Abstract
Hair cells are the mechanotransducing cells of the inner ear that are essential for hearing and balance. POU4F3 – a POU-domain transcription factor selectively expressed by these cells – has been shown to be essential for hair cell differentiation and survival in mice and its mutation in humans underlies late-onset progressive hearing loss (DFNA15). The downstream targets of POU4F3 are required for hair cell differentiation and survival. We aimed to identify such targets in order to elucidate the molecular pathways involved in hair cell production and maintenance. The orphan thyroid nuclear receptor Nr2f2 was identified as a POU4F3 target using a subtractive hybridization strategy and EMSA analysis showed that POU4F3 binds to two sites in the Nr2f2 5′ flanking region. These sites were shown to be required for POU4F3 activation as their mutation leads to a reduction in the response of an Nr2f2 5′ flanking region reporter construct to POU4F3. Immunocytochemistry was carried out in the developing and adult inner ear in order to investigate the relevance of this interaction in hearing. NR2F2 expression in the postnatal mouse organ of Corti was shown to be detectable in all sensory epithelia examined and characterised. These data demonstrate that Nr2f2 is a direct target of POU4F3 in vitro and that this regulatory relationship may be relevant to hair cell development and survival.
Introduction
POU4F3 is a member of the POU family of transcription factors that regulate a wide array of neuroendocrine developmental pathways [1]. This family is characterised by the presence of a bipartite DNA binding domain known as the POU domain which comprises a POU-homeodomain and a POU-specific domain separated by a linker [2]. All of these components are required for sequence-specific DNA binding [3], [4].
Though known to be expressed in the retina, spinal cord, dorsal root ganglia and Merkel cells, the investigation of POU4F3 expression and function has focused on its role in the development and maintenance of hair cells – the sensory cells of the inner ear [5]–[8]. Homozygous Pou4f3 mutant mice demonstrate deafness and balance deficits due to the failure of nascent hair cells – cells that express hair cell markers – to develop into morphologically recognisable hair cells and subsequent death of these cells [6]–[9]. Furthermore, heterozygous mutation of human POU4F3 is the cause of autosomal dominant late-onset progressive hearing loss (DFNA15) in several families [10]–[13].
As the molecular pathways involved in differentiation and maintenance of hair cells are largely unknown, identification of the downstream targets of POU4F3 would greatly improve current understanding of the transcription factors and target genes that mediate hair cell differentiation and maturation. Such an understanding may also aid in the explanation of the mechanism by which POU4F3 mutation causes hearing loss in humans.
To date, the only known direct targets of POU4F3 are BDNF, NT-3 and Caprin-1 [14], [15], though the expression of other genes is strongly associated with POU4F3 expression [14], [16], [17]. In previous work we identified Caprin-1 as a direct target of POU4F3 in a subtractive hybridization screen that was carried out in UB/OC-2 cells – a conditionally immortal cell line, derived from the developing mouse inner ear sensory epithelium, that displays expression of hair cell markers [18]– that were manipulated to differentially express POU4F3 [15]. The Caprin-1 5′ flanking sequence contains POU4F3 binding sites which mediate repression of Caprin-1 expression by POU4F3 [15]. We showed that Caprin-1, which is known to promote stress granule formation [19]–[21], is expressed in both the hair cells and supporting cells of the organ of Corti as well as being involved in the hair cell response to stress [15].
The same subtractive hybridization screen also returned the orphan thyroid nuclear receptor Nr2f2 as a putative target of POU4F3. Nr2f2 is an orphan steroid/thyroid hormone nuclear receptor which is expressed in a range of organs in the developing embryo [22] as well as adult tissues [23]. Its most essential roles in development are in angiogenesis, heart development and remodelling the primitive capillary plexus into large and small microcapillaries; as demonstrated by knocking out the Nr2f2 gene in mice. This change results in lethality at around E10 that is likely to be due to haemorrhage and oedema in the brain and heart [24]. Further study of the function of NR2F2 in the vascular system has revealed its importance in designating venous identity via its suppression of the Notch signalling pathway. In addition to its previously reported roles in generation of venous identity, these studies show a role for NR2F2 in cell fate determination [25].
Despite having a general role in embryogenesis, NR2F2 also demonstrates more specific actions in organogenesis. In the brain, it regulates cell migration [26]; it is essential for correct stomach patterning [27]; and has been proposed to be involved in dorso-ventral patterning of the mammalian retina [28]. Both NR2F2 and its closely related family member NR2F1 have been shown to be variably expressed in the developing mouse inner ear. NR2F2 is expressed in the distal tip of the elongating cochlear epithelium at E10 to E13.5; homogeneous expression is seen across cell types of both the greater and lesser epithelial ridges (GER & LER) in the apical coil at E15.5; and predominantly LER expression is seen in the apical-to-middle coil at the same age [29].
In contrast to NR2F2, NR2F1 expression in the developing inner ear between E14.5 and E15.5 becomes more confined to the GER in the apical cochlea. In the basal cochlea, its expression extends to the LER and the level of expression appears to decrease [29]. This decrease in basal expression correlates with the wave of hair cell differentiation from base to apex [30]. Consistent with a role in cochlear development, Nr2f1 −/− mice display shortened cochlear ducts with an increased number of hair cells in the mid-to-apical cochlear turns; an anomaly which appears consistent with a role in Notch regulation of organ of Corti differentiation [31].
Though NR2F2 has been implicated in a number of developmental processes, its expression is known to be maintained in postnatal vertebrates. Post-natal NR2F2 expression is perhaps best-characterised in the liver where it is known to downregulate the apolipoprotein A1 (apoA1) gene [32]. However, its expression is maintained in the adult mouse brain and in a wide array of postnatal human tissue types, suggesting additional uncharacterised roles in cell maintenance [23], [33].
In this paper, we present evidence for the direct regulation of Nr2f2 by POU4F3. Furthermore, we show that NR2F2 expression is maintained postnatally in the mouse cochlear sensory epithelium. These data add to our understanding of the downstream effects of POU4F3 signalling and widen the array of possible functions of NR2F2 to the postnatal inner ear.
Materials and Methods
Subtractive hybridization
POU4F3-regulated genes were identified as previously described [15]. Briefly, two populations of UB/OC-2 cells [18] were created. In one population, POU4F3 expression was increased by stable transfection of a Pou4f3 expression construct. In the other, POU4F3 expression was reduced by stable transfection of an antisense Pou4f3 construct. cDNA was prepared from these two cell populations and used in a subtractive hybridization screen using the PCR-Select cDNA Subtraction kit (Clontech). Differentially expressed cDNA sequences identified in this analysis were verified by a series of hybridization experiments with cDNA from the original analysis and virtual northern blot experiments with cDNA from transiently transfected cells to increase the stringency of the analysis [15]. Clones that displayed differential expression were selected for further analysis.
BLAST analysis
Following identification of differentially expressed transcripts by subtractive hybridization, cloned cDNA was subjected to Sanger sequencing. This was followed by BLASTN analysis of transcript sequences against the ENSEMBL mouse cDNA database (NCBI m35) with a ‘near-exact match’ stringency. Matches were confirmed by alignment of clone sequence to the cDNA sequence in the ENSEMBL database using the BioEdit software package [34].
Nr2f2 5′ flanking region Genomatix analysis
The Genomatix Gene2Promoter software [35] was used to identify predicted Nr2f2 promoters for analysis. The promoter that corresponded to the transcript that best matched the Nr2f2 transcript identified in the BLAST analysis was selected for further investigation (GXP_158616). To identify putative POU4F3 binding sites in this predicted promoter, the selected region was interrogated using the Genomatix MatInspector [36], [37] and ModelInspector [38] programs. Binding sites for analysis were selected on the basis of search stringency, proximity to the predicted transcriptional start site and promoter architecture at the relevant site. Two predicted binding sites were selected for functional analysis and were named POU recognition element-1 (PRE1) and POU recognition element-2 (PRE2).
Electrophoretic mobility-shift assay (EMSA) analysis
Both in vitro translated protein and UB/OC-2 cell nuclear protein extract were used. Pou4f3 was cloned into the pGEM-T Easy vector (Promega) under the control of the T7 or SP6 promoter and in vitro translated protein was generated using either the TNT-T7 Coupled Reticulocyte Lysate System (Promega) or the TNT-SP6 Quick Coupled Transcription/Translation System (Promega).
Double-stranded probe oligonucleotides were labelled in a standard T4 kinase reaction with γ32P γATP (GE Healthcare) using 50 ng probe per reaction. Assay reactions containing 10 µl 2x Parker buffer (16% Ficoll, 40 mM HEPES at pH 7.9, 100 mM KCl, 2 mM EDTA at pH 8.0 and 1 mM DTT), 3 µg poly(dI·dC), 2–4 µl nuclear protein extract or in vitro translated protein and 0–1000 ng non-radiolabelled competition probe were made up to a total volume of 20 µl and incubated on ice for ten minutes. Assay reactions were subsequently incubated at room temperature for 15–30 minutes following the addition of 1 ng labelled probe.
Reactions were loaded onto a 4% polyacrylamide (29∶1), 0.25x TBE gel and electrophoresed in 0.25x TBE at 200 V for one to three hours at 4°C. The polyacrylamide gel was subsequently dried and autoradiographs were produced by exposure of dried gels to X-Ray film at −80°C or room temperature for varying durations.
Luciferase assays
The 4.2 kb-Nr2f2-Luc Nr2f2 5′ flanking region luciferase reporter construct was kindly provided by Dr M Vasseur-Cognet [39]. The PRE1-Luc and PRE2-Luc POU recognition element reporter constructs were cloned in our laboratory by inserting a single copy of PRE1 and PRE2 into the pGL4.23 [luc2/minP] reporter construct (Promega). The Dreidel expression construct was created by subcloning the Dreidel sequence from a pHM6 vector, kindly provided by Professor K Avraham [40], into pSi (Promega).
For transient transfections, ND7 cells [41] were plated at approximately 2×105 cells/well density in six-well plates and cultured at 37°C with 5% carbon dioxide in L-glutamine-containing L-15 media supplemented with 10% heat-inactivated foetal bovine serum, 0.32% sodium bicarbonate, 0.25% glucose and 0.85% penicillin-streptomycin. The next morning, the culture medium was changed to DMEM with 10% heat-inactivated foetal bovine serum for at least one hour prior to transfection. Cells were transfected using a standard calcium precipitation method [42] with 100–200 ng reporter construct, 1–3 µg expression vector and 10 ng pRL-null vector (Promega). The total amount of DNA used per transfection was kept constant using pSi (Promega). Cells were then incubated for a minimum of six hours followed by washing and returning to normal culture media. Cells were harvested at least 24 hours later and assayed using the Dual-Luciferase kit (Promega).
Site-directed mutagenesis
The 4.2 kb-Nr2f2-Luc-Mut reporter construct was generated by overlap extension PCR [43] using the following mutagenic primers (mutated bases underlined): PRE1 forward mutagenic primer, CTTTTTAGCCGATTTGATCACTTTGATT; PRE1 reverse mutagenic primer, AATCAAAGTGATCAAATCGGCTAAAAAG; PRE1 forward flanking primer, AAGCCTCCGGGTCGGGCCCGGAG; PRE1 reverse flanking primer, TCCGCGCTCCGGGGTCCAC; PRE2 forward mutagenic primer, GATAAAGTTGAGAGGAATTTATTTTAATTGCAGGGTAACAATGAGGTGAAGTCTGGTGTT; PRE2 reverse mutagenic primer, AACACCAGACTTCACCTCATTGTTACCCTGCAATTAAAATAAATTCCTCTCAACTTTATC; PRE2 forward flanking primer, GCTTAATGAATTCCCATCACTTGC; PRE2 reverse flanking primer, GGAATTCTCACAATCAACTAGCGG. The purified mutated fragment of PRE2 was cloned into 4.2 kb-Nr2f2-Luc, replacing the wild type sequence. The purified mutated fragment of PRE1 was cloned into this plasmid to create a double-mutant 4.2 kb-Nr2f2-Luc-Mut. Further subcloning was carried out to correct presumed PCR errors though a c to t missense mutation remained 288 bp downstream from the intended PRE1 mutagenesis site. The double-mutant probe was used in luciferase assays as described above.
Transcription factor binding site evolutionary conservation analysis
The ECR Browser [44] was used to navigate to the region of the human genome (build “hg19”) that corresponded to the mouse sequence for PRE1 (CTTTTTAGCATATTTGATCACTTTGATT) or PRE2 (GGAATTTATTTTAATTGCATCATAACAATGAGGTGA). The view obtained was expanded to include approximately 70 bp of flanking sequence and adjusted to include only mouse, rat and chimpanzee alignments. This view was submitted to the Mulan software and the phylogenetic tree obtained was not adjusted. The results of the Mulan analysis were submitted to MultiTF using the ‘vertebrates’ and ‘optimised for function’ settings against the TRANSFAC Professional Version 10.2 library to identify evolutionarily conserved POU domain transcription factor binding sites. Positional weight matrices (PWMs) for POU-domain transcription factors were included in this analysis as POU family binding sites are clearly related [4] despite there being subtle sequence-specific differences in the recognition elements of different POU4 family members [45]: OCT1_B, OCT1_Q5, OCT1_Q6, OCT4, OCT_C, OCT_Q6, POU1F1_Q6, POU3F2 and POU6F1.
Cryosectioning and immunocytochemistry
Adult (i.e. >P21) C57BL/6J mice, adult Sprague Dawley rats and P1 C57BL/6J mice were killed in accordance with Schedule I of the UK Animals (Scientific Procedures) Act (1986). This study was approved by the UCL Biological Services Ethical Review Committee. Inner ears were isolated and the apical bone of the cochlea was perforated to facilitate fixation in 4% paraformaldehyde solution in PBS for one to two hours at room temperature or overnight at 4°C. Cryoprotection was carried out by incubation of samples in 30% sucrose in PBS at 4°C overnight. Tissue was mounted in 1% low gelling agarose (Sigma-Aldrich) in PBS which had been heated to 140°C and allowed to cool to 37°C. Once set, a block was cut, attached to a cryostat chuck with OCT compound and rapidly frozen by immersion in liquid nitrogen prior to cryosectioning at −20 to −25°C. Sections of 15 µm were cut using a CM1850 cryostat (Leica).
Prior to immunolabelling, sections were permeabilised and blocked for one hour at room temperature with blocking solution (10% goat serum in PBS) containing 0.1–0.5% Triton-X. Slides were then incubated in blocking solution with an antibody raised against NR2F2 (a kind gift from Dr M Studer) 1∶1000 at 4°C overnight [26]. Following three five-minute PBS washes, 2 µg/ml Alexa 488-conjugated anti-rabbit secondary antibody, 1 Unit/ml Alexa 633-conjugated Phalloidin and 5 µM DAPI in PBS or blocking solution was added for two hours at room temperature. Following three further five-minute PBS washes, samples were mounted in Fluoromount G and visualised using a Zeiss LSM 510 Meta microscope. Relative expression comparisons were made between apical to basal cochlear turns within individual slides.
Results
Identification of Nr2f2 as a putative target of POU4F3
In order to identify target genes of POU4F3 that may be involved in hair cell survival and maintenance, a subtractive hybridization was carried out in UB/OC-2 cells followed by serial virtual Northern analysis as described previously [15]. One putative upregulatory target of POU4F3 identified by this screen was Clone D8. This clone was sequenced and subjected to BLAST analysis against the ENSEMBL mouse cDNA database (based on NCBI m35 data) to identify known mouse cDNA sequences to which it corresponded. Clone D8 matched the 3′ region of the related Nr2f1 and Nr2f2 cDNA sequences which are highly homologous in this region. However, the match to Nr2f2 (ENSMUST00000032768) was more extensive (343 bp, 100% sequence similarity: p = 8.6×10−273) than the match to Nr2f1 (116 bp, 87% sequence similarity: p = 3.3×10−25). Therefore, this analysis identified Nr2f2 as an upregulated transcript from the POU4F3 forward subtractive hybridization library.
As Nr2f2 was identified as a putative upregulatory target of POU4F3 in the subtractive hybridization screen, its expression in UB/OC-2 cells was investigated as these cells constitutively express POU4F3 in their proliferating state and are derived from the mouse inner ear at E13, i.e. after the onset of NR2F2 expression in the developing ear [18], [29]. Nr2f2 mRNA and protein were found to be present in proliferating UB/OC-2 cells by reverse transcriptase PCR, western blot and immunohistochemistry (Figure S1). NR2F2 expression was localised to UB/OC-2 cell nuclei, consistent with its reported subcellular localisation [26], [29].
POU4F3 binds two sites in the Nr2f2 5′ flanking region identified by Genomatix software analysis
For POU4F3 to directly regulate a gene, it must bind to the target gene promoter via a sequence specific binding site. To establish whether Nr2f2 is a direct or indirect target of POU4F3 we performed bioinformatic and functional analysis of the Nr2f2 5′ flanking region to identify any such binding sites. The Genomatix MatInspector and ModelInspector programmes were used to interrogate the most proximal 5 kb of the predicted Nr2f2 promoter (GXP_158616, identified by the Gene2Promoter software) [35]–[38].
Using MatInspector software at the highest stringency (0.05) a number of putative POU4F3 binding sites were identified. Of these, the two most proximal to the Nr2f2 transcriptional start site were selected for further analysis and designated POU regulation element (PRE) 1 and 2 (Figure 1a). PRE2 also overlapped with a SORY OCT1 module identified using the Genomatix ModelInspector software i.e. PRE2 is within an experimentally verified functional promoter subunit that contains predicted binding sites for both OCT1 and SORY.
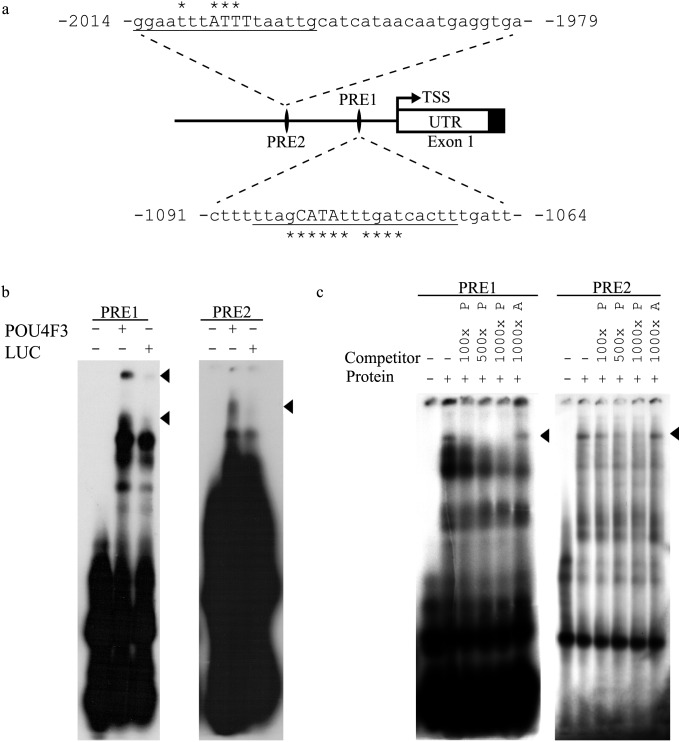
Figure 1. Identification and verification of POU4F3 recognition elements in the Nr2f2 5′ flanking region.
a. Schematic diagram of the location of two putative POU4F3 recognition elements (PREs) identified in the Nr2f2 5′ flanking region and used in EMSA analysis. Underlined bases correspond to POU4F3 binding sites predicted by MatInspector software with capital letters denoting matches to the core sequence. The asterisks signify matrix position conservation >60/100 (TSS, transcriptional start site and UTR, un-translated region). b. Sequences shown in a were used as radiolabelled probes in EMSA analysis with in vitro translated POU4F3 or a Luciferase control. Bandshifts due to protein-specific binding by POU4F3 are indicated by arrowheads. c. The same probes were used in EMSA analysis with UB/OC-2 cell nuclear protein extract. Reactions were incubated either alone, with UB/OC-2 cell nuclear protein extract or with the nuclear protein extract and an excess of non-radiolabelled competitor oligonucleotide as indicated. The ‘P’ suffix refers to non-radiolabelled POU4F3 binding sequence whereas the ‘A’ suffix indicates a non-radiolabelled AP4 binding sequence. POU4F3-sequence-specific shifts are indicated by arrowheads.
The ability of POU4F3 to bind these putative recognition elements was tested by electrophoretic mobility-shift assay (EMSA) analysis. In vitro translated POU4F3 produced bandshifts for both PRE1 and PRE2 that were not reproduced by in vitro translated luciferase protein (Figure 1b), demonstrating POU4F3-specific binding to PRE1 and PRE2. In addition, endogenous UB/OC-2 cell POU4F3 was shown to bind to these two sequences in EMSA competition analysis (Figure 1c). Hence, UB/OC-2 cell nuclear protein extract binding was reduced with increasing amounts of unlabelled POU4F3 consensus sequence but not with an excess of an unrelated transcription factor binding site, demonstrating the presence of POU4F3 in the shifted complex (Figure 1c).
POU4F3 activates the 4.2 kb Nr2f2 5′ flanking region containing PRE1 and PRE2
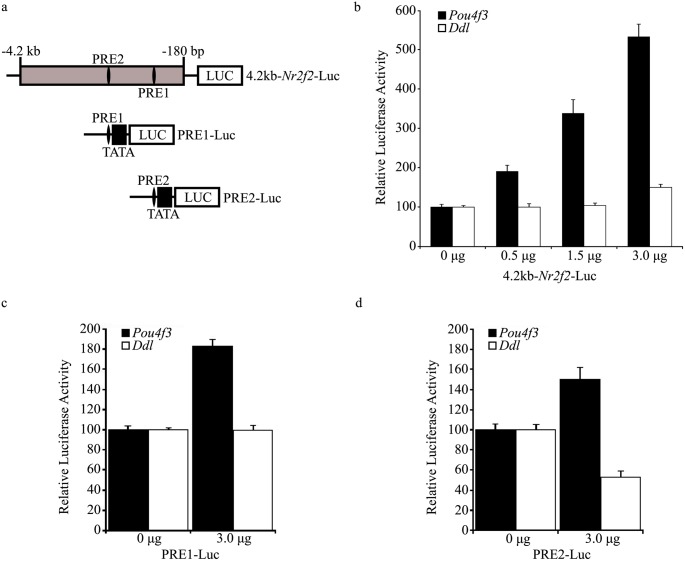
Having demonstrated the ability of POU4F3 to bind to PRE1 and PRE2, we tested whether POU4F3 can regulate the Nr2f2 5′ flanking region that contains these sites. A luciferase reporter construct containing 4.2 kb of the Nr2f2 5′ flanking region (4.2 kb-Nr2f2-Luc, Figure 2a) was used in co-transfection studies in ND7 cells. This analysis demonstrated a dose-dependent increase in 4.2 kb-Nr2f2-Luc activity in response to increasing POU4F3 levels. Compared to basal activity, promoter activity was five times higher at the maximal amount of POU4F3 expression construct used (3 µg) and was not replicated with a non-DNA-binding POU4F3 mutant (dreidel), showing that this activation is dependent on POU4F3-specific DNA binding (Figure 2b).
Figure 2. POU4F3-mediated activation of PRE1, PRE2 and a 4.2 kb Nr2f2 5′ flanking sequence.
a. Schematic diagram of reporter constructs used in luciferase reporter assays in ND7 cells. POU4F3 recognition element (PRE) 1 and 2 are shown in an Nr2f2 5′ flanking region fragment (grey box) cloned upstream of a luciferase reporter gene (LUC). The location of this fragment relative to the Nr2f2 transcriptional start site is shown. b. Evaluation of the response of 4.2 kb-Nr2f2-Luc to increasing levels of POU4F3 or dreidel mutant POU4F3 (Ddl) expression construct. The luciferase activity of the reporter is normalised to its response to the empty expression vector and results are expressed relative to this. c. Response of the PRE1-Luc reporter construct in co-transfection experiments with POU4F3 and dreidel expression constructs. d. Evaluation of the response of PRE2-Luc in similar experiments to c. Error bars represent the s.e.m in b, c and d (n = 6 for each data point).
These experiments were also conducted in UB/OC-2 cells but gave inconsistent results (data not shown). Experiments in this cell type may have been complicated by a number of factors. For example, proliferating UB/OC-2 cells endogenously express POU4F3 which may have been sufficiently expressed to mask the variation induced by the co-transfected POU4F3 expression construct. UB/OC-2 cells also vary their POU4F3 expression depending on differentiation status; it is, therefore, possible that POU4F3 expression varies more dynamically than is currently known. Given these possibilities, ND7 cells were used as POU4F3 activation of Nr2f2 constructs was consistent in this assay.
To investigate whether POU4F3 activates 4.2 kb-Nr2f2-Luc via PRE1, PRE2 or a combination of both, each was cloned upstream of a minimal promoter in a luciferase vector to generate PRE1-Luc and PRE2-Luc (Figure 2a) and used in co-transfection studies with increasing amounts of POU4F3. In ND7 cells, co-transfection with a POU4F3 expression construct produced a 1.8-fold upregulation of PRE1-Luc (Figure 2c) and a 1.5-fold upregulation of PRE2-Luc (Figure 2d). In both cases, this activation required functional POU4F3 as dreidel mutant protein did not upregulate either binding site (Figure 2c & d).
Activation of 4.2 kb-Nr2f2-Luc is dependent on PRE1 and PRE2
To confirm that POU4F3-mediated regulation of the Nr2f2 promoter is dependent on PRE1 and PRE2, both sites were mutated to attenuate POU4F3 binding. Mutant versions of PRE1 and PRE2 were synthesised and tested in EMSA analysis which showed effective attenuation of in vitro translated POU4F3 binding (Figure 3a). Both mutations were then introduced into the 4.2 kb-Nr2f2-Luc construct by overlap extension PCR to produce 4.2 kb-Nr2f2-Luc-Mut and the effect of these mutations on POU4F3-specific activation was investigated in co-transfection studies. Regulation of 4.2 kb-Nr2f2-Luc-Mut was attenuated to 1.3-fold compared to a two-fold activation of 4.2 kb-Nr2f2-Luc at the maximal amount of POU4F3 expression construct used (1 µg). This suggests that PRE1 and PRE2 are required for activation of the Nr2f2 5′ flanking region by POU4F3 (Figure 3b).
Figure 3. POU4F3 recognition element mutation attenuates POU4F3 binding and activation of the Nr2f2 5′ flanking region.
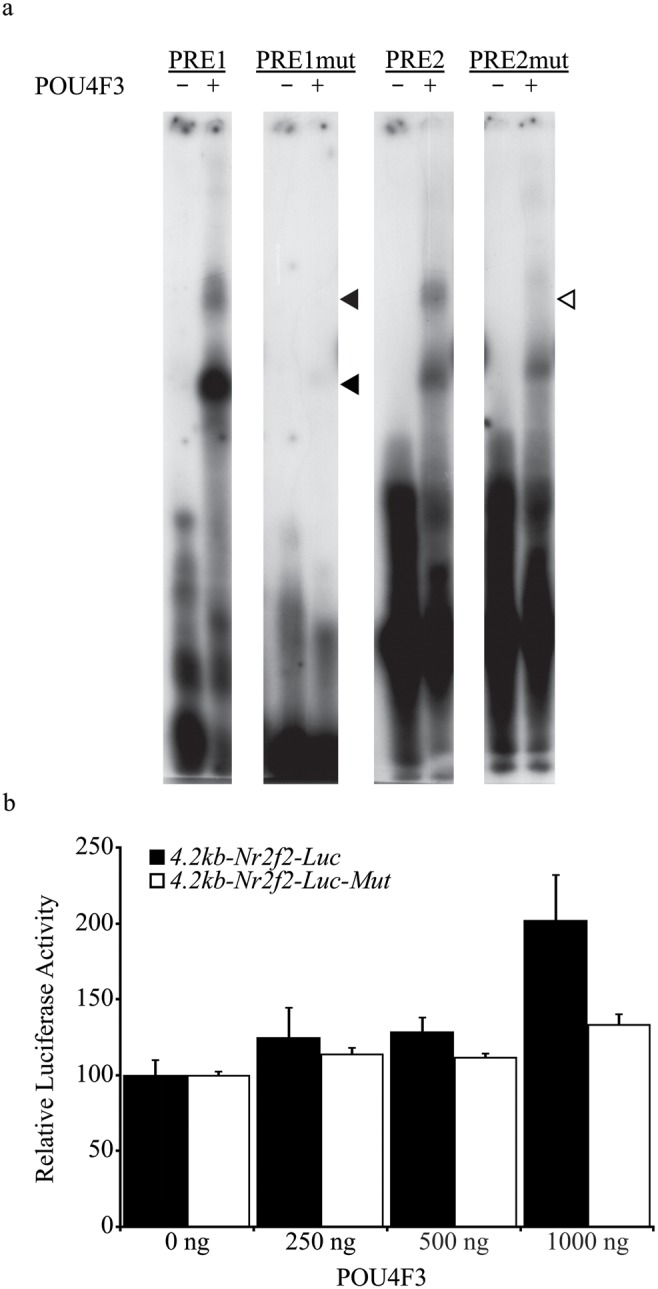
a. EMSA analysis of POU4F3 recognition element mutations on POU4F3 binding. Probes were either incubated alone or with in vitro translated POU4F3. Closed arrowheads indicate POU4F3-specific PRE1 bandshifts and the open arrowhead indicates the POU4F3-specific PRE2 bandshift. The mutation of each site severely compromises the ability of POU4F3 to bind to each sequence. b. Luciferase reporter assay upon co-transfection of POU4F3 with the 4.2 kb-Nr2f2-Luc-Mut reporter construct. Error bars represent the s.e.m (n = 6 for each data point).
Evolutionary Conservation of PRE1 and PRE2
Similar to protein-coding regions of the genome, functionally important regulatory elements are conserved across species [46]–[48]. Therefore, the evolutionary conservation of the POU4F3 binding sites PRE1 and PRE2 within the Nr2f2 5′ flanking sequence was investigated to indicate the potential functional importance of these sites in vivo. The Mulan program performs an analysis of sequences from multiple species; constructs phylogenetic trees; creates alignments; and identifies evolutionarily conserved regions. The output of this analysis is used as input for the MultiTF program which identifies evolutionarily conserved transcription factor binding sites [48].
The MultiTF program relies on the TRANSFAC Professional Version 10.2 library [49], however, no PWM is available for POU4F3 in this database. Therefore, PWMs for closely related transcription factor binding site families (i.e. OCT and POU) were used as POU family binding sites are closely related [4]. This analysis revealed two evolutionarily conserved binding sites in the human region of NR2F2 that correspond to the PRE1 locus in the mouse Nr2f2 5′ flanking region. The overlapping binding sites identified correspond to OCT1 and OCT_C PWMs (Figure 4a). No such evolutionarily conserved binding sites were identified for PRE2 which is less well-conserved than PRE1 across the species assessed (Figure 4b). This conservation suggests that PRE1 is more likely to play an important role in the regulation of Nr2f2 expression in vivo over recent evolution.
Figure 4. Evolutionary conservation of transcription factor binding sites in the Nr2f2 5′ flanking region.
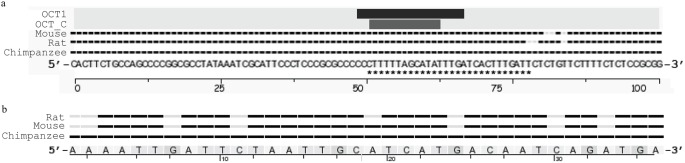
Schematic representation of the conservation of the region surrounding the human PRE1 sequence (a) and the human PRE2 sequence (b) compared to corresponding mouse, rat and chimpanzee sequences. a. The well-conserved PRE1 sequence is indicated by asterisks with evolutionarily conserved binding sites for the POU domain transcription factors OCT1 and OCT_C indicated by horizontal bars. Conserved bases are represented by black boxes with mismatches in grey. b. PRE2 is less well conserved with eight mismatched bases and no corresponding POU domain transcription factor binding sites in the TRANSFAC database. Conserved bases are again represented by black boxes with mismatches in grey.
Expression of NR2F2 in the embryonic rat inner ear
The inner ear’s function is primarily achieved by mechanosensory hair cells which are surrounded by support cells and extracellular matrix that form a complicated microanatomy which is required to achieve the special senses of hearing and balance. Accordingly, the development of this organ requires a complex co-ordination of cellular proliferation, differentiation and morphogenetic movement [50].
POU4F3 and NR2F2 are both expressed in the embryonic mouse inner ear. In the developing cochlea, Pou4f3 expression begins in the base and extends to include all hair cells of the inner ear by birth. This expression is unique to hair cells and correlates with the wave of hair cell differentiation [6]–[8], [51]. NR2F2 is more widely expressed in the developing apical organ of Corti with its expression level gradually reducing to become more refined to the LER in the basal cochlea [29].
The putative interaction of POU4F3 and Nr2f2 in the inner ear was investigated by further characterising the expression profile of NR2F2 in the E18 rat inner ear. NR2F2 expression was seen throughout the developing cochlea at this age with strongest expression seen in the greater epithelial ridge, lesser epithelial ridge and supero-medial wall of the apical cochlear duct (Figure 5). Expression decreased in an apical-to-basal direction with NR2F2 expression being limited to the lesser epithelial ridge of the basal cochlear duct (Figure 5). This embryonic expression profile is in keeping with the previously reported expression profile of NR2F2 in the embryonic mouse inner ear [29].
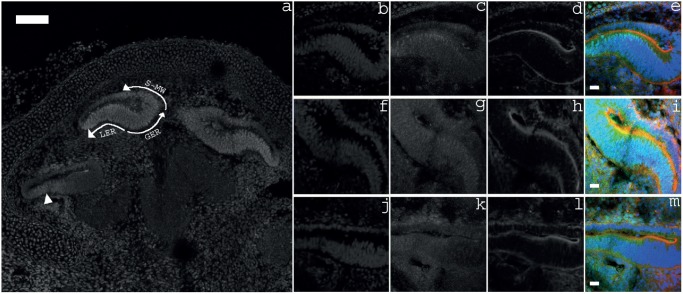
Figure 5. Expression of NR2F2 in the embryonic rat cochlea.
a. Overview of NR2F2 expression throughout the cochlea at E18. NR2F2 is most strongly expressed in the cochlear duct. In the apical duct, strong expression is seen in the super-medial wall (S-MW), greater epithelial ridge (GER), and lesser epithelial ridge (LER). This expression is reduced to the lesser epithelial ridge in the basal duct (arrowhead). b–e. Apical cochlear duct; f–i. Middle cochlear duct; and j–m. Basal cochlear duct. b, f & j, cell nuclei stained with DAPI; c, g & k, NR2F2 labelling; d, h & l, filamentous actin labelled by Phalloidin; and e, i & m, Merge of DAPI (blue), NR2F2 (green) and Phalloidin (red), scale bars: 100 µm in a and 20 µm in e, i & m.
NR2F2 expression in the postnatal mouse inner ear
The subtractive hybridization experiment that identified Nr2f2 as a putative target of POU4F3 was conducted to elucidate the pathway through which POU4F3 affects hair cell differentiation and survival. In the murine model, hair cell differentiation requires POU4F3 in the embryonic and early postnatal period [8]. However POU4F3 is also required for hair cell survival and is expressed postnatally [10]–[12], [17]. It must therefore influence its targets postnatally – though the targets required for hair cell maturation will not necessarily be identical to those required for maintenance. As NR2F2 is expressed postnatally in a number of organs, we investigated whether it is expressed in the postnatal inner ear to ascertain whether it is has the potential to play a role in hair cell maintenance as a POU4F3 target.
As in the embryonic cochlea, NR2F2 expression was widespread in the postnatal mouse cochlea both within and around the sensory epithelium. Throughout the P1 cochlea a typical nuclear pattern of NR2F2 expression was seen in hair cells as well as a number of supporting cells of the sensory epithelium i.e. Hensen cells, Claudius cells and inner sulcus cells (Figure 6). Expression appeared stronger in the apical coil including the hair cells as compared to the basal coil and hair cells. This apical-to-basal expression gradient was maintained in the adult cochlea where NR2F2 expression in the basal cochlea further decreased so that NR2F2 expression was no longer observed in the nuclei of basal hair cells. As in the cochlea, NR2F2 expression is widespread though variable in the postnatal cristae where its expression is greatest within the sensory epithelium, including hair cells (Figure 6).
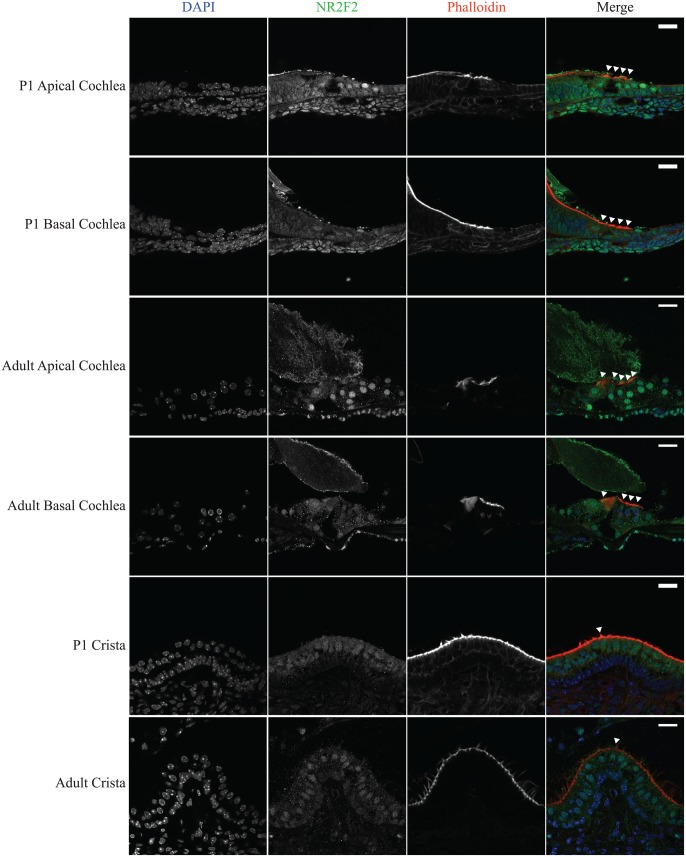
Figure 6. Expression of NR2F2 in the postnatal mouse inner ear.
Cryotome sections of P1 and adult mouse inner ears were subjected to immunohistochemistry in order to characterise their postnatal NR2F2 expression pattern. NR2F2 labelling is seen throughout the sensory epithelia of the cochlea and cristae. In the cochlea, NR2F2 labelling is of greater intensity in the apex than the base. NR2F2 is expressed in the nuclei of apical hair cells at all ages examined. In P1 mice, NR2F2 expression is reduced in basal hair cells. In adult mice, the expression of NR2F2 in basal hair cell nuclei appears further reduced. NR2F2 is also expressed in the hair cells of the cristae of the semicircular canals and this expression is maintained into adulthood. In merged images DAPI is blue, NR2F2 is green, Phalloidin is red, arrowheads indicate hair cells and scale bars: 20 µm.
Discussion
POU4F3 expression is essential for hair cell differentiation and maintenance [6]–[8], [10]–[12]. However, it’s only known direct targets (BDNF, NT-3 and Caprin-1) and downstream targets (Gfi1 and Lhx3) do not sufficiently explain the link between POU4F3 and the hair cell phenotype [14]–[16], [40]. In this paper, we furthered the understanding of this pathway by identifying and verifying Nr2f2 as a new regulatory target of POU4F3.
A combination of bioinformatic analysis, EMSA and reporter gene assays demonstrated that POU4F3 binds to two sites in the 4.2 kb Nr2f2 5′ flanking region and that the most proximal binding site (PRE1) is well-conserved through recent evolution. The sites identified were shown to be required for POU4F3-mediated upregulation of this region as their mutation attenuates this effect and were sufficient to confer POU4F3 activation upon a heterologous promoter (Figure 2). These results suggest that Nr2f2 is a direct regulatory target of POU4F3.
NR2F2 in the developing inner ear
Experiments in embryonic rats confirmed expression of NR2F2 in the developing cochlea (Figure 5) [29]. The widespread expression of NR2F2 indicates the presence of otic regulators of this gene. However, our data reveal that hair cell-selective Nr2f2 expression could be regulated by POU4F3 given its hair cell-specific expression pattern in the inner ear [6], [8].
As well as the POU4F3 binding sites described here, the Nr2f2 5′ flanking region is known to contain response elements for retinoic acid, sonic hedgehog, and Notch [52]–[54] which are all involved in inner ear development [50], [55], [56]. Furthermore, NR2F2 is known to mediate signalling between mesenchymal and endothelial compartments and it is thought that organs which require the differentiation of mesenchyme to epithelium display NR2F2 expression in the mesenchyme [57]. Epithelial-mesenchymal interactions give rise to most of the otic capsule and retinoic acid (which interacts with NR2F2) is thought to orchestrate mesenchyme-epithelial interactions in inner ear morphogenesis [54], [57], [58]. Though their relevance in the inner ear is not yet known, NR2F2 could act as an integrator of these important developmental pathways.
NR2F2 in the postnatal inner ear
The potential role of NR2F2 in hair cell survival and maintenance was investigated in the adult inner ear. In the postnatal mouse cochlea, NR2F2 expression is highest in the apex and falls to undetectable levels in basal hair cells. This pattern correlates with a previous report of higher apical POU4F3 expression in the postnatal rat cochlea [17]. The potentially matching expression pattern of these two transcription factors, if verified, supports the possibility that their interaction is active in the postnatal cochlea.
Apical hair cells are responsible for the detection of low frequency sounds and are the first hair cells of the cochlea to exit the cell cycle at approximately E13 in the developing mouse [59]. However, these hair cells are the last to complete their differentiation as the wave of hair cell differentiation proceeds from base to apex [30]. Though the gradient of NR2F2 expression seen in development mirrors hair cell differentiation this pattern persists despite full differentiation, i.e. after the onset of hearing. The nuclear expression pattern in these cells suggests that NR2F2 is acting as a transcription factor. However, as the postnatal role of NR2F2 is currently poorly understood, further work is required to determine its role in the hair cells of the inner ear.
The postnatal cochlear expression pattern of NR2F2 reported here is contrary to previously published data where it was not detected in the postnatal mouse organ of Corti [29]. This may be explained by several factors. For example, different antibodies were used in these experiments and tissue was obtained from different mouse strains [26], [29], [60]. As transcription factors typically have low expression levels, a small change in the sensitivity of the differing immunohistochemistry assays could account for this discrepancy.
In addition to its cochlear expression, NR2F2 expression was detected in the nuclei of hair cells and supporting cells of the postnatal mouse crista. The cristae form part of the vestibular apparatus and are required for the detection of rotational acceleration. A role for NR2F2 in this system would, therefore, imply a role in balance function. This is the first report of NR2F2 expression in the vestibular system and further work to improve the understanding of its relative expression and function at this site, as compared to the cochlea, may assist in elucidation of the molecular basis for phenotypic differences between cochlear and vestibular hair cells.
NR2F2 function in the inner ear
Given the diversity of genes and pathways in which NR2F2 is involved, it is possible that NR2F2 could play a novel role in cochlear development and maintenance. However, despite knowledge of its regulation, the function of NR2F2 in the inner ear remains unknown. Furthermore, the complex regulation of Nr2f2 and its various targets [61] make it difficult to differentiate any effects of POU4F3 from other activators of Nr2f2 which are known to be important in inner ear differentiation e.g. sonic hedgehog [62]. The hair-cell-specific expression of POU4F3 suggests that its interaction with Nr2f2 represents a regulatory mechanism that may be restricted to these sensory cells.
It is notable that a number of other reported regulatory targets of POU4F3 are transcription factors, including Lhx3 and Gfi-1 [16], [40] suggesting that it lies at the head of a complex and diverse regulatory pathway and may act as master-regulator of hair cell function. This is certainly consistent with the evidence provided from both transgenic mice with targeted deletion of Pou4f3 and humans with POU4F3 mutations, which exhibit loss of hearing due to loss of hair cell function.
Postnatally, mitochondrial biogenesis has been implicated in hair cell survival following the administration of ototoxic medications and noise exposure [63], [64]. NR2F2 has been shown to be involved in white adipose tissue mitochondrial biogenesis and may therefore influence this otoprotective function in postnatal hair cells [65]. Also, established roles of NR2F2 such as angiogenesis may be relevant to other cochlear cells that do not express POUF43 but do express NR2F2 [65].
Embryonic lethality at E10 makes it hard to assess the effect of Nr2f2 knockout on the developing mouse inner ear and no relevant conditional knockout mouse exists [24]. In the adult Nr2f2 knockout mouse, there is no overt phenotype, though hearing has yet to be assessed in these animals [66]. However, postnatal expression of NR2F2 in a number of organs – including the inner ear – suggests that it plays a functional role beyond development. Current evidence suggests that NR2F2 is most important in regeneration and de-differentiation in pathological conditions [65].
Therefore, beyond the verification of POU4F3 regulation of Nr2f2 in vivo, the characterisation of NR2F2 function in development and under pathological conditions in mature animals is essential to clarify the putative role of this versatile orphan nuclear receptor in the development and maintenance of hearing and balance.
Supporting Information
Nr2f2 expression in UB/OC-2 cells. Nr2f2 expression was investigated in proliferating UB/OC-2 cells. a, cDNA was generated from proliferating UB/OC-2 cells and subjected to PCR which demonstrated amplification of the predicted Nr2f2 fragment (213 bp). Integrity of the cDNA was demonstrated by amplification of Gapdh (450 bp) with sizes of amplified fragments measured against a 1 kb marker (M). Adjacent lanes have been removed for presentation purposes. b, The specificity of an anti-NR2F2 antibody was verified by western blot. A predominant 50 kDa band (arrow) with the expected molecular weight of NR2F2 is detected. Immunohistochemistry was carried out with (c) and without (d, secondary antibody alone) an anti-NR2F2 antibody in UB/OC-2 cells. This analysis showed NR2F2 expression localised to OC-2 cell nuclei. i, nuclei were stained DAPI; ii, NR2F2 expression; iii, Phalloidin labelling; and iv, shows a merged image of i, ii and iii with DAPI in blue, NR2F2 in green and Phalloidin in red. Scale bars: 100 µm.
(TIF)
Acknowledgments
We would like to thank Dr M Vasseur-Cognet, Dr M Studer and Professor K Avraham for their kind gifts of reagents. We would also like to thank Dr L Nolan for her general advice and technical assistance with the above experiments.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by grants from: Action on hearing loss, http://actiononhearingloss.org.uk/ (grant number 483 to JEG and SJD); the Wellcome Trust, http://www.wellcome.ac.uk/ (grant number 091092/Z/09/Z to JEG and SJD); and the original subtractive hybridisation screen was supported by a grant from the Wellcome Trust (grant number 064599 to SJD). CT’s research was undertaken as part of a studentship that was funded by GlaxoSmithKline (http://www.gsk.com/uk) via the UCL MB PhD programme (http://www.ucl.ac.uk/mbphd). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Andersen B, Rosenfeld MG (2001) POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr Rev 22: 2–35. [DOI] [PubMed] [Google Scholar]
- 2. Scott MP, Tamkun JW, Hartzell III GW (1989) The structure and function of the homeodomain. Biochim Biophys Acta - Rev Cancer 989: 25–48. [DOI] [PubMed] [Google Scholar]
- 3. Aurora R, Herr W (1992) Segments of the POU domain influence one another’s DNA-binding specificity. Mol Cell Biol 12: 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Verrijzer CP, Van der Vliet PC (1993) POU domain transcription factors. Biochim Biophys Acta 1173: 1–21. [DOI] [PubMed] [Google Scholar]
- 5. Badea TC, Williams J, Smallwood P, Shi M, Motajo O, et al. (2012) Combinatorial expression of Brn3 transcription factors in somatosensory neurons: genetic and morphologic analysis. J Neurosci 32: 995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Erkman L, McEvilly RJ, Luo L, Ryan AK, Hooshmand F, et al. (1996) Role of transcription factors a Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature 381: 603–606. [DOI] [PubMed] [Google Scholar]
- 7. Xiang M, Gan L, Li D, Chen ZY, Zhou L, et al. (1997) Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc Natl Acad Sci U S A 94: 9445–9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiang, Gao WQ, Hasson T, Shin JJ, Xiang M (1998) Requirement for Brn-3c in maturation and survival, but not in fate determination of inner ear hair cells. Development 125: 3935–3946. [DOI] [PubMed] [Google Scholar]
- 9. Keithley EM, Erkman L, Bennett T, Lou L, Ryan AF (1999) Effects of a hair cell transcription factor, Brn-3.1, gene deletion on homozygous and heterozygous mouse cochleas in adulthood and aging. Hear Res 134: 71–76. [DOI] [PubMed] [Google Scholar]
- 10. Collin RWJ, Chellappa R, Pauw R-J, Vriend G, Oostrik J, et al. (2008) Missense mutations in POU4F3 cause autosomal dominant hearing impairment DFNA15 and affect subcellular localization and DNA binding. Hum Mutat 29: 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee HK, Park H-J, Lee K-Y, Park R, Kim U-K (2010) A novel frameshift mutation of POU4F3 gene associated with autosomal dominant non-syndromic hearing loss. Biochem Biophys Res Commun 396: 626–630. [DOI] [PubMed] [Google Scholar]
- 12. Vahava O, Morell R, Lynch ED, Weiss S, Kagan ME, et al. (1998) Mutation in Transcription Factor POU4F3 Associated with Inherited Progressive Hearing Loss in Humans. Science 279: 1950–1954. [DOI] [PubMed] [Google Scholar]
- 13. Kim H-J, Won H-H, Park K-J, Hong SH, Ki C-S, et al. (2013) SNP linkage analysis and whole exome sequencing identify a novel POU4F3 mutation in autosomal dominant late-onset nonsyndromic hearing loss (DFNA15). PLoS One 8: e79063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clough RL, Sud R, Davis-Silberman N, Hertzano R, Avraham KB, et al. (2004) Brn-3c (POU4F3) regulates BDNF and NT-3 promoter activity. Biochem Biophys Res Commun 324: 372–381. [DOI] [PubMed] [Google Scholar]
- 15. Towers ER, Kelly JJ, Sud R, Gale JE, Dawson SJ (2011) Caprin-1 is a target of the deafness gene Pou4f3 and is recruited to stress granules in cochlear hair cells in response to ototoxic damage. J Cell Sci 124: 1145–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hertzano R, Dror AA, Montcouquiol M, Ahmed ZM, Ellsworth B, et al. (2007) Lhx3, a LIM domain transcription factor, is regulated by Pou4f3 in the auditory but not in the vestibular system. Eur J Neurosci 25: 999–1005. [DOI] [PubMed] [Google Scholar]
- 17. Gross J, Angerstein M, Fuchs J, Stute K, Mazurek B (2011) Expression analysis of prestin and selected transcription factors in newborn rats. Cell Mol Neurobiol 31: 1089–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rivolta MN, Grix N, Lawlor P, Ashmore JF, Jagger DJ, et al. (1998) Auditory hair cell precursors immortalized from the mammalian inner ear. Proc Biol Sci 265: 1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shiina N, Shinkura K, Tokunaga M (2005) A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J Neurosci 25: 4420–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shiina N, Yamaguchi K, Tokunaga M (2010) RNG105 deficiency impairs the dendritic localization of mRNAs for Na+/K+ ATPase subunit isoforms and leads to the degeneration of neuronal networks. J Neurosci 30: 12816–12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Solomon S, Xu Y, Wang B, David MD, Schubert P, et al. (2007) Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol Cell Biol 27: 2324–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsai SY, Tsai MJ (1997) Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocr Rev 18: 229–240. [DOI] [PubMed] [Google Scholar]
- 23. Suzuki T, Moriya T, Darnel a D, Takeyama J, Sasano H (2000) Immunohistochemical distribution of chicken ovalbumin upstream promoter transcription factor II in human tissues. Mol Cell Endocrinol 164: 69–75. [DOI] [PubMed] [Google Scholar]
- 24. Pereira FA, Qiu Y, Zhou G, Tsai M-J, Tsai SY (1999) The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev 13: 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. You L, Lin F, Lee C, DeMayo F, Tsai M (2005) Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature 435: 98–104. [DOI] [PubMed] [Google Scholar]
- 26. Tripodi M, Filosa A, Armentano M, Studer M (2004) The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development 131: 6119–6129. [DOI] [PubMed] [Google Scholar]
- 27. Takamoto N, You L-R, Moses K, Chiang C, Zimmer WE, et al. (2005) COUP-TFII is essential for radial and anteroposterior patterning of the stomach. Development 132: 2179–2189. [DOI] [PubMed] [Google Scholar]
- 28. McCaffery P, Wagner E, O’Neil J, Petkovich M, Dräger UC (1999) Dorsal and ventral retinal territories defined by retinoic acid synthesis, break-down and nuclear receptor expression. Mech Dev 82: 119–130. [DOI] [PubMed] [Google Scholar]
- 29. Tang LS, Alger HM, Lin F, Pereira FA (2005) Dynamic expression of COUP-TFI and COUP-TFII during development and functional maturation of the mouse inner ear. Gene Expr Patterns 5: 587–592. [DOI] [PubMed] [Google Scholar]
- 30.Lim DJ, Anniko M (1985) Developmental morphology of the mouse inner ear. A scanning electron microscopic observation. Acta Otolaryngol Suppl 422: 1–69. [PubMed]
- 31. Tang LS, Alger HM, Pereira FA (2006) COUP-TFI controls Notch regulation of hair cell and support cell differentiation. Development 133: 3683–3693. [DOI] [PubMed] [Google Scholar]
- 32. Ladias JAA, Karathanasis SK (1991) Regulation of the Apolipoprotein AI Gene by ARP-1, a Novel Member of the Steroid Receptor Superfamily. Science 251: 561–565. [DOI] [PubMed] [Google Scholar]
- 33. Lopes da Silva S, Cox JJ, Jonk LJ, Kruijer W, Burbach JP (1995) Localization of transcripts of the related nuclear orphan receptors COUP-TF I and ARP-1 in the adult mouse brain. Brain Res Mol Brain Res 30: 131–136. [DOI] [PubMed] [Google Scholar]
- 34. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 35. Scherf M, Klingenhoff A, Werner T (2000) Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: a novel context analysis approach. J Mol Biol 297: 599–606. [DOI] [PubMed] [Google Scholar]
- 36. Quandt K, Frech K, Karas H, Wingender E, Werner T (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23: 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, et al. (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942. [DOI] [PubMed] [Google Scholar]
- 38. Frech K, Werner T (1996) Specific Modelling of Regulatory Units in DNA Sequences. Nucleic Acids Res 24: 1212–1219. [PubMed] [Google Scholar]
- 39. Perilhou A, Tourrel-Cuzin C, Zhang P, Kharroubi I, Wang H, et al. (2008) The MODY1 gene for hepatocyte nuclear factor 4alpha and a feedback loop control COUP-TFII expression in pancreatic beta cells. Mol Cell Biol 28: 4588–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hertzano R, Montcouquiol M, Rashi-Elkeles S, Elkon R, Yücel R, et al. (2004) Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet 13: 2143–2153. [DOI] [PubMed] [Google Scholar]
- 41. Wood JN, Bevan SJ, Coote PR, Dunn PM, Harmar A, et al. (1990) Novel cell lines display properties of nociceptive sensory neurons. Proc Biol Sci 241: 187–194. [DOI] [PubMed] [Google Scholar]
- 42.Kingston RE, Chen CA, Okayama H (2001) Calcium phosphate transfection. Curr. Protoc. Immunol. p. Unit 10.13. [DOI] [PubMed]
- 43. Ling MM, Robinson BH (1997) Approaches to DNA mutagenesis: an overview. Anal Biochem 254: 157–178. [DOI] [PubMed] [Google Scholar]
- 44. Ovcharenko I, Nobrega MA, Loots GG, Stubbs L (2004) ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res 32: W280–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gruber CA, Rhee JM, Gleiberman A, Turner EE (1997) POU domain factors of the Brn-3 class recognize functional DNA elements which are distinctive, symmetrical, and highly conserved in evolution. Mol Cell Biol 17: 2391–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, et al. (2000) Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288: 136–140. [DOI] [PubMed] [Google Scholar]
- 47. Elnitski L, Li J, Noguchi CT, Miller W, Hardison R (2001) A negative cis-element regulates the level of enhancement by hypersensitive site 2 of the beta-globin locus control region. J Biol Chem 276: 6289–6298. [DOI] [PubMed] [Google Scholar]
- 48. Ovcharenko I, Loots GG, Giardine BM, Hou M, Ma J, et al. (2005) Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res 15: 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matys V, Kel-Margoulis O V, Fricke E, Liebich I, Land S, et al. (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bok J, Chang W, Wu DK (2007) Patterning and morphogenesis of the vertebrate inner ear. Int J Dev Biol 51: 521–533. [DOI] [PubMed] [Google Scholar]
- 51. Raz Y, Kelley MW (1999) Retinoic acid signaling is necessary for the development of the organ of Corti. Dev Biol 213: 180–193. [DOI] [PubMed] [Google Scholar]
- 52. Diez H, Fischer A, Winkler A, Hu C-J, Hatzopoulos AK, et al. (2007) Hypoxia-mediated activation of Dll4-Notch-Hey2 signaling in endothelial progenitor cells and adoption of arterial cell fate. Exp Cell Res 313: 1–9. [DOI] [PubMed] [Google Scholar]
- 53. Krishnan V (1997) Mediation of Sonic Hedgehog-Induced Expression of COUP-TFII by a Protein Phosphatase. Science 278: 1947–1950. [DOI] [PubMed] [Google Scholar]
- 54. Qiu Y, Krishnan V, Pereira FA, Tsai SY, Tsai MJ (1996) Chicken ovalbumin upstream promoter-transcription factors and their regulation. J Steroid Biochem Mol Biol 56: 81–85. [DOI] [PubMed] [Google Scholar]
- 55. Romand R, Dolle P, Hashino E (2006) Retinoid Signaling in Inner Ear Development. J Neurobiol 66: 687–704. [DOI] [PubMed] [Google Scholar]
- 56. Kelley MW (2006) Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci 7: 837–849. [DOI] [PubMed] [Google Scholar]
- 57. Pereira FA, Tsai MJ, Tsai SY (2000) COUP-TF orphan nuclear receptors in development and differentiation. Cell Mol Life Sci 57: 1388–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fekete DM (1999) Development of the vertebrate ear: insights from knockouts and mutants. Trends Neurosci 22: 263–269. [DOI] [PubMed] [Google Scholar]
- 59.Ruben RJ (1967) Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta Otolaryngol: Suppl 220: 1–44. [PubMed]
- 60. Zhou C, Qiu Y, Pereira FA, Crair MC, Tsai SY, et al. (1999) The nuclear orphan receptor COUP-TFI is required for differentiation of subplate neurons and guidance of thalamocortical axons. Neuron 24: 847–859. [DOI] [PubMed] [Google Scholar]
- 61. Park J-I, Tsai SY, Tsai M-J (2003) Molecular mechanism of chicken ovalbumin upstream promoter-transcription factor (COUP-TF) actions. Keio J Med 52: 174–181. [DOI] [PubMed] [Google Scholar]
- 62. Krishnan V, Elberg G, Tsai MJ, Tsai SY (1997) Identification of a novel sonic hedgehog response element in the chicken ovalbumin upstream promoter-transcription factor II promoter. Mol Endocrinol 11: 1458–1466. [DOI] [PubMed] [Google Scholar]
- 63. Hyde GE, Rubel EW (1995) Mitochondrial role in hair cell survival after injury. Otolaryngol Head Neck Surg 113: 530–540. [DOI] [PubMed] [Google Scholar]
- 64. Choi C-H, Chen K, Vasquez-Weldon A, Jackson RL, Floyd RA, et al. (2008) Effectiveness of 4-hydroxy phenyl N-tert-butylnitrone (4-OHPBN) alone and in combination with other antioxidant drugs in the treatment of acute acoustic trauma in chinchilla. Free Radic Biol Med 44: 1772–1784. [DOI] [PubMed] [Google Scholar]
- 65. Lin F-J, Qin J, Tang K, Tsai SY, Tsai M-J (2011) Coup d’Etat: an orphan takes control. Endocr Rev 32: 404–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Qin J, Tsai M-J, Tsai SY (2008) Essential roles of COUP-TFII in Leydig cell differentiation and male fertility. PLoS One 3: e3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nr2f2 expression in UB/OC-2 cells. Nr2f2 expression was investigated in proliferating UB/OC-2 cells. a, cDNA was generated from proliferating UB/OC-2 cells and subjected to PCR which demonstrated amplification of the predicted Nr2f2 fragment (213 bp). Integrity of the cDNA was demonstrated by amplification of Gapdh (450 bp) with sizes of amplified fragments measured against a 1 kb marker (M). Adjacent lanes have been removed for presentation purposes. b, The specificity of an anti-NR2F2 antibody was verified by western blot. A predominant 50 kDa band (arrow) with the expected molecular weight of NR2F2 is detected. Immunohistochemistry was carried out with (c) and without (d, secondary antibody alone) an anti-NR2F2 antibody in UB/OC-2 cells. This analysis showed NR2F2 expression localised to OC-2 cell nuclei. i, nuclei were stained DAPI; ii, NR2F2 expression; iii, Phalloidin labelling; and iv, shows a merged image of i, ii and iii with DAPI in blue, NR2F2 in green and Phalloidin in red. Scale bars: 100 µm.
(TIF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.