Abstract
Summary: Recently, several high profile studies collected cell viability data from panels of cancer cell lines treated with many drugs applied at different concentrations. Such drug sensitivity data for cancer cell lines provide suggestive treatments for different types and subtypes of cancer. Visualization of these datasets can reveal patterns that may not be obvious by examining the data without such efforts. Here we introduce Drug/Cell-line Browser (DCB), an online interactive HTML5 data visualization tool for interacting with three of the recently published datasets of cancer cell lines/drug-viability studies. DCB uses clustering and canvas visualization of the drugs and the cell lines, as well as a bar graph that summarizes drug effectiveness for the tissue of origin or the cancer subtypes for single or multiple drugs. DCB can help in understanding drug response patterns and prioritizing drug/cancer cell line interactions by tissue of origin or cancer subtype.
Availability and implementation: DCB is an open source Web-based tool that is freely available at: http://www.maayanlab.net/LINCS/DCB
Contact: avi.maayan@mssm.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Patient-derived cancer cell lines have long been used as experimental models for tumor biology in cancer research. These cell lines preserve the genomic, transcriptomic and proteomic features of tumor cells and emulate tumor cells’ response to stimuli. A drug that substantially inhibits the growth of a cancer cell line would potentially suppress the progression of its tumor of origin. Cell viability assays measure the number of live cells in response to treatment by drugs applied in different concentrations. This type of assay is used to test the efficacy of a drug for inhibiting the growth or killing cancer cell lines. A cancer cell line is considered sensitive to a drug if it has low viability when the drug is applied at low concentration. Screening many cancer cell lines with many drugs applied in many concentrations can be used to match tumors with the most likely effective drugs. These drugs could potentially be used to eventually treat individual patients (Duan et al., 2013).
Aberrant tumor growth through rapid cell proliferation could be attributed to malfunction of many pathways (Hanahan and Weinberg, 2011). The regulatory networks that regulate cell proliferation and tumor growth in cancer are wired differently from cell line to cell line, and such intricacies imply that different drugs, targeting different pathways, would have different effects on different cell lines. Yet, until recently, efforts to screen drugs against cell lines in high throughput were impeded by the amount of cell viability assays that could be performed in a reasonable time. In recent years, however, the technology for high-throughput cell viability assays has been developed, and several datasets consisting of tens of thousands cell viability experiments have been published (Barretina et al., 2012; Yang et al., 2013). These datasets include cell viability assays of dozens of drugs applied to hundreds of cell lines. Analysis and visualization of such data can potentially uncover informative patterns that would have direct therapeutic potential. Here, we present Drug/Cell-line Browser (DCB), a HTML5 Web-based application that uses canvas clustering (Tan et al., 2013) to visualize three recent cell viability assay datasets: from the Cancer Cell Line Encyclopedia (CCLE) project (Barretina et al., 2012), from the Genomics of Drug Sensitivity (GDS) in cancer project (Yang et al., 2013) and from an anticancer compound study for breast cancer (Heiser et al., 2012).
2 METHODS
Cancer cell line canvas: Each tile on the canvas represents a cancer cell line. The tiles are colored by tissue of origin or by cluster fitness using basal gene expression vector, mutation status or overall drug response. To construct the canvases, adjacency matrices were created where cell lines are connected if they share the tissue of origin or cancer subtype, display similar basal expression profile, mutation status profile or respond similarly to the vector of drugs. These matrices are fed into the Network2Canvas Python script (Tan et al., 2013) to generate the coordinates of the tiles in each cancer cell line canvas. Such information is stored in a JavaScript Object Notation (JSON) format. DCB visualizes each cancer cell line canvas as an Scalable Vector Graphics (SVG) rect element using the D3 JavaScript library (Bostock et al., 2011). The edges of the canvas fold on themselves forming a torus. Drug canvas: Each tile on the drug canvas represents a drug. Each tile is colored by its fitness based on similarity with its eight surrounding neighboring drugs. Fitness is defined as the similarity of a drug’s sensitivity profile vector, chemical structure similarity, drug targets and changes in expression in cell lines after treatment with the drug. Details for computing cell line/cell line similarity are provided in the Supplementary File.
The drug similarity adjacency matrices were created from distance tables that measure similarity between drugs.
Then, similarly to the way the cancer cell line canvas was processed, the matrix is fed into the Network2Canvas Python script, and the DCB application visualizes the drugs JSON canvases with SVG elements using D3. The drug canvas is also continuous where the edges fold on themselves forming a torus. To fill all the tiles on the canvas, some drugs appear twice. Tissue panel: The tissue panel shows the tissue of origin in lexicographical order. Cancer cell lines are grouped by tissue of origin in a JavaScript object so that tissues of origin and cancer cell lines map to each other. Tissues are then sorted in lexicographical order and visualized as SVG rect elements on the panel. An event listener function listens to click events on the drug canvas to adjust each tissue rect. Clicking on a drug tile, the function computes the significance of a tissue’s sensitivity to that drug using Fisher’s exact test based on the number of highly sensitive cancer cell lines for that tissue. The number of highly sensitive cell lines per drug can be filtered using a slider.
3 RESULTS
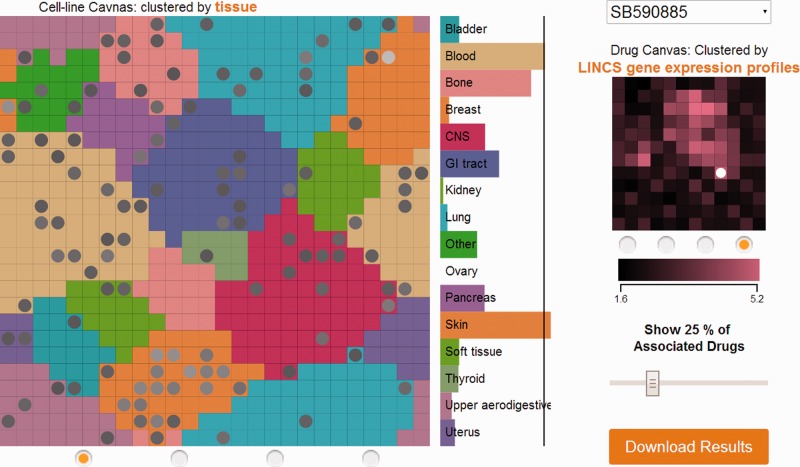
DCB comprises two canvases, one in the middle to visualize cancer cell lines and the other on the right to visualize drugs (Fig. 1). On the cancer cell line canvas, each tile represents a cancer cell line, and the tiles are colored by tissue of origin or by level of clustering where brighter colors mean high neighborhood similarity. Cell lines are clustered by tissue type, basal gene expression similarity, mutation status and response to drugs vector. On the drug canvas, each title represents a drug and the tiles are colored by how similar they are based on their sensitivity profile, structure, shared targets and gene expression signatures induced by the drugs. The tissue bar graph panel on the right lists tissues in lexicographical order. The colors of tissues match to the colors of cancer cell line tiles in the tissue view. Mouse hovering on a cancer cell line tile displays its name and highlights its tissue of origin on the tissue panel. Mouse hovering on a drug tile shows its name. Scrolling with the mouse wheel on the cancer cell line canvas enables zooming in and out, and dragging the canvases allows panning. Tiles on the cancer cell line canvas can be labeled by the Label Cell Lines selection on the left. Users may need to zoom in to read the cell line name labels. Users can focus on a specific tissue using the Filter by Cell Type selection. If a tissue is selected, the cancer cell line canvas will show only cancer cell lines of the selected tissue. Clicking on a cancer cell line on the canvas shows the top 25% most-associated (sensitive) drugs on the drug canvas. A slider can be used to adjust the percentage of drugs that are shown. Similarly, clicking on a drug on the drug canvas shows the top 25% most-associated (sensitive) cancer cell lines for the drug on the cancer cell line canvas. A slider can be used to adjust the percentage of cell lines that are shown. Additionally, the tissue panel shows the significance of sensitivity of each tissue to that drug. The black line on the panel delineates significance threshold of P < 0.05. The significance is calculated based on the number of sensitive cancer cell lines for a particular tissue for a drug using the Fisher’s exact test. Besides clicking on the drug canvas, drugs can be chosen from a drop-down menu. DCB visualizes three datasets: GDS, CCLE and Heiser et al. In the Heiser dataset, all cell lines are derived from breast cancer and are colored by subtypes instead of tissue of origin. Consequently, the tissue panel shows subtypes instead of tissues. Overall, DCB can be used to identify opportunities for cancer drug discovery and drug repurposing.
Fig. 1.
Visualization of cancer cell lines from GDS and their sensitivity to the drug SB590885, a Raf inhibitor. Only top 25% of the most-associated (sensitive) cell lines are highlighted on the cell line canvas with circles. The tissue panel shows that the tissue that is most significantly sensitive to SB590885 is skin. This is consistent with the known role of B-Raf is many melanomas
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Drs Pillai and Larkin from NHGRI and Dr Shamu from HMS for useful suggestions.
Funding: This work was supported in part by grants from the NIH: R01GM098316-01A1, R01DK088541-01A1, U54HG006097-02S1 and U54CA189201 to A.M.
Conflict of interest: none declared.
REFERENCES
- Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock M, et al. D-3: data-driven documents. IEEE T. Vis. Comput. Gr. 2011;17:2301–2309. doi: 10.1109/TVCG.2011.185. [DOI] [PubMed] [Google Scholar]
- Duan Q, et al. Metasignatures identify two major subtypes of breast cancer. CPT Pharmacometrics Syst. Pharmacol. 2013;2:e35. doi: 10.1038/psp.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Heiser LM, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl Acad. Sci. USA. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CM, et al. Network2Canvas: network visualization on a canvas with enrichment analysis. Bioinformatics. 2013;29:1872–1878. doi: 10.1093/bioinformatics/btt319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.