Abstract
To study the effect of melanogenesis on HIF-1α expression and attendant pathways, we used stable human and hamster melanoma cell lines in which the amelanotic vs melanotic phenotypes are dependent upon the concentration of melanogenesis precursors in the culture media. The induction of melanin pigmentation led to significant up-regulation of HIF-1α, but not HIF-2α, protein in melanized cells for both lines. Similar upregulation of nuclear HIF-1α was observed in excisions of advanced melanotic vs. amelanotic melanomas. In cultured cells, melanogenesis also significantly stimulated expression of classical HIF-1-dependent target genes involved in angiogenesis and cellular metabolism, including glucose metabolism and stimulation of activity of key enzymes in the glycolytic pathway. Several other stress related genes containing putative HRE consensus sites were also upregulated by melanogenesis, concurrently with modulation of expression of HIF-1-independent genes encoding for steroidogenic emzymes, cytokines and growth factors. Immunohistochemical studies using a large panel of pigmented lesions revealed that higher levels of HIF-1α and GLUT-1 were detected in advanced melanomas in comparison to melanocytic nevi or thin melanomas localized to the skin. However, the effects on overall or disease free survival in melanoma patients were modest or absent for GLUT-1 or for HIF-1α, respectively. In conclusion, induction of the melanogenic pathway leads to robust upregulation of HIF-1-dependent and independent pathways in cultured melanoma cells, suggesting a key role for melanogenesis in regulation of cellular metabolism.
Keywords: melanoma, melanogenesis, HIF-1, metabolism, glycolysis, melanoma progression
Introduction
Cutaneous melanoma is the most rapidly increasing malignancy in Caucasian populations, and causes a high mortality rate due to onset of therapeutic resistance in stages III and IV of the disease [1-3]. Recently, multiple strategies have been proposed or implemented that target the hallmark features of cancer progression [4], with an expectation of finding a cure for this devastating disease [1-3, 5-8]. Unfortunately, efficacies of these modalities last only a few months before the disease re-emerges, leading to death of the patients. Overall, most therapies lead only to modest improvements in overall survival (OS) and are accompanied by toxic side effects and very high costs of treatment [2, 5, 7, 9-11]. Therefore, there is an urgent need for additional research in order to better define the molecular mechanisms that underline the ability of melanomas to re-wire metabolic and homeostatic responses that are permissive to development of resistance to targeted therapies [11]. Melanoma behavior is driven by several growth factors and cytokines that interact with their corresponding receptors [1, 7, 11], as well as various hormones, neuropeptides and neurotransmitters [12, 13], These include proopiomelanocortin (POMC)-derived peptides ([14, 15] corticotropin releasing hormone (CRH) [16], steroids [17], biogenic amines and melatonin [18, 19].
Normal and malignant melanocytes are armed with an unique metabolic pathway that begins with tyrosinase mediated oxidation of L-tyrosine to L-DOPA [20, 21], followed by a tightly regulated series oxidoreduction reactions that involve production of several intermediates, including toxic compounds such as quinones, semiquinones and reactive oxygen species (ROS) [21-26]. The melanin synthesis apparatus serves not only as a differentiation marker but also, and more importantly, protects normal and malignant melanocytes and epidermal keratinocytes containing melanin from a myriad of physical and chemical insults [12, 25]. These latter properties can lead to melanotic melanomas that are relatively resistant to chemo-, radio- and phototherapy [12, 25, 27, 28]. Melanogenesis also influences the behavior of normal and malignant melanocytes and their surrounding microenvironment [26, 27, 29] through the action of intermediates of melanogenesis [30-32], oxygen consumption and stimulation of aerobic glycolysis [26, 33] or interaction with other metabolic pathways [12, 34-36].
There is an abundance of evidence that malignant neoplasms rely on aerobic glycolysis, known as the “Warburg effect” [37] to provide energy to the tumor [4, 38-41]. Changes in cellular metabolism, including stimulation of glycolysis, play a central role in tumor development, progression and therapeutic resistance. In fact, targeting dysregulated tumor metabolism has emerged as a promising approach to prevent drug resistance [42, 43]. Key master regulators of cellular metabolism and therapeutic resistance are the Hypoxia-Inducible Factors (HIF)-1α and HIF-2α, oxygen-responsive, basic helix-loop-helix-PAS domain proteins expressed in all metazoan organisms [44]. In partnership with aryl hydrocarbon nuclear receptor translocator (ARNT), HIF-1α or HIF-2α, from the HIF-1 or HIF-2 transcription factors, respectively, which regulate transcription of hundreds of direct gene targets by binding to hypoxic response elements (HREs) in gene regulatory regions [45]. Protein stability of the HIFα subunits is regulated by hydroxylation of two key proline residues in the C-terminus by a family of prolyl hydroxylase (PHD) enzymes. Hydroxylation of the oxygen-dependent degradation (ODD) domain of the HIFα subunits promotes direct interaction with the von Hippel-Lindau protein (VHL), which functions as an E3 ubiquitin ligase, leading to HIFα destruction by the 26S proteasome [38, 39, 44]. HIF-1 and HIF-2 regulate gene transcription in a cell type-specific and microenvironment context dependent manner [46-49]. Some of the classic HIF target genes include multiple enzymes in the glycolytic pathway, such as lactate dehydrogenase A (LDH-A), pyruvate dehydrogenase kinase, isozyme 1 (PDK1), aldolase A, fructose-bisphosphate (ALDOA), hexokinase 2 (HK2), BNIP3 (BCL2/adenovirus E1B 19kDa interacting protein 3), sodium hydrogen exchanger (NHEI), monocarboxylate transporter 4 (MCT4), glucose transporters 1 and 4 (GLUT-1 and GLUT-4) and angiogenic factors, including vascular endothelial growth factor A (VEGF), growth factors and transferrin receptor 1 [38, 50-54]. HIF-1α is over-expressed in several types of human cancers, including melanoma [55, 56] and direct impacts several hallmarks of malignancy [39].
Since melanogenesis has the potential to regulate tumor behavior [26, 27], we have tested the effects of stimulation of melanogenesis on the expression of HIF-1α and HIF-2α proteins, the genes regulated downstream of the HIF transcription factors, as well as the activity of select enzymes in the glycolytic pathway. For these studies, we used human melanoma and hamster melanoma lines whose amelanotic versus melanotic phenotypes are regulated by the concentration of melanin precursors added to culture medium, as previously described [30, 32, 57, 58]. These studies were complemented by pathological analysis of clinical samples to examine the relationships between HIF-1α or GLUT-1 expression and the progression of melanocytic lesions and the clinical outcome of metastatic disease.
Materials and Methods
Patients
We analyzed 106 samples obtained from 84 patients treated in the Oncology Centre - Prof. Franciszek Łukaszczyk Memorial Hospital, Bydgoszcz, Poland. Samples of normal skin (n=5) were obtained from patients who underwent surgery not related to skin disorders. The characteristics of the patients and pigmented lesions are presented in Table 1. This study was approved by the Committee of Ethics of Scientific Research of Collegium Medicum of Nicolaus Copernicus University, Poland.
Table 1.
Sequences of primers used for qPCR
| Genes | Description | Primers |
|---|---|---|
| CYCB | cyclophillin B | L 5′tgtggtgtttggcaaagttc-3′ R 5′-gtttatcccggctgtctgtc-3′ |
| β-actin | β-actin | L 5′-ccaaccgcgagaagatga-3′ R 5′-ccagaggcgtacagggatag-3′ |
| ALDOA | aldolase A, fructose-bisphosphate | L 5′- cgggaagaaggagaacctg-3′ R 5′-gaccgctcggagtgtacttt-3′ |
| PDK1 | pyruvate dehydrogenase kinase, isozyme 1 | L 5′-ccgctctccatgaagcagtt-3′ R 5′-ttgccgcagaaacataaatgag-3′ |
| HK2 | hexokinase 2 | L 5′-caaagtgacagtgggtgtgg-3′ R 5′-gccaggtccttcactgtctc-3′ |
| MCT2 | monocarboxylate transporter 2 | L 5′-atcctgggcttcattgacat-3′ R 5′-atcctgggcttcattgacat-3′ |
| MCT4 | monocarboxylate transporter 4 | L 5’atcctgggcttcattgacat-3’ R 5’-atggagaagctgaagaggta-3’ |
| GLUT1 | glucose transporter 1 | L 5′-ccattggctccggtatcgt-3′ R 5′-tgctcgctccaccacaaac-3′ |
| NHEI | sodium hydrogen exchanger | L 5′-tctgccgtctccactgtctcca-3′ R 5′-cccttcagctcctcattcacca-3′ |
| LDHA | lactate dehydrogenase A | L 5′-acccagtttccaccatgatt-3′ L 5′-cccaaaatgcaaggaacact-3′ |
| VEGFA | vascular endothelial growth factor A | L 5′-tggaattggattcgccattt-3′ R 5′-tgggtgggtgtgtctacagga-3′ |
| TYR | tyrosinase | L 5′-ggctgttttgtactgcctgct-3′ R 5′-aggagacacaggctctagggaa-3′ |
| Cyp11A1, exon 4 | cytochrome P450, family 11, subfamily A, polypeptide 1, exon 4 | L 5′ ccagacctgttccgtctgtt-3′ R 5′ aaaatcacgtcccatgcag-3′ |
| Cyp11B1 | cytochrome P450, family 11, subfamily B, polypeptide 1 | L 5′-aggtggacagcctgcatc-3′ R 5′-ccattcaggcccattcag-3′ |
| Cyp17A1 | cytochrome P450, family 17, subfamily A, polypeptide 1 | L 5′-gcatcatagacaacctgagcaa-3′ R 5′-gggttttgttggggaaaatc-3′ |
| Cyp21A2v2 | cytochrome P450, family 21, subfamily A, polypeptide 2,variant2 | L 5′-gcttggcctgactcagaaat-3′ R 5′-gacaccagcttgtcttgcag-3′ |
| HSD11B1 | hydroxysteroid (11-beta) dehydrogenase 1 | L 5′-caatggaagcattgttgtcg-3′ R 5′-ggcagcaaccattggataag-3′ |
| HSD11B2 | hydroxysteroid (11-beta) dehydrogenase 2 | L 5′-gtcaaggtcagcatcatcca-3′ R 5′-cactgacccacgtttctcac-3′ |
| 7DHCR | 7-dehydrocholesterol reductase | L 5′-ttgtaaaagaaattgcctgtgaat-3′ R 5′-gccatggtcaagggctac-3′ |
| 3βHSD | 3β-hydroxysteroid dehydrogenase | L 5′-cttggacaaggccttcagac-3′ R 5′-tcaagtacagtcagcttggtcct-3′ |
| CRH | corticotropin releasing hormone | L 5′-ctccgggaagtcttggaaat-3′ R 5′-gttgctgtgagcttgctgtg-3′ |
| CRH, exon2 | corticotropin releasing hormone, exon 2 | L 5′-actcagagaccaagtc ca-3′ R cttcccaggcgcttcgcaggt-3′ |
| CRHR1 | corticotropin releasing hormone receptor 1 | L 5′-tggatgttcatctgcattgg-3′ R 5′-tgccaaaccagcacttctc-3′ |
| POMC, exon3 | proopiomelanocortin, exon 3 | L 5′-_agcctcagcctgcctggaa-3′ R 5′- cagcaggttgctttccgtggtg-3′ |
| MC1 | melanocortin receptor 1 | L 5′-actccgtctgctccaatgac-3′ R 5′-agagctggcagcaaagatg-3′ |
| MC2 | melanocortin receptor 2 | L 5′-tcttcagcctgtctgtgattg-3′ R 5′-ggcacaggatgaagaccag-5′ |
| PCSK1 | proprotein convertase subtilisin/kexin type 1 | L 5′-tgatcccacaaacgagaaca-3′ R 5′-tgtgattatttgcttgcatgg-3′ |
| PCSK2 | proprotein convertase subtilisin/kexin type 2 | L 5′-agcatacaactccaaggttgc-3′ R 5′-aggcctcgatgatgtctgtc-3′ |
| NQO2 | NAD(P)H dehydrogenase, quinone2 | L 5′-ttgacatcccaggattctacg-3′ R 5′-ccgtggttacggaaaggag-3′ |
| EGFR | epidermal growth factor receptor | L 5′-acacagaatctatacccaccagagt-3′ R 5′-atcaactcccaaacggtcac -3′ |
| ICAM1 | intercellular adhesion molecule 1 | L 5′-ccttcctcaccgtgtactgg-3′ R 5′-agcgtagggtaaggttcttgc-3′ |
| IFNα1 | interferon α1 | L 5′-ccctctctttatcaacaaacttgc-3′ L 5′-ttgttttcatgttggaccaga-3′ |
| IFNβ1 | interferon β1 | L 5′-gatgagtacaaaagtcctgatcca-3′ R 5′-ctgcagccactggttctgt-3′ |
| IL1α | interleukin 1α | L 5′-ggttgagtttaagccaatcca-3′ R 5′-tgctgacctaggcttgatga-3′ |
| IL1β | interleukin 1β | L 5′-crgtcctgcgtgttgaaaga-3′ R 5′-ttgggtaatttttgggatctaca-3′ |
| IL2 | interleukin 1 | L 5′-aagttttacatgcccaagaagg-3′ R 5′-aagtgaaagtttttgctttgagcta-3′ |
| IL4 | interleukin 3 | L 5′-caccgagttgaccgtaacag-3′ R 5′-gccctgcagaaggtttcc-3′ |
| IL5 | interleukin 5 | L 5′-ggtttgttgcagccaaagat-3′ R 5′-tcttggccctcattctcact-3′ |
| IL6 (IFNβ2) | interleukin 6 | L 5′-gaagctctatctcgcctcca-3′ R 5′-agcaggcaacaccaggag-3′ |
| IL8 (CXCL-8) | interleukin 8, chemokine ligand 8 | L 5′-agacagcagagcacacaagc-3′ R 5′-atggttccttccggtggt-3′ |
| IL10 | interleukin 10 | L 5′-tgggggagaacctgaagac-3′ R 5′-ccttgctcttgttttcacagg-3′ |
| IL17A | interleukin 17A | L 5′-tgggaagacctcattggtgt-3′ R 5′-ggatttcgtgggattgtgat-3′ |
| TNFα | tumor necrosis factor alpha | L 5′-cagcctcttctccttcctgat-3′ R 5′-gccagagggctgattagaga-3′ |
| IFNγ | interferon gamma | L 5′-ggcattttgaagaattggaaag-3′ R 5′-tttggatgctctggtcatctt-3′ |
| PDGFA | platelet-derived growth factor alpha polypeptide | L 5′-gcagtcagatccacagcatc-3′ R 5′-tccaaagaatcctcactcccta-3′ |
| TGFA | transforming growth factor, alpha | L 5′-ttgctgccactcagaaacag-3′ R 5′-atctgccacagtccacctg-3′ |
| TGFB1 | transforming growth factor, beta 1 | L 5′-gcagcacgtggagctgta-3′ R 5′-cagccggttgctgaggta-3′ |
| TGFB2 | transforming growth factor, beta 2 | L 5′-ccaaagggtacaatgccaac-3′ R 5′-cagatgcttctggatttatggtatt-3′ |
| RANTES | CCL5 chemokine (C-C motif) ligand 5 | L 5′-tgcccacatcaaggagtattt-3′ R 5′-tttcgggtgacaaagacga-3′ |
| TRANSFERRIN RC | transferrin receptor | L 5′-ttgagaaaacaatgcaaaatgtg-3′ R 5′-cccagttgctgtcctgatataga-3′ |
| NPL | N-acetylneuraminate pyruvate lyase | L 5′-ccagagatttatcaactttgttgtca-3′ R 5′-gaggctttctgcagtggaag-3′ |
Cell culture
Human SKMEL-188 [59] and AbC1 hamster [57] melanoma cells were cultured in Ham's F10 supplemented with 5% fetal bovine serum (FBS) and 1% antibiotics (penicillin/streptomycin/amphotericin, Sigma-Aldrich, St. Louis, MO) at 37°C in 5% CO2 and the media were changed every second day. Although Ham's F-10 contains a low concentration of melanin precursor (10 μM of L-tyrosine), cells cultured in this medium remained amelanotic [30, 57]. To induce production of melanin pigment, the medium was changed to Dulbecco's Modified Eagle's Medium (DMEM) containing 400 μM of L-tyrosine or Ham's F-10 plus 400 μM of L-tyrosine and cells were cultured for 2, 3 or 6 days and collected as described previously [30, 57]. Non-pigmented cells were grown in parallel in Ham's F10 medium [30, 57] and processed using the same procedures as the pigmented cells.
Western Blot analyses
The expression of HIF-1α and HIF-2α was compared in non-pigmented and pigmented melanoma cells, with Caki cells as a positive control for HIF-2α [60], and Hif1a wild type (WT) and knockout (KO) mammary tumor epithelial cells (MTECs) derived from the polyoma virus middle T transgenic mouse (MMTV-PyMT) [61] as controls for HIF-1α. Cells were grown to 80% confluence and harvested after culture for 6 hours at either normoxia (ambient air; 5% CO2) or hypoxia (0.5% O2, 5% CO2). Cell extracts were prepared using a modified RIPA buffer to produce a whole cell extract (WCE) comprised of cytoplasmic and nuclear proteins. Insoluble material remaining after preparation of WCE was then re-extracted in high-salt (HS) (400 mM NaCl) buffer containing the deubiquitinase inhibitor N-ethylmaleimide (NEM, 0.5 μM) to enrich for nuclear proteins. HS-WCE was resolved on 3% to 8% Tris-acetate gels (10 μg/lane; Invitrogen) and transferred onto polyvinylidene fluoride (PVDF) membrane. Membranes were blocked in 5% milk prior to addition of anti-HIF1A (1:5,000, Novus Biologicals, NB100-479) or anti-HIF2A (1:2,000, Novus Biologicals, NB100-122) primary antibodies, which were incubated overnight at 4°C followed by incubation in anti-rabbit igG-HRP (Jackson Laboratories, 1;40,000) for 45 minutes and detection using enhanced chemiluminescence (ECL).
Reverse transcription (RT) and quantitative polymerase chain reaction (qPCR)
RNA was harvested from SKMEL-188 melanoma cells cultured for two or three days in F10 media (non-pigmented) or in DMEM media (pigmented), supplemented with 5% FBS, using the Absolutely RNA Miniprep kit (Stratagen) as previously described [62]. Reverse transcription was performed using the Transcriptor First Strand cDNA Synthesis kit (Roche). Primers were designed using either the Universal Probe Library software (UPL Roche, Germany) or as previously reported in [63-74]. Primer sequences are listed in Table 1. Real-time PCR (qPCR) data were generated from input cDNA using a SYBR Green Master Mix (n=3 technical/replicates/gene) that was amplified using standard settings on a Roche LC480 LightCycler machine. Gene expression (expressed as crossing point values, Cp) was normalized using either cyclophillin B or ß-actin by the ΔΔCp method. Changes in gene expression are presented as a relative quantities based on the mean difference between target gene and reference gene in the cycle of appearance in time ± SD. For each gene, data were compared between pigmented and non-pigmented cells using a Student's t-test. It must be noted that the lower is the number presented in tables or figures the higher is relative gene expression level. Since the Cp value represents the cycle at which signal is first detected above a threshold, increases in the relative abundance of the PCR product detected are reflected by a lower Cp numeric value for each particular gene (PCR product) of interest
Enzyme assays
Enzyme assays were performed using protocols described in Methods in Enzymology (Volume 9) [75-77]. Briefly, the cell pellets were suspended in cold 50 mM potassium phosphate buffer containing 10% glycerol, 10 mM DTT and a protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO). Suspensions were sonicated followed by centrifugation at 12,000g and the supernatants were collected for enzyme assays. Extracts were resuspended in 100 mM potassium phosphate buffer (pH 7.0) containing 1 mM Sodium pyruvate and 0.13 mM NADH for LDH (lactate dehydrogenase) assays, or 20 mM Tris-HCl buffer (pH 7.6) containing 15 mM glucose, 20 mM MgCl2, 0.13 mM NADP, 0.01mM EDTA, 1 mM ATP and 0.2 units of glucose-6-phosphate dehydrogenase (Sigma-Aldrich, St. Louis, MO) for the hexokinase assay. An aldolase assay was performed by resuspending extracts in a buffer containing 50 mM glycylglycine (pH 7.5) containing 0.2 mM NADH, 10 mM FDP and 15 units of glycerophosphate dehydrogenase (Sigma-Aldrich, St Louis, MO). Changes in optical density were measured at room temperature at a wavelength of 340 nm and the units of activity were reported as the changes in optical density per minute per 1 mg protein, as in [75-77]. Total protein content per extract was measured using Coomassie brilliant blue (Sigma-Aldrich, St. Louis, MO).
NAD and NADH ratios
NAD and the NADH ratio were measured using a commercial kit (Sigma-Aldrich, St. Louis, MO) per manufacturer instructions. Briefly, SKMEL-188 cells were grown in Ham's F10 media + 5% FBS and 400 μM L-tyrosine for 6 days or in DMEM/Ham's F10 (75/25) +10% FBS for 2 days to generate pigmented cells. For the non-pigmented cells, L-tyrosine was either omitted, or Ham's F10 + 5% FBS was used. At harvest, cells were washed with PBS and total NAD and NADH were extracted using the buffer provided in the kit, followed by filtration using a 10 kDa cut-off spin filter (Abcam, Cambridge, MA). NAD was decomposed by heating at 60°C for 30 min for detection of NADH only. After the reaction with an enzyme mix and buffer provided in the kit and addition of the NADH developer, the absorbance was measured at 450 nM.
Immunohistochemistry staining and analysis
Immunohistochemistry was performed on 4 μm sections of formalin-fixed paraffin-embedded samples of human melanocytic lesions following protocols described previously [78]. The characteristics of this large panel of pigmented lesions isolated from different patients are listed in Table 2. After antigen retrieval in high pH buffer in a PT-Link device (Dako, Carpinteria, CA), endogenous peroxidase was blocked in H2O2-based blocking reagent (Dako, Carpinteria, CA). For GLUT-1 immunostaining, sections were incubated with a monoclonal anti-GLUT-1 antibody (Abcam, Cambridge, UK) at a dilution of 1:75 for 1 hour. After excessive washing and incubation (30 minutes) with anti-mouse EnVision+System-peroxidase labeled polymer (Dako, Carpinteria, CA), antibody-antigen interactions were visualized using a peroxidase substrate, ImmPACT NovaRED (Vector Laboratories Inc., Burlingame, CA). For HIF1α immunostaining, a TSA (tyramide signal amplification) kit (Life Technologies, Carlsbad, CA) was used per the manufacturer's protocol. A mouse monoclonal anti-HIF1A antibody (Santa Cruz Biotechnology, sc-53546) was applied at a dilution of 1:750 for 1 hour at RT, followed by overnight incubation at 4°C. After washing and incubation for 1 hour with HRP-conjugated goat anti-mouse secondary antibody (1:75, Abcam, Cambridge, UK), antibody complexes were visualized using Vector Red substrate (Vector Laboratories Inc., Burlingame, CA). All sections were counterstained with Harris hematoxylin and mounted in non-aqueous medium (Consul Mount; Thermo Fisher Scientific Inc. Waltham, MA, USA).
Table 2.
Clinical, pathological and morphometry characteristic of patient samples.
| Features | Number of cases |
|---|---|
| Nevi | 26 |
| Junctional | 6 |
| Intradermal | 7 |
| Compound | 8 |
| Dysplastic | 5 |
| Melanomas | 75 |
| Superficial spreading | 36 |
| Nodular | 37 |
| Lentiginous | 2 |
| Age | |
| Nevi | Median 38,2 years (range 20-85) |
| ≤20 years | 2 |
| 21–40 years | 15 |
| 41–60 years | 7 |
| 61–80 years | 1 |
| >80 years | 1 |
| Melanomas | Median 58,0 years (range 25-100) |
| ≤20 years | 0 |
| 21–40 years | 5 |
| 41–60 years | 36 |
| 61–80 years | 28 |
| >80 years | 6 |
| Female/male ratio | |
| Nevi | |
| Female/Male | 18/8 |
| Melanomas | |
| Female/Male | 36/39 |
| pT | |
| pT1 | 21 |
| pT2 | 11 |
| pT3 | 15 |
| pT4 | 28 |
| pN | |
| pN0 | 38 |
| pN1 | 16 |
| pN2 | 11 |
| pN3 | 10 |
| pM | |
| pM0 | 66 |
| pM1 | 9 |
| overall stage | |
| 1 | 22 |
| 2 | 15 |
| 3 | 29 |
| 4 | 9 |
Both HIF-1α and GLUT-1 immunostaining were scored semi-quantitatively as described previously [78, 79]. Briefly, the level of GLUT-1 was calculated based on the portion of positive cells and the mean staining intensity evaluated from 0 to 3 arbitrary units (A.U.) with 0 as negative (0), weak (1), moderate (2) and strong (3). Staining intensity was evaluated with reference to immunostaining of erythrocytes, which scored as strong. The level of HIF-1α was also assessed according to the above formulation. The percentage of HIF-1α-positive cell nuclei per field was also analyzed with reference to clinical pathology morphological data.
Bioinformatics analysis for putative Hypoxic Response Elements (HREs)
The promoter regions of a panel of genes involved in melanogenesis were scanned for the presence of putative HIF binding sites (HREs). The publicly available web server, Transcription Factor Matrix (TFM) Explorer [80] was used for analysis of a total of 2,500 base pairs (bp) of sequence [2000 bp upstream of the transcription start site (TSS) and 500 base pairs after the TSS]. Each gene was scanned using weight matrices obtained from JASPER [81] and TRANSFAC® [82] for the HRE consensus sequence 5’-RCGTG-3’, where R is any purine A or G. A list of genes and their identified putative HRE locations is summarized in Table 3.
Table 3.
Indentification of putative HREs in genes related to stress response, steroidogenesis, immunity and cell growth.
| Gene name or symbol | Accession | HREs | Location relative to TSS | Strand | p-value |
|---|---|---|---|---|---|
| CRH | NM_000756 | 3 | + 260 | forward | <.001 |
| + 280 | forward | <.001 | |||
| + 338 | forward | <.001 | |||
| CRHR1 | NM_004382 | 0 | NA | NA | NA |
| POMC | NM_000939 | 2 | − 248 | reverse | NS |
| + 382 | reverse | NS | |||
| PCSK1 | NM_000439 | 2 | − 1582 | forward | <.005 |
| − 1294 | reverse | NS | |||
| PCSK2 | NM_002594 | 2 | − 1292 | forward | <.05 |
| − 1287 | forward | <.005 | |||
| MC2R | NM_000529 | 2 | − 1530 | forward | <.05 |
| − 829 | reverse | <.05 | |||
| NQO2 | NM_000904 | 0 | NA | NA | NA |
| TGFB1 | 000660 | 2 | − 936 | forward | NS |
| − 133 | reverse | NS | |||
| TGFB2 | NM_003238 | 5 | − 790 | reverse | <.001 |
| − 767 | reverse | <.001 | |||
| − 466 | reverse | <.001 | |||
| − 762 | forward | <.05 | |||
| + 42 | forward | <.05 | |||
| CYP11A1 | NM_000781 | 0 | NA | NA | NA |
| CYP11B1 | NM_000497 | 1 | − 75 | reverse | NS |
| HSD3B1 | NM_000862 | 0 | NA | NA | NA |
| CYP17A1 | NM_000102 | 0 | NA | NA | NA |
| CYP21A2 | NM_000500 | 0 | NA | NA | NA |
| HSD11B1 | NM_005525 | 0 | NA | NA | NA |
| HSD11B2 | NM_000196 | 0 | NA | NA | NA |
| DHCR7 | NM_001360 | 5 | − 901 | forward | <.05 |
| − 760 | forward | <.05 | |||
| − 637 | forward | <.05 | |||
| − 622 | forward | <.005 | |||
| − 251 | forward | <.001 | |||
| IL1A | NM_000575 | 0 | NA | NA | NA |
| IL1B | NM_000576 | 2 | − 1123 | forward | NS |
| − 296 | forward | <.05 | |||
| IL5 | NM_000879 | 0 | NA | NA | NA |
| IL10 | NM_000572 | 0 | NA | NA | NA |
| IL17A | NM_002190 | 0 | NA | NA | NA |
| PDGFA | NM_002607 | 5 | − 1972 | reverse | <.05 |
| − 1549 | reverse | <.05 | |||
| − 1432 | forward | <.05 | |||
| − 1401 | forward | <.005 | |||
| − 574 | forward | <.005 | |||
| TFRC | NM_003234 | 3 | − 1112 | forward | <.05 |
| − 327 | reverse | NS | |||
| − 19 | forward | <.05 | |||
| IFNA1 | NM_024013 | 0 | NA | NA | NA |
| IFNB1 | NM_024013 | 0 | NA | NA | NA |
| TGFA | NM_003236 | 0 | NA | NA | NA |
| EGFR | NM_005228 | 0 | NA | NA | NA |
| CCL5 | NM_002985 | 2 | + 32 | forward | NS |
| + 468 | forward | NS | |||
| NPL | NM_030769 | 0 | NA | NA | NA |
The promoter regions of a panel of genes involved in melanogensis were scanned for the presence of putative HIF binding sites (HREs) as described in the methods. A summary of each genes profile, its accession number at the National Center for Biotechnology Information (NCBI), and the identified putative HRE locations are listed; NA-not applicable (no binding sites identified), NS-not significant.
Statistical analysis
Statistical analysis was performed with Prism 5.0 (GraphPad Software, San Diego, CA). To test the significance of differences between cohorts, either one way ANOVA or Student's t-tests were performed as appropriate. To examine the relationship between two variables, Pearson's correlation was used. Survival was compared using Kaplan-Meier plots and the log-rank test. In all tests, a p-value of <0.05 was considered statistically significant.
Results and Discussion
Medium rich in melanin precursors stimulates melanogenesis in melanoma cells
Medium containing high concentrations of melanin precursors induces rapid melanization of Bomirski hamster amelanotic AbC1 melanoma [57] and SKMEL-188 human melanoma [28, 30]. When cells are switched from Ham's F10 (containing 10 μM of L-tyrosine) to DMEM media (containing 400 μM L-tyrosine), the production of melanin pigment is observed after two days of culture, with an increase in melanization levels observed on day 3 that is accompanied by rounding of heavily melanized cells and their detachment from the culture surface into the media as described previously [57, 83, 84]. In contrast, melanoma cells cultured in Ham's F10 medium (control medium) remain amelanotic and are tightly attached to the culture surface with multipolar morphology during the entire culture period.
Melanogenesis stimulates expression of HIF-1α in melanoma cells
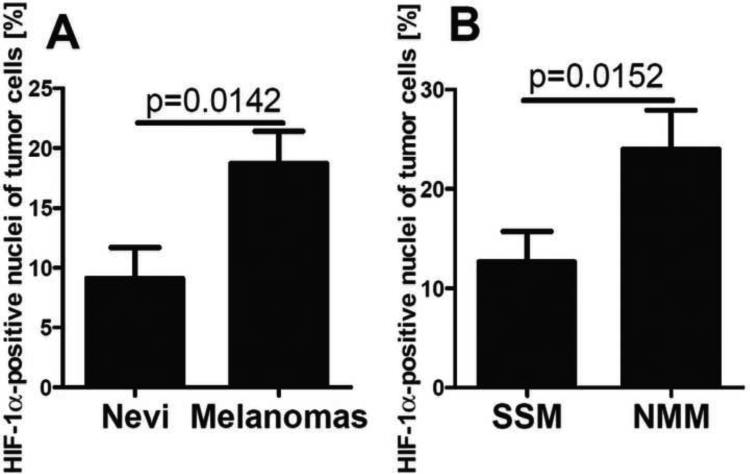
Expression of HIF-1α, but not HIF-2α, protein is dramatically up-regulated in response to induction of melanogenesis in melanoma cells (Fig. 1; data representative of two independent experiments). The specificity of the HIF-1α and HIF-2α bands detected by western blotting was confirmed using both positive (HIF-1 WT MTECs, Caki) and negative control cell lines (HIF-1 KO MTECs) (Fig. 1). To evaluate whether this relationship is also observed in vivo in human melanoma clinical specimens, we performed immunocytochemical analyses on 75 excisional biopsies or excisions of human melanomas, including 34 amelanotic and 41 melanotic tumors. Melanin production in these specimens was heterogeneous, with higher melanin levels detected in less advanced melanomas (pT1-2, 66%) relative to advanced tumors (pT3-4, 47%, Chi square test: χ2=7.341, p=0.0067). We next compared nuclear HIF-1α immunostaining in pigmented vs. amelanotic melanomas and observed significantly higher levels of HIF-1α expression in melanotic tumors (Fig. 2A-C). When samples were stratified according to the pT, this relationship was even more pronounced in advanced melanomas (pT3-4), with loss of correlation for melanoma at pT1-2. It should be also noted that high levels of nuclear HIF-1α immunostaining were observed in cells located close to the areas of necrosis, as expected (data not shown).
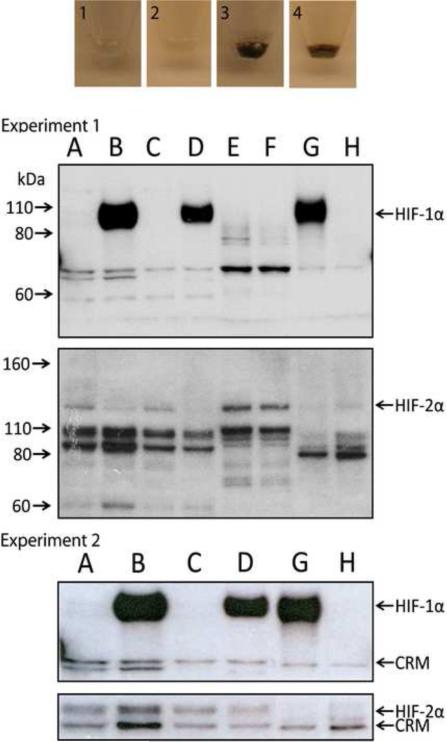
Figure 1.
The induction of melanin pigmentation is associated with up-regulation of HIF-1α, but not HIF-2α protein. For each independent experiment, experiment 1 (upper panels) and experiment 2 (lower panels), respectively, images of cell pellets obtained from two stable melanoma cell lines [Bomirski hamster AbC1 (1, 3) and human SKMEL-188 (2, 4)] that were cultured as described in the Materials and Methods and western blots for HIF-1α and HIF-2α are shown. For the western blots, lanes correspond to the following HS-WCE: A and C: amelanotic (non-pigmented) AbC1 and SKMEL-188 cells, respectively; B and D: melanotic (pigmented) AbC1 and SK-MEL 188 cells, respectively; E, Caki cells cultured 6h at hypoxia; F, Caki cells cultured at normoxia; G and H, HIF-1 WT (G) and HIF-1 KO (H) PyMT MTECs cultured at hypoxia (6h at 0.5% O2). Arrows to the left indicate the molecular weight (MW, kDa), whereas arrows to the right point to either HIF-1α, HIF-2α or cross-reactive material (CRM).
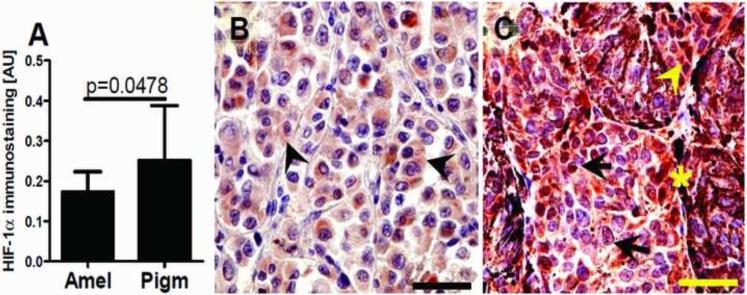
Figure 2.
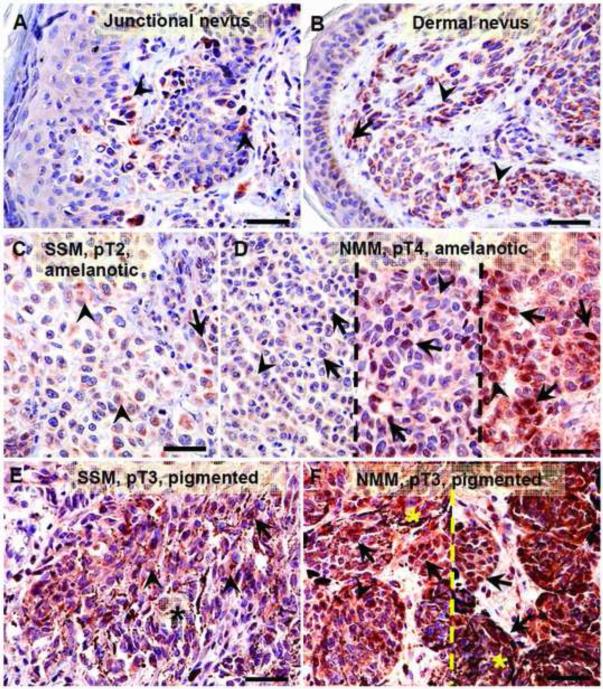
Relationship between maximal HIF-1α expression and melanin production (A). Statistically significant differences are denoted using p values as determined by Student's t-test. Amel: amelanotic melanomas, Pigm: pigmented melanomas. Representative immunostaining in amelanotic (B) and pigmented (C) melanomas. Arrows show nuclear HIF-1α immunostaining, whereas arrowheads show cytoplasmic HIF-1α immunostaining, asterisks indicate melanin; scale bar: 50 μm.
Therefore, these combined in vitro and in vivo data demonstrate that stimulation of melanogenesis increases the overall expression of HIF-1α and its nuclear localization. It is possible that HIF-1α is induced by the production of intermediates of melanogenesis, including ROS and the consumption of intracellular oxygen by the melanogenic process [12, 26]. Both ROS and hypoxic environments are known to efficiently stimulate nuclear HIF-1α localization in other tumors [38, 44]. Further support for our hypothesis is that melanogenesis induces a hypoxic intracellular oxidative environment is supported by the significant increase in the NAD/NADH ratio in melanized vs. non-melanized cells (Table 4).
Table 4.
NAD/NADH ratio in pigmented and non-pigmented human melanoma cells.
| Media used for pigmentation | NAD / NADH ratio |
|
|---|---|---|
| Pigmented SKMEL-188 | Non-pigmented SKMEL-188 | |
| Ham's F10 with L-tyrosine | 3.39 ± 0.23 | 2.73 ± 0.06** |
| DMEM/Ham's F10 (75/25) | 4.11 ± 0.15 | 3.56 ± 0.19** |
p < 0.01 by Student's t-test
Melanogenesis stimulates the expression of glycolysis related genes, glycolytic enzyme activity and VEGFA
Next we investigated the effect of melanogenesis on the expression of genes known to be regulated by HIF-1 as well as the enzymatic activity of key enzymes in the glycolytic pathway (Table 5 and Fig. 3). Table 5 shows that induction of melanogenesis significantly stimulates the expression of genes coding vascular endothelial growth factor A (VEGFA), glucose transporter 1 (GLUT-1), monocarboxylate transporter 2 (MCT2), sodium hydrogen exchanger (NHEI), which regulates intracellular acidity, and the glycolytic enzymes pyruvate dehydrogenase kinase, isozyme 1 (PDK1), aldolase A, fructose-bisphosphate (ALDOA), lactate dehydrogenase A (LDHA) and hexokinase 2 (HK2) (p <0.0001). These changes were also associated with increased expression of tyrosinase (Tyr) mRNA, a HIF-1-independent marker that indicates induction of the melanogenic pathway. As results are based on relative crossing point (Cp) values, it should be noted that a lower Cp value corresponds to higher gene expression, because all data are presented as the mean ΔΔCp between the target and reference genes (refer to Materials and Methods).
Table 5.
Melanogenesis stimulates expression of several genes regulated by HIF-1.
| Mean Cp value ± S.D. | ||
|---|---|---|
| Gene | Non-pigmented cells | Pigmented cells |
| VEGF | 6.04 ±0.31 | 0.85±0.18 |
| GLUT1 | 6.55 ±0.28 | 0.11±0.31 |
| Tyr | 4.68 ±0.22 | 3.05±0.10 |
| LDHA | 9.34 ±0.16 | 3.05±0.10 |
| MCT2 | 3.52 ±0.17 | 0.16±0.15 |
| NHEI | 9.34 ±0.16 | −3.09±0.26 |
| PDK1 | −2.12 ±0.33 | −8.25±0.16 |
| ALDOA | 0.04 ±0.35 | −5.11±0.31 |
| HK2 | 0.38 ±0.16 | −4.77±0.32 |
Expression of mRNAs in non-pigmented versus pigmented SKMEL-188 melanoma cells grown as described as in the methods was compared to β-actin using a comparative CP method. Relative gene expression data were calculated using the ΔΔCp method. Changes in gene expression are presented as a relative quantities using the mean ΔCp (normalized target) ± S.D. to caculate the difference between the target gene and the reference gene by comparing the first cycle of appearance above threshold (crossing point, Cp) using standardized algorithms in the Roche LC480 1.5 software package. All data (pigmented vs non-pigmented cells) are statistically significant, p <0.0001. VEGFA (vascular endothelial growth factor A); GLUT-1 (glucose receptor 1); Tyr (Tyrosinase); LDHA (lactate dehydrogenase A); MCT2 (monocarboxylate transporter 2); NHEI (sodium hydrogen exchanger); PDK1 (pyruvate dehydrogenase kinase, isozyme 1); ALDOA (aldolase A, fructose-bisphosphate); HK2 (hexokinase 2). A lower Cp numeric value reflects a more abundant signal for each particular gene (PCR product) of interest.
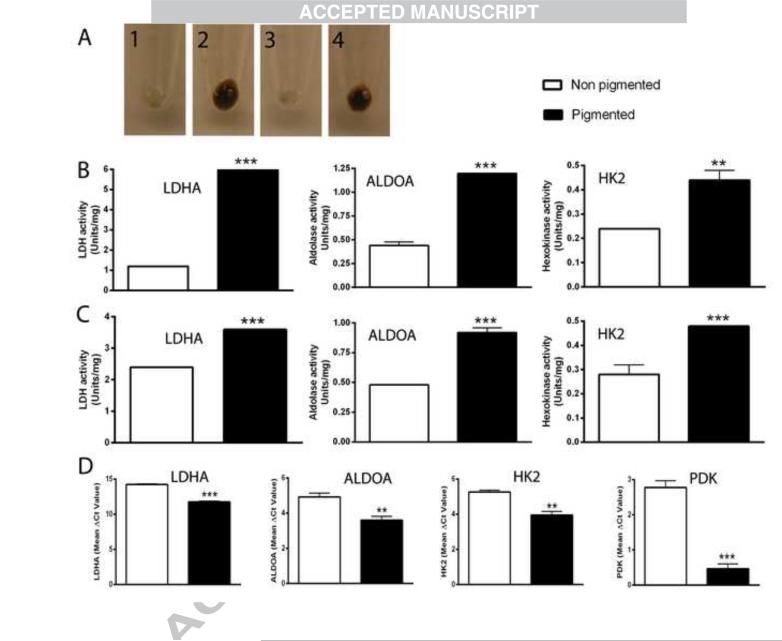
Figure 3.
Melanogenesis induces gene expression associated with increased HIF-1α expression. SKMEL-188 cells were grown in Ham's F10 containing 5% FBS, followed by incubation in fresh Ham's F10 containing 5% FBS for non-pigmentation or 75% DMEM and 25% Ham's F10 containing 5% FBS for pigmentation. The cells were harvested after 2 days (B) or 3 days (C) for enzyme assays and after 2 days for qPCR (D). A-1, non-pigmented cells at day 2; A-2, pigmented cells at day 2; A-3, non-pigmented cells at day 3; A-4, pigmented cells at day 3. Panels B and C summarize the enzyme activities as determined using whole cell protein extracts as described in the methods. Panel D shows the mean values of ΔCp for each gene (with lower values indicative or relatively higher gene expression levels). Data are presented as the mean ± SE (n = 3). **p < 0.01; ***p < 0.001 by Student's t-test.
The induction of melanogenesis is accompanied by cell rounding, followed by the detachment of heavily pigmented cells from the culture dish. Therefore, we also analyzed the time dependent expression of selected genes in pigmented vs. non-pigmented cells (Table 6). Again, we observed that the stimulation of HIF-1-dependent genes (VEGFA, GLUT-1, LDHA, PDK1, ALDOA, HK2 and MCT4) was highly reproducible for all-time points and conditions tested. It was previously shown that increases in HIF-1 dependent mRNAs for genes in the glycolytic pathway are accompanied by increases in total protein levels [85]. Therefore, changes in mRNA levels were confirmed at the protein level by measuring enzymatic activities of several glycolytic enzymes, including LDHA, ALDOA, HK2 and PDK1 enzymes activities, all of which increased in melanized vs. amelanotic melanomas (Fig. 3A-C). These data are also in agreement with previous observations showing that one intermediate of melanogenesis, L-DOPA, can stimulate lactate production in melanotic but not amelanotic melanomas [33]. In summary, stimulation of melanogenesis leads to the increased transcription of classical target genes regulated by HIF-1, and is accompanied by increased activities of several key enzymes that mediate glycolysis. Therefore, as is the case for other tumors, increased HIF-1 activity in melanized melanomas leads to up-regulation of the glycolytic pathway [38, 41, 44].
Table 6.
Time- and pigmentation-dependent changes in gene expression in human melanoma cells.
| 2 days culture, mean Cp ± S.D. | 3 days culture, mean Cp ± S.D. | ||||
|---|---|---|---|---|---|
| Gene | Non-pigmented cells | Pigmented cells | Non-pigmented cells | Pigmented cells | Pigmented and detached cells |
| ALDOA | 12.95±0.27 | 11.57±0.22 ** | 11.49±0.51 | 4.71±0.44 **** | 2.91±0.95 *** |
| PDK1 | 5.5±0.18 | 3.24±0.18 **** | 4.79±0.43 | −1.19±0.22 **** | −2.41±0.34 **** |
| HK2 | 11.17±0.21 | 10.25±0.28 ** | 10.13±0.53 | 5.54±0.22 *** | 3.27±0.37 **** |
| GLUT1 | 17.02±0.11 | 13.56±0.22 **** | 14.68±0.46 | 9.61±0.37 *** | 8.29±0.41 **** |
| LDHA | 14.91±0.22 | 10.22±0.19 **** | 13.4±0.36 | 8.85±0.17 **** | 6.37±0.16 **** |
| VEGFA | 17.02±0.11 | 6.39±0.14 **** | 17.42±0.36 | 11.87±0.16 **** | 11.85±0.43 **** |
| Tyr | 9.61±0.21 | 0.26±0.31 **** | 3.23±0.39 | 2.83±0.18 | ND |
| MCT4 | 8.48±0.102 | 7.1±0.11 **** | 7.89±0.16 | 6.24±0.11 **** | 8.48±0.102 |
Relative mRNA levels present in of Sk-Mel-188 non-pigmented and pigmented cells grown for 2 or 3 days after stimulation of melanogenesis were compared to reference the gene Cyclophillin B. Relative gene expression data were calculated using ΔΔCp method. Changes in gene expression are presented as a relative quantities using mean ΔCp (normalized target) ± S.D. as the difference between target gene and reference gene in cycle of appearance in time (Cp). Data were compared between pigmented and non-pigmented cells using Student's t-test
* p <0.05
p <0.01
p <0.001
p <0.0001
ND-not detected. A lower Cp numeric value reflects a more abundant signal for each particular gene (PCR product) of interest.
Yet, the inhibition of expression of Tyr (Table 6) following melanization requires a separate explanation. After 3 days of culture, when melanoma cells became heavily pigmented, the relative expression of Tyr mRNA decreased, approaching values of the control amelanotic cells. Tyr expression levels became undetectable in detached pigmented cells (Table 5). The decrease of Tyr may be explained by a feedback-dependent inhibition of enzyme activity and gene expression in heavily melanized cells, as described and discussed previously [86-89].
Differential effect of melanogenesis on expression of stress related, steroidogenic, immune and growth related genes
As melanin pigmentation leads to increased HIF-1α protein levels and HIF-1 transcriptional activity, a panel of genes involved in stress response, regulation of melanogenesis, immunity, angiogenesis and cell growth were screened for the presence of putative Hypoxia Response Elements (HRE; 5’-RCGTC, where R is any purine, A or G) within each gene's regulatory regions using publicly available bioinformatic tools (Table 3). This screen identified several potential HIF target genes that are known to be involved in stress-responses, the HPA axis, steroidogenesis and the regulation of melanogenic activity, as well as genes involved in immunity, angiogenesis and cell proliferation. The two most striking genes identified as putative HIF target genes were CRH and DHCR7. There are three putative HRE sites identified within the regulatory region of CRH, each with a significant matrix score (p <0.001, based on TRANSFAC and JASPAR transcription factor binding matrix models) [81, 82]. Multiple putative HREs were also identified for DHCR7: five HRE sites were identified within the first 1000 bp of the transcriptional start site (TSS; p <0.05 to p <0.001). Additional HPA axis genes, including PCKS1, PCSK2, MC2, and the previously identified POMC and MC1 each contained at least two putative HREs within the regulatory regions [56]. In agreement with this analysis, we discovered that expression of these genes is also stimulated by melanogenesis, as shown in Table 7, which suggests involvement of HIF-1α. It must be noted that the PCSK1 and PCSK2 convertase enzymes differentially process pro-CRH and POMC to CRH peptide and POMC-derived ACTH, α-MSH and β-endorphin, respectively [14]. Each of these factors can stimulate melanogenesis through activation of their corresponding receptors [12, 14-16, 90, 91]. Collectively, these observations strongly suggest that HIF-1 is involved in regulation of the stress responses through the activation of pro-hormones, convertases, CRH and the MC1 and MC2 coding genes, which in a context-dependent manner could regulate either the HPA or melanogenic activity following schemes previously outlined [12, 13, 16]. Additional mechanisms of induction of pro-melanogenic molecules could include a direct action of intermediates of melanogenesis on POMC and MC1 [26], since stimulation of POMC by L-DOPA [92] and of MSH receptors by L-tyrosine and L-DOPA have already been reported [58, 59, 93]. It is possible that induction of these classes of genes (see Table 7) could be secondary to HIF-1 transcriptional activity and/or may alternatively represent the cellular response to intermediates of melanogenesis [26] or of oxidative stress.
Table 7.
Pigmentation-dependent gene expression in human SKMEL-188 melanoma cells.
| Mean Cp ± S.D. | ||
|---|---|---|
| Gene | Non Pigmented Cells | Pigmented Cells |
| CRH | 4.51±0.37 | 3.14±0.63 * |
| CRH, exon 2 | 5.24±0.28 | 3.14±0.63 ** |
| CRHR1 | 3.45±0.36 | 1.53±0.72 * |
| POMC, exon 3 | 5.24±0.28 | 2.02±0.66 ** |
| MC1 | 5.24±0.28 | −3.01±0.69 **** |
| MC2 | 3.07±0.31 | 2.02±0.67 |
| PCSK1 | 5.24±0.28 | 3.14±0.63 ** |
| PCSK2 | 5.24±0.28 | −31.86±0.001 **** |
| NQO2 | 5.24±0.28 | −2.89±0.65 **** |
| Cyp11A1, exon 4 | 5.24±0.26 | 3.14±0.63 ** |
| Cyp11B1 | 4.16±0.31 | 0.17±0.64 *** |
| 3βHSD | 5.24±0.28 | 3.14±0.63 ** |
| Cyp17A1 | −10.70±0.3 | 0.15±0.63 **** |
| Cyp21A2v2 | −29.76±0.001 | 3.14±0.063 **** |
| HSD11B1 | −12.86±0.42 | −4.38±0.64 **** |
| HSD11B2 | −13.48±0.45 | −4.04±0.7 **** |
| 7DHCR | −1.87±0.34 | 3.14±0.63 *** |
| ICAM1 | 11.41±0.19 | 11.78±0.25 |
| IFNα1 | −0.07±0.20 | 0.25±0.26 |
| IFNβ1 | 7.41±0.31 | 7.83±0.26 |
| IFNγ | −23.59±0.1 | −23.22±0.1 |
| TNFα | 7.02±0.35 | 7.12±0.32 |
| IL1α | 11.41±0.19 | 11.78±0.25 |
| IL1β | 11.38±0.20 | 11.41±0.41 |
| IL2 | 11.41±0.19 | 11.78±0.25 |
| IL4 | 9.47±0.22 | 9.77±0.81 |
| IL5 | −6.13±0.36 | 11.14±0.41 **** |
| IL6 | 7.28±0.21 | 7.86±0.32 |
| IL8 (CXCL-8) | 11.41±0.19 | 11.78±0.25 |
| IL10 | 10.57±0.41 | 11.56±0.46 * |
| IL17A | −4.89±0.47 | 7.78±0.25 **** |
| RANTES | 11.41±0.19 | 11.78±0.25 |
| PDGFA | −0.61±0.21 | 6.84±0.30 **** |
| TGFA | 3.59±0.34 | 4.29±0.40 |
| TGFB1 | 9.33±0.24 | 9.82±0.27 |
| TGFB2 | 0.37±0.20 | −23.22±0.001 **** |
| EGFR | 7.10±0.34 | 7.57±0.28 |
| TRANSFERRIN RC | −11.69±0.37 | −5.61±0.42 **** |
| NPL | 11.41±0.19 | 11.78±0.25 |
Changes in gene expression of human SKMEL-188 melanoma cells grown for three days in culture in F10 media (non-pigmented) or in DMEM media (pigmented) as described in the methods, after normalization to the reference gene cyclophillin B. Changes in gene expression are presented as a relative quantities using mean ΔCp (normalized target) ± S.D. as the difference between target gene and reference gene in cycle of appearance in time (Cp). Data were compared between pigmented and non-pigmented cells using Student's t-test. Genes profiled include: CYP11A1 (cytochrome P450, family 11, subfamily A, polypeptide 1) exon 4; CYP11B1 (cytochrome P450, family 11, subfamily B, polypeptide 1); CYP17A1 (cytochrome P450, family 17, subfamily A, polypeptide 1); CYP21 A2v2 (cytochrome P450, family 21, subfamily A, polypeptide 2); HSD11B1 (hydroxysteroid (11-beta) dehydrogenase 1); HSD11B1 (hydroxysteroid (11-beta) dehydrogenase 2); 7DHCR (7-dehydrocholesterol reductase); 3βHSD (3-beta-hydroxysteroid dehydrogenase); CRH (corticotropin releasing hormone); CRHR1 (corticotropin releasing hormone receptor 1); POMC (proopiomelanocortin); MC (melanocortin receptor); PCSK (proprotein convertase subtilisin/kexin type); NQO2 (NAD(P)H dehydrogenase, quinone 2); EGFR (epidermal growth factor receptor); ICAM1 (intercellular adhesion molecule 1); IFN (interferon); IL (interleukin); CXCL (chemokine ligand); TNF (tumor necrosis factor); PDGFA (platelet-derived growth factor alpha polypeptide); TGFB (transforming growth factor, beta); RANTES (CCL5 chemokine (C-C motif) ligand 5); TRANSFERRIN RC (transferrin receptor); NPL (N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
p <0.05
p <0.01
p <0.001
p <0.0001.
A lower Cp numeric value reflects a more abundant signal for each particular gene (PCR product) of interest.
Excluding 7DHCR described above (which encodes for the 7Δreductase that transforms 7-dehydrocholesterol, 7DHC, to cholesterol), many genes involved in steroidogenesis did not contain any putative HREs using our search parameters, including CYP11A1, HSD3B1, CYP17A1, CYP21A2, HSD11B1, and HSD11B2. Only one putative HRE was identified for CYP11B1, but it was detected below the matrix significance level (p >0.05; Table 3). Interestingly, we observed melanogenesis-dependent stimulation of CYP11A1, CYP11B1, 3βHSD, but inhibition of CYP17A1, CYP21A2, HSD11B1, HSD11B2 and 7DHCR (Table 7). Together, these results imply that there is HIF-1 independent regulation of steroidogenesis in pigment cells, similar to the complexity of the pathways operating in the skin [reviewed in [13, 17, 18]]. In the context of downregulation of 7DHCRF, which contains several putative HREs, we speculate that HIF-1 might be involved in the inhibition of cholesterol production by attenuation of reduction of unsaturated B ring and the corresponding accumulation of 7DHC. In the skin, UVB exposure will transform 7DHC to vitamin D3 [94], which can be further hydroxylated by CYP27A1 and CYP27B1 [95] to its active form, 1,25-dihydroxyvitamin D3 (calcitriol). Of note, both of these CYP genes were previously shown to contain putative HRE elements [56].
Genes for which no HREs were identified included CRHR1 and NQO2 (Table 3). Therefore, their stimulation (see Table 7) could be secondary to the action of intermediates of melanogenesis or oxidative stress. Since CRHR1 is a part of the HPA axis, this relationship could link its activity with oxidative stress. Stimulation of NQO2 by melanogenesis could result from a protective role of the protein product quinone reductase 2 (originally considered as a melatonin receptor type 3 and ubiquitously expressed in the skin) against oxidative damage and quinone compounds [96, 97], since both ROS and quinones are produced during melanogenesis [12].
TGFB2 and PDGFA, genes involved in regulation of cell growth and immunity, each contained five putative HRE sites (p <0.05) (Table 3). Whereas TGFB2 expression was stimulated by melanogenesis, PDGFA was inhibited by melanogenesis (see Table 7). In addition, TFGB1, TFRC, IL1B and CCL5 contained at least two HREs (Table 3), of which the expression of only TFRC was downregulated by melanogenesis, with other enzymes showing steady rate of expression of mRNA (see Table 7). Additional genes in this category, including IL5, IL10, IL17A, IFNA1, IFNB1, TFGA, EGFR, and NPL did not contain any putative HREs (Table 3). Interestingly, IL-5, IL-10 and IL17A expression levels were stimulated by melanogenesis, but there was no effect on the expression of other variety of growth factors or cytokines, including ICAM1, INFA, INFB, INFG, TGFA, TGFB1, EGFR, IL1A, IL1B, IL2, IL2, IL4, IL6, IL8, IL10, RANTES or NPL (see Table 7). The above pattern suggests limited effects of melanogenesis on expression of growth factors or cytokines in melanoma cells and that there are selective as well as both HIF-1 -dependent and independent effects on gene expression for these classes of genes.
Concluding remarks for cell culture studies
In conclusion, induction of melanogenesis affects the expression of multiple genes involved in the regulation of melanocyte/melanoma behavior, including the metabolic switch to glycolysis, coordinated by HIF-1, with accompanying changes in stress related genes. However, it is not yet clear if these genes are exclusively HIF-dependent since analysis is by complicated by potential secondary effects on gene expression, and since the HRE sites have not been shown to be functional using chromatin immunoprecipitation or other assays. The proposed interactions on the cellular level of these complex biochemical and biological networks are outlined in Fig. 4. Similar studies using normal human melanocytes remain to be performed to evaluate whether similar relationships are operating under physiological conditions of normal skin.
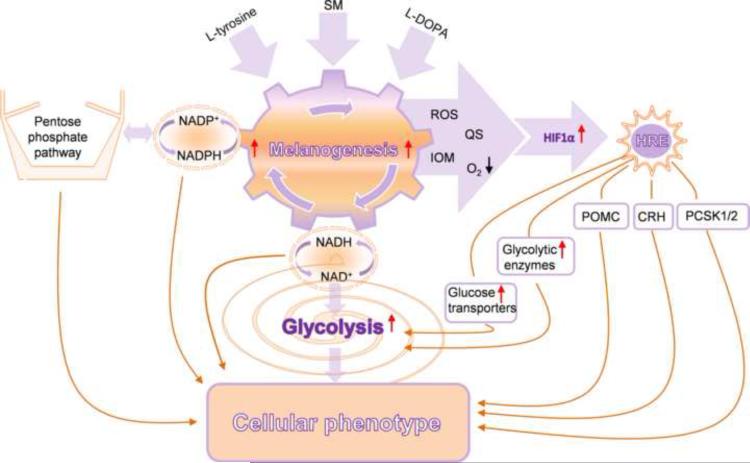
Figure 4.
The proposed interactions between melanogenesis, glucose metabolism and potential HIF-1 dependent genes/pathways that culminate to regulate cellular metabolism. SM: stimulators of melanogenesis, ROS: reactive oxygen species, QS: quinones and semiquinones, IOM: intermediates of melanogenesis, POMC: proopiomelanocortin, CRH: corticotropin releasing hormone, PCSK1/2: proprotein convertase subtilisin/kexin types 1 and 2.
Changes in expression of HIF-α during progression of melanocytic lesions: clinico-pathological analyses
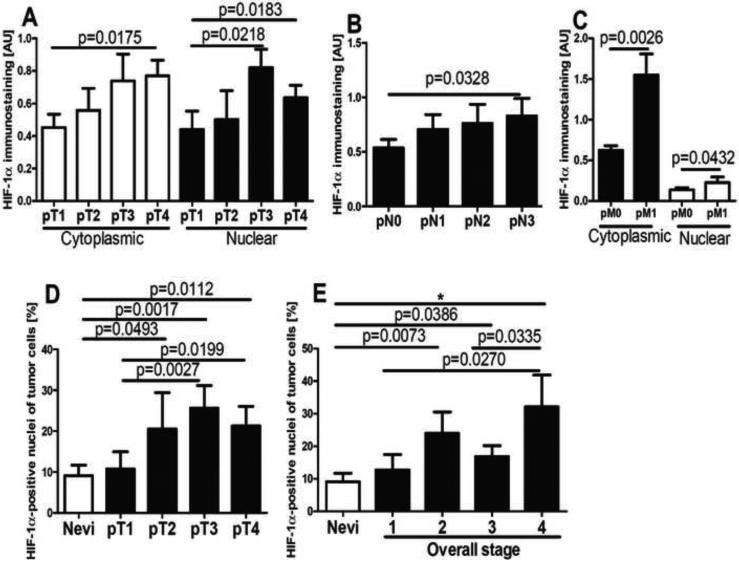
In the context of above studies and the literature on the constitutive expression of HIF-1α in melanomas [56, 98-100], we have evaluated a relationship between HIF-1α expression and the progression of melanocytic lesions in a large panel of pigmented lesions obtained from patients listed in Table 2. Representative immunostaining of HIF-1α in melanocytic nevi and melanomas is shown in Fig 5. In normal skin, not involved in pathology, expression of HIF-1α was below the detection limits of our protocols. In melanocytic nevi and melanomas, HIF-1α was expressed both in the cytoplasm and nuclei (Fig. 6), with significant differences in expression between these groups found for nuclear immunoreactivity. There was no difference in immunostaining between different types of nevi including the dysplastic type. Cells with HIF-1α-positive nuclei were found more frequently in malignant lesions (Chi square test: χ2=6.697, p=0.0097) in comparison to melanocytic nevi. The nuclear HIF-1α-positivity was found in 60% of melanocytic nevi and in 77% of malignant melanomas. The percentage of HIF-1α-positive nuclei was 18.7% in melanomas and 9.1% in melanocytic nevi (Fig. 5A-F, 6A). When melanomas where stratified according to different types, a higher portion of HIF1α-positive nuclei was found in nodular (24.0%) in comparison to superficial spreading type (12.7%) melanomas (Fig. 5C-F, 6B). Stratification of melanomas according pTNM stage revealed a positive correlation between cytoplasmic HIF-1α and the pT (r=0.2705, p=0.0128), pN (r=0.2029, p=0.0407) or pM (r=0.2872, p=0.0131) stages. The increases for both mean cytoplasmic and nuclear HIF-1α levels with the pT or pM stages were observed, with the highest levels of HIF-1α detected in melanomas at the advanced stages (pT3-4, pM1) (Fig. 5C-F, 7A-C). For melanomas stratified according pN stage, only cytoplasmic HIF-1α expression increased with pN advancement (Fig. 7B). The percentage of HIF-1α-positive cell nuclei correlated also with pT (r=0.2511, p=0.0195, data not shown). In addition, thorough analysis of portion of HIF-1α-positive cell nuclei in melanomas stratified according pT and overall stage revealed no differences between pT1 and stage 1 melanomas and nevi (Fig. 7D,E). There was also lack of correlation between OS and DFS of melanoma patients and tumor HIF-1α levels (data not shown).
Figure 5.
Representative immunostaining with antibodies against HIF-1α in nevi (A, B) and melanomas (C-F). Dashed lines separates cases with these same pathomorphological features, arrows show nuclear HIF-1α immunostaining, arrow-head shows cytoplasmic HIF-1α immunostaining, asterisks indicates melanin; scale bar 50 μm.
Figure 6.
Differences in HIF-1α immunostaining between nevi (n=26) and melanomas (n=75) (A) and between superficial spreading (SSM) (n=36) or nodular (NMM) (n=37) melanomas (B). Statistically significant differences are denoted with p values as determined by Student's t-test.
Figure 7.
Expression of cytoplasmic (A,B,C) and nuclear (A,C) HIF-1α antigen in melanomas stratified according pT (A), pN (B) and pM (C) stage. The percentage of HIF-1α-positive tumor cell nuclei in nevi and melanomas stratified according pT (D) and overall stage (E). Statistically significant differences are denoted with p values determined by Student's t-test and with asterisks by ANOVA (*p <0.05). Refer to Table 1 for information regarding the number of cases per melanoma stage.
Thus, analysis of 26 cases of melanocytic nevi and 75 cases of melanoma demonstrates distinct and significant increases of HIF-1α expression in melanomas at advanced stages as indicated by higher expression in nodular vs. superficial spreading melanomas, or in thicker (pT3 and pT4) vs. thinner (pT1) melanomas or localized (pN0 or pM0) vs. metastatic melanomas that involve multiple lymph nodes (pN3) or visceral sites (pM1). This was further supported by higher expression in highly advanced stages (stage 4) vs. localized (stage 1) disease. These results are in agreement with studies by Mills et al., 2009 [100] who showed the highest expression of HIF-1α in metastatic melanomas and a higher expression in the vertical growth phase vs. radial growth phase. Our data are also in agreement with a study by Giatrmanolaki et al., who demonstrated a contribution of HIF-1α to melanoma progression and/or metastasis [101]. Other investigators have not detected a clear correlation between the expression of HIF-1α and any clinicopathological variables, including patient prognosis or survival [99]. Overall, our studies are in partial agreement with these analyses as we also found no significant correlation between HIF-1α expression and either OS or DFS. However, a significant correlation was observed between progression and HIF-1α expression in highly advanced melanomas, with higher HIF-1α levels in thicker (≥pT2) but not thin (pT1) melanomas in comparison to melanocytic nevi. A similar trend was reported previously by other investigators [56]. Despite the limitations listed above, our pathologic data support the hypothesis that overexpression of HIF-1α can contribute to the increased aggressiveness of melanoma, which is in agreement to prior reports in the literature on melanomas [100, 102, 103] and other tumor types [38, 44]. However, it is unlikely that HIF-1α acts alone in driving melanoma progression.
Changes in expression of GLUT-1 during progression of melanocytic lesions: clinico-pathological analyses
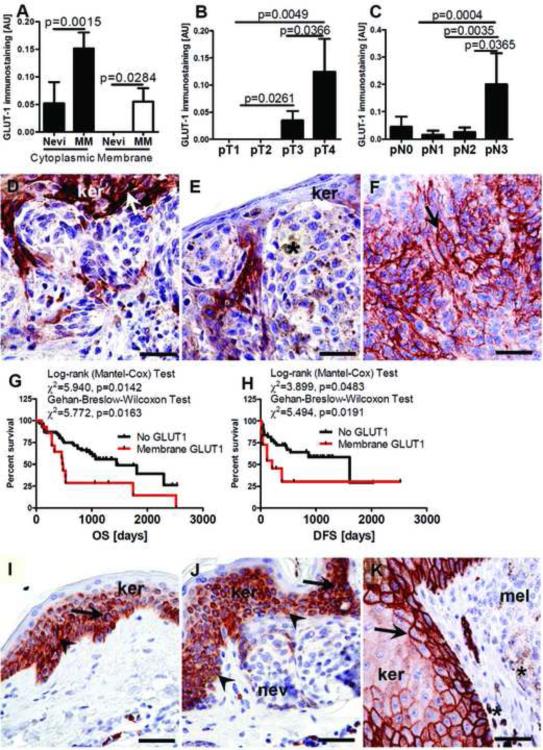
Comparisons of GLUT-1 expression in the same specimens revealed both cytoplasmic and membranous patterns of localization (Fig. 8A, J). The membrane-localized GLUT-1 was observed only in melanomas (55%). Cytoplasmic GLUT-1 was more frequently observed in malignant (49%) vs. benign (20%) melanocytic lesions (Chi square test: χ2=18.61, p<0.0001). GLUT-1 immunostaining in melanomas was heterogeneous, being clustered in patches of melanomas cells in various areas of tumor. A high level of membrane-localized GLUT-1 immunostaining was observed around necrotic areas (data not shown). Comprehensive analysis of GLUT-1 levels in reference to pathomorphological features revealed significantly higher membranous GLUT-1 expression in pT3-4 stages and pN3 melanomas as compared to less advanced tumors (Fig. 8B-F). The OS and DFS in melanomas with membranous GLUT-1 expression was shortened (Fig. 8G,H). However, there was lack of such correlation for cytoplasmic GLUT-1 staining. In normal skin, GLUT-1 was localized predominantly to the keratinocytes’ cytoplasm, while in keratinocytes immediately surrounding melanomas GLUT-1 expression increased having predominant membranous localization (Fig. 8 I-K). There were no significant differences in GLUT-1 immunostaining levels and either melanin level, pM or overall melanoma stage. We also observed that OS and DFS in melanomas expressing membrane-localized GLUT-1 was shorter. Other laboratories have reported trends between GLUT-1 expression and some melanoma features of increased tumor aggressiveness [104, 105]. However, GLUT-1's role in melanoma progression requires further investigation using large number of specimens as other investigators have reported a decreased frequency of GLUT-1 in aggressive melanomas as compared to melanocytic nevi [106], or have reported variable effects on glucose uptake in melanomas [107-109].
Figure 8.
GLUT-1 expression in nevi and melanomas (A), membrane GLUT-1 expression and melanoma stratified according pT (B) and pN (C) classification. Representative GLUT-1 immunostaining in pT1 (D), pT2 (E) and pT4 (F) melanomas. Relationship between OS (G) or DFS (H) and membrane GLUT-1 levels. Representative immunostaining of GLUT-1 in normal skin (I), in the epidermis above the nevus (J) or in melanoma (K). Statistically significant differences are denoted with p values as determined by Student's t-test. Arrows show membrane GLUT-1 immunostaining, whereas arrowheads indicate cytoplasmic GLUT-1 immunostaining, and asterisks indicate melanin, MM: melanomas, ker: keratinocytes, nev: nevus cells, mel: melanoma cells; scale bar 50 μm. Refer to Table 2 for information regarding the number of cases per melanoma stage.
Final concluding remarks
Induction of melanogenesis in melanoma cells is associated with dramatic increases of nuclear HIF1-α expression accompanied by upregulation of HIF-1 dependent multiple genes involved in the regulation of glucose metabolism, angiogenesis and stress responses. It is, however, unclear whether these regulatory pathways are exclusively or predominantly coordinated by HIF-1, since melanogenesis had several secondary effects on gene expression that are HIF-1 independent. Therefore, we propose a complex set of interactions between melanogenesis, glucose metabolism and HIF-1-dependent- or independent pathways that can regulate both cellular metabolism and cell behavior (Fig. 4). The immunocytochemical analyses revealing higher levels of HIF-1α and of GLUT-1 in advanced melanomas suggest that HIF-1 might also be involved in melanoma progression at local levels.
Highlights.
Melanogenesis stimulates expression of HIF-1α and downstream regulators of metabolism
Melanogenesis affects expression of stress related genes
Advanced melanomas show increased expression of HIF-1α protein
Advanced melanomas show increased expression of membrane-bound GLUT-1
Acknowledgements
This work was supported by grants from NIH/NIAMS (2R06AR052190 and 1R01AR056666 to AS), NIH/NCI (1R01CA138488 to TS) and the West Clinic Cancer Foundation awards to AS and TS. The paper is dedicated to Dr. Andrzej Bomirski, who established Bomirski melanomas that are transplantable to Syrian golden hamsters [110]. We thank Dr. Zbigniew Pawlowicz, Director of Oncology Centre - Prof. Franciszek Łukaszczyk Memorial Hospital for generously supporting collaborations advantageous for our research.
Nonstandard abbreviations
- VEGFA
vascular endothelial growth factor A
- GLUT-1
glucose transporter 1
- Tyr
Tyrosinase
- LDHA
lactate dehydrogenase A
- MCT
monocarboxylate transporter
- NHEI
sodium hydrogen exchanger
- PDK1
pyruvate dehydrogenase kinase, isozyme 1
- ALDOA
aldolase A, fructose-bisphosphate
- HK2
hexokinase 2
- Cyp11A1
(cytochrome P450, family 11, subfamily A, polypeptide 1) exon 4
- Cyp11B1
cytochrome P450, family 11, subfamily B, polypeptide 1
- Cyp17A1
cytochrome P450, family 17, subfamily A, polypeptide 1
- Cyp21 A2v2
cytochrome P450, family 21, subfamily A, polypeptide 2
- HSD11B1
hydroxysteroid (11-beta) dehydrogenase 1
- HSD11B1
hydroxysteroid (11-beta) dehydrogenase 2
- 7DHCR
7-dehydrocholesterol reductase
- 3βHSD
3-beta-hydroxysteroid dehydrogenase
- CRH
corticotropin releasing hormone
- CRHR1
corticotropin releasing hormone receptor 1
- POMC
proopiomelanocortin
- MC
melanocortin receptor
- PCSK
proprotein convertase subtilisin/kexin type
- NQO2
NAD(P)H dehydrogenase, quinone 2
- EGFR
epidermal growth factor receptor
- ICAM1
intercellular adhesion molecule 1
- IFN
interferon
- IL
interleukin
- CXCL
chemokine ligand
- TNF
tumor necrosis factor
- PDGFA
platelet-derived growth factor alpha polypeptide
- TGFB
transforming growth factor, beta
- RANTES
CCL5 chemokine (C-C motif) ligand 5
- TRANSFERRIN RC
transferrin receptor
- NPL
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
- HIF
Hypoxia-Inducible Factor
- HRE
HIF responsive element
- SM
stimulators of melanogenesis
- ROS
reactive oxygen species
- QS
quinones and semiquinones
- IOM
intermediates of melanogenesis
- OS
overall survival
- DFS
disease free survival
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levine D, Fisher DE. Methods Mol Biol. 2014;1102:177–197. doi: 10.1007/978-1-62703-727-3_11. [DOI] [PubMed] [Google Scholar]
- 2.Schadendorf D, Hauschild A. Nat Rev Clin Oncol. 2014 doi: 10.1038/nrclinonc.2013.246. [DOI] [PubMed] [Google Scholar]
- 3.Kwong LN, Davies MA. Oncogene. 2014;33:1–9. doi: 10.1038/onc.2013.34. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Robert C, Dummer R, Gutzmer R, Lorigan P, Kim KB, Nyakas M, Arance A, Liszkay G, Schadendorf D, Cantarini M, Spencer S, Middleton MR. Lancet Oncol. 2013;14:733–740. doi: 10.1016/S1470-2045(13)70237-7. [DOI] [PubMed] [Google Scholar]
- 6.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, Schuchter L, Cebon J, Ibrahim N, Kudchadkar R, Burris HA, 3rd, Falchook G, Algazi A, Lewis K, Long GV, Puzanov I, Lebowitz P, Singh A, Little S, Sun P, Allred A, Ouellet D, Kim KB, Patel K, Weber J. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah DJ, Dronca RS. Mayo Clinic Proc. 2014;89:504–519. doi: 10.1016/j.mayocp.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber JS, Kudchadkar RR, Yu B, Gallenstein D, Horak CE, Inzunza HD, Zhao X, Martinez AJ, Wang W, Gibney G, Kroeger J, Eysmans C, Sarnaik AA, Chen YA. J Clin Oncol. 2013;31:4311–4318. doi: 10.1200/JCO.2013.51.4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, Rutkowski P, Blank CU, Miller WH, Jr., Kaempgen E, Martin-Algarra S, Karaszewska B, Mauch C, Chiarion-Sileni V, Martin AM, Swann S, Haney P, Mirakhur B, Guckert ME, Goodman V, Chapman PB. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 10.Johnson DB, Wallender EK, Cohen DN, Likhari SS, Zwerner JP, Powers JG, Shinn L, Kelley MC, Joseph RW, Sosman JA. Cancer Immunol Res. 2013;1:373. doi: 10.1158/2326-6066.CIR-13-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slominski AT, Carlson JA. Mayo Clinic proceedings. 2014;89:429–433. doi: 10.1016/j.mayocp.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slominski A, Tobin DJ, Shibahara S, Wortsman J. Physiol Rev. 2004;84:1155–1228. doi: 10.1152/physrev.00044.2003. [DOI] [PubMed] [Google Scholar]
- 13.Slominski AT, Zmijewski MA, Skobowiat C, Zbytek B, Slominski RM, Steketee JD. Adv Anat Embryol Cell Biol. 2012;212:1–115. doi: 10.1007/978-3-642-19683-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slominski A, Wortsman J, Luger T, Paus R, Solomon S. Physiol Rev. 2000;80:979–1020. doi: 10.1152/physrev.2000.80.3.979. [DOI] [PubMed] [Google Scholar]
- 15.Bohm M, Luger TA, Tobin DJ, Garcia-Borron JC. J Invest Dermatol. 2006;126:1966–1975. doi: 10.1038/sj.jid.5700421. [DOI] [PubMed] [Google Scholar]
- 16.Slominski AT, Zmijewski MA, Zbytek B, Tobin DJ, Theoharides TC, Rivier J. Endocr Rev. 2013;34:827–884. doi: 10.1210/er.2012-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slominski A, Zbytek B, Nikolakis G, Manna PR, Skobowiat C, Zmijewski M, Li W, Janjetovic Z, Postlethwaite A, Zouboulis CC, Tuckey RC. J Steroid Biochem Mol Biol. 2013;137:107–123. doi: 10.1016/j.jsbmb.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slominski AT, Zmijewski MA, Semak I, Zbytek B, Pisarchik A, Li W, Zjawiony J, Tuckey RC. Anticancer Agents Med Chem. 2014;14:77–96. doi: 10.2174/18715206113139990308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slominski A, Wortsman J, Tobin DJ. FASEB J. 2005;19:176–194. doi: 10.1096/fj.04-2079rev. [DOI] [PubMed] [Google Scholar]
- 20.Korner A, Pawelek J. Science. 1982;217:1163–1165. doi: 10.1126/science.6810464. [DOI] [PubMed] [Google Scholar]
- 21.Lerner AB, Fitzpatrick TB. Physiol Rev. 1950;30:1–126. doi: 10.1152/physrev.1950.30.1.91. [DOI] [PubMed] [Google Scholar]
- 22.Pawelek JM, Korner AM. Am Sci. 1982;70:136–145. [PubMed] [Google Scholar]
- 23.Pawelek J, Korner A, Bergstrom A, Bologna J. Nature. 1980;286:617–619. doi: 10.1038/286617a0. [DOI] [PubMed] [Google Scholar]
- 24.Prota G. Fortschr Chem Org Naturst. 1995;64:93–148. doi: 10.1007/978-3-7091-9337-2_2. [DOI] [PubMed] [Google Scholar]
- 25.Wood JM, Jimbow K, Boissy RE, Slominski A, Plonka PM, Slawinski J, Wortsman J, Tosk J. Exp Dermatol. 1999;8:153–164. doi: 10.1111/j.1600-0625.1999.tb00365.x. [DOI] [PubMed] [Google Scholar]
- 26.Slominski A, Zmijewski MA, Pawelek J. Pigment Cell Melanoma Res. 2012;25:14–27. doi: 10.1111/j.1755-148X.2011.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slominski A, Paus R, Mihm MC. Anticancer Res. 1998;18:3709–3715. [PubMed] [Google Scholar]
- 28.Brozyna AA, VanMiddlesworth L, Slominski AT. Int J Cancer. 2008;123:1448–1456. doi: 10.1002/ijc.23664. [DOI] [PubMed] [Google Scholar]
- 29.Slominski A, Paus R, Schadendorf D. J Theor Biol. 1993;164:103–120. doi: 10.1006/jtbi.1993.1142. [DOI] [PubMed] [Google Scholar]
- 30.Slominski A, Zbytek B, Slominski R. Int J Cancer. 2009;124:1470–1477. doi: 10.1002/ijc.24005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slominski A, Friedrich T. Pigment Cell Res. 1992;5:396–399. doi: 10.1111/j.1600-0749.1992.tb00569.x. [DOI] [PubMed] [Google Scholar]
- 32.Slominski A, Moellmann G, Kuklinska E. Pigment Cell Res. 1989;2:109–116. doi: 10.1111/j.1600-0749.1989.tb00170.x. [DOI] [PubMed] [Google Scholar]
- 33.Scislowski PW, Slominski A, Bomirski A. Int J Biochem. 1984;16:327–331. doi: 10.1016/0020-711x(84)90107-1. [DOI] [PubMed] [Google Scholar]
- 34.Scislowski PW, Slominski A, Bomirski A, Zydowo M. Neoplasma. 1985;32:593–598. [PubMed] [Google Scholar]
- 35.Li W, Slominski R, Slominski AT. Anal Biochem. 2009;386:282–284. doi: 10.1016/j.ab.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scislowski PW, Slominski A. Neoplasma. 1983;30:239–243. [PubMed] [Google Scholar]
- 37.Warburg O. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 38.Kroemer G, Pouyssegur J. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Semenza GL. Drug Discov Today. 2007;12:853–859. doi: 10.1016/j.drudis.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 40.Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, Saavedra E. FEBS J. 2007;274:1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, Cairns R, Papandreou I, Koong A, Denko NC. PLoS One. 2009;4:e7033. doi: 10.1371/journal.pone.0007033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey KM, Wojtkowiak JW, Hashim AI, Gillies RJ. Adv Pharmacol. 2012;65:63–107. doi: 10.1016/B978-0-12-397927-8.00004-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dang NH, Singla AK, Mackay EM, Jirik FR, Weljie AM. Curr Pharm Des. 2013 doi: 10.2174/13816128113199990489. [DOI] [PubMed] [Google Scholar]
- 44.Semenza GL. Sci STKE 2007. 2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 45.Schodel J, Mole DR, Ratcliffe PJ. Biol Chem. 2013;394:507–517. doi: 10.1515/hsz-2012-0351. [DOI] [PubMed] [Google Scholar]
- 46.Sowter HM, Raval RR, Moore JW, Ratcliffe PJ, Harris AL. Cancer Res. 2003;63:6130–6134. [PubMed] [Google Scholar]
- 47.Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. Blood. 2011;117:e207–217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu J, Stiehl DP, Setzer C, Wichmann D, Shinde DA, Rehrauer H, Hradecky P, Gassmann M, Gorr TA. Mol Cancer Res. 2011;9:1520–1536. doi: 10.1158/1541-7786.MCR-11-0090. [DOI] [PubMed] [Google Scholar]
- 49.Stiehl DP, Bordoli MR, Abreu-Rodriguez I, Wollenick K, Schraml P, Gradin K, Poellinger L, Kristiansen G, Wenger RH. Oncogene. 2012;31:2283–2297. doi: 10.1038/onc.2011.417. [DOI] [PubMed] [Google Scholar]
- 50.Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Nature. 2003;425:307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- 51.Rolfs A, Kvietikova I, Gassmann M, Wenger RH. J Biol Chem. 1997;272:20055–20062. doi: 10.1074/jbc.272.32.20055. [DOI] [PubMed] [Google Scholar]
- 52.Haase VH. Am J Physiol Renal Physiol. 2006;291:F271–281. doi: 10.1152/ajprenal.00071.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, Chi JT, Jeffrey SS, Giaccia AJ. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 55.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 56.Zbytek B, Peacock DL, Seagroves TN, Slominski A. Dermatoendocrinol. 2013;5:239–251. doi: 10.4161/derm.22678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slominski A, Moellmann G, Kuklinska E, Bomirski A, Pawelek J. J Cell Sci. 1988;89(Pt 3):287–296. doi: 10.1242/jcs.89.3.287. [DOI] [PubMed] [Google Scholar]
- 58.Slominski A, Jastreboff P, Pawelek J. Biosci Rep. 1989;9:579–586. doi: 10.1007/BF01119801. [DOI] [PubMed] [Google Scholar]
- 59.Slominski A, Ermak G, Wortsman J. In Vitro Cell Develop Biol. 1999;35:564–565. doi: 10.1007/s11626-999-0093-6. [DOI] [PubMed] [Google Scholar]
- 60.Shinojima T, Oya M, Takayanagi A, Mizuno R, Shimizu N, Murai M. Carcinogenesis. 2007;28:529–536. doi: 10.1093/carcin/bgl143. [DOI] [PubMed] [Google Scholar]
- 61.Schwab LP, Peacock DL, Majumdar D, Ingels JF, Jensen LC, Smith KD, Cushing RC, Seagroves TN. Breast Cancer Res. 2012;14:R6. doi: 10.1186/bcr3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Janjetovic Z, Brozyna AA, Tuckey RC, Kim TK, Nguyen MN, Jozwicki W, Pfeffer SR, Pfeffer LM, Slominski AT. Br J Cancer. 2011;105:1874–1884. doi: 10.1038/bjc.2011.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen PJ, Huang YS. EMBO J. 2012;31:959–971. doi: 10.1038/emboj.2011.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng Q, Yu X, Xiao L, Hu Z, Luo X, Tao Y, Yang L, Liu X, Chen H, Ding Z, Feng T, Tang Y, Weng X, Gao J, Yi W, Bode AM, Dong Z, Liu J, Cao Y. Cell Death Dis. 2013;4:e804. doi: 10.1038/cddis.2013.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Esteve PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, Jacobsen SE, Pradhan S. Proc Natl Acad Sci U S A. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaber T, Schellmann S, Erekul KB, Fangradt M, Tykwinska K, Hahne M, Maschmeyer P, Wagegg M, Stahn C, Kolar P, Dziurla R, Lohning M, Burmester GR, Buttgereit F. J Immunol. 2011;186:764–774. doi: 10.4049/jimmunol.0903421. [DOI] [PubMed] [Google Scholar]
- 67.Grisouard J, Timper K, Radimerski TM, Frey DM, Peterli R, Kola B, Korbonits M, Herrmann P, Krahenbuhl S, Zulewski H, Keller U, Muller B, Christ-Crain M. Biochem Pharmacol. 2010;80:1736–1745. doi: 10.1016/j.bcp.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 68.Kobayashi M, Fujita I, Itagaki S, Hirano T, Iseki K. Biol Pharm Bull. 2005;28:1197–1201. doi: 10.1248/bpb.28.1197. [DOI] [PubMed] [Google Scholar]
- 69.Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. PLoS One. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lindsey JD, Jones HL, Hewitt EG, Angert M, Weinreb RN. Arch Ophthalmol. 2001;119:853–860. doi: 10.1001/archopht.119.6.853. [DOI] [PubMed] [Google Scholar]
- 71.Liu L, Schlesinger PH, Slack NM, Friedman PA, Blair HC. J Cell Physiol. 2011;226:1702–1712. doi: 10.1002/jcp.22501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murai M, Toyota M, Suzuki H, Satoh A, Sasaki Y, Akino K, Ueno M, Takahashi F, Kusano M, Mita H, Yanagihara K, Endo T, Hinoda Y, Tokino T, Imai K. Clin Cancer Res. 2005;11:1021–1027. [PubMed] [Google Scholar]
- 73.Shi Q, Nguyen AT, Angell Y, Deng D, Na CR, Burgess K, Roberts DD, Brunicardi FC, Templeton NS. Gene Ther. 2010;17:1085–1097. doi: 10.1038/gt.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spriet LL, Tunstall RJ, Watt MJ, Mehan KA, Hargreaves M, Cameron-Smith D. J Appl Physiol. 1985;2004;96:2082–2087. doi: 10.1152/japplphysiol.01318.2003. [DOI] [PubMed] [Google Scholar]
- 75.Joshi MD, Jagannathan V. Methods in enzymology. 1966;9:371–375. [Google Scholar]
- 76.Rutter WJ, Hunsley JR, Groves WE, Calder J, Rajkumar TV, Woodfin BM. Methods in enzymology. 1966;9:479–498. [Google Scholar]
- 77.Stolzenbach F. Methods in enzymology. 1966;9:278–288. [Google Scholar]
- 78.Brozyna AA, Jozwicki W, Janjetovic Z, Slominski AT. Hum Pathol. 2011;42:618–631. doi: 10.1016/j.humpath.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brozyna AA, Jozwicki W, Janjetovic Z, Slominski AT. Hum Pathol. 2013;44:374–387. doi: 10.1016/j.humpath.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tonon L, Touzet H, Varre JS. Nucleic Acids Res. 2010;38:W286–292. doi: 10.1093/nar/gkq473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. Nucleic Acids Res. 2008;36:D102–106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Slominski A. J Cancer Res Clin Oncol. 1985;109:29–37. doi: 10.1007/BF01884251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Slominski A. Biosci Rep. 1983;3:189–194. doi: 10.1007/BF01121951. [DOI] [PubMed] [Google Scholar]
- 85.Seagroves TN, Ryan HE, Lu H, Wouters BG, Knapp M, Thibault P, Laderoute K, Johnson RS. Mol Cell Biol. 2001;21:3436–3444. doi: 10.1128/MCB.21.10.3436-3444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Slominski A, Costantino R. Life Sci. 1991;48:2075–2079. doi: 10.1016/0024-3205(91)90164-7. [DOI] [PubMed] [Google Scholar]
- 87.Slominski A, Costantino R, Howe J, Moellmann G. Anticancer Res. 1991;11:257–262. [PubMed] [Google Scholar]
- 88.Slominski A, Scislowski PW, Bomirski A. Biosci Rep. 1983;3:1027–1034. doi: 10.1007/BF01121029. [DOI] [PubMed] [Google Scholar]
- 89.Slominski A, Costantino R. Experientia. 1991;47:721–724. doi: 10.1007/BF01958826. [DOI] [PubMed] [Google Scholar]
- 90.Kauser S, Thody AJ, Schallreuter KU, Gummer CL, Tobin DJ. J Invest Dermatol. 2004;123:184–195. doi: 10.1111/j.0022-202X.2004.22724.x. [DOI] [PubMed] [Google Scholar]
- 91.Kauser S, Schallreuter KU, Thody AJ, Gummer C, Tobin DJ. J Invest Dermatol. 2003;120:1073–1080. doi: 10.1046/j.1523-1747.2003.12242.x. [DOI] [PubMed] [Google Scholar]
- 92.Slominski A. FEBS Lett. 1991;291:165–168. doi: 10.1016/0014-5793(91)81274-c. [DOI] [PubMed] [Google Scholar]
- 93.Slominski A, Pawelek J. Biosci Rep. 1987;7:949–954. doi: 10.1007/BF01122128. [DOI] [PubMed] [Google Scholar]
- 94.Holick MF. J Cell Biochem. 2003;88:296–307. doi: 10.1002/jcb.10338. [DOI] [PubMed] [Google Scholar]
- 95.Bikle DD. Experimental Dermatology. 2011;20:7–13. doi: 10.1111/j.1600-0625.2010.01202.x. [DOI] [PubMed] [Google Scholar]
- 96.Nosjean O, Ferro M, Coge F, Beauverger P, Henlin JM, Lefoulon F, Fauchere JL, Delagrange P, Canet E, Boutin JA. J Biol Chem. 2000;275:31311–31317. doi: 10.1074/jbc.M005141200. [DOI] [PubMed] [Google Scholar]
- 97.Slominski A, Fischer TW, Zmijewski MA, Wortsman J, Semak I, Zbytek B, Slominski RM, Tobin DJ. Endocrine. 2005;27:137–148. doi: 10.1385/ENDO:27:2:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kuphal S, Winklmeier A, Warnecke C, Bosserhoff AK. Eur J Cancer. 2010;46:1159–1169. doi: 10.1016/j.ejca.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 99.Valencak J, Kittler H, Schmid K, Schreiber M, Raderer M, Gonzalez-Inchaurraga M, Birner P, Pehamberger H. Clin Exp Dermatol. 2009;34:e962–964. doi: 10.1111/j.1365-2230.2009.03706.x. [DOI] [PubMed] [Google Scholar]
- 100.Mills CN, Joshi SS, Niles RM. Mol Cancer. 2009;8:104. doi: 10.1186/1476-4598-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Giatromanolaki A, Sivridis E, Kouskoukis C, Gatter KC, Harris AL, Koukourakis MI. Melanoma Res. 2003;13:493–501. doi: 10.1097/00008390-200310000-00008. [DOI] [PubMed] [Google Scholar]
- 102.Victor N, Ivy A, Jiang BH, Agani FH. Clin Exp Metastasis. 2006;23:87–96. doi: 10.1007/s10585-006-9024-z. [DOI] [PubMed] [Google Scholar]
- 103.Iervolino A, Trisciuoglio D, Ribatti D, Candiloro A, Biroccio A, Zupi G, Del Bufalo D. FASEB J. 2002;16:1453–1455. doi: 10.1096/fj.02-0122fje. [DOI] [PubMed] [Google Scholar]
- 104.Mihic-Probst D, Ikenberg K, Tinguely M, Schraml P, Behnke S, Seifert B, Civenni G, Sommer L, Moch H, Dummer R. PLoS One. 2012;7:e33571. doi: 10.1371/journal.pone.0033571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee JH, Gulec SA, Kyshtoobayeva A, Sim MS, Morton DL. Ann Surg Oncol. 2009;16:2834–2839. doi: 10.1245/s10434-009-0583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Parente P, Coli A, Massi G, Mangoni A, Fabrizi MM, Bigotti G. J Exp Clin Cancer Res. 2008;27:34. doi: 10.1186/1756-9966-27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wachsberger PR, Gressen EL, Bhala A, Bobyock SB, Storck C, Coss RA, Berd D, Leeper DB. Melanoma Res. 2002;12:35–43. doi: 10.1097/00008390-200202000-00006. [DOI] [PubMed] [Google Scholar]
- 108.Park SG, Lee JH, Lee WA, Han KM. Nucl Med Biol. 2012;39:1167–1172. doi: 10.1016/j.nucmedbio.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 109.Yamada K, Brink I, Bisse E, Epting T, Engelhardt R. J Dermatol. 2005;32:316–334. doi: 10.1111/j.1346-8138.2005.tb00903.x. [DOI] [PubMed] [Google Scholar]
- 110.Bomirski A, Slominski A, Bigda J. Cancer Metastasis Rev. 1988;7:95–118. doi: 10.1007/BF00046481. [DOI] [PubMed] [Google Scholar]