Abstract
Background
LKB1 is a serine/threonine kinase that functions as a tumor suppressor, which regulates cell polarity, proliferation and metabolism. Mutations in LKB1 are associated with Peutz-Jeghers syndrome, as well as sporadic cervical and lung cancers. While LKB1-null mice develop invasive endometrial cancers, the role and regulation of LKB1 in the pathogenesis of human endometrial cancer are not well defined, and are the focus of these studies.
Methods
LKB1 protein and mRNA expression levels were evaluated in high and low-grade endometrioid endometrial cancer (EEC) and cell lines by RT-PCR, western blot analysis and immunohistochemistry. Mutational and promoter analyses of the LKB1 (STK11) gene were performed to identify mechanisms contributing to the loss of LKB1 in high-grade EEC.
Results
Analysis of the LKB1 gene in low and high-grade EECs revealed no genetic mutations, suggesting that alterations in LKB1 transcription may be responsible for LKB1 protein loss in high-grade EEC. Analysis of the LKB1 promoter revealed four putative p53 binding sites. Quantitative chromatin immunoprecipitation demonstrated that p53 bound directly to one of these sites and increased LKB1 promoter activity 140-fold. LKB1 promoter activity, mRNA, and protein levels were suppressed following silencing of p53 with siRNA, and elevated in cells over-expressing p53. P53 mRNA and protein expression were decreased in high-grade EEC, and positively correlated with LKB1 protein levels (Spearman correlation, r = 0.601, p<0.001).
Conclusions
LKB1 is a direct transcriptional target of p53. The loss of wild-type p53 in high-grade EEC may contribute to LKB1 loss seen in these more aggressive tumors.
Keywords: Endometrial Cancer, LKB1, p53, transcriptional regulation, serine threonine kinase
Introduction
Endometrial carcinoma is the most common gynecologic malignancy in the United States. Approximately 80% of endometrial carcinomas are endometrioid tumors (EEC), which are typically low-grade, early stage disease with good outcomes. The 5-year survival rate for women with Grade 1 EEC exceeds 90%, whereas it is only 69% for women with Grade 3 EEC1. Unlike well-differentiated low-grade tumors, poorly differentiated high-grade tumors are usually associated with poor prognosis due to high risk of relapse and distant metastases. High-grade tumors demonstrate a higher extent of myometrial wall infiltration, lymph-vascular space invasion and higher proliferative rate when compared with Grade 1 EEC 2. The complex molecular mechanisms contributing to the development of high-grade EEC remain unresolved.
Liver kinase B1 (LKB1) is a multifunctional serine/threonine kinase that regulates cell polarity, proliferation, apoptosis, cell cycle progression and energy metabolism through phosphorylation and activation of AMP-dependent kinase (AMPK) and other substrates. Germline mutations of LKB1 gene (also known as STK11) are responsible for Peutz-Jeghers syndrome, which is characterized by hamartomatous polyps and increased incidence of several cancers including endometrial cancer 3.
Genetically engineered LKB1 mutant mice demonstrated a critical role of LKB1 in endometrial carcinogenesis. 53% of LKB1−/+ female mice spontaneously developed endometrial adenocarcinomas with myometrial invasion, while 100% of female mice with homozygous endometrial-specific LKB1 inactivation developed aggressive and invasive endometrial cancers which spread to adjacent organs including ovary, cervix and bladder 4, 5. Consistent with these mouse studies, our lab has previously shown that 5 of 26 (19%) of human endometrial cancer cell lines exhibited loss of LKB1 protein by western blot analysis 6. Among these 5 cases, 4 originated from grades 2/3 endometrioid adenocarcinoma. The absence of LKB1 is thought to contribute to a loss of AMPK and tuberous sclerosis complex-2 activities. The subsequent hyper-activation of mTOR signaling is associated with endometrial carcinogenesis 7. We therefore hypothesized that the loss of LKB1 in high-grade endometrial cancers may contribute to the development of more aggressive phenotype.
In the current study, we sought to extend our preliminary observations and identify potential clinical consequences resulting from LKB1 loss in endometrial cancer. We further investigated potential molecular mechanisms responsible for regulating LKB1 expression in endometrial tumors.
Materials and Methods
Clinical Specimens
Following IRB approval, tissue from a total of 74 endometrioid endometrial adenocarcinomas cases were obtained for analysis from the MD Anderson Gynecologic Cancer Translational Research Tissue Bank, including 22 grade 1; 24 grade 2 and 28 grade 3 tumors. Tumor grade was assessed using the revised International Federation of Gynecology and Obstetrics (FIGO) system. All slides were examined by an expert gynecologic pathologist for confirmation of the histologic type and grade.
Cell culture and reagents
Human endometrial carcinoma cell lines, ECC-1, Ishikawa, Hec-1A, Hec-1B and KLE cells (ATCC, Manassas, VA) were cultured in RPMI-1640 supplemented with 10% fetal bovine serum and 2 mM L-glutamine (Invitrogen, Carlsbad, CA) and grown in humidified incubator with 5% CO2 at 37°C.
siRNA transfection and retroviral infection
The siGENOME smartpool of siRNA against p53 (M-003329-03) and control siRNA (RISC-free 1, D-001220-01) were purchased from Dharmacon (Thermo Scientifics, Chicago, IL). Cells were transfected using RNAiMax (Invitrogen, Carlsbad, CA) based on the manufacturer’s instruction. A retroviral plasmid encoding full-length p53 gene was a generous gift from Dr. Kwong-Kwok Wong (UT MD Anderson Cancer Center). P53 and control retroviral particles were incubated with cells for 4 h, complete culture medium were then added to the cells and incubated for 48 h.
Quantitative RT-PCR
Total RNA was extracted by lysing cells with TRI reagent (Molecular Research Center, Cincinnati, OH), and 0.5 μg total RNA was used to synthesize first strand cDNA by ImProm-II™ Reverse Transcription System (Promega, Madison, WI). Real-time PCR was performed using the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA) and the TaqMan® Gene Expression Master Mix (Applied Biosystems, Foster City, CA) under the standard thermal-cycling condition. The LKB1 and GAPDH specific Taqman probes and primers were from Applied Biosystems TaqMan® Gene Expression Assay. The Relative Standard Curve Method (2−ΔΔCt) was used to determine relative mRNA expression, using GAPDH as the reference.
Western blot analysis
The cells were washed thrice with ice-cold PBS and lysed in RIPA buffer (20 mM sodium phosphate, 150 mM NaCl (pH 7.4), 1% Nonidet P-40, 0.1% SDS, and 0.5% deoxycholic acid) containing complete protease inhibitor mixture (Roche). The proteins were separated on SDS-polyacrylamide gels and electrophoretically transferred to Immobilon PVDF membrane (Millipore, Billerica, MA). The membranes were incubated with primary antibodies against LKB1 (Santa Cruz Biotechnology, Santa Cruz, CA), p53 (Cell Signaling, Beverly MA) and β-actin (Sigma-Aldrich, St. Louis, MO) for overnight at 4°C followed by appropriate horseradish peroxidase-conjugated secondary antibodies at 1:10,000 dilution (GE Healthcare Biosciences, Pittsburgh, PA) for 1 h at room temperature. Signals were developed by using ECL chemiluminescence detection reagents (GE Healthcare Biosciences) and visualized on X-ray film (Fuji Photo Film, Tokyo, Japan). The experiment was repeated three times.
Cell proliferation and apoptotic cell death assays
2×104 ECC-1 or KLE cells were seeded into each well of 96-well plate one day before transfection. ECC-1 cells were transfected with LKB1 siRNA (VHS50411, Invitrogen) using RNAiMax (Invitrogen) based on the manufacturer’s instruction. KLE cells were transfected with 0.2 μg of full length LKB1 expression vector, LKB1-pCMV using Lipofectamine 2000 (Invitrogen) and pCMV empty vector was used as a negative control. 72 h after transfection, the cells were subjected to MTT and apoptotic cell death assays. For MTT assay, the cells were incubated with 50 μl of 1 mg/ml 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma) in PBS for 3 h. The formazan formed was then solubilized by adding 150 μl dimethyl sulfoxide (DMSO). The absorbance was read at 570 nm using a FLUOstar Galaxy plate reader. For apoptotic cell death assay, the cells was evaluated by a Cell Death Detection ELISA Plus kit purchased from Roche Molecular Biochemicals (Indianapolis, IN), following the manufacturer’s instructions. The absorbance was read at 405 nm using a FLUOstar Galaxy plate reader. The data was expressed as relative percentage of apoptosis induction.
Direct sequencing of LKB1 coding exons
Laser-capture microdissection was used to collect the pure population of cancer cells from 20 Grade 3 endometrioid endometrial tumors and DNA extraction was performed using QIAamp DNA Micro Kit (Qiagen). 5 ng genomic DNA from each patient was amplified using the following intron-specific primers flanking each LKB1 exon: Exon 1, forward 5′-ACAAGGAAGGACCGCTCACC-3′, reverse 5′-CGACCCCAGCAAGCCATACT-3′; Exon 2, forward 5′-ATCCTGACGTTGGGTCGGCT-3′, reverse 5′-ACAATGGCTGACTTCCGGGG-3′; Exon 3, forward 5′-TCCAGAGCCCCTTTTCTGGC-3′, reverse 5′-TGTGGCCTCACGGAAAGGA-3′; Exon 4/5, forward 5′-TGGGCCTGTGGTGTTTGGGA-3′, reverse 5′-AGTGTGCGTGTGGTGAGTGC-3′; Exon 6, forward 5′-AGGGCGTCAACCACCTTGACT-3′, reverse 5′-TTCTGCACAAAAGCCCCGCC-3′; Exon 7, forward 5′-AGGGCCTGACAACAGAGGCT-3′, reverse 5′-CGGTAACAGGACACTGCCCA-3′; Exon 8, forward 5′-TCGGAAAACTGGACCGCCCT-3′, reverse 5′-ACGTGGGATTGGCCACCAGA-3′; Exon 9, forward 5′-TGTAAGTGCGTCCCCGTGGT-3′, reverse 5′-TCCAGGCGTTGTCCCCACAT-3′. The amplified DNA fragments were subjected to Sanger sequencing.
Promoter constructs and site-directed mutagenesis
The DNA fragments containing the putative p53 binding sites in upstream sequence of LKB1/STK11 were amplified from ECC-1 genomic DNA using the following primers: LKB1(−1726/−1625): forward 5′-TCTTACGCGTAAGCAGTTCTCCTGCCTCAG-3′, reverse 5′-AGATCTCGAGTGGTGAAACCCCATCTCTAC-3′; LKB1(−937/−858): forward 5′-TCTTACGCGTCTCCTCCCTCAGCCTTCTG-3′, reverse 5′-AGATCTCGAGCCCGGCTAATTTTTGTATTTTT-3′; LKB1(−664/−578): forward 5′-TCTTACGCGTGGCAACTCTTGTTTTTCACGA-3′, reverse 5′-AGATCTCGAGTCTGCCCTATCGGAACTCAT-3′; LKB1(−164/−1): forward 5′-TCTTACGCGTAACGCTCCAATCGTCAGC-3′, reverse 5′-AGATCTCGAGGCCGCCATCTTGTTTACCTC-3′. PCR products were subsequently cloned into the MluI and XhoI sites of the luciferase reporter vector pGL3-Basic (Promega) and the integrity of constructs was confirmed by DNA sequencing. Site-directed mutagenesis was carried out to introduce mutations into the putative p53 binding site from LKB1(−164/−1) using the QuikChange II site-directed kit (Stratagene, La Jolla, CA) and the mutation-containing primers are 5′-CGCGTCAGCGGCGGCGGGGCGGGCAGAGGGCCGGGGATGGCAGGTTCAACCAACCGGTGGGGACCTCGTCCTCGCGAGGAGGCGTGCCCTGCGGCCGGGCGTGCGGTGTC-3′ (sense) and 5′-TCGAGACACCGCACGCCCGGCCGCAGGGCACGCCTCCTCGCGAGGACGAGGTCCCCACCGGTTGGTTGAACCTGCCATCCCCGGCCCTCTGCCCGCCCCGCCGCCGCTGA-3′ (antisense).
Transient transfection and luciferase reporter gene activity assay
1×105 ECC-1 cells were seeded into 24-well plates one day before transfection. The cells were transfected with 0.8 μg of luciferase-reporter vector containing the LKB1 upstream genomic sequence using Lipofectamine 2000 (Invitrogen) and pGL3 empty vector was used as a negative control. 1:20 of pRL-TK, which encoding Renilla luciferase, was included in all transfections to normalize transfection efficiency. 24 h after transfection, the cells were washed and lysed with the passive lysis buffer from the Dual-Luciferase Reporter Assay System (Promega). Luciferase activity was measured in each cell lysate using a FLUOstar Omega microplate reader (BMG LABTECH, Germany). Firefly luciferase activity was normalized to Renilla luciferase and the data are expressed as fold induction relative to that of the empty pGL3 Basic plasmid.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using MAGnify™ Chromatin Immunoprecipitation System based on the manufacturer’s instruction (Invitrogen). Briefly, ECC1 cells were grown to confluence, crosslinked, lysed, and sheared. The cell supernatants were diluted with Dilution Buffer and aliquots of samples were saved as the input DNA for quantization of the amount of total DNA. Cross-linked p53 protein-DNA complexes were immunoprecipitated with a p53-specific antibody (Invitrogen) or normal rabbit IgG. Thereafter, the immunoprecipitated DNAs were amplified by quantitative PCR with the primers indicated below. LKB1 promoter region (−174/−89): forward 5′-GCCGGGTCCAAACGCTCCAAT-3′; reverse 5′-GACGTGCCCACCCGTTGGTT-3′. intron region between exon 1 and 2 of LKB1 (+2338/+2423): forward 5′-TCGAGGGACTGGCAAGGACA-3′; reverse 5′-TGAGAGCAGACACCGCAGCA-3′; p21 promoter with known p53-binding site: forward 5′-CCCTTCCTCACCTGAAAACA-3′; reverse 5′-GTGGCTCTGATTGGCTTTCTG-3′.
Immunohistochemistry
Immunohistochemical staining was performed based on the protocol and reagents from Biocare Medical (Concord, CA). Briefly, 4-μm thick sections of formalin-fixed paraffin embedded tissue were deparaffinized, rehydrated, and exposed to heated antigen decloaking with pH6.0 citrate buffer or DakoCytomation Target Retrieval Solution High pH for LKB1 or p53 staining respectively. The specimens were blocked with BACKGROUNDsniper to reduce nonspecific background staining, and incubated with polyclonal rabbit anti-LKB1 antibody (dilution 1:60, clone HPA017254, Sigma) and monoclonal mouse anti-p53 antibody (dilution 1:100, clone DO-7, Dako) for 1 h at room temperature. MACH 3 Rabbit/Mouse AP Polymer Detection and Vulcan Fast Red Chromogen Kit were used according to the manufacturer’s protocol. The images of the immunostained slides were examined and captured using a computerized image analysis system comprised of a high definition Leica microscope and Image Pro Plus software (Media Cybernetics, Silver Spring, MD). Five random microscope fields per section were evaluated at 20x magnification and the integral optical density (IOD) of every visual field was calculated. The data was normalized to that of the sample with the lowest expression, of which the expression is designated as 1.0.
Results
LKB1 loss in high-grade endometrial cancer
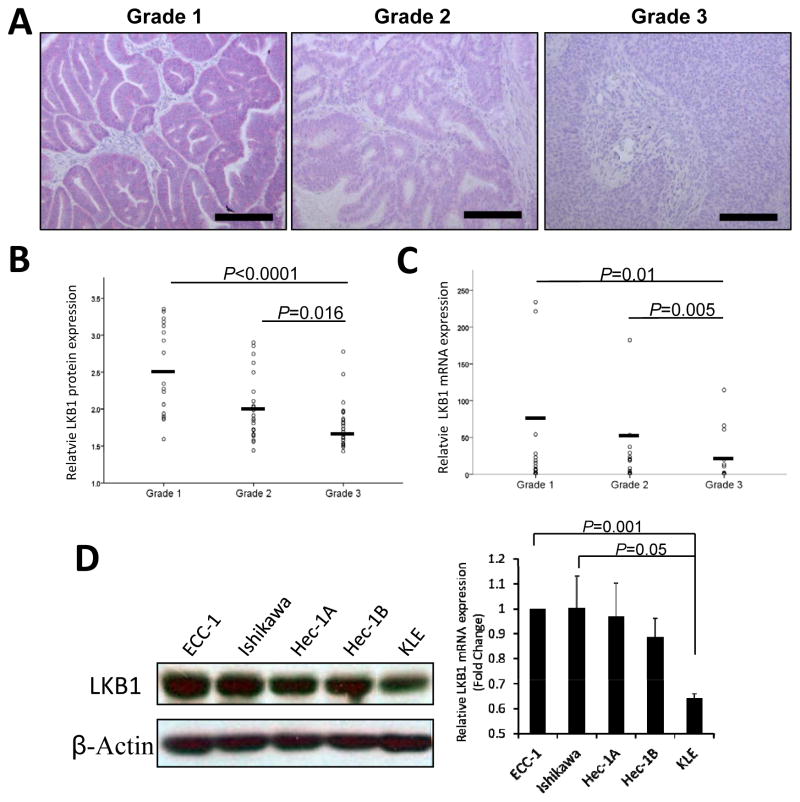
LKB1 protein expression levels were evaluated in different grades of human endometrial cancer by immunohistochemistry. A total of 17 Grade 1, 24 Grade 2, and 28 Grade 3 human endometrioid endometrial adenocarcinomas were examined (Fig 1). The well-differentiated glandular Grade 1 tumors showed strong LKB1 staining. LKB1 staining intensity decreased in the moderately differentiated Grade 2 tumors. LKB1 protein expression was significantly lower in the poorly differentiated Grade 3 tumors when compared with Grade 1 (P<0.0001) and Grade 2 (P=0.016) tumors (Fig. 1A and 1B). To further test whether LKB1 mRNA expression is also downregulated in high-grade endometrial cancer cells, we isolated total RNA of cancer cells from fresh frozen endometrioid endometrial tissues using laser-capture microdissection. LKB1 mRNA expression was significantly downregulated in 23 Grade 3 endometrial cancer cells when compared with 22 Grade 1 (P=0.01) and 21 Grade 2 (P=0.005) (Fig. 1C). Levels were normalized by relative quantitation of LKB1 expression as compared to GAPDH expression.
Figure 1. LKB1 protein and mRNA expression levels in human endometrial carcinomas.
A) Immunohistochemical staining of LKB1. Formalin-fixed, paraffin-embedded sections of 17 Grade 1, 24 Grade 2, and 28 Grade 3 human endometrioid endometrial adenocarcinomas were immunostained with anti-LKB1 antibody. The slides were counter-stained with Hematoxylin. The staining intensity of LKB1 (red color) was computationally calculated and shown in the dot plot (B). Horizontal bars represent the mean intensities. C) LKB1 mRNA expression in Grade 1, 2 and 3 endometrial carcinoma. Laser-capture microdissection was used to isolate tumor tissue from 22 Grade 1, 21 Grade 2 and 23 Grade 3 fresh frozen endometrioid endometrial tissues, from which RNA was prepared. LKB1 mRNA expression levels in microdissected tumor samples were quantified by TaqMan quantitative PCR. D) LKB1 mRNA and protein levels in well-differentiated (ECC-1 and Ishikawa), moderately-differentiated (Hec-1A and Hec-1B) and poorly differentiated (KLE) endometrial cancer cell lines. The relative LKB1 mRNA and protein levels were measured by quantitative RT-PCR and Western blot analyses respectively. A representative experiment is shown, and similar results were obtained from three independent experiments.
We further examined the expression of LKB1 in human endometrial cancer cell lines. ECC-1 and Ishikawa cells are derived from primary well-differentiated endometrial adenocarcinomas and ECC-1 cells formed tumors with glandular structures following transplantation to athymic nude mice. Hec-1A and Hec-1B cells were derived from moderately differentiated, Grade 2 endometrial adenocarcinomas. KLE cells were generated from a poorly-differentiated, high-grade endometrial carcinoma. By quantitative RT-PCR and western blotting analyses, the well-differentiated ECC-1 and Ishikawa cells expressed elevated levels of LKB1 mRNA and protein. In contrast, poorly differentiated KLE cells expressed low levels of LKB1 while Hec-1A and Hec-1B demonstrated moderate LKB1 expression (Fig. 1D). These findings confirm that LKB1 is expressed at lower levels in both poorly-differentiated, high-grade endometrial primary tumors and cell lines derived from high-grade tumors, as compared with low-grade tumors.
LKB1 expression modulates cell proliferation and apoptosis in endometrial cancer cell lines
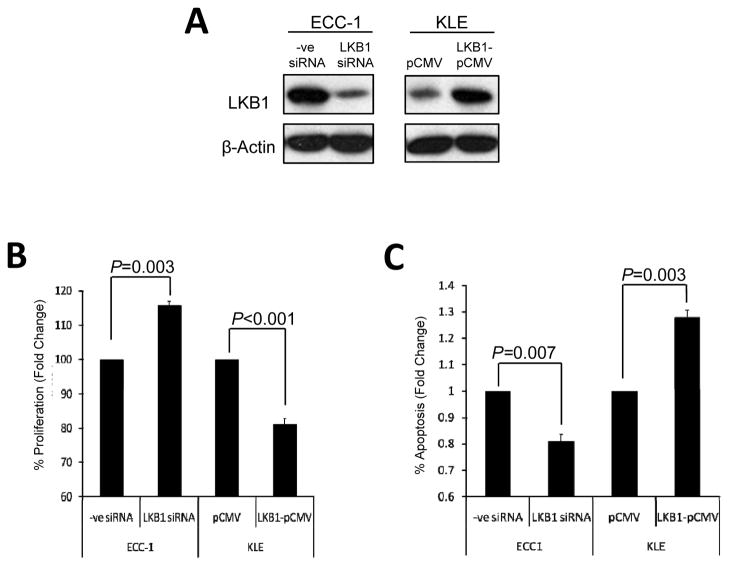
To evaluate the effect of LKB1 levels on cellular proliferation and apoptosis, ECC-1, which express comparatively high levels of LKB1, were transfected with LKB1-silencing siRNA, while KLE cells, which express low levels of LKB1 were transfected with a vector expressing full-length LKB1 (LKB1-pCMV) (Fig. 2A). After 72 h, MTT assay and cytoplasmic DNA fragmentation ELISA were used to measure the proliferation rate and apoptosis induction respectively. Our in vitro findings confirmed that elevated levels of LKB1 expression were associated with a suppression of cell proliferation (Fig. 2B) and increased apoptosis (Fig. 2C).
Figure 2. LKB1 loss in endometrial cancers confer increased cell proliferation and decreased apoptosis.
A) ECC-1 and KLE cells were transfected with LKB1 siRNA and vector expressing full-length LKB1 (LKB1-pCMV) respectively. After 72 h, B) MTT assay and C) cytoplasmic DNA fragmentation ELISA were used to measure the proliferation rate and apoptosis induction respectively.
Absence of LKB1 mutations in high-grade endometrial cancer
Because somatic LKB1 mutations have been found in several sporadic human cancers, we initially performed direct sequence analysis of LKB1 coding exons using genomic DNA from 20 microdissected Grade 3 endometrial cancers to investigate whether LKB1 is inactivated by allelic deletions and/or somatic mutations. As illustrated in Table 1, 4 samples had synonymous mutation p.Tyr175=, 3 samples had synonymous mutation p.Ile161= and 1 sample had both p.Tyr175= and p.Ile161= synonymous mutations. No non-synonymous mutations were identified, indicating that mutational alterations of LKB1 are not the major mechanism leading to the loss of LKB1 in high-grade endometrial cancer.
Table 1. Direct genomic DNA sequencing of LKB1 coding exons on 20 microdissected Grade 3 endometrioid endometrial tumors.
Total number of mutations found in each exon for each patient is listed in the table. No somatic mutations leading to amino acid substitutions or frameshifts were found.
| Patient ID | Exon 1 | Exon 2 | Exon 3 | Exon 4 | Exon 5 | Exon 6 | Exon 7 | Exon 8 | Exon 9 |
|---|---|---|---|---|---|---|---|---|---|
| #1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #2 | 0 | 0 | 0 | 1 (NP_000446.1:p.Tyr175=) | 0 | 0 | 0 | 0 | 0 |
| #3 | 0 | 0 | 0 | 1 (NP_000446.1:p.Ile161=) | 0 | 0 | 0 | 0 | 0 |
| #4 | 0 | 0 | 0 | 1 (NP_000446.1:p.Tyr175=) | 0 | 0 | 0 | 0 | 0 |
| #5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #10 | 0 | 0 | 0 | 1 (NP_000446.1:p.Tyr175=) | 0 | 0 | 0 | 0 | 0 |
| #11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #12 | 0 | 0 | 0 | 1 (NP_000446.1:p.Tyr175=) | 0 | 0 | 0 | 0 | 0 |
| #13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #17 | 0 | 0 | 0 | 2 (NP_000446.1:p.Tyr175= & p.Ile161=) | 0 | 0 | 0 | 0 | 0 |
| #18 | 0 | 0 | 0 | 1 (NP_000446.1:p.Ile161=) | 0 | 0 | 0 | 0 | 0 |
| #19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #20 | 0 | 0 | 0 | 1 (NP_000446.1:p.Ile161=) | 0 | 0 | 0 | 0 | 0 |
The LKB1 gene is infrequently hypermethylated in EEC
To evaluate the role of hypermethylation on LKB1 expression in EEC, we examined LKB1 promoter methylation in the 270 EEC cases in the TCGA dataset for which both methylation and clinical data were available8. In total, 132 of the cases were grade 1 and 2, while 138 of these tumors were grade 3. Methylation status was evaluated using the Illumina Infinium HumanMethylation450 BeadChip microarray. 485,577 methylation sites are covered on the HumanMethylation450 Platform, of which three probes mapped to the LKB1 promoter and putative CpG island as predicted by the UCSC Genome Browser9. For these three genomic loci, the mean beta value was 0.030 ± 0.008; beta values range from 0 to 1, with a value below 0.3 indicating no methylation. These values are comparable to the mean beta value for 46 normal tissue samples from the TCGA dataset (0.032 ± 0.011). Taken together, these data suggest that loss of LKB1 protein expression observed in high-grade EEC is likely not due to epigenetic silencing.
Hypermethylation of the LKB1 gene has previously been examined in cervical, lung, colon, head and neck, pancreatic and breast cancers 10,11,12,13. Epigenetic silencing of the LKB1 gene also does not appear to be the major cause of LKB1 loss in these tumor types, which like EEC, is predominantly observed in high grade tumors.
Regulation of LKB1 transcription by p53
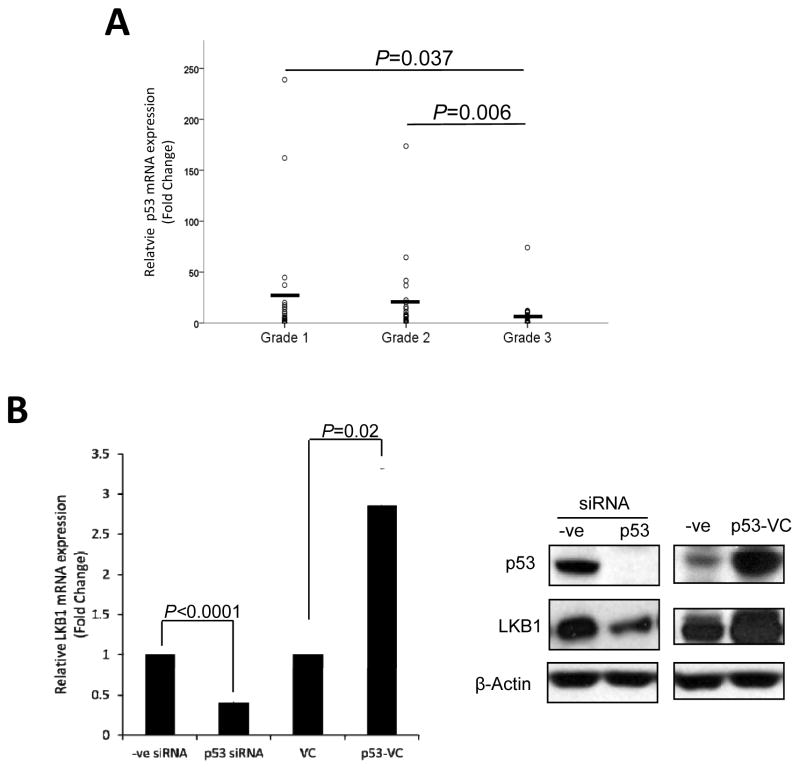
Based on the observation that both LKB1 mRNA and protein levels were decreased in high-grade endometrial tumors, we further investigated the transcriptional regulation of LKB1 expression. Computational sequence analysis of the LKB1 5′-flanking region using MatInspector and TESS (Transcription Element Search Software) revealed a number of binding sites for several potential transcription factors, including AP-1 (activator proteins 1), STAT5 (signal transducer and activator of transcription 5), C/EBP (CCAAT/Enhancer Binding Protein) and others. Notable were four potential binding sites for p53, which is reported to be both an interaction partner and phosphorylation target of LKB1 14, 15. Since LKB1 is downregulated in high-grade endometrial cancers, we first examined the expression of p53 in human endometrial tumor samples to see whether its expression is correlated with that of LKB1. By quantitative RT-PCR, p53 mRNA expression was significantly downregulated in microdissected Grade 3 endometrioid endometrial tumor tissue (n=24) compared with Grade 1 (P=0.037; n=22) and Grade 2 (0.006; n=21) tumors, and a positive correlation was observed between p53 and LKB1 mRNA levels in the tissues using Spearman correlation analysis (correlation coefficient=0.362, P=0.001; Table 2). Furthermore, by quantitative RT-PCR and western blot analyses, gene silencing of p53 by siRNA suppressed the LKB1 mRNA and protein expression; retroviral infection of full-length p53 increased LKB1 mRNA and protein level in ECC-1 cells (Fig. 3B), indicating that LKB1 expression is induced by p53.
Table 2. Correlation between LKB1 and p53 mRNA expression.
mRNA expression of p53 and LKB1 were examined in 24 micro-dissected Grade 3 and 22 micro-dissected Grade 1 endometrioid endometrial tumor tissues and their correlation was calculated by Spearman correlation analysis.
| p53 mRNA level | |||
|---|---|---|---|
| Spearman’s rho | LKB1 mRNA level | Correlation Coefficient | 0.382 |
| Sig. (2-tailed) | 0.001 | ||
| N | 67 |
Figure 3. LKB1 expression is regulated by p53.
A) p53 mRNA expression in microdissected Grade 1 (n=22), Grade 2 (n = 21) and Grade 3 (n= 24) fresh frozen endometrioid endometrial tissues as quantified by quantitative PCR. The correlations between the mRNA expression of LKB1 and p53 were carried out using Spearman correlation analysis. B) The effect of p53 knockdown and overexpression on the mRNA and protein levels of LKB1. ECC-1 cells were transfected with p53 siRNA or infected with retrovirus encoding full-length p53 (p53-VC) for 24 h, after which total RNA and protein were extracted and subjected to quantitative RT-PCR and Western blot analyses.
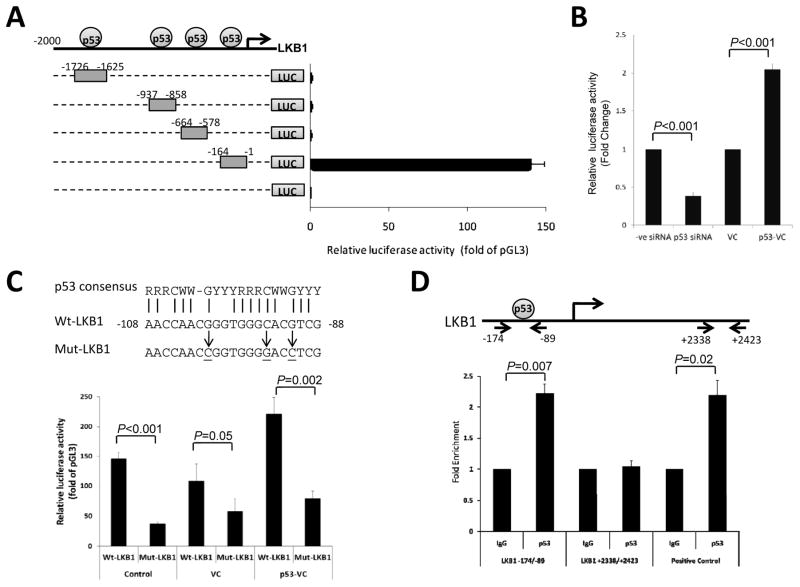
To elucidate the transcriptional regulation of LKB1 by p53, primers were designed to amplify the DNA fragments (−1726/−1625, −937/−858, −664/−578 and −164/−1) containing the putative p53 binding sites in the upstream promoter region of the LKB1 gene. These fragments were cloned into the pGL3 basic luciferase vector for transfection and luciferase reporter assays. The luciferase activity of ECC-1 cells transfected with LKB1(−164/−1) was induced by 147 fold while there was no significant increase in luciferase activity for the other three constructs, as compared to the vector control (Fig. 4A). To further confirm the role of p53 in the regulation of LKB1 transcriptional activity, ECC-1 cells were transfected with p53 siRNA or infected with full-length p53 retrovirus to knock down or overexpress p53 proteins in the cells, respectively. Knockdown of p53 significantly reduced LKB1(−164/−1) luciferase activity in ECC-1 cells while p53 overexpression increased the luciferase activity by 2-fold (Fig. 4B), suggesting p53 regulates the activity of the LKB1 promoter within the −164-bp region. We further compared the candidate DNA elements in the LKB1(−164/−1) with the p53 consensus binding sequence as described by El-Deiry et al. 16. A 20 bp sequence located at −108 to −88 bp of LKB promoter shared 75% identity with the human p53 consensus site. Site-directed mutagenesis was carried out to introduce mutations into the putative p53 binding site in the LKB1 promoter. We found that LKB1 promoter activity was greatly decreased upon mutation of the p53 binding site and the p53 activation of LKB1 promoter activity was blocked by the mutation (Fig. 4C). To show direct binding of p53 to the LKB1 promoter region at −108 to −88 bp in vivo, a quantitative chromatin immunoprecipitation assay was performed in ECC-1 cells using anti-p53 antibody. P53 bound chromatin was immunoprecipitated and subjected to quantitative PCR analysis using primers covering the p53 binding site in the −108 to −88 bp of LKB1 promoter. Fig. 4D showed that p53 bound to the LKB1 promoter and the same p53 binding result was observed in the positive control, known p53 binding site in p21 promoter. In contract, no binding of p53 was observed in the intron region between exon 1 and 2 of LKB1 (+2338 to +2423 bp). The result confirms that LKB1 transcription activity is directly regulated by p53 at −108 to −88 bp of LKB1 promoter.
Figure 4. Transcriptional regulation of LKB1 by p53.
A) Schematic representation of the putative p53 binding sites in upstream sequence of LKB1 and the DNA fragments cloned into the upstream of the firefly luciferase reporter gene (LUC). Each reporter construct was transiently transfected into ECC-1 cells and the cells were harvested after 24 h for the dual luciferase assay. The promoter activity of each construct was measured using the firefly luciferase activity that was first normalized to Renilla activity and then calculated as fold induction relative to that of the empty pGL3 Basic plasmid. B) The effect of p53 knockdown and overexpression on the promoter activity of LKB1. After transfection with p53 siRNA or infection with retrovirus encoding full-length p53 (p53-VC) for 6 h, ECC-1 cells were then transfected with LKB1(−164/−1) reporter construct and subjected to the dual luciferase assay. C) The sequence of the p53-binding site in LKB1 genomic sequence is shown and compared to the p53 consensus binding. R, G or A; W, A or T; Y, C or T. The sequence of the mutant version of the p53-binding site is also shown. ECC-1 cells were transduced with p53-VC or control retroviral particles (VC) for 6 h and then transfected with LKB1(−164/−1) (Wt–LKB1) or mutated construct (MutLKB1). D) Chromatin immunoprecipitation assay was performed to determine the association of p53 with the LKB1 promoter. Normal rabbit IgG was included as the negative control (IgG). The recovered chromatin was subjected to quantitative PCR analysis using primers shown in the diagram. PCR amplification of the p53 site in the p21 promoter was included as positive control. Results are given as fold enrichment relative to IgG.
Wild-type p53 controls LKB1 expression
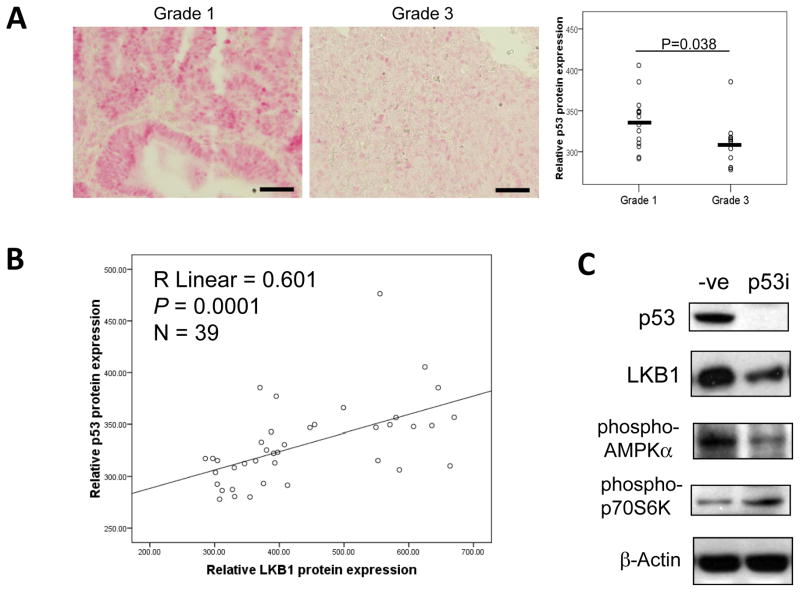
Since we have shown that p53 could bind to the LKB1 promoter in vivo and transactivates the LKB1 promoter, we further examined p53 protein expression in human endometrial tumor samples and studied its correlation with LKB1 expression. While P53 mutations are observed in up to 90% of non-endometrioid endometrial cancers (NEEC), P53 mutations are seen in only 10–20% of EECs, most of which are high grade tumors17–19. We therefore evaluated the expression status of full-length, wild type p53 protein in grade 3 versus grade 1 EECs, and were compared p53 expression status to relative levels of LKB1 expression observed in each sample. Immunohistochemical staining showed a significant decrease in wild type p53 protein levels in Grade 3 tumors (n=16) when compared with those observed in Grade 1 (P=0.038; n=13) tumors (Fig. 5A). As predicted, protein levels of p53 positively correlated with those of LKB1 (Spearman correlation coefficient=0.601, P=0.0001) (Fig. 5B), in support of a direct role of p53 in the regulation of LKB1 expression levels. Since LKB1 is known to downregulate mTOR signaling by activating the AMPK signaling pathway7, we further investigated the effect of p53 on LKB1 regulated AMPK/mTOR pathway. By Western blot analyses, p53 silencing by siRNA transfection decreased the protein expression of LKB1 and phospho-AMPKα; and increased the phosphorylation of p70S6K, the downstream target of mTOR (Fig. 5C).
Figure 5. Wild type p53 protein expression is correlated with LKB1.
A) Immunohistochemical staining of wild type p53 on FFPE sections of Grade 1 (n=13) and Grade 3 (n=16) human endometrioid endometrial adenocarcinomas. The staining intensity was computationally calculated and shown in the dot plot. B) The protein expression of LKB1 and p53 were evaluated in 39 EEC samples with wild type p53 using immunohistochemistry and their correlation is calculated by Spearman correlation analysis. C) The effect of p53 on the LKB1/AMPK pathway. An siRNA pool for p53 was transfected into ECC-1 cells for 48 h, cells were then lysed for protein extraction and subjected to Western blot analyses. The experiment was repeated three times and a representative experiment is shown.
Discussion
Our current studies confirm our original observations that LKB1 mRNA and protein expression are significantly decreased in high-grade, poorly differentiated endometrioid endometrial tumors and cell lines. Furthermore, siRNA mediated knockdown of LKB1 in ECC-1 cells increased cell proliferation and decreased apoptosis, while restoration of LKB1 expression in KLE cells by gene transfection decreased proliferation and increased apoptosis. Collectively, these findings suggest that the loss of the tumor suppressing effect of LKB1 in endometrial tumors may lead to progression of endometrial cancer towards a more aggressive, high-grade phenotype.
Given these observations, we sought to identify the underlying mechanisms responsible for LKB1 modulation in endometrial tumors. LKB1 (STK11) gene mutation has been extensively investigated in a variety of cancers. Germline mutations in LKB1 not only cause Peutz-Jeghers syndrome, but are also associated with the development of some sporadic tumors. LKB1 is the third most-commonly mutated gene in non-small cell lung carcinoma (NSCLC); nearly 90% in 124 NSCLC cases tested had either loss of heterozygosity or homozygous deletion of the LKB1 gene 20. Point mutation and/or deletion of the LKB1 gene was also observed in 20% of the primary cervical tumor samples and in the cervical cancer cell lines, HeLa and SiHa 21. Sporadic LKB1 mutations are less common in other tumor types 22.
Curiously, unlike cervical cancer and lung cancer, we demonstrated the loss of LKB1 in endometrial tumors does not result from direct mutations and deletions of the LKB1 gene. Neither is it a consequence of DNA methylation. Instead, we have identified a previously undescribed and direct role for p53 in the regulation of LKB1 expression. The increased incidence of p53 mutations has been reported in Grade 3 versus Grade 1 and 2 EEC 23, suggesting that loss of functional p53 may account, in part, for decreased LKB1 expression that contributes to a more aggressive phenotype and poor outcome for those patients with high-grade EEC. Interestingly, a recent report in non-small cell lung cancers also demonstrated that alterations in LKB1 expression often occurred simultaneously with mutations in p5324.
Mutational analysis revealed that the p53-binding site at −108 to −88 bp site in the LKB1 gene is required for p53-mediated activation of the LKB1 promoter. P53 was confirmed to bind to this region in vivo using a quantitative chromatin immunoprecipitation assay. Furthermore, overexpression of p53 increased LKB1 promoter activity, which was reflected by an elevation both LKB1 mRNA and protein levels. While we did not sequence the LKB1 promoter region ourselves, we queried the LKB1 promoter in 247 EECs included in the Catalogue of Somatic Mutations in Cancer (COSMIC) data set25, 26. No obvious mutations were observed in 165 bp region which contains the p53 binding site, suggesting that mutations in this sequence are an extremely rare event
This is the first report describing the modulation of LKB1 mRNA and protein expression by direct binding p53 to the LKB1 gene promoter. Previous studies have identified important but independent roles for both p53 and LKB1 in the regulation of cellular metabolism. In normal tissues, the expression of p53 and LKB1 are each associated with tumor suppression. In addition to its known role in cell cycle blockade and growth arrest in response to DNA damage and hypoxia, additional roles for p53 in the regulation of energy metabolism in response to stress have been recently described27, 28. P53 activates AMPK expression and AMPK and TSC1/2 activity, thereby suppressing cell growth, reducing energy-consuming processes such as protein synthesis, and promoting ATP producing processes such as glycolysis 29. Similarly, in conditions of ATP and nutrient depletion, LKB1 is known to directly phosphorylate AMPK and activate the AMPK signaling pathway. By Western blot analyses, we showed that p53 silencing not only decreased LKB1 protein expression, thereby downregulating AMPKα activity and increasing expression of phospho-p70S6K. Our findings demonstrate a direct link between p53 and LKB1 expression, and provide an additional mechanism by which p53 regulates AMPK-dependent pathways to maintain cellular homeostasis.
Acknowledgments
This work was supported by a grant from the National Cancer Institute (5 P50 CA098258).
Footnotes
Conflict of Interest
There are no competing financial interests in relation to the work described in this manuscript.
References
- 1.Creasman WT, Kohler MF, Odicino F, Maisonneuve P, Boyle P. Prognosis of papillary serous, clear cell, and grade 3 stage I carcinoma of the endometrium. Gynecol Oncol. 2004;95:593–596. doi: 10.1016/j.ygyno.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 2.Zannoni GF, Vellone VG, Arena V, et al. Does high-grade endometrioid carcinoma (grade 3 FIGO) belong to type I or type II endometrial cancer? A clinical-pathological and immunohistochemical study. Virchows Arch. 457:27–34. doi: 10.1007/s00428-010-0939-z. [DOI] [PubMed] [Google Scholar]
- 3.van Lier MG, Wagner A, Mathus-Vliegen EM, Kuipers EJ, Steyerberg EW, van Leerdam ME. High cancer risk in Peutz-Jeghers syndrome: a systematic review and surveillance recommendations. Am J Gastroenterol. 105:1258–1264. doi: 10.1038/ajg.2009.725. author reply 1265. [DOI] [PubMed] [Google Scholar]
- 4.Contreras CM, Akbay EA, Gallardo TD, et al. Lkb1 inactivation is sufficient to drive endometrial cancers that are aggressive yet highly responsive to mTOR inhibitor monotherapy. Dis Model Mech. 3:181–193. doi: 10.1242/dmm.004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contreras CM, Gurumurthy S, Haynie JM, et al. Loss of Lkb1 provokes highly invasive endometrial adenocarcinomas. Cancer Res. 2008;68:759–766. doi: 10.1158/0008-5472.CAN-07-5014. [DOI] [PubMed] [Google Scholar]
- 6.Lu KH, Wu W, Dave B, et al. Loss of tuberous sclerosis complex-2 function and activation of mammalian target of rapamycin signaling in endometrial carcinoma. Clin Cancer Res. 2008;14:2543–2550. doi: 10.1158/1078-0432.CCR-07-0321. [DOI] [PubMed] [Google Scholar]
- 7.Korets SB, Czok S, Blank SV, Curtin JP, Schneider RJ. Targeting the mTOR/4E-BP pathway in endometrial cancer. Clin Cancer Res. 2011;17:7518–7528. doi: 10.1158/1078-0432.CCR-11-1664. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research N. Kandoth C, Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karolchik D, Hinrichs AS, Kent WJ. The UCSC Genome Browser. Curr Protoc Bioinformatics. 2012;Chapter 1(Unit1 4) doi: 10.1002/0471250953.bi0104s28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteller M, Avizienyte E, Corn PG, et al. Epigenetic inactivation of LKB1 in primary tumors associated with the Peutz-Jeghers syndrome. Oncogene. 2000;19:164–168. doi: 10.1038/sj.onc.1203227. [DOI] [PubMed] [Google Scholar]
- 11.Ekizoglu S, Dalay N, Karaman E, Akdeniz D, Ozaydin A, Buyru N. LKB1 downregulation may be independent of promoter methylation or FOXO3 expression in head and neck cancer. Transl Res. 2013;162:122–129. doi: 10.1016/j.trsl.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Trojan J, Brieger A, Raedle J, Esteller M, Zeuzem S. 5′-CpG island methylation of the LKB1/STK11 promoter and allelic loss at chromosome 19p13.3 in sporadic colorectal cancer. Gut. 2000;47:272–276. doi: 10.1136/gut.47.2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Wilde RF, Ottenhof NA, Jansen M, et al. Analysis of LKB1 mutations and other molecular alterations in pancreatic acinar cell carcinoma. Mod Pathol. 2011;24:1229–1236. doi: 10.1038/modpathol.2011.83. [DOI] [PubMed] [Google Scholar]
- 14.Zeng PY, Berger SL. LKB1 is recruited to the p21/WAF1 promoter by p53 to mediate transcriptional activation. Cancer Res. 2006;66:10701–10708. doi: 10.1158/0008-5472.CAN-06-0999. [DOI] [PubMed] [Google Scholar]
- 15.Cheng H, Liu P, Wang ZC, et al. SIK1 couples LKB1 to p53-dependent anoikis and suppresses metastasis. Sci Signal. 2009;2:ra35. doi: 10.1126/scisignal.2000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 17.Catasus L, Gallardo A, Cuatrecasas M, Prat J. Concomitant PI3K-AKT and p53 alterations in endometrial carcinomas are associated with poor prognosis. Mod Pathol. 2009;22:522–529. doi: 10.1038/modpathol.2009.5. [DOI] [PubMed] [Google Scholar]
- 18.Dedes KJ, Wetterskog D, Ashworth A, Kaye SB, Reis-Filho JS. Emerging therapeutic targets in endometrial cancer. Nat Rev Clin Oncol. 2011;8:261–271. doi: 10.1038/nrclinonc.2010.216. [DOI] [PubMed] [Google Scholar]
- 19.Yeramian A, Moreno-Bueno G, Dolcet X, et al. Endometrial carcinoma: molecular alterations involved in tumor development and progression. Oncogene. 2013;32:403–413. doi: 10.1038/onc.2012.76. [DOI] [PubMed] [Google Scholar]
- 20.Gill RK, Yang SH, Meerzaman D, et al. Frequent homozygous deletion of the LKB1/STK11 gene in non-small cell lung cancer. Oncogene. 30:3784–3791. doi: 10.1038/onc.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wingo SN, Gallardo TD, Akbay EA, et al. Somatic LKB1 mutations promote cervical cancer progression. PLoS One. 2009;4:e5137. doi: 10.1371/journal.pone.0005137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez-Cespedes M. A role for LKB1 gene in human cancer beyond the Peutz-Jeghers syndrome. Oncogene. 2007;26:7825–7832. doi: 10.1038/sj.onc.1210594. [DOI] [PubMed] [Google Scholar]
- 23.Kihana T, Hamada K, Inoue Y, et al. Mutation and allelic loss of the p53 gene in endometrial carcinoma. Incidence and outcome in 92 surgical patients. Cancer. 1995;76:72–78. doi: 10.1002/1097-0142(19950701)76:1<72::aid-cncr2820760110>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Lee SM, Choi JE, Na YK, et al. Genetic and epigenetic alterations of the LKB1 gene and their associations with mutations in TP53 and EGFR pathway genes in Korean non-small cell lung cancers. Lung Cancer. 2013;81:194–199. doi: 10.1016/j.lungcan.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Forbes SA, Bhamra G, Bamford S, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008;Chapter 10(Unit 10):11. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 28.Puzio-Kuter AM. The Role of p53 in Metabolic Regulation. Genes Cancer. 2011;2:385–391. doi: 10.1177/1947601911409738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Z, Hu W, de Stanchina E, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]