Abstract
Despite an intensive vaccine program influenza infections remain a major health problem, due to the viruses’ ability to change its envelope glycoprotein hemagglutinin (HA), through shift and drift, permitting influenza to escape protection induced by current vaccines or natural immunity. Recently a new variant, H7N9, has emerged in China causing global concern. First, there have been more than 130 laboratory-confirmed human infections resulting in an alarmingly high death rate (32.3%). Second, genetic changes found in H7N9 appear to be associated with enabling avian influenza viruses to spread more effectively in mammals, thus transmitting infections on a larger scale. Currently, no vaccines or drugs are effectively able to target H7N9. Here, we report the rapid development of a synthetic consensus DNA vaccine (pH7HA) to elicit potent protective immunity against the H7N9 viruses. We show that pH7HA induces broad antibody responses that bind to divergent HAs from multiple new members of the H7N9 family. These antibody responses result in high-titer HAI against H7N9. Simultaneously, this vaccine induces potent polyfunctional effector CD4 and CD8 T cell memory responses. Animals vaccinated with pH7HA are completely protected from H7N9 virus infection and any morbidity associated with lethal challenge. This study establishes that this synthetic consensus DNA vaccine represents a new tool for targeting emerging infection, and more importantly, its design, testing and development into seed stock for vaccine production in a few days in the pandemic setting has significant implications for the rapid deployment of vaccines protecting against emerging infectious diseases.
Keywords: H7N9, hemagglutinin (HA), DNA vaccine
1. Introduction
In recent decades, both high pathogenicity avian influenza (HPAI) and low pathogenicity avian influenza (LPAI) H7 viruses have caused large poultry outbreaks in Italy [1], the Netherlands [2, 3], Canada [4], the United Kingdom [5], the United States of America [6], and Mexico [7]. During viral circulation in poultry, with the exception of the 1999 H7N1 outbreak in Italy, these H7 viruses have acquired the ability to be transmitted from poultry to humans and cause disease and mortality [8, 9]. The outbreak of the highly pathogenic H7N7 influenza virus in Netherlands in 2003 resulted in an estimated 450 human cases and one death [3]. The newly emerged Chinese avian influenza A (H7N9) virus is one subgroup among the larger group of H7 viruses [10]. As of July 4th, 2013, there have been a total of 133 laboratory-confirmed cases and 43 people have died, indicating a very high mortality rate (32.3%) [11]. The H7N9 virus exhibits several genetic features of mammalian influenza viruses, including the specificity of their HA protein binding to mammalian cellular receptors [12, 13]; a deletion in NA stalk associated with increased virulence in mammals [14]; and an important mutation in the PB2 protein that is essential for the efficient replication of avian viruses in mammalian species [15]. The efficient transmission of H7N9 virus in ferrets suggested that human-to-human transmission of this virus might be possible under appropriate conditions [16]. As a result, the emergence of the novel H7N9 has raised concerns about its pandemic potential, as well as that of related influenza viruses.
As there is no vaccine currently available for the prevention of avian influenza H7N9 infection in humans [17], there remains an urgent need to develop a protective vaccine against H7N9 infection. New synthetic DNA vaccines have emerged as an attractive approach against various infectious diseases and cancers. Conceptually, DNA vaccines have many useful attributes over traditional vaccination strategies, such as live-attenuated vaccines, protein/peptide-based vaccines [18-20]. While DNA vaccines have been shown to be capable of eliciting balanced CD4+ and CD8+ T cell responses as well as humoral immune responses in small-animal models [21], their progress in the clinic historically has been hampered by difficulties in generating sufficiently potent T cell and humoral responses in humans [22, 23]. Until recently, the DNA platform has been used primarily in prime-boost strategies along with viral vectors and proteins, thus creating an inordinately long testing and development window for addressing emerging pandemics rapidly. In order to address the technical hurdles associated with weak vaccine-induced immunity, we have recently applied many synthetic DNA design strategies, including codon/RNA optimization, the addition of highly efficient immunoglobulin leader sequences [24-26], use of ‘centralized’ immunogens to broaden immunity and remove dependence on any individual viral sequence [27, 28], new formulations [29] combined with highly efficient DNA delivery methods such as in vivo electroporation (EP) [30, 31], to improve the induction of immune responses induced by DNA vaccines in small animals, macaques [31, 32], and most recently and importantly, in humans [33, 34]. Here we present the first adaptation of this newly developed synthetic platform deployed to approach the feasibility of developing a protective vaccine against a rapidly emerging pathogen in real time.
We developed a synthetic H7N9 HA DNA vaccine using a combination of strategies in gene optimization. The DNA vaccine was delivered by EP and its immunogenicity was evaluated in mice. We observed that this vaccine was capable of inducing strong H7 HA-specific polyfunctional CD4 and CD8 effector T cell memory responses. In addition, the vaccine was able to elicit protective levels of HAI titers to the H7N9 strain A/Anhui/1/2013. Challenge of mice with a lethal dose of A/Anhui/1/2013 virus confirmed 100% protection from influenza-driven mortality. The rapid development of potential seed stock production, in just a few days, combined with the improved immune profile of this platform support the further study of synthetic H7N9 HA vaccine antigens in combination with electroporation delivery against diverse emerging pathogens.
2. Materials and methods
2.1. Influenza H7N9 Hemagglutinin DNA Vaccine Design and Development
To design a H7N9 hemagglutinin DNA vaccine, the hemagglutinin (HA) sequences of the first four identified H7N9 human isolates were retrieved from The Global Initiative on Sharing All Influenza Data (GISAID). All HA sequences were aligned using MegAlign (DNASTAR, Madison, WI) and a consensus HA sequence (H7HA) was developed, codon/RNA optimized and subsequently synthesized by GenScript. The synthesized H7HA was cloned into the expression vector pGX0001, which is under the control of the cytomegalovirus immediate-early promoter. This construct was named pH7HA.
2.2. Phylogenetic Analysis of H7N9 HA protein sequences
Twenty-four primary HA protein sequences of human H7N9 virus isolates were retrieved from GISAID. The alignment applied in the phylogenetic study was performed using Clustal × (version 2.0) and a phylogenetic tree was constructed based on Neighbor-joining evaluation of the alignment.
2.3. Indirect Immunofluorescent assay
An indirect immunofluorescent assay was utilized to confirm H7HA gene expression as described previously [35]. Briefly, human rhabdomyosarcoma (RD) cells were plated on two-well chamber slides (BD Biosciences), at a density to obtain 60-70% confluency the next day in complete DMEM medium with 10% FBS (GIBCO) and allowed to adhere overnight. The cells were transfected with pH7HA and the control plasmid pGX0001 (1 ug/well) using TurboFectin™8.0 Transfection Reagent (OriGene) according to the manufacturer’s instructions. Forty-eight hours later, the cells were washed gently three times with 1XPBS and fixed on slides using ice cold methanol for 10 min. The cells were incubated with anti-H7N9 HA mouse monoclonal antibody (Sino Biological Inc., Cat# 11082-MM04) at a 1:400 dilution for 2h at room temperature. The slides were then incubated with the Alexa 555-conjugated anti-mouse secondary antibody (Cell Signaling Technology) for 60 min in the dark, and analyzed by fluorescent microscopy (Leica DM4000B, Leica Microsystems Inc, USA) using the SPOT Advanced software program (SPOT™ Diagnostic Instruments, Inc).
2.4. Immunization of Mice
Female 8-week-old BALB/c mice were purchased from Jackson Laboratory. Their care was in accordance with the guidelines of the National Institutes of Health and the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC).
Mice were immunized with 25 ug of pH7HA by intramuscular injection (IM) into the quadriceps muscle followed by in vivo electroporation (EP) using the CELLECTRA® adaptive constant current electroporation device (Inovio Pharmaceuticals Inc.). Two 0.1 Amp constant current square-wave pulses were delivered through a triangular 3-electrode array consisting of 26-gauge solid stainless steel electrodes. Each pulse was 52 milliseconds in length with a 1 second delay between pulses. The mice received two immunizations, three weeks apart. Serum samples were collected pre-immunization and two weeks after the second vaccination time point for the ELISA and Hemagglutination Inhibition (HAI) assays. Four weeks after the second immunization, the mice were sacrificed for analysis of cellular immune responses.
2.5. H7N9 HA-specific Antibody Determination
The measurement of IgG antibodies specific for H7HA was performed by ELISA in both immunized and control mice. The plates were coated with 0.5 μg/ml of H7N9 A/Shanghai/1/2013 influenza HA protein, A/Anhui/1/2013 influenza HA1 protein and A/Hangzhou/1/2013 HA1 protein (Sino Biological Inc.), respectively, and standard ELISA was carried out [27]. Endpoint titers were determined as previously described [36]. The mathematical formula used to calculate the endpoint titer cutoffs is expressed as the standard deviation multiplied by a factor that was based on the number of negative controls (naïve sera) (n = 10) and the confidence level (95%). The endpoint titer is reported as the reciprocal of the last dilution that remained above the endpoint cutoff.
2.6. Hemagglutination Inhibition Assay
Sera samples were treated with receptor-destroying enzyme (RDE, 1:3 ratio) at 37°C overnight for 18-20 hours followed by complement inactivation at 56°C for 45 minutes. Starting with a 1:10 dilution in PBS, twofold serial dilutions of RDE-treated serum were serially diluted down on 96-well V-bottom microtiter plates. Four hemagglutinating dose of A/Anhui/1/13 was added to each well and the serum-virus mixture were incubated at room temperature for 1.5 h. Following incubation, 50 μl horse red blood cells (1% cells + 0.5% Bovine Serum Albumin Fraction V in 0.85% saline solution) were added to each well and incubated for 1.25 h at room temperature. The HAI antibody titer was scored as the reciprocal of the highest dilution that did not exhibit agglutination of red blood cells. Each assay was performed in duplicate.
2.7. IFN-γELISpot assay
Mouse IFN-γ ELISpot assay was performed as described previously [35]. A set of peptides spanning the entire consensus H7HA protein, each containing 15 amino acid residues overlapping by 8 amino acids, were synthesized from GenScript (Piscataway, NJ). The entire set of peptides was pooled at a concentration of 2 ug/ml/peptide into 4 pools as antigens for specific stimulation of the IFN-γ release. Concavalin A (Sigma–Aldrich, St. Louis, MO), at 5 ug/ml, and complete culture medium were used as positive and negative control, respectively. The average number of spot forming cells (SFC) was adjusted to 1 × 106 splenocytes.
2.8. Intracellular Cytokine stain for Flow Cytometry
Splenocytes were added to a 96-well plate (1×106/well) and were stimulated with H7 peptide for 5-6 hours at 37°C/5% CO2 in the presence of Protein Transport Inhibitor Cocktail (Brefeldin A and Monensin) (ebioscience) according to the manufacturers’ instructions. The Cell Stimulation Cocktail (plus protein transport inhibitors) (phorbol 12-myristate 13-acetate (PMA), ionomycin, brefeldin A and monensin) (ebioscience) was used as a positive control and R10 media for as negative control. In cultures being used to measure degranulation, anti-CD107a (FITC; clone 1D4B; Biolegend) was added at this time to enhance staining. The cells were then fixed and stained as described as elsewhere [37]. Briefly, the cells were washed in FACS buffer (PBS containing 0.1% sodium azide and 1% FCS) before surface staining with flourochrome-conjugated antibodies. Cells were washed with FACS buffer fixation and permeabilization using the BD Cytofix/Ctyoperm™ (BD, San Diego, CA, USA) according to the manufacturer’s protocol followed by intracellular staining. The following antibodies were used for surface staining: LIVE/DEAD Fixable Violet Dead Cell stain kit (Invitrogen), CD19 (V50; clone 1D3; BD Biosciences) CD4 (V500; clone RM4-5; BD Biosciences), CD8 (PE-TexasRed; clone 53-6.7; Abcam), CD44 (A700; clone IM7; Biolegend). For intracellular staining the following antibodies were used: IFN-γ (APC; clone XMG1.2; Biolegend), TNF-α (PE; clone MP6-XT22; ebioscience), IL-2 (PeCy7; clone JES6-SH4; ebioscience), CD3 (PerCP/Cy5.5; clone 145-2C11; Biolegend). All data were collected using a LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star, Ashland, OR) and SPICE v5.2 (free available from http://exon.niaid.nih.gov/spice/). Boolean gating was performed using FlowJo software to examine the polyfunctionality of the T cells from vaccinated animals. For flow cytometry, cells were gated on singlets using FSC-H by FSC-A followed by gating on LIVE-DEAD (dump channel), CD3+ CD4+ CD8− T and CD3+ CD8+ CD4− T cells to examine the CD4+ and CD8+ T-cell populations.
2.9. Virus Challenge in Mice
Twenty female BALB/c mice were divided into two groups (n=10): the naïve and immunized group. The mice in the immunized group were immunized with 25 μg of pH7HA twice, three weeks apart. Four weeks after the second immunization, the mice were anesthetized with isoflurane and subsequently challenged by intranasal administration (bolus delivery into the nostrils using a standard micropipette) of 100 × LD50 of A/Anhui/1/13 influenza virus in 50 μl Dulbecco’s Modified Eagle Medium (DMEM) plus 2% Fetal Bovine Serum (FBS). After challenge, the animals were weighed daily for 14 days and monitored for clinical signs of influenza infection using an approved scoring sheet. All surviving animals were monitored for a total of 28 days. All procedures and the scoring method were approved by the Institutional Animal Care Committee at the National Microbiology Laboratory (NML) of the Public Health Agency of Canada (PHAC) according to the guidelines of the Canadian Council on Animal Care. All infectious work was performed in the ‘Biosafety Level 4’ (BSL4) facility.
2.10. Statistical analysis
Standard and paired student’s t-tests were applied to analyze statistical significance of all quantitative data produced in this study. A p < 0.05 was considered statistically significant.
3. Results
3.1. Development of a novel H7N9 HA DNA immunogen
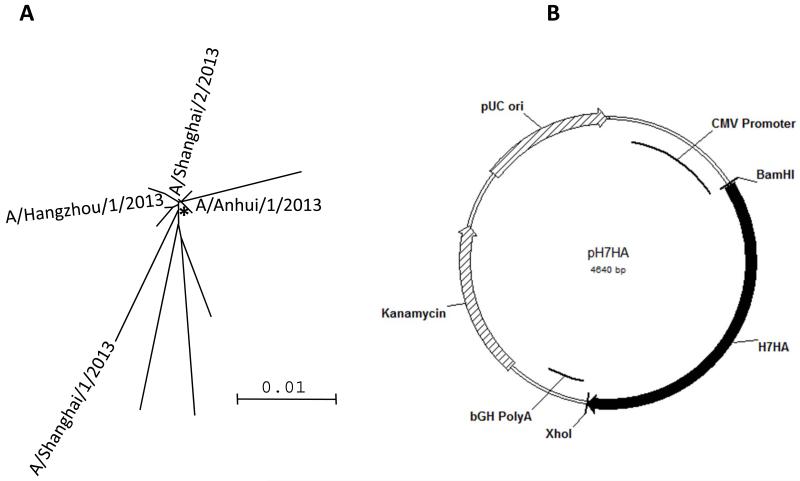
The H7N9 HA consensus sequence was generated from the first four H7N9 HA sequences isolated from humans published in GISAID (The Global Initiative on Sharing All Influenza Data). Phylogenetic analysis indicated that the sequence identity between any two H7 HA proteins of these human isolates could be as low as 95.9%, while the identities between the consensus HA and primary HA proteins were 97.5% and above. The consensus approach to antigen development for influenza, therefore, is not dependent upon any one viral isolate that may not represent the totality of the pandemic in real time. The relevant placement of the consensus H7 HA sequence is indicated in Fig. 1A. After generating the consensus HA sequence, codon and RNA optimizations were performed, as previously described [27]. The synthetic H7HA gene, which is 1683 bp in length, was synthesized, sequence verified, and was subcloned into the expression vector pGX0001 at BamHI and NotI sites and named pH7HA (Fig. 1B) in just a few days.
Figure 1.
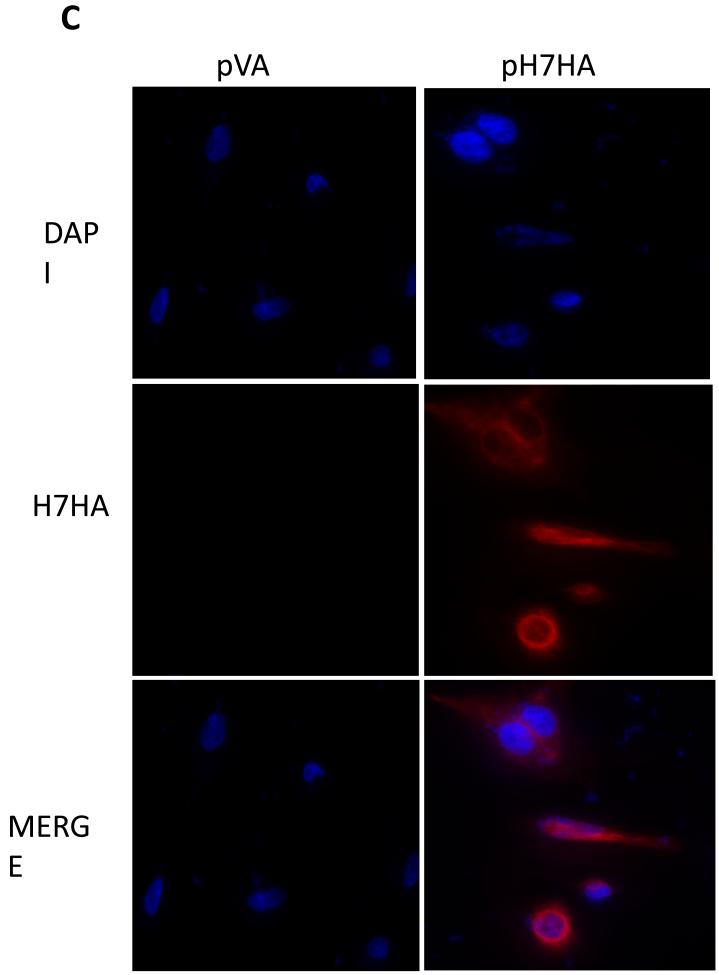
The H7N9 HA DNA vaccine design and its expression (A) Phylogenetic tree based on neighbor-joining evaluation of H7HA alignment. Asterisks indicate location of consensus sequence. The strains used to generate the consensus HA are indicated. (B) The plasmid map of the H7HA plasmid. (C) Immunofluorescence assay of pH7HA. The transfected RD cells expressing H7HA protein showed typical red fluorescence. An anti-H7N9 HA mouse monoclonal antibody served as the source of primary antibody.
3.2. Expression of pH7HA DNA vaccine
To determine the expression of the H7HA construct, an indirect immunofluorescent assay was carried out using an anti-H7N9 HA mouse monoclonal antibody. High membrane expression was observed by fluorescent microscopy in the pH7HA-transfected cells (Fig. 1C), supporting the idea that the expressed HA protein was a surface protein and the protein exhibited a relatively native conformation. As a control, expression was not detected in pGX0001 (null vector)-transfected cells.
3.3. Induction of cross-reactive antibodies by H7HA DNA immunogen
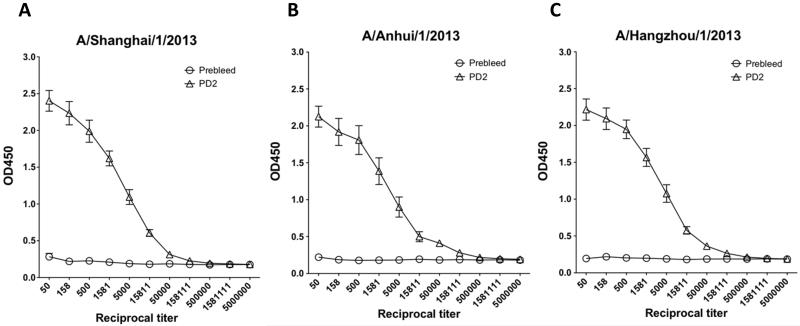
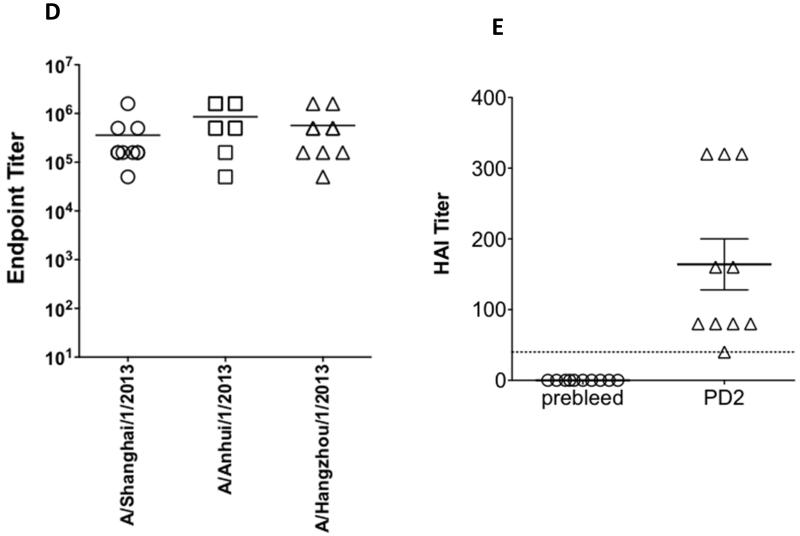
Upon confirming expression of the pH7HA construct, we first investigated whether this immunogen was capable of driving cross-reactive antibody responses. Mice were immunized twice, three weeks apart, with pH7HA and the serum samples collected at pre-vaccination and at two weeks post the second immunization were utilized for the ELISA assay. As shown in Fig. 2A, 2B and 2C, we were able to detect high-titer vaccine-induced antibody responses against three different HA proteins in the immunized mice. The average HA-specific antibody endpoint titers against A/Shanghai/1/2013 HA, A/Anhui/1/2013 HA1 and A/Hangzhou/1/2013 HA1 were 222,074 (range 50,000-1,581,111), 368,019 (range 50,000-1,581,111) and 315,542 (range 50,000-1,581,111), respectively (Fig. 2D). These data indicated that vaccination with pH7HA elicited strong and broad antibody responses.
Figure 2.
Robust HA-specific IgG antibody titers and hemagglutination-inhibition titers in the sera of the immunized mice. (A) IgG antibodies against H7N9 A/Shanghai/1/2013 influenza HA protein. (B) IgG antibodies against H7N9 A/Anhui/1/2013 influenza HA protein. (C) IgG antibodies against H7N9 A/Hangzhou/1/2013 influenza HA protein. (D) HA-specific IgG endpoint titers. (E) Hemagglutination-inhibition titers against A/Anhui/1/2013. Each BALB/c mouse was immunized intramuscularly followed by electroporation with 25 μg of pH7HA DNA twice, three weeks apart. Mice (n=10) were bled before and two weeks after second immunization. IgG antibody titers and hemagglutination-inhibition titers were measured by endpoint enzyme-linked immunosorbent assay (ELISA) and hemagglutination-inhibition assay, respectively. Error bars represent 1 standard deviation from the mean.
3.4. Induction of protective antibodies by H7HA DNA immunogen
In order to test whether the cross-reactive antibodies may be protective, we next employed a standard Hemagglutination Inhibition assay (HAI) to test the ability of the pH7N9 plasmid to generate functional antibodies with HAI activity. Due to the timing of the emergence of the H7N9 strains and our animal experiments, only the A/Anhui/1/2013 H7N9 strain was quantified at the resource lab and available for this test. Accordingly it was used to determine a serum HAI antibody titer induced by the vaccination. As depicted in Fig. 2E, two weeks after two immunizations, pH7HA induced protective HAI titers (≥1:40) in every immunized animal (n=10) with a GMT titer of 1:130 in the pH7HA-immunized mice.
3.5. Induction of potent antigen-specific effector T cell memory responses
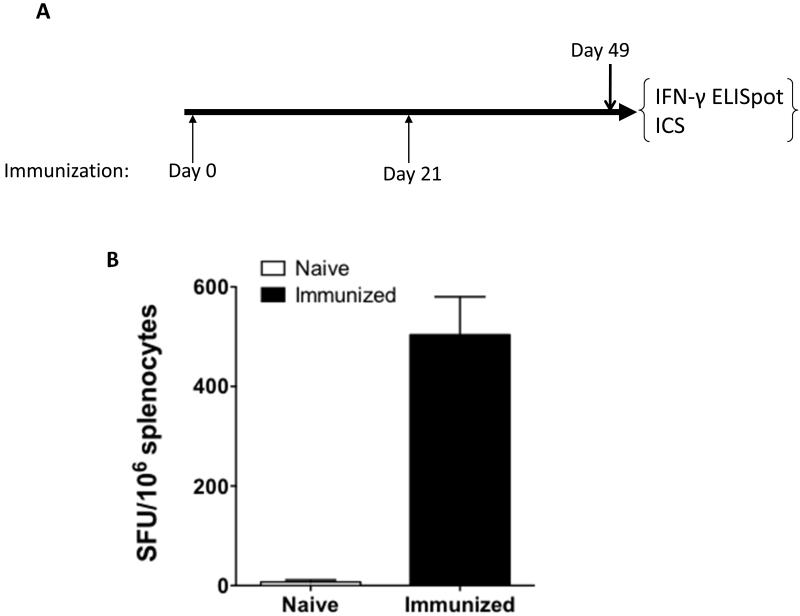
It is suggested that cell mediated immunity is crucial to virus clearance, with the number of IFN-γ secreting cells correlating with the efficacy of live, attenuated influenza vaccine in children [38]. Therefore, after confirming pH7HA elicited protective antibody responses, we explored pH7HA’s ability to induce antigen-specific cellular immune responses. C57BL/6 mice were immunized twice with pH7HA, sacrificed four weeks post the second immunization (Fig. 3A), and the IFN-γ ELISpot assay was performed. As shown in Fig. 3B, the average response against four pools of H7HA overlapping peptides in the mice immunized with pH7HA was 504 ± 132 SFU/106 splenocytes, while minimal background spots were observed in naïve mice. Clearly, strong IFN-γ responses were induced by vaccination with pH7HA.
Figure 3.
HA-specific cellular immune response induced in the immunized mice. (A) Immunization schedule. (B) Total IFN-γ responses induced by pH7HA. Frequencies of HA-specific IFN-γ-secreting cells per million splenocytes after two DNA immunizations with pH7HA were determined by IFN-γ ELISpot assay. The splenocytes were isolated from each mouse (n=5), stimulated in vitro with four overlapping HA peptide pools for 24 h and IFN-γ secreting cells were determined by ELISpot assay. Naïve mice were included as a negative control. The values are the means ± standard error of the means.
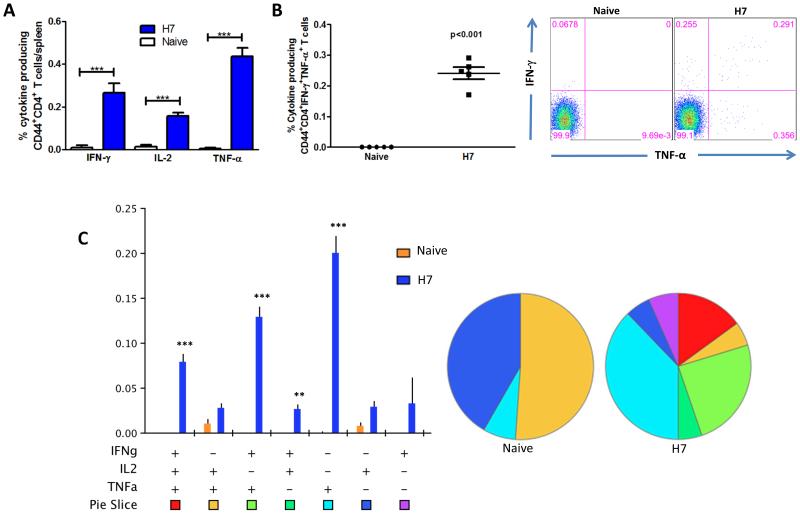
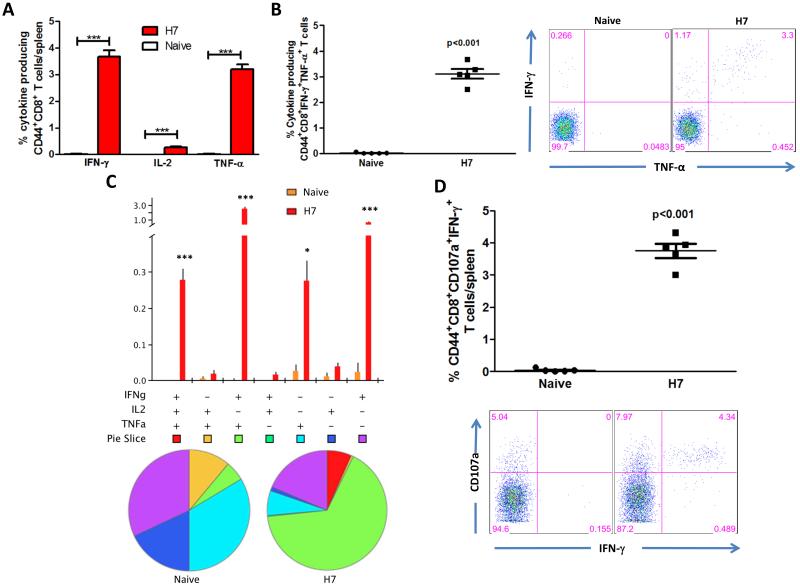
Subsequently, we characterized the phenotype and cytokine profile production of the memory T cells generated. Herein, we studied the longevity and quality of CD4 and CD8 effector T cell memory induced by the H7HA DNA vaccine. Four weeks after the last vaccination, mice were sacrificed and splenocytes were stimulated in vitro with H7HA pooled peptides and the production of IFN-γ, TNF-α, and IL-2 by CD4+ CD44+ and CD8+ CD44+ T cells was analyzed. The H7HA DNA vaccine elicited significant HA-specific CD4+ T cells producing all three cytokines (Fig. 4A), and a significant number of these cells were double positive (67%) and triple-positive (7%) (Figs. 4B-C). The triple positive and TNFα+IL2+ double positive T cell phenotypes normally represent effector memory and central memory T cells (2), indicating the induction of a memory like CD4 T cell immune response by our HA vaccine. In terms of CD8 T cells, we found that vaccination with pH7HA elicited substantially higher frequencies of HA-specific IFNγ+CD8 T cells, with up to 3.7% of total splenic CD44+CD8+ T cells (Fig. 5A) producing either IFN-γ+ alone (0.76%), dual IFNγ+/TNFα+ (2.7%) or triple IFNγ+/IL-2+/TNFα+ (0.28%) (Figs. 5A-C). To further characterize the vaccine-induced T cells, we next analyzed the cytotoxic potential of the induced CD8+ T cells undergoing degranulation. Therefore, cultures were stained with an antibody to CD107a, which is a marker for degranulation, and we observed that CD8 T cells from the vaccinated mice showed a significant increase of antigen-specific (IFNγ+CD107a+: 3.8%) degranulation compared to naïve mice. These results are indicative of the vaccine’s potent ability to induce cytolytic T cell responses with the potential ability to clear H7N9 influenza infected cells. In addition, the vaccine also induced a high frequency of TNF-α producing HA-specific CD4 (0.4%) and CD8 T cells (3.2%), with a modest but significant increase in IL-2 responses (0.2% and 0.3%, respectively) compared to the naïve group (Fig. 4A and 5A). Interestingly, the proportional order of effector CD4 and CD8 T cell subpopulations in response to H7 HA stimulation was similar, with IFNγ+/TNFα+ being greater than IFNγ+/IL2+/TNFα+. The high frequencies of effector cells secreting IFNγ (CD8: 3.7%; CD4: 0.3%), IFNγ+/TNFα+ (CD8: 3.1%; CD4: 0.24%), and IFNγ+/IL2+/TNFα+ among the HA-specific CD4 and CD8 T cell populations are indicative of strong vaccine potency induced by pH7HA vaccination (Figs. 4 & 5). Overall, the frequency hierarchy of effector CD4 and CD8 memory T cells expressing one (1+), any combination of two (2+), or all three cytokine (3+) by flow cytometry were CD4+CD44+: 1+ (26%), 2+ (67%), 3+ (7%) and CD8+CD44+: 1+ (50%), 2+ (35%), 3+ (15%). Taken together, we observed that strong effector and memory T cells, as well as protective antibody responses induced by this vaccination approach. The magnitude of the T cell responses are similar to those induced by viral infection rather than weak responses which are generated by traditional DNA immunization [39].
Figure 4.
Cytokine frequencies and phenotypic profiles of specific CD4 T cells after pH7HA immunization. Cytokine recall responses to H7 HA were measured 4 weeks after last immunization by ICS and flow cytometry. CD4 T cells were identified by CD3 expression and further gated as CD44+. (A) The percentage of total CD44+CD4 T cells expressing IFN-γ in response to H7 HA stimulation. (B) Average percentage of HA-specific CD44+CD4 double-positive-producing cells. (C) Multiparameter flow cytometry was used to determine the percentages of multifunctional CD4 T cell cytokine profile of H7HA. The bar chart shows the percentage of specific CD44+CD4 T cells displayed as triple, double, or single positive CD4 T cells. Pie charts show the relative proportion of each cytokine subpopulation to H7 HA stimulation. Background staining from cells stimulated with medium alone has been subtracted. Data represent the mean±SEM of five mice per group with ***P < 0.001, **P < 0.01, *P < 0.05 using Student’s t-test. Figure 5. Cytokine frequencies and phenotypic profiles of specific CD8 T cells after pH7HA immunization.
Figure 5.
Cytokine frequencies and phenotypic profiles of specific CD8 T cells after pH7HA immunization. Cytokine recall responses to H7 HA were measured 4 weeks after last immunization by ICS and flow cytometry. CD8 T cells were identified by CD3 expression and further gated as CD44+. (A) The percentage of total CD44+CD8 T cells expressing IFN-γ in response to H7 HA stimulation. (B) Average percentage of HA-specific CD44+CD8 double-positive-producing cells. (C) Multiparameter flow cytometry was used to determine the percentages of multifunctional CD8 T cell cytokine profile of H7HA. The bar chart shows the percentage of specific CD44+CD8 T cells displayed as triple, double, or single positive CD8 T cells. Pie charts show the relative proportion of each cytokine subpopulation to H7 HA stimulation. (D) Antigen-specific cytolytic degranulation T cells were measured by degranulation marker expression, CD107a and IFN-γ. Background staining from cells stimulated with medium alone has been subtracted. Data represent the mean±SEM of five mice per group with ***P < 0.001, **P < 0.01, *P < 0.05 using Student’s t-test.
3.6. Vaccination with pH7HA elicits complete protection against H7N9 virus challenge
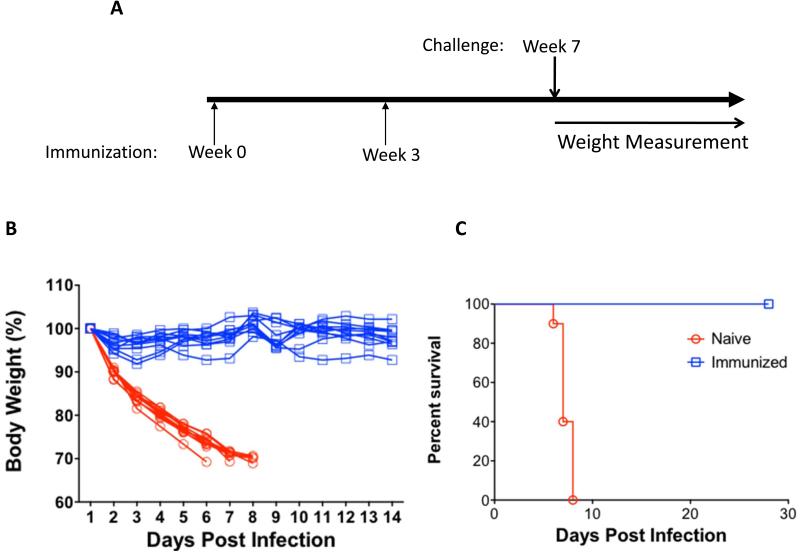
To determine if vaccination with pH7HA could elicit protective immunity to a lethal viral challenge, mice were immunized with pH7HA twice, three weeks apart, then challenged with a lethal dose of the H7N9 virus A/Anhui/1/2013 four weeks post the final immunization, and then monitored for weight loss for 14 days (Fig. 6A). According to the protocol established prior to the study, animals falling below the threshold of 70% of their initial body weight were humanely euthanized. As shown in Fig. 6B and 6C, in naïve group, one mouse lost 30% of its body weight by day 6 post-challenge, five mice exhibited 30% weight loss by day 7 post-challenge, and the rest of four mice lost 30% of their body weight by day 8. In contrast, all vaccinated mice (n=10) survived to day 28 post challenge with no observed pathogenic effects or weight loss at which time the study was ended. These data support that the synthetic DNA vaccine-induced immune responses could protect against influenza H7N9 infection.
Figure 6.
Protection from H7N9 A/Anhui/1/2013 virus challenge in the immunized mice. (A) Experimental schedule of challenge study. The mice (n=10) were immunized with 25 μg of pH7HA twice, three weeks apart. Four weeks after the second immunization, the mice were challenged intranasally with a lethal dose of A/Anhui/1/2013 H7N9 virus and monitored daily for weight loss and mortality. (B) Weight loss of each individual surviving mice in both naïve and immunized groups. The data are plotted as percentage of the weight on day 1. (C) Kaplan-Meier survival curve showing the percent survival following challenge. All surviving animals were monitored for a total of 28 days.
4. Discussion
The emergence of deadly H7N9 influenza virus in Eastern China invokes questions about the origins of influenza pandemics, the risk to humans of avian influenza viruses, and their potential ability to infect humans and be stably transmitted by humans. In the current situation the entire population is at risk and the potential for a severe pandemic is of great concern. Our study details the rapid development of a synthetic H7N9 HA DNA vaccine efficiently delivered in vivo by EP capable of eliciting not only strong effector T cell memory responses, but also broadly reactive antibody responses. More importantly, we confirmed that immunization with the pH7HA DNA vaccine induced protective HAI titers in all immunized animals and resulted in 100% protection from mortality and morbidity due to influenza H7N9 infection. To date, this is the first report demonstrating that an H7N9 HA DNA vaccine can protect against this newly emergent influenza subtype.
Current vaccination platforms require significant development and production timelines to address pandemics. Given the mortality rate of H7N9 and its potential to infect a large number of humans, it is important to develop strategies that can thwart a potential growing pandemic. An ideal influenza vaccine platform for rapid deployment should include: 1) technologies that can be rapidly and straightforwardly utilized to design an effective vaccine against multiple known and new strains, 2) a delivery mechanism that can quickly induce cellular and humoral immune responses against new antigens in humans, 3) an approach that is rapid from design to scale up and mass production, with a relevant clinical track record and 4) an approach that is very stable for distribution purposes. Here we show that the enhanced DNA approach can exhibit these properties using the H7N9 pandemic as a demonstration platform. From computer immunogen design to having the immunogen in hand for animal studies is as short as 2-3 days under a pandemic setting. Immunization data is available in three weeks of this receipt. Seed stock is generated in just 3-4 days without any amplification or selection required thus limiting potential mutations being induced in the process of immunogen development. Distribution for production can begin as early as 5 days post pathogen detection. Using this approach, millions of doses could be produced at scale within a few months of identification of the pathogen [29, 40]. In addition the stability of DNA and the lack of requirement of any viral amplification step ensure fidelity in the process. Furthermore the growing safety record of DNA in the clinic in adults, the elderly and children all are important in suggesting the potential of this improved potency platform [41].
The surface HA protein of the influenza virus contains two structural elements (head and stalk) that differ in their potential utility as vaccine targets. Antibodies targeting areas of the HA globular head region (HA1) can drive HAI activity and viral neutralization. In this study, due to the limited availability of H7N9 virus strains, we tested HAI titers against A/Anhui/1/2013. However, high-titer cross-reactive binding antibodies against A/Anhui/1/2013 HA1 and A/Hangzhou/1/2013 HA1 regions were confirmed in all vaccinated mice. High titer binding was also observed against A/Shanghai/2013. Therefore, vaccination with pH7HA DNA vaccine appears to induce broadly reactive antibodies against multiple H7N9 strains. Efforts to perform additional HAI assays against more H7N9 virus strains are ongoing as stocks become available. However, from our past research on the H5 influenza model system [42], and the data presented for the H7 vaccine, it appears that breath of HAI would be similar.
HAI antibody is the major correlate of protection for flu vaccines. Although all immunized mice exhibited greater than 1:40 protective HAI titer in this study. Older designed DNA vaccines generally did not elicit workable levels of antibody in humans [22, 23]. Recently, by using a similar combination of approaches in gene optimization and delivery, we have successfully demonstrated that all human subjects immunized with a novel HPV E6/E7 DNA vaccine could induce strong de novo anti-HPV antibody responses, and the antibody titers remained high even 6 months after the last immunization [33]. The strong and long-lasting antibodies measured in this clinical study are encouraging for the development of HAI antibody dependent influenza DNA vaccines. Furthermore, these studies suggest that a potential human dose could be in the range of just 500 μg to 1 mg per immunization.
Cellular immune responses play an important role in influenza virus infection [43]. Many studies have shown that CD8+ T cells are induced following infection with influenza virus and play a critical role in rapid clearance of influenza virus, thus limiting pathogenesis [44, 45]. CD4+ T cells are also induced following influenza virus infection and play a role in maintaining CD8+ T cell memory response and providing help for B cells for rapid antibody production [46, 47]. Previous studies have identified 113 CD4 T cell and 35 CD8 T cell epitopes within the HA antigen [48] and some of these epitopes have been shown to confer protection in humans [49, 50]. The current study showed pH7HA could induce both potent polyfunctional effector CD4 and CD8 T cell memory responses. The percentage of antigen-specific CD8 cells secreting IFNγ+CD107a+ increased significantly, indicating potential ability of this vaccine to induce cytolytic T cell responses to clear influenza H7N9 infected cells. These data support the idea that the synthetic vaccine could induce both antigen-specific antibody and T cell responses, which would be important for protection during a pandemic.
Herein we report on the development of a highly potent, synthetic, H7N9 HA DNA vaccine which in combination with an adaptive constant current electroporation delivery platform is capable of eliciting robust cellular immune responses, broadly cross-reactive antibody responses and generating complete protection from lethal challenge with just a few week development and vaccination regime. The potent synthetic DNA vaccine platform eliminates many of hurdles that limited the rapid development and deployment of a vaccine against an emerging pandemic. The lack of a requirement for any recombination or in vivo production, as with some recent competing technologies, both quickens the production time line as well as eliminates a major potential source of errors in seed development. New production methods for DNA are further shortening this production timeline with a platform exhibiting a growing safety record in the clinic. Further study of this new platform for development of vaccines to combat pandemic infections, including influenza is warranted.
Acknowledgments
DBW notes funding from NIH including Flu (R01 AI092843) and HVDDT (U19 AI078675) and SRA funding to DBW laboratory for Consensus Immunogen Design. J.N.W. notes funding from Ruth L. Kirschstein National Research Service Award (5T32CA115299-07).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Puzelli S, Di Trani L, Fabiani C, Campitelli L, De Marco MA, Capua I, et al. Serological analysis of serum samples from humans exposed to avian H7 influenza viruses in Italy between 1999 and 2003. J Infect Dis. 2005;192:1318–22. doi: 10.1086/444390. [DOI] [PubMed] [Google Scholar]
- [2].Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–93. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- [3].Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101:1356–61. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pasick J, Berhane Y, Hooper-McGrevy K. Avian influenza: the Canadian experience. Rev Sci Tech. 2009;28:349–58. doi: 10.20506/rst.28.1.1875. [DOI] [PubMed] [Google Scholar]
- [5].Nguyen-Van-Tam JS, Nair P, Acheson P, Baker A, Barker M, Bracebridge S, et al. Outbreak of low pathogenicity H7N3 avian influenza in UK, including associated case of human conjunctivitis. Euro Surveill. 2006;11:E060504 2. doi: 10.2807/esw.11.18.02952-en. [DOI] [PubMed] [Google Scholar]
- [6].Trock SC, Huntley JP. Surveillance and control of avian influenza in the New York live bird markets. Avian Dis. 2010;54:340–4. doi: 10.1637/8728-032409-ResNote.1. [DOI] [PubMed] [Google Scholar]
- [7].Kapczynski DR, Pantin-Jackwood M, Guzman SG, Ricardez Y, Spackman E, Bertran K, et al. Characterization of the 2012 Highly Pathogenic Avian Influenza H7N3 virus isolated from poultry in an outbreak in Mexico: Pathobiology and Vaccine Protection. J Virol. 2013 doi: 10.1128/JVI.00666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Belser JA, Bridges CB, Katz JM, Tumpey TM. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg Infect Dis. 2009;15:859–65. doi: 10.3201/eid1506.090072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Morens DM, Taubenberger JK, Fauci AS. H7N9 Avian Influenza A Virus and the Perpetual Challenge of Potential Human Pandemicity. MBio. 2013;4 doi: 10.1128/mBio.00445-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381:1916–25. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wu S, Wu F, He J. Emerging risk of H7N9 influenza in China. Lancet. 2013;381:1539–40. doi: 10.1016/S0140-6736(13)60767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog. 2010;6:e1001081. doi: 10.1371/journal.ppat.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Xiong X, Martin SR, Haire LF, Wharton SA, Daniels RS, Bennett MS, et al. Receptor binding by an H7N9 influenza virus from humans. Nature. 2013 doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- [14].Matsuoka Y, Swayne DE, Thomas C, Rameix-Welti MA, Naffakh N, Warnes C, et al. Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza H5N1 viruses for mice. J Virol. 2009;83:4704–8. doi: 10.1128/JVI.01987-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–2. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- [16].Zhu H, Wang D, Kelvin DJ, Li L, Zheng Z, Yoon SW, et al. Infectivity, Transmission, and Pathology of Human-Isolated H7N9 Influenza Virus in Ferrets and Pigs. Science. 2013 doi: 10.1126/science.1239844. [DOI] [PubMed] [Google Scholar]
- [17].Zhou J, Wang D, Gao R, Zhao B, Song J, Qi X, et al. Biological features of novel avian influenza A (H7N9) virus. Nature. 2013 doi: 10.1038/nature12379. [DOI] [PubMed] [Google Scholar]
- [18].Martin T, Parker SE, Hedstrom R, Le T, Hoffman SL, Norman J, et al. Plasmid DNA malaria vaccine: the potential for genomic integration after intramuscular injection. Hum Gene Ther. 1999;10:759–68. doi: 10.1089/10430349950018517. [DOI] [PubMed] [Google Scholar]
- [19].Chattergoon M, Boyer J, Weiner DB. Genetic immunization: a new era in vaccines and immune therapeutics. Faseb J. 1997;11:753–63. doi: 10.1096/fasebj.11.10.9271360. [DOI] [PubMed] [Google Scholar]
- [20].Hokey DA, Weiner DB. DNA vaccines for HIV: challenges and opportunities. Springer Semin Immunopathol. 2006;28:267–79. doi: 10.1007/s00281-006-0046-z. [DOI] [PubMed] [Google Scholar]
- [21].Ferraro B, Cisper NJ, Talbott KT, Philipson-Weiner L, Lucke CE, Khan AS, et al. Co-delivery of PSA and PSMA DNA vaccines with electroporation induces potent immune responses. Hum Vaccin. 2011;7(Suppl):120–7. doi: 10.4161/hv.7.0.14574. [DOI] [PubMed] [Google Scholar]
- [22].Kennedy JS, Co M, Green S, Longtine K, Longtine J, O’Neill MA, et al. The safety and tolerability of an HIV-1 DNA prime-protein boost vaccine (DP6-001) in healthy adult volunteers. Vaccine. 2008;26:4420–4. doi: 10.1016/j.vaccine.2008.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Trimble CL, Peng S, Kos F, Gravitt P, Viscidi R, Sugar E, et al. A phase I trial of a human papillomavirus DNA vaccine for HPV16+ cervical intraepithelial neoplasia 2/3. Clin Cancer Res. 2009;15:361–7. doi: 10.1158/1078-0432.CCR-08-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Deml L, Bojak A, Steck S, Graf M, Wild J, Schirmbeck R, et al. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J Virol. 2001;75:10991–1001. doi: 10.1128/JVI.75.22.10991-11001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Muthumani K, Zhang D, Dayes NS, Hwang DS, Calarota SA, Choo AY, et al. Novel engineered HIV-1 East African Clade-A gp160 plasmid construct induces strong humoral and cell-mediated immune responses in vivo. Virology. 2003;314:134–46. doi: 10.1016/s0042-6822(03)00459-8. [DOI] [PubMed] [Google Scholar]
- [26].Schneider R, Campbell M, Nasioulas G, Felber BK, Pavlakis GN. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Revindependent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yan J, Yoon H, Kumar S, Ramanathan MP, Corbitt N, Kutzler M, et al. Enhanced cellular immune responses elicited by an engineered HIV-1 subtype B consensusbased envelope DNA vaccine. Mol Ther. 2007;15:411–21. doi: 10.1038/sj.mt.6300036. [DOI] [PubMed] [Google Scholar]
- [28].Laddy DJ, Yan J, Corbitt N, Kobasa D, Kobinger GP, Weiner DB. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine. 2007;25:2984–9. doi: 10.1016/j.vaccine.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cai Y, Rodriguez S, Rameswaran R, Draghia-Akli R, Juba RJ, Jr., Hebel H. Production of pharmaceutical-grade plasmids at high concentration and high supercoiled percentage. Vaccine. 2010;28:2046–52. doi: 10.1016/j.vaccine.2009.10.057. [DOI] [PubMed] [Google Scholar]
- [30].Sardesai NY, Weiner DB. Electroporation delivery of DNA vaccines: prospects for success. Curr Opin Immunol. 2011;23:421–9. doi: 10.1016/j.coi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hirao LA, Wu L, Khan AS, Satishchandran A, Draghia-Akli R, Weiner DB. Intradermal/subcutaneous immunization by electroporation improves plasmid vaccine delivery and potency in pigs and rhesus macaques. Vaccine. 2008;26:440–8. doi: 10.1016/j.vaccine.2007.10.041. [DOI] [PubMed] [Google Scholar]
- [32].Hirao LA, Draghia-Akli R, Prigge JT, Yang M, Satishchandran A, Wu L, et al. Multivalent smallpox DNA vaccine delivered by intradermal electroporation drives protective immunity in nonhuman primates against lethal monkeypox challenge. J Infect Dis. 2011;203:95–102. doi: 10.1093/infdis/jiq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bagarazzi ML, Yan J, Morrow MP, Shen X, Parker RL, Lee JC, et al. Immunotherapy against HPV16/18 generates potent TH1 and cytotoxic cellular immune responses. Sci Transl Med. 2012;4:155ra38. doi: 10.1126/scitranslmed.3004414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kalams SA, Parker SD, Elizaga M, Metch B, Edupuganti S, Hural J, et al. Safety and Comparative Immunogenicity of an HIV-1 DNA Vaccine in Combination with Plasmid Interleukin 12 and Impact of Intramuscular Electroporation for Delivery. J Infect Dis. 2013 doi: 10.1093/infdis/jit236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yan J, Yoon H, Kumar K, Ramanathan MP, Corbitt N, Kutzler M, et al. Enhanced diversity and magnitude of cellular immune responses elicited by a novel engineered HIV-1 subtype B consensus-based envelope DNA vaccine. Molecular Therapy. 2007:411–21. doi: 10.1038/sj.mt.6300036. [DOI] [PubMed] [Google Scholar]
- [36].Frey A, Di Canzio J, Zurakowski D. A statistically defined endpoint titer determination method for immunoassays. J Immunol Methods. 1998;221:35–41. doi: 10.1016/s0022-1759(98)00170-7. [DOI] [PubMed] [Google Scholar]
- [37].Shedlock DJ, Aviles J, Talbott KT, Wong G, Wu SJ, Villarreal DO, et al. Induction of broad cytotoxic T cells by protective DNA vaccination against marburg and ebola. Mol Ther. 2013;21:1432–44. doi: 10.1038/mt.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Forrest BD, Pride MW, Dunning AJ, Capeding MR, Chotpitayasunondh T, Tam JS, et al. Correlation of cellular immune responses with protection against cultureconfirmed influenza virus in young children. Clin Vaccine Immunol. 2008;15:1042–53. doi: 10.1128/CVI.00397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shedlock DJ, Talbott KT, Cress C, Ferraro B, Tuyishme S, Mallilankaraman K, et al. A highly optimized DNA vaccine confers complete protective immunity against high-dose lethal lymphocytic choriomeningitis virus challenge. Vaccine. 2011;29:6755–62. doi: 10.1016/j.vaccine.2010.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cai Y, Rodriguez S, Hebel H. DNA vaccine manufacture: scale and quality. Expert Rev Vaccines. 2009;8:1277–91. doi: 10.1586/erv.09.84. [DOI] [PubMed] [Google Scholar]
- [41].Sheets RL, Stein J, Manetz TS, Andrews C, Bailer R, Rathmann J, et al. Toxicological safety evaluation of DNA plasmid vaccines against HIV-1, Ebola, Severe Acute Respiratory Syndrome, or West Nile virus is similar despite differing plasmid backbones or gene-inserts. Toxicol Sci. 2006;91:620–30. doi: 10.1093/toxsci/kfj170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Laddy DJ, Yan J, Khan AS, Andersen H, Cohn A, Greenhouse J, et al. Electroporation of synthetic DNA antigens offers protection in nonhuman primates challenged with highly pathogenic avian influenza virus. J Virol. 2009;83:4624–30. doi: 10.1128/JVI.02335-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Thomas PG, Keating R, Hulse-Post DJ, Doherty PC. Cell-mediated protection in influenza infection. Emerg Infect Dis. 2006;12:48–54. doi: 10.3201/eid1201.051237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Brincks EL, Katewa A, Kucaba TA, Griffith TS, Legge KL. CD8 T cells utilize TRAIL to control influenza virus infection. J Immunol. 2008;181:4918–25. doi: 10.4049/jimmunol.181.7.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Topham DJ, Tripp RA, Doherty PC. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J Immunol. 1997;159:5197–200. [PubMed] [Google Scholar]
- [46].Swain SL, Agrewala JN, Brown DM, Jelley-Gibbs DM, Golech S, Huston G, et al. CD4+ T-cell memory: generation and multi-faceted roles for CD4+ T cells in protective immunity to influenza. Immunol Rev. 2006;211:8–22. doi: 10.1111/j.0105-2896.2006.00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Brown DM, Roman E, Swain SL. CD4 T cell responses to influenza infection. Semin Immunol. 2004;16:171–7. doi: 10.1016/j.smim.2004.02.004. [DOI] [PubMed] [Google Scholar]
- [48].Bui HH, Peters B, Assarsson E, Mbawuike I, Sette A. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc Natl Acad Sci U S A. 2007;104:246–51. doi: 10.1073/pnas.0609330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lee LY, Ha do LA, Simmons C, de Jong MD, Chau NV, Schumacher R, et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest. 2008;118:3478–90. doi: 10.1172/JCI32460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gelder C, Davenport M, Barnardo M, Bourne T, Lamb J, Askonas B, et al. Six unrelated HLA-DR-matched adults recognize identical CD4+ T cell epitopes from influenza A haemagglutinin that are not simply peptides with high HLA-DR binding affinities. Int Immunol. 1998;10:211–22. doi: 10.1093/intimm/10.2.211. [DOI] [PubMed] [Google Scholar]