Abstract
Ninety seven Rhizoctonia isolates were collected from different Brassica species with typical Rhizoctonia symptoms in different provinces of Vietnam. The isolates were identified using staining of nuclei and sequencing of the rDNA-ITS barcoding gene. The majority of the isolates were multinucleate R. solani and four isolates were binucleate Rhizoctonia belonging to anastomosis groups (AGs) AG-A and a new subgroup of A-F that we introduce here as AG-Fc on the basis of differences in rDNA-ITS sequence. The most prevalent multinucleate AG was AG 1-IA (45.4% of isolates), followed by AG 1-ID (17.5%), AG 1-IB (13.4%), AG 4-HGI (12.4%), AG 2-2 (5.2%), AG 7 (1.0%) and an unknown AG related to AG 1-IA and AG 1-IE that we introduce here as AG 1-IG (1.0%) on the basis of differences in rDNA-ITS sequence. AG 1-IA and AG 1-ID have not been reported before on Brassica spp. Pathogenicity tests revealed that isolates from all AGs, except AG-A, induced symptoms on detached leaves of several cabbage species. In in vitro tests on white cabbage and Chinese cabbage, both hosts were severely infected by AG 1-IB, AG 2-2, AG 4-HGI, AG 1-IG and AG-Fc isolates, while under greenhouse conditions, only AG 4-HGI, AG 2-2 and AG-Fc isolates could cause severe disease symptoms. The occurrence of the different AGs seems to be correlated with the cropping systems and cultural practices in different sampling areas suggesting that agricultural practices determine the AGs associated with Brassica plants in Vietnam.
Introduction
Vietnam is a country in Southeast Asia in which the agricultural sector accounts for more than 22% of the GDP, 30% of export and 52% of all employment. Vietnam is not only one of the world leaders in rice and coffee export, but also the third world's largest vegetable producer. Brassicas are among the main vegetables produced for both local consumption and export [1]. Vegetables in Vietnam are mainly produced by poor households living in the Red River and Mekong River delta (see Figure 1) in intensive cultivation systems or in rotation with other crops. Due to the lack of knowledge in crop management, limited availability of technology and land fragmentation, farmers are suffering heavy yield losses year after year. Among the limiting factors in vegetable production is the occurrence of Rhizoctonia diseases, which has been recognized as one of the most important threats.
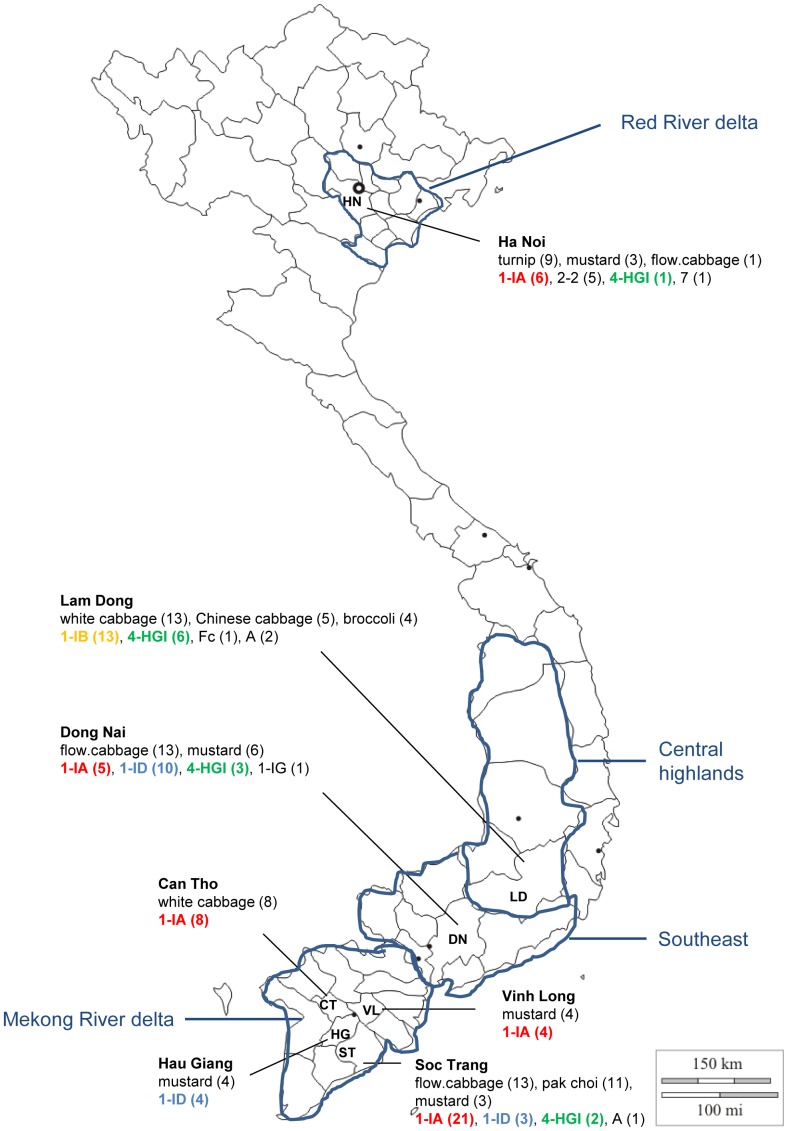
Figure 1. Location of sites for collection of Rhizoctonia isolates from Brassica spp. in Vietnam.
The seven provinces sampled are: Ha Noi (districts of Gia Lam, Thanh Tri and Dong Anh), Lam Dong (Da Lat city and Duc Trong district), Dong Nai (Bien Hoa city), Vinh Long (Binh Tan district), Can Tho (Cai Rang district), Hau Giang (Phung Hiep district) and Soc Trang (Soc Trang city and My Xuyen district). In each city or district of one province, one to two wards were surveyed and these wards are marked with a start . Different colors are used to highlight the most important AGs found in our survey including AG 1-IA, AG 1-IB, AG 1-ID and AG 4-HGI.
. Different colors are used to highlight the most important AGs found in our survey including AG 1-IA, AG 1-IB, AG 1-ID and AG 4-HGI.
Rhizoctonia is a genus of basidiomycete fungi causing many important plant diseases. Based on differences in the number of nuclei per cell, Rhizoctonia isolates have been differentiated into uninucleate Rhizoctonia, binucleate Rhizoctonia (teleomorphs: Ceratobasidium spp. and Tulasnella spp.) and multinucleate Rhizoctonia (teleomorphs: Thanatephorus spp. and Waitea spp.) [2]. Rhizoctonia species can also be classified using biochemical and molecular techniques. Among those, rDNA-ITS sequence analysis appears to be the most convenient and reliable method [3]. Currently, isolates of R. solani, the most widely recognized species within the multinucleate Rhizoctonia group, have been divided into 13 anastomosis groups (AGs), while 16 AGs of binucleate Rhizoctonia have been recognized [2]–[7]. Due to the considerable genetic diversity, several AGs have been further divided into subgroups based on phylogenetic differences. These phylogenetic differences can be associated with differences in morphology, ecology, pathogenicity and biochemical characteristics, although this is not necessarily the case [8].
Compared to binucleate Rhizoctonia, multinucleate R. solani AGs usually have a wider host range and higher virulence. R. solani can survive for a long period in plant debris, contaminated seeds, or infested soils as mycelium or sclerotia [9], [10]. Under favorable conditions, sclerotia geminate and form delicate hyphae that will grow toward the host plants [11]. Brassica vegetables can be attacked by several different AGs of R. solani resulting in the development of various diseases such as foliar blight, wirestem and damping off. In previous studies, AGs 1-IB, 1-IC, 2-1, 2-2 IIIB, 3, 4-HGI, 4-HGII, 4-HGIII, 5, 7, 9 and 10 were shown to be pathogenic on Brassica crops grown in Canada [12], [13], Australia [14], Japan [15], [16], North America [17]–[19], Brazil [20], China [21], Belgium [22], and the UK [23]. Although Rhizoctonia diseases occur severely and frequently on leafy vegetables cultivated in Vietnam, there have been no reports about the AGs and subgroups that attack Brassica crops. Therefore, our research aimed at (i) identifying the species and AGs of Rhizoctonia present on Brassica plants in different vegetable producing regions in Vietnam, and (ii) verifying the susceptibility of Brassica vegetables to Rhizoctonia isolates collected. Our story revealed that Rhizoctonia AGs such as AG 1-IA and AG 1-ID, which have not been reported before on Brassica spp., are predominant in Vietnam, which is presumably linked with the cultural practices and cropping systems in the different sampling areas. Moreover, we describe two new AGs that were previously unknown.
Materials and Methods
Field sampling and pathogen isolation
Sampling was done on private farms by Gia Khuong Hoang Hua (Vietnamese citizen) with permission from the farmer. In general, permission by the authorities was not required since the sampling studies were carried out on private farms. Only cabbage plants showing symptoms of the undesired Rhizoctonia fungus were sampled, hence the field studies did not involve endangered or protected species. The GPS coordinates of the locations where samples were collected are presented in Table S1 in File S1.
The survey was conducted from September to October 2011 on various fields in the Red River delta (Ha Noi; an important vegetable production area of the North), the Mekong River delta (Vinh Long, Can Tho, Hau Giang and Soc Trang; main vegetable production areas of the South), the Central Highlands (Lam Dong; vegetables are mainly produced for export) and the Southeast (Dong Nai; vegetables are mainly produced for domestic consumption) (Figure 1). These regions were chosen because of their importance in vegetable production and because they offer a good representation of the different climatic and topographic conditions in Vietnam and, therefore, a good representation of the distribution of Rhizoctonia spp. on Brassica spp. in Vietnam can be obtained. Total production area, climatic conditions and main agricultural activities of these regions are listed in Table 1.
Table 1. Sampling locations and their relevant characteristics [53]–[59].
| Sampling location | Agricultural area (1000 ha) | Average temperature (°C) | Main crop |
| Red River Delta | |||
| Ha Noi | 152.24 | 24 | - Cabbage, tomato, cucumber, radish |
| - Rice | |||
| Central highlands region | |||
| Lam Dong | 279.00 | High land: 14 | - Lettuce, cabbage, carrot, potato |
| Low land: 21 | - Coffee, tea, cashew-nut tree, cotton | ||
| Southeast region | |||
| Dong Nai | 289.02 | 27 | - Coffee, cotton, black pepper |
| - Durian, grapefruit, mango | |||
| Mekong River Delta | |||
| Vinh Long | 116.18 | 27 | - Rice |
| - Mango, orange, grapefruit, durian | |||
| - Cabbage, cucumber, bean | |||
| Can Tho | 115.00 | 27 | - Rice |
| - Cabbage, cucumber | |||
| Hau Giang | 139.07 | 27 | - Durian, pineapple, grapefruit |
| - Rice | |||
| Soc Trang | 278.15 | 27 | - Rice |
| - Grapefruit, mango, durian |
A total of 142 Brassica plants with Rhizoctonia-like symptoms were sampled. Infected root and leaf tissues were washed in running tap water, surface-disinfected in 1% sodium hypochlorite solution for two min and then rinsed twice in sterile water before placing on 1% water agar medium supplemented with streptomycin (0.05 g L−1). After 24 h of incubation, Rhizoctonia-like hyphal tips growing out of these tissues were transferred to fresh potato dextrose agar (PDA; Difco) plates and incubated for two to four days at 28°C.
Ninety seven Rhizoctonia isolates were recovered and subjected to nuclei staining and sequencing of the ITS-rDNA region. All isolates are listed in Table 2.
Table 2. Characterization of Rhizoctonia isolates collected from diseased Brassica crops grown in Vietnam by sequencing the ITS-region.
| AG/Subgroup | Host plant | Isolateab | Genbank accession numbers |
| 1-IA | B. parachinensis (Chinese flowering cabbage) | STST03-1, STST03-3, STST03-4, STST04-2, STMX04-1, STMX04-2, STMX04-3, STMX04-4, STMX04-5 | KF907702, KF907703, KF907704, KF907705 |
| B. juncea (Mustard cabbage) | DNBH01-1, DNBH01-2, DNBH01-3, DNBH02-2, DNBH02-3 | ||
| HNGL01-1, HNGL01-2, HNGL01-3 | KF907706 | ||
| STST02-1, STST02-2 | |||
| VLBT01-1, VLBT01-2, VLBT01-3, VLBT01-4 | |||
| B. chinensis (Pak choi) | STMX01-1, STMX01-2, STMX01-4, STMX01-5, STMX02-1, STMX02-2, STMX02-3, STMX03-1, STMX03-2, STMX03-3 | KF907707, KF907708, KF907709, KF907710, KF907711 | |
| B. oleraceae (Turnip cabbage) | HNDD01-1, HNDD01-2, HNDD01-3 | KF907712 | |
| B. oleraceae (White cabbage) | CTCR01-1, CTCR01-2, CTCR01-3, CTCR02-1, CTCR02-2, CTCR02-3, CTCR03-1, CTCR03-2 | KF907713, KF907714, KF907715 | |
| 1-IB | B. chinensis (Chinese cabbage) | LDDT03-1 | |
| B. oleraceae (Broccoli) | LDDL01-1, LDDL01-2, LDDL01-3, LDDL01-4 | KF907716 | |
| B. oleraceae (White cabbage) | LDDL04-1, LDDL04-2, LDDL04-3, LDDL04-4, LDDL04-5, LDDL05-1, LDDL05-2, LDDL05-3 | KF907717, KF907718, KF907719 | |
| 1-ID | B. parachinensis (Chinese flowering cabbage) | DNBH03-1, DNBH03-2, DNBH03-3, DNBH03-4, DNBH03-5, DNBH05-1-1, DNBH05-1-3, DNBH05-2-2, DNBH05-3-1, DNBH05-4 | KF907720, KF907721, KF907722, KF907723, KF907724 |
| STST04-1, STST04-3 | |||
| B. juncea (Mustard cabbage) | HGPH01-1, HGPH01-2, HGPH01-3, HGPH01-4 | KF907725 | |
| STST02-3 | |||
| 1-IG | B. parachinensis (Chinese flowering cabbage) | DNBH05-1-2 | KF907730 |
| 2-2 | B. parachinensis (Chinese flowering cabbage) | HNTT01-1 | KF907726 |
| B. oleraceae (Turnip cabbage) | HNDA01-1, HNDA01-2, HNDA01-3, HNDA01-4 | KF907727, KF907728, KF907729 | |
| 4-HGI | B. chinensis (Chinese cabbage) | LDDT01-1, LDDT01-2, LDDL02-2 | KF907731 |
| B. parachinensis (Chinese flowering cabbage) | DNBH05-2-1, DNBH05-3-2 | ||
| STST01-1, STST03-2 | KF907732 | ||
| B. juncea (Mustard cabbage) | DNBH02-1 | ||
| B. oleraceae (Turnip cabbage) | HNDD01-4 | KF907733 | |
| B. oleraceae (White cabbage) | LDDT02-1, LDDT02-2, LDDT02-3 | ||
| 7 | B. oleraceae (Turnip cabbage) | HNDA02-1 | KF907734 |
| A | B. chinensis (Pak choi) | STMX01-3 | |
| B. oleraceae (White cabbage) | LDDL03-1, LDDL03-2 | KF907735 | |
| Fc | B. chinensis (Chinese cabbage) | LDDL02-1 | KF907736 |
The first two letters represent provinces in which the samples were collected (i.e. CT: Can Tho, VL: Vinh Long, HG: Hau Giang, ST: Soc Trang, DN: Dong Nai, LD: Lam Dong and HN: Ha Noi).
Isolates in bold (unique sequences) are submitted to Genbank.
Nuclei staining
Rhizoctonia isolates were cultured on sterile glass slides covered by PDA for two days at 28°C. Actively growing fungal hyphae were stained with 10 µg mL−1 4, 6-diamino-2-phenyl indole (DAPI; Sigma-Aldrich) and the number of nuclei per hyphal cell was determined using an Olympus BX51 microscope [22].
DNA extraction, PCR and sequencing of the rDNA-ITS region
The rDNA-ITS region of all collected isolates was sequenced for identification to the AG and subgroup level. The usefulness of the rDNA-ITS region for identification of unknown Rhizoctonia spp. has been clearly shown by Sharon et al. [2], [3].
Rhizoctonia isolates were grown on potato dextrose broth at 28°C for one week. Mycelial mats were harvested by filtration and ground in liquid nitrogen to produce a fine powder. Total genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen). The rDNA-ITS fragment including the 5.8 S gene was amplified using primers ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) and ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) [24]. The PCR amplification reactions were performed by adding 2 µL genomic DNA (5–10 ng µL−1) to 23 µL of reaction mixture containing 2.5 µL PCR buffer (10×; Qiagen), 5 µL Q-solution (Qiagen), 0.5 µL dNTPs (10 mM; Fermentas GmbH), 1.75 µL of each primer (10 µM), 0.15 µL Taq DNA polymerase (5 units µL−1; Fermentas GmbH) and 11.35 µL ultrapure sterile water. Amplification was performed using a Flexcycler PCR Thermal Cycler (Analytik Jena) programmed for an initial denaturation step at 94°C for 10 min followed by 35 cycles at 94°C for 1 min, 55°C for 1 min and 72°C for 1 min. Cycling ended with a final extension step at 72°C for 10 min. Amplification products were separated in 1% agarose gels in TAE-buffer at 100 V for 30 min and visualized by ethidium bromide staining on a UV transilluminator. The sequences of both strands were determined by LGC Genomics GmbH (Berlin, Germany) using Sanger sequencing.
Identification using BLAST and phylogenetic analysis
Consensus sequences for all 97 isolates were created with BioEdit version 7.1.11. To determine the AG of the isolates, the rDNA-ITS consensus sequences obtained were compared to those in Genbank using the BLASTn tool.
However, since Genbank is an uncurated database, it can contain inaccurately designated Rhizoctonia spp., as has been previously shown by Sharon et al. [3]. Therefore, comparison of rDNA-ITS sequences of unknown isolates to a curated database of sequences containing representative rDNA-ITS sequences of all known uninucleate, binucleate and multinucleate Rhizoctonia AG and subgroups provides a more reliable identification. Such a database of representative sequences is available from Sharon et al. [2] and was provided by Michal Sharon to us. However, not all known AGs were present in this database, therefore we added representative isolates of the following AGs: AG 1-1E, AG 1-1F, AG 2-2 WB [25] and AG 13 [4]. The total number of sequences in the database was 129 and the Genbank accession numbers of all these sequences can be found in Table S2 in File S2.
Multiple alignments for multinucleate and binucleate Rhizoctonia isolates were constructed using MUSCLE which is implemented in MEGA 6 [26] and checked manually afterwards. The resulting alignments had a length of 717 bp (multinucleate Rhizoctonia isolates) and 768 bp (binucleate Rhizoctonia isolates).
Separate phylogenetic trees were constructed for multinucleate Rhizoctonia and binucleate Rhizoctonia isolates. For the binucleate tree, one representative isolate for each known binucleate AG was included together with the binucleate Rhizoctonia isolates obtained in this work.
For the multinucleate tree, reference isolates from the curated database (Table S2 in File S2) were added for all AGs present in our collection from Vietnam. Another 32 isolates from a characterization study in Vietnam that has not been published (Thuan et al., unpublished; Genbank accession numbers with prefix ‘EF’ in Table 3), and found on a range of crops and belonging to AGs AG 1-IA, AG 1-ID and AG 4-HGI were also added to the multinucleate Rhizoctonia alignment for phylogenetic analysis. Phylogenetic trees were built using the neighbour joining algorithm with 1000 bootstrap repeats using MEGA 6 [26]. Model testing was done using the software implemented in MEGA 6 and the K2+ G DNA substitution model was chosen.
Table 3. Multinucleate and binucleate Rhizoctonia isolates derived from Genbank included in the phylogenetic analysis for comparison.
| AG/Subgroup | Isolate | Host plant | Origin | Genbank accession number | Reference |
| 1-IA | L31-1, L66-1, L73, L59, L38, L52, L62-1 | Rice | Vietnam | EF206342, EF429208, EF429211, EF429212, EF429210, EF429209, EF429207 | unpublished |
| RM61 | Water spinach | Vietnam | EF429216 | unpublished | |
| LB71 | Water hyacinth | Vietnam | EF429215 | unpublished | |
| DP38 | Peanut | Vietnam | EF429214 | unpublished | |
| CLV72-2 | Barnyard grass | Vietnam | EF429213 | unpublished | |
| BV71-2, BV61-2, BV50-1 | Cotton | Vietnam | EF429206, EF429205, EF206341 | unpublished | |
| CC72 | Bermuda grass | Vietnam | EF429204 | unpublished | |
| B34-1 | Corn | Vietnam | EF429203 | unpublished | |
| 1-IG | RMPG28 | Chickpea | India | JF701750 | [29] |
| 1-ID | BV62-1, BV61-6, BV61-5, BV61-4, BV61-1 | Cotton | Vietnam | EF197803, EF197804, EF197802, EF197801, EF197800 | unpublished |
| SR61, SR650 | Durian | Vietnam | EF197798, EF197797 | [30] | |
| B61-1 | Corn | Vietnam | EF197796 | unpublished | |
| CCD61-1 | Sugar beet | Vietnam | EF197799 | unpublished | |
| 4-HGI | XL4 | Cauliflower | Vietnam | EF203247 | unpublished |
| CB63, CB34-2 | Cabbage | Vietnam | EF203251, EF203245 | unpublished | |
| CP50-2 | Coffee | Vietnam | EF203250 | unpublished | |
| BV68-1, BV68-2 | Cotton | Vietnam | EF203249, EF203248 | unpublished | |
| KT63-1 | Potato | Vietnam | EF203246 | unpublished | |
| Fc | BS-YT-06-5-14, YT, BS-J-06-6-3, DL-jiang-06-2-4, DL-YT-06-4-10, DL-YT-06-4-9, DL-YT-06-3-4 | Taro, Ginger | China | HM623619, HM623631, HM623615, HM623622, HM623625, HM623624, HM623623 | unpublished |
Aggressiveness of Rhizoctonia isolates towards detached leaves of cabbages, rice and water spinach
Nine isolates (representing the nine different Rhizoctonia AGs collected) were randomly selected and tested for virulence towards several Brassica crops in two independent experiments (Figures 2, 3 and 4). To confirm their pathogenicity, tests were repeated for AGs 1-IA, 1-IB, 1-ID, 2-2, 4-HGI and A since each of these AGs consists of more than one isolate. Results of additional tests are shown in Tables S3, S4 and S5 in File S1.
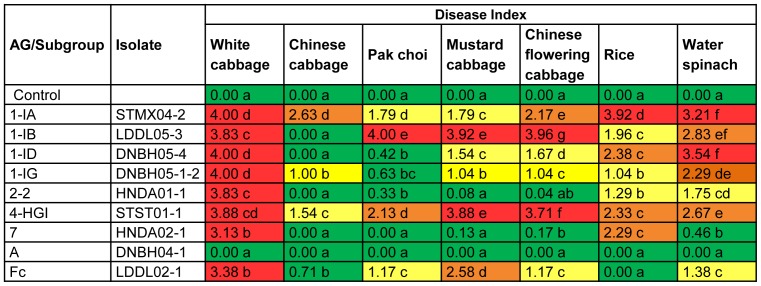
Figure 2. Aggressiveness of Rhizoctonia isolates towards detached leaves of white cabbage, Chinese cabbage, pak choi, mustard cabbage, Chinese flower cabbage, rice and water spinach.
Leaves were scored using a scale ranging from 0 (no disease symptoms) to 4 (lesions covered more than 75% of leaf surface or dead leaf). For rapid visual evaluation of the data, a coloring scale with green (0<DI≤1), yellow (1<DI≤2), orange (2<DI≤3) and red (3<DI≤4) was used. The experiment was conducted twice and each treatment consisted of 12 leaves or leaf pieces. The data of the two experiments were pooled before Mann-Whitney comparisons were applied at p = 0.05. Within columns, disease severities followed by the same letter are not significantly different.
Figure 3. Pathogenic potential of Rhizoctonia isolates on seedlings of white cabbage and Chinese cabbage in in vitro bio-assays.
Disease severity was assessed on a scale ranging from 0 (no symptoms) to 4 (lesions covering more than 75% of root, hypocotyl or leaf surface or dead plant). For rapid visual evaluation of the data, a coloring scale with green (0<DI≤1), yellow (1<DI≤2), orange (2<DI≤3) and red (3<DI≤4) was used. The experiment was done twice with 12 seedlings maintained in two square Petri plates for one treatment. The data of the two experiments were pooled before Mann-Whitney comparisons were applied at p = 0.05. Within columns, disease severities followed by the same letter are not significantly different.
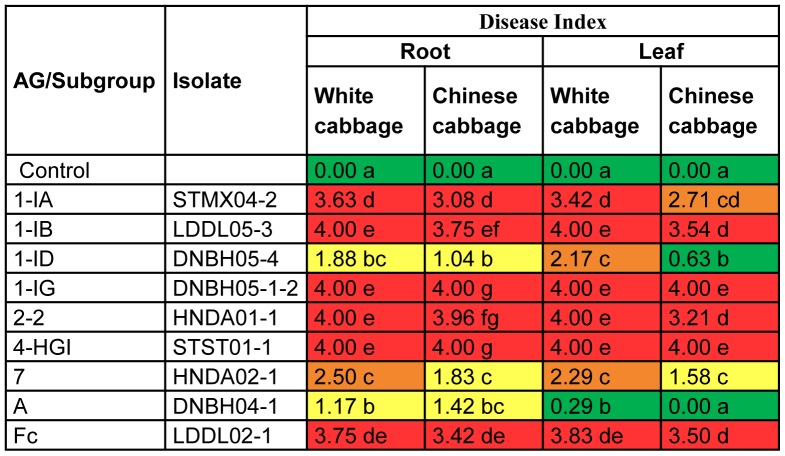
Figure 4. Pathogenic potential of Rhizoctonia isolates on roots and hypocotyls of white cabbage and Chinese cabbage in in vivo experiment.
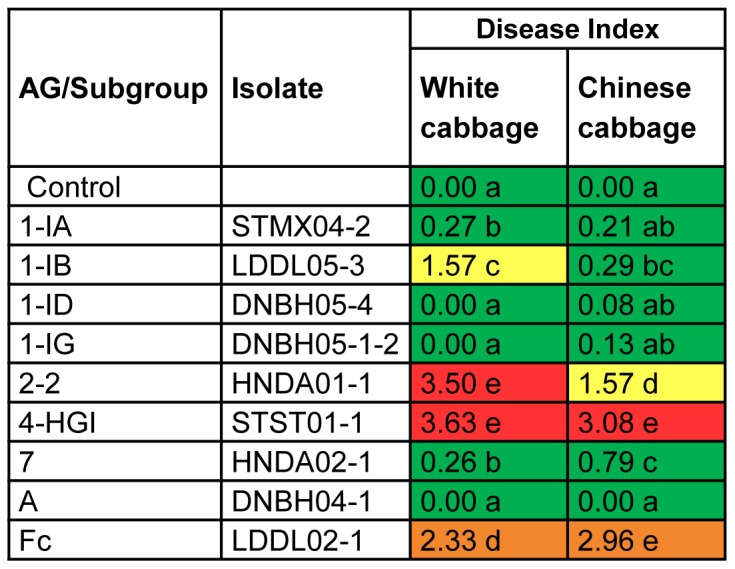
Disease severity on roots was assessed on a scale ranging from 0 (no symptoms) to 4 (seedling dead). For rapid visual evaluation of the data, a coloring scale with green (0<DI≤1), yellow (1<DI≤2), orange (2<DI≤3) and red (3<DI≤4) was used. The experiment was performed twice with 12 seedlings cultivated in two plastic boxes per treatment. The data of the two experiments were pooled before Mann-Whitney comparisons were applied at p = 0.05. Within columns, disease severities followed by the same letter are not significantly different.
Leaves of white cabbage (Brassica oleracea), Chinese cabbage (B. chinensis), pak choi (B. chinensis), mustard cabbage (B. juncea) and Chinese flowering cabbage (B. parachinensis) were cut into pieces (3×3 cm). The soil substrate used in our experiments was a mixture (w/w) of 50% potting soil (Structural; Snebbout, Kaprijke, Belgium) and 50% sand (Cobo garden; Belgium). Sets of six leaf discs were placed in a plastic box (16×11×6 cm) containing 400 g of soil substrate. Inoculum of Rhizoctonia spp. was produced according to the method described by Scholten et al. [27]. Briefly, water-soaked wheat kernels were autoclaved for 25 min on two successive days and then inoculated with three fungal discs (diameter 5 mm) cut at the edge of a 3-day-old Rhizoctonia colony cultured on PDA. Flasks containing the inoculated kernels were incubated for 14 days at 28°C and shaken every 3–4 days to avoid coagulation. Two Rhizoctonia-infected kernels which were comparable in size were buried 2 cm below each leaf disc. Leaf discs inoculated with sterile wheat kernels served as a control. All boxes were incubated in a growth chamber at 22°C.
The detached leaf bio-assay was also conducted to investigate the pathogenicity of the Rhizoctonia isolates on rice and water spinach, two important hosts of Rhizoctonia spp. in tropical countries. Surface-sterilized seeds of rice (Oryza sativa cv. CO39) and water spinach (Ipomoea aquatic cv. Trang Nong) were sown in plastic trays (45×45×10 cm) filled with 4 kg of soil substrate and kept in a growth chamber at 28°C for four weeks before their leaves were detached. Rice leaves were then cut into pieces (8 cm long) and six rice leaf pieces or six water spinach leaves were put in one square Petri dish containing a sterile filter paper moistened with sterile water. Two sterile glass slides were placed in the middle of each Petri dish to keep the leaves away from water. A 5-mm plug harvested from 3-day-old cultures of Rhizoctonia spp. on PDA was placed at the center of each leaf or leaf piece and the Petri dishes were incubated at 28°C.
After four days of incubation, disease severity was scored based on the following disease scale: 0 = no symptoms observed; 1 = lesions covered less than 25% of leaf surface; 2 = lesions covered 25–50% of leaf surface; 3 = lesions covered 50–75% of leaf surface; 4 = lesions covered more than 75% of leaf surface or dead leaf. The experiment had a completely randomized design. Each treatment consisted of 12 leaves or leaf pieces equally divided into two boxes or Petri dishes.
In vitro pathogenic potential of Rhizoctonia spp. on seedlings of white cabbage and Chinese cabbage
The same nine Rhizoctonia isolates were studied for their in vitro pathogenic potential using the method described by Keijer et al. [28]. Six surface-sterilized seeds of white cabbage (B. oleracea cv. TN180) or Chinese cabbage (B. chinensis cv. Elton) were germinated on Gamborg B5 medium (Gamborg B5 medium including vitamins; Duchefa) in a square Petri dish. Two mycelial disks (5 mm in diameter) from 3-day-old Rhizoctonia cultures grown on PDA were placed between seeds. In the control dishes, sterile PDA discs were used for inoculation. The Petri dishes were incubated at 22°C in the dark for two days for seed germination. Then, the second halves of the Petri dishes were covered with aluminum foil to protect the roots from light and placed in an upright position in a growth chamber (22°C, 12 h light). The disease severity was recorded for root and hypocotyl or for leaves after six days of incubation using the following disease scale: 0 = healthy, no symptoms; 1 = lesions covering less than 25% of the root, hypocotyl or leaf surface; 2 = lesions covering between 25% and 50% of the root, hypocotyl or leaf surface; 3 = wilted plant with lesions covering between 50% and 75% of the root, hypocotyl or leaf surface; 4 = lesions covering more than 75% of root, hypocotyl or leaf surface or dead plant. In this test, a complete randomized design was applied with two Petri dishes (six seedlings each) per treatment and this experiment was done twice.
In vivo pathogenic potential of Rhizoctonia spp. on roots and hypocotyls of white cabbage and Chinese cabbage
Surface-sterilized seeds of white cabbage and Chinese cabbage were germinated on wet filter paper in Petri dishes at 22°C one day before sowing into 600 g of soil substrate. Four days after sowing, each perforated plastic box (22×15×6 cm) with six seedlings was inoculated by placing a row of 12 Rhizoctonia-colonized wheat kernels in the middle of the box. The kernels used for inoculation had equivalent sizes and were produced as described previously. Control seedlings were similarly treated with sterile wheat kernels. All plants were incubated at 22°C. Disease severity on root and hypocotyl was evaluated 14 days after inoculation using the same disease scale described for the in vitro experiment. A completely randomized design was used with 12 seedlings cultivated in two experimental boxes per treatment and this test was performed twice with the same nine isolates that were used in the previous tests.
Statistical analysis
The severity of Rhizoctonia diseases on roots and leaves of Brassica seedlings are presented in Figures 2–4 and Tables S3–S5 in File S1 as Disease index (DI). DI was calculated using the following formula:
Pathogenicity data for the two experiments with nine isolates of nine different AGs were always very similar and no significant interaction was found between the experiments. Therefore, statistical analysis was done on pooled data for the different repeats. The non-parametric Kruskal-Wallis test for k independent samples was used, after which pair-wise comparisons were performed for all treatments using Mann-Whitney tests at a confidence level of p = 0.05.
To determine the correlation between the distributions of AGs and sampling locations and between the distributions of AGs and sampled Brassica spp., contingency tables were constructed using Excel. Then, potential significant differences between the variables were revealed with Fisher's Exact Tests. All statistical analyses were conducted in SPSS 22.0 (SPSSinc, Illinois, USA).
Results
Molecular characterization and phylogenetic analysis of Rhizoctonia isolates
A total of 142 Brassica plants with Rhizoctonia-like symptoms were sampled in various important vegetable producing regions in Vietnam (see Figure 1). Of the 97 Rhizoctonia isolates recovered, only four were binucleate with two nuclei per hyphal cell. The other 93 isolates had multinucleate cells (data not shown).
Analysis of the rDNA-ITS region using the BLASTn tool (against Genbank and against the curated database) revealed that three binucleate Rhizoctonia isolates belonged to AG-A while the fourth isolate (LDDL02-1) could not be assigned to any known AG.
Pairwise sequence similarity scores of isolate LDDL02-1 against all isolates in the curated database (representing all known AGs) were determined. Highest pairwise sequence similarities were found with isolates of multinucleate AG 6 (94%) and binucleate AG-Fb (93%) (Table 4). When blasting to Genbank, several isolates from taro and ginger from Yunnan Province in China were found to be nearly identical to isolate LDDL02-1.
Table 4. Pairwise sequence similarities of unknown isolates LDDL02-1 and DNBH05-1-2 to all known AGs from the curated database in Table S2 in File S2.
| LDDL02-1 | DNBH05-1-2 | |
| AG 1-IA | 0.88 | 0.92 |
| AG 1-IB | 0.84–0.87 | 0.84–0.87 |
| AG 1-IC | 0.89–0.90 | 0.89–0.90 |
| AG 1-ID | 0.85–0.86 | 0.85 |
| AG 1-IE | 0.89 | 0.94 |
| AG 1-IF | 0.84 | 0.85 |
| AG 2-1 | 0.87–0.90 | 0.82–0.85 |
| AG 2-2 | 0.86–0.87 | 0.84–0.85 |
| AG 2-3 | 0.89–0.90 | 0.85 |
| AG 3 | 0.89–0.90 | 0.84–0.85 |
| AG 4 | 0.86–0.88 | 0.85–0.88 |
| AG 5 | 0.91 | 0.86–0.87 |
| AG 6 | 0.89–0.94 | 0.88–0.90 |
| AG 7 | 0.91 | 0.89 |
| AG 8 | 0.92–0.93 | 0.89–0.90 |
| AG 9 | 0.9 | 0.86 |
| AG 10 | 0.89–0.90 | 0.84–0.85 |
| AG 11 | 0.88–0.89 | 0.84 |
| AG 12 | 0.89–0.90 | 0.87–0.88 |
| AG 13 | 0.90 | 0.90 |
| AG 2-BI | 0.85–0.86 | 0.81–0.82 |
| AG-A | 0.84 | 0.84 |
| AG-K | 0.84 | 0.84 |
| AG-Bb | 0.79–0.80 | 0.78–0.79 |
| AG-Q | 0.80 | 0.79 |
| AG-Bo | 0.83 | 0.83 |
| AG-Ba | 0.83–0.84 | 0.83–0.84 |
| AG-C | 0.80–0.82 | 0.82 |
| AG-H | 0.81–0.82 | 0.81 |
| AG-I | 0.81 | 0.80–0.81 |
| AG-D | 0.78–0.81 | 0.77–0.79 |
| AG-G | 0.85 | 0.85 |
| AG-L | 0.85 | 0.85–0.86 |
| AG-O | 0.86 | 0.86 |
| AG-Fb | 0.93 | 0.89 |
| AG-P | 0.86–0.89 | 0.86–0.90 |
| AG-R | 0.88 | 0.85 |
| AG-S | 0.88 | 0.88 |
| AG-Fa | 0.91 | 0.91 |
| AG-E | 0.90–0.91 | 0.89 |
| UNR1 | 0.82–0.83 | 0.84 |
| UNR2 | 0.80 | 0.82 |
| AG-N | 0.67 | 0.66 |
| W. circinata | 0.66–0.67 | 0.64–0.66 |
LDDL02-1 shows most similarity to AG 6 and AG-Fb. DNBH05-1-2 shows highest pairwise sequence similarity to AG 1-IA and AG 1-IE.
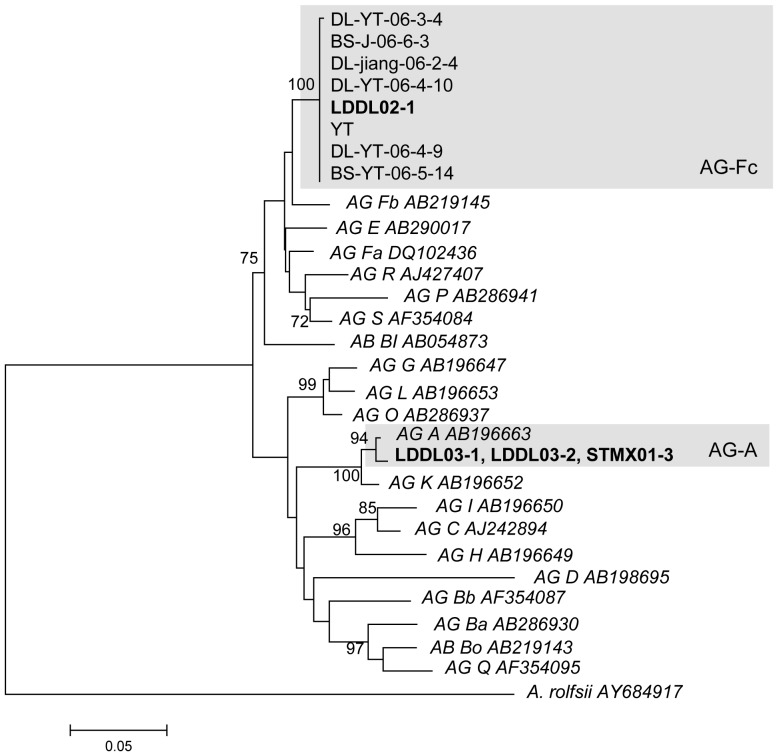
From the binucleate phylogenetic tree (Figure 5), it is clear that the unknown isolate LDDL02-1 clusters together with the isolates from Yunnan province, forming a clade with high bootstrap support that is different from any known binucleate AG, with the closest related AG being AG-Fb. So far, nothing has been published about the Chinese isolates. Pairwise sequence similarity within AG-F is 90–100% [2], and the closest related AGs are AG-Fb (93%) and AG-Fa (91%). Therefore, we propose to assign these isolates as a new AG-F subclade, namely AG-Fc.
Figure 5. rDNA-ITS phylogeny of binucleate Rhizoctonia spp. sampled from Brassica spp. in Vietnam.
Neighbour joining tree derived from the alignment of 31 binucleate Rhizoctonia isolates and the outgroup Athelia rolfsii (AY684917). Isolates in bold are the 4 isolates derived from Brassica spp. in Vietnam during this study. For each known binucleate Rhizoctonia AG, a representative isolate (in italics) from the curated database (Table S2 in File S2) is included. Bootstraps are only given for those branches with bootstrap support higher than 70. The tree was made using only isolates with unique sequences. Isolates with identical sequences were added afterwards on the same line.
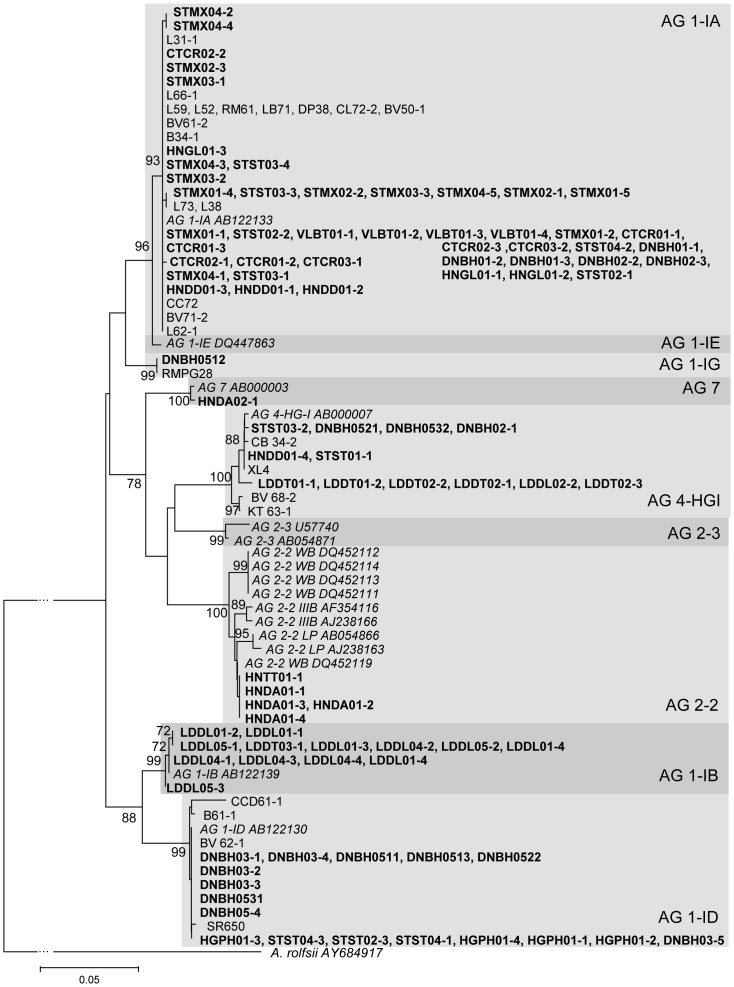
In the multinucleate tree, all our Vietnamese isolates form clades with high bootstrap supports together with representative isolates from the curated database (Figure 6). The largest group belonged to AG 1-IA (44 isolates), followed by AG 1-ID (17 isolates), AG 1-IB (13 isolates) and AG 4-HGI (12 isolates). Five isolates belonged to AG 2-2. AG 2-2 is further divided into four groups: AG 2-2 IV, AG 2-2 LP, AG 2-2 IIIB and AG 2-2 WB. The similarity scores between our isolates and AG 2-2 IIIB and AG 2-2 LP were comparable (between 97 and 98%). Also, our isolates show 99% similarity with isolate Barranca (DQ452119), which is assigned as AG 2-2 WB by Godoy-Lutz et al. [25]. However, with other isolates from AG 2-2 WB (DQ452111-114), pairwise sequence similarity is lower (between 96 and 97%). Therefore, we decided that it is impossible at this point to assign our isolates to any of the AG 2-2 phylogenetic subgroups. One isolate belonged to AG 7 and the last isolate (DNBH05-1-2) could not be assigned to any of the known multinucleate AG groups in our reference database. The isolate showed the highest pairwise sequence similarity with AG 1-IE (94%, see Table 4). A BLASTn search on Genbank identified an isolate from chickpea in India (RMPG28, [29]) assigned to AG 2–3 to be>99% similar (1 bp substitution) to DNBH05-1-2. However, when comparing these isolates to the AG 2–3 isolates in the curated database, we found low pairwise sequence similarity (85%) (Table 4). Also in the phylogenetic tree (see Figure 6), isolate DNBH05-1-2 clusters together with isolate RMPG28, but not with the representative AG 2–3 isolates from the curated database, indicating that isolate RMPG28 is wrongly designated as AG 2–3 in Genbank. Hence, these isolates represent a new subgroup of AG 1 and we propose the name AG 1-IG.
Figure 6. rDNA-ITS phylogeny of multinucleate Rhizoctonia spp. sampled from Brassica spp. in Vietnam.
Neighbour joining tree derived from the alignment of 128 multinucleate Rhizoctonia isolates and the outgroup Athelia rolfsii (AY684917). Isolates in bold are the isolates derived from Brassica spp. in Vietnam during this study. For each of the multinucleate Rhizoctonia AG subgroups present in our sampling, representative isolates (in italics) from the curated database (Table S2 in File S2) are included. Bootstraps are only given for those branches with bootstrap support higher than 70. The tree was made using only isolates with unique sequences. Isolates with identical sequences were added afterwards on the same line. Only half of the length of the outgroup branch is shown to increase clarity.
For most of the AGs we detected in Vietnam there was very little variation in ITS sequence, except for AG 4-HGI where six variable positions were found between the isolates from Lam Dong and the isolates from the other three provinces where this AG was detected (Soc Trang, Dong Nai and Ha Noi).
The AG 1-IA isolates showed identical or nearly identical (max. 4 SNPs) sequences to AG 1-IA isolates previously isolated from rice, water spinach, water hyacinth and other crops in Vietnam (Table 3). The AG 1-ID isolates showed high similarity (identical sequence or one SNP) to isolates detected on coffee, cotton, durian [30], corn and sugar beet (Table 3) in Vietnam.
Relationship between the AGs found and sampling locations and between AGs found and Brassica species
There seems to be a relationship between the AGs present and the sampling areas (Figure 1 and Table 5). Thirty three out of 44 isolates belonging to AG 1-IA were recovered from samples collected from Soc Trang, Can Tho and Vinh Long in the Mekong River delta, the main rice production region of Vietnam. On the other hand, no AG 1-IA isolates were detected in Lam Dong although the expected frequency of this group in this province was high (9.98). The observed numbers were also significantly higher than the expected numbers for AG 1-IB and AG 4-HGI in Lam Dong as well as for AG 1-ID in Hau Giang and Dong Nai.
Table 5. Contingency table with observed and expected frequencies of anastomosis groups (AGs) of Rhizoctonia spp. obtained from Brassica fields in different provinces of Vietnam.
| Province | AG/Subset | Total/province | ||||||||
| Multinucleate Rhizoctonia | Binucleate Rhizoctonia | |||||||||
| 1-IA | 1-IB | 1-ID | 2-2 | 1-IG | 4-HGI | 7 | A | Fc | ||
| Ha Noi | 6 (5.90) | 0 (1.74) | 0 (2.28) | 5 (0.67) | 0 (0.13) | 1 (1.61) | 1 (0.13) | 0 (0.40) | 0 (0.13) | 13 |
| Lam Dong | 0 (9.98)* | 13 (2.95)* | 0 (3.86) | 0 (1.13) | 0 (0.45) | 6 (2.72)* | 0 (0.23) | 2 (0.68) | 1 (0.23) | 22 |
| Dong Nai | 5 (8.62) | 0 (2.55) | 10 (3.33)* | 0 (0.98) | 1 (0.20) | 3 (2.35) | 0 (0.20) | 0 (0.59) | 0 (0.20) | 19 |
| Vinh Long | 4 (1.81) | 0 (0.54) | 0 (0.70) | 0 (0.21) | 0 (0.04) | 0 (0.49) | 0 (0.04) | 0 (0.12) | 0 (0.04) | 4 |
| Can Tho | 8 (3.63) | 0 (1.07) | 0 (1.40) | 0 (0.41) | 0 (0.08) | 0 (0.99) | 0 (0.08) | 0 (0.25) | 0 (0.08) | 8 |
| Hau Giang | 0 (1.81) | 0 (0.54) | 4 (0.70)* | 0 (0.21) | 0 (0.04) | 0 (0.49) | 0 (0.04) | 0 (0.12) | 0 (0.04) | 4 |
| Soc Trang | 21 (12.25)* | 0 (3.62) | 3 (4.73) | 0 (1.39) | 0 (0.28) | 2 (3.34) | 0 (0.28) | 1 (0.84) | 0 (0.28) | 27 |
| Total/AG | 44 | 13 | 17 | 5 | 1 | 12 | 1 | 3 | 1 | 97 |
Data show actual numbers of isolates collected among the different provinces. According to Fisher's exact test (p = 0.05), the AGs found are related to the sampling locations. An asterisk* indicates significant differences between observed and expected numbers. Values in parentheses represent the expected numbers.
The relationship between the occurrence of different AGs and the host plants is presented in Table 6. Significant AG-host correlations were identified in the following combinations: AG 1-IB and white cabbage, AG 1-IB and broccoli, AG 1-ID and Chinese flowering cabbage, and AG 2-2 and turnip cabbage.
Table 6. Contingency table with observed and expected frequencies of anastomosis groups (AGs) of Rhizoctonia spp. obtained from field-grown Brassica crops in Vietnam.
| Host plant | AG/Subseta | Total/host plant | ||||||||
| Multinucleate Rhizoctonia | Binucleate Rhizoctonia | |||||||||
| 1-IA | 1-IB | 1-ID | 2-2 | 1-IG | 4-HGI | 7 | A | Fc | ||
| Mustard cabbage | 14 (9.07) | 0 (2.68) | 5 (3.51) | 0 (1.03) | 0 (0.21) | 1 (2.47) | 0 (0.21) | 1 (0.62) | 0 (0.21) | 20 |
| White cabbage | 8 (9.53) | 8 (2.81)* | 0 (3.68) | 0 (1.08) | 0 (0.22) | 3 (2.60) | 0 (0.22) | 2 (0.65) | 0 (0.22) | 21 |
| Chinese flowering cabbage | 9 (12.25) | 0 (3.62) | 12 (4.73)* | 1 (1.39) | 1 (0.28) | 4 (3.34) | 0 (0.28) | 0 (0.84) | 0 (0.28) | 27 |
| Chinese cabbage | 0 (2.27) | 1 (0.67) | 0 (0.88) | 0 (0.26) | 0 (0.05) | 3 (0.62) | 0 (0.05) | 0 (0.15) | 1 (0.05) | 5 |
| Pak choi | 10 (4.99) | 0 (1.47) | 0 (1.93) | 0 (0.57) | 0 (0.11) | 0 (1.36) | 0 (0.11) | 1 (0.34) | 0 (0.11) | 11 |
| Broccoli | 0 (1.81) | 4 (0.54)* | 0 (0.70) | 0 (0.21) | 0 (0.04) | 0 (0.49) | 0 (0.04) | 0 (0.12) | 0 (0.04) | 4 |
| Turnip cabbage | 3 (4.08) | 0 (1.21) | 0 (1.58) | 4 (0.46)* | 0 (0.09) | 1 (1.11) | 1 (0.09) | 0 (0.28) | 0 (0.09) | 9 |
| Total/AG | 44 | 13 | 17 | 5 | 1 | 12 | 1 | 3 | 1 | 97 |
Data show actual numbers of isolates collected among the different Brassica crops. According to Fisher's exact test (p = 0.05), the AGs found are related to the crops. An asterisk* indicates significant differences between observed and expected numbers. Values in parentheses represent the expected numbers.
Aggressiveness of Rhizoctonia isolates towards detached leaves
Nine Rhizoctonia isolates, randomly selected from each identified AG, were tested for their pathogenicity on detached leaves. Due to practical reasons, experiments with Brassica crops were conducted in a growth chamber specifically built for Brassica spp. (22°C, RH = 60%, 12 h photoperiod) and experiments with rice and water spinach were done in a growth chamber specifically built for rice (28°C, RH = 60%, 16 h photoperiod). Although the temperature and humidity in these chambers are slightly lower than those of Vietnam, they are still suitable for the growth of host plants and Rhizoctonia isolates involved in our study.
As shown in Figure 2, leaves of white cabbage, Chinese cabbage, pak choi, mustard cabbage, Chinese flowering cabbage, rice, and water spinach responded differently to the infection of Rhizoctonia spp. although each host was affected by at least four AGs. For each plant species, a wide variation in symptom severity induced by different AGs was observed and the only AG that could not cause disease on any of the plants tested was AG-A. White cabbage leaves were most severely infected by all AGs (except AG-A) with a DI varying from 3.13 to 4.00. Compared to other hosts, Chinese cabbage appeared to be most resistant to Rhizoctonia isolates. No disease symptoms were observed on Chinese cabbage leaves challenged with AG 1-IB, AG 1-ID, AG 2-2 and AG 7, while other AGs were weak to moderately aggressive (DI varied from 0.71 in AG-Fc to 2.63 in AG 1-IA). Inoculation of pak choi with AG 1-IB resulted in a complete decay of all leaf discs (DI = 4.00). For mustard cabbage and Chinese flowering cabbage, a strong disease pattern was obtained on leaves confronted with AG 1-IB (DI≥3.92) and AG 4-HGI (DI≥3.71). Rice leaves were very susceptible to AG 1-IA and they were severely destroyed within four days of inoculation (DI = 3.92). Disease induced by other R. solani isolates on rice was also significantly different from the control although the two binucleate Rhizoctonia isolates tested were not pathogenic on rice. For water spinach, a wide variation in aggressiveness of R. solani isolates was observed, resulting in DI values ranging from 0.46 to 3.54. Large lesions (DI≥3.21) developed on water spinach leaves inoculated with AG 1-IA and AG 1-ID isolates. The difference in pathogenicity of these AGs towards the different Brassica species was confirmed when additional isolates from AGs 1-IA, 1-IB, 1-ID, 2-2, 4-HGI and A were tested in a detached leaf assay on the same series of plants (see supplementary information, Table S3 in File S1).
In vitro pathogenic potential of Rhizoctonia spp. isolates on seedlings of white cabbage and Chinese cabbage
The results of the detached leaf assay revealed that white cabbage leaves were remarkably more susceptible than other cabbage species, while only some AGs could attack the leaves of Chinese cabbage. Therefore, these two plant species were selected as hosts for in vitro and in vivo pathogenicity tests using the same nine Rhizoctonia isolates as mentioned above. Under in vitro conditions (Figure 3), all AGs could cause disease and symptoms were observed on both roots and leaves. Isolates of AG 1-IA, AG 1-IB, AG 2-2, AG 1-IG, AG 4-HGI and AG-Fc were most aggressive towards these hosts (for white cabbage: DI on roots ≥3.63 and DI on leaves ≥3.42; for Chinese cabbage: DI on roots ≥3.08 and DI on leaves ≥2.71). Fewer symptoms were recorded on seedlings inoculated with isolates belonging to AG 1-ID, AG 7 and AG-A. The virulence of AG 1-IA, AG 1-IB, AG 2-2 and AG 4-HGI towards white cabbage and Chinese cabbage was confirmed when in vitro assays were repeated with additional isolates of these AGs (Table S4 in File S1). Towards white cabbage, DI on roots varied from 2.17 to 4.00 and DI on leaves fluctuated between 2.33 and 4.00. DI on roots and leaves of Chinese cabbage ranged from 2.08 to 4.00 and from 1.92 to 4.00, respectively.
In vivo pathogenic potential of Rhizoctonia spp. isolates on roots and hypocotyls of white cabbage and Chinese cabbage
The results displayed in Figure 4 demonstrate that severe Rhizoctonia-induced damage on white cabbage roots could only be seen for AG 2-2 (DI = 3.50) and AG 4-HGI (DI = 3.63). Moderate infection (DI = 2.33) was observed in response to AG-Fc. Towards Chinese cabbage, severe disease symptoms were incited by AG 4-HGI (DI = 3.08) and AG-Fc (DI = 2.96). No symptoms were detected on roots and hypocotyls of seedlings challenged with AG-A. In addition, the inoculation of isolates belonging to AG 1-IA, AG 1-IB, AG 1-ID, AG 1-IG and AG 7 did not result in the formation of large lesions on roots of white cabbage and Chinese cabbage seedlings (DI≤1.57). Data obtained from the tests conducted with additional Rhizoctonia isolates belonging to AGs 1-IA, 1-IB, 1-ID, 2-2, 4-HGI and AG-A are presented in Table S5 in File S1. These data confirm the high aggressiveness of AG 2-2 on white cabbage (DI = 4.00) and that of AG 4-HGI on both white cabbage and Chinese cabbage (DI≥3.50).
Discussion
Rhizoctonia is an important fungal ‘form genus’ occurring worldwide and including many important plant pathogenic strains as well as mycorrhizal fungi and hypovirulent or avirulent strains among which there are strains that are capable to protect plants against pathogenic Rhizoctonia and other pathogens as well as increase plant growth. Plants can be infected by different Rhizoctonia AGs from the time of sowing resulting in the development of both foliar and root diseases. This is the first time Rhizoctonia species that attack field-grown Brassica crops in Vietnam were isolated and characterized. Ninety seven isolates of Rhizoctonia were recovered from symptomatic plant tissues of seven brassicaceous hosts (mustard cabbage, white cabbage, Chinese flowering cabbage, pak choi, turnip cabbage, Chinese cabbage and broccoli). Of all the isolates collected, 4% were binucleate Rhizoctonia and 96% were multinucleate Rhizoctonia. Molecular characterization by sequencing of the rDNA-ITS region showed that the binucleate isolates found belonged to AG-A (3 isolates) and an unknown AG introduced here as AG-Fc (1 isolate). Comparison to a curated sequence database of all known Rhizoctonia multinucleate, binucleate and uninucleate AGs did not reveal high homology of this isolate to a known AG. Pairwise sequence similarities to all known multinucleate, binucleate and uninucleate Rhizoctonia AGs showed highest similarity to AG-Fb, but also to AG 6, which is a multinucleate AG. Close relationships of some binucleate groups with multinucleate groups has been previously noted by Sharon et al. [2] who did a combined ITS sequence analysis of multinucleate, binucleate and uninucleate groups. These authors stated that the clustering of binucleate and uninucleate groups close to certain multinucleate clusters may indicate a possible evolutionary bridge between multinucleate and binucleate groups and our results support this hypothesis.
We conducted pathogenicity assays on detached leaves, in vitro seedlings and plants grown in vivo. In general, pathogenicity was highest on roots and leaves under in vitro conditions, which is probably due to the high humidity and the young age of the plants in this system. It should be mentioned that we only checked in vivo pathogenicity towards roots and hypocotyls, but it should be borne in mind that some of our isolates are mainly leaf pathogens.
Although AG-A isolates were obtained from symptomatic plants, they were unable to induce disease on detached leaves or on seedlings in vivo, and were only slightly pathogenic on cabbage seedlings under in vitro conditions. This low virulence may be due to the differences in humidity between our experimental conditions and the field situation in Vietnam. Alternatively, AG-A isolates might be avirulent, but since they grow very fast in vitro, they may have masked the presence of slower growing, virulent AGs. In contrast to AG-A, AG-Fc appeared to be moderately to highly virulent in all experiments. This also concurs with previous studies that some binucleate Rhizoctonia species are highly virulent [31]–[33], whereas others are weakly virulent or avirulent [34]–[36].
Among the multinucleate AGs, AG 1-IA was the dominant group, followed by AG 1-ID, AG 1-IB, AG 4-HGI, AG 2-2, AG 7 and an unknown AG that we introduced here as AG 1-IG. The occurrence of AG 1-IB and AG 4 on Brassica spp. is well known from previous studies. According to Pannecoucque et al. [22], AG 1-IB is one of the causal agents of wirestem in Belgian cauliflower fields. AG 1-IB isolates are also highly virulent on lettuce in Belgium [37]. In our study, AG 1-IB isolates were only recovered from Lam Dong, a province in the cool Central Highlands of Vietnam and the main lettuce production region of Vietnam, suggesting that the presence of this AG is associated with cool climates. The presence of AG 4 has also been reported on Brassica oleracea [21] and B. rapa subsp. chinensis [38] in China, B. oleracea in the UK [23], B. napus L. and B. campestris L. in Canada [39], and B. oleracea [18] and B. napus L. in the US [40]. AG 4-HGI occurred in most regions sampled. This AG has a wide host range due to its ability to adapt to temperature variation and cropping patterns [41]. AG 4 isolates are able to induce disease on all plant parts and in our pathogenicity trials, the highest disease ratings on cabbage in all bioassays were shown for the AG 4-HGI isolates. For white cabbage, severe disease symptoms were observed on seedlings inoculated with AG 2-2 and AG 4-HGI isolates in vivo or on detached leaves challenged with isolates of all AGs. Although R. solani AG 2-1 is considered the most dominant and damaging anastomosis group attacking Brassica spp. [22], [23], [42], isolates belonging to this group were not detected in our survey.
The predominance of R. solani isolates of AG 1-IA and also the presence of isolates belonging to AG 1-ID in our collection were not anticipated because these AGs have not been described on Brassica crops before. The presence of unusual AGs in our sampling is probably due to three aspects: (i) alternative hosts of Rhizoctonia present in the sampling locations; (ii) poor cultural practices and (iii) high temperature. AG 1-IA isolates were mainly recovered from samples collected in provinces of the Mekong delta including Vinh Long, Can Tho and Soc Trang. This is a low-lying coastal region of Vietnam, characterized by high temperature and humidity and prone to flooding every rainy season. Due to water availability and soil type, the agricultural production in this area is dominated by rice [43], [44]. As previously reported, sheath blight, caused by R. solani AG 1-IA, is a major disease of rice cultivated in intensive production systems [45]–[47]. With the ability to float and to survive in water [48], sclerotia of AG 1-IA isolates may easily spread from the rice paddy fields to vegetable fields through irrigation or flood water. The recovery of AG 1-IA in the hot region located in the South of Vietnam is consistent with the findings of Harikrishnan and Yang [41] that temperature can influence growth rate, sclerotia production of Rhizoctonia spp. and the distribution of Rhizoctonia isolates belonging to different anastomosis groups. AG 1 is a high temperature group [8] and its vegetative growth as well as sclerotia production and survival are inhibited at low temperatures. The occurrence of AG 1-IA on Brassica crops is probably even increased due to farmers' lack of knowledge about crop protection. AG 1 (not specific) has been considered as one of the causal agents of foliar diseases on water hyacinth (Eichhornia crassipes) [49], water lettuce (Pistia stratioites) and anchoring hyacinth (E. azurea) [50]. In this study, water spinach (Ipomoea aquatic) was found to be susceptible to the AG 1-IA isolates that we collected from Brassica spp. Vietnamese farmers commonly use these aquatic plants as cover materials in vegetable production and use irrigation water from sources where these plants are present, thus bringing the fungus from these alternative hosts to Brassica spp. (Figure 7). Isolates of AG 1-IA appeared to be very pathogenic towards leaves of cabbages but they could not induce severe disease on roots, especially under in vivo conditions. This finding is in agreement with results reported previously by Yang and Li [51] that AG 1-IA isolates have a tendency to attack aerial parts of plants. These data also support our hypothesis about the spread of AG 1-IA isolates from rice and water spinach to vegetables. In other words, rice and water spinach that are infected by R. solani could be an important source of inoculum that may contribute to the disease caused by AG 1-IA on Brassica spp.
Figure 7. Rhizoctonia-infected water hyacinth is introduced as cover material to a white cabbage field in Vietnam.
Rhizoctonia-infected water hyacinth is taken from a nearby water ditch and used as cover material on the white cabbage field. Via this common practice, Vietnamese farmers unintentionally introduce the Rhizoctonia fungus to their crops.
In our assays AG 1-ID was mainly pathogenic on leaves of white cabbage. Rhizoctonia AG 1-ID was previously found to be pathogenic on durian [30] and coffee [52]. Interestingly, ten out of 17 AG 1-ID isolates were collected from Dong Nai, a province where durian, coffee and cotton are widely cultivated [53], again suggesting a correlation between cropping patterns and Rhizoctonia distribution in Vietnam.
Collectively, it seems that the distribution of Rhizoctonia AGs in Vietnam is correlated with the cropping patterns and climatic conditions. However, it is difficult to draw a strong conclusion about the influence of cropping patterns and climatic conditions on the occurrence of Rhizoctonia spp. in Vietnam because the sampling regime for R. solani isolates comprises two variable parameters, namely the plant species infected by R. solani and the sampling site. Due to the variation in soil and climatic conditions, different Brassica species are grown in different geographic regions and there is a possibility that particular plant species might be more susceptible to infections by specific R. solani AGs than others. Therefore, another field survey with a systematic sampling regime needs to be conducted to confirm our hypothesis.
Our research also points towards the need to have good extension programs to improve the farmers' knowledge about crop protection. Additionally, knowing which AGs are responsible for Rhizoctonia diseases on Brassica spp. in Vietnam is an essential prerequisite for developing successful disease management strategies in this country.
Supporting Information
Contains the following files: Table S1. GPS co-ordinates of the wards in each province of Vietnam where Rhizoctonia-infected Brassica crops were sampled. Table S3. Aggressiveness of Rhizoctonia isolates towards white cabbage, Chinese cabbage, pak choi, mustard cabbage, Chinese flower cabbage, rice and water spinach in detached leaf bio-assays. Leaves were scored using a scale ranging from 0 (no disease symptoms) to 4 (lesions covered more than 75% of leaf surface or dead leaf). For rapid visual evaluation of the data, a coloring scale with green (0<DI≤1), yellow (1<DI≤2), orange (2<DI≤3) and red (3<DI≤4) was used. The test was done once with 12 leaves or leaf pieces per treatment. All data were statistically analyzed and within columns, disease severities followed by the same letter are not significantly different. Table S4. Aggressiveness of Rhizoctonia isolates towards roots and leaves of white cabbage and Chinese cabbage seedlings in in vitro bio-assays. Disease severity on roots or leaves was assessed on a scale ranging from 0 (no symptoms) to 4 (lesions covering more than 75% of root, hypocotyl or leaf surface or dead plant). For rapid visual evaluation of the data, a coloring scale with green (0<DI≤1), yellow (1<DI≤2), orange (2<DI≤3) and red (3<DI≤4) was used. Experiment was conducted once with 12 seedlings maintained in two square Petri plates for one treatment. All data were statistically analyzed and within columns, disease severities followed by the same letter are not significantly different. Table S5. Aggressiveness of Rhizoctonia isolates towards roots of white cabbage and Chinese cabbage seedlings in in planta experiment. Disease severity on roots was assessed on a scale ranging from 0 (no symptoms) to 4 (seedling dead). For rapid visual evaluation of the data, a coloring scale with green (0<DI≤1), yellow (1<DI≤2), orange (2<DI≤3) and red (3<DI≤4) was used. Experiment was performed once. Each treatment consisted of 12 seedlings cultivated in two plastic boxes. Data were statistically analyzed and within columns, disease severities followed by the same letter are not significantly different.
(DOC)
Table S2. Curated database of sequences containing representative rDNA-ITS sequences of all known uninucleate, binucleate and multinucleate Rhizoctonia AG and subgroups.
(XLSX)
Acknowledgments
The authors wish to thank Dr. Tran Thi Thu Thuy and Dr. Nguyen Thi Thu Nga (Can Tho University) and Dr. Ha Viet Cuong (Ha Noi University of Agriculture) for helpful advice and Dr. Michal Sharon for providing the rDNA-ITS sequence alignment of UNR, BNR and MNR reference isolates.
Funding Statement
This work was supported by a scholarship from the Special Research Fund of Ghent University (BOF) given to Gia Khuong Hoang Hua. Lien Bertier was funded by a PhD grant of the Agency of Innovation by Science and Technology in Flanders (IWT). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vietnam trade promotion agency (2008) Report on Vietnamese vegetable and fruit sector. Available: http://www.aseankorea.org/aseanZone/downloadFile2.asp?boa_filenum=1575. Accessed 2014 Jan 24.
- 2. Sharon M, Kuninaga S, Hyakumachi M, Naito S, Sneh B (2008) Classification of Rhizoctonia spp. using rDNA-ITS sequence analysis supports the genetic basis of the classical anastomosis grouping. Mycoscience 49: 93–114. [Google Scholar]
- 3. Sharon M, Kuninaga S, Hyakumachi M, Sneh B (2006) The advancing identification and classification of Rhizoctonia spp. using molecular and biotechnological methods compared with the classical anastomosis grouping. Mycoscience 47: 299–316. [Google Scholar]
- 4. Carling DE, Baird RE, Gitaitis RD, Brainard KA, Kuninaga S (2002) Characterization of AG-13, a newly reported anastomosis group of Rhizoctonia solani . Phytopathology 92: 893–899. [DOI] [PubMed] [Google Scholar]
- 5. Carling DE, Pope EJ, Brainard KA, Carter DA (1999) Characterization of mycorrhizal isolates of Rhizoctonia solani from an orchid, including AG-12, a new anastomosis group. Phytopathology 89: 942–946. [DOI] [PubMed] [Google Scholar]
- 6. Carling DE, Kuninaga S, Brainard KA (2002) Hyphal anastomosis reactions, rDNA-internal transcribed spacer sequences, and virulence levels among subsets of Rhizoctonia solani anastomosis group-2 (AG-2) and AG-BI. Phytopathology 92: 43–50. [DOI] [PubMed] [Google Scholar]
- 7. Hyakumachi M, Priyatmojo A, Kubota M, Fukui H (2005) New anastomosis groups, AG-T and AG-U, of binucleate Rhizoctonia spp. causing root and stem rot of cut-flower and miniature roses. Phytopathology 95: 784–792. [DOI] [PubMed] [Google Scholar]
- 8.Sneh B, Burpee L, Ogoshi A (1991). Identification of Rhizoctonia species. St. Paul Minnesota: APS Press. 133 p. [Google Scholar]
- 9.Schwartz HF, Gent DH, Franc GD, Harveson RM (2007) Dry bean-Rhizoctonia root rot. Available: http://www.scarab.msu.montana.edu/HpIPMSearch/AuthorSearch.exe?Complete=true&sort=asc. Accessed 2014 Jan 24.
- 10.Wharton P, Kirk W, Berry D, Snapp S (2007) Rhizoctonia stem canker and black scurf of potato. Available: http://www.potatodiseases.org/pdf/rhizoctonia-bulletin.pdf. Accessed 2014 Jan 24.
- 11.Keijer J (1996) The initial steps of the infection process in Rhizoctonia solani In: Sneh B, Jabaji-Hare S, Neate S, Dijst G, editors. Rhizoctonia species: Taxonomy, molecular biology, ecology, pathology and disease control. Dordrecht: Springer Netherlands. pp. 149–162. [Google Scholar]
- 12. Verma PR (1996) Biology and control of Rhizoctonia solani on rapeseed: A review. Phytoprotection 77: 99–111. [Google Scholar]
- 13. Yang J, Kharbanda PD, Wang H (1996) Characterization, virulence, and genetic variation of Rhizoctonia solani AG-9 in Alberta. Plant Dis 80: 513–518. [Google Scholar]
- 14. Khangura RK, Barbetti MJ, Sweetingham MW (1999) Characterization and pathogenicity of Rhizoctonia species on canola. Plant Dis 83: 714–721. [DOI] [PubMed] [Google Scholar]
- 15. Sayama A (2000) Occurrence of damping-off disease caused by Rhizoctonia solani AG 2-2-IIIB on cabbage plug seedlings. Annu Rep Soc Plant Protect N Jpn 51: 54–57. [Google Scholar]
- 16. Homma Y, Yamashita Y, Ishii M (1983) A new anastomosis group AG-7 of Rhizoctonia solani Kuhn from Japanese radish fields. Ann Phytopathol Soc Jpn 49: 184–190. [Google Scholar]
- 17. Keinath AP, Farnham MW (1997) Differential cultivars and criteria for evaluating resistance to Rhizoctonia solani in seedling Brassica oleracea . Plant Dis 81: 946–952. [DOI] [PubMed] [Google Scholar]
- 18. Rollins PA, Keinath AP, Farnham MW (1999) Effect of inoculum type and anastomosis group of Rhizoctonia solani causing wirestem of cabbage seedlings in a controlled environment. Can J Plant Pathol 21: 119–124. [Google Scholar]
- 19. Paulitz TC, Okubara PA, Schillinger WF (2006) First report of damping-off of canola caused by Rhizoctonia solani AG 2-1 in Washington State. Plant Dis 90: 829–829. [DOI] [PubMed] [Google Scholar]
- 20. Kuramae E, Buzeto A, Ciampi M, Souza N (2003) Identification of Rhizoctonia solani AG 1-IB in lettuce, AG 4 HG-I in tomato and melon, and AG 4 HG-III in broccoli and spinach, in Brazil. Eur J Plant Pathol 109: 391–395. [Google Scholar]
- 21. Yang GH, Chen JY, Pu WQ (2007) First report of head rot of cabbage and web blight of snap bean caused by Rhizoctonia solani AG-4 HGI. Plant Pathol 56: 351–351. [Google Scholar]
- 22. Pannecoucque J, Van Beneden S, Höfte M (2008) Characterization and pathogenicity of Rhizoctonia isolates associated with cauliflower in Belgium. Plant Pathol 57: 737–746. [Google Scholar]
- 23. Budge GE, Shaw MW, Lambourne C, Jennings P, Clayburn R, et al. (2009) Characterization and origin of infection of Rhizoctonia solani associated with Brassica oleracea crops in the UK. Plant Pathol 58: 1059–1070. [Google Scholar]
- 24.White TJ, Bruns TD, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfland DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to methods and applications. New York: Academic Press. pp. 315–322. [Google Scholar]
- 25. Godoy-Lutz G, Kuninaga S, Steadman JR, Powers K (2008) Phylogenetic analysis of Rhizoctonia solani subgroups associated with web blight symptoms on common bean based on ITS-5.8S rDNA. J Gen Plant Pathol 74: 32–40. [Google Scholar]
- 26. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scholten OE, Panella LW, De Bock TSM, Lange W (2001) A greenhouse test for screening sugar beet (Beta vulgaris) for resistance to Rhizoctonia solani . Eur J Plant Pathol 107: 161–166. [Google Scholar]
- 28. Keijer J, Korsman MG, Dullemans AM, Houterman PM, De Bree J, et al. (1997) In vitro analysis of host plant specificity in Rhizoctonia solani . Plant Pathol 46: 659–669. [Google Scholar]
- 29. Dubey SC, Tripathi A, Upadhyay BK (2012) Molecular diversity analysis of Rhizoctonia solani isolates infecting various pulse crops in different agro-ecological regions of India. Folia Microbiol (Praha) 57: 513–524. [DOI] [PubMed] [Google Scholar]
- 30. Thuan TTM, Tho N, Tuyen BC (2008) First report of Rhizoctonia solani subgroup AG 1-ID causing leaf blight on durian in Vietnam. Plant Dis 92: 648–648. [DOI] [PubMed] [Google Scholar]
- 31. Martin B (1988) Identification, isolation, frequency, and pathogenicity of anastomosis groups of binucleate Rhizoctonia spp. from strawberry roots. Phytopathology 78: 379–384. [Google Scholar]
- 32. Demirci E, Eken C, Zengin H (2002) First report of Rhizoctonia solani and binucleate Rhizoctonia from Johnsongrass in Turkey. Plant Pathol 51: 391–391. [Google Scholar]
- 33. Babiker EM, Hulbert SH, Schroeder KL, Paulitz TC (2013) Evaluation of Brassica species for resistance to Rhizoctonia solani and binucleate Rhizoctonia (Ceratobasidum spp.) under controlled environment conditions. Eur J Plant Pathol 136: 763–772. [Google Scholar]
- 34. Herr LJ (1995) Biological control of Rhizoctonia solani by binucleate Rhizoctonia spp. and hypovirulent R. solani agents. Crop Prot 14: 179–186. [Google Scholar]
- 35. Poromarto SH, Nelson BD, Freeman TP (1998) Association of binucleate Rhizoctonia with soybean and mechanism of biocontrol of Rhizoctonia solani . Phytopathology 88: 1056–1067. [DOI] [PubMed] [Google Scholar]
- 36. Ross RE, Keinath AP, Cubeta MA (1998) Biological control of wirestem on cabbage using binucleate Rhizoctonia spp. Crop Prot 17: 99–104. [Google Scholar]
- 37. Van Beneden S, Pannecoucque J, Debode J, De Backer G, Höfte M (2008) Characterisation of fungal pathogens causing basal rot of lettuce in Belgian greenhouses. Eur J Plant Pathol 124: 9–19. [Google Scholar]
- 38. Yang GH, Chen XQ, Chen HR, Naito S, Ogoshi A, et al. (2004) First report of foliar blight in Brassica rapa subsp. chinensis caused by Rhizoctonia solani AG-4. Plant Pathol 53: 260–260. [Google Scholar]
- 39. Yitbarek SM, Verma PR, Morrall RAA (1987) Anastomosis groups, pathogenicity, and specificity of Rhizoctonia solani isolates from seedling and adult rapeseed/canola plants and soils in Saskatchewan. Can J Plant Pathol 9: 6–13. [Google Scholar]
- 40. Baird RE (1996) First report of Rhizoctonia solani AG-4 on canola in Georgia. Plant Dis 80: 104–104. [DOI] [PubMed] [Google Scholar]
- 41. Harikrishnan R, Yang XB (2004) Recovery of anastomosis groups of Rhizoctonia solani from different latitudinal positions and influence of temperatures on their growth and survival. Plant Dis 88: 817–823. [DOI] [PubMed] [Google Scholar]
- 42. Ohkura M, Abawi GS, Smart CD, Hodge KT (2009) Diversity and aggressiveness of Rhizoctonia solani and Rhizoctonia-like fungi on vegetables in New York. Plant Dis 93: 615–624. [DOI] [PubMed] [Google Scholar]
- 43.Ministry of Natural Resources and Environment Sub-Institute of Hydrometeorology and Environment of South Vietnam (2010) Climate change in Mekong delta: Climate scenario's, sea level rise, other effects. Available: http://wptest.partnersvoorwater.nl/wp…/CLIMATE-CHANGE-final-draft.pdf. Accessed 2014 Jan 24.
- 44.International Federation of Red Cross and Red Crescent Societies (2013) Emergency appeal final report Viet Nam: Mekong delta floods. Available: http://reliefweb.int/sites/reliefweb.int/files/resources/MDRVN009fr.pdf. Accessed 2014 Jan 24.
- 45. Lee FN, Rush MC (1983) Rice sheath blight: A major rice disease. Plant Dis 67: 829–832. [Google Scholar]
- 46. Ogoshi A (1987) Ecology and pathogenicity of anastomosis and intraspecific groups of Rhizoctonia solani Kuhn. Annu Rev Phytopathol 25: 125–143. [Google Scholar]
- 47. Taheri P, Gnanamanickam S, Höfte M (2007) Characterization, genetic structure, and pathogenicity of Rhizoctonia spp. associated with rice sheath diseases in India. Phytopathology 97: 373–383. [DOI] [PubMed] [Google Scholar]
- 48. Hashiba T, Yamaguchi T, Mogi S (1972) Biological and ecological studies on the sclerotium of Pellicularia sasakii (Shirai) S. Ito. I. Floating on the water surface of sclerotium. Ann Phytopathol Soc Jpn 38: 414–425. [Google Scholar]
- 49. Freeman TE, Charudattan R, Cullen RE (1982) Rhizoctonia blight on waterhyacinth in the United States. Plant Dis 66: 861–862. [Google Scholar]
- 50. Zettler FW, Freeman TE (1972) Plant pathogens as biocontrols of aquatic weeds. Annu Rev Phytopathol 10: 455–470. [Google Scholar]
- 51.Yang G, Li C (2012) General description of Rhizoctonia species complex. In: Cumagun CJR, editor. Plant Pathology. Croatia: InTech. pp. 41–52. [Google Scholar]
- 52. Priyatmojo A, Escopalao VE, Tangonan NG, Pascual CB, Suga H, et al. (2001) Characterization of a new subgroup of Rhizoctonia solani anastomosis group 1 (AG-1-ID), causal agent of a necrotic leaf spot on coffee. Phytopathology 91: 1054–1061. [DOI] [PubMed] [Google Scholar]
- 53.People's committee of Dong Nai province (2011) Program to develop main crops, domestic animals and build up trade name of agricultural products, duration 2011–2015. Available: http://laws.dongnai.gov.vn/2011_to_2020/2011/201109/201109260003/lawdocument_view. Accessed 2014 Jan 24.
- 54.Commerce (2014) Can Tho province. Available: http://www.thuongmai.vn/thuong-mai/du-an-keu-goi-dau-tu/54004-can-tho.html. Accessed 2014 Jan 24.
- 55.Department of Culture, Sport and Tourism of Soc Trang (2014) Overview about Soc Trang province. Available: http://www.sovhttdl.soctrang.gov.vn/wps/portal/!ut/p/c4/04_SB8K8xLLM9MSSzPy8xBz9CP0os3gLR1dvZ09LYwOL4GAnA08TRwsfvxBDI4swc_2CbEdFAM7mSnc!/. Accessed 2014 Jan 24.
- 56.Department of Planning and Investment of Vinh Long (2014) Overview about Vinh Long province. Available: http://www.skhdt.vinhlong.gov.vn/Default.aspx?tabid=36. Accessed 2014 Jan 24.
- 57.Department of Propaganda and Training of Hau Giang (2014) Overview about Hau Giang province. Available: http://www.haugiang.gov.vn/Portal/Default.aspx?pageindex=2&pageid=2979&siteid=59#. Accessed 2014 Jan 24.
- 58.Integrated Water Resources Management (2014) Lam Dong province. Available: http://www.iwrm.vn/index.php?page=2&id=21. Accessed 24 January 2014.
- 59.Viet's life (2014) Ha Noi capital. Available: http://www.cuocsongviet.com.vn/index.asp?act=dp&id_tinh=27. Accessed 2014 Jan 24.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains the following files: Table S1. GPS co-ordinates of the wards in each province of Vietnam where Rhizoctonia-infected Brassica crops were sampled. Table S3. Aggressiveness of Rhizoctonia isolates towards white cabbage, Chinese cabbage, pak choi, mustard cabbage, Chinese flower cabbage, rice and water spinach in detached leaf bio-assays. Leaves were scored using a scale ranging from 0 (no disease symptoms) to 4 (lesions covered more than 75% of leaf surface or dead leaf). For rapid visual evaluation of the data, a coloring scale with green (0<DI≤1), yellow (1<DI≤2), orange (2<DI≤3) and red (3<DI≤4) was used. The test was done once with 12 leaves or leaf pieces per treatment. All data were statistically analyzed and within columns, disease severities followed by the same letter are not significantly different. Table S4. Aggressiveness of Rhizoctonia isolates towards roots and leaves of white cabbage and Chinese cabbage seedlings in in vitro bio-assays. Disease severity on roots or leaves was assessed on a scale ranging from 0 (no symptoms) to 4 (lesions covering more than 75% of root, hypocotyl or leaf surface or dead plant). For rapid visual evaluation of the data, a coloring scale with green (0<DI≤1), yellow (1<DI≤2), orange (2<DI≤3) and red (3<DI≤4) was used. Experiment was conducted once with 12 seedlings maintained in two square Petri plates for one treatment. All data were statistically analyzed and within columns, disease severities followed by the same letter are not significantly different. Table S5. Aggressiveness of Rhizoctonia isolates towards roots of white cabbage and Chinese cabbage seedlings in in planta experiment. Disease severity on roots was assessed on a scale ranging from 0 (no symptoms) to 4 (seedling dead). For rapid visual evaluation of the data, a coloring scale with green (0<DI≤1), yellow (1<DI≤2), orange (2<DI≤3) and red (3<DI≤4) was used. Experiment was performed once. Each treatment consisted of 12 seedlings cultivated in two plastic boxes. Data were statistically analyzed and within columns, disease severities followed by the same letter are not significantly different.
(DOC)
Table S2. Curated database of sequences containing representative rDNA-ITS sequences of all known uninucleate, binucleate and multinucleate Rhizoctonia AG and subgroups.
(XLSX)