ABSTRACT
The emergence in Japan of field isolates of type 1 porcine reproductive and respiratory syndrome virus (PRRSV) suggests problems with control. We therefore developed a one-step real-time reverse transcription polymerase chain reaction (qRT-PCR) with improved sensitivity that detects as little as 1 × 10−2 TCID50/ml of viral RNA. We tested serum samples collected in January and September 2008, October 2009 and January 2011 from a farm with an outbreak and found infected pigs between January and September 2008, but not in January 2011. Further, between 2008 and 2011, we did not detect infection in pigs at 8 nearby farms or in 2,052 serum samples collected from pigs from 74 farms in 12 prefectures. This assay should help prevent future outbreaks.
Keywords: epidemiological survey, PRRS, real-time RT-PCR, type 1 PRRSV
Porcine reproductive and respiratory syndrome (PRRS) is characterized by reproductive failure in sows and respiratory disease in piglets [12]. The PRRS virus (PRRSV) is classified into type 1 (European) and type 2 (North American) genotypes according to genetic, antigenic and pathogenic differences [1, 2, 5, 8, 11]. The first isolate of PRRSV in Japan in 1994 was type 2 [9], which rapidly spread through the country during the following two decades and has since markedly diverged [4]. We first isolated type 1 PRRSV from diseased pigs in 2009 [3]. Here, we describe the development of a SYBR® Green one-step real-time RT-PCR assay to detect type 1 PRRSV RNA. We applied this method to detect the transmission of type 1 PRRSV in pig farms.
Viral RNA extracted using a QIAamp Viral RNA Mini Kit (QIAGEN, Tokyo, Japan) was used as a template for one-step real-time RT-PCR (qRT-PCR) with TaKaRa One Step SYBR® PrimeScript® RT-PCR Kit II (Perfect Real Time) (TaKaRa, Otsu, Japan). We modified the sequences of published primers [6] to amplify a broad range of type 1 PRRSV strains, including the European prototype strain Lelystad (GenBank accession number: M96262), field isolates from the United States and Japanese type 1 PRRSV isolates. Briefly, a primer pair (forward, 5′-GCACCACCTCACCCAAAC-3′ and reverse, 5′-CAGTTCCTGCGCCTTGAT-3′; the modified nucleotide is underlined) was used to detect part of the ORF7 gene (77 nucleotides) without using a dual-labeled probe, which was designed for TaqMan qRT-PCR [6]. Fluorescence data were analyzed using the PE 7500 Sequence Detection System Software (Version 1.4; Life Technologies Inc., Carlsbad, CA, U.S.A.).
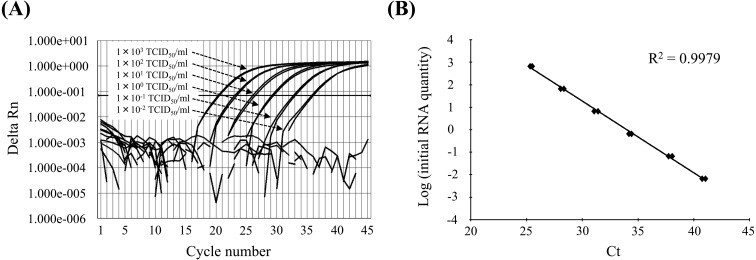
To determine the detection limit of the qRT-PCR assay, the equivalent of 1 × 103 tissue culture infectious doses (TCID)50/ml of Japanese type 1 PRRSV RNA was extracted from the culture supernatant, and serial 10-fold dilutions were analyzed. The results of the qRT-PCR assay were compared with those of a previously established nested PCR assay [7]. Positive signals were observed with 1 × 103 to 1 × 10−2 TCID50/ml of the diluted type 1 PRRSV RNA (Fig. 1A). In contrast, the detection limits of conventional RT-PCR and subsequent nested-PCR methods were 1 × 102 TCID50/ml and 1 × 10−1 TCID50/ml per sample, respectively. Further, a linear standard curve was generated in each qRT-PCR run with a series of serial dilutions (R2=0.9979). The threshold cycle value (Ct) indicates the amount of target gene that produces a signal that exceeds a preset threshold value, which is obtained from a calibration curve (Fig. 1B). The Ct value is valid only between the minimum and maximum values obtained using the standard RNAs. The amplification and dissociation curves for Lelystad (data not shown) and Japanese type 1 PRRSV RNA were indistinguishable. To test the specificity of the method, RNAs were prepared from 1 × 103 TCID50/ml of other type 2 PRRSV RNAs (EDRD1, M96262; RespPRRS MLV vaccine strain, AF095499; Jpn5-37, AB546125). Amplification of these RNAs was not detected (data not shown). Moreover, 100 sera collected from healthy pigs were also tested; however, viral sequences were undetectable.
Fig. 1.
Quantitative detection of PRRSV using a SYBR® Green qPCR assay. (A) Amplification of serially diluted PRRSV RNA (duplicates) containing 1 × 103 to 1 × 10−4 TCID50/ml. Amplification plots are shown from 1 × 103 to 1 × 10−2 TCID50/ml. (B) Standard curve for quantification of the partial ORF7 gene of PRRSV. The standard curve plots Ct values against the log of 10-fold dilutions of viral RNA equivalent to 1 × 103 to 1 × 10−4 TCID50/ml.
We next evaluated the prevalence of type 1 PRRSV over time in a farm with an outbreak. Animals housed in the farm that tested positive for type 1 PRRS were analyzed using conventional nested RT-PCR in 2009 and were subsequently subjected to annual inspections. We determined the prevalence of type 1 PRRSV by testing serum samples taken during January and September 2008, October 2009 and January 2011 (Table 1). Viral RNA was undetectable in all 35 samples collected in January 2008 using either the qRT-PCR or nested PCR assays. After 8 months, 19/35 (54.3%) and 13/35 (37%) samples were positive using the qRT-PCR and nested PCR assays, respectively. In October 2009, 5 (14.2%) and 4 (11%) positive samples were detected using the respective assays, and all samples from January 2011 were negative using both methods. These results indicate that type 1 PRRSV infected the pigs housed on the farm during January to September 2008 and that the virus became gradually undetectable after spreading through the farm between 2009 and 2011, suggesting that the virus had been transmitted to most of the pigs and that the pigs had then developed immunity that inhibited further virus replication. This inference is also supported by the results of serological testing using an ELISA (Table 2). A PRRSV-specific antibody was evaluated using a commercially available ELISA (HerdCheck PRRS ELISA, IDEXX Laboratories). The highest mortality rate was observed in September 2008 (Table 3), and the rate may have increased with further spread of the virus throughout the farm.
Table 1. Comparison of the real-time PCR assay with the nested PCR assay for detection of type 1 PRRSV from serum samples collected from pigs living at the outbreak farm.
| Age (Days old) |
January 2008 | September 2008 | October 2009 | January 2011 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Real-time PCR (%) |
Nested PCR ((%) |
Real-time PCR ((%) |
Nested PCR ((%) |
Real-time PCR ((%) |
Nested PCR ((%) |
Real-time PCR ((%) |
Nested PCR ((%) |
|||||||||
| + | – | + | – | + | – | + | – | + | – | + | – | + | – | + | – | |
| < 60 | 0 | 10 (100) | 0 | 10 (100) | 9 (90) | 1 (10) | 6 (60) | 4 (40) | 0 | 10 (100) | 0 | 10 (100) | 0 | 5 (100) | 0 | 5 (100) |
| 60 < 120 | 0 | 10 (100) | 0 | 10 (100) | 8 (80) | 2 (20) | 6 (60) | 4 (40) | 5 (50) | 5 (50) | 4 (40) | 6 (60) | 0 | 10 (100) | 0 | 10 (100) |
| 120 < | 0 | 10 (100) | 0 | 10 (100) | 2 (20) | 8 (80) | 1 (10) | 9 (90) | 0 | 10 (100) | 0 | 10 (100) | 0 | 5 (100) | 0 | 5 (100) |
| Sow | 0 | 5 (100) | 0 | 5 (100) | 0 | 5 (100) | 0 | 5 (100) | 0 | 10 (100) | 0 | 5 (100) | 0 | 5 (100) | 0 | 5 (100) |
| Total | 0 (0) | 35 (100) | 0 (0) | 35 (100) | 19 (54.3) | 16 (45.7) | 13 (37) | 22 (63) | 5 (14.2) | 30 (85.7) | 4 (11) | 31 (89) | 0 (0) | 25 (100) | 0 (0) | 25 (100) |
Table 2. Distribution of the prevalence of type 1 PRRSV-specific antibodies from serum samples collected from pigs living at the outbreak farm.
| Age (Days old) |
January 2008 | September 2008 | October 2009 | January 2011 | ||||
|---|---|---|---|---|---|---|---|---|
| + | – | + | – | + | – | + | – | |
| < 60 | 4 (40%) | 6 (60%) | 10 (100%) | 0 (0%) | 7 (70%) | 3 (30%) | 3 (60%) | 2 (40%) |
| 60 < 120 | 4 (40%) | 6 (60%) | 8 (80%) | 2 (20%) | 3 (30%) | 7 (70%) | 4 (40%) | 6 (60%) |
| 120 < | 6 (60%) | 4 (40%) | 10 (100%) | 0 (0%) | 10 (100%) | 0 (0%) | 5 (100%) | 0 (0%) |
| Sow | 3 (60%) | 2 (40%) | 5 (100%) | 0 (0%) | 5 (100%) | 0 (0%) | 4 (100%) | 1 (0%) |
| Total | 17 (48.6%) | 18 (51.4%) | 33 (94.3%) | 2 (5.7%) | 25 (71.4%) | 10 (28.6%) | 16 (64%) | 9 (36%) |
Table 3. Comparison of the mortality rate (%) in each stage on farm A.
| Age | January 2008 |
September 2008 |
October 2009 |
January 2011 |
|---|---|---|---|---|
| < 60 | 20 | 20 | 3 | 20 |
| 60 < 120 | 10 | 30 | 30 | 3 |
| 120 < | 3 | 30 | 20 | 3 |
We tested 70 animals housed at 8 other pig farms in the same area using the qRT-PCR and nested PCR assays. However, no positive animals were detected. We further investigated a total of 2,052 serum samples from 74 pig farms in 12 prefectures, which were collected between 2008 and 2011 by the Livestock Hygiene Service Centers of each prefecture. The virus was undetectable in all of these samples, leading us to conclude that type 1 PRRSV was not widely spread across the country.
Here, we describe the development of a SYBR® Green one-step qRT-PCR assay for detecting type 1 PRRSV and show that the assay is highly sensitive for detecting the type 1 PRRSV ORF7 gene. Amplification of PRRSV RNA is a powerful tool for detecting PRRS during the early phase of infection and in carrier animals [10]. The specificity of the primers without use of a dual-labeled probe targeting the partial ORF7 gene of type 1 PRRSV was proven by successful amplification of the PRRSV RNAs in our laboratory’s collection and by the positive results compared with using the conventional nested PCR method. Although the qRT-PCR and nested PCR assays are useful for analysis of clinical specimens and may achieve high sensitivity, the major advantages of the qRT-PCR assay are its wide range of detection (starting from 1 × 10−2 TCID50/ml of viral RNA) and its ability to quantify the infection load of clinical specimens. Further, the adaptability of this technique to a high-throughput 96-well format significantly reduces the overall time and costs per sample in a clinical laboratory that processes a large number of samples. We therefore believe that the qRT-PCR assay developed here can be implemented as a diagnostic tool to detect type 1 PRRSV in field samples. This conclusion is supported by our ability to follow, for the first time to our knowledge, type 1 PRRSV transmission over time in pigs raised on a farm in Japan. We intend to apply this assay to future viral epidemiological studies to compare the effects of different drug regimens for prevention and treatment, to routine monitoring of herds and to diagnosis of PRRS infections.
Acknowledgments
We thank the following members of the Working Group for the Diagnosis of PMWS for conducting microbiological and histopathological examinations of pigs and investigations of mortality on farms: Okamoto K., Sugawara K., Takamori H., Oikawa T., Onuki A., Sawada N., Shimizu H., Ashizawa T., Sato T., Terasaki T., Wakuda T., Totsuka T., Sakai Y., Nishi D., Nakashima D. and Nitta Y. We thank Nachiko Hattori for technical support and gratefully acknowledge the financial support provided by the National Agriculture and Food Research Organization and the Ministry of Agriculture, Forestry and Fisheries of Japan.
REFERENCES
- 1.Forsberg R., Storgaard T., Nielsen H. S., Oleksiewicz M. B., Cordioli P., Sala G., Hein J., Bøtner A.2002. The genetic diversity of European type PRRSV is similar to that of the North American type but is geographically skewed within Europe. Virology 299: 38–47. doi: 10.1006/viro.2002.1450 [DOI] [PubMed] [Google Scholar]
- 2.Goldberg T. L., Hahn E. C., Weigel R. M., Scherba G.2000. Genetic, geographical and temporal variation of porcine reproductive and respiratory syndrome virus in Illinois. J. Gen. Virol. 81: 171–179. [DOI] [PubMed] [Google Scholar]
- 3.Iseki H., Takagi M., Kawashima K., Shibahara T., Kuroda Y., Tsunemitsu H.2012. Type I porcine reproductive and respiratory syndrome virus emerged in Japan. The 22nd International Pig Veterinary Society Congress. Abstract II: p978.
- 4.Iseki H., Takagi M., Miyazaki A., Katsuda K., Mikami O., Tsunemitsu H.2011. Genetic analysis of ORF5 in porcine reproductive and respiratory syndrome virus in Japan. Microbiol. Immunol. 55: 211–216. doi: 10.1111/j.1348-0421.2010.00303.x [DOI] [PubMed] [Google Scholar]
- 5.Kapur V., Elam M. R., Pawlovich T. M., Murtaugh M. P.1996. Genetic variation in porcine reproductive and respiratory syndrome virus isolates in the midwestern United States. J. Gen. Virol. 77: 1271–1276. doi: 10.1099/0022-1317-77-6-1271 [DOI] [PubMed] [Google Scholar]
- 6.Kleiboeker S. B., Schommer S. K., Lee S. M., Watkins S., Chittick W., Polson D.2005. Simultaneous detection of North American and European porcine reproductive and respiratory syndrome virus using real-time quantitative reverse transcriptase-PCR. J. Vet. Diagn. Invest. 17: 165–170. doi: 10.1177/104063870501700211 [DOI] [PubMed] [Google Scholar]
- 7.Kono Y., Kanno T., Shimizu M., Yamada S., Ohashi S., Nakamine M., Shirai J.1996. Nested PCR for detection and typing of porcine reproductive and respiratory syndrome (PRRS) virus in pigs. J. Vet. Med. Sci. 58: 941–946. doi: 10.1292/jvms.58.10_941 [DOI] [PubMed] [Google Scholar]
- 8.Mateu E., Martín M., Vidal D.2003. Genetic diversity and phylogenetic analysis of glycoprotein 5 of European-type porcine reproductive and respiratory virus strains in Spain. J. Gen. Virol. 84: 529–534. doi: 10.1099/vir.0.18478-0 [DOI] [PubMed] [Google Scholar]
- 9.Murakami Y., Kato A., Tsuda T., Morozumi T., Miura Y., Sugimura T.1994. Isolation and serological characterization of porcine reproductive and respiratory syndrome (PRRS) viruses from pigs with reproductive and respiratory disorders in Japan. J. Vet. Med. Sci. 56: 891–894. doi: 10.1292/jvms.56.891 [DOI] [PubMed] [Google Scholar]
- 10.Reiner G., Fresen C., Bronnert S., Willems H.2009. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) infection in wild boars. Vet. Microbiol. 136: 250–258. doi: 10.1016/j.vetmic.2008.11.023 [DOI] [PubMed] [Google Scholar]
- 11.Ropp S. L., Wees C. E., Fang Y., Nelson E. A., Rossow K. D., Bien M., Arndt B., Preszler S., Steen P., Christopher-Hennings J., Collins J. E., Benfield D. A., Faaberg K. S.2004. Characterization of emerging European-like porcine reproductive and respiratory syndrome virus isolates in the United States. J. Virol. 78: 3684–3703. doi: 10.1128/JVI.78.7.3684-3703.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossow K. D.1998. Porcine reproductive and respiratory syndrome. Vet. Pathol. 35: 1–20. doi: 10.1177/030098589803500101 [DOI] [PubMed] [Google Scholar]