Abstract
Tumor-targeted antibody therapy has had a major impact on reducing morbidity and mortality in a wide range of cancers. Antibodies mediate their antitumor activity in part by activating immune effector cells; however, the tumor microenvironment (TME) is enriched with cellular and soluble mediators that actively suppress generation of antitumor immunity. Here, we investigate the potential of prospectively identifying and neutralizing an immunomodulatory soluble mediator within the TME to enhance therapeutic efficacy of the HER2-directed antibody trastuzumab. Using the D5-HER2 cell line and an immunocompetent human HER2 transgenic animal (hmHER2Tg) in which human HER2 is a self-antigen, we determined that IL4 was present in the TME and produced by both tumor and stromal cells. A siRNA-based screening approach identified Stat5a as a novel negative regulator of IL4 production by D5-HER2 tumor cells. Furthermore, IL4 neutralization using the anti-IL4 antibody 11B11 enhanced the efficacy of trastuzumab and modulated the TME. For example, IL4 neutralization resulted in reduced levels of myeloid chemoattractants CCL2, CCL11, and CXCL5 in the TME. Combination therapy with 11B11 and trastuzumab resulted in a reduction of tumor-infiltrating CD11b+CD206+ myeloid cells compared to monotherapy. These data suggest that IL4 neutralization enhances the efficacy of trastuzumab by influencing the phenotype of myeloid cells within the TME and provides further rationale for combining tumor-targeted antibody therapy with agents that neutralize factors in the TME that suppress generation of productive antitumor immune responses.
Keywords: Antibody, Tumor Immunology, Tumor Microenvironment, Macrophage, Cytokine/Chemokine
INTRODUCTION
Monoclonal antibody (mAb) therapy has revolutionized treatment of both hematologic malignancies and solid tumors (1). In the setting of solid tumors, growth factor receptors, including epidermal growth factor receptor (EGFR) family members EGFR and HER2, have proven to be useful targets for antibody therapy. HER2, which is overexpressed and gene amplified in approximately 30% of invasive breast cancers, confers a poor prognosis and is the target of the mAb trastuzumab (Herceptin®)(2). Trastuzumab showed activity alone and in combination with chemotherapy as a first or second line agent in patients with HER2+ breast cancer and is currently FDA approved for use in the adjuvant setting in patients with node positive, HER2 overexpressing breast cancer (3, 4). In 2010, trastuzumab was also approved for use in patients with HER2+ metastatic gastric or gastroesophageal junction tumors based on data showing an approximately 2.7 month improvement in survival when trastuzumab was added to a regimen of chemotherapy (5).
The clinical utility of antibody therapy has been attributed to multi-modal mechanisms of action that include perturbation of signal transduction through cell-surface receptors and activation of immune effectors through, for example, interactions between antibody Fc domains and Fc-gamma receptors (FcγR) expressed on immune cells. Tumors overexpressing growth factor receptors are particularly prone to signaling perturbation via antibody blockade of ligand binding, which leads to disruption of the cell cycle and induction of apoptosis. However, a growing body of evidence suggests that activation of immune effectors also contributes to efficacy of tumor-targeted antibodies in vivo. Deletion of the inhibitory FcγRIIB in mice resulted in enhanced antitumor activity of trastuzumab and the CD20-targeted antibody rituximab (6). Clinically, patients with activating FcγR polymorphisms have a more favorable prognosis in the setting of non-Hodgkin’s lymphoma, colorectal cancer and breast cancer (7–9). Engagement of activating Fc-receptors can lead to induction of antibody-dependent cell mediated cytotoxicity (ADCC), complement fixation or phagocytosis, all of which can lead to tumor cell death and liberation of tumor antigens. These tumor antigens can be processed and presented to T cells by antigen-presenting cells, such as macrophages and dendritic cells (DC), and result in generation of an antibody-initiated, tumor-directed adaptive immune response. Our group and others have demonstrated that antibody-initiated adaptive immunity plays an important role in the therapeutic activity of HER2-directed antibodies (10, 11). The phenotype of this immune response is critical and is shaped by the composition of the tumor microenvironment (TME). Cellular components, such as myeloid derived suppressor cells (MDSC) and T-regulatory cells (T reg), together with soluble factors, such as IL10 and TGF-β, collectively serve to limit infiltration of effector T cells and inhibit Th1 polarization, which is associated with a more favorable prognosis in a wide range of cancers (12). In addition, tumor-infiltrating macrophages are often “alternatively activated” and serve to drive tumor growth and dissemination (13).
We previously reported results using a unique model system to study and optimize tumor-targeted antibody therapy. This model employs a human HER2 transgenic (hmHER2Tg) mouse that ubiquitously expresses a kinase-inactivated human HER2, and the murine melanoma cell line D5, which has been engineered to overexpress full length human HER2 (D5-HER2)(11). hmHER2Tg animals are immunologically tolerant to human HER2 and do not mount a xenogenic immune response to D5-HER2; therefore, this model permits direct study of the impact of trastuzumab on the induction of T cell-based anti-HER2 immune responses. Using this model, trastuzumab monotherapy prevented tumor growth in approximately 30% of animals, and protected those animals against tumor re-challenge (11). In the current study we have used this model system to investigate the potential of enhancing trastuzumab’s efficacy by targeting and neutralizing immunomodulatory soluble mediators in the TME in order to skew polarization of immune effectors and ultimately drive productive antitumor immunity.
MATERIALS AND METHODS
Cell Lines
Generation of D5-HER2 has previously been described (11). Briefly, D5 cells (a subclone of the poorly immunogenic B16/BL6 murine melanoma; gift from Dr. Suyu Shu; Cleveland Clinic Foundation, Cleveland, OH) were transfected with full length human HER2 cDNA expressed under control of the CMV promoter. D5-HER2 cells were grown and maintained in RPMI supplemented with 10% fetal bovine serum (FBS) and 2mM L-glutamine. HEK293T (gift from Dr. Anton Wellstein; Georgetown University; Washington, DC), EO771 (gift from Dr. Peter Goedegeburre; Washington University in St. Louis; St. Louis, MO) and EO771-HER2 cell lines were maintained in DMEM supplemented with 10% FBS and 2mM L-glutamine. All cell lines were tested and determined to be free of Mycoplasma and other rodent pathogens; no other authentication assay was performed.
Generation of EO771-HER2 cell line
Human ERBB2 cDNA (DNASU Plasmid repository (14); plasmid: HsCD0002235(15)) was cloned into the entry vector pENTR4 (Life Technologies; Carlsbad, CA). LR Clonase enzyme mix (Life Technologies) was used to facilitate recombination of HER2 cDNA from pENTR4-ERBB2 into the destination vector pLENTI CMV Blast DEST (Addgene plasmid 17451 submitted by Dr. Eric Campeau (16)). To generate functional virions, HEK293T cells were transfected at approximately 70% confluence with 15μg pLENTI-ERBB2, 20μg psPAX2 (Addgene plasmid 12260 submitted by Dr. Didier Trono) and 3ug VSV-G (gift from Dr. Todd Waldman; Georgetown University; Washington, DC) plasmids using 18μL of FuGENE6 transfection reagent (Promega; Madison, Wisconsin). Media was replaced with normal growth media 24 hours after transfection. Virus containing media was harvested 48 hours post-transfection, centrifuged, filtered using a 0.45 micron syringe filter and stored at −80°C until use. To generate EO771-HER2 cells, EO771 cells transduced with 3mL virus containing media, 1ml normal growth media and 3.2μg polybrene (Sigma; St. Louis, MO). Media was replaced 24 hours post transduction with normal growth media and at 48 hours with media containing blasticidin. After 7 days of selection, single clones were identified by limiting dilution assay. Surface expression of human HER2 was assessed on individual clones using flow cytometry (not shown).
Luminex-based Cytokine Analysis
Tumors were harvested when they reached approximately 10–15mm in diameter, and homogenized in five volumes of PBS+0.5% Tween-20 with protease inhibitors (Roche; Penzberg, Bavaria, Germany). Homogenates were centrifuged and the supernatant was immediately stored at −80°C. Protein concentration was determined using the BCA assay (Bio-Rad; Hercules, CA). For cell culture analysis, supernatant was harvested from D5-HER2 cells grown in 96-well plates for 72 hours (75–80% confluence). Media was centrifuged and stored at −80°C. Samples were sent to Eve Technologies (Calgary, Alberta, Canada) and analyzed in duplicate using the mouse 32-plex cytokine/chemokine Discovery Assay.
Creation of IL4 siRNA library
Pathway Studio (Ariadne Genomics; Elsevier; Amsterdam, Netherlands) was used to search the literature and curate a list of proteins that positively regulate IL4 expression (supplemental table 1). Search criteria included proteins that bound the IL4 promoter and/or positively regulated IL4 transcription. The resultant gene list was used to create a custom siRNA library in 96-well plate format (Qiagen; Venlo, Netherlands). Each well of the plate contained two pooled siRNAs targeting a single gene.
Primary siRNA Screen and identification of hits
Library plates containing siRNAs were resuspended in siRNA suspension buffer (Qiagen) and used in conjunction with RNAiMax transfection reagent (Life Technologies) to create siRNA/lipid complexes. D5-HER2 cells were harvested at 75–80% confluence and reverse transfected with siRNA/lipid complexes in a total volume of 110μL (15nM siRNA). A non-targeting control siRNA (siNEG) and a murine death control siRNA (siDEATH) were used to verify the fidelity of transfection for each screen. Supernatant was harvested at 72 hours post-transfection to determine IL4 protein concentration and cells were stained with crystal violet (CV) as a surrogate measurement of viability. Genes whose knockdown resulted in greater than a 50% reduction in viability were excluded from analysis. ELISA and CV data were normalized to values obtained from transfection with siNEG and the ELISA:CV ratios were generated. These ratios were log-transformed and a hit was identified as having a z-score of at least +/−1.20. All screens were performed in duplicate and repeated three times.
Validation of hits obtained from primary siRNA screen
For each hit identified in the primary screen, four unique siRNAs targeting each gene were used in validation studies (two siRNAs used in primary screen and two additional siRNAs). Each siRNA was placed in a single well of a 96-well pate and used to transfect D5-HER2 cells as previously described. A hit was considered validated if two or more siRNAs targeting the gene recapitulated the phenotype observed in the primary screen. All screens were performed in duplicate and repeated three times.
ELISA
Cell culture supernatants and tumor homogenates were prepared as described above. IL4 protein concentration was determined using the standard mouse IL-4 ELISA MAX kit (Biolegend; San Diego, CA) according to the manufacturer’s recommendations.
Real-Time PCR
RNA was extracted from cell pellets using the PureLink RNA Mini Kit (Life Technologies). RNA concentration was determined spectrophotometrically using NanoDrop 1000 (Thermo Scientific; Waltham, MA). 1μg of RNA was reverse-transcribed using Omniscript RT (Qiagen) in a total volume of 25μL. Quantitative PCR was performed using 2μL of cDNA, Quantitect SYBR Green (Qiagen) and pre-designed primer pairs to IL4 and GAPDH (Quantitect Primer Assay; Qiagen) in a total volume of 25μL according to manufacturer’s recommendation. Reactions were performed using the 7900HT Real-Time PCR System (Thermo Scientific) and analyzed using the ΔΔCT method using GAPDH as the endogenous control.
Western Blotting
D5-HER2 cells were lysed using radio-immunoprecipitation assay (RIPA) buffer (Boston Bioproducts; Ashland, MA) and protein concentration was determined by the BCA assay (Bio-Rad). Approximately 16–20μg of protein was run on tris-glycine gels under reducing conditions. Protein was transferred to a nitrocellulose membrane and blocked for two hours using non-fat milk (Bio-Rad). The following primary antibody dilutions were used: Stat5a (ab32403; 1:1000; Abcam; Cambridge, England, United Kingdom), anti-phospho Y694 Stat5a (ab30648; 1:500; Abcam), GAPDH (D16H11; 1:1000; Cell Signaling; Danvers, MA). Primary antibodies were diluted in PBS-T +5% non-fat milk and incubated overnight at 4°C. Membranes were incubated for one hour at room temperature with secondary antibody (HRP labeled anti-rabbit IgG; 1:10,000; GE Healthcare; Little Chalfont, Bucking hamshire, United Kingdom) diluted in PBS-T. For Stat5a and phospho-Stat5a westerns, Supersignal West Femto high sensitivity substrate (Thermo Scientific) was used for visualization while Supersignal West Pico (Thermo Scientific) was used for GAPDH westerns.
Stat5a Overexpression
Stat5a cDNA from pMXc-neo-Stat5a WT-FLAG and pMXc-neo-Stat5a1*6-FLAG constructs (gift from Dr. Toshio Kitamura; University of Tokyo) were cloned into pCMV-SPORT6 mammalian expression vectors (Life Technologies). Stat5a1*6 is a constitutively active mutant of Stat5a, which contains a H229R point mutation in the N-terminal domain and a S711F point mutation in the transactivation domain (17). For transient transfections, 3000 D5-HER2 cells were plated on day 0 and transfected on day 1 with either pCMV-SPORT5-Stat5aWT or pCMV-SPORT6-Stat5a1*6 using Lipofectamine 2000 transfection reagent (Life Technologies). Media was collected 72hrs post transfection for ELISA and cells were collected and prepared for western blotting.
Mice
Generation of human HER2 transgenic mice (hmHER2Tg) was previously described (11). Abrogation of HER2 kinase activity was accomplished by replacement of a lysine residue at position 753 with a methionine residue. The resultant HER2 transgene was transferred to FVB donor zygotes and animals were backcrossed to C57Bl/6 animals for 14 generations. IL4−/− animals (C57BL/6-Il4tm1Nnt/J) were purchased from Jackson Laboratories (Bar Harbor, Maine) and crossed to hmHER2Tg animals to create hmHER2Tg:IL4−/− animals.
Subcutaneous inoculation, treatment and monitoring of D5-HER2 and EO771-HER tumors
Cohorts of 8 to 12-week old female hmHER2Tg or hmHER2Tg:IL4−/− mice were injected in the flank with 3×103 D5-HER2 cells or 1×106 EO771-HER2 cells subcutaneously on day 0. Animals were randomized on day 1 to receive intraperitoneal injections (i.p.) of either PBS, 200μg trastuzumab (Herceptin®; Genentech, South San Francisco, CA), 1mg anti-IL4 (clone 11B11; BioXcell; West Lebanon, NH), 1mg GL113 (rat IgG1 isotype control), or trastuzumab + 11B11 (or GL113) combination therapy. The GL113 antibody was produced from a hybridoma (gift from Dr. Fred Finkelman; University of Cincinnati) and purified using Protein G agarose beads (Thermo Scientific). Trastuzumab was dosed twice a week for four weeks and 11B11/GL113 was dosed every five days for a total of six doses. Tumor growth was followed every 2–3 days and animals were sacrificed when tumors reached 2cm in the largest diameter. All animal experiments were performed in accordance with the Georgetown University Institutional Animal Care and use Committee.
Animal re-challenge experiments
Approximately 120 days after the initiation of primary challenge experiments, tumor-free animals were re-challenged with 1.5×104 D5-HER2 tumor cells subcutaneously in the opposite flank. No additional treatments were administered. Tumors were measured every 2–3 days and animals were sacrificed when tumors reached 2cm in the largest diameter.
Flow Cytometry
Tumors were harvested when they reached approximately 10–15mm in the largest diameter and were mechanically disrupted and subjected to enzymatic digest using a 1mg/mL collagenase D solution (Roche). Red blood cells were lysed using a 0.84% NH4Cl solution. Cells were washed twice with RPMI+10%FBS and passed through a 70-μm cell strainer. The resultant cell suspension was washed twice with ice cold PBS and stained with LIVE/DEAD-Violet dye according to manufacturer’s recommendation (Life Technologies). Fc-receptors were blocked using 1μg anti-CD16/CD32 (Biolegend; TruStain FcX). Staining for surface antigens was performed in PBS+1%BSA using the following antibodies: CD45 (Biolegend; clone 30-F11), F4/80 (Biolegend; clone BM8), MHCII (Biolegend; clone M5/114.15.2), CD80 (BD Biosciences; San Jose, CA; clone 16-10A1), CD86 (BD Biosciences; clone GL1), Gr-1 (Biolegend; clone RB6-8C5), CD11b (Biolegend; clone M1/70), NK1.1 (eBioscience; San Diego, CA; clone PK136), CD3 (Biolegend; clone 145-2C11), CD4 (eBioscience; clone RM4-5), CD8a (Biolegend; clone 53-6.7), CD69 (Biolegend; clone H1.2F3), CD206 (Biolegend; clone C068C2), FoxP3 (eBioscience; clone NRRF-30), and HER2 (BD Bioscience; clone Neu 24.7). Intracellular staining for Foxp3 was performed using fixation and permeabilization buffers according to the manufacturer’s recommended protocol (eBioscience).
Statistical Analysis
All statistical analysis was performed using GraphPad Prism version 6 (GraphPad Software; La Jolla, CA). Log transformation and z-score analysis was performed using Microsoft Excel.
RESULTS
IL4 is present in the tumor microenvironment and is expressed by D5-HER2 cells
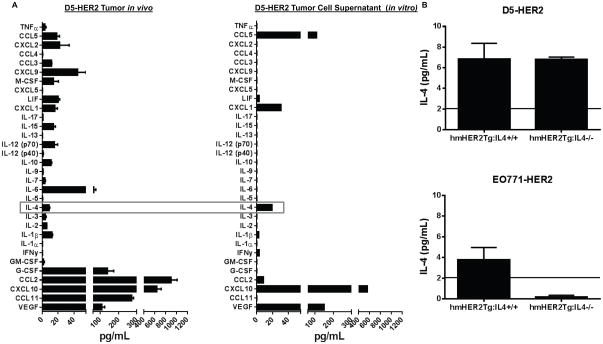
In order to determine which immunomodulatory soluble mediators may be relevant targets in our model system, a Luminex assay was performed to determine the concentration of 32 cytokines and chemokinesin the D5-HER2 TME(Fig. 1A). Various cytokines and chemokines, including CCL2, CXCL10, CCL11 and IL4, were expressed in the microenvironment of D5-HER2 tumors. D5-HER2 cells grown in vitro also expressed high levels of CXCL10 in addition to CCL5, VEGF and IL4 (Fig. 1A). This analysis revealed several putative targets for therapeutic neutralization; we ultimately chose IL4 for further study due to the pleiotropic effects of this cytokine on the phenotype and activation of both myeloid and lymphoid cells, and the fact that it was both present in the TME and produced by D5-HER2 cells in vitro (Fig. 1A). To determine if IL4 is produced by D5-HER2 tumor cells in vivo or is the product of the stromal cells surrounding the tumor, we assessed the concentration of IL4 in the TME of D5-HER2 tumors grown in hmHER2Tg:IL4−/− and IL4+/+animals. Tumors grown in hmHER2Tg:IL4−/− animals had a similar concentration of IL4 compared to tumors grown in hmHER2Tg:IL4+/+ animals, suggesting that D5-HER2 cells do indeed express IL4 in vivo (Fig. 1B). In contrast, the medullary breast cancer cell line EO771-HER2, which does not express IL4 in vitro (data not shown), contained low levels of IL4 in tumors grown in hmHER2Tg:IL4+/+ and no IL4 in tumors grown in hmHER2Tg:IL4−/− animals, suggesting that the IL4 from hmHER2Tg:IL4+/+ animals is derived exclusively from the stromal compartment (Fig. 1B).
Figure 1. Characterization of cytokines/chemokines expressed by D5-HER2 in vitro and in vivo.
A. Expression of 32 cytokines/chemokines in the microenvironment of D5-HER2 tumors was analyzed using a luminex-based approach. n=5 animals. B. A representative characterization of the cytokine/chemokine expression profile of D5-HER2 cells grown in vitro. Cell supernatant was harvested after 72 hours of growth and analyzed using a luminex-based approach. C. Evaluation of IL4 production by tumors grown in hmHER2Tg:IL4+/+ or hmHER2Tg:IL4−/− animals. 3×103 D5-HER2 or 1×106 EO771-HER2 cells were inoculated subcutaneously in the flank of indicated animals. Tumors were harvested, homogenized and IL4 was quantified using ELISA. Solid line represents the limit of detection of the assay. n=5 animals per group. Error bars represent standard error of the mean (SEM).
siRNA-based screen reveals positive and negative regulators of IL4 in D5-HER2 cells
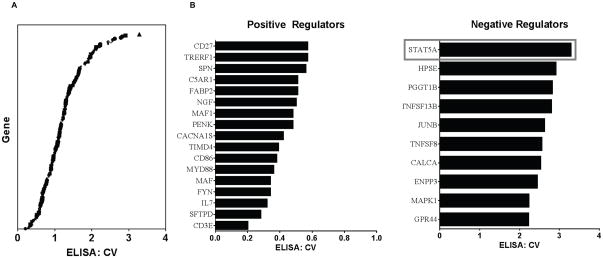
Given the importance of tumor cell-derived IL4 on the growth and survival of cancer cells (18, 19), we sought to determine the mechanism by which tumor cells regulate the expression of IL4. D5-HER2 cells served as an appropriate model to address this question since these cells, unlike many murine and human epithelial cancer cell lines, express detectable quantities of IL4 protein in vitro. Pathway Studio was used to curate the literature and identify 193 genes that positively regulate IL4 expression (Supplementary Table 1). Based on this gene list, a siRNA library was constructed and used to screen D5-HER2 cells to identify genes that positively regulate IL4 expression without a significant impact on viability. A number of genes from the primary screen were candidate positive regulators (genes whose knockdown results in an ELISA: crystal violet (CV) ratio 1) (Fig. 2A). Surprisingly, the knockdown of many of these genes led to ELISA: CV values of greater than 1, suggesting that these genes were in fact negative regulators of IL4 production (Fig. 2A). Applying z-score cutoffs of +/−1.2, 17 genes were identified in the primary screen as positive regulators and 10 genes were identified as negative regulators (Fig. 2B).
Figure 2. Identification of positive and negative regulators of IL4 production in D5-HER2 cells using a siRNA-based screening approach.
A. Plot of the ELISA: crystal violet (CV) ratios for all genes included in the screen. ELISA and CV values from each screen were normalized to siNEG. Mean ELISA and CV values from three independent screens were used to generate the ELISA:CV ratios. Ratios were log-transformed, and positive and negative regulators (B) were identifying using a z-score cutoff of −1.2 and +1.2, respectfully. All screens were performed in duplicate and repeated three times.
Stat5a is a negative regulator of IL4 expression in D5-HER2 cells
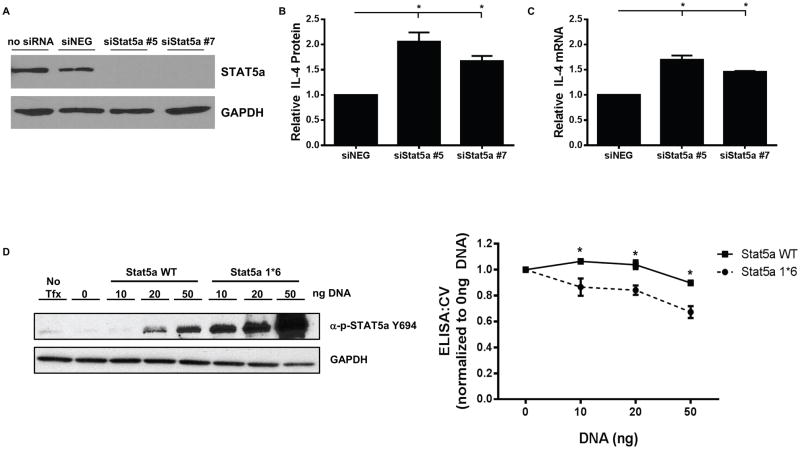
Further validation of hits identified in the primary screen (described in Materials and Methods) revealed Stat5a as a negative regulator of IL4 production. Knockdown of Stat5a resulted in an approximately 2-fold increase in IL4 protein production (Fig. 3A–B) and a 1.5 fold increase in IL4 mRNA (Fig. 3C), suggesting that Stat5a regulates IL4 at the transcriptional level. Overexpression of a constitutively active mutant of Stat5a (Stat5a1*6) resulted in hyper-phosphorylation of Stat5a and reduced IL4 protein expression compared to overexpression of wild type Stat5a in D5-HER2 cells (Fig. 3D). Modulation of Stat5a is not sufficient to drive expression of IL4 in an IL4-negative cell line as knockdown of Stat5a in EO771-HER2 cells does not result in production of IL4 mRNA or protein (data not shown). Collectively, these data reveal a previously undescribed function of Stat5a as a negative regulator of IL4 expression in D5-HER2 tumor cells.
Figure 3. Stat5a is a negative regulator of IL4 production in D5-HER2 cells.
A. Efficiency of Stat5a knockdown was evaluated by western blotting. D5-HER2 cells were reverse transfected with indicated siRNAs and incubated for 72 hours. A representative western blot is shown. B. D5-HER2 cells were transfected with indicated siRNAs and the concentration of IL4 in the cell culture supernatant was determined by ELISA. IL4 concentrations from siStat5a samples were normalized to IL4 concentrations from siNEG. C. The relative abundance of IL4 mRNA was determined using quantitative real-time PCR. Values from siStat5a were normalized to values obtained from siNEG. D. D5-HER2 cells were transfected with either WT Stat5a or a constitutively active mutant (Stat5a1*6) for 72 hours and phosphorylation of Stat5a was determined using western blotting. The concentration of IL4 in the supernatant from transfected cells was determined by ELISA and depicted in the line graph. Data were transformed into ELISA:CV ratios and normalized to the 0ng DNA transfection condition. Each experiment was performed at least three times. Error bars represent SEM of replicate experiments. *p<0.05. Statistical significance was determined by one way ANOVA using Dunnett multiple comparison analysis.
Neutralization of IL4 enhances the efficacy of trastuzumab during primary challenge
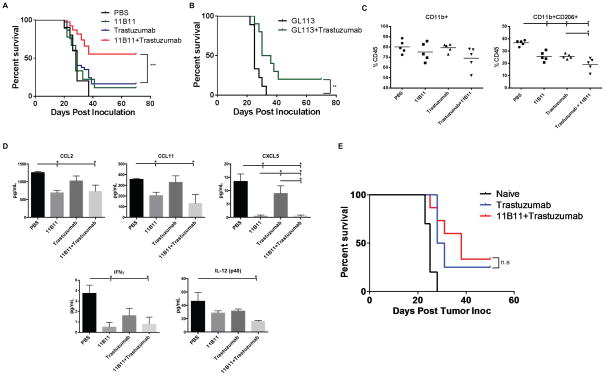
To determine if neutralizing IL4 could augment the therapeutic benefit of trastuzumab in our model system, we used the anti-IL4 antibody 11B11 alone or in combination with trastuzumab, which we have previously shown to be an effective therapeutic agent in the D5-HER2/hmHER2Tg model (11). Combination treatment with 11B11 and trastuzumab significantly enhanced the survival of hmHER2Tg animals compared to either agent alone (Fig. 4A). Treatment with GL113, the isotype control for 11B11, did not impact survival when used as monotherapy and performed similarly to trastuzumab monotherapy when combined with trastuzumab (Fig. 4B).
Figure 4. IL4 neutralization enhances the efficacy of trastuzumab during primary challenge and changes the composition of the tumor microenvironment.
hmHER2Tg animals were subcutaneously inoculated with 3000 D5-HER2 cells on day 0 and randomized to receive PBS, 200μg trastuzumab b.i.w., 1mg anti-IL4 (11B11), or isotype control (GL113) every five days, or combination therapy on day 1. A. Kaplan-Meier survival curve of animals treated with PBS (n=10), 11B11 (n=9), trastuzumab (37), or 11B11+trastuzumab (n=38). Statistical significance was determined by the log-rank test. B. Kaplan-Meier survival curve of animals treated with GL113 (n=9) or GL113+trastuzumab (n=10). Statistical significance determined by the log-rank test. C. Tumor-infiltrating cells were quantified using flow cytometry. Events were first gated on live cells using the LIVE/DEAD fixable dye. n=5 animals per group. Statistical significance was determined by one-way ANOVA using Tukey’s test. D. Cytokine and chemokine analysis from tumor homogenates was performed using a Luminex-based assay. n=5 animals per group. Statistical significance determined by one-way ANOVA using Tukey’s test. E. Kaplan-Meier plot of tumor-free animals from the primary challenge that were re-challenged with 1.5×104 D5-HER2 cells with no additional treatment. n=10 for naïve animals, n=4 for trastuzumab-treated, and n=14 for trastuzumab+11B11. Statistical significance was determined by the log-rank test. N.S., not significant. *p≤0.05, **p<0.01, ***p<0.001. Error bars represent SEM.
Neutralization of IL4 changes the composition of the tumor microenvironment
To elucidate a potential mechanism by which 11B11 enhanced trastuzumab efficacy, we investigated the impact of IL4 neutralization on the composition of the TME. Tumor-infiltrating immune cells were characterized using flow cytometry and the cytokine/chemokine profile of tumors from different treatment groups was determined. We chose to harvest tumors that were approximately 10–15mm in their largest diameters to facilitate acquisition of sufficient numbers of LIVE events to allow for multi-color flow cytometry-based immunophenotyping. We previously determined that smaller D5-HER2 tumors (5–8mm largest diameter) show no changes in the cellular compartment of the TME compared to larger tumors (data not shown). While no difference in recruitment of CD11b+ myeloid cells was observed, monotherapy with 11B11 or trastuzumab significantly reduced the recruitment of CD11b+CD206+ myeloid cells to the TME (Fig. 4C). This difference in recruitment was enhanced with combination therapy suggesting a synergistic effect of trastuzumab and 11B11 in reducing recruitment of CD11b+CD206+ myeloid cells to the TME (Fig. 4C). No differences in recruitment of T cells, T regs, NK cells, MDSCs, or F4/80 macrophages were observed between the treatment groups (not shown). IL4 neutralization reduced the concentration of potent myeloid chemoattractants CCL2, CCL11 and CXCL5and the differentiation factor GM-CSF in the TME (Fig. 4D and Supplemental Fig. 1A). Combination treatment reduced the concentrations of IL1α, IL6, and IL3 compared to monotherapy (Supplemental Fig. 1A). Surprisingly, IL4 neutralization seemed to reduce the concentrations of cytokines important for T-cell activation and proliferation: treatment with 11B11 reduced levels of IFNγ and combination treatment reduced levels of IL-12(p40) (Fig. 4D).
IL4 neutralization does not enhance trastuzumab-initiated protective adaptive immunity
To determine if IL4 neutralization enhanced generation of trastuzumab-initiated adaptive immunity during primary challenge, tumor-free animals were re-challenged with D5-HER2 with no additional treatments. Approximately 30% of animals initially treated with trastuzumab remained tumor-free following re-challenge, which is consistent with our previous findings (11). However, despite changes observed in the TME during primary challenge, addition of 11B11 to trastuzumab therapy during primary challenge did not enhance survival during re-challenge and thus did not potentiate generation of trastuzumab-initiated protective adaptive immunity (Fig. 4E).
Host-derived IL4 contributes to tumor control in the D5-HER2 model but does not impact efficacy of trastuzumab
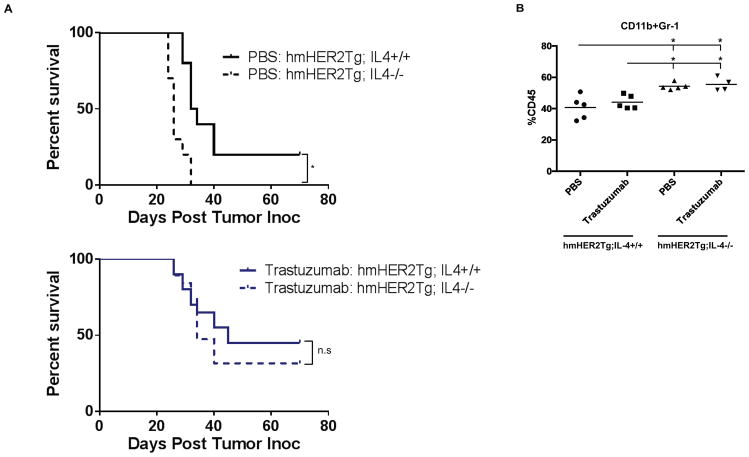
Since IL4 is produced by both D5-HER2 tumor cells and by the stroma, we sought to understand the relative roles of tumor-derived and stromal-derived IL4 in modulating the therapeutic activity of trastuzumab and the composition of the TME. Unexpectedly, D5-HER2 tumors grown in hmHER2Tg:IL4−/− animals grew more quickly compared to tumors grown in hmHER2Tg:IL4+/+ animals (data not shown), and tumor-bearing hmHER2Tg:IL4−/− animals displayed reduced survival compared to tumor-bearing hmHER2Tg:IL4+/+ animals (Fig. 5A). Tumors grown in hmHER2Tg:IL4−/− animals showed a marginally reduced level of CCL2 and contained more CD11b+Gr-1+ MDSCs compared to tumors in hmHER2Tg:IL4+/+animals (Supplemental Fig. 2A and Fig. 5B). Despite enhanced growth of tumors in hmHER2Tg:IL4−/− animals, knockout of host-derived IL4 did not impact the therapeutic efficacy of trastuzumab (Fig. 5A). Cytokine/chemokine analysis of animals treated with trastuzumab revealed a reduction of CCL2 in the microenvironment of tumors grown in hmHER2Tg:IL4−/− compared to hmHER2Tg:IL4+/+ animals, which is consistent with data from untreated animals (Supplemental Fig. 2B). Collectively, these data show that modulation of host-derived IL4 does not impact the antitumor activity of trastuzumab, but may aid in control of D5-HER2 tumor growth.
Figure 5. Differential role of host- versus tumor-derived IL4.
A. Kaplan-Meier plots from cohorts of hmHER2Tg:IL4+/+ or hmHER2Tg:IL4−/− animals that were challenged with 3×103 D5-HER2 tumor cells on day 0 and randomized to receive either PBS (n=5) or 200μg trastuzumab (n=10) b.i.w. on day 1. Statistical significance determine by the log-rank test. N.S, not significant. B. Tumor-infiltrating cells were analyzed using flow cytometry. All events were gated on live cells using the LIVE/DEAD fixable dye. n=5 animals per group. *p<0.05. Statistical significance was determined using one-way ANOVA using Tukey’s test.
DISCUSSION
A growing body of clinical and preclinical data suggests that activation of immune effectors is important for the maximal therapeutic efficacy of tumor-targeted antibodies. However, the tumor microenvironment is composed of cells and soluble mediators that collectively serve to limit generation of antitumor immunity and drive tumor growth and invasion. Thus, we sought to investigate the therapeutic potential of identifying and neutralizing a soluble factor in the microenvironment to enhance trastuzumab-initiated activation of immune effectors and subsequently enhance efficacy of trastuzumab. We identified many potential soluble mediators in the D5-HER2 TME that might suppress trastuzumab-initiated activation of antitumor immunity. Many of these mediators, including CCL2 and VEGF are highly expressed in the TME and determining the therapeutic impact of neutralizing these mediators in the context of trastuzumab therapy is the subject of future investigation. We chose to study IL4 for a number of reasons. IL4 is important for development of Th2 responses and limits production of IFNγ and the subsequent Th1 polarization, which is of relevance because patients with a more Th1-polarized TME have a more favorable prognosis (20, 21). IL4 also promotes the alternative activation of macrophages, leading to suppression of T-cell function via increased arginase activity, and suppresses generation of tumoricidal, classically activated M1 macrophages (22, 23). IL4 directly influences activity of monocytes and macrophages by downregulating expression of activating FcγR and upregulating expression of inhibitory FcγR, suggesting that IL4 can modulate the capacity of effectors to mediate ADCC(24, 25).
Using the D5-HER2/hmHER2Tg animal model, we demonstrated that IL4 is expressed by both tumor and stromal cells in the TME. This is consistent with published data demonstrating that a majority of primary human colorectal, breast and lung tumor cells express IL4 and that tumor-derived IL4 helps protect tumor cells against the effects of chemotherapy by inducing expression of anti-apoptotic proteins (19). Apoptosis induction is important for immune effector functions elicited by trastuzumab; our study provides additional rationale for targeting IL4 in our model system. Similarly, endogenous tumor-derived IL4 from human pancreatic cancer and prostate cancer cell lines serves as an autocrine growth factor and neutralization of IL4 resulted in diminished proliferation (18, 26).
Given the importance of tumor-derived IL4 on tumor cell biology and its potential to modulate the phenotype of immune effectors in response to antibody therapy, we sought to understand how IL4 is regulated in D5-HER2 tumor cells. Using a gene library of known positive regulators of IL4 expression, we conducted an siRNA screen against D5-HER2 that revealed Stat5a to be a negative regulator of IL4 production. To our knowledge, this is the first attempt to understand how tumor cells regulate the expression of IL4. In D5-HER2 cells, knockdown of Stat5a resulted in increased IL4 protein production, and overexpression of a constitutively active Stat5a mutant resulted in decreased IL4 production. We also observed that Stat5a knockdown resulted in increased expression of IL4 mRNA, suggesting that Stat5a negatively regulates IL4 transcription, either directly or indirectly. This finding is contrary to published data, which reported that Stat5a maintains the IL4 locus in a demethylated state, rendering it accessible to the transcriptional machinery (27). The IL4 gene has an intronic STAT5 binding site that serves as an enhancer element to maintain IL4 locus accessibility in mast cells (28). Therefore, this work raises the possibility of differential regulation of gene expression in tumor versus normal cells. Whether Stat5a directly regulates IL4 gene expression in D5-HER2, or indirectly via modulation of another transcription factor is unknown at this time. Future experiments aimed at understanding how Stat5a-mediated regulation of IL4 impacts the composition of the TME will shed light on whether Stat5a impacts the antitumor immune response via modulation of tumor-derived IL4.
Next, we sought to determine whether neutralization of IL4 could improve the in vivo efficacy of trastuzumab. Combination therapy with trastuzumab and 11B11 resulted in enhanced survival compared to either agent alone. IL4 neutralization changed the composition of the TME by decreasing expression of the myeloid chemoattractants CCL2, CCL11, and CXCL5. The dramatic reduction in CXCL5 levels is of note as this chemokine is associated with recruitment of tumor-promoting neutrophils and is associated with a poor prognosis (29, 30). Similarly, CCL2-mediated recruitment of pro-tumorigenic myeloid cells to the TME is well documented and treatment with CCL2 neutralizing antibodies has been shown to reduce growth of established tumors (31, 32). Treatment with 11B11 and trastuzumab combination therapy also resulted in a decreased recruitment of CD11b+CD206+ myeloid cells to the microenvironment. CD206 (mannose receptor) is upregulated on M2 alternatively activated macrophages and is associated with a poor prognosis in patients with renal cell carcinoma (33). Accumulation of CD206+ cells within tumors and draining lymph nodes may also be partially responsible for failure of vaccine therapy following surgery (34). CD206 is expressed by both macrophages and DCs. However, given the abundance of CD11b+CD206+ cells we identified in the TME of PBS-treated control animals, we do not believe these are DCs. Dendritic cells are present at a lower frequency in the D5-HER2 TME compared to the roughly 38% abundance of the CD11b+CD206+ cells (data not shown). Considering tumor-associated macrophages express CD11b in addition to F4/80, we believe the CD11b+CD206+ cells that were identified are in fact alternatively activated macrophages (35).
These data provide evidence that treatment with 11B11 can alter the composition of the TME and limit the generation of alternatively activated macrophages to enhance the efficacy of trastuzumab. Despite changes observed in the myeloid cell compartment, we cannot exclude the possibility that the enhanced antitumor activity elicited by combination therapy is mediated in part by T cells. Future studies aimed at elucidating the relative contributions between the myeloid and lymphoid compartments during primary challenge are currently underway.
Despite changes induced in the myeloid cell compartment, addition of 11B11 did not enhance generation of trastuzumab-initiated adaptive immunity. Tumor-free animals originally treated with combination therapy during primary challenge did not receive any further protection from re-challenge compared to animals originally receiving trastuzumab. This can be explained partially because combination therapy resulted in decreased IFNγ and IL12p40in the TME during re-challenge. The decrease was modest, but could explain failure of combination therapy to enhance generation of HER2-specific memory T cells. These data reveal an important role of IL4 in the context of tumor-targeted antibody therapy: neutralization of IL4 primarily affects the myeloid-cell compartment without significantly altering the T-cell compartment. Evidence of a similar finding was observed in the setting of a CD40 agonist antibody for the treatment of pancreatic cancer. Beatty and colleagues demonstrated that the antitumor activity of a CD40 agonist antibody was T-cell independent and instead dependent on the re-programming of tumor-infiltrating macrophages from tumor promoting to tumoricidal (36). Therefore, combining strategies primarily aimed at modulating the myeloid-cell compartment, such as IL4 neutralization, with strategies that target T-cell activation, including the checkpoint inhibitors anti-CTLA-4 and anti-PD-1, may be a rational approach to maximally liberate and activate the antitumor immune response.
Finally, we sought insight into the relative role between tumor-derived IL4 and stromal-derived IL4. Surprisingly, D5-HER2 tumors grew more rapidly in hmHER2Tg:IL4−/− animals compared to that in hmHER2Tg:IL4+/+ animals and removal of host-derived IL4 did not impact the efficacy of trastuzumab. These data suggest an intriguing duality of IL4 function; host-derived IL4 seems to contribute to tumor control, while tumor-derived IL4 contributes to tumor growth. Other groups have investigated the role of tumor-derived IL4 in shaping the TME and impacting tumor growth, however, these models involved expression of IL4 under control of viral promoters that results in supraphysiological levels of IL4 (37, 38). In contrast, we provide the first reported insight into the relative contribution of host- versus tumor-derived IL4 using a model system that expresses endogenous levels of IL4. We also observed an increased recruitment of MDSCs in tumors from hmHER2Tg:IL4−/− animals, which may help explain why these tumors grew more rapidly.
In summary, we utilized a unique model system to identify Stat5a as a negative regulator of IL4 expression in D5-HER2 tumor cells and provide evidence that IL4 neutralization enhances the therapeutic efficacy of trastuzumab via modulation of the myeloid-cell compartment. Our results collectively demonstrate that prospectively identifying and targeting soluble factors in the TME is a rational strategy to improve the efficacy of tumor-targeted antibody therapy.
Supplementary Material
Acknowledgments
Grant Support
This work is supported by NIH/NCI grant RO1CA050633 (L.W.) and by a seed grant from the American Medical Association Foundation (R.S.). The Flow Cytometry & Cell Sorting Shared Resource is partially supported by NIH/NCI grant P30-CA051008.
The authors would like to thank Karen Creswell from the Flow Cytometry and & Cell Sorting Shared Resource for technical assistance, Yong Tang for technical assistance with antibody production and purification, and Todd Waldman for assistance with production of lentivirus.
Footnotes
Conflicts of interest: None
References
- 1.Weiner LM, Surana R, Wang S. Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat Rev Immunol. 2010;10:317–27. doi: 10.1038/nri2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 3.Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639–48. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 4.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 5.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 6.Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytoxicity against tumor targets. Nat Med. 2000;6:443–6. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 7.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. 2002;99:754–8. doi: 10.1182/blood.v99.3.754. [DOI] [PubMed] [Google Scholar]
- 8.Bibeau F, Lopez-Crapez E, Di Fiore F, Thezenas S, Ychou M, Blanchard F, et al. Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J Clin Oncol. 2009;27:1122–9. doi: 10.1200/JCO.2008.18.0463. [DOI] [PubMed] [Google Scholar]
- 9.Tamura K, Shimizu C, Hojo T, Akashi-Tanaka S, Kinoshita T, Yonemori K, et al. FcgammaR2A and 3A polymorphisms predict clinical outcome of trastuzumab in both neoadjuvant and metastatic settings in patients with HER2-positive breast cancer. Ann Oncol. 2011;22:1302–7. doi: 10.1093/annonc/mdq585. [DOI] [PubMed] [Google Scholar]
- 10.Park S, Jiang Z, Mortenson ED, Deng L, Radkevich-Brown O, Yang X, et al. The therapeutic effect of anti-HER2/neu antibody depends on both innate and adaptive immunity. Cancer Cell. 2010;18:160–70. doi: 10.1016/j.ccr.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Astsaturov IA, Bingham CA, McCarthy KM, von Mehren M, Xu W, et al. Effective antibody therapy induces host-protective antitumor immunity that is augmented by TLR4 agonist treatment. Cancer Immunol Immunother. 2012;61:49–61. doi: 10.1007/s00262-011-1090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 13.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 14.Seiler CY, Park JG, Sharma A, Hunter P, Surapaneni P, Sedillo C, et al. DNASU plasmid and PSI: Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 2014;42:D1253–60. doi: 10.1093/nar/gkt1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witt AE, Hines LM, Collins NL, Hu Y, Gunawardane RN, Moreira D, et al. Functional proteomics approach to investigate the biological activities of cDNAs implicated in breast cancer. J Proteome Res. 2006;5:599–610. doi: 10.1021/pr050395r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PloS one. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onishi M, Nosaka T, Misawa K, Mui AL, Gorman D, McMahon M, et al. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol Cell Biol. 1998;18:3871–9. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prokopchuk O, Liu Y, Henne-Bruns D, Kornmann M. Interleukin-4 enhances proliferation of human pancreatic cancer cells: evidence for autocrine and paracrine actions. Br J Cancer. 2005;92:921–8. doi: 10.1038/sj.bjc.6602416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todaro M, Lombardo Y, Francipane MG, Alea MP, Cammareri P, Iovino F, et al. Apoptosis resistance in epithelial tumors is mediated by tumor-cell-derived interleukin-4. Cell Death Differ. 2008;15:762–72. doi: 10.1038/sj.cdd.4402305. [DOI] [PubMed] [Google Scholar]
- 20.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 21.Oldford SA, Robb JD, Codner D, Gadag V, Watson PH, Drover S. Tumor cell expression of HLA-DM associates with a Th1 profile and predicts improved survival in breast carcinoma patients. Int Immunol. 2006;18:1591–602. doi: 10.1093/intimm/dxl092. [DOI] [PubMed] [Google Scholar]
- 22.Liu Q, Zhang C, Sun A, Zheng Y, Wang L, Cao X. Tumor-educated CD11bhighIalow regulatory dendritic cells suppress T cell response through arginase I. J Immunol. 2009;182:6207–16. doi: 10.4049/jimmunol.0803926. [DOI] [PubMed] [Google Scholar]
- 23.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 24.Tridandapani S, Siefker K, Teillaud JL, Carter JE, Wewers MD, Anderson CL. Regulated expression and inhibitory function of Fcgamma RIIb in human monocytic cells. J Biol Chem. 2002;277:5082–9. doi: 10.1074/jbc.M110277200. [DOI] [PubMed] [Google Scholar]
- 25.te Velde AA, Huijbens RJ, de Vries JE, Figdor CG. IL-4 decreases Fc gamma R membrane expression and Fc gamma R-mediated cytotoxic activity of human monocytes. J Immunol. 1990;144:3046–51. [PubMed] [Google Scholar]
- 26.Lee SO, Pinder E, Chun JY, Lou W, Sun M, Gao AC. Interleukin-4 stimulates androgen-independent growth in LNCaP human prostate cancer cells. Prostate. 2008;68:85–91. doi: 10.1002/pros.20691. [DOI] [PubMed] [Google Scholar]
- 27.Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–48. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- 28.Hural JA, Kwan M, Henkel G, Hock MB, Brown MA. An intron transcriptional enhancer element regulates IL-4 gene locus accessibility in mast cells. J Immunol. 2000;165:3239–49. doi: 10.4049/jimmunol.165.6.3239. [DOI] [PubMed] [Google Scholar]
- 29.Zhou SL, Dai Z, Zhou ZJ, Chen Q, Wang Z, Xiao YS, et al. CXCL5 contributes to tumor metastasis and recurrence of intrahepatic cholangiocarcinoma by recruiting infiltrative intratumoral neutrophils. Carcinogenesis. 2014;35:597–605. doi: 10.1093/carcin/bgt397. [DOI] [PubMed] [Google Scholar]
- 30.Kawamura M, Toiyama Y, Tanaka K, Saigusa S, Okugawa Y, Hiro J, et al. CXCL5, a promoter of cell proliferation, migration and invasion, is a novel serum prognostic marker in patients with colorectal cancer. Eur J Cancer. 2012;48:2244–51. doi: 10.1016/j.ejca.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 31.Siveen KS, Kuttan G. Role of macrophages in tumour progression. Immunol Lett. 2009;123:97–102. doi: 10.1016/j.imlet.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 32.Li M, Knight DA, LAS, Smyth MJ, Stewart TJ. A role for CCL2 in both tumor progression and immunosurveillance. Oncoimmunology. 2013;2:e25474. doi: 10.4161/onci.25474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu L, Zhu Y, Chen L, An H, Zhang W, Wang G, et al. Prognostic Value of Diametrically Polarized Tumor-Associated Macrophages in Renal Cell Carcinoma. Ann Surg Oncol. 2014;21:3142–50. doi: 10.1245/s10434-014-3601-1. [DOI] [PubMed] [Google Scholar]
- 34.Predina J, Eruslanov E, Judy B, Kapoor V, Cheng G, Wang LC, et al. Changes in the local tumor microenvironment in recurrent cancers may explain the failure of vaccines after surgery. Proc Natl Acad Sci U S A. 2013;110:E415–24. doi: 10.1073/pnas.1211850110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beatty GL, Chiorean EG, Fishman MP, Saboury B, Teitelbaum UR, Sun W, et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331:1612–6. doi: 10.1126/science.1198443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang J, Wang Z, Li Z, Zhang J, Wang C, Xu X, Qin Z. Early exposure of high-dose interleukin-4 to tumor stroma reverses myeloid cell-mediated T-cell suppression. Gene Ther. 2010;17:991–9. doi: 10.1038/gt.2010.54. [DOI] [PubMed] [Google Scholar]
- 38.Kajiwara A, Doi H, Eguchi J, Ishii S, Hiraide-Sasagawa A, Sakaki M, et al. Interleukin-4 and CpG oligonucleotide therapy suppresses the outgrowth of tumors by activating tumor-specific Th1-type immune responses. Oncol Rep. 2012;27:1765–71. doi: 10.3892/or.2012.1723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.