Abstract
The Golgi apparatus lies at the heart of the secretory pathway where it receives, modifies, and sorts protein cargo to the proper intracellular or extracellular location. Although this secretory function is highly conserved throughout the eukaryotic kingdom, the structure of the Golgi complex is arranged very differently among species. In particular, Golgi membranes in vertebrate cells are integrated into a single compact entity termed the Golgi ribbon that is normally localized in the perinuclear area and in close vicinity to the centrosomes. This organization poses a challenge for cell division when the single Golgi ribbon needs to be partitioned into the two daughter cells. To ensure faithful inheritance in the progeny, the Golgi ribbon is divided in three consecutive steps in mitosis, namely disassembly, partitioning, and reassembly. However, the structure of the Golgi ribbon is only present in higher animals and Golgi disassembly during mitosis is not ubiquitous in all organisms. Therefore, there must be unique reasons to build up the Golgi in this particular conformation and to preserve it over generations. In this review, we first highlight the diversity of the Golgi architecture in different organisms and revisit the concept of the Golgi ribbon. Following on, we discuss why the ribbon is needed and how it forms in vertebrate cells. Lastly, we conclude with likely purposes of mitotic ribbon disassembly and further propose mechanisms by which it regulates mitosis.
Keywords: Golgi, stacks, ribbon, perinuclear, secretion, glycosylation, polarity, mitosis, spindle, disassembly, partitioning, organelle inheritance, golgin, GRASP
What is the Golgi ribbon?
The basic function of the Golgi apparatus is to post-translationally modify secretory and membrane proteins received from the endoplasmic reticulum (ER) and to sort them to the appropriate cellular destinations. Although this function is highly conserved across the eukaryotic kingdom, the morphological organization of the Golgi apparatus varies among different cell types and species (Table 1). In the vast majority of eukaryotes, the Golgi complex is composed of disk-shaped, flattened cisternae that are piled on top of each other and aligned in parallel into compact stacks. In protozoa, fungi (such as Schizosaccharomyces pombe and Pichia pastoris), plants and invertebrates, stacks are dispersed throughout the cytoplasm and frequently reside near the ER exit sites. However, in certain unicellular eukaryotes including some parasitic protists and the yeast Saccharomyces cerevisiae, Golgi membranes appear as single, isolated cisternae randomly distributed throughout the cytoplasm (1–3). Phylogenetic analysis of Golgi morphology indicates that ancient eukaryotes likely possessed a stacked Golgi and that the emergence of stack-lacking organisms is probably due to multiple independent incidents of unstacking in the evolutionary history (1).
Table 1.
Structure and localization of the Golgi apparatus in eukaryotes.
| Organism | Interphase | Mitosis | |||
|---|---|---|---|---|---|
| Structure | Localization | Cisternae and stacks | Localization | ||
| Protozoa | Toxoplasma gondii | Single stack (71, 104) | Apical perinuclear region (104) | Intact (71) | |
| Trypanosoma brucei | Single stack (105, 106) | Near the flagellar pocket (105) | Intact (106) | ||
| Trichomonas vaginalis | Single stack (70) | Intact (70) | |||
| Tritrichomonas foetus | Single stack (70) | Intact (70) | |||
| Plasmodium falciparum | Single cisterna (107) | ||||
| Dictyostelium discoideum | Stacks (108) | Perinuclear (109, 110) | Dispersed (110) | ||
| Fungi | Saccharomyces cerevisiae | Cisternae (3); tubular networks (2) | Dispersed (3) | Intact (3) | Dispersed in the cytoplasm and clustered at the site of bud emergence (3) |
| Schizosaccharomyces pombe | Stacks (111) | Dispersed (111) | Intact (112) | Dispersed (112) | |
| Pichia pastoris | Stacks (113) | Dispersed (72, 113) | |||
| Plants | Nicotiana tabacum | Stacks (114, 115) | Dispersed (33, 115) | Intact (115) | Dispersed in the cytoplasm and accumulated near spindle poles (33) |
| Arabidopsis thaliana | Stacks (116) | Dispersed (116) | Intact (116) | Dispersed (116) | |
| Invertebrates | Caenorhabditis elegans | Stacks (117) | Dispersed (118) | Dispersed (119) | |
| Drosophila melanogaster | Stacks (22) | Dispersed (22, 120) | Disassembled; intact in embryos (120) | Dispersed (120) | |
| Aedes albopictus (mosquito) | Stacks (121) | Dispersed (121) | |||
| Lytechinus variegates (sea urchin) | Stacks (122) | Dispersed in early embryo; perinuclear after the 9th division (122) | Dispersed in the cytoplasm and accumulated at the spindle poles (122) | ||
| Vertebrates | Danio rerio (fish) | Stacks (123) | Perinuclear ribbon (124) | ||
| Xenopus laevis (frog) | Stacks (125) | Perinuclear ribbon (125, 126) | Disassembled in oocytes (127) | ||
| Potorous tridactylus (PtK) | Stacks (42) | Perinuclear ribbon (42, 81) | Disassembled (128) | Dispersed in the cytoplasm and accumulated around spindle poles (42, 81, 128) | |
| Rattus norvegicus (NRK) | Stacks (4) | Perinuclear ribbon (4) | Disassembled (128) | Dispersed in the cytoplasm and accumulated around spindle poles (15) | |
| Homo sapiens (HeLa) | Stacks (129) | Perinuclear ribbon (129) | Disassembled (129) | Dispersed in the cytoplasm and accumulated around spindle poles (81) | |
In contrast to simple eukaryotes that have lost structural characteristics, vertebrates have instead gained a more complex level of Golgi organization. A typical Golgi apparatus in vertebrate cells, ranging from fish to mammals, exhibits a twisted ribbon-like network, which is situated in the juxtanuclear and most often pericentriolar region of the cell. This higher-ordered structure, the Golgi ribbon, is named after its appearance under the light microscope. At the ultrastructural level, the Golgi ribbon is composed of compact stacks that are connected by noncompact regions of tubular membranes. These tubules join and laterally link equivalent cisternae in adjacent stacks, thereby generating an interconnected reticulum (4).
The term Golgi ribbon used to describe the structure in most studies consists of two distinct but often intimately coupled features: structural continuity and perinuclear localization. Experiments to determine the molecular requirements for ribbon formation rely primarily on one scenario, in which loss or inhibition of a particular factor converts the Golgi ribbon into mini-stacks that are scattered throughout the cytoplasm. In these cases, loss of structural continuity often coincides with disappearance of perinuclear positioning. Therefore, although various advanced techniques such as electron tomography (4) and fluorescence recovery after photobleaching (5, 6) have been developed to examine structural continuity, the simplest criterion for a Golgi ribbon remains with the initial definition at the level of the light microscope as a higher-ordered, continuous Golgi structure.
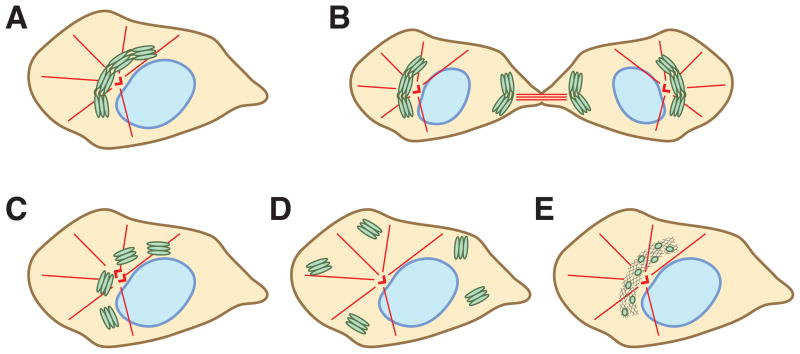
However, it is not always obvious whether the Golgi is assembled into a ribbon. In principle, the Golgi in vertebrate cells can be organized in at least five different configurations (Figure 1). First, as observed in the majority of cell types, the Golgi complex appears in the perinuclear region as a twisted ribbon-like structure made of a collection of interconnected stacks. Second, the continuous Golgi ribbon can be positioned in a cellular location different from the perinuclear area. During cytokinesis, for instance, a small Golgi ribbon is localized next to the midbody and flanks the intracellular bridge between the two daughter cells (7, 8). As another example, a non-perinuclear ribbon can be found in terminally differentiated post-mitotic cells such as neurons where a set of specialized Golgi structures called Golgi outposts often localize at branch points of dendrites (9). A third possibility is unlinked Golgi stacks that cluster around the major microtubule-organizing center (MTOC) in the perinuclear region, a conformation resembling the perinuclear recycling endosomes (10). Under physiological conditions, Golgi ribbon unlinking is observed in late G2 phase when Golgi stacks lose mutual connectivity but remain concentrated in the perinuclear region (11). This is in contrast to unlinked Golgi stacks that are scattered all over the cytoplasm. Dispersion of Golgi stacks can be induced by interfering with the microtubule cytoskeleton, Golgi structural proteins or membrane dynamics (12). Lastly, Golgi structural proteins including the golgin and GRASP families can form a ribbon, regardless of the presence of Golgi enzymes or the organization of Golgi membranes (13). These proteins, collectively referred to as the Golgi matrix (14), form a dynamic scaffold which enzymes and membranes normally populate (13).
Figure 1. Golgi organization in vertebrate cells.
A) A typical interphase Golgi ribbon is composed of interconnected stacks and is localized in the perinuclear region in close vicinity to the centrosome. B) The Golgi ribbon can be positioned in a cellular location different from the perinuclear area. During cytokinesis, a small Golgi ribbon is situated next to the midbody and flanks the intracellular bridge between the two daughter cells. C) In late G2 phase, Golgi stacks lose mutual connectivity but remain clustered in the perinuclear region around the centrosomes. D) Unlinked Golgi stacks are scattered throughout the cytoplasm. E) Golgi matrix proteins, in particular the golgin and GRASP families, are sufficient to assemble and maintain a higher-ordered ribbon structure in the perinuclear region, regardless of the presence of Golgi enzymes or the organization of Golgi membranes.
The matrix-based Golgi ribbon has been generated in two ways (13, 15). In cells pretreated with Brefeldin A (BFA) to relocate Golgi enzymes into the ER, inhibiting ER-to-Golgi transport by microinjecting dominant negative Sar1 protein (dnSar1) followed by BFA washout regenerates a Golgi ribbon in the perinuclear region. Similarly, a perinuclear ribbon has been reconstituted in cells directly microinjected with dnSar1. In both cases, a continuous ribbon structure positive for Golgi matrix markers but negative for Golgi-resident enzymes can be identified by light microscopy. Moreover, ultrastructural analysis revealed that no cisternae but vesicles and vacuoles are present near the nucleus in these cells (13). In this regard, the structural continuity of the ribbon is conferred by the Golgi matrix proteins rather than the membranes. In sum, the Golgi matrix alone is sufficient to organize a higher-ordered ribbon structure. While membrane continuity is not absolutely required for a ribbon, it is often considered characteristic under most physiologically relevant conditions.
Although the perinuclear ribbon seems to be the default structure in most vertebrate cells, morphological variations do occur during differentiation. For instance, during skeletal myogenesis when undifferentiated myoblasts fuse to form myotubes, the compact Golgi ribbon is progressively fragmented and scattered between multiple nuclei (16). Similar Golgi dispersion is observed in differentiating megakaryocytes where the perinuclear ribbon starts to disassemble when proplatelet extension forms (17).
Why does the Golgi ribbon form in interphase?
It is quite puzzling why the Golgi apparatus in vertebrate cells is arranged in such a sophisticated manner. Evolutionarily, stacks are the more ancient structure (1), indicating that natural selection favors a ribbon in higher animals. Therefore, the organisms should benefit at least in part from maintaining this particular organization. Furthermore, it is mechanistically more complicated and energetically unfavorable to divide a single perinuclear ribbon via a disassembly-reassembly process than to simply apportion dispersed stacks during cell division. So why is the vertebrate Golgi organized as a ribbon? Does the ribbon provide advantages compared to unlinked stacks? In other words, does ribbon dispersion compromise Golgi function, such as secretion, glycosylation or cell polarization?
Membrane trafficking, the most fundamental function of the Golgi complex, does not require a ribbon structure. Disruption of ribbon continuity by either down-regulating Golgi-localized proteins (5, 18) or depolymerizing microtubules (19, 20) blocks neither intra-Golgi transport nor overall secretion to the plasma membrane. In fact, trafficking through a perinuclear Golgi ribbon to the cell surface occurs with similar kinetics to that through dispersed individual stacks (5, 19). In addition, secretion remains functional through disorganized stacks (21, 22) or even through individual, unstacked cisternae (3).
A continuous ribbon allows lateral diffusion of its glycosylation enzymes between constituent stacks (5, 6, 23, 24). However, due to opposing results it remains unclear whether ribbon disruption affects glycosylation efficiency and/or oligosaccharide pattern of cargo (5, 25). This may be due to differences in the experimental setups and cargo molecules of interest, as well as the current limited detection techniques for sugar modification. A systematic analysis of the relationship between ribbon integrity and glycosylation function is perhaps needed.
In contrast to the above functions, the presence of a Golgi ribbon is generally recognized as a requirement for cell polarization. Proper orientation of the Golgi ribbon is essential for specialized cells that rely on polarized secretion for their advanced functions, such as directional cell migration of fibroblasts (26, 27), determination of apical-basal axis of the epithelium (28), polarized secretion of lytic granules in cytotoxic T-cells (29), and regulated dendritic growth in neurons (30). An intact Golgi ribbon is necessary to provide polarity cues that are asymmetrically delivered to specific domains on the cell surface. Impairing ribbon integrity by either altering the microtubule cytoskeleton or modulating Golgi-localized proteins results in polarization defects and failure in directed cell migration (26, 31, 32). In addition, overexpression of the Golgi structural protein GRASP65, which leads to dispersion of the somatic Golgi ribbon, has been shown to suppress asymmetric dendritic growth in hippocampal neurons (30).
Presumably driven by distinct organizations of the Golgi membranes, eukaryotic cells utilize very diverse mechanisms to achieve polarized secretion. For example, in the budding yeast Saccharomyces cerevisiae, single Golgi cisternae frequently cluster to the site of bud emergence as well as at the cell septum (3). This suggests that polarized positioning of the organelle itself facilitates asymmetric deposition of membranes. Likewise, temporary relocation of Golgi membranes is also observed during cell division in higher plants where the Golgi stacks are brought in close proximity to the phragmoplast, an equatorial array of microtubules at the cell center, to facilitate cytokinesis (33). An alternative mode is to target cargo mRNA to the ER exit sites near the secretion site. In Drosophila melanogaster, secreted cargo can be locally translated from mRNA and then directly transported from the ER exit sites through closely apposed Golgi stacks to the plasma membrane (34). These two ways of polarized secretion are in sharp contrast to that used in vertebrate cells where cargo is synthesized in the ER, processed by the centralized, perinuclear Golgi ribbon, and then sorted from the trans-Golgi network (TGN). To direct polarized trafficking, the ribbon is oriented towards the secretion site, which allows cargo to be efficiently and directionally transported along microtubules to the cell periphery (28, 35). Notably, the above mechanisms are not mutually exclusive. Instead, they can act cooperatively to exert more specialized or dynamic function. In neurons, for instance, mRNAs encoding post-synaptic receptors and secreted cargo are actively delivered to the dendrites where they are translated and transported through local Golgi outposts (36).
How does the Golgi ribbon form in interphase?
Although it is not fully understood why the vertebrate Golgi is organized into a perinuclear ribbon, various factors involved in maintaining its architecture have been explored including the cytoskeleton, Golgi structural proteins and membrane trafficking molecules. The current understanding of mechanisms of Golgi ribbon formation is largely derived from analyses of ribbon assembly upon nocodazole washout and from RNAi-mediated loss-of-function studies. Mechanistically, the ribbon is formed by two processes. One is microtubule-dependent clustering of Golgi stacks in the cell periphery followed by positioning in the perinuclear area. The other involves lateral association and homotypic fusion of Golgi stacks into a continuous ribbon.
The requirement for microtubules in Golgi ribbon formation is demonstrated by the effects of nocodazole that depolymerizes microtubules and accordingly disperses the ribbon into numerous mini-stacks throughout the cytoplasm. Upon nocodazole washout, the dispersed Golgi stacks undergo two stages of microtubule-driven and CLASP-dependent coalesce (23). First they are locally clustered in the cell periphery by Golgi-derived microtubules and then moved towards the centrosomes in the perinuclear area by radial centrosomal microtubules. Indispensable for both clustering and positioning stages is the minus end-directed motor dynein, which gathers and transports Golgi stacks along microtubules into the perinuclear region (23). Perturbation of microtubule integrity, microtubule dynamic instability, or dynein function scatters the Golgi throughout the cytoplasm (19, 37–40).
Interestingly, although the centrosome is the major MTOC that efficiently nucleates and organizes microtubules into a radial array, it is neither necessary nor sufficient for Golgi ribbon formation. In fact, Golgi membranes, including nocodazole-induced mini-stacks in interphase cells and mitotic membranes in post-metaphase cells, self-organize into a single continuous unit in the absence of centrosomes (41, 42). In support of this notion, the Golgi itself has been reported as an MTOC to nucleate microtubules (35, 43). Microtubule nucleation requires a ring-like multi-subunit structure known as the γ-tubulin ring complex (γ-TuRC). Several recent studies have revealed that γ-TuRC anchors on the Golgi membrane via either direct or indirect association with cis-Golgi or TGN proteins such as GMAP210, GCC185 (via CLASP) and GM130 (via AKAP450) (35, 44, 45). These findings thus provide a molecular basis for autonomy of the Golgi as well as dispensability of centrosomes in building the ribbon.
Once present in close proximity, the Golgi stacks can be further linked together. Lateral, homotypic linking of cisternae between stacks requires not only proper orientation of polarized stacks but also precise recognition of cisternal identity. It is not completely clear how such specificity is achieved, but Golgi-derived microtubules may partially contribute to it. Since these microtubule filaments seem to nucleate from either the cis-side of the Golgi or the TGN (35, 45), Golgi-derived microtubules may preferentially select or promote the line-up of stacks to be linked due to geometrical constraints. Another level of specificity comes from stack-linking factors that are specifically associated with a particular cisterna. In analogy to heterotypic tethering of vesicles to Golgi cisternae, homotypic linking of two cisternae can also be achieved by specific membrane proteins. The best-studied case is the stacking factor GRASP65, which holds cisternae together into stacks and maintains ribbon integrity (5, 46, 47). By forming anti-parallel homo-oligomers in trans that bring cisternae in close vicinity, GRASP65 is important for stacking as well as lateral linking of cisternae (47–49). In sharp contrast to cisternal stacking where a precise distance between cisternae is maintained, lateral linking is followed by fusion of equivalent cisternae. So far the molecules and mechanisms that regulate this downstream homotypic fusion event remain to be uncovered. In addition, other Golgi structural proteins, mostly golgins, have been widely implicated in the maintenance of an intact Golgi ribbon (5, 18, 25, 32, 50–53), but the molecular mechanisms are largely unknown.
To maintain ribbon integrity, it is critical to balance the flux of membranes passing through the Golgi. Disturbing the dynamic equilibrium between membrane inflow and outflow leads to a disrupted Golgi structure (21, 25, 54–59). Moreover, other important regulators including kinases, phosphoinositide phosphatases and cholesterol transfer proteins also contribute to maintaining this dynamic organelle (60–64). Notably, while likely sharing some common components, de novo ribbon assembly in post-mitotic cells may involve additional players other than those required for maintaining the interphase ribbon.
Why does the Golgi ribbon disassemble in mitosis?
During cell division, the single Golgi ribbon must be divided into the two daughter cells. To prepare for proper segregation, the ribbon first unlinks into individual stacks, which further undergo unstacking and vesiculation, and then eventually disassembles into a collection of vesicles and tubules. These mitotic Golgi membranes are then partitioned between the two daughter cells where they reassemble into a functional Golgi in late mitosis (65).
Mechanistically, disassembly of the Golgi ribbon in mitosis is an outcome of imbalanced membrane trafficking. Budding of COPII vesicles out of the ER is blocked upon mitotic entry due to disassembly of the ER exit sites (66, 67) and fusion of COPI vesicles with Golgi cisternae is also inhibited due to impaired vesicle tethering (68). Therefore, membrane input into the Golgi is ceased at the levels of both ER-to-Golgi and intra-Golgi transport. Furthermore, membrane flow out of the Golgi not only persists but increases, as formation of COPI vesicles is accelerated due to cisternal unstacking, which exposes a greater surface area from which vesicles can bud (69). Consequently, Golgi membranes are rapidly converted into numerous vesicles.
Intriguingly, disassembly of the Golgi complex during cell division is not a conserved phenomenon among eukaryotes (Table 1). In plants, the Golgi does not vesiculate, but instead maintains the stacked organization throughout the entire cell cycle (33). Similarly, partitioning of the single Golgi stack in simple protozoa is achieved by binary fission without further disintegration (70, 71). This indicates that mitotic disassembly is not a general requirement for Golgi inheritance. So why would vertebrates develop a disassembly-reassembly mechanism for Golgi partitioning, especially given the fact that coordination of this process with the intrinsic mitotic program is not a trivial effort? What could be the possible purposes or advantages, for disassembly of the Golgi ribbon during cell division?
One reason for mitotic Golgi disassembly is to facilitate its own inheritance. Theoretically, there is no need for receiving Golgi membranes from the mother cell as long as de novo assembly can take place in the progeny. However, while Golgi stacks have the ability to form de novo (72, 73), the template inheritance is the dominant route in most cases once the template is present (74). In this scenario, there are two ways by which the single Golgi ribbon can be separated into the two daughter cells. One is to simply split the single Golgi ribbon into two smaller ribbons, each of which is then segregated into one of the daughter cells, a process similar to binary fission in protozoa (75). In reality, vertebrate cells adopt a more complex approach by which the single Golgi ribbon is first disassembled into numerous vesicles and tubules, which are then fused back into a functional organelle after partitioning (76). Therefore, there appear to be additional reasons to disassemble the Golgi ribbon during mitosis.
Second, inhibition of intracellular trafficking and resulting Golgi disassembly is perhaps intended to prevent unregulated activation of biochemical pathways during mitosis. This is best exemplified by SREBP and ATF6 (77), which are transcription factors that reside as silent precursors in the ER and await proper stimulation (low cholesterol for SREBP and ER stress for ATF6) to be transported to the Golgi (78–80). Upon arrival, they are cleaved and activated by Golgi-resident proteases. To avoid potential activation induced by content mixing between two membrane compartments, it is conceivable to temporarily shut down membrane exchange in mitosis. This maintains a discrete boundary between the two organelles, so that transcriptional activation is only induced in the presence of the appropriate signals (7).
As the third reason, mitotic Golgi disassembly may physically segregate different pools of Golgi molecules with distinct tasks. This is exemplified by our recent study on the requirement of the mitotic spindle for Golgi inheritance (42). Intrigued by the observation that disassembled Golgi membranes are concentrated around the spindle poles (81), we dissected the role of the spindle in Golgi partitioning. By inducing asymmetrical cell division where the spindle is segregated into only one of the daughter cells (82), we unexpectedly discovered two levels of regulation governing Golgi inheritance. On one hand, Golgi stacks, which are functional in basic secretion, reform independently of the spindle. On the other hand, the ribbon, as a structural prerequisite for polarized secretion, requires the mitotic spindle to ensure its integrity after mitosis. This demonstrates that the factors necessary for post-mitotic ribbon formation (ribbon determinants) are associated and partitioned with the spindle machinery (83). More importantly, it further implies that mitotic disassembly of the ribbon separates different subsets of Golgi proteins into distinct pools prior to partitioning. Similar mechanisms have been reported for the nuclear pore complex, which disintegrates into several sub-complexes upon mitotic entry (84). As the nuclear membrane continues to resolve, most nucleoporins are dispersed into the cytoplasm while the membrane proteins are left behind in the ER. Notably, certain nucleoporins are localized in part to the kinetochores and/or spindle microtubules and are partitioned together with the spindle (85–87). Likewise, through the disassembly process, Golgi ribbon determinants may be selectively separated from the rest of the membranes or dissociated from the interphase protein complexes and then hook up to the spindle for subsequent division.
The fourth possibility suggests that the very initial step of Golgi disassembly may act as a signaling trigger for entry into mitosis. Golgi ribbon unlinking, which severs the lateral connections between adjacent stacks, has been recently reported to occur in the late G2 phase (88). This subtle change in Golgi morphology correlates with mitotic entry, as prevention of ribbon unlinking delays cells from progressing through the G2/M border. In this sense, Golgi disassembly (unlinking by a more precise definition) actively promotes mitotic entry rather than being a passive consequence of mitosis. This has led to the hypothesis that a Golgi-based checkpoint exists and acts as a part of or in parallel with the intrinsic G2/M checkpoint. However, although the membrane fission factor BARS (89, 90) as well as MEK/ERK kinases and their Golgi targets GRASP65/55 (11, 91–93) contribute to ribbon unlinking, the mechanism by which the morphological change is sensed and transmitted to cell cycle machinery still awaits elucidation.
In addition to promoting mitotic entry, Golgi disassembly may also have a vital role in mediating other important processes during cell division. These key events include nuclear envelope breakdown, spindle formation, spindle assembly checkpoint signaling, cytokinesis and cell fate determination. Below we suggest potential mechanisms to achieve such regulation.
How does Golgi ribbon disassembly regulate mitosis?
Once the ribbon is disassembled, Golgi-localized proteins are exposed and become accessible, both physically and biochemically, to binding partners that are involved in mitotic regulation. In this respect, GRASP65 has been shown to contribute to spindle assembly and mitotic progression, as its downregulation by RNAi results in aberrant multipolar spindles and mitotic arrest (93). In mitosis, transient switch of protein functions is often mediated by post-translational modifications such as phosphorylation, which, in the case of GRASP65, disrupts its self-oligomerization and accordingly unstacks Golgi cisternae (46, 47). GRASP65 phosphorylation may thus change the conformation and/or free binding sites for spindle assembly factors. GM130 down-regulation shows a similar mitotic defect (94), implying an additional role of GM130 in regulating spindle formation. Interestingly, since GM130 is localized to the more central region of Golgi cisternae (18), it is likely that mitotic disassembly relieves the steric hindrance around the molecules. Moreover, GM130 is also mitotically phosphorylated (68), which releases GM130 from its interphase interactor p115 (95, 96). This may allow GM130 to associate with other mitotic regulators and thus mediate specific mitotic events (Wei and Seemann, unpublished data).
Furthermore, factors associated with Golgi membranes are librated after mitotic disassembly into the cytoplasm or translocated to other intracellular locations where they acquire novel functions. In support of this idea, several Golgi peripheral proteins have been found in various locations at different mitotic stages that regulate diverse aspects of cell division. For instance, COPI is recruited by the nucleoporin Nup153 to the nuclear envelope where it promotes nuclear envelope breakdown (97). ZW10 and RINT-1, which mediate transport between the ER and the Golgi during interphase (58, 98), are targeted to the kinetochores where they assist the termination of the spindle assembly checkpoint (99, 100). In addition, clathrin directly binds to spindle microtubules where it stabilizes kinetochore fibers and facilitates chromosome congression to the metaphase plate (101). Similarly, through recruitment to the cleavage furrow and midbody, Nir2 regulates cytokinesis by interacting with Plk1 and RhoA (102). Moreover, during neurogenesis when stem cells undergo asymmetric cell division, ACBD3 is briefly released into the cytoplasm and interacts with Numb to defend the progenitor fate (103). In sum, the Golgi complex functions as a molecular reservoir for mitotic regulators and Golgi disassembly is utilized as a switch that triggers their actions in a precise spatiotemporal manner.
Future perspectives
Considerable progress has been made in elucidating the factors required for the Golgi ribbon structure. Among them, golgins have attracted particular attention due to their dual abilities to facilitate vesicle tethering as well as to form a structural support for Golgi membranes. Complexity of the ribbon structure might be attributed to the golgins that are uniquely expressed in higher eukaryotes. Furthermore, accumulating evidence shows that golgins can link membranes to the cytoskeleton, making them the most versatile players for ribbon formation. Although the significance of these proteins has been clearly demonstrated, a coherent view is still lacking as to how they interact with the cytoskeleton and the membrane fusion machinery to orchestrate the process. In addition, many other questions remain unanswered. For instance, what are the molecules responsible for post-mitotic ribbon reassembly? Do they share common components with interphase ribbon factors? How does ribbon disassembly participate in regulating other mitotic events? Moreover, future efforts should also be made to better understand the regulation of ribbon dynamics under different biological contexts, for example, during animal development and disease pathogenesis.
Acknowledgments
We thank Katie Zhang for critical reading of the manuscript and Nobuhiro Nakamura for advice. J.S. is a Virginia Murchison Lithicum Scholar in Medical Research and supported by grants from the NIH (GM096070) and the American Heart Association (8065090F).
References
- 1.Mowbrey K, Dacks JB. Evolution and diversity of the Golgi body. FEBS Lett. 2009;583:3738–3745. doi: 10.1016/j.febslet.2009.10.025. [DOI] [PubMed] [Google Scholar]
- 2.Rambourg A, Jackson CL, Clermont Y. Three dimensional configuration of the secretory pathway and segregation of secretion granules in the yeast Saccharomyces cerevisiae. J Cell Sci. 2001;114:2231–2239. doi: 10.1242/jcs.114.12.2231. [DOI] [PubMed] [Google Scholar]
- 3.Preuss D, Mulholland J, Franzusoff A, Segev N, Botstein D. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol Biol Cell. 1992;3:789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J Cell Biol. 1999;144:1135–1149. doi: 10.1083/jcb.144.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- 6.Cole NB, Smith CL, Sciaky N, Terasaki M, Edidin M, Lippincott-Schwartz J. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 1996;273:797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- 7.Bartz R, Sun LP, Bisel B, Wei JH, Seemann J. Spatial separation of Golgi and ER during mitosis protects SREBP from unregulated activation. EMBO J. 2008;27:948–955. doi: 10.1038/emboj.2008.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaietta GM, Giepmans BN, Deerinck TJ, Smith WB, Ngan L, Llopis J, Adams SR, Tsien RY, Ellisman MH. Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proc Natl Acad Sci U S A. 2006;103:17777–17782. doi: 10.1073/pnas.0608509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J Neurosci. 2003;23:6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feinstein TN, Linstedt AD. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol Biol Cell. 2007;18:594–604. doi: 10.1091/mbc.E06-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutterlin C, Colanzi A. The Golgi and the centrosome: building a functional partnership. J Cell Biol. 2010;188:621–628. doi: 10.1083/jcb.200910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seemann J, Jokitalo E, Pypaert M, Warren G. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature. 2000;407:1022–1026. doi: 10.1038/35039538. [DOI] [PubMed] [Google Scholar]
- 14.Ramirez IB, Lowe M. Golgins and GRASPs: holding the Golgi together. Semin Cell Dev Biol. 2009;20:770–779. doi: 10.1016/j.semcdb.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Seemann J, Pypaert M, Taguchi T, Malsam J, Warren G. Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science. 2002;295:848–851. doi: 10.1126/science.1068064. [DOI] [PubMed] [Google Scholar]
- 16.Lu Z, Joseph D, Bugnard E, Zaal KJ, Ralston E. Golgi complex reorganization during muscle differentiation: visualization in living cells and mechanism. Mol Biol Cell. 2001;12:795–808. doi: 10.1091/mbc.12.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White JG, Key NS, King RA, Vercellotti GM. The White platelet syndrome: a new autosomal dominant platelet disorder. Platelets. 2004;15:173–184. doi: 10.1080/09537100410001682805. [DOI] [PubMed] [Google Scholar]
- 18.Diao A, Rahman D, Pappin DJ, Lucocq J, Lowe M. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J Cell Biol. 2003;160:201–212. doi: 10.1083/jcb.200207045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trucco A, Polishchuk RS, Martella O, Di Pentima A, Fusella A, Di Giandomenico D, San Pietro E, Beznoussenko GV, Polishchuk EV, Baldassarre M, Buccione R, Geerts WJ, Koster AJ, Burger KN, Mironov AA, et al. Secretory traffic triggers the formation of tubular continuities across Golgi sub-compartments. Nat Cell Biol. 2004;6:1071–1081. doi: 10.1038/ncb1180. [DOI] [PubMed] [Google Scholar]
- 21.Zolov SN, Lupashin VV. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J Cell Biol. 2005;168:747–759. doi: 10.1083/jcb.200412003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondylis V, Rabouille C. A novel role for dp115 in the organization of tER sites in Drosophila. J Cell Biol. 2003;162:185–198. doi: 10.1083/jcb.200301136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188:237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marra P, Salvatore L, Mironov A, Jr, Di Campli A, Di Tullio G, Trucco A, Beznoussenko G, Mironov A, De Matteis MA. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol Biol Cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bisel B, Wang Y, Wei JH, Xiang Y, Tang D, Miron-Mendoza M, Yoshimura S, Nakamura N, Seemann J. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J Cell Biol. 2008;182:837–843. doi: 10.1083/jcb.200805045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preisinger C, Short B, De Corte V, Bruyneel E, Haas A, Kopajtich R, Gettemans J, Barr FA. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3zeta. J Cell Biol. 2004;164:1009–1020. doi: 10.1083/jcb.200310061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 29.Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. Centrosome polarization delivers secretory granules to the immunological synapse. Nature. 2006;443:462–465. doi: 10.1038/nature05071. [DOI] [PubMed] [Google Scholar]
- 30.Horton AC, Racz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Magdalena J, Millard TH, Machesky LM. Microtubule involvement in NIH 3T3 Golgi and MTOC polarity establishment. J Cell Sci. 2003;116:743–756. doi: 10.1242/jcs.00288. [DOI] [PubMed] [Google Scholar]
- 32.Yadav S, Puri S, Linstedt AD. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nebenfuhr A, Frohlick JA, Staehelin LA. Redistribution of Golgi stacks and other organelles during mitosis and cytokinesis in plant cells. Plant Physiol. 2000;124:135–151. doi: 10.1104/pp.124.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herpers B, Rabouille C. mRNA localization and ER-based protein sorting mechanisms dictate the use of transitional endoplasmic reticulum-golgi units involved in gurken transport in Drosophila oocytes. Mol Biol Cell. 2004;15:5306–5317. doi: 10.1091/mbc.E04-05-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, Yates JR, 3rd, Maiato H, Khodjakov A, Akhmanova A, Kaverina I. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanus C, Ehlers MD. Secretory outposts for the local processing of membrane cargo in neuronal dendrites. Traffic. 2008;9:1437–1445. doi: 10.1111/j.1600-0854.2008.00775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wehland J, Henkart M, Klausner R, Sandoval IV. Role of microtubules in the distribution of the Golgi apparatus: effect of taxol and microinjected anti-alpha-tubulin antibodies. Proc Natl Acad Sci U S A. 1983;80:4286–4290. doi: 10.1073/pnas.80.14.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Y, Yu W, Li Y, Yang Z, Yan X, Huang Q, Zhu X. Nudel functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein. J Cell Biol. 2004;164:557–566. doi: 10.1083/jcb.200308058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corthesy-Theulaz I, Pauloin A, Pfeffer SR. Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J Cell Biol. 1992;118:1333–1345. doi: 10.1083/jcb.118.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelletier L, Jokitalo E, Warren G. The effect of Golgi depletion on exocytic transport. Nat Cell Biol. 2000;2:840–846. doi: 10.1038/35041089. [DOI] [PubMed] [Google Scholar]
- 42.Wei JH, Seemann J. The mitotic spindle mediates inheritance of the Golgi ribbon structure. J Cell Biol. 2009;184:391–397. doi: 10.1083/jcb.200809090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, Pous C. The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rios RM, Sanchis A, Tassin AM, Fedriani C, Bornens M. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell. 2004;118:323–335. doi: 10.1016/j.cell.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 45.Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barr FA, Puype M, Vandekerckhove J, Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Seemann J, Pypaert M, Shorter J, Warren G. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sengupta D, Truschel S, Bachert C, Linstedt AD. Organelle tethering by a homotypic PDZ interaction underlies formation of the Golgi membrane network. J Cell Biol. 2009;186:41–55. doi: 10.1083/jcb.200902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Satoh A, Warren G. Mapping the functional domains of the Golgi stacking factor GRASP65. J Biol Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Derby MC, Lieu ZZ, Brown D, Stow JL, Goud B, Gleeson PA. The trans-Golgi network golgin, GCC185, is required for endosome-to-Golgi transport and maintenance of Golgi structure. Traffic. 2007;8:758–773. doi: 10.1111/j.1600-0854.2007.00563.x. [DOI] [PubMed] [Google Scholar]
- 51.Short B, Preisinger C, Korner R, Kopajtich R, Byron O, Barr FA. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J Cell Biol. 2001;155:877–883. doi: 10.1083/jcb.200108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu L, Tai G, Hong W. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-golgi network. Mol Biol Cell. 2004;15:4426–4443. doi: 10.1091/mbc.E03-12-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feinstein TN, Linstedt AD. GRASP55 regulates Golgi ribbon formation. Mol Biol Cell. 2008;19:2696–2707. doi: 10.1091/mbc.E07-11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shestakova A, Zolov S, Lupashin V. COG complex-mediated recycling of Golgi glycosyltransferases is essential for normal protein glycosylation. Traffic. 2006;7:191–204. doi: 10.1111/j.1600-0854.2005.00376.x. [DOI] [PubMed] [Google Scholar]
- 56.Sohda M, Misumi Y, Yoshimura S, Nakamura N, Fusano T, Sakisaka S, Ogata S, Fujimoto J, Kiyokawa N, Ikehara Y. Depletion of vesicle-tethering factor p115 causes mini-stacked Golgi fragments with delayed protein transport. Biochem Biophys Res Commun. 2005;338:1268–1274. doi: 10.1016/j.bbrc.2005.10.084. [DOI] [PubMed] [Google Scholar]
- 57.Suga K, Hattori H, Saito A, Akagawa K. RNA interference-mediated silencing of the syntaxin 5 gene induces Golgi fragmentation but capable of transporting vesicles. FEBS Lett. 2005;579:4226–4234. doi: 10.1016/j.febslet.2005.06.053. [DOI] [PubMed] [Google Scholar]
- 58.Sun Y, Shestakova A, Hunt L, Sehgal S, Lupashin V, Storrie B. Rab6 regulates both ZW10/RINT-1 and conserved oligomeric Golgi complex-dependent Golgi trafficking and homeostasis. Mol Biol Cell. 2007;18:4129–4142. doi: 10.1091/mbc.E07-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu Y, Martin S, James DE, Hong W. GS15 forms a SNARE complex with syntaxin 5, GS28, and Ykt6 and is implicated in traffic in the early cisternae of the Golgi apparatus. Mol Biol Cell. 2002;13:3493–3507. doi: 10.1091/mbc.E02-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bejarano E, Cabrera M, Vega L, Hidalgo J, Velasco A. Golgi structural stability and biogenesis depend on associated PKA activity. J Cell Sci. 2006;119:3764–3775. doi: 10.1242/jcs.03146. [DOI] [PubMed] [Google Scholar]
- 61.Diaz Anel AM, Malhotra V. PKCeta is required for beta1gamma2/beta3gamma2-and PKD-mediated transport to the cell surface and the organization of the Golgi apparatus. J Cell Biol. 2005;169:83–91. doi: 10.1083/jcb.200412089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y, Boukhelifa M, Tribble E, Morin-Kensicki E, Uetrecht A, Bear JE, Bankaitis VA. The Sac1 phosphoinositide phosphatase regulates Golgi membrane morphology and mitotic spindle organization in mammals. Mol Biol Cell. 2008;19:3080–3096. doi: 10.1091/mbc.E07-12-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun KH, de Pablo Y, Vincent F, Johnson EO, Chavers AK, Shah K. Novel genetic tools reveal Cdk5’s major role in Golgi fragmentation in Alzheimer’s disease. Mol Biol Cell. 2008;19:3052–3069. doi: 10.1091/mbc.E07-11-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ngo M, Ridgway ND. Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol Biol Cell. 2009;20:1388–1399. doi: 10.1091/mbc.E08-09-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei JH, Seemann J. Mitotic division of the mammalian Golgi apparatus. Semin Cell Dev Biol. 2009;20:810–816. doi: 10.1016/j.semcdb.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 66.Farmaki T, Ponnambalam S, Prescott AR, Clausen H, Tang BL, Hong W, Lucocq JM. Forward and retrograde trafficking in mitotic animal cells. ER-Golgi transport arrest restricts protein export from the ER into COPII-coated structures. J Cell Sci. 1999;112 (Pt 5):589–600. doi: 10.1242/jcs.112.5.589. [DOI] [PubMed] [Google Scholar]
- 67.Kano F, Tanaka AR, Yamauchi S, Kondo H, Murata M. Cdc2 kinase-dependent disassembly of endoplasmic reticulum (ER) exit sites inhibits ER-to-Golgi vesicular transport during mitosis. Mol Biol Cell. 2004;15:4289–4298. doi: 10.1091/mbc.E03-11-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin DJ, Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, Wei JH, Bisel B, Tang D, Seemann J. Golgi Cisternal Unstacking Stimulates COPI Vesicle Budding and Protein Transport. PLoS ONE. 2008;3:e1647. doi: 10.1371/journal.pone.0001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benchimol M. Trichomonads under Microscopy. Microsc Microanal. 2004;10:528–550. doi: 10.1017/S1431927604040905. [DOI] [PubMed] [Google Scholar]
- 71.Pelletier L, Stern CA, Pypaert M, Sheff D, Ngo HM, Roper N, He CY, Hu K, Toomre D, Coppens I, Roos DS, Joiner KA, Warren G. Golgi biogenesis in Toxoplasma gondii. Nature. 2002;418:548–552. doi: 10.1038/nature00946. [DOI] [PubMed] [Google Scholar]
- 72.Bevis BJ, Hammond AT, Reinke CA, Glick BS. De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat Cell Biol. 2002;4:750–756. doi: 10.1038/ncb852. [DOI] [PubMed] [Google Scholar]
- 73.Puri S, Linstedt AD. Capacity of the golgi apparatus for biogenesis from the endoplasmic reticulum. Mol Biol Cell. 2003;14:5011–5018. doi: 10.1091/mbc.E03-06-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Emr S, Glick BS, Linstedt AD, Lippincott-Schwartz J, Luini A, Malhotra V, Marsh BJ, Nakano A, Pfeffer SR, Rabouille C, Rothman JE, Warren G, Wieland FT. Journeys through the Golgi--taking stock in a new era. J Cell Biol. 2009;187:449–453. doi: 10.1083/jcb.200909011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He CY. Golgi biogenesis in simple eukaryotes. Cell Microbiol. 2007;9:566–572. doi: 10.1111/j.1462-5822.2006.00874.x. [DOI] [PubMed] [Google Scholar]
- 76.Lowe M, Barr FA. Inheritance and biogenesis of organelles in the secretory pathway. Nat Rev Mol Cell Biol. 2007;8:429–439. doi: 10.1038/nrm2179. [DOI] [PubMed] [Google Scholar]
- 77.Bartz R, Seemann J. Mitotic regulation of SREBP and ATF6 by separation of the Golgi and ER. Cell Cycle. 2008;7:2100–2105. doi: 10.4161/cc.7.14.6327. [DOI] [PubMed] [Google Scholar]
- 78.Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annual review of genetics. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 79.Goldstein JL, Debose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 80.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Molecular cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 81.Shima DT, Cabrera-Poch N, Pepperkok R, Warren G. An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J Cell Biol. 1998;141:955–966. doi: 10.1083/jcb.141.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei JH, Seemann J. Induction of asymmetrical cell division to analyze spindle-dependent organelle partitioning using correlative microscopy techniques. Nat Protoc. 2009;4:1653–1662. doi: 10.1038/nprot.2009.160. [DOI] [PubMed] [Google Scholar]
- 83.Wei JH, Seemann J. Spindle-dependent partitioning of the Golgi ribbon. Commun Integr Biol. 2009;2:406–407. doi: 10.4161/cib.2.5.8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol. 2010;2:a000539. doi: 10.1101/cshperspect.a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, Ellenberg J, Doye V. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol. 2002;156:595–602. doi: 10.1083/jcb.200110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Salina D, Enarson P, Rattner JB, Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol. 2003;162:991–1001. doi: 10.1083/jcb.200304080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rabouille C, Kondylis V. Golgi ribbon unlinking: an organelle-based G2/M checkpoint. Cell Cycle. 2007;6:2723–2729. doi: 10.4161/cc.6.22.4896. [DOI] [PubMed] [Google Scholar]
- 89.Colanzi A, Hidalgo Carcedo C, Persico A, Cericola C, Turacchio G, Bonazzi M, Luini A, Corda D. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. EMBO J. 2007;26:2465–2476. doi: 10.1038/sj.emboj.7601686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hidalgo Carcedo C, Bonazzi M, Spano S, Turacchio G, Colanzi A, Luini A, Corda D. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 2004;305:93–96. doi: 10.1126/science.1097775. [DOI] [PubMed] [Google Scholar]
- 91.Duran JM, Kinseth M, Bossard C, Rose DW, Polishchuk R, Wu CC, Yates J, Zimmerman T, Malhotra V. The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis. Mol Biol Cell. 2008;19:2579–2587. doi: 10.1091/mbc.E07-10-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shaul YD, Seger R. ERK1c regulates Golgi fragmentation during mitosis. J Cell Biol. 2006;172:885–897. doi: 10.1083/jcb.200509063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sutterlin C, Polishchuk R, Pecot M, Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kodani A, Sutterlin C. The Golgi protein GM130 regulates centrosome morphology and function. Mol Biol Cell. 2008;19:745–753. doi: 10.1091/mbc.E07-08-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakamura N. Emerging new roles of GM130, a cis-Golgi matrix protein, in higher order cell functions. J Pharmacol Sci. 2010;112:255–264. doi: 10.1254/jphs.09r03cr. [DOI] [PubMed] [Google Scholar]
- 96.Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- 97.Liu J, Prunuske AJ, Fager AM, Ullman KS. The COPI complex functions in nuclear envelope breakdown and is recruited by the nucleoporin Nup153. Dev Cell. 2003;5:487–498. doi: 10.1016/s1534-5807(03)00262-4. [DOI] [PubMed] [Google Scholar]
- 98.Hirose H, Arasaki K, Dohmae N, Takio K, Hatsuzawa K, Nagahama M, Tani K, Yamamoto A, Tohyama M, Tagaya M. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 2004;23:1267–1278. doi: 10.1038/sj.emboj.7600135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chan GK, Jablonski SA, Starr DA, Goldberg ML, Yen TJ. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat Cell Biol. 2000;2:944–947. doi: 10.1038/35046598. [DOI] [PubMed] [Google Scholar]
- 100.Xiao J, Liu CC, Chen PL, Lee WH. RINT-1, a novel Rad50-interacting protein, participates in radiation-induced G(2)/M checkpoint control. J Biol Chem. 2001;276:6105–6111. doi: 10.1074/jbc.M008893200. [DOI] [PubMed] [Google Scholar]
- 101.Royle SJ, Bright NA, Lagnado L. Clathrin is required for the function of the mitotic spindle. Nature. 2005;434:1152–1157. doi: 10.1038/nature03502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Litvak V, Argov R, Dahan N, Ramachandran S, Amarilio R, Shainskaya A, Lev S. Mitotic phosphorylation of the peripheral Golgi protein Nir2 by Cdk1 provides a docking mechanism for Plk1 and affects cytokinesis completion. Mol Cell. 2004;14:319–330. doi: 10.1016/s1097-2765(04)00214-x. [DOI] [PubMed] [Google Scholar]
- 103.Zhou Y, Atkins JB, Rompani SB, Bancescu DL, Petersen PH, Tang H, Zou K, Stewart SB, Zhong W. The mammalian Golgi regulates numb signaling in asymmetric cell division by releasing ACBD3 during mitosis. Cell. 2007;129:163–178. doi: 10.1016/j.cell.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 104.Hager KM, Striepen B, Tilney LG, Roos DS. The nuclear envelope serves as an intermediary between the ER and Golgi complex in the intracellular parasite Toxoplasma gondii. J Cell Sci. 1999;112 (Pt 16):2631–2638. doi: 10.1242/jcs.112.16.2631. [DOI] [PubMed] [Google Scholar]
- 105.Duszenko M, Ivanov IE, Ferguson MA, Plesken H, Cross GA. Intracellular transport of a variant surface glycoprotein in Trypanosoma brucei. J Cell Biol. 1988;106:77–86. doi: 10.1083/jcb.106.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.He CY, Ho HH, Malsam J, Chalouni C, West CM, Ullu E, Toomre D, Warren G. Golgi duplication in Trypanosoma brucei. J Cell Biol. 2004;165:313–321. doi: 10.1083/jcb.200311076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bannister LH, Hopkins JM, Fowler RE, Krishna S, Mitchell GH. Ultrastructure of rhoptry development in Plasmodium falciparum erythrocytic schizonts. Parasitology. 2000;121 (Pt 3):273–287. doi: 10.1017/s0031182099006320. [DOI] [PubMed] [Google Scholar]
- 108.Becker B, Melkonian M. The secretory pathway of protists: spatial and functional organization and evolution. Microbiol Rev. 1996;60:697–721. doi: 10.1128/mr.60.4.697-721.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kinseth MA, Anjard C, Fuller D, Guizzunti G, Loomis WF, Malhotra V. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 2007;130:524–534. doi: 10.1016/j.cell.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 110.Schneider N, Schwartz JM, Kohler J, Becker M, Schwarz H, Gerisch G. Golvesin-GFP fusions as distinct markers for Golgi and post-Golgi vesicles in Dictyostelium cells. Biol Cell. 2000;92:495–511. doi: 10.1016/s0248-4900(00)01102-3. [DOI] [PubMed] [Google Scholar]
- 111.Chappell TG, Warren G. A galactosyltransferase from the fission yeast Schizosaccharomyces pombe. J Cell Biol. 1989;109:2693–2702. doi: 10.1083/jcb.109.6.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Johnson BF, Yoo BY, Calleja GB. Cell division in yeasts: movement of organelles associated with cell plate growth of Schizosaccharomyces pombe. J Bacteriol. 1973;115:358–366. doi: 10.1128/jb.115.1.358-366.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rossanese OW, Soderholm J, Bevis BJ, Sears IB, O’Connor J, Williamson EK, Glick BS. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dupree P, Sherrier DJ. The plant Golgi apparatus. Biochim Biophys Acta. 1998;1404:259–270. doi: 10.1016/s0167-4889(98)00061-5. [DOI] [PubMed] [Google Scholar]
- 115.Staehelin LA, Kang BH. Nanoscale architecture of endoplasmic reticulum export sites and of Golgi membranes as determined by electron tomography. Plant Physiol. 2008;147:1454–1468. doi: 10.1104/pp.108.120618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Segui-Simarro JM, Staehelin LA. Cell cycle-dependent changes in Golgi stacks, vacuoles, clathrin-coated vesicles and multivesicular bodies in meristematic cells of Arabidopsis thaliana: a quantitative and spatial analysis. Planta. 2006;223:223–236. doi: 10.1007/s00425-005-0082-2. [DOI] [PubMed] [Google Scholar]
- 117.Wolf N, Hirsh D, McIntosh JR. Spermatogenesis in males of the free-living nematode, Caenorhabditis elegans. J Ultrastruct Res. 1978;63:155–169. doi: 10.1016/s0022-5320(78)80071-9. [DOI] [PubMed] [Google Scholar]
- 118.Rolls MM, Hall DH, Victor M, Stelzer EH, Rapoport TA. Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol Biol Cell. 2002;13:1778–1791. doi: 10.1091/mbc.01-10-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Andrews R, Ahringer J. Asymmetry of early endosome distribution in C. elegans embryos. PLoS ONE. 2007;2:e493. doi: 10.1371/journal.pone.0000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stanley H, Botas J, Malhotra V. The mechanism of Golgi segregation during mitosis is cell type-specific. Proc Natl Acad Sci U S A. 1997;94:14467–14470. doi: 10.1073/pnas.94.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rolls MM, Marquardt MT, Kielian M, Machamer CE. Cholesterol-independent targeting of Golgi membrane proteins in insect cells. Mol Biol Cell. 1997;8:2111–2118. doi: 10.1091/mbc.8.11.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Terasaki M. Dynamics of the endoplasmic reticulum and golgi apparatus during early sea urchin development. Mol Biol Cell. 2000;11:897–914. doi: 10.1091/mbc.11.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ho SY, Lorent K, Pack M, Farber SA. Zebrafish fat-free is required for intestinal lipid absorption and Golgi apparatus structure. Cell Metab. 2006;3:289–300. doi: 10.1016/j.cmet.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schlager MA, Kapitein LC, Grigoriev I, Burzynski GM, Wulf PS, Keijzer N, de Graaff E, Fukuda M, Shepherd IT, Akhmanova A, Hoogenraad CC. EMBO J. 2010. Pericentrosomal targeting of Rab6 secretory vesicles by Bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Le Bot N, Antony C, White J, Karsenti E, Vernos I. Role of xklp3, a subunit of the Xenopus kinesin II heterotrimeric complex, in membrane transport between the endoplasmic reticulum and the Golgi apparatus. J Cell Biol. 1998;143:1559–1573. doi: 10.1083/jcb.143.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roghi C, Allan VJ. Dynamic association of cytoplasmic dynein heavy chain 1a with the Golgi apparatus and intermediate compartment. J Cell Sci. 1999;112 (Pt 24):4673–4685. doi: 10.1242/jcs.112.24.4673. [DOI] [PubMed] [Google Scholar]
- 127.Colman A, Jones EA, Heasman J. Meiotic maturation in Xenopus oocytes: a link between the cessation of protein secretion and the polarized disappearance of Golgi apparati. J Cell Biol. 1985;101:313–318. doi: 10.1083/jcb.101.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jokitalo E, Cabrera-Poch N, Warren G, Shima DT. Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J Cell Biol. 2001;154:317–330. doi: 10.1083/jcb.200104073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shima DT, Haldar K, Pepperkok R, Watson R, Warren G. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J Cell Biol. 1997;137:1211–1228. doi: 10.1083/jcb.137.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]