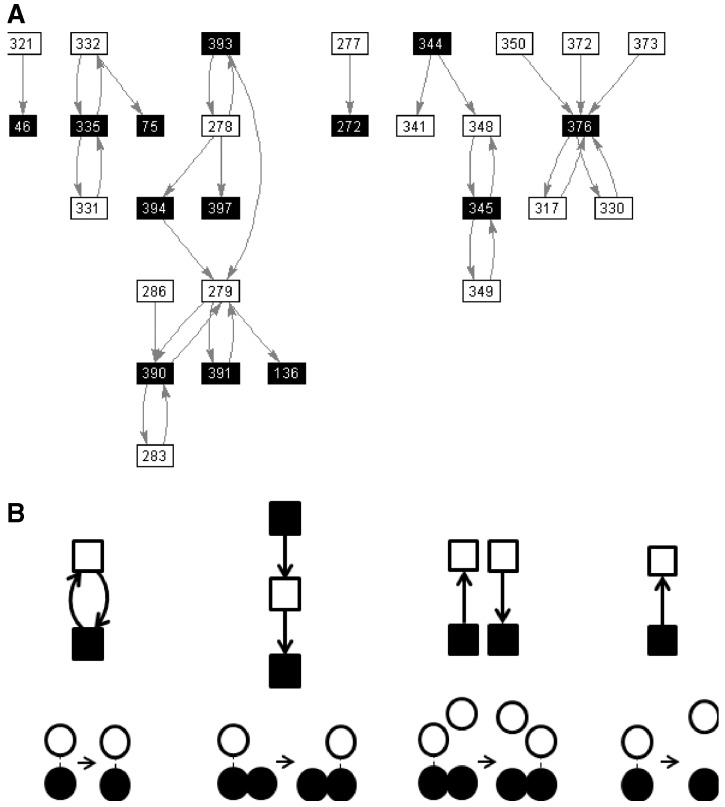
Fig. 2.
DCG and decomposition. (A) The DCG for the domain movement between conformation 1 (PDB accession code: 1CTS) and conformation 2 (PDB accession code: 1CSH) in citrate synthase. A filled square corresponds to a residue in domain A, and an open square corresponds to a residue in domain B with the residue number written in the square. An arrow from a residue in A to a residue in B indicates a contact between the residues in conformation 1. An arrow from a residue in B to a residue in A indicates a contact between the residues in conformation 2. (B) The elemental DCGs for ‘maintained’, ‘exchanged-partner’, ‘exchanged-pair’ and ‘new’ that represent the pairwise residue contact changes depicted underneath each graph. The DCG in (A) is decomposed into these elemental DCGs to give Nmaint = 10, Nexchpart = 2, Nexchpair = 2 and Nnew = 6. The prediction value for this domain movement is 0.55, which puts it in the Mixed class