and EFR-mediated responses to brassinosteroids, flagellin and elongation factor Tu (EF-Tu), respectively. Here, crystal structures of the LRR domains of OsSERK2 and a D128N OsSERK2 mutant, expressed as hagfish variable lymphocyte receptor (VLR) fusions, are reported. These structures suggest that the aspartate mutation does not generate any significant conformational change in the protein, but instead leads to an altered interaction with partner receptors.
1. Introduction
Leucine-rich repeat (LRR) domains are commonly used for the detection of molecules in all life forms and are critical for many innate immune responses. Some of the best known examples of LRR domain-containing proteins are the animal Toll-like receptors (TLRs) and plant LRR-receptor kinases (LRR-RKs). TLRs form either a homodimer or a heterodimer with another TLR in response to a specific microbial-associated molecular pattern (MAMP) so that immune signaling is triggered. For example, TLR5 homodimerizes in response to bacterial flagellin (Hayashi et al., 2001 ▶), whereas bacterial lipopeptide induces the dimerization of TLR1 and TLR2 (Takeuchi et al., 2002 ▶). In plants, immune response often involves the dimerization of LRR-RKs with common somatic embryogenesis receptor kinases (SERKs; Schulze et al., 2010 ▶).
The best characterized SERK protein is Arabidopsis thaliana SERK3 (AtSERK3). AtSERK3 is the coreceptor for the brassinosteroid receptor brassinosteroid insensitive-1 (BRI1); thus, AtSERK3 is commonly called BRI1-associated kinase 1 (BAK1; Li et al., 2002 ▶). BAK1 is critical for immune signaling through the pattern-recognition receptors (PRRs) FLS and EFR (Heese et al., 2007 ▶). FLS2 and EFR detect the MAMPs flagellin and elongation factor Tu (EF-Tu), respectively. BAK1 participates in these diverse signaling pathways through direct interaction with the receptors. Ligand binding induces rapid heterodimerization of BAK1 with BRI1, FLS2 or EFR, which leads to transphosphorylation between the kinase domains of BAK1 and the binding partner (Schulze et al., 2010 ▶). BAK1 is also required for response to the Arabidopsis damage-associated molecular pattern peptide 1, which is detected by the LLR-RKs PEPR1 and PEPR2 (Krol et al., 2010 ▶). BAK1 is involved in responding to a variety of other MAMPs, including peptidoglycan, lipopolysaccharide and HrpZ, although the primary receptors for these MAMPs are unknown (Shan et al., 2008 ▶). Collectively, these and other studies suggest that BAK1 is a common co-receptor involved in signaling by numerous LRR-RKs in Arabidopsis.
A mutant A. thaliana line with an elongated (elg) phenotype owing to aberrant brassinosteroid signaling has a mutation on BAK1 (Whippo & Hangarter, 2005 ▶). The mutation, a substitution of Asp122 by asparagine, lies within the LRR domain of BAK1 (Whippo & Hangarter, 2005 ▶). The mutant form of BAK1 is impaired in its interaction with FLS2, but aberrantly dimerizes with BRI1 in the absence of ligand (Jaillais et al., 2011 ▶). Thus, the LRR domain of BAK1 is essential for its association with FLS2 and BRI1.
Recently, the structures of A. thaliana SERK1 (AtSERK1) and BAK1 have been determined (Santiago et al., 2013 ▶; Sun, Han et al., 2013 ▶; Sun, Li et al., 2013 ▶). AtSERK1 and BAK1 were crystallized in complexes with BRI1 and brassinosteroid (Santiago et al., 2013 ▶; Sun, Han et al., 2013 ▶). The structure of a BAK1–FLS2–flagellin peptide complex has also been determined (Sun, Li et al., 2013 ▶). These three structures have revealed for the first time how SERK proteins recognize ligand-bound receptors.
Rice has two members of the SERK family: OsSERK1 and OsSERK2 (Chen et al., 2014 ▶; Ito et al., 2005 ▶). We have recently shown that OsSERK2 positively regulates immune defenses mediated by rice FLS2 (OsFLS2) and by the immune receptors XA21 and XA3, which confer resistance to the rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae (Chen et al., 2014 ▶). OsSERK1 is not required for XA21-mediated resistance. This may be owing to the differential expression of the OsSERK2 proteins. OsSerk2 is expressed primarily in leaves, whereas OsSerk1 is primarily expressed in flowers and stems (Chen et al., 2014 ▶).
Like BAK1, OsSERK2 also interacts with the rice brassinosteroid receptor OsBRI1 and is critical for brassinosteroid-regulated development (Park et al., 2011 ▶; Chen et al., 2014 ▶). The kinase domain of OsSERK2 transphosphorylates the kinase domains of OsBRI1, XA21 and XA3 (Chen et al., 2014 ▶). These results indicate that OsSERK2 is a functional homolog of BAK1 in rice, serving as a common mediator of various LRR-RK signaling pathways.
Here, we present the crystal structures of parts of the extracellular domains of OsSERK2 and a D128N mutant of OsSERK2 that were obtained by expressing the proteins as hagfish VLR fusions. These structures reveal how the OsSERK2 aspartate mutation, which corresponds to the BAK1 D122N mutation that causes the elg phenotype, may cause altered interaction between SERK proteins and their partner LRR receptors.
2. Materials and methods
2.1. Recombinant protein expression and purification
Recombinant OsSERK2 was produced in High Five insect cells (Invitrogen, Carlsbad, California, USA) using a recombinant baculovirus expression vector, AcOsSERK2. AcOsSERK2 was generated using the Bac-to-Bac Expression System (Invitrogen) following the manufacturer’s protocol. Briefly, a recombinant donor plasmid, pFastBac-HT-OsSERK2, was constructed and transformed into competent DH10Bac Escherichia coli cells. The expression cassette in pFastBac-HT-OsSERK2 consisted of sequences encoding an N-terminal GP67 signal sequence for secretion, amino-acid residues 32–173 of OsSERK2 from Nipponbare rice (Os04g38480), amino-acid residues 126–200 of hagfish VLR B.61 at the C-terminus of OsSERK2 for improved expression and crystallization, a thrombin cleavage site, a Strep-Tactin II tag and a six-histidine tag, as described previously for zebrafish TLR5 (Yoon et al., 2012 ▶). Expression is driven by a polyhedrin promoter. Subsequently, recombinant bacmid DNA was isolated from the transformed DH10Bac E. coli cells and 2 µg was used to transfect insect Sf-9 cells in order to generate AcOsSERK2. AcOsSERK2 was subjected to one round of amplification in Sf-9 cells to increase the titer prior to the inoculation of High Five cells for protein expression.
For protein expression, High Five cells were inoculated with AcOsSERK2 at a multiplicity of infection of 0.5 and cultured in suspension (100 rev min−1) at 28°C in 500 ml ESF921 medium (Expression Systems, Davis, California, USA) in a 1 l round-bottom flask. The infected High Five cells and culture supernatant were separated 66 h post-inoculation by centrifugation (2000g, 15 min, 5°C). The proteins in the supernatant were then concentrated using an Amicon filtration device (10 kDa MWCO) and diluted twofold in 500 mM NaCl, 60 mM Tris pH 8. OsSERK2-VLR was purified by Ni–NTA affinity chromatography followed by gel-filtration chromatography in 50 mM NaCl, 25 mM Tris pH 8. The total yield of purified protein was approximately 4 mg per litre of infected High Five cells.
2.2. Crystallization
Purified wild-type and mutant OsSERK2 were concentrated to 10 mg ml−1 in 20 mM Tris pH 8.0, 100 mM NaCl. Crystallization screening was carried out with a Phoenix robot (Art Robbins Instruments, Sunnyvale, California, USA) using the sparse-matrix screening method (Jancarik & Kim, 1991 ▶). OsSERK2 was crystallized by the sitting-drop vapor-diffusion method with drops consisting of a 1:1 ratio of protein solution and 100 mM bis-tris pH 6.5, 200 mM MgCl2, 23%(w/v) PEG 3350. Plate-like crystals were observed within one week. For data collection, crystals were flash-cooled in liquid nitrogen in a buffer containing a 1:9 ratio of glycerol to crystallization buffer.
2.3. X-ray data collection and structure determination
The X-ray data set for OsSERK2 was collected at the Berkeley Center for Structural Biology on beamline 8.2.1 of the Advanced Light Source at Lawrence Berkeley National Laboratory. Diffraction data were recorded using an ADSC Q315R detector (Area Detector Systems Corporation, San Diego, California, USA). Processing of image data was performed using the HKL-2000 suite of programs (Otwinowski & Minor, 1997 ▶). Phases were calculated by molecular replacement with Phaser (McCoy et al., 2007 ▶) using residues 377–495 of brassinosteroid receptor BRI1 (PDB entry 3rgx; She et al., 2011 ▶) as a search model. Automated model building was conducted using AutoBuild (Terwilliger et al., 2008 ▶) from the PHENIX suite of programs (Adams et al., 2010 ▶), resulting in a model that was 85% complete. Manual building using Coot (Emsley & Cowtan, 2004 ▶) was alternated with reciprocal-space refinement using phenix.refine (Afonine et al., 2012 ▶). Water molecules were automatically placed using PHENIX and manually added or deleted with Coot according to peak height (3.0σ in the F o − F c map) and distance from a potential hydrogen-bonding partner (<3.5 Å). TLS refinement (Painter & Merritt, 2006a ▶) using ten groups chosen using the TLSMD web server (Painter & Merritt, 2006b ▶) was used in later rounds of refinement. The OsSERK2 D128N structure was refined and built in the same manner as the wild-type model. All data-collection, phasing and refinement statistics are summarized in Table 1 ▶. Atomic coordinates and experimental structure factors have been deposited in the Worldwide Protein Data Bank as PDB entries 4q3g for OsSERK2 and 4q3i for OsSERK2 D128N.
Table 1. X-ray data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| OsSERK2 | D128N mutant | |
|---|---|---|
| PDB code | 4q3g | 4q3i |
| Resolution () | 502.78 (2.802.78) | 502.35 (2.392.35) |
| Space group | P212121 | P21 |
| Unit-cell parameters (, ) | a = 49.9, b = 81.7, c = 126.5, = = = 90 | a = 67.9, b = 50.6, c = 83.2, = = 90, = 113.3 |
| Total reflections | 43101 | 80877 |
| Unique reflections | 13469 | 22466 |
| Average multiplicity | 3.2 (3.0) | 3.6 (2.5) |
| Completeness (%) | 93.2 (94.5) | 95.0 (71.1) |
| I/(I) | 7.75 (1.52) | 10.4 (1.34) |
| R merge † | 0.13 (0.69) | 0.11 (0.52) |
| R factor | 0.24 | 0.24 |
| R free | 0.28 | 0.29 |
| Geometry statistics | ||
| R.m.s. deviations from ideal geometry‡ | ||
| Bonds () | 0.003 | 0.002 |
| Angles () | 0.9 | 0.6 |
| MolProbity analysis§ | ||
| Ramachandran plot | ||
| Favored (%) | 95 | 94 |
| Outliers (%) | 0 | 0.2 |
| Rotamer outliers (%) | 0.8 | 0.3 |
| Clashscore | 8.6 | 6.0 |
3. Results
OsSERK2 is a multidomain integral membrane protein (Fig. 1 ▶ a). The OsSERK2 gene encodes a protein with an N-terminal secretion signal which targets the protein to the secretory pathway (Petersen et al., 2011 ▶). The mature extracellular domain consists of five LRRs flanked by an N-terminal LRR-capping domain (LRRNT) and a C-terminal proline-rich domain containing the SPP (Ser-Pro-Pro) motif. The protein spans the membrane once, and the intracellular portion of the protein consists of juxtamembrane and kinase domains.
Figure 1.
Structure of the OsSERK2 ectodomain. (a) OsSERK2 encodes a 628-amino-acid protein containing a secretion signal sequence (Sig), an N-terminal LRR capping domain (LRRNT), five LRRs, a proline-rich domain (SPP), a transmembrane sequence (Trans), a juxtamembrane domain (Juxt) and kinase domains. (b) The crystallized protein contains the LRRNT, the first four LRRs and part of the fifth LRR from OsSERK2 fused to the C-terminus of VLR B.61 with C-terminal Strep-Tag II and 6×His purification tags (Str-His). (c) The five LRRs of OsSERK2 have the plant-specific LRR consensus sequence LxxLxxLxLxxNxLSGxIPxxLGx. (d) The structure of the OsSERK2-VLR fusion protein is shown colored as in (b). The structure was determined by X-ray crystallography at 2.78 Å resolution.
To facilitate the expression and crystallization of the OsSERK2 ectodomain, we fused the OsSERK2 ectodomain to the hagfish variable lymphocyte receptor (VLR; Fig. 1 ▶ b). OsSERK2 amino acids 32–173, containing the LRRNT, the first four LRRs and part of the fifth LRR, were fused to the VLR (Figs. 1 ▶ b, 1c and Supplementary Fig. S1a 1). The chimeric protein was expressed in insect cells as a soluble secreted protein. The structure was determined by X-ray crystallography at 2.78 Å resolution (Table 1 ▶, Fig. 1 ▶ d, Supplementary Figs. S1b and 1c). The asymmetric unit consists of two nearly identical molecules (r.m.s.d. of 0.29 Å over the backbone Cα atoms), which both contain the entire sequence of the OsSERK2 (amino acids 32–173) and VLR fragments (Supplementary Fig. S1c). From here on, we refer only to the OsSERK2 portion of the structure (chain A).
As predicted, the OsSERK2 ectodomain contains an N-terminal LRRNT followed by five LRRs (the fifth LRR is only partially represented in this structure). The LRRs form a short curved solenoid structure (Fig. 1 ▶ d, Supplementary Fig. S2). The OsSERK2 ectodomain is highly similar to the recently determined structures of AtSERK1 and BAK1 (Fig. 2 ▶). The OsSERK2 structure aligns with BAK1 (Santiago et al., 2013 ▶) with an r.m.s.d. of 1.67 Å over 121 backbone Cα atoms (Fig. 2 ▶ a). The ectodomain of OsSERK2 is 70 and 67% identical to the ectodomains of AtSERK1 and BAK1, respectively.
Figure 2.
The OsSERK2 ectodomain (cyan) is similar in structure to the Arabidopsis LRRs (a) BAK1 (yellow), (b) TMK1 (magenta) and (c) BRI1 (orange). OsSERK2 closely aligns with BAK1. The structures of TMK1 and BRI1 are long twisted helical solenoids characteristic of plant LRRs. OsSERK2 only aligns with a portion of these structures.
Three-dimensional homology searches reveal that the OsSERK2 ectodomain is also similar in structure to the Arabidopsis proteins BRI1 (PDB entry 3rgx; She et al., 2011 ▶), a portion of which was used as a search model for molecular replacement, and TMK1 (PDB entry 4hq1; Liu et al., 2013 ▶) (Fig. 2 ▶). TMK1 has two solenoids containing ten and three LRRs, respectively, which are oriented nearly perpendicular to each other in the shape of a ‘7’ (Liu et al., 2013 ▶). OsSERK2 aligns most closely with the first solenoid of TMK1, but also aligns with the second solenoid of TMK1. OsSERK2 is also similar in structure to polygalacturonase-inhibiting protein 2 (PGIP2) from Phaseolus vulgaris, the first plant LRR to be structurally determined (Di Matteo et al., 2003 ▶). BRI1, TMK1, PGIP2 and the SERK proteins all share the LRR consensus motif LxxLxxLxLxxNxLSGxIPxxLGx (Fig. 1 ▶ c). This consensus sequence is unique to plants and causes a twisted or helical horseshoe structure (Hothorn et al., 2011 ▶; She et al., 2011 ▶; Di Matteo et al., 2003 ▶). The OsSERK2 LRRs form a similarly twisted solenoid (Supplementary Fig. S2). The LRRNT of OsSERK2 is also highly similar to the LRRNTs of BRI1, TMK1, BAK1 and AtSERK1. The LRRNT is stabilized by a single disulfide between Cys63 and Cys70.
There are six predicted N-glycosylation sites in the OsSERK2 ectodomain: Asn109, Asn120, Asn133, Asn155, Asn168 and Asn181 (Chauhan et al., 2012 ▶; Zhang et al., 2004 ▶). The first four of these predicted glycosylation sites are conserved in OsSERK1 and BAK1. Evidence of glycosylation is seen in the electron-density map, particularly for Asn120 (Fig. 3 ▶ a). N-Acetyl glucosamine is modeled into the additional density at this residue. The predicted glycosylated residues are located to either side of the convex surface of the OsSERK2 ectodomain (Fig. 3 ▶ b).
Figure 3.

OsSERK2 glycosylation and the concave surface. (a) 2mF o − DF c electron-density map (contoured at 1.0σ) showing density indicating glycosylation of Asn120. N-Acetylglucosamine (NAG) is modeled into the density. (b) SERK proteins have a conserved concave surface which may be involved in receptor binding. Asp128 is on the concave surface of the third LRR (shown as purple sticks). Predicted glycosylated residues are shown as red sticks. (c) Asp128 forms a salt bridge with Arg152 and hydrogen bonds to Ser126. (d) The sequences of SERK proteins from rice, wheat, Z. mays and A. thaliana were aligned and scored according to the extent of sequence variation. Scores were displayed on the surface of the OsSERK2 as a heat map with conserved residues colored red and with the most variable residues colored dark blue. The left panel, which is in the same orientation as in (b), shows that the concave surface where Asp128 is located is highly conserved. The position of Asp128 is indicated by an asterisk. In contrast, the heat map shown in the right panel shows that the convex surface is not highly conserved.
The concave surface of the OsSERK2 ectodomain is largely conserved among SERK proteins (Santiago et al., 2013 ▶; Fig. 3 ▶ c, Supplementary Fig. S3). Sequences of OsSERK1, OsSERK2, the five A. thaliana SERK proteins and SERK proteins from Zea mays and Triticum aestivum were aligned and scored for conservation according to a Risler matrix (Gouet et al., 1999 ▶). The homology score was mapped onto the surface of the OsSERK2 structure, with the most highly conserved residues colored red and divergent residues colored blue (Fig. 3 ▶ d). This analysis reveals that a large portion of the concave surface of the LRRs is conserved. The crystal structures of AtSERK1 and BAK1 in complex with BRI1 and FLS2 reveal that the conserved, concave surface of SERK proteins is involved in receptor binding (Santiago et al., 2013 ▶; Sun, Han et al., 2013 ▶; Sun, Li et al., 2013 ▶).
The phenotype of the Arabidopsis elongated (elg) mutant has been shown to be owing a single substitution of Asp122 to asparagine on BAK1 (Whippo & Hangarter, 2005 ▶). The mutant BAK1 has altered interactions with the receptors BRI1 and FLS2 (Jaillais et al., 2011 ▶). This aspartate residue is conserved among all of the A. thaliana and rice SERK proteins (Supplementary Fig. S3). In OsSERK2, the corresponding residue, Asp128, maps onto the concave surface of the ectodomain on LRR-3 (Figs. 3 ▶ b and 3 ▶ c). Asp128 forms a salt bridge to Arg152 on LRR-4 and hydrogen bonds to Ser126 (Fig. 3 ▶ c). These residues are also strictly conserved within the Arabidopsis and rice SERK families (Supplementary Fig. S3).
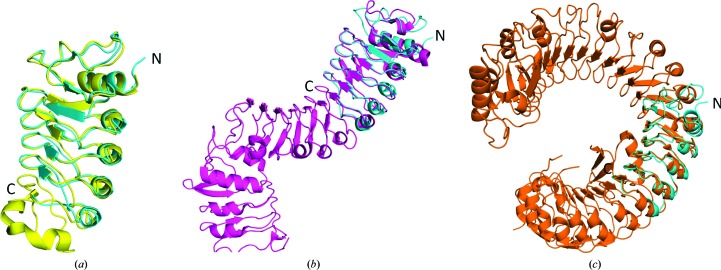
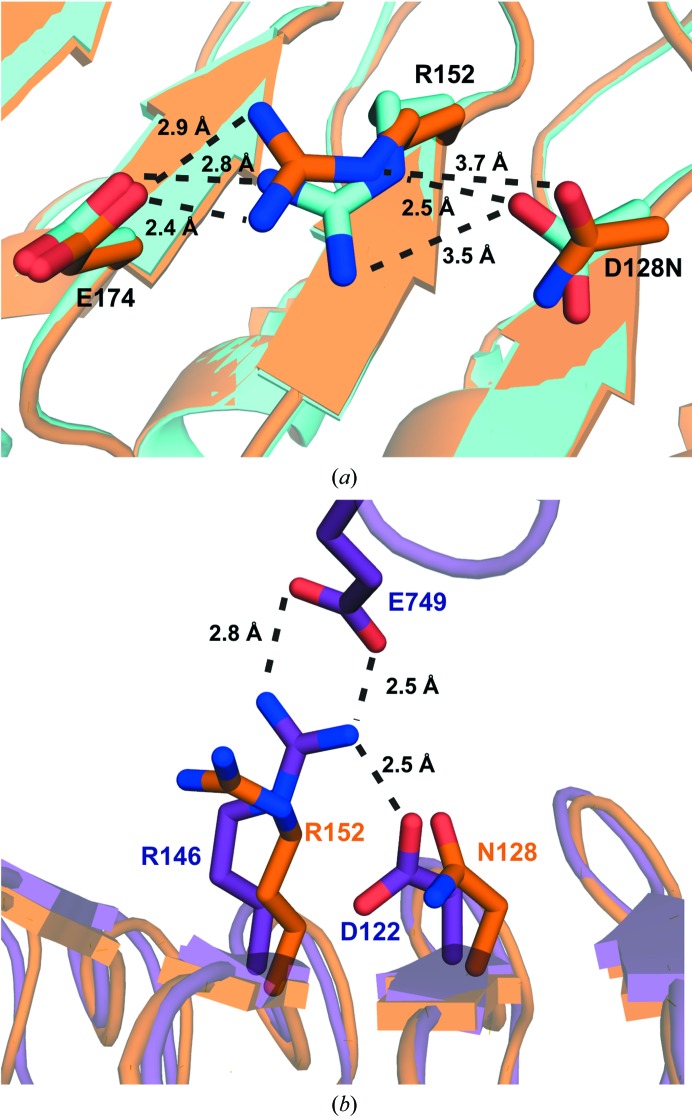
Crystal structures of the BAK1–BRI1 complex show that Asp122 is not directly involved in binding to BRI1 (Santiago et al., 2013 ▶; Sun, Han et al., 2013 ▶). To examine the role of Asp122 in BAK1 co-receptor binding, we mutated the corresponding aspartate in OsSERK2 (Asp128) to asparagine and solved the structure using crystallographic methods. We observe that the D128N mutant disrupts the formation of a salt bridge between Asp128 and Arg152. Instead, in the mutant, Arg152 forms a salt bridge with the nearby residue Glu174 (Fig. 4 ▶ a). In Fig. 4 ▶(b), an overlay of OsSERK D128N and the BRI1–BAK1 complex (PDB entry 4lsx; Santiago et al., 2013 ▶) shows that the analogous arginine in BAK1 (Arg146) forms a salt bridge with Glu749 from BRI1. BAK1 Asp122 forms a salt bridge with BAK1 Arg146 and helps to position BAK1 Arg146 for binding to BRI1. Our structure shows that OsSERK2 D128N is no longer able to make the analogous interaction, thus leading to altered binding to BRI1 and the elg phenotype.
Figure 4.
(a) Overlay of wild-type OsSERK2 (cyan) and OsSERK2 D128N (orange). In the wild-type protein, Asp128 forms a salt bridge to Arg152. The D128N mutant breaks this interaction. Instead, Arg152 of the OsSERK2 D128N mutant forms a salt bridge with Glu174. (b) Overlay of OsSERK D128N (orange) and the BRI1–BAK1 complex (PDB entry 4lsx; purple). The analogous arginine in BAK1 (Arg146) forms a salt bridge with Glu749 from BRI1. BAK1 Asp122 forms a salt bridge with BAK1 Arg146 and helps to position BAK1 Arg146 for binding to BRI1. Our structure shows that OsSERK2 D128N is no longer able to make the analogous interaction, therefore leading to altered binding to BRI1 and the elg phenotype.
4. Discussion
SERK proteins play a central role in immune and developmental signaling pathways in plants. Brassinosteroid receptors and several PRRs heterodimerize with a common SERK coreceptor. The recent structures of SERK–receptor complexes have significantly advanced our understanding of plant immune and hormone signaling (Santiago et al., 2013 ▶; Sun, Han et al., 2013 ▶; Sun, Li et al., 2013 ▶). Prior to these structures, SERK co-receptors were not thought to be involved in ligand binding directly but rather to bind to the ligand-bound LRR-RK (Chinchilla et al., 2007 ▶; Hothorn et al., 2011 ▶; She et al., 2011 ▶). However, these structures show that in the cases of the brassinosteroid and flagellin, the N-termini of SERK proteins directly participate in ligand binding (Santiago et al., 2013 ▶; Sun, Han et al., 2013 ▶; Sun, Li et al., 2013 ▶). A key question remaining in SERK signaling is how a single SERK coreceptor can interact with such a large set of diverse LRR–ligand complexes.
We have performed structural studies in order to better understand the role of the OsSERK2 coreceptor in signaling with its partner receptors OsBRI1, XA21, XA3 and OsFLS2. The OsSERK2 ectodomain has an N-terminal LRR capping domain followed by five LRRs (Fig. 1 ▶ a). A proline-rich SPP domain lies between the LRRs and the transmembrane region. The function of the proline-rich domain is unknown, but it is considered to be one of the defining features of SERKs. It has been suggested that the SPP domain may act as a flexible hinge and/or interact with the cell wall (Hecht et al., 2001 ▶).
In order to facilitate protein expression and crystallization, we fused OsSERK2 within the fifth LRR to the hagfish VLR. Without the VLR, we were unable to express sufficient quantities of OsSERK for crystallization trials. VLR is a stable, readily crystallizable protein that has been used to obtain structures of several LRR proteins including Toll receptor, Toll-like receptor 1 (TLR1), TLR2, TLR4, TLR5 and TLR6 (Gangloff et al., 2013 ▶; Yoon et al., 2012 ▶; Kang et al., 2009 ▶; Kim et al., 2007 ▶; Jin et al., 2007 ▶). This study reveals that VLR fusion is a useful tool for plant LRRs as well as animal LRRs. This was not necessarily expected because plant LRRs such as OsSERK2 have the consensus sequence LxxLxxLxLxxNxLSGxIPxxLGx as opposed to the canonical animal LRR consensus sequence LxxLxxLxLxxNxLxxLpxxoFxx (Kajava, 1998 ▶). The plant consensus sequence causes proteins to have a twisted horseshoe structure distinct from the planar horseshoe structures seen for animal LRRs (Hothorn et al., 2011 ▶; She et al., 2011 ▶; Di Matteo et al., 2003 ▶).
Sequence analysis reveals that much of the concave surface surrounding Asp128 is conserved among a diverse set of SERK proteins from O. sativa, A. thaliana, Z. mays and T. aestivum (Fig. 3 ▶ c, Supplementary Fig. S4). The conservation extends across the β-sheet portion of the LRRs as well as much of the LRRNT. The surface residues of the concave surface are not conserved in the structurally similar proteins TMK1 and BRI1, and are thus not conserved for structural reasons. The residues are also not conserved in the closely related class of rice SERK-like proteins. The surface residues on the concave surface of OsSERK2, however, are more diverse (Fig. 3 ▶ c). The predicted glycosylated residues are also on the nonconserved surfaces of the protein, which are unlikely to be involved in receptor binding (Fig. 3, Supplementary Fig. S3). The structures of the AtSERK1–BRI1, BAK1–BRI1 and BAK1–FLS2 complexes reveal that the conserved surface of the SERK proteins is largely involved in binding partner receptors (Santiago et al., 2013 ▶; Sun, Han et al., 2013 ▶; Sun, Li et al., 2013 ▶).
BAK1 D122N has altered interaction with the partner receptors FLS2 and BRI1 (Jaillais et al., 2011 ▶). The recent structures of the BAK1–BRI1 and BAK1–FLS2 complexes reveal that Asp122 of BAK1 does not directly contact amino-acid residues of the partner receptor (Sun, Li et al., 2013 ▶; Santiago et al., 2013 ▶; Sun, Han et al., 2013 ▶). In the BAK1 D122N mutant, Asn122 is not predicted to be glycosylated (Chauhan et al., 2012 ▶; Zhang et al., 2004 ▶). This suggests that the D122N mutation indirectly affects binding by altering the position of the amino acids near residue 122.. To investigate this hypothesis, we generated OsSERK2 D128N, an OsSERK2 mutation which corresponds to the D122N mutation of BAK1.
Our structure of the OsSERK2 D128N mutant helps to explain why the BAK1 D122N mutant is impaired in its interaction with the receptors BRI1 and FLS2. The aspartate-to-asparagine mutation causes Arg152 to lose its interaction with Asp128. Instead, Arg152 forms a salt bridge with Glu174. The structures of the BRI1–SERK1 and the BRI1–BAK1 complexes (Santiago et al., 2013 ▶; Sun, Han et al., 2013 ▶) reveal that the analogous residue in BAK1 (Arg146) forms a salt bridge with the neighboring co-receptor. Changes to the positioning of Arg146 could therefore directly impact co-receptor interaction. Thus, our structure suggests that the effects of the D122N mutation are not through a direct interaction between Asp122 and BRI1, but indirectly through Arg146. The structures of the OsSERK2 ectodomain and the D128N mutant presented in this study provide additional resources for structure–function studies to answer this key question of how SERK proteins recognize a diverse range of LRR-RK signaling partners.
Supplementary Material
PDB reference: OsSERK2, 4q3g
PDB reference: OsSERK2, D128N mutant, 4q3i
Supporting Information.. DOI: 10.1107/S1399004714021178/kw5096sup1.pdf
Acknowledgments
We thank the Wilson laboratory for providing the TLR5-N14VLR-pAcGP67 vector which was used to construct the OsSERK2 expression vector. This work was part of the DOE Joint BioEnergy Institute (JBEI), which is funded by the US Department of Energy, Office of Science, Office of Biological and Environmental Research through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy. This work was also funded by NIH grant No. GM55962 to PCR and NIEHS grant R01 ES002710 to BDH. DM was supported by the Department of Biotechnology, Government of India Overseas Fellowship. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences of the US Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: KW5096).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Chauhan, J. S., Bhat, A. H., Raghava, G. P. & Rao, A. (2012). PLoS One, 7, e40155. [DOI] [PMC free article] [PubMed]
- Chen, X., Zuo, S., Schwessinger, B., Chern, M., Canlas, P. E., Ruan, D., Zhou, X., Wang, J., Daudi, A., Petzold, C. J., Heazlewood, J. L. & Ronald, P. C. (2014). Mol. Plant, 7, 874–892. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., Jones, J. D., Felix, G. & Boller, T. (2007). Nature (London), 448, 497–500. [DOI] [PubMed]
- Di Matteo, A., Federici, L., Mattei, B., Salvi, G., Johnson, K. A., Savino, C., De Lorenzo, G., Tsernoglou, D. & Cervone, F. (2003). Proc. Natl Acad. Sci. USA, 100, 10124–10128. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Engh, R. A. & Huber, R. (1991). Acta Cryst. A47, 392–400.
- Gangloff, M., Arnot, C. J., Lewis, M. & Gay, N. J. (2013). Structure, 21, 143–153. [DOI] [PMC free article] [PubMed]
- Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., Eng, J. K., Akira, S., Underhill, D. M. & Aderem, A. (2001). Nature (London), 410, 1099–1103. [DOI] [PubMed]
- Hecht, V., Vielle-Calzada, J.-P., Hartog, M. V., Schmidt, E. D. L., Boutilier, K., Grossniklaus, U. & de Vries, S. C. (2001). Plant Physiol. 127, 803–816. [PMC free article] [PubMed]
- Heese, A., Hann, D. R., Gimenez-Ibanez, S., Jones, A. M., He, K., Li, J., Schroeder, J. I., Peck, S. C. & Rathjen, J. P. (2007). Proc. Natl Acad. Sci. USA, 104, 12217–12222. [DOI] [PMC free article] [PubMed]
- Hothorn, M., Belkhadir, Y., Dreux, M., Dabi, T., Noel, J. P., Wilson, I. A. & Chory, J. (2011). Nature (London), 474, 467–471. [DOI] [PMC free article] [PubMed]
- Ito, Y., Takaya, K. & Kurata, N. (2005). Biochim. Biophys. Acta, 1730, 253–258. [DOI] [PubMed]
- Jaillais, Y., Belkhadir, Y., Balsemão-Pires, E., Dangl, J. L. & Chory, J. (2011). Proc. Natl Acad. Sci. USA, 108, 8503–8507. [DOI] [PMC free article] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411.
- Jin, M. S., Kim, S. E., Heo, J. Y., Lee, M. E., Kim, H. M., Paik, S.-G., Lee, H. & Lee, J.-O. (2007). Cell, 130, 1071–1082. [DOI] [PubMed]
- Kajava, A. V. (1998). J. Mol. Biol. 277, 519–527. [DOI] [PubMed]
- Kang, J. Y., Nan, X., Jin, M. S., Youn, S.-J., Ryu, Y. H., Mah, S., Han, S. H., Lee, H., Paik, S.-G. & Lee, J.-O. (2009). Immunity, 31, 873–884. [DOI] [PubMed]
- Kim, H. M., Park, B. S., Kim, J.-I., Kim, S. E., Lee, J., Oh, S. C., Enkhbayar, P., Matsushima, N., Lee, H., Yoo, O. J. & Lee, J.-O. (2007). Cell, 130, 906–917. [DOI] [PubMed]
- Krol, E., Mentzel, T., Chinchilla, D., Boller, T., Felix, G., Kemmerling, B., Postel, S., Arents, M., Jeworutzki, E., Al-Rasheid, K. A., Becker, D. & Hedrich, R. (2010). J. Biol. Chem. 285, 13471–13479. [DOI] [PMC free article] [PubMed]
- Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E. & Walker, J. C. (2002). Cell, 110, 213–222. [DOI] [PubMed]
- Liu, P., Hu, Z., Zhou, B., Liu, S. & Chai, J. (2013). Cell Res. 23, 303–305. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol, 276, 307–326. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2006a). Acta Cryst. D62, 439–450. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2006b). J. Appl. Cryst. 39, 109–111.
- Park, H. S., Ryu, H. Y., Kim, B. H., Kim, S. Y., Yoon, I. S. & Nam, K. H. (2011). Mol. Cells, 32, 561–569. [DOI] [PMC free article] [PubMed]
- Petersen, T. N., Brunak, S., von Heijne, G. & Nielsen, H. (2011). Nature Methods, 8, 785–786. [DOI] [PubMed]
- Santiago, J., Henzler, C. & Hothorn, M. (2013). Science, 341, 889–892. [DOI] [PubMed]
- Schulze, B., Mentzel, T., Jehle, A. K., Mueller, K., Beeler, S., Boller, T., Felix, G. & Chinchilla, D. (2010). J. Biol. Chem. 285, 9444–9451. [DOI] [PMC free article] [PubMed]
- Shan, L., He, P., Li, J., Heese, A., Peck, S. C., Nürnberger, T., Martin, G. B. & Sheen, J. (2008). Cell Host Microbe, 4, 17–27. [DOI] [PMC free article] [PubMed]
- She, J., Han, Z., Kim, T.-W., Wang, J., Cheng, W., Chang, J., Shi, S., Wang, J., Yang, M., Wang, Z.-Y. & Chai, J. (2011). Nature (London), 474, 472–476. [DOI] [PMC free article] [PubMed]
- Sun, Y., Han, Z., Tang, J., Hu, Z., Chai, C., Zhou, B. & Chai, J. (2013). Cell Res. 23, 1326–1329. [DOI] [PMC free article] [PubMed]
- Sun, Y., Li, L., Macho, A. P., Han, Z., Hu, Z., Zipfel, C., Zhou, J.-M. & Chai, J. (2013). Science, 342, 624–628. [DOI] [PubMed]
- Takeuchi, O., Sato, S., Horiuchi, T., Hoshino, K., Takeda, K., Dong, Z., Modlin, R. L. & Akira, S. (2002). J. Immunol. 169, 10–14. [DOI] [PubMed]
- Terwilliger, T. C., Grosse-Kunstleve, R. W., Afonine, P. V., Moriarty, N. W., Zwart, P. H., Hung, L.-W., Read, R. J. & Adams, P. D. (2008). Acta Cryst. D64, 61–69. [DOI] [PMC free article] [PubMed]
- Whippo, C. W. & Hangarter, R. P. (2005). Plant Physiol. 139, 448–457. [DOI] [PMC free article] [PubMed]
- Yoon, S.-I., Kurnasov, O., Natarajan, V., Hong, M., Gudkov, A. V., Osterman, A. L. & Wilson, I. A. (2012). Science, 335, 859–864. [DOI] [PMC free article] [PubMed]
- Zhang, M., Gaschen, B., Blay, W., Foley, B., Haigwood, N., Kuiken, C. & Korber, B. (2004). Glycobiology, 14, 1229–1246. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: OsSERK2, 4q3g
PDB reference: OsSERK2, D128N mutant, 4q3i
Supporting Information.. DOI: 10.1107/S1399004714021178/kw5096sup1.pdf