Abstract
Background
Specific pathogenic bacteria have been implicated in recurrent aphthous stomatitis (RAS), a chronic inflammatory condition characterised by ulcerations in the oral mucosa. However, the aetiology behind this condition still remains unclear.
Objective
The buccal microbiota of patients with RAS was compared to that of control subjects to investigate its potential role for this condition.
Design
Buccal swabs were obtained from non-ulcerative areas of 60 patients, of whom 42 patients had lesions at the time of sampling, and 60 healthy age- and gender-matched controls. Bacterial DNA was extracted and analysed by Terminal-Restriction Fragment Length Polymorphism, using enzymatic digestion of the polymerase chain reaction-amplified 16S rRNA gene, yielding a series of peaks, each representing a bacterial taxon.
Results
Two peaks, 60 and 489, were more prevalent in patients with RAS than controls. Conversely, peaks 58 and 490 were less common in patients than controls. When the patients were divided into subgroups, we found that the observed differences in peak-pattern were related to the presence of lesions during sampling.
Conclusions
The microbiota of the non-inflamed buccal mucosa differed between patients and controls. The differences were most pronounced in patients who presented with lesions during sampling, suggesting that a disturbance in the normal buccal microbiota triggers the presence of lesions or that presence of lesions alters the microbiota.
Keywords: oral mucosa, oral mucosal disease, oral bacteria, Terminal-Restriction Fragment Length Polymorphism, oral ulceration, oral cavity
Recurrent aphthous stomatitis (RAS) is a chronic inflammation of the oral mucosa characterised by recurrent ulcerations that affect primarily the non-keratinised mucosa. RAS is one of the most common oral mucosal disorders, with a prevalence of approximately 2%, although geographical variations in prevalence have been identified (1). Currently, no curative therapy is available and treatment strategies are aimed at reducing the discomfort caused by ulcerations. The aetiology of RAS remains unknown. Viruses and bacteria (2) as well as genetic susceptibility (3) have been implicated in this disease.
Bacteria may contribute to the pathogenesis of RAS, acting either as pathogens or as the source of antigens that induce the production of antibodies, which can cross-react with oral mucosal keratinocytes. Barile et al. proposed Streptococcus sanguinis 2A as a causative agent of RAS (2), where the unstable L-form of this bacterium would convert to a transient pathogenic form responsible for disease progression while the stable L-form would be inactive. This could explain the recurrent nature of aphthous stomatitis. Other streptococcal species, such as Streptococcus mitis (4) and Streptococcus oralis (5), have been suspected to provoke development of RAS. Furthermore, a 65-kDa heat shock protein released by various streptococcal strains was shown to cross-react with peptides in the oral epithelium, suggesting that streptococci can trigger lesions based on an autoimmune reaction (6). Helicobacter pylori has also been found in RAS lesions (7). H. pylori causes duodenal ulcers with clinical features similar to those observed in RAS. However, the pathogenic roles of these different bacteria in RAS have not been resolved to date, and it remains to be proven that this condition has a microbiological aetiology (8).
The microbial communities in the oral cavity form a complex commensal microbiota. Disruption of the gut microbiota has been implicated in the development of gut mucosal inflammation in inflammatory bowel diseases (9, 10). To date, the contribution of the normal oral microbiota in RAS has not been investigated.
Traditionally, culture-based methods have been used to study bacterial communities. More recently, molecular-based methods have enabled the analysis of non-cultivable bacteria. Terminal-Restriction Fragment Length Polymorphism (T-RFLP) is a technique that is based on variations in the 16S rRNA gene, as revealed by restriction enzyme digestion patterns (11). The method provides a culturing-independent, rapid, sensitive and reproducible fingerprint of complex microbial communities.
The aim of this study was to characterise the oral microbiota of patients with RAS using T-RFLP. In contrast to previous studies, in which the microbiota was sampled in the ulcerative lesion (12, 13) we focused on the microbiota of the non-inflamed buccal mucosa in patients with RAS. Patients with or without ulcerations at the time of sampling were included, to determine whether the microbiota changes in relation to disease activity. The present study is the first to investigate if the profile of the oral microbiota in patients with RAS is different compared to healthy control subjects.
Subjects and methods
Study population
In this case-control study, 60 patients with RAS were enrolled, together with 60 healthy, age-and gender-matched controls. All the patients were selected from the referral population at the Clinic of Oral Medicine, Public Dental Health, Gothenburg, the Region Västra Götaland of Sweden, while the control subjects were recruited from local Public Dental Health clinics across the Region, in the period 2010–2012. Most of the patients visited the clinic regularly for at least six months for check-ups. As all of the control subjects visited their local Public Dental Health clinics for annual examinations, all of the subjects in the study were fully characterised. Patients and controls were not related or shared a common household.
The patients were selected randomly in that all individuals who fulfilled the inclusion criteria were offered to participate; the episodes of aphthous ulcers should occur at least once a month. Exclusion criteria were use of antibiotics or antibacterial mouth rinses during the previous month or drugs for the treatment of ulcers in the previous six months. Further exclusion criteria were other types of oral mucosal lesions, smoking or excessive consumption of alcohol (alcohol intake more than three times weekly). Of the 60 patients with RAS, 24 used some form of medication regularly, for other conditions than their mouth ulcers. Characteristics of the patients are listed in Table 1 and Supplementary Table 1. Overall dental health was not examined in detail, although no patient suffered from any severe caries or periodontal disease. All patients were categorised as ‘minor RAS’ or ‘major RAS’ (N=30 per group), based on whether the lesion had a diameter of<10 mm (minor RAS) or ≥10 mm (major RAS), in accordance with the standard nomenclature. The control subjects did not suffer from any systemic or oral disease, were not taking any medication, had not used antibacterial mouth rinses or antibiotics in the month prior to sampling, did not suffer from allergies, did not smoke, and consumed alcohol in moderation (alcohol intake of maximum three times weekly) or not at all. All the clinical data concerning the patients were recorded as single data entries in the web-based Medview programme (14).
Table 1.
Clinical characteristics of patients with recurrent aphthous stomatitis (RAS) and healthy gender- and age-matched controls
| Characteristic | RAS patients (N=60) | Controls (N=60) | ||
|---|---|---|---|---|
| Age (years) | ||||
| Median age | 23 | 26 | ||
| Range | 6–68 | 7–70 | ||
| N | % | N | % | |
| Gender | ||||
| Females | 36 | 60 | 36 | 60 |
| Lesion size | ||||
| Minor RASa | 30 | 50 | 0 | 0 |
| Major RASb | 30 | 50 | 0 | 0 |
| Lesion at sampling | ||||
| Patients with lesion at samplingc | 42 | 70 | 0 | 0 |
| Minor RAS patients with lesion at sampling | 25 | 83 | 0 | 0 |
| Major RAS patients with lesion at sampling | 17 | 57 | 0 | 0 |
| Medication at sampling | ||||
| Patients on medication at samplingd | 24 | 40 | 0 | 0 |
Lesion diameter<10 mm.
Lesion diameter ≥10 mm.
RAS exhibits a recurrent pathology and some patients presented with a lesion at the time of sampling.
Includes all types of medication, apart from those used to treat RAS.
The guidelines of the World Medical Association Declaration of Helsinki were followed. The Ethical Review Board in Gothenburg approved the study, and written informed consent was obtained from all the patients and control subjects (Dr no: 386-10).
Sampling procedures
Sampling of the buccal mucosa was carried out with a sterile swab (Isohelix/Cell Projects Ltd, Kent, UK). Sampling was performed when the patients had the possibility to visit the clinic. For this reason, some patients had active ulcers at the time of sampling whereas others did not. Thus, one or several aphthous ulcers were present in the oral mucosa in 42 cases (25 minor RAS and 17 major RAS) while 12 patients had no ulcers at sampling (five minor RAS and seven major RAS). For six of the patients in the major RAS group, it was not clear from their oral medical journals whether or not they presented with an ulcer upon sampling. The samples were taken from the buccal mucosa at a location distant from the ulcers. The participants were requested not to have brushed their teeth or consumed any food or beverage within an hour before sampling. Two dentists participated in the collection of samples using a standardised technique, according to the protocol of the manufacturer (Isohelix/Cell Projects Ltd, Kent, UK).
T-RFLP analysis
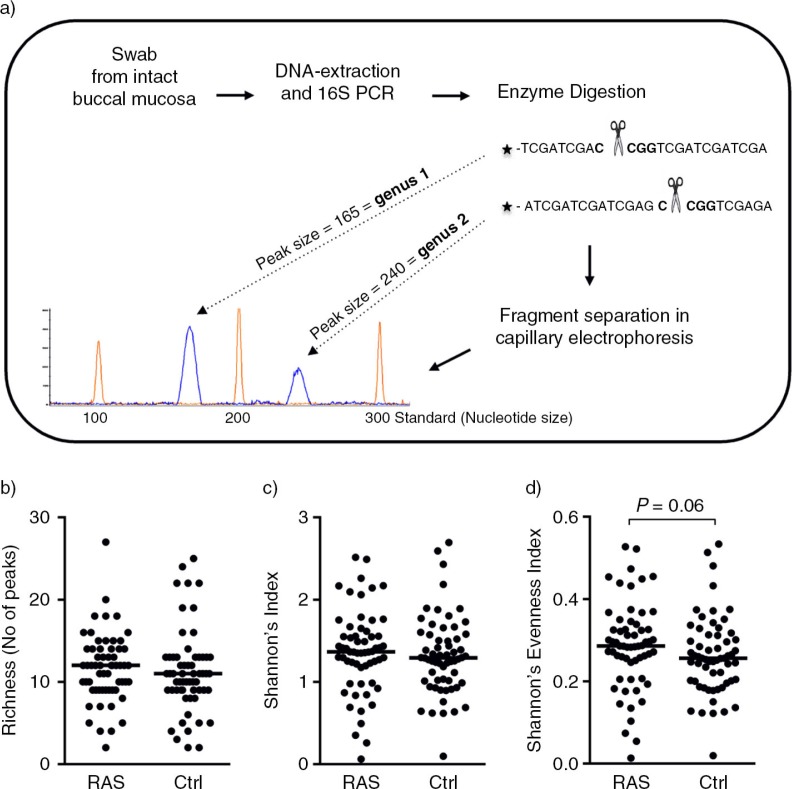
This method is widely used to study bacterial communities in complex environments, such as the oral cavity, using variations in 16S rRNA genes (Fig. 1a). The 16S rRNA gene contains sequences that are highly conserved among bacteria, and variable regions that differ in sequence between bacterial species. In T-RFLP, the 16S rRNA gene is amplified and the terminal fragment fluorescently labelled during the polymerase chain reaction (PCR) reaction, thereby facilitating its detection. The PCR products are then digested with a specific restriction enzyme, and the fragments are detected. Each fragment generates an individual peak of a certain size, a T-RF, which represents a specific bacterial taxon, while the pattern of peaks reveals the bacterial profile of an individual. Generally, T-RFLP permits differentiation to the genus level and sometimes to the species level (15). At least eight streptococcal species can be distinguished using T-RFLP.
Fig. 1.
No difference in bacterial diversity between patients with recurrent aphthous stomatitis (RAS) and control subjects. (a) The Terminal-Restriction Fragment Length Polymorphism (T-RFLP) method is based on variations in 16S rRNA genes within bacterial populations. Bacterial DNA was prepared from swabs taken from the intact buccal mucosa of the study subjects and used as template for PCR amplification of 16S rRNA genes using universal primers. The PCR products were digested with a restriction enzyme that yields a fragment fingerprint representative of a specific bacterial species, and the cleaved PCR products were detected with a fragment analyser. The size of each fragment/height of each peak is representative of an individual bacterial genus/species. Richness, calculated as (b) the number of peaks in each individual or (c) by Shannon's Index. (d) Evenness, calculated by Shannon's Evenness Index. Horizontal bars indicate median values and each symbol represents a single individual. The data was analysed with a Mann-Whitney U-test.
DNA extraction
Buccal swabs were stored at room temperature after sampling. Thereafter bacterial DNA extraction was performed by LGC Genomics, Germany, using the sbeadex® Forensic Kit for the preparation of nucleic acids (LGC Ltd, Middlesex, UK).
Amplification
The T-RFLP analysis was set-up and performed as described previously (15) (Fig. 1a). In brief, bacterial 16S rRNA genes were amplified using the universal primers ENV 1 (5′-6-FAM-AGA GTT TGA TII TGG CTC AG-3′ (for Escherichia coli nucleotides (nt) 8-27) and ENV 2 (5′-CGG ITA CCT TGT TAC GAC TT-3′ (for E. coli nt 1511-1492) (16), with the forward primer being fluorescently labelled with 6-FAM at the 5′ -end. The PCR mixture contained 100 ng DNA, 25 µl Hot Start Taq Master Mix (2.5 U Taq DNA polymerase, 1.5 mM MgCl2, 200 µM dNTP) (Qiagen, Hilden, Germany), 0.3 µM primer, 1 mM MgCl2, and H2O to a final volume of 50 µl for each buccal swab sample. The PCR reaction was performed in an Eppendorf Mastercycler Gradient (Eppendorf, Hamburg, Germany) using the following steps: initial activation of Taq polymerase at 95°C for 15 min; 25 cycles of 94°C for 1 min; 50°C for 45 s; 72°C for 2 min; and a final extension step at 72°C for 7 min. Negative controls that contained all the PCR reagents were included throughout the analysis, to detect potential DNA contamination. Each buccal swab sample was analysed in three independent PCRs and then the triplicate PCR products were pooled.
Restriction enzyme digestion and fragment analysis
PCR products were purified using the QIAquick PCR Purification kit (Qiagen, Hilden, Germany). For each sample, 100 ng of the 16S amplicon were digested with 16 U MspI (New England Biolabs, Ipswich, replace with state) in a final volume of 5 µl at 37°C for 5 hours. The reaction was stopped by incubating the samples at 65°C for 20 min. The digested, fluorescently labelled fragments were detected using the ABI PRISM 310 genetic analyser (Applied Biosystems, Carlsbad, CA) with an injection time of 5 s and separation for 50 min at 15 kV in a Performance Optimized Polymer (Pop) 4 gel. A mixture of 1 µl of digested 16S amplicon, 9 µl of formamide, and 0.5 µl of GeneScan LIZ 1200 size standard (Applied Biosystems) was denatured at 95°C for 3 min and then placed immediately on ice. Each sample was analysed three times. The fragment lengths were determined with the GeneMapper ver. 4 software (Applied Biosystems) using the Local Southern Method.
Identification of bacterial taxa (genus/species level) associated with RAS
The In silico software (http://insilico.ehu.es/) was used for the identification of peaks that were previously correlated with RAS (17). This online database was used for simulation of enzymatic digestion, so as to identify the RAS-correlated bacterial taxa from the peaks generated by the T-RFLP, that is, T-RFs. The positions of the cleavage sites and the sizes of the fragments generated by the MspI enzyme from the 16S rRNA genes were determined.
Statistical analysis
The thresholds employed to identify ‘true’ T-RFs from the background ‘noise’ were set statistically within a fragment length in the range of 28–1,000 bp (18). In brief, the data were standardised by dividing the area of each peak by the total peak area of that particular sample. The standard deviation (SD) of the dataset was then computed assuming that the true mean was zero. Peaks with an area greater than the mean plus three SDs were considered true signals, and therefore retained for further analyses. This process was reiterated until no additional ‘true’ peaks were identified. Fragments that differed by no more than±1 bp in different analysis were considered to be identical. The analysis was performed with the Perl (http://www.perl.org/get.html) and R (http://cran.r-project.org/bin/windows/base/old/2.9.2/) software packages. Samples were run in triplicate, and only those fragments that were represented in at least 2/3 runs of the same sample were considered; the peak sizes and areas from the duplicate/triplicate runs were averaged.
Multivariate analysis (SIMCA P+ ver. 13; Umetrics AB, Umeå, Sweden) was used to project associations for the microbial patterns. This was achieved using a Partial Least Squares-discriminant analysis (PLS-DA). Partial Least Squares regression (PLS) is used for pattern recognition between two matrices, for example, X and Y (19); in the present case, X corresponds to the individual T-RFs, and Y denotes the diagnosis (RAS versus controls, major RAS versus minor RAS). The variables are plotted along an orthogonal axis. The importance of each X-variable to the Y-variable is presented as column bars in a corresponding column loading plot. The final PLS-DA loading column plots in the results section are models based on X-variables with Variable Influence of Projection (VIP) values ≥1. These VIP-values can be used to discriminate between important and unimportant predictors for the overall model. X-variables with VIP- values ≥1 were kept while those with lower VIP-values were discarded. The quality of the multivariate analysis was assessed based on the parameters R2Y, that is, how well the variation of the Y-variable is explained by the model, and Q2, that is, how well a variable can be predicted by the model. Univariate data analysis was performed on, according to the SIMCA analysis, relevant X-variables using Fisher's exact test (GraphPad Prism 6 software; GraphPad Inc., San Diego, CA). A P≤0.05 was considered to be statistically significant (*P≤0.05; ** P≤0.01). Adjustment for mass-significances was not performed, since the univariate analysis was based on SIMCA results where only the most important peaks were further analysed.
Diversity within a bacterial community can be described using the concepts of ‘richness’ (the total number of peaks, each peak reflecting a bacterial taxon) and ‘evenness’ (the peak size, reflecting the amount of each taxon). Shannon's Index () was used to estimate the diversity (richness and evenness). This index quantifies the uncertainty regarding the species identity of an individual who is picked at random from the dataset (20). A high value of H represents a diverse and equally distributed bacterial community, whereas a low value characterises a bacterial community with lower levels of diversity. A value of 0 represents a community with just one taxon. Evenness within a group was calculated with Shannon's Evenness Index (E=H/lnS), where E assumes a value between 0 and 1 and where 1 represents complete evenness. S equals the total number of species, and pi corresponds to the proportion of S made up of the ith species in the community. Univariate data analysis was performed using Mann-Whitney U-test (GraphPad Prism 6 software).
Results
Bacterial diversity in the oral microbiota is similar in patients with RAS and control subjects
The richness (the total number of T-RFs reflecting the number of different taxa within the community) and evenness (diversity including a measure of T-RF size, reflecting the relative amount of each different taxon) of the buccal microbiota are indicators of bacterial diversity. The richness of the microbiota of an individual was estimated by counting the number of T-RFs detected for that particular individual. In total, 192 different T-RFs could be identified within the 120 individuals, 156 were identified in the control group and 118 in the RAS group. While 82 species were common to both groups, 74 were found exclusively in the control group and 36 exclusively in the RAS group. The richness showed no differences between the RAS and control groups (median values of 12 and 11 peaks, respectively) (Fig. 1b). The same pattern was observed for diversity estimated with the Shannon's index (Fig. 1c). There was a tendency towards a higher Shannon's Evenness Index in the patients with RAS than in the control subjects (P =0.06) (Fig. 1d), suggesting that variation in population levels between different taxa is lower within the RAS group than the control group.
Differences in oral microbiota compositions between patients with RAS and control subjects
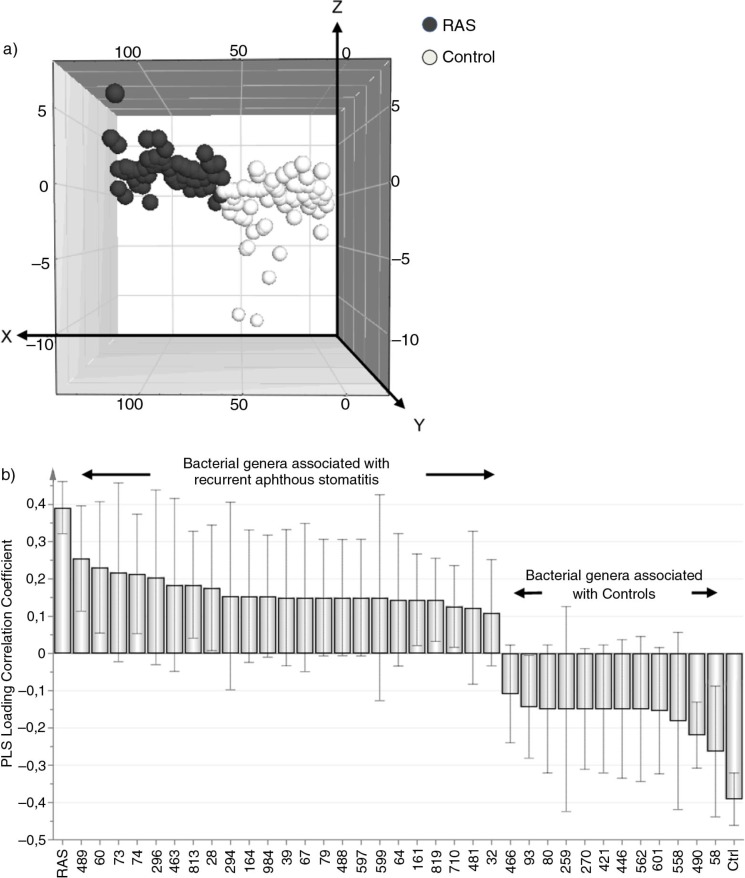
A multivariate analysis was performed to compare the buccal microbiota of the patients with RAS and control subjects using a PLS-DA. The results are illustrated in a 3D-score scatter plot (Fig. 2a). The relationships between the X-variables, that is, the buccal microbiota (identified as T-RFs), and the Y-variable (i.e. the diagnosis of RAS or control subject) are depicted. The patients with RAS were clustered, indicating similarities in their buccal microbiotas, whereas the controls were separate from the patients, indicating a different microbiota profile. The related column-loading plot reveals the T-RFs that contributed to the difference between the two groups (Fig. 2b). Only the X-variables of significance for the model, that is, VIP values ≥1, are projected. These are the X-variables that make the largest contribution to the separation. The X-variables that showed little or no difference between the two groups were assigned VIP-values ≤1 (Supplementary Fig. 1). These variables were therefore excluded, as they did not contribute to the separation of the two groups.
Fig. 2.
Multivariate analysis reveals differences in the oral microbiota between patients with recurrent aphthous stomatitis (RAS) and control subjects. (a) The Partial Least Squares-discriminant analysis (PLS-DA) 3D-score scatter plot depicts the relationship between the X-variables, that is, the buccal microbiota (identified as Terminal-Restriction Fragment Length Polymorphism (T-RFLP) peaks) and the Y-variables, that is, the study population (patients with RAS and control subjects). Each symbol represents the peak pattern of a single individual. Individuals with similar buccal microbiotas cluster together, whereas individuals with different microbiotas are separated. (b) A column-loading plot depicting only the X-variables that are of significance for the model, that is, having Variable Influence of Projection (VIP) values ≥1. X-variables projected on the same side are associated with Y-variables, and X-variables projected on the opposite side are inversely associated with Y. The closer the X-variable is to the Y-variable, the higher the bar and the lower the spread, therefore the stronger the association. R2Y indicates how well the variation of Y is explained, while Q2 indicates how well Y can be predicted. For this model: R2Y=0.33 and Q2=0.21.
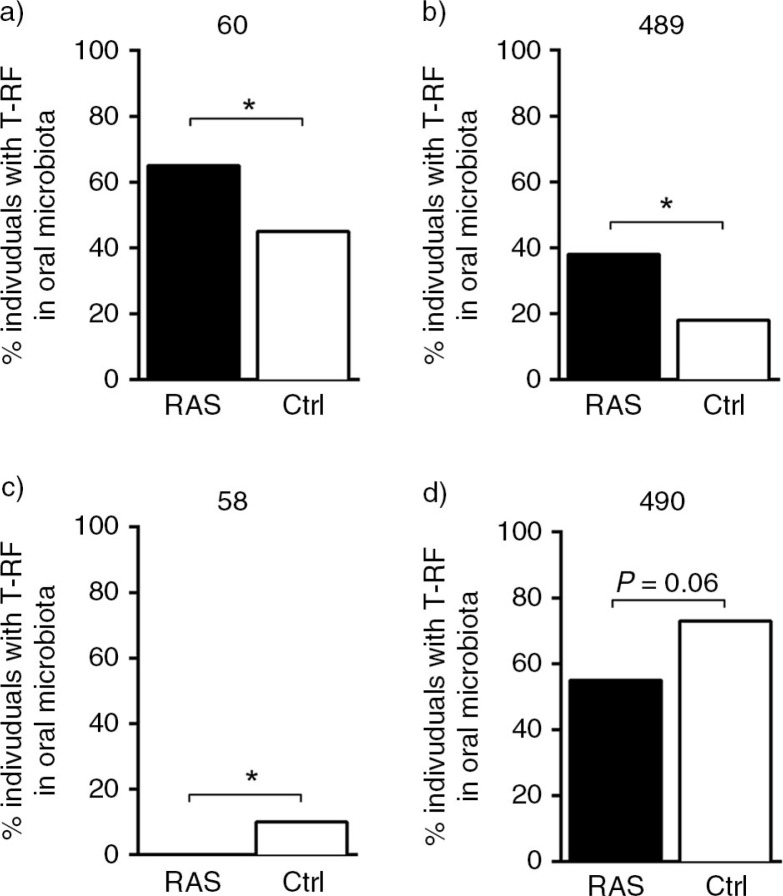
As shown in the column-loading plot (Fig. 2b) several peaks were associated with the patients with RAS and others were associated with the controls. These peaks were selected and subjected to univariate analysis (Fig. 3). Peaks 60 and 489 were more prevalent within the buccal microbiota of the patients with RAS than in the controls (P=0.04 and P=0.03, respectively) (Fig. 3a and b). Whereas peak 58 was associated with the controls, as none of the patients with RAS presented with this peak (P=0.03) (Fig. 3c). A tendency toward a difference was noted for peak 490 (P=0.06) (Fig. 3d), which was more frequently observed in the control subjects than in the patients with RAS.
Fig. 3.
Patients with recurrent aphthous stomatitis (RAS) have an altered prevalence of certain bacterial genus/species in their oral mucosa, as compared to control subjects. (a–d) Four relevant peaks, 58, 60, 489 and 490, were identified in the multivariate analysis as differentiating between patients with RAS and control subjects. These peaks were selected for further statistical analysis with Fisher's exact test (*P≤0.05). Each bar represents the proportion of individuals within the group that have the indicated peak.
Differences in the oral microbiota persist when the patients with RAS are divided into subgroups
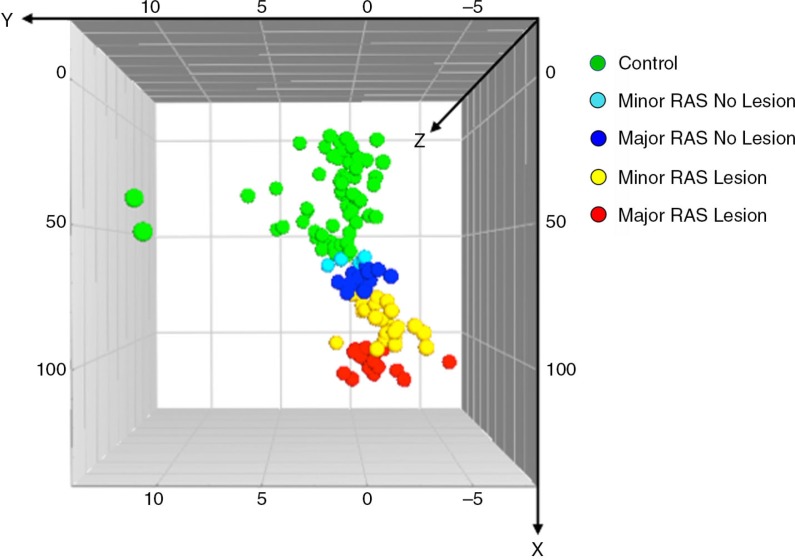
The patients with RAS were categorised according to lesion size (major RAS/minor RAS), presence of lesion when sampling (RAS Lesion/RAS No Lesion), and whether or not they were on a regular medication (RAS Medication/RAS No Medication). This was done to see if the observed differences in the buccal microbiota, between the patients with RAS and controls, were in any way linked to these parameters. For this purpose, we compared the buccal microbiota of the subdivided patients with RAS to controls by a PLS-DA analysis. The results are illustrated in the 3D-score scatter plot (Fig. 4). The patients with RAS clustered together, which indicates similarities in their buccal microbiotas, whereas the controls were separated from the patients, demonstrating a different microbial pattern. The patients who had lesions at the time of sampling clustered furthest away from the controls, indicating that their oral microbiotas differed the most from the controls. The patients with major RAS lesions clustered further away from the controls than the patients with minor lesions. Hence, patients with major RAS that had lesions at time of sampling clustered furthest from the controls, whereas patients with minor RAS that did not have lesions at the time of sampling clustered closest to the controls. Further subdivision, taking into account the usage of medication, was not performed since the number of patients became too low.
Fig. 4.
Multivariate analysis reveals that the presence of lesions during sampling, contributes to the observed differences in oral microbiota between patients with recurrent aphthous stomatitis (RAS) and control subjects. The patients with RAS were divided into subgroups based on: 1) the size of the lesion (minor RAS < 10 mm and major RAS ≥10 mm), and 2) presenting with or not presenting with a lesion at the time of sampling (RAS Lesion/RAS No Lesion). A Partial Least Squares-discriminant analysis (PLS-DA) 3D-score scatter plot depicts the relationship between the X-variables, that is, the buccal microbiota (identified as Terminal-Restriction Fragment Length Polymorphism (T-RFLP) peaks) and the Y-variables, that is, the study population (identified as minor RAS patients with and without lesions, major RAS patients with and without lesions, and control subjects). Each symbol represents the peak pattern of one individual. Individuals with similar buccal microbiotas cluster together, whereas individuals that have different microbiotas are separated. The six major RAS patients for whom it was not known if they presented with a lesion upon sampling or not were excluded from this part of the analysis. R2Y indicates how well the variation of Y is explained, while Q2 indicates how well Y can be predicted. For this model: R2Y = 0.35 and Q2 = 0.03.
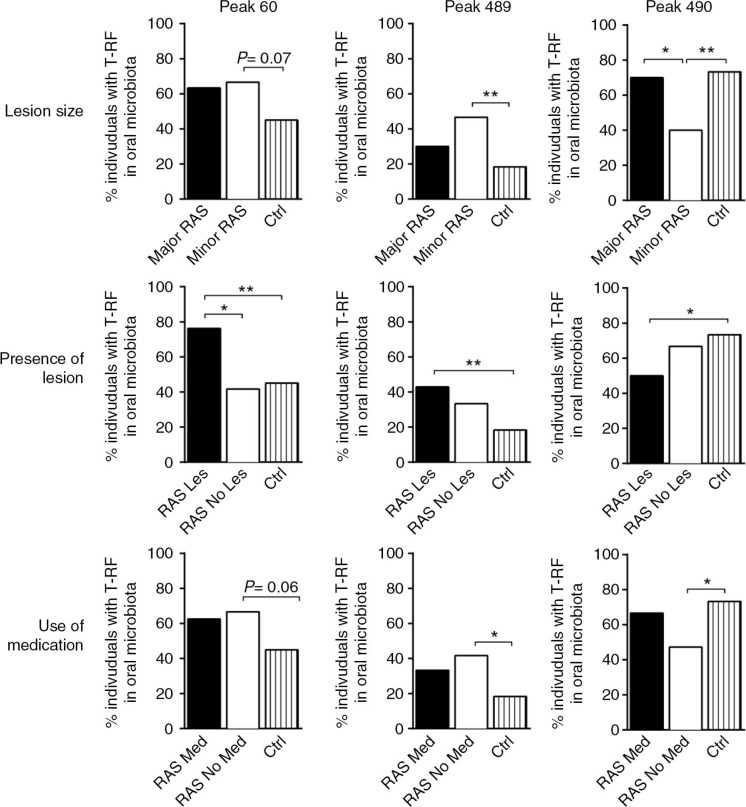
The three T-RFs, present in the patients with RAS that differentiated from the controls (60, 489, and 490; Fig. 3), were studied according to their presence in the different RAS patient subgroups (Fig. 5). Peaks 60 and 489 were both detected more frequently in patients who presented with a lesion upon sampling, as compared with patients who lacked lesions. These two peaks also tended to be present more often in patients with minor RAS lesions and in patients who did not receive medication. In contrast, peak 490 was less prevalent in the patients with RAS who presented with a lesion upon sampling as compared to controls. This peak was also less often found in the patients with RAS who had minor lesions as compared to patients with major RAS lesions or to controls and in patients who did not receive medication compared to controls.
Fig. 5.
The differences in oral microbiota between patients with recurrent aphthous stomatitis (RAS) and control subjects are most prominent in the patients who had lesions during sampling. Peaks 60, 489, and 490 were also found to be of importance in the multivariate analysis when patients with RAS were further divided into the subgroups: minor and major RAS (minor RAS<10 mm and major RAS ≥10 mm), Lesion and No Lesion (RAS Les/RAS No Les) and Medication and No Medication (RAS Med/RAS No Med). They were further analysed statistically with Fisher's exact test (*P≤0.05, * P≤0.01). Each bar represents the proportion of individuals within the group that has the indicated peak.
Identification of RAS-associated bacterial species
T-RFLP does not yield immediate information with regards to the genus/species identity. Such information can be obtained by comparison of observed T-RFs with theoretical sizes acquired by in silico digestion. This method calculates, from known restriction cleavage sites in a particular species, the sizes of the fragment that should be generated. We used this methodology to analyse whether the three species, S. sanguinis, H. pylori and S. oralis were present in the microbiota and, if so, differed in abundance between the patients with RAS and controls as suggested by previous studies (2, 5, 7). H. pylori was calculated to generate a fragment of 270–272 bp, S. oralis gave fragments of 549 bp or 553 bp, and S. sanguinis gave fragments of 545, 546, 548, 555 or 557 bp. T-RFs corresponding to these fragment lengths were not found to be relevant in our model separating patients with RAS from control subjects based on microbiota composition.
Discussion
RAS takes different clinical forms and may be present either on its own or as part of a broad spectrum of systemic disorders (21). Currently, it is unclear if RAS qualifies as a disease per se or is merely a manifestation of different underlying systemic disorders. Thus, RAS may be considered as a pathological reaction pattern of the oral mucosa rather than as a specific disease.
The present study focuses on the commensal oral microbiota as a potential aetiological factor in RAS, by sampling the buccal mucosa. Previous studies have indicated that the oral microbiota in the healthy oral cavity differs from that observed in various oral diseases (22–26). The oral microbiota seems to be more stable both within and between individuals, than microbes colonising other niches within the body (27). Using T-RFLP, we found suggestive evidence of differences in oral microbiota profiles of patients with RAS and healthy age- and gender-matched controls. Certain T-RFs were more prevalent in patients with RAS than controls, while other T-RFs showed the opposite pattern. Differences were also evident when the patients with RAS were further subdivided on the bases of presenting with a lesion upon sampling and the actual size of the lesion. The microbiotas of the patients differed to the greatest extent from those of the controls when a lesion was present at the time of sampling, although the buccal swab was not applied to the lesion itself.
Several different bacterial species have been associated with RAS. Since the study by Barile et al. (2) additional studies have supported a possible relationship between S. sanguinis 2A and this condition (28–30). However, conflicting results have been reported (31, 32) and a direct association has therefore not been confirmed. H. pylori has been found in RAS ulcers and implicated in the aetiology (7, 33). The effect of eradication of this bacterium during the clinical course of RAS has been studied with positive results (34, 35). However, other studies (36–38) have failed to find an association between H. pylori infection and RAS or other oral mucosal lesions, indicating that this bacterium is not to be of aetiological significance for the development of RAS.
The result of this study indicates that either the presence of a lesion alters the microbiota of the entire oral cavity or that a changed microbiota triggers the development of lesions. To resolve this issue, further studies are required in which the patients are followed for a longer period of time to study the microbiota at different intervals both during the presence of lesions and between episodes of lesions. It is also important to acknowledge possible differences in the resident bacterial species within and outside of the lesions. This approach enables to elucidate the extent to which the microbiota of the lesions affect the non-inflamed tissue and vice versa. The multivariate analysis suggests that there are differences in the buccal microbiota between patients with minor and major RAS, that is, different lesion sizes. However, as lesions were more frequently seen in the minor RAS group at the time of sampling, we cannot exclude the possibility that the differences observed between these two groups were simply due to the presence or absence of a lesion. Therefore, a larger study population is required. Nevertheless, we can conclude from the present study that the presence of lesions has greater impact on microbiota composition than the actual size of the ulcer. It is reasonable to assume that certain types of medication affect the prevalence or composition of the oral bacterial community. However, as we did not observe any large differences between the patients on medications and those who had no prescribed medications, we exclude medication as a confounding factor.
A gut microbiota of low diversity during early infancy is known to be a risk factor for the development of allergy (39–41). Therefore, we investigated whether low-level diversity of the buccal microbiota is related to RAS. We found no differences in microbiota richness at the individual level between the patients with RAS and the control subjects. Therefore, a low level of complexity of the oral microbiota does not seem to underlie the aetiology of RAS. However, at the group level, fewer bacterial species were represented in the RAS patient group than in the equally sized control group (118 species versus 156 species). Accordingly, 74 species were found exclusively in the control group, whereas only 36 species were found exclusively in the patients with RAS.
The T-RFLP methodology was chosen because it is an excellent method for fingerprinting the microbiota and revealing overall differences in microbiota pattern. However, a limitation is its inability to identify genus/species that generate a particular T-RF. Databases are needed that allow conversions of each T-RF size to a genus/species. To identify genus composition, additional molecular-based methods are required, e.g. cloning and sequencing of the 16S rRNA genes. However, these methods are labour intensive and not suitable for larger sample sizes. The T-RF size of a known bacterium can be calculated based on knowledge of the specific enzyme digestion site (‘in silico’ simulation). However, this strategy is not suitable for unknown bacteria, since different species of bacteria may generate the same T-RFs of identical or near identical size. In addition, a discrepancy between the actual peak size and the observed experimental peak size has previously been reported (15, 42). High-throughput sequencing, such as pyro-sequencing, has greatly facilitated the sequencing of genomes. However, it is currently expensive to conduct this type of study with a large number of samples. Furthermore, bacterial identification, based on the 16S rRNA gene, remains a challenge, since bacteria with a close phylogenetic relationship cannot be separated, e.g. the various Streptococcus species that dominate the microbiota in the human oral cavity that are highly similar at the 16S rRNA sequence level.
We have assessed the microbiota of the buccal mucosa rather than that of the ulcer itself, which has been the main focus in previous studies. The rationale for this approach was our belief that it is more relevant to study the preconditions for the development of RAS than studying the effect of inflammation in already established ulcerations. Further research using larger cohorts, in which the patients are allocated so as to create more homogenous subgroups, are required to reveal the role of the oral microbiota in this complex condition.
Supplementary Material
Acknowledgements
We thank the Genomics Core Facility of the University of Gothenburg and LGC Genomics, Germany for the DNA extraction and Gunnar Dahlén for valuable discussions. This study was supported by grants from the Swedish National Graduate School in Odontological Science and the Swedish Dental Society.
Footnotes
†These two authors contributed equally to the study.
Conflict of interest and funding
The authors report no conflicts of interest related to this study.
References
- 1.Jurge S, Kuffer R, Scully C, Porter SR. Mucosal disease series. Number VI. Recurrent aphthous stomatitis. Oral Dis. 2006;12:1–21. doi: 10.1111/j.1601-0825.2005.01143.x. [DOI] [PubMed] [Google Scholar]
- 2.Barile MF, Graykowski EA, Driscoll EJ, Riggs DB. L Form of bacteria isolated from recurrent aphthous stomatitis lesions. Oral Surg Oral Med Oral Pathol. 1963;16:1395–402. doi: 10.1016/0030-4220(63)90416-x. [DOI] [PubMed] [Google Scholar]
- 3.Ship II. Inheritance of aphthous ulcers of the mouth. J Dent Res. 1965;44:837–44. doi: 10.1177/00220345650440051501. [DOI] [PubMed] [Google Scholar]
- 4.Hoover CI, Greenspan JS. Immunochemical comparison of cell-wall antigens of various viridans streptococci, including strain 2A2 + 3 hot from recurrent oral aphthous ulceration in man. Arch Oral Biol. 1983;28:917–22. doi: 10.1016/0003-9969(83)90087-0. [DOI] [PubMed] [Google Scholar]
- 5.Narikawa S, Suzuki Y, Takahashi M, Furukawa A, Sakane T, Mizushima Y. Streptococcus oralis previously identified as uncommon Streptococcus sanguis in Behcet's disease. Arch Oral Biol. 1995;40:685–90. doi: 10.1016/0003-9969(95)00042-n. [DOI] [PubMed] [Google Scholar]
- 6.Lehner T, Lavery E, Smith R, van der Zee R, Mizushima Y, Shinnick T. Association between the 65-kilodalton heat shock protein, Streptococcus sanguis, and the corresponding antibodies in Behcet's syndrome. Infect Immun. 1991;59:1434–41. doi: 10.1128/iai.59.4.1434-1441.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leimola-Virtanen R, Happonen RP, Syrjanen S. Cytomegalovirus (CMV) and Helicobacter pylori (HP) found in oral mucosal ulcers. J Oral Pathol Med. 1995;24:14–17. doi: 10.1111/j.1600-0714.1995.tb01123.x. [DOI] [PubMed] [Google Scholar]
- 8.Greenspan JS, Gadol N, Olson JA, Hoover CI, Jacobsen PL, Shillitoe EJ, et al. Lymphocyte function in recurrent aphthous ulceration. J Oral Pathol. 1985;14:592–602. doi: 10.1111/j.1600-0714.1985.tb00536.x. [DOI] [PubMed] [Google Scholar]
- 9.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berry D, Reinisch W. Intestinal microbiota: a source of novel biomarkers in inflammatory bowel diseases? Best Pract Res Clin Gastroenterol. 2013;27:47–58. doi: 10.1016/j.bpg.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Liu WT, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–22. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donatsky O, Justesen T, Lind K, Vestergaard BF. Microorganisms in recurrent aphthous ulcerations. Scand J Dent Res. 1977;85:426–33. doi: 10.1111/j.1600-0722.1977.tb00576.x. [DOI] [PubMed] [Google Scholar]
- 13.Marchini L, Campos MS, Silva AM, Paulino LC, Nobrega FG. Bacterial diversity in aphthous ulcers. Oral Microbiol Immunol. 2007;22:225–31. doi: 10.1111/j.1399-302X.2006.00345.x. [DOI] [PubMed] [Google Scholar]
- 14.Jontell M, Mattsson U, Torgersson O. MedView: an instrument for clinical research and education in oral medicine. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2005;99:55–63. doi: 10.1016/j.tripleo.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Sjoberg F, Nowrouzian F, Rangel I, Hannoun C, Moore E, Adlerberth I, et al. Comparison between Terminal-Restriction Fragment Length Polymorphism (T-RFLP) and quantitative culture for analysis of infants’ gut microbiota. J Microbiol Methods. 2013;94:37–46. doi: 10.1016/j.mimet.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Gutell RR. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures: 1994. Nucleic Acids Res. 1994;22:3502–7. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bikandi J, San Millan R, Rementeria A, Garaizar J. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR and endonuclease restriction. Bioinformatics. 2004;20:798–9. doi: 10.1093/bioinformatics/btg491. [DOI] [PubMed] [Google Scholar]
- 18.Abdo Z, Schuette UM, Bent SJ, Williams CJ, Forney LJ, Joyce P. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ Microbiol. 2006;8:929–38. doi: 10.1111/j.1462-2920.2005.00959.x. [DOI] [PubMed] [Google Scholar]
- 19.Westerhuis JA, van Velzen EJ, Hoefsloot HC, Smilde AK. Multivariate paired data analysis: multilevel PLSDA versus OPLSDA. Metabolomics. 2010;6:119–28. doi: 10.1007/s11306-009-0185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuomisto H. A consistent terminology for quantifying species diversity? Yes, it does exist. Oecologia. 2010;164:853–60. doi: 10.1007/s00442-010-1812-0. [DOI] [PubMed] [Google Scholar]
- 21.Baccaglini L, Lalla RV, Bruce AJ, Sartori-Valinotti JC, Latortue MC, Carrozzo M, et al. Urban legends: recurrent aphthous stomatitis. Oral Dis. 2011;17:755–70. doi: 10.1111/j.1601-0825.2011.01840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakamoto M, Takeuchi Y, Umeda M, Ishikawa I, Benno Y. Application of terminal RFLP analysis to characterize oral bacterial flora in saliva of healthy subjects and patients with periodontitis. J Med Microbiol. 2003;52:79–89. doi: 10.1099/jmm.0.04991-0. [DOI] [PubMed] [Google Scholar]
- 23.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang JG, Kim SH, Ahn TY. Bacterial diversity in the human saliva from different ages. J Microbiol. 2006;44:572–6. [PubMed] [Google Scholar]
- 25.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–43. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69:137–43. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graykowski EA, Barile MF, Lee WB, Stanley HR., Jr. Recurrent aphthous stomatitis. Clinical, therapeutic, histopathologic, and hypersensitivity aspects. JAMA. 1966;196:637–44. doi: 10.1001/jama.196.7.637. [DOI] [PubMed] [Google Scholar]
- 29.Donatsky O, Bendixen G. In vitro demonstration of cellular hypersensitivity to strep 2A in recurrent aphthous stomatitis by means of the leucocyte migration test. Acta Allergol. 1972;27:137–44. doi: 10.1111/j.1398-9995.1972.tb01410.x. [DOI] [PubMed] [Google Scholar]
- 30.Donatsky O, Dabelsteen E. An immunofluorescence study on the humoral immunity to Strep. 2A in recurrent aphthous stomatitis. Acta Pathol Microbiol Scand Section B: Microbiol Immunol. 1974;82:107–12. doi: 10.1111/j.1699-0463.1974.tb02300.x. [DOI] [PubMed] [Google Scholar]
- 31.Oppenheim JJ, Francis TC. The role of delayed hypersensitivity in immunological processes and its relationship to aphthous stomatitis. J Periodontol. 1970;41:205–9. doi: 10.1902/jop.1970.41.4.205. [DOI] [PubMed] [Google Scholar]
- 32.Martin DK, Nelms DC, Mackler BF, Peavy DL. Lymphoproliferative responses induced by streptococcal antigens in recurrent aphthous stomatitis and Behcet's syndrome. Clin Immunol Immunopathol. 1979;13:146–55. doi: 10.1016/0090-1229(79)90058-8. [DOI] [PubMed] [Google Scholar]
- 33.Birek C, Grandhi R, McNeill K, Singer D, Ficarra G, Bowden G. Detection of Helicobacter pylori in oral aphthous ulcers. J Oral Pathol Med. 1999;28:197–203. doi: 10.1111/j.1600-0714.1999.tb02024.x. [DOI] [PubMed] [Google Scholar]
- 34.Albanidou-Farmaki E, Giannoulis L, Markopoulos A, Fotiades S, Aggouridaki X, Farmakis K, et al. Outcome following treatment for Helicobacter pylori in patients with recurrent aphthous stomatitis. Oral Dis. 2005;11:22–6. doi: 10.1111/j.1601-0825.2004.01053.x. [DOI] [PubMed] [Google Scholar]
- 35.Tas DA, Yakar T, Sakalli H, Serin E. Impact of Helicobacter pylori on the clinical course of recurrent aphthous stomatitis. J Oral Pathol Med. 2013;42:89–94. doi: 10.1111/j.1600-0714.2012.01197.x. [DOI] [PubMed] [Google Scholar]
- 36.Porter SR, Barker GR, Scully C, Macfarlane G, Bain L. Serum IgG antibodies to Helicobacter pylori in patients with recurrent aphthous stomatitis and other oral disorders. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1997;83:325–8. doi: 10.1016/s1079-2104(97)90237-7. [DOI] [PubMed] [Google Scholar]
- 37.Mravak-Stipetic M, Gall-Troselj K, Lukac J, Kusic Z, Pavelic K, Pavelic J. Detection of Helicobacter pylori in various oral lesions by nested polymerase chain reaction (PCR) J Oral Pathol Med. 1998;27:1–3. doi: 10.1111/j.1600-0714.1998.tb02081.x. [DOI] [PubMed] [Google Scholar]
- 38.Mansour-Ghanaei F, Asmar M, Bagherzadeh AH, Ekbataninezhad S. Helicobacter pylori infection in oral lesions of patients with recurrent aphthous stomatitis. Med Sci Monit. 2005;11:CR576–9. [PubMed] [Google Scholar]
- 39.Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol. 2008;121:129–34. doi: 10.1016/j.jaci.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 40.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129:434–40, 440. e1–2. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 41.Ismail IH, Oppedisano F, Joseph SJ, Boyle RJ, Licciardi PV, Robins-Browne RM. Reduced gut microbial diversity in early life is associated with later development of eczema but not atopy in high-risk infants. Pediatr Allergy Immunol. 2012;23:674–81. doi: 10.1111/j.1399-3038.2012.01328.x. [DOI] [PubMed] [Google Scholar]
- 42.Pandey J, Ganesan K, Jain RK. Variations in T-RFLP profiles with differing chemistries of fluorescent dyes used for labeling the PCR primers. J Microbiol Methods. 2007;68:633–8. doi: 10.1016/j.mimet.2006.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.