Abstract
Background
SMO (the official symbol for “smoothened, frizzled family receptor”) is an important component of the hedgehog signaling pathway, which has been implicated in various human carcinomas. However, clinical, molecular, and prognostic associations of SMO expression in colorectal cancer remain unclear.
Methods
Using a database of 735 colon and rectal cancers in the Nurse’s Health Study and the Health Professionals Follow-up Study, we examined the relationship of tumor SMO expression (assessed by immunohistochemistry) to prognosis, and to clinical, pathological and tumor molecular features, including mutations of KRAS, BRAF and PIK3CA, microsatellite instability, CpG island methylator phenotype (CIMP), LINE-1 methylation, and expression of phosphorylated AKT and CTNNB1.
Results
SMO expression was detected in 370 (50%) tumors. In multivariate logistic regression analysis, SMO expression was independently inversely associated with phosphorylated AKT expression [odds ratio (OR), 0.48; 95% confidence interval (CI), 0.34–0.67] and CTNNB1 nuclear localization (OR, 0.48; 95% CI, 0.35–0.67). SMO expression was not significantly associated with colorectal cancer-specific or overall survival. However, in CIMP-high tumors, but not CIMP-low/0 tumors, SMO expression was significantly associated with better colorectal cancer-specific survival (log-rank P = 0.012; multivariate hazard ratio, 0.36; 95% CI, 0.13–0.95; Pinteraction = 0.035, for SMO and CIMP status).
Conclusions
Our data reveal novel potential associations between the hedgehog, the WNT/CTNNB1, and the PI3K (phosphatidylinositol-4,5-bisphosphonate 3-kinase)/AKT pathways, supporting pivotal roles of SMO and hedgehog signaling in pathway networking. SMO expression in colorectal cancer may interact with tumor CIMP status to affect patient prognosis, although confirmation by future studies is needed.
Keywords: colon carcinoma, rectal cancer, hedgehog, HH, molecular pathology, clinical outcome
INTRODUCTION
Colorectal cancers represent a heterogeneous group of complex multifactorial diseases, which are influenced by host and environmental factors.1 Molecular classification (e.g. by KRAS, BRAF, and MSI status) has become essential in both research and clinical practice to better predict tumor progression and behavior.2–5
The hedgehog signaling pathway plays a role in patterning, growth, and differentiation in various tissues, including the gastrointestinal tract.6–8 In mammals, hedgehog signaling is initiated through binding of one of three ligands [sonic hedgehog (SHH), indian hedgehog (IHH) and desert hedgehog (DHH)] to the trans-membrane receptor patched 1 (PTCH1), leading to release of the suppressed transmembrane protein smoothened, fizzled family receptor (SMO) and subsequent activation of GLI transcription factors.6 Hedgehog signaling has been implicated in the pathogenesis of various human cancers, either through hedgehog ligand-dependent activation, or through ligand-independent activation, i.e., by loss of function mutations in PTCH1, or gain of function mutations in the proto-oncogene SMO.9–11 Consequently, the hedgehog pathway is viewed as a potential therapeutic target.9,12
Although evidence supporting a role of the hedgehog pathway in colorectal neoplasia has tended to be inconsistent,13–21 accumulating experimental data demonstrate that the hedgehog signaling pathway cooperates with other molecular alternations and signaling pathways, such as WNT signaling,22,23 and phosphatidylinositol-4,5-biphosphonate 3-kinase (PI3K) /AKT,24–26 in multiple tumorigenic contexts, even in the absence of hedgehog ligand-dependent pathway activation.22,25
Given evidence of cross-talk between hedgehog and other signaling pathways in human carcinogenesis, we hypothesized that SMO expression in colorectal cancer might be associated with other important tumor characteristics, such as CTNNB1 and phosphorylated AKT expression. We therefore utilized a molecular pathological epidemiology database,27,28 derived from colorectal cancers arising in two U.S. nationwide prospective cohort studies, to examine SMO expression status in colorectal cancer, and to assess the relationships between SMO expression and other important molecular features, including: microsatellite instability (MSI); CpG island methylation phenotype (CIMP); long interspersed nucleotide element-1 (LINE-1) methylation; and KRAS, BRAF, and PIK3CA mutations. We also sought to evaluate the prognostic association of SMO expression, and to explore the potential for its interaction with other tumor features in survival analyses.
MATERIALS AND METHODS
Study Group
We used the database of two prospective cohort studies, the Nurses’ Health Study (NHS, N = 121,700 women observed since 1976) and the Health Professionals Follow-Up Study (HPFS, N = 51,500 men observed since 1986).29 Participants were sent follow-up biennial questionnaires to update information on diet and lifestyle factors, and to identify newly diagnosed cancers and other diseases. In this population-based study, besides medications that a given patient took by themselves, treatment modality was chosen by treating physicians, and detailed treatment data were not available. After confirmation of colorectal cancer, we requested paraffin embedded tissue blocks from hospitals across the U.S., where participants had undergone resection of primary tumors. We were able to obtain colorectal cancer specimens for 1443 cases out of 3019 colorectal cancer cases recorded up to June 2006. Diagnostic biopsy specimens from rectal cancer patients who received pre-operative therapy were collected in order to avoid treatment-related artifact. Tumor location was categorized [cecum; ascending colon (including hepatic flexure); transverse colon; descending colon (including splenic flexure); sigmoid colon; rectum] based on medical records.30 All colorectal cancer cases were confirmed through review of histology by a pathologist (S.O.) blinded to exposure data. Tumor grade was categorized as high (≤50% glandular area) or low (> 50% glandular area). Based on the availability of SMO expression data and survival data, a total of 735 colorectal cancer cases diagnosed up to 2006 were included in this study. Patients were observed until death, or January 2011, whichever came first. Death of a participant was ascertained through the National Death Index, or by reporting by family members or postal authorities. The cause of death was assigned by study physicians. Written informed consent was obtained from all study subjects. Human Subjects Committees at Harvard School of Public Health and Brigham and Women’s Hospital approved this study.
Immunohistochemistry for SMO, Phosphorylated AKT and CTNNB1
Tissue microarray blocks were constructed as previously described.31 Methods of immunohistochemical staining and interpretations for phosphorylated AKT (at amino acid position Ser 473) and CTNNB1 have been described previously.32–34 For SMO immunostaining, deparaffinized tissue sections were heated in a microwave for 15 minutes in Antigen Retrieval Citra Solution, pH 6 (BioGenex Laboratories, San Ramon, CA). Tissue sections were incubated with Dual Endogenous Enzyme Block (DAKO, Carpinteria, CA), then Serum Free Protein Block (DAKO), each for 15 minutes. Slides were incubated at room temperature for one hour with a primary antibody against SMO (1:100, rabbit polyclonal; Santa Cruz, sc-13943, San Diego, CA). Envision™ anti-rabbit HRP-labeled polymer (DAKO) was applied to the sections for 30 minutes, followed by visualization using the chromogen 3,3-diaminobenzidine (DAKO), and hematoxylin counterstain. The specificity of the SMO antibody was confirmed by previous studies in different tissues and cells.35–37 Positive and negative controls were included in each panel of immunohistochemistry for all markers.33,34 Known positive prostate carcinoma was used as a positive control for SMO.38 Sections processed with replacement of primary antibody by Tris-buffered saline were used as a negative control.
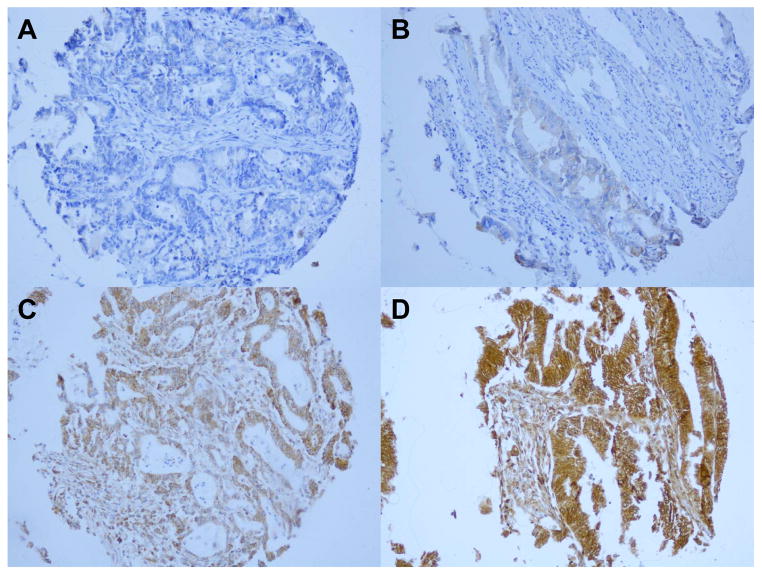
For each case, cytoplasmic SMO status was recorded as absent, weak, moderate or intense staining. SMO expression was defined as the presence of weak to intense staining (Figure 1). Immunostained tissue for each marker was scored by a single pathologist (SMO by X.L.; phosphorylated AKT by Y.B.; and CTNNB1 by T.M.) blinded to other data. A subset sample of over 100 cases for each marker was scored independently by a second pathologist (SMO by T.M.; phosphorylated AKT by K.S.; and CTNNB1 by S.O.) unaware of other data. The concordance between the two observers (all P < 0.0001) was 0.91 (κ = 0.79, N = 118) for SMO, 0.81 (κ = 0.59, N = 132) for phosphorylated AKT, and 0.90 (κ= 0.80, N = 292) for nuclear CTNNB1 localization, indicating good to substantial agreement.
Figure 1.
SMO expression in colorectal cancer. No expression (A), weak expression (B), moderate expression (C), and intense expression (D) in colorectal cancer cells.
Sequencing of KRAS, BRAF and PIK3CA Mutation, and Analysis for Microsatellite Instability
Genomic DNA was extracted from paraffin-embedded tissue. PCR and pyrosequencing targeted at KRAS [codons 12 and 13 (since 90% of KRAS mutations occur in these two codons)],32,39 BRAF (codon 600),40,41 and PIK3CA exons 9 and 20,42 were performed as previously described. Microsatellite instability was assessed using a panel of 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67 and D18S487).41 MSI-high was defined as the presence of instability in 30% or more of the markers, and MSI-low/microsatellite stability (MSS) as instability 0–29% of the markers.41
Real-time PCR for CpG Island Methylation and Pyrosequencing to Measure LINE-1 Methylation
Sodium bisulfite treatment of DNA, and real-time PCR assays (MethyLight) were performed as previously described.41,43,44 We quantified promoter methylation at eight CIMP-specific loci: CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1. CIMP-high was defined as ≥ 6 (of 8) methylated promoters, and CIMP-low/0 as 0–5 (of 8) methylated promoters. To accurately quantify methylation level in LINE-1, a PCR-pyrosequencing assay was employed.45,46
Statistical Analysis
All statistical analyses were performed using SAS software (version 9.2; SAS Institute Inc, Cary, NC). All P values were two-sided. When multiple hypothesis testing was performed, the P value for significance was adjusted to P = 0.0033 (= 0.05/15) by Bonferroni correction. For categorical data, the chi-square test or Fisher’s exact test was performed. To compare mean age and mean LINE-1 methylation levels, a t-test, assuming equal variances, was performed.
The Kaplan-Meier method and the log-rank test were performed for survival analyses. Deaths from causes other than colorectal cancer were censored in colorectal cancer-specific mortality analyses. To control for confounding, we used Cox proportional hazards models to calculate hazard ratio (HR) of death according to tumor SMO expression status. The model initially included age at diagnosis (continuous), sex, year of diagnosis (continuous), body mass index, tumor location (proximal vs. distal colon vs. rectum), tumor grade, MSI (high vs. low/MSS), CIMP (high vs. low/0), LINE-1 methylation (continuous), BRAF mutation, KRAS mutation, and PIK3CA mutation, in addition to CTNNB1 and phosphorylated AKT expression. To minimize residual confounding and overfitting, disease stage (I, II, III, IV, or unknown) was used as a stratifying variable using the “strata” option in the SAS “proc phreg” command. To avoid overfitting, variables in the final model were selected using backward stepwise elimination with a threshold of P = 0.05. Interaction was assessed using the Wald test on the cross-product of SMO and another variable of interest (excluding cases missing data) in a multivariate Cox model. To improve efficiency of the models, cases with missing data in any of the categorical variables [CIMP (1.8%), MSI (2.0%), BRAF (1.3%), KRAS (1.1%), PIK3CA (9.8%), CTNNB1 (4.7%) and phosphorylated AKT (6.9%)], were included in the majority category for that variable. We confirmed that excluding cases with missing information in any of the covariates did not substantially alter the results (data not shown).
To assess whether associations between SMO expression and the variables in Table 1 were independent of other variables, a multivariate logistic regression analysis was conducted for cross sectional analyses. To calculate adjusted odds ratios (OR), the model initially included variables as in Cox proportional hazards models. To avoid overfitting, a backward stepwise elimination with a threshold of P = 0.05 was used to select variables in the final model. After the variables in the final logistic regression model were selected, we employed a missing indicator method for those cases with missing data in a given variable, to obtain a more accurate effect estimate in the given variable.
Table 1.
Clinical, pathological and molecular features of colorectal cancer according to SMO expression status
| Feature | Total | SMO non-expression | SMO expression | P value | |||
|---|---|---|---|---|---|---|---|
| Total No. | 735 | 365 | 370 | ||||
| Sex | 0.093 | ||||||
| Male (HPFS) | 271 | 37% | 146 | 40% | 125 | 34% | |
| Female (NHS) | 464 | 63% | 219 | 60% | 245 | 66% | |
| Mean age at diagnosis (years) ± SD | 67.2 ± 8.4 | 67.4 ± 8.1 | 67.0 ± 8.7 | 0.60 | |||
| Family history of colorectal cancer in first-degree relatives | 0.20 | ||||||
| Absent | 587 | 80% | 299 | 82% | 288 | 78% | |
| Present | 148 | 20% | 66 | 18% | 82 | 22% | |
| Body mass index (kg/m2) | 0.31 | ||||||
| <30 | 591 | 81% | 299 | 82% | 292 | 79% | |
| ≥30 | 142 | 19% | 65 | 18% | 77 | 21% | |
| Tumor location | 0.60 | ||||||
| Cecum | 129 | 18% | 64 | 18% | 65 | 17% | |
| Ascending colon | 156 | 21% | 78 | 22% | 78 | 21% | |
| Transverse colon | 75 | 10% | 41 | 11% | 34 | 9% | |
| Descending colon | 59 | 8% | 34 | 9% | 25 | 7% | |
| Sigmoid colon | 165 | 23% | 77 | 21% | 88 | 24% | |
| Rectum | 148 | 20% | 68 | 19% | 80 | 22% | |
| Disease stage | 0.46 | ||||||
| I | 158 | 21% | 82 | 22% | 76 | 20% | |
| II | 227 | 31% | 108 | 30% | 119 | 32% | |
| III | 207 | 28% | 96 | 26% | 111 | 30% | |
| IV | 102 | 14% | 55 | 15% | 47 | 13% | |
| Unknown | 41 | 6% | 24 | 7% | 17 | 5% | |
| Tumor grade | 0.044 | ||||||
| Low | 664 | 90% | 321 | 88% | 343 | 93% | |
| High | 70 | 10% | 43 | 12% | 27 | 7% | |
| MSI status | 0.087 | ||||||
| MSI-low/MSS | 602 | 84% | 288 | 81% | 314 | 86% | |
| MSI-high | 118 | 16% | 67 | 19% | 51 | 14% | |
| CIMP status | 0.0035 | ||||||
| CIMP-low/0 | 604 | 84% | 284 | 80% | 320 | 88% | |
| CIMP-high | 118 | 16% | 73 | 20% | 45 | 12% | |
| BRAF status | 0.0026 | ||||||
| Wild-type | 616 | 85% | 293 | 81% | 323 | 89% | |
| Mutant | 109 | 15% | 69 | 19% | 40 | 11% | |
| KRAS status | 0.0027 | ||||||
| Wild-type | 461 | 63% | 248 | 69% | 213 | 58% | |
| Mutant | 266 | 37% | 112 | 31% | 154 | 42% | |
| PIK3CA status | 0.92 | ||||||
| Wild-type | 556 | 84% | 275 | 84% | 281 | 84% | |
| Mutant | 107 | 16% | 54 | 16% | 53 | 16% | |
| Mean LINE-1 methylation level (%) ± SD | 61.3 ± 9.4 | 61.7 ± 10.0 | 61.0 ± 8.8 | 0.33 | |||
| CTNNB1 nuclear localization | 0.0005 | ||||||
| Negative | 372 | 53% | 159 | 46% | 213 | 60% | |
| Positive | 329 | 47% | 184 | 54% | 145 | 40% | |
| Phosphorylated AKT expression | < 0.0001 | ||||||
| Negative | 253 | 37% | 99 | 29% | 154 | 45% | |
| Positive | 431 | 63% | 240 | 71% | 191 | 55% | |
The % number indicates the proportion of patients with a specific clinical, pathological or molecular feature among all patients, or patients with specific tumor SMO expression status.
CIMP, CpG island methylator phenotype; HPFS, Health Professionals Follow-up Study; LINE-1, long interspersed nucleotide element-1; MSI, microsatellite instability; MSS, microsatellite stable; NHS, Nurses’ Health Study; SD, standard deviation.
RESULTS
SMO Expression in Colorectal Cancer
Among 735 colorectal cancer cases diagnosed up to 2006 with SMO expression data, we observed SMO expression in 370 tumors (50%) by immunohistochemistry (Figure 1). SMO expression was positively associated with KRAS mutation (P = 0.0027), and inversely associated with phosphorylated AKT expression (P < 0.0001), BRAF mutation (P = 0.0026), CTNNB1 nuclear localization (P = 0.0005) and CIMP-high status (P = 0.0035) (Table 1).
Multivariate Logistic Regression Analysis to Assess Associations with SMO Expression in Colorectal Cancer
Multivariate logistic regression analysis was performed to assess independent relationships between SMO expression and other factors. Phosphorylated AKT expression [multivariate OR = 0.48; 95% confidence interval (CI), 0.34–0.67; P < 0.0001] and CTNNB1 nuclear localization (OR = 0.48; 95% CI, 0.35–0.67; P < 0.0001) remained significantly associated with SMO expression in the final model.
In addition, BRAF-mutation/KRAS-wild-type (vs. BRAF-wild-type/KRAS-wild-type) and CIMP-high remained in the final model [(OR = 0.49; 95% CI, 0.28–0.85; P = 0.011) and (OR = 0.59; 95% CI, 0.35–0.98; P = 0.043), respectively], but these associations were not statistically significant given multiple hypothesis testing (Table 2).
Table 2.
Multivariate logistic regression analysis to calculate adjusted odds ratio (OR) for the association of a given variable (in the left column) with SMO expression (as an outcome variable)
| Variable in the final multivariate model | Multivariate OR (95% CI) | P value |
|---|---|---|
| Phosphorylated AKT expression | 0.48 (0.34–0.67) | < 0.0001 |
| CTNNB1 nuclear localization | 0.48 (0.35–0.67) | < 0.0001 |
| BRAF/KRAS status | ||
| BRAF-mutation/KRAS-wild-type (vs. BRAF-wild-type/KRAS-wild-type) | 0.49 (0.28–0.85) | 0.011 |
| BRAF-wild-type/KRAS-mutation (vs. BRAF-wild-type/KRAS-wild-type) | 1.35 (0.96–1.89) | 0.082 |
| CIMP- high (vs. low/0) | 0.59 (0.35–0.98) | 0.043 |
The multivariate logistic regression model initially included age, sex, year of diagnosis, body mass index, tumor location, family history, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF and PIK3CA mutation, LINE-1 methylation, CTNNB1 nuclear localization and phosphorylated AKT expression. A backward elimination with threshold of P = 0.05 was used to select variables in the final model. When multiple hypothesis testing was performed, the P value for significance was adjusted to P = 0.0033 (= 0.05/15) by Bonferroni correction.
CI, confidence interval; OR, odds ratio.
SMO Expression and Patient Survival in Colorectal Cancer
During follow-up of 735 patients with survival data (median follow-up time 14.1 years for censored cases), there were 373 deaths, including 216 deaths due to colorectal cancer. In Kaplan-Meier analyses, SMO expression was not significantly associated with colorectal cancer-specific survival (log-rank P = 0.85) or overall survival (log-rank P = 0.72).
We performed Cox proportional hazards regression models to assess mortality according to SMO status, but did not observe a significant association between SMO expression and survival in univariate, stage-stratified, or multivariate stage-stratified analyses (data not shown).
Interactions between SMO Expression and other Variables in Colorectal Cancer Survival Analysis
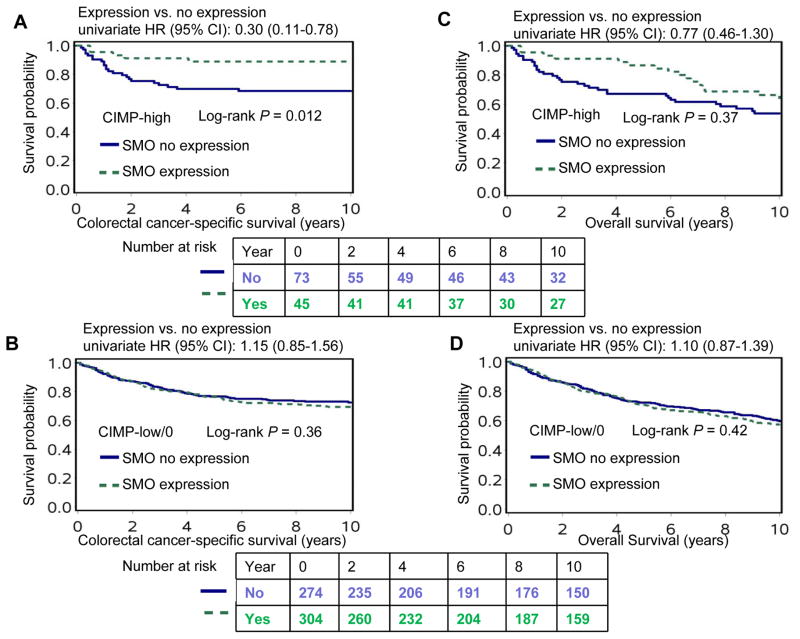
We examined whether any clinical, pathological, or molecular variables significantly modified the association of SMO expression with patient survival. We observed a borderline significant interaction between SMO expression and CIMP status in colorectal cancer-specific survival (Pinteraction = 0.035, given multiple testing significance level was adjusted to P = 0.0033). For patients with CIMP-high tumor, SMO positivity was significantly associated with better colorectal cancer-specific survival (multivariate HR, 0.36, 95% CI, 0.13–0.95); whereas, for patients with CIMP-low/0 tumor, SMO positivity was not significantly associated with colorectal cancer-specific survival (Table 3). The differential effect of SMO expression on colorectal cancer-specific survival according to CIMP status was also evident in Kaplan-Meier analyses (Figure 2).
Table 3.
SMO expression status in colorectal cancer and patient mortality in strata of CpG island methylator phenotype status
| Colorectal cancer-specific mortality
|
Overall mortality
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tumor characteristics | Total No. | No. of events | Univariate HR (95% CI) | Stage- stratified HR (95% CI) | Multivariate stage-stratified HR (95% CI) | No. of events | Univariate HR (95% CI) | Stage-stratified HR (95% CI) | Multivariate stage-stratified HR (95% CI) |
| CIMP-low/0 | |||||||||
| No expression | 274 | 75 | 1 (referent) | 1 (referent) | 1 (referent) | 131 | 1 (referent) | 1 (referent) | 1 (referent) |
| Expression | 304 | 95 | 1.15 (0.85–1.56) | 1.11 (0.82–1.51) | 1.08 (0.79–1.47) | 156 | 1.10 (0.87–1.39) | 1.09 (0.86–1.38) | 1.02 (0.80–1.29) |
| CIMP-high | |||||||||
| No expression | 73 | 23 | 1 (referent) | 1 (referent) | 1 (referent) | 39 | 1 (referent) | 1 (referent) | 1 (referent) |
| Expression | 45 | 5 | 0.30 (0.11–0.78) | 0.37 (0.14–0.98) | 0.36 (0.13–0.95) | 23 | 0.77 (0.46–1.30) | 0.87 (0.51–1.47) | 0.73 (0.43–1.23) |
| P for interaction (SMO expression and CIMP status) | 0.0090 | 0.035 | 0.035 | 0.22 | 0.44 | 0.26 | |||
The multivariate, stage-stratified Cox regression model initially included age, sex, year of diagnosis, body mass index, tumor location, tumor grade, microsatellite instability, KRAS mutation, BRAF mutation, PIK3CA mutation, LINE-1 methylation, CTNNB1 nuclear localization and phosphorylated AKT expression. A backward elimination with a threshold of P = 0.05 was used to select variables in the final models.
CI, confidence interval; HR, hazard ratio.
Figure 2.
Colorectal cancer-specific and overall survival in patients with colorectal cancer according to SMO expression status in strata of CIMP status. CI, confidence interval; HR, hazard ratio.
The association of SMO expression with cancer-specific mortality did not significantly differ according to any of the other variables.
DISCUSSION
In this study, the unique resource of a molecular pathological epidemiology database,27,28 containing a large number of colorectal cancers, and prospectively collected data from two cohort studies, enabled us to comprehensively evaluate the associations of SMO expression with clinical, pathological and tumor molecular features. We observed that SMO was expressed in around one half of colorectal cancers. In a multivariate logistic regression model, SMO expression was significantly inversely associated with phosphorylated AKT expression and CTNNB1 nuclear localization. An inverse association was also observed between SMO expression and BRAF-mutant/KRAS wild-type; however, the association was of borderline significance, when multiple testing was taken into account.
Recent studies have demonstrated that colorectal cancers constitute a group of heterogeneous tumors at the molecular level.47,48 The development and progression of colorectal neoplasia is attributable to the accumulation of genetic and epigenetic changes and the complex interaction of aberrations in various signaling pathways.49–52 Each tumor has its own unique characteristics in terms of molecular phenotype, tumor microenvironment, and interactomes within and between neoplastic and host cells.1,53 Therefore, tumor biomarker testing contributes to personalized medicine research and, ultimately, to clinical practice.50,54–56
Experimental data suggest a link between SMO expression and the PI3K/AKT pathway. AKT is a major downstream effector of PI3K, and plays a crucial role in regulating a wide variety of cellular process, including cellular metabolism as well as cell proliferation and survival.57 Riobo et al. have previously shown that PI3K and AKT are essential for SHH signaling.24 Furthermore, SMO activity is required for cooperation between SHH and insulin-like growth factor in promoting myogenic proliferation and differentiation via the MAPK/ERK and PI3K/AKT pathways.25 In our dataset, SMO expression was inversely associated with phosphorylated AKT expression in colorectal cancer, suggesting that SMO activation may tend to be mutually exclusive with AKT activation in colorectal cancer development.
Crosstalk between the hedgehog and WNT signaling pathways in intestinal tumorigenesis remains controversial.7,20,22,50,58 Several groups have reported possible negative regulation of the WNT pathway by hedgehog signaling.7,58 In one study, overexpression of IHH resulted in down-regulation of intestinal CTNNB1.58 Nuclear expression of CTNNB1 has been found to be inversely associated with GLI1 staining in colorectal cancers, suggesting that GLI1 plays an inhibitory role in the development of colorectal cancer driven by WNT signaling.20 However, Arimura et al. have shown that reduced SMO expression inhibits WNT signaling by down-regulating nuclear CTNNB1 expression, independent of GLI-mediated hedgehog signaling.22 Our current findings suggest that tumors with SMO expression are inversely associated with CTNNB1 nuclear expression, favoring a negative regulation of the WNT pathway by hedgehog signaling.
While SMO expression was not associated with colorectal cancer-specific survival or overall survival, our data suggest a possible interaction with CIMP status in patient prognosis. CIMP constitutes an epigenomic phenomenon characterized by widespread promoter methylation, which leads to tumor suppressor gene silencing.59 CIMP status has been extensively investigated in colorectal cancer.60–65 In our current study, we observed that SMO expression was inversely associated with CIMP-high status. Moreover, our data suggest that, within CIMP-high cancers (but not within CIMP-low/0 cancers), patients with SMO-expressing tumors may expect better cancer-specific survival compared to those with SMO-nonexpression tumors. Given multiple hypotheses testing, and the exploratory nature of our interaction analyses, these findings need confirmation by additional independent studies.
Interestingly, we observed a possible inverse association between SMO expression and BRAF-mutation/KRAS-wild-type in colorectal cancers, independent of other molecular variables. BRAF mutation is present in 10–15% of colorectal cancers and associated with inferior prognosis.66–69 Nonetheless, our results need to be confirmed by independent studies.
There are also limitations in the present study. Firstly, data on treatment were limited. We speculated that chemotherapy administration did not substantially differ by tumor SMO expression, since the data were not available for treating physicians. Nevertheless, our regression analyses were adjusted for TNM stage, on which treatment decisions are largely based. Secondly, despite quite high agreement of readings of the two pathologists for SMO immunohistochemistry there was still a 9% discordance rate.
In conclusion, our large cohort study has shown that SMO expression in colorectal cancer is inversely associated with phosphorylated AKT expression and CTNNB1 nuclear localization. SMO expression in colorectal cancer may interact with tumor CIMP status to affect patient prognosis, although confirmation by future studies is needed. Our data are compatible with literature supporting a role for SMO in pathway networking in colorectal carcinogenesis.
Synopsis.
Our data reveal novel potential associations between the hedgehog, the WNT/CTNNB1, and the PI3K/AKT pathways, supporting pivotal roles of SMO and hedgehog signaling in pathway networking. SMO expression in colorectal cancer may interact with tumor CIMP status to affect patient prognosis, although confirmation by future studies is needed.
Acknowledgments
Funding: This work was supported by USA National Institute of Health (NIH) [P01 CA87969 (to SE Hankinson), UM1 CA167552 and P01 CA55075 (to WC Willett), R01 CA137178 (to ATC), P50 CA127003 (to CSF), and R01 CA151993 (to SO)]; the Bennett Family Fund for Targeted Therapies Research; and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. PL was supported by a Harvard University Frank Knox Memorial Fellowship and a fellowship from the Chief Scientist Office, Scotland. ATC is a Damon Runyon Clinical Investigator. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. Funding agencies did not have any role in the design of the study; the collection, analysis, or interpretation of the data; the decision to submit the manuscript for publication; or the writing of the manuscript.
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-Up Study for their valuable contributions, as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. In addition, this study was approved by the Connecticut Department of Public Health (DPH) Human Investigations Committee. Certain data used in this publication were obtained from the DPH. The authors assume full responsibility for analyses and interpretation of these data.
Abbreviations
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- HPFS
the Health Professionals Follow-Up Study
- HR
hazard ratio
- MSI
microsatellite instability
- MSS
microsatellite stable
- NHS
the Nurses’ Health Study
- OR
odds ratio
- PI3K
phosphatidylinositol-4,5-biphosphonate 3-kinase
- SD
standard deviation
Footnotes
Conflict of Interest Statement:
No potential conflicts of interest exist.
Use of standardised official symbols: We use HUGO (Human Genome Organisation)-approved official symbols for genes and gene products, including BRAF; SMO (smoothened, frizzled family receptor); PIK3CA, AKT and KRAS; all of which are described at www.genenames.org.
References
- 1.Ogino S, Fuchs CS, Giovannucci E. How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn. 2012;12:621–628. doi: 10.1586/erm.12.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Funkhouser WK, Jr, Lubin IM, Monzon FA, et al. Relevance, pathogenesis, and testing algorithm for mismatch repair-defective colorectal carcinomas: a report of the association for molecular pathology. J Mol Diagn. 2012;14:91–103. doi: 10.1016/j.jmoldx.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. International journal of molecular sciences. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers. 2013;5:676–713. doi: 10.3390/cancers5020676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JH, Kang GH. Molecular and prognostic heterogeneity of microsatellite-unstable colorectal cancer. World journal of gastroenterology : WJG. 2014;20:4230–4243. doi: 10.3748/wjg.v20.i15.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berman DM, Karhadkar SS, Maitra A, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 7.Kolterud A, Grosse AS, Zacharias WJ, et al. Paracrine Hedgehog signaling in stomach and intestine: new roles for hedgehog in gastrointestinal patterning. Gastroenterology. 2009;137:618–628. doi: 10.1053/j.gastro.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fendrich V, Oh E, Bang S, et al. Ectopic overexpression of Sonic Hedgehog (Shh) induces stromal expansion and metaplasia in the adult murine pancreas. Neoplasia. 2011;13:923–930. doi: 10.1593/neo.11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng JM, Curran T. The Hedgehog’s tale: developing strategies for targeting cancer. Nat Rev Cancer. 2011;11:493–501. doi: 10.1038/nrc3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teglund S, Toftgard R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 2010;1805:181–208. doi: 10.1016/j.bbcan.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Michael LE, Westerman BA, Ermilov AN, et al. Bmi1 is required for Hedgehog pathway-driven medulloblastoma expansion. Neoplasia. 2008;10:1343–1349. 1345–1349. doi: 10.1593/neo.81078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Laterra J, Pomper MG. Hedgehog pathway inhibitor HhAntag691 is a potent inhibitor of ABCG2/BCRP and ABCB1/Pgp. Neoplasia. 2009;11:96–101. doi: 10.1593/neo.81264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatel G, Ganeff C, Boussif N, et al. Hedgehog signaling pathway is inactive in colorectal cancer cell lines. Int J Cancer. 2007;121:2622–2627. doi: 10.1002/ijc.22998. [DOI] [PubMed] [Google Scholar]
- 14.Oniscu A, James RM, Morris RG, Bader S, Malcomson RD, Harrison DJ. Expression of Sonic hedgehog pathway genes is altered in colonic neoplasia. J Pathol. 2004;203:909–917. doi: 10.1002/path.1591. [DOI] [PubMed] [Google Scholar]
- 15.Qualtrough D, Buda A, Gaffield W, Williams AC, Paraskeva C. Hedgehog signalling in colorectal tumour cells: induction of apoptosis with cyclopamine treatment. Int J Cancer. 2004;110:831–837. doi: 10.1002/ijc.20227. [DOI] [PubMed] [Google Scholar]
- 16.Alinger B, Kiesslich T, Datz C, et al. Hedgehog signaling is involved in differentiation of normal colonic tissue rather than in tumor proliferation. Virchows Arch. 2009;454:369–379. doi: 10.1007/s00428-009-0753-7. [DOI] [PubMed] [Google Scholar]
- 17.Mazumdar T, DeVecchio J, Shi T, Jones J, Agyeman A, Houghton JA. Hedgehog signaling drives cellular survival in human colon carcinoma cells. Cancer Res. 2011;71:1092–1102. doi: 10.1158/0008-5472.CAN-10-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varnat F, Duquet A, Malerba M, et al. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol Med. 2009;1:338–351. doi: 10.1002/emmm.200900039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saqui-Salces M, Merchant JL. Hedgehog signaling and gastrointestinal cancer. Biochim Biophys Acta. 2010;1803:786–795. doi: 10.1016/j.bbamcr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akiyoshi T, Nakamura M, Koga K, et al. Gli1, downregulated in colorectal cancers, inhibits proliferation of colon cancer cells involving Wnt signalling activation. Gut. 2006;55:991–999. doi: 10.1136/gut.2005.080333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding YL, Wang QS, Zhao WM, Xiang L. Expression of smoothened protein in colon cancer and its prognostic value for postoperative liver metastasis. Asian Pac J Cancer Prev. 2012;13:4001–4005. doi: 10.7314/apjcp.2012.13.8.4001. [DOI] [PubMed] [Google Scholar]
- 22.Arimura S, Matsunaga A, Kitamura T, Aoki K, Aoki M, Taketo MM. Reduced level of smoothened suppresses intestinal tumorigenesis by down-regulation of Wnt signaling. Gastroenterology. 2009;137:629–638. doi: 10.1053/j.gastro.2009.04.059. [DOI] [PubMed] [Google Scholar]
- 23.Morris JPt, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riobo NA, Lu K, Ai X, Haines GM, Emerson CP., Jr Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci U S A. 2006;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madhala-Levy D, Williams VC, Hughes SM, Reshef R, Halevy O. Cooperation between Shh and IGF-I in promoting myogenic proliferation and differentiation via the MAPK/ERK and PI3K/Akt pathways requires Smo activity. J Cell Physiol. 2012;227:1455–1464. doi: 10.1002/jcp.22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo YA, Kang MH, Lee HJ, et al. Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 2011;71:7061–7070. doi: 10.1158/0008-5472.CAN-11-1338. [DOI] [PubMed] [Google Scholar]
- 27.Ogino S, Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: the evolving field of molecular pathological epidemiology. J Natl Cancer Inst. 2010;102:365–367. doi: 10.1093/jnci/djq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 32.Kawasaki T, Nosho K, Ohnishi M, et al. IGFBP3 promoter methylation in colorectal cancer: relationship with microsatellite instability, CpG island methylator phenotype, and p53. Neoplasia. 2007;9:1091–1098. doi: 10.1593/neo.07760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baba Y, Nosho K, Shima K, et al. Phosphorylated AKT expression is associated with PIK3CA mutation, low stage, and favorable outcome in 717 colorectal cancers. Cancer. 2011;117:1399–1408. doi: 10.1002/cncr.25630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morikawa T, Kuchiba A, Yamauchi M, et al. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011;305:1685–1694. doi: 10.1001/jama.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dierks C, Grbic J, Zirlik K, et al. Essential role of stromally induced hedgehog signaling in B-cell malignancies. Nature medicine. 2007;13:944–951. doi: 10.1038/nm1614. [DOI] [PubMed] [Google Scholar]
- 36.Riobo NA, Saucy B, Dilizio C, Manning DR. Activation of heterotrimeric G proteins by Smoothened. Proc Natl Acad Sci U S A. 2006;103:12607–12612. doi: 10.1073/pnas.0600880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerber AN, Wilson CW, Li YJ, Chuang PT. The hedgehog regulated oncogenes Gli1 and Gli2 block myoblast differentiation by inhibiting MyoD-mediated transcriptional activation. Oncogene. 2007;26:1122–1136. doi: 10.1038/sj.onc.1209891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao X, Siu MK, Au CW, et al. Aberrant activation of hedgehog signaling pathway in ovarian cancers: effect on prognosis, cell invasion and differentiation. Carcinogenesis. 2009;30:131–140. doi: 10.1093/carcin/bgn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imamura Y, Morikawa T, Liao X, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer Res. 2012;18:4753–4763. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao X, Morikawa T, Lochhead P, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18:2257–2268. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogino S, Kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10:13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D, Xia D, Dubois RN. The Crosstalk of PTGS2 and EGF Signaling Pathways in Colorectal Cancer. Cancers. 2011;3:3894–3908. doi: 10.3390/cancers3043894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertrand FE, Angus CW, Partis WJ, Sigounas G. Developmental pathways in colon cancer: Crosstalk between WNT, BMP, Hedgehog and Notch. Cell Cycle. 2012;11:4344–4351. doi: 10.4161/cc.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088–2100. doi: 10.1053/j.gastro.2009.12.066. [DOI] [PubMed] [Google Scholar]
- 52.Rosty C, Hewett DG, Brown IS, Leggett BA, Whitehall VL. Serrated polyps of the large intestine: current understanding of diagnosis, pathogenesis, and clinical management. Journal of gastroenterology. 2013;48:287–302. doi: 10.1007/s00535-012-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Domingo E, Ramamoorthy R, Oukrif D, et al. Use of multivariate analysis to suggest a new molecular classification of colorectal cancer. J Pathol. 2013;229:441–448. doi: 10.1002/path.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayashi Y, Molina JR, Hamilton SR, Georgescu MM. NHERF1/EBP50 is a new marker in colorectal cancer. Neoplasia. 2010;12:1013–1022. doi: 10.1593/neo.10780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soerjomataram I, Thong MS, Korfage IJ, et al. Excess weight among colorectal cancer survivors: target for intervention. Journal of gastroenterology. 2012;47:999–1005. doi: 10.1007/s00535-012-0567-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu KL, Huang EY, Jhu EW, et al. Overexpression of galectin-3 enhances migration of colon cancer cells related to activation of the K-Ras-Raf-Erk1/2 pathway. Journal of gastroenterology. 2013;48:350–359. doi: 10.1007/s00535-012-0663-3. [DOI] [PubMed] [Google Scholar]
- 57.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 58.van den Brink GR, Bleuming SA, Hardwick JC, et al. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat Genet. 2004;36:277–282. doi: 10.1038/ng1304. [DOI] [PubMed] [Google Scholar]
- 59.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beggs AD, Jones A, El-Bahrawy M, Abulafi M, Hodgson SV, Tomlinson IP. Whole-genome methylation analysis of benign and malignant colorectal tumours. J Pathol. 2013;229:697–704. doi: 10.1002/path.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Q, Dong Y, Wu W, et al. Detection and differential diagnosis of colon cancer by a cumulative analysis of promoter methylation. Nature communications. 2012;3:1206. doi: 10.1038/ncomms2209. [DOI] [PubMed] [Google Scholar]
- 62.Dahlin AM, Palmqvist R, Henriksson ML, et al. The Role of the CpG Island Methylator Phenotype in Colorectal Cancer Prognosis Depends on Microsatellite Instability Screening Status. Clin Cancer Res. 2010;16:1845–1855. doi: 10.1158/1078-0432.CCR-09-2594. [DOI] [PubMed] [Google Scholar]
- 63.Zlobec I, Bihl M, Foerster A, Rufle A, Lugli A. Comprehensive analysis of CpG Island Methylator Phenotype (CIMP)-high, -low, and -negative colorectal cancers based on protein marker expression and molecular features. J Pathol. 2011;225:336–343. doi: 10.1002/path.2879. [DOI] [PubMed] [Google Scholar]
- 64.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hughes LA, Khalid-de Bakker CA, Smits KM, et al. The CpG island methylator phenotype in colorectal cancer: Progress and problems. Biochim Biophys Acta. 2012;1825:77–85. doi: 10.1016/j.bbcan.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 66.Price TJ, Hardingham JE, Lee CK, et al. Impact of KRAS and BRAF Gene Mutation Status on Outcomes From the Phase III AGITG MAX Trial of Capecitabine Alone or in Combination With Bevacizumab and Mitomycin in Advanced Colorectal Cancer. J Clin Oncol. 2011;29:2675–2682. doi: 10.1200/JCO.2010.34.5520. [DOI] [PubMed] [Google Scholar]
- 67.Ogino S, Shima K, Meyerhardt JA, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. Clin Cancer Res. 2012;18:890–900. doi: 10.1158/1078-0432.CCR-11-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 69.Lochhead P, Kuchiba A, Imamura Y, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]