Abstract
Biological questions are increasingly being addressed using a wide range of quantitative analytical tools to examine protein complex composition. Knowledge of the absolute number of proteins present provides insights into organization, function, and maintenance and is used in mathematical modeling of complex cellular dynamics. In this chapter, we outline and describe three microscopy-based methods for determining absolute protein numbers—fluorescence correlation spectroscopy, stepwise photobleaching, and ratiometric comparison of fluorescence intensity to known standards. In addition, we discuss the various fluorescently labeled proteins that have been used as standards for both stepwise photobleaching and ratiometric comparison analysis. A detailed procedure for determining absolute protein number by ratiometric comparison is outlined in the second half of this chapter. Counting proteins by quantitative microscopy is a relatively simple yet very powerful analytical tool that will increase our understanding of protein complex composition.
INTRODUCTION
The intersection of physics and computational, molecular, and cellular biology reflects major changes in our approach to basic cell biological questions in the post-genome era. New strategies to beat the resolution limit in live cells, examine dynamic processes with speed and accuracy, and perform these genome-wide challenge cell biologists to make quantitatively accurate measurements. Determining the protein composition of complex dynamic structures is needed for a complete understanding of cellular function. Quantitative analysis of fluorescence microscopy images can provide absolute protein numbers and information regarding stoichiometry of protein complexes. Knowledge of the number of proteins present in a given complex is crucial for the development of structural and dynamic models of cellular processes. Here, we discuss three methods for determining absolute protein numbers using quantitative fluorescence microscopy and provide a step-by-step protocol for counting molecules by ratiometric comparison of fluorescence intensity.
19.1 METHODS FOR COUNTING MOLECULES
19.1.1 IMAGING AND MEASUREMENT CONSIDERATIONS
In order to obtain reliable and quantifiable images for analysis, some general considerations should be kept in mind. General microscope alignment and sample preparation concerns are discussed in greater detail elsewhere (Rottenfusser, 2013; Salmon et al., 2013; Waters, 2013). In order to accurately measure fluorescence intensity, it is essential to maximize the signal-to-noise ratio while also minimizing photobleaching. Microscope alignment, the objective lens, and the sample preparation contribute in large part to image quality. Proper alignment ensures even illumination across the field of view. The objective lens should have a high numerical aperture (NA) and be corrected for optical aberrations at a magnification level appropriate for the sample to obtain the greatest image intensity. For quantitative image acquisition in budding yeast, we acquire images on a widefield microscope with a 100× objective with an NA of at least 1.4. For proteins of interest in thicker specimens, it may be preferable to use a confocal microscope or total internal reflection (TIRF) microscopy to reduce out-of-focus light (Hallworth & Nichols, 2012; Joglekar, Bouck, et al., 2008; Ulbrich & Isacoff, 2007). The sample should be fluorescently labeled in a manner that ensures a consistent ratiometric relationship between fluorescent signal intensity and number of proteins of interest. This can be most easily achieved using a genetically encoded fluorophore that is both bright and stable (Douglass & Vale, 2008; Johnson & Straight, 2013; Xia, Li, & Fang, 2013). Imaging parameters should minimize sample photobleaching, and all methods discussed are very sensitive to loss of signal intensity due to unintended photobleaching during image acquisition (Coffman & Wu, 2012; Johnson & Straight, 2013). The detailed protocol that follows includes specific guidelines for optimization of image acquisition.
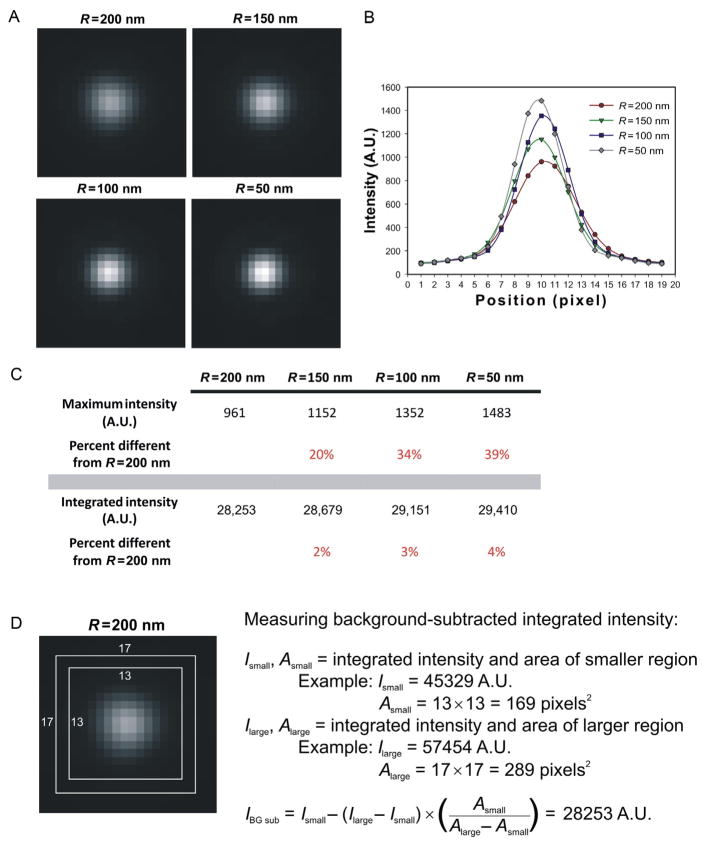
The details of postacquisition image analysis vary by method, but proper quantification of image intensity is universally important. The fluorescence intensity of a two-dimensional image can be measured from either the peak intensity of the spot (brightest pixel intensity) or the integrated intensity of the whole spot. We use integrated intensity for intensity quantification since this method does not assume a constant volume. When comparing multiple structures that differ in size and/or shape, measurement by integrated intensity will more accurately describe the intensity independent of fluorophore density (Fig. 19.1). Brightest pixel measurements will show a reduced signal intensity if a structure increases in size (reducing fluorophore density) and can result in misleading analysis of the number of fluorophores. It may also be necessary to sum intensity values of multiple z-planes if the structure of interest is larger than the resolution limit in z. For relatively small structures, such as yeast kinetochore spots, we acquire sufficiently closely spaced z-planes (with respect to the objective) to capture the in-focus image plane for analysis (Joglekar, Salmon, & Bloom, 2008). For larger structures, it may be necessary to use the sum intensity of multiple z-planes to fully capture the intensity (Coffman & Wu, 2012; Wu & Pollard, 2005). In addition to using integrated intensity measurements, it is important to correct for background fluorescence (Hoffman et al., 2001). This is done by measuring total integrated intensity of the region of interest and that of a slightly larger region and obtaining the background intensity value (Fig. 19.1D). This value is then subtracted to calculate the intensity of the spot of interest.
FIGURE 19.1.
Methods for measuring fluorescence intensity. (A) Simulated and convolved spheres of known subresolution diameters populated with a constant number of fluorophores (N=50) shown on the same intensity scale (generated using FluoroSim; Quammen et al., 2008). (B) Linescans through the brightest pixel of the simulated sphere images. The maximum intensity decreases as the size of the sphere is increased. (C) Comparison of maximum intensity and integrated intensity measurements. Integrated intensity values show a 4% difference between values measured for the largest and smallest spheres. For comparison, the maximum intensity values show an almost 40% difference. (D) The procedure for measuring background=corrected integrated intensity. Briefly, two square regions are drawn around the signal of interest and the integrated intensity values of these are recorded. Using the areas and integrated intensities of these squares, the final background=corrected integrated intensity can be calculated (Example shown is for the R=200 nm simulated sphere image from (A).).
19.1.2 FLUORESCENCE CORRELATION SPECTROSCOPY
Fluorescence correlation spectroscopy (FCS) is a microscopy method in which the fluorescence intensity arising from molecules within a small volume is collected over time and correlated to obtain information regarding dynamics and concentrations. This method can be applied in vivo and, like other fluorescence microscopy techniques, is nondestructive. FCS measurements are highly sensitive and can be done at the single-molecule level (Chen, Muller, Ruan, & Gratton, 2002). In principle, FCS measures the small changes in fluorescence intensity arising when a molecule enters the observation volume and the corresponding drop when it leaves (Braeckmans, Deschout, Demeester, & De Smedt, 2011; Bulseco & Wolf, 2013; Gosch & Rigler, 2005; Levin & Carson, 2004). Correlation analysis of the measured fluorescence intensity over time should reveal the concentration and diffusion rate of particles through the observation volume.
FCS experiments require a more specialized optical setup than stepwise photo-bleaching or ratiometric comparison of fluorescence intensity (Bacia & Schwille, 2003; Bulseco & Wolf, 2013; Haustein & Schwille, 2007). Recent advances in microscope detector sensitivity (photomultiplier tube or avalanche photodiode (APD)) have allowed for greater sensitivity and analysis in FCS experiments (Ries & Schwille, 2012; Tian, Martinez, & Pappas, 2011; Vukojevic et al., 2005). In contrast to a standard laser scanning confocal microscope, for FCS experiments, the laser beam position remains constant and the fluorescence intensity within the observation volume is measured over time. The confocal FCS observation volume is defined by the focusing of laser excitation light, and, as with typical confocal microscopes, apertures are used to reduce out-of-focus light (Bulseco & Wolf, 2013). For aligned and optimized confocal FCS microscope systems, the observation volume is approximately 0.5 fL and 600 nm in diameter (Bulseco & Wolf, 2013; Slaughter & Li, 2010).
FCS relies on the dynamic diffusion of particles through the observation volume, and this method is limited to measuring diffusion rates and molecule numbers for mobile samples. The length of observation is determined by the speed of particle diffusion and, as with the other techniques described here, it is important to consider and minimize photobleaching effects when choosing fluorophores and during image acquisition (Bacia & Schwille, 2003; Ries & Schwille, 2012). In addition to being limited to measuring mobile samples, FCS is best applied to certain concentration ranges (~1 fluorescent particle per observation volume), and concentrations that are too low or too high require very long observation times for reliable analysis (Enderlein, Gregor, Patra, & Fitter, 2004; Levin & Carson, 2004; Slaughter & Li, 2010).
Photon counting histogram (PCH) analysis (and fluorescence intensity distribution analysis) can be applied to the data to measure the absolute number of particles (Thompson, Lieto, & Allen, 2002). PCH analysis utilizes the fluorescence measurements observed within the observation volume and mathematically relates this intensity distribution to the number of molecules present (Chen, Muller, Berland, & Gratton, 1999; Chen, Muller, So, & Gratton, 1999; Kask, Palo, Ullmann, & Gall, 1999). FCS imaging within a small observation volume and PCH analysis have been used to generate a calibration curve relating brightness and absolute number of particles and compare these to experimental structures in vivo (Shivaraju et al., 2012; Slaughter, Huff, Wiegraebe, Schwartz, & Li, 2008).
19.1.3 STEPWISE PHOTOBLEACHING
The measurement of protein counts by observation of photobleaching dynamics has been applied to a wide range of biological systems to determine number and stoichiometry of protein subunits. This method captures the irreversible photobleaching of fluorophores fused to the protein of interest at single-molecule resolution. In addition to imaging considerations previously discussed, the experimental setup should be optimized to minimize photobleaching multiple fluorophores in the same event (Coffman & Wu, 2012; Hallworth & Nichols, 2012). This includes those considerations discussed in the preceding text and the incorporation of well-characterized control structures. A range of structures have been used as controls to assess the reliability of detection and analysis, including various membrane-bound channels and receptors, cytosolic fluorophores, or the bacterial flagellar motor MotB (Coffman, Wu, Parthun, & Wu, 2011; Leake et al., 2006; Padeganeh et al., 2013; Ulbrich & Isacoff, 2007).
There are two general approaches for the measurement of the number of proteins present in a given structure or complex by stepwise photobleaching—direct counting of photobleaching steps and the determination of the step size of a single photo-bleaching event to extrapolate the total number of fluorophores. The direct counting of photobleaching steps is most relatable for low protein numbers. The maximum number of steps that can be measured without additional mathematical extrapolation ranges from 5–7 to 15 steps (Das, Darshi, Cheley, Wallace, & Bayley, 2007; Ulbrich & Isacoff, 2007). The raw data can be further processed to enable detection of a larger number of photobleaching steps using a Chung–Kennedy filter or methods to detect changes in intensity state such as hidden Markov modeling (Chung & Kennedy, 1991; Engel, Ludington, & Marshall, 2009; McKinney, Joo, & Ha, 2006; Watkins & Yang, 2005). Alternatively, it is possible to measure the intensity drop corresponding to the photobleaching of one fluorophore and compare this value to the unbleached spot intensity to extrapolate the number of fluorophores in the structure. This approach has been used to determine the composition of various complexes including the bacterial flagellar motor and yeast centromeres (Coffman et al., 2011; Leake et al., 2006). A combination of photobleaching and brightness analysis has been used to measure the subunit composition of mammalian membrane proteins (Madl et al., 2010).
19.1.4 RATIOMETRIC COMPARISON OF FLUORESCENCE INTENSITY TO KNOWN STANDARDS
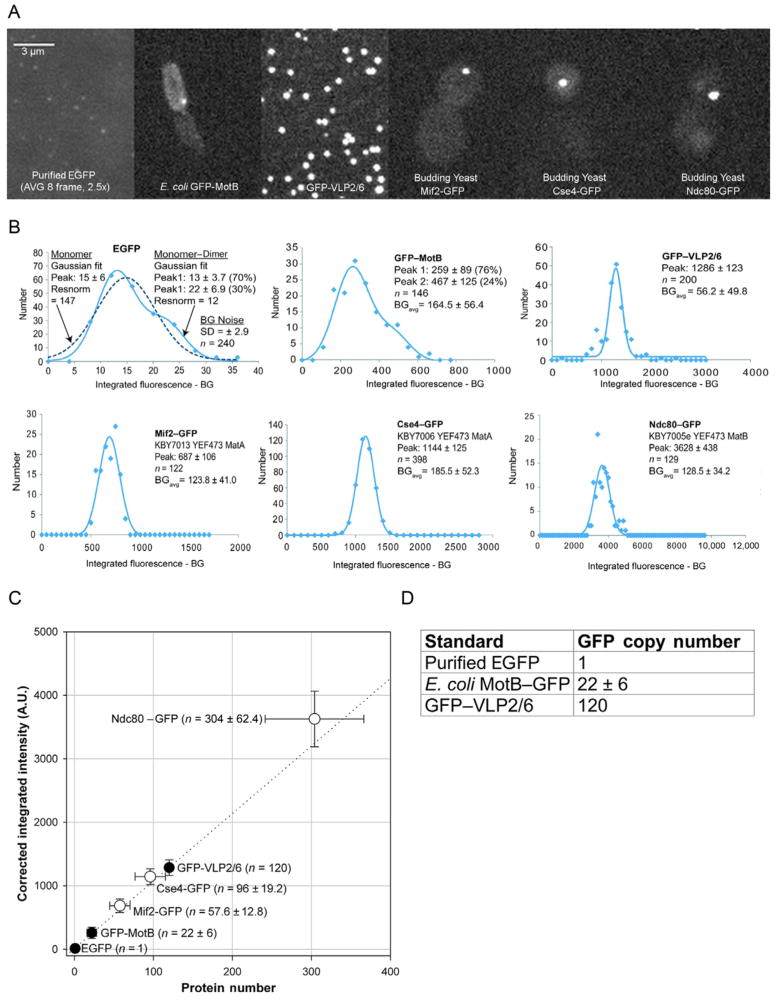
The stepwise photobleaching method can thus be applied in a manner that compares the fluorescence drop upon bleaching a single fluorophore to the total fluorescence of the sample. This is, in principle, similar to the ratiometric comparison of fluorescence intensity to determine the absolute protein number and allows for the measurement of a greater number of protein counts than direct stepwise photobleaching experiments. This method works by building a standard curve relating fluorescence intensity to number of molecules through careful and consistent measurement of fluorescence intensity of one or more fluorescence standards (Fig. 19.2). Alternatively, one can measure the ratio of bulk to single-molecule intensity of a standard and compare this to the bulk intensity of the labeled protein of interest to determine the intensity of a single fluorescent protein of interest (Graham, Johnson, & Marko, 2011). Fluorescence standards are discussed in greater detail later in the text and should be characterized using biochemical or electron microscopy assays to confirm their composition. The fluorescence of an experimental spot can then be measured under identical conditions and compared to the standard curve to determine protein count (Fig. 19.2C).
FIGURE 19.2.
Generating a standard curve. (A) Representative images of standards used in Lawrimore, Bloom, and Salmon (2011) and yeast strains in which anaphase copy numbers were measured. Purified EGFP (top left panel) was imaged with 2.5=fold longer exposure time (1500 vs. 600 ms) than other specimens and image shown is an average of eight images. (B) Gaussian fits of depth= and photobleaching=corrected integrated fluorescence intensity for standards and anaphase GFP spots in yeast strains. Peak intensities of each Gaussian fit are provided with standard deviation. EGFP and GFP–MotB can be fitted with two Gaussian curves (peak 1 and peak 2). BG noise is the average background intensity corrected for in each sample. (C) Standard curve generated from EGFP=, GFP–MotB=, and GFP–VLP2/6=corrected integrated fluorescence intensity versus protein number (black circles) with GFP spots from yeast strains (white circles). The dotted line represents a linear regression of the three standards (black circles). Values±standard deviation. (D) Table of GFP copy numbers for three fluorescence standards used to generate the standard curve in (C).
Ratiometric comparison of fluorescence intensities can be applied to a range of biological questions, including measurements of the budding yeast kinetochore to examine transcription dynamics in Escherichia coli (Taniguchi et al., 2010; Yu, Xiao, Ren, Lao, & Xie, 2006) and the organization of the kinetochore–microtubule attachment in budding yeast (Joglekar, Bouck, et al., 2008; Joglekar, Bouck, Molk, Bloom, & Salmon, 2006; Joglekar, Salmon, et al., 2008). More recently, ratiometric comparison of fluorescence intensity has been applied to understand the γ-tubulin microtubule nucleation structure (Erlemann et al., 2012). Fission yeast cytokinetic contractile ring proteins have been measured by comparing fluorescence intensity to a quantitative immunoblotting standard curve (McCormick, Akamatsu, Ti, & Pollard, 2013; Wu & Pollard, 2005). Overall, ratiometric comparison of fluorescence intensities is a powerful method of determining protein counts in a variety of systems that does not require highly specialized equipment. This method, like the FCS-based counting and stepwise photobleaching, requires rigorous quantification of known fluorescence standards to validate and calibrate the experiment.
19.1.5 FLUORESCENCE STANDARDS
For all the methods discussed, it is essential to validate the acquisition and analysis methodology using a range of known fluorescence standards to determine the relationship between fluorescence intensity and number of molecules (Fig. 19.2). The protein composition and stoichiometry of these fluorescence standards have been characterized using a range of different procedures. The most straightforward standard, though technically challenging to image, is soluble GFP either in vitro or cytosolic (Lawrimore et al., 2011; Padeganeh et al., 2013). The typical E. coli flagellar motor is composed of 11 stators, and each contains two copies of the MotB protein, as determined by electron microscopy and biochemical analysis (Khan, Dapice, & Reese, 1988; Kojima & Blair, 2004). Fluorescence imaging and stepwise photo-bleaching analysis have shown that GFP–MotB clusters contain approximately 22 times the intensity of a single GFP molecule (Leake et al., 2006). Subsequent studies have confirmed the composition of this structure by stepwise photobleaching and ratiometric comparison of fluorescence intensity (Coffman et al., 2011; Lawrimore et al., 2011). The virus-like particles (VLP), formed by proteins GFP–VP2/6, contains 120 GFPs as determined by electron tomography and an extinction coefficient predicted for 120 GFPs per virus capsid and has been used as a fluorescence standard for ratiometric comparison (Charpilienne et al., 2001; Lawrimore et al., 2011).
The centromere-specific histone H3 variant in budding yeast, Cse4p, has been used as a fluorescence standard, and recent studies have further clarified the composition of Cse4p clusters in vivo. Given the sequence-specific nature of the budding yeast centromere, it was thought that each chromosome contained only one Cse4p-containing nucleosome, making this an attractive fluorescent standard (Joglekar, Bouck, et al., 2008; Joglekar et al., 2006; Johnston et al., 2010). The 16 budding yeast kinetochores are clustered together into two close to diffraction-limited spots during M phase. These clusters have been shown to appear anisotropic during metaphase and more compact during anaphase (Haase, Stephens, Verdaasdonk, Yeh, & Bloom, 2012). The peak intensity value of these clusters is increased during anaphase as the spots are more compacted, but there is no change in integrated intensity between metaphase and anaphase (Fig. 19.1).
The single nucleosome concept was derived from chromatin immunoprecipitation demonstrating that Cse4p was concentrated at the centromere DNA (Furuyama & Biggins, 2007; Verdaasdonk & Bloom, 2011). However, the single Cse4p nucleosome standard failed to match protein numbers estimated from biochemistry. Various groups have measured the number of Cse4p proteins in each kinetochore cluster. Coffman et al. and Lawrimore et al. each generated a standard curve of fluorescence intensity versus number of molecules using some of the fluorescence standards described in the preceding text by stepwise photobleaching or ratiometric comparison of fluorescence intensity, respectively (Coffman et al., 2011; Lawrimore et al., 2011). Lawrimore et al. reported ~5 Cse4p per chromosome for a total of 80 per haploid cluster and Coffman et al. found ~7–8 Cse4p per chromosome for a total of 122 per cluster (Coffman et al., 2011; Lawrimore et al., 2011). These studies show that there are extra Cse4p molecules incorporated at random positions over 20–50 kb of DNA flanking the centromere. This anisotropy of Cse4p clusters is abolished in the mRNA processing pat1Δ or xrn1Δ mutants, and the number of Cse4p molecules associated with chromatin is also reduced (Haase et al., 2013; Maresca, 2013). These findings support the presence of extra Cse4p molecules per chromosome and show that these are not essential for chromosome segregation.
Using FCS of soluble GFP to calibrate APD confocal imaging, Shivaraju et al. found 1–2 Cse4p per chromosome (depending on cell cycle stage) (Shivaraju et al., 2012). The FCS imaging methodology used in this study examines fluorescence in a defined volume that may be excluding fluorescence resulting from the extra Cse4p incorporated away from the centromere in anisotropic fluorescence clusters. Previous work has shown that Cse4p clusters change size/shape throughout the cell cycle (Haase et al., 2012), and thus, the use of maximum intensity instead of integrated intensity measurements could account for the variation in Cse4p intensity between metaphase and anaphase observed by Shivaraju et al. (2012). Therefore, it is possible that Shivaraju et al. had very accurately measured the Cse4p content at the centromere (~2 Cse4p per chromosome) while excluding the fluorescence intensity from the extra Cse4p molecules observed by Coffman et al. (2011), Lawrimore et al. (2011), and Shivaraju et al. (2012). The result that the centromere nucleosome contains 2 Cse4p proteins is consistent with TIRF stepwise photobleaching of single nucleosomes in mammalian cells (Padeganeh et al., 2013) and BiFC complementation experiments (Aravamudhan, Felzer-Kim, & Joglekar, 2013).
Analysis of whole fluorescent clusters of Cse4p yields a number of 5–6 Cse4p per chromosome (80–96 molecules per cluster). Using these values, the ratiometric comparison of fluorescence intensity approach is consistent with independent protein measurements for cytokinesis (McCormick et al., 2013; Wu & Pollard, 2005), γ-tubulin small complex (Erlemann et al., 2012), and Cnp1 in fission yeast (Lando et al., 2012).
19.2 PROTOCOL FOR COUNTING MOLECULES BY RATIOMETRIC COMPARISON OF FLUORESCENCE INTENSITY
This protocol uses the protein copy number of Cse4–GFP (anaphase) published in Lawrimore et al. (2011) to calculate protein copy numbers of other GFP-fused proteins. As discussed earlier in the text, Cse4–GFP intensity has been compared to a range of other fluorescence standards of known composition to validate its use as a standard. Either GFP(S65T) or EGFP(S65T, F64L) can be used as they have similar emission spectra and other properties (Patterson, Knobel, Sharif, Kain, & Piston, 1997). In addition to yeast, this protocol has been used to count the number of molecules in DT40 cells (Johnston et al., 2010). The protocol in the succeeding text is primarily designed for imaging punctate spots in budding yeast cells; however, these methods can be adapted for imaging larger GFP signals or in other cell types. Since this method is based on comparing the intensity of a known standard (Cse4–GFP) to other samples, consistency during the experiment is crucial.
19.2.1 MINIMIZING INSTRUMENT ERROR
Before undertaking any quantitative fluorescence measurements, it is essential to understand how the specifications and setup of an imaging system will affect the precision of the measurements. The following steps will help minimize any potential systematic errors:
Camera: Ensure that the camera you are using has high quantum efficiency for the EGFP emission spectrum (Tsien, 1998). The lower the quantum efficiency, the more variation will occur in all of the fluorescence intensity measurements. In addition, use a camera with the smallest possible pixel size. Images can be binned to increase signal if needed. Suggested pixel size of the images is 130 nm.
Objective: Only use the highest NA and magnification objectives. An NA of 1.4 or higher and a magnification of 100× are required.
Stage: Since fluorescence intensity reduces as a function of sample depth, a stage that allows accurate and consistent Z-steps should be used.
Light source: The consistency of the light used in quantitative measurements is essential. No matter the light source used, the intensity of the light should be checked regularly. Measure the intensity of the light every 20 min after allowing 30 min of warm-up to ensure the light source is stable. Arc lamps are less stable than laser and LED-based lighting systems and thus should be used with caution. However, frequent light intensity readings and allowing proper warm-up time will mitigate variation in light intensity.
Imaging environment: Any ambient light will cause increased variation in fluorescence intensity measurements. All imaging should be performed in the darkest and most consistent conditions possible.
19.2.2 MEASURING INSTRUMENT VARIATION
The steps in the succeeding text directly measure the precision of an imaging system and are intended to quantify the amount of variation resulting from different imaging components that will influence fluorescence intensity measurements. Note that the sources of variation are additive in the order they are given. It is strongly suggested that the following steps be performed in the order given:
-
Dark noise
Turn on the imaging system and allow for proper warm-up of all components.
Take five, full-chip images with the camera shutter closed.
Measure the mean intensity of several regions across the full-chip image. Note any intensity variations present in the dark images and take it into account when selecting a region of interest for imaging.
Measure the mean intensity and standard deviation of your region of interest to be used during imaging.
Average the five mean intensity and standard deviation measurements together to calculate the noise due to electronic noise.
-
Light leakage
Repeat the steps earlier in the text but with the camera shutter open but with no light from the light source allowed in the camera path to test for any possible light leakage.
-
Light noise
Repeat the steps in the first section but allow excitation light through the objective. The light coming from the light source should be measured by carefully removing the objective or rotating the microscope turret to an empty slot and using a light meter to measure the intensity of the light. Suggested light intensity is 0.5 mW of 488 nm light. Any increase in the standard deviation will reflect the variation from the light source.
-
Sample buffer noise
Repeat the steps in the first section with a slide filled with imaging buffer/media. For yeast, use a synthetic media. Autoclaved rich yeast media containing sugar is highly autofluorescent and should not be used.
19.2.3 BUDDING YEAST IMAGING PROTOCOL
This section outlines the procedure for growth and imaging of the yeast strain expressing Cse4–GFP (KBY7006). To minimize protein count variation due to different health conditions of yeast, all yeast should be grown to an optical density (λ=660 nm) of at least 0.4 twice before starting an imaging culture. Image each strain until a sample size of 100 is obtained. Do not analyze any images where the GFP spot is moving:
Grow yeast in YPD media at 24 °C in 50 mL or greater flasks until reaching mid-logarithmic phase (OD660 = 0.4–0.8).
Thirty minutes prior to imaging, turn on all imaging components.
Spin down 1 mL yeast culture for 1 min at 4000 rpm.
Aspirate supernatant and resuspend in 1 mL synthetic media.
Spin down 1 mL yeast culture for 1 min at 4000 rpm.
Aspirate supernatant and resuspend in 20–100 μL synthetic media depending on the size of the pellet.
Pipet yeast resuspension on a concanavalin A-coated coverslip, place coverslip on slide, and seal edges with VALAP (1:1:1 mix of vaseline/lanolin/paraffin).
Immediately prior to imaging, measure the light intensity by removing the objective or moving turret to blank slot. Place light meter where slide will rest during imaging. Set light intensity to 0.5 mW.
Obtain Z-series image stacks with 40, 200 nm step sizes of yeast in anaphase. Anaphase yeast will have large buds and the GFP spots will be separated by 4–5 μm. The objective should be focused above/below the coverslip so that the Z-series will pass through the coverslip before focusing on yeast. If the pixel size of the image is near 65 nm, use 2×2 binning.
Note the frame where the coverslip is in focus. There will be an autofluorescent residue on the coverslip surface to indicate when the coverslip is in focus.
Note the frame where the GFP spot is in focus.
After 20 min of imaging, remove the slide and check the image intensity. If the intensity has drifted, do not analyze the z-stacks acquired. Do not image a slide for longer than 20 min as the yeast viability will deteriorate over time.
19.2.4 MEASURING BACKGROUND-SUBTRACTED, INTEGRATED INTENSITY
The image analysis described here entails measuring the integrated intensity (summing of all pixel values in a region of interest) of a larger and a smaller region of interest around the in-focus GFP spot and subtracting the integrated intensity of the surrounding background. Different imaging systems and specimens will require different regions of interest sizes. Ensure that region size and shape selected are large enough to capture the entire signal of the GFP spot. For most punctate GFP spots, a 5×5 pixel square (where 1 pixel=135 nm) is sufficient to encompass the GFP spot. For GFP signals that are not punctate, draw a region large enough to encompass the whole signal.
In order to measure the background of a GFP spot, draw a second region of interest centered on the region encompassing the GFP spot. For the punctate spots within a 5×5 pixel region, a larger 7×7 (where 1 pixel=135 nm) pixel square was used. The following equation describes how to calculate the background-subtracted, integrated intensity from the two concentric regions (adapted from Hoffman et al., 2001):
where Ismall is the integrated intensity of the smaller region, Ilarge is the integrated intensity of the larger region, Asmall is the area in pixels of the smaller region, and Alarge is the area in pixels of the larger region (Fig. 19.1D). To minimize error, the area of the larger region should be close to twice the size of the smaller region. However, regions of any size and shape can be used. In yeast, the nucleus is present during mitosis and has a slightly higher background than the cytoplasm. In cases where the GFP spot is against the nuclear envelope, the larger region can be shifted to capture more of the nuclear background. However, the larger region must fully encompass the smaller region.
For specimens where the GFP signal background cannot be measured as described in the preceding text, regions distal to the GFP signal can be used if the background intensity is similar to the region proximal to the GFP signal. Alternatively, specimens lacking the GFP signal can be measured to calculate an average background. However, this method will introduce measurement error if the background intensities of the specimen lacking GFP differ or are highly variable. For these methods, use the same region sizes for the sample and the background and directly subtract the background-subtracted, integrated intensity from the sample’s integrated intensity.
19.2.5 DEPTH CORRECTION
The further away a GFP spot is from the coverslip surface, the lower the integrated intensity will be. To calculate the depth of a GFP spot, subtract the frame number of the coverslip from the frame number of the in-focus GFP spot. Plot the background-subtracted, integrated intensity against the depth. Perform a linear regression on the data and calculate the slope of the line. For each background-subtracted, integrated intensity, use the following equation to correct for depth variation:
where Idepth is the background-subtracted, depth-corrected, integrated intensity; fspot is the frame number of the in-focus GFP spot; fcs is the frame number of the coverslip; m is the slope of the linear regression; and IBGsub,PC is the background-subtracted, photobleach-corrected, integrated intensity. Plot the depth-corrected data against the depth and perform another linear regression. Ensure the slope of the depth-corrected intensities is now zero.
19.2.6 CALCULATING PHOTOBLEACHING CORRECTION FACTOR
As a consequence of taking multiple pictures per Z-series, a small amount of photo-bleaching will occur. In order to minimize the variation that results from differing rates of photobleaching, each experimental strain should have a photobleaching curve constructed. Take five consecutive Z-series with the same settings used for normal image acquisition. A sample size of at least 10 GFP spots should be obtained. Measure the background-subtracted, integrated intensity of each in-focus GFP spot as described previously in the text. Use the slope of the background-subtracted, integrated intensity versus depth plot to correct for depth variation.
To calculate the photobleaching correction factor, calculate the four percent differences for each of the five timelapses taken. Then, average all of the percent difference together and divided by two. This step is summarized in the following equation:
where CF is the photobleaching correction factor; Idepthx is the background-subtracted, depth-corrected, integrated intensity of a particular timelapse; and Idepthx+1 is the background-subtracted, depth-corrected, integrated intensity of the next sequential timelapse.
Multiply each background-subtracted, depth-corrected, integrated intensity by this factor to calculate the amount of integrated intensity lost due to photobleaching during image acquisition. Add this amount to the background-subtracted, depth-corrected, integrated intensity to correct for photobleaching. This step is summarized in the following equation:
where Idepth,photo is the background-subtracted, depth- and photobleach-corrected, integrated intensity; Idepth is the background-subtracted, depth-corrected, integrated intensity of a spot; and CF is the photobleaching correction factor.
19.2.7 GAUSSIAN FITTING AND RATIOMETRIC COMPARISON TO DETERMINE PROTEIN COUNT
Perform a least-squares fit to a Gaussian curve on the background-subtracted, depth-and photobleach-corrected, integrated intensities to calculate the mean and standard deviation of each data set. To determine the intensity to copy number conversion factor, divide the mean and standard deviation of the experimental data set by the number of Cse4–GFP molecules/cluster(=96±19.2, anaphase) (Fig. 19.2C; Lawrimore et al., 2011). This conversion factor can be used to calculate the copy number of other proteins tagged with GFP.
CONCLUSIONS
The methods discussed in this chapter provide a starting point for researchers wishing to determine the absolute number of their protein of interest. A broad range of biological questions can benefit from knowledge of protein numbers, such as examining protein complex organization throughout the cell cycle, how protein composition is maintained, or allowing for mathematical modeling of protein complex architecture and behavior. The ratiometric comparison of fluorescence intensity to known standards allows for measurement of a broad range of protein numbers using standard high-end microscopy equipment. We encourage scientists to consider protein counting as another tool to address their research questions.
References
- Aravamudhan P, Felzer-Kim I, Joglekar AP. The budding yeast point centromere associates with two Cse4 molecules during mitosis. Current Biology. 2013;23(9):770–774. doi: 10.1016/j.cub.2013.03.042. http://dx.doi.org/10.1016/j.cub.2013.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacia K, Schwille P. A dynamic view of cellular processes by in vivo fluorescence auto- and cross-correlation spectroscopy. Methods. 2003;29(1):74–85. doi: 10.1016/s1046-2023(02)00291-8. [DOI] [PubMed] [Google Scholar]
- Braeckmans K, Deschout H, Demeester J, De Smedt SC. Optical fluorescence microscopy: From the spectral to the nano dimension. Berlin, Heidelberg: Springer-Verlag; 2011. Measuring molecular dynamics by FRAP, FCS, and SPT; pp. 153–163. [Google Scholar]
- Bulseco DA, Wolf DE. Fluorescence correlation spectroscopy: Molecular complexing in solution and in living cells. Methods in Cell Biology. 2013;114:489–524. doi: 10.1016/B978-0-12-407761-4.00021-X. http://dx.doi.org/10.1016/B978-0-12-407761-4.00021-X. [DOI] [PubMed] [Google Scholar]
- Charpilienne A, Nejmeddine M, Berois M, Parez N, Neumann E, Hewat E, et al. Individual rotavirus-like particles containing 120 molecules of fluorescent protein are visible in living cells. The Journal of Biological Chemistry. 2001;276(31):29361–29367. doi: 10.1074/jbc.M101935200. http://dx.doi.org/10.1074/jbc.M101935200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Muller JD, Berland KM, Gratton E. Fluorescence fluctuation spectroscopy. Methods. 1999;19(2):234–252. doi: 10.1006/meth.1999.0854. http://dx.doi.org/10.1006/meth.1999.0854. [DOI] [PubMed] [Google Scholar]
- Chen Y, Muller JD, Ruan Q, Gratton E. Molecular brightness characterization of EGFP in vivo by fluorescence fluctuation spectroscopy. Biophysical Journal. 2002;82(1 Pt 1):133–144. doi: 10.1016/S0006-3495(02)75380-0. http://dx.doi.org/10.1016/S0006-3495(02)75380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Muller JD, So PT, Gratton E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophysical Journal. 1999;77(1):553–567. doi: 10.1016/S0006-3495(99)76912-2. http://dx.doi.org/10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SH, Kennedy RA. Forward-backward non-linear filtering technique for extracting small biological signals from noise. Journal of Neuroscience Methods. 1991;40(1):71–86. doi: 10.1016/0165-0270(91)90118-j. [DOI] [PubMed] [Google Scholar]
- Coffman VC, Wu JQ. Counting protein molecules using quantitative fluorescence microscopy. Trends in Biochemical Sciences. 2012;37(11):499–506. doi: 10.1016/j.tibs.2012.08.002. http://dx.doi.org/10.1016/j.tibs.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Wu P, Parthun MR, Wu JQ. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. The Journal of Cell Biology. 2011;195(4):563–572. doi: 10.1083/jcb.201106078. http://dx.doi.org/10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SK, Darshi M, Cheley S, Wallace MI, Bayley H. Membrane protein stoichiometry determined from the step-wise photobleaching of dye-labelled subunits. Chembiochem: A European Journal of Chemical Biology. 2007;8(9):994–999. doi: 10.1002/cbic.200600474. http://dx.doi.org/10.1002/cbic.200600474. [DOI] [PubMed] [Google Scholar]
- Douglass AD, Vale RD. Single-molecule imaging of fluorescentproteins. Methods in Cell Biology. 2008;85:113–125. doi: 10.1016/S0091-679X(08)85006-6. http://dx.doi.org/10.1016/S0091-679X(08)85006-6. [DOI] [PubMed] [Google Scholar]
- Enderlein J, Gregor I, Patra D, Fitter J. Art and artefacts of fluorescence correlation spectroscopy. Current Pharmaceutical Biotechnology. 2004;5(2):155–161. doi: 10.2174/1389201043377020. http://dx.doi.org/10.2174/1389201043377020. [DOI] [PubMed] [Google Scholar]
- Engel BD, Ludington WB, Marshall WF. Intraflagellar transport particle size scales inversely with flagellar length: Revisiting the balance-point length control model. The Journal of Cell Biology. 2009;187(1):81–89. doi: 10.1083/jcb.200812084. http://dx.doi.org/10.1083/jcb.200812084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlemann S, Neuner A, Gombos L, Gibeaux R, Antony C, Schiebel E. An extended gamma-tubulin ring functions as a stable platform in microtubule nucleation. The Journal of Cell Biology. 2012;197(1):59–74. doi: 10.1083/jcb.201111123. http://dx.doi.org/10.1083/jcb.201111123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(37):14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosch M, Rigler R. Fluorescence correlation spectroscopy of molecular motions and kinetics. Advanced Drug Delivery Reviews. 2005;57(1):169–190. doi: 10.1016/j.addr.2004.07.016. http://dx.doi.org/10.1016/j.addr.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Graham JS, Johnson RC, Marko JF. Counting proteins bound to a single DNA molecule. Biochemical and Biophysical Research Communications. 2011;415(1):131–134. doi: 10.1016/j.bbrc.2011.10.029. http://dx.doi.org/10.1016/j.bbrc.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J, Mishra PK, Stephens A, Haggerty R, Quammen C, Taylor RM, 2nd, et al. A 3D map of the yeast kinetochore reveals the presence of core and accessory centromere-specific histone. Current Biology. 2013;23(19):1939–1944. doi: 10.1016/j.cub.2013.07.083. http://dx.doi.org/10.1016/j.cub.2013.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J, Stephens A, Verdaasdonk J, Yeh E, Bloom K. Bub1 kinase and Sgo1 modulate pericentric chromatin in response to altered microtubule dynamics. Current Biology. 2012;22(6):471–481. doi: 10.1016/j.cub.2012.02.006. http://dx.doi.org/10.1016/j.cub.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallworth R, Nichols MG. Single molecule imaging approach to membrane protein stoichiometry. Microscopy and Microanalysis: The Official Journal of Microscopy Society of America, Microbeam Analysis Society, Microscopical Society of Canada. 2012;18(4):771–780. doi: 10.1017/S1431927612001195. http://dx.doi.org/10.1017/S1431927612001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haustein E, Schwille P. Fluorescence correlation spectroscopy: Novel variations of an established technique. Annual Review of Biophysics and Biomolecular Structure. 2007;36:151–169. doi: 10.1146/annurev.biophys.36.040306.132612. http://dx.doi.org/10.1146/annurev.biophys.36.040306.132612. [DOI] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Molecular Biology of the Cell. 2001;12(7):1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck D, Finley K, Liu XK, Wan YK, Berman J, et al. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. Journal of Cell Biology. 2008;181(4):587–594. doi: 10.1083/jcb.200803027. http://dx.doi.org/10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nature Cell Biology. 2006;8(6):581–585. doi: 10.1038/ncb1414. http://dx.doi.org/10.1038/Ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Salmon ED, Bloom KS. Counting kinetochore protein numbers in budding yeast using genetically encoded fluorescent proteins. Methods in Cell Biology. 2008;85:127–151. doi: 10.1016/S0091-679X(08)85007-8. http://dx.doi.org/10.1016/S0091-679X(08)85007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WL, Straight AF. Fluorescent protein applications in microscopy. Methods in Cell Biology. 2013;114:99–123. doi: 10.1016/B978-0-12-407761-4.00005-1. http://dx.doi.org/10.1016/B978-0-12-407761-4.00005-1. [DOI] [PubMed] [Google Scholar]
- Johnston K, Joglekar A, Hori T, Suzuki A, Fukagawa T, Salmon ED. Vertebrate kinetochore protein architecture: Protein copy number. The Journal of Cell Biology. 2010;189(6):937–943. doi: 10.1083/jcb.200912022. http://dx.doi.org/10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kask P, Palo K, Ullmann D, Gall K. Fluorescence-intensity distribution analysis and its application in biomolecular detection technology. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(24):13756–13761. doi: 10.1073/pnas.96.24.13756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Dapice M, Reese TS. Effects of mot gene expression on the structure of the flagellar motor. Journal of Molecular Biology. 1988;202(3):575–584. doi: 10.1016/0022-2836(88)90287-2. [DOI] [PubMed] [Google Scholar]
- Kojima S, Blair DF. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry. 2004;43(1):26–34. doi: 10.1021/bi035405l. http://dx.doi.org/10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- Lando D, Endesfelder U, Berger H, Subramanian L, Dunne PD, McColl J, et al. Quantitative single-molecule microscopy reveals that CENP-A(Cnp1) deposition occurs during G2 in fission yeast. Open Biology. 2012;2(7):120078. doi: 10.1098/rsob.120078. http://dx.doi.org/10.1098/rsob.120078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. Journal of Cell Biology. 2011;195(4):573–582. doi: 10.1083/jcb.201106036. http://dx.doi.org/10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443(7109):355–358. doi: 10.1038/nature05135. http://dx.doi.org/10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- Levin MK, Carson JH. Fluorescence correlation spectroscopy and quantitative cell biology. Differentiation; Research in Biological Diversity. 2004;72(1):1–10. doi: 10.1111/j.1432-0436.2004.07201002.x. http://dx.doi.org/10.1111/j.1432-0436.2004.07201002.x. [DOI] [PubMed] [Google Scholar]
- Madl J, Weghuber J, Fritsch R, Derler I, Fahrner M, Frischauf I, et al. Resting state Orai1 diffuses as homotetramer in the plasma membrane of live mammalian cells. The Journal of Biological Chemistry. 2010;285(52):41135–41142. doi: 10.1074/jbc.M110.177881. http://dx.doi.org/10.1074/jbc.M110.177881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ. Chromosome segregation: Not to put too fine a point (centromere) on it. Current Biology. 2013;23(19):R875–R878. doi: 10.1016/j.cub.2013.08.049. http://dx.doi.org/10.1016/j.cub.2013.08.049. [DOI] [PubMed] [Google Scholar]
- McCormick CD, Akamatsu MS, Ti SC, Pollard TD. Measuring affinities of fission yeast spindle pole body proteins in live cells across the cell cycle. Biophysical Journal. 2013;105(6):1324–1335. doi: 10.1016/j.bpj.2013.08.017. http://dx.doi.org/10.1016/j.bpj.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophysical Journal. 2006;91(5):1941–1951. doi: 10.1529/biophysj.106.082487. http://dx.doi.org/10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padeganeh A, Ryan J, Boisvert J, Ladouceur AM, Dorn JF, Maddox PS. Octameric CENP-A nucleosomes are present at human centromeres throughout the cell cycle. Current Biology. 2013;23(9):764–769. doi: 10.1016/j.cub.2013.03.037. http://dx.doi.org/10.1016/j.cub.2013.03.037. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophysical Journal. 1997;73(5):2782–2790. doi: 10.1016/S0006-3495(97)78307-3. http://dx.doi.org/10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quammen CW, Richardson AC, Haase J, Harrison BD, Taylor RM, 2nd, Bloom KS. FluoroSim: A visual problem-solving environment for fluorescence microscopy. Eurographics Workshop on Visual Computing for Biomedicine. 2008;2008:151–158. doi: 10.2312/VCBM/VCBM08/151-158. http://dx.doi.org/10.2312/VCBM/VCBM08/151-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries J, Schwille P. Fluorescence correlation spectroscopy. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology. 2012;34(5):361–368. doi: 10.1002/bies.201100111. http://dx.doi.org/10.1002/bies.201100111. [DOI] [PubMed] [Google Scholar]
- Rottenfusser R. Proper alignment of the microscope. Methods in Cell Biology. 2013;114:43–67. doi: 10.1016/B978-0-12-407761-4.00003-8. http://dx.doi.org/10.1016/B978-0-12-407761-4.00003-8. [DOI] [PubMed] [Google Scholar]
- Salmon ED, Shaw SL, Waters JC, Waterman-Storer CM, Maddox PS, Yeh E, et al. A high-resolution multimode digital microscope system. Methods in Cell Biology. 2013;114:179–210. doi: 10.1016/B978-0-12-407761-4.00009-9. http://dx.doi.org/10.1016/B978-0-12-407761-4.00009-9. [DOI] [PubMed] [Google Scholar]
- Shivaraju M, Unruh JR, Slaughter BD, Mattingly M, Berman J, Gerton JL. Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell. 2012;150(2):304–316. doi: 10.1016/j.cell.2012.05.034. http://dx.doi.org/10.1016/j.cell.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter BD, Huff JM, Wiegraebe W, Schwartz JW, Li R. SAM domain-based protein oligomerization observed by live-cell fluorescence fluctuation spectroscopy. PLoS One. 2008;3(4) doi: 10.1371/journal.pone.0001931. http://dx.doi.org/10.1371/Journal.Pone.0001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter BD, Li R. Toward quantitative “in vivo biochemistry” with fluorescence fluctuation spectroscopy. Molecular Biology of the Cell. 2010;21(24):4306–4311. doi: 10.1091/mbc.E10-05-0451. http://dx.doi.org/10.1091/mbc.E10-05-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329(5991):533–538. doi: 10.1126/science.1188308. http://dx.doi.org/10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson NL, Lieto AM, Allen NW. Recent advances in fluorescence correlation spectroscopy. Current Opinion in Structural Biology. 2002;12(5):634–641. doi: 10.1016/s0959-440x(02)00368-8. [DOI] [PubMed] [Google Scholar]
- Tian Y, Martinez MM, Pappas D. Fluorescence correlation spectroscopy: A review of biochemical and microfluidic applications. Applied Spectroscopy. 2011;65(4):115A–124A. doi: 10.1366/10-06224. http://dx.doi.org/10.1366/10-06224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien RY. The green fluorescent protein. Annual Review of Biochemistry. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. http://dx.doi.org/10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nature Methods. 2007;4(4):319–321. doi: 10.1038/NMETH1024. http://dx.doi.org/10.1038/nmeth1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk JS, Bloom K. Centromeres: Unique chromatin structures that drive chromosome segregation. Nature Reviews Molecular Cell Biology. 2011;12(5):320–332. doi: 10.1038/nrm3107. http://dx.doi.org/10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukojevic V, Pramanik A, Yakovleva T, Rigler R, Terenius L, Bakalkin G. Study of molecular events in cells by fluorescence correlation spectroscopy. Cellular and Molecular Life Sciences. 2005;62(5):535–550. doi: 10.1007/s00018-004-4305-7. http://dx.doi.org/10.1007/s00018-004-4305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC. Live-cell fluorescence imaging. Methods in Cell Biology. 2013;114:125–150. doi: 10.1016/B978-0-12-407761-4.00006-3. http://dx.doi.org/10.1016/B978-0-12-407761-4.00006-3. [DOI] [PubMed] [Google Scholar]
- Watkins LP, Yang H. Detection of intensity change points in time-resolved single-molecule measurements. The Journal of Physical Chemistry B. 2005;109(1):617–628. doi: 10.1021/jp0467548. http://dx.doi.org/10.1021/jp0467548. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310(5746):310–314. doi: 10.1126/science.1113230. http://dx.doi.org/10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Xia T, Li N, Fang X. Single-molecule fluorescence imaging in living cells. Annual Review of Physical Chemistry. 2013;64:459–480. doi: 10.1146/annurev-physchem-040412-110127. http://dx.doi.org/10.1146/annurev-physchem-040412-110127. [DOI] [PubMed] [Google Scholar]
- Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311(5767):1600–1603. doi: 10.1126/science.1119623. http://dx.doi.org/10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]