Abstract
Melanoma is frequently lethal and its global incidence is steadily increasing. Despite the rapid development of different modes of targeted treatment, durable clinical responses remain elusive. A complete understanding of the molecular mechanisms that drive melanomagenesis is required, both genetic and epigenetic, in order to improve prevention, diagnosis, and treatment. There is increased appreciation of the role of microRNAs (miRNAs) in melanoma biology, including in proliferation, cell cycle, migration, invasion, and immune evasion. Data are also emerging on the role of long non-coding RNAs (lncRNAs), such as SPRY4-IT1, BANCR, and HOTAIR, in melanomagenesis. Here we review the data on the miRNAs and lncRNAs implicated in melanoma biology. An overview of these studies will be useful for providing insights into mechanisms of melanoma development and the miRNAs and lncRNAs that might be useful biomarkers or future therapeutic targets.
Introduction
Melanoma is the leading cause of skin cancer deaths in the United States [1]. Melanoma survival rates are good when the disease is detected early; precise diagnostic tests for early melanoma detection would therefore be useful, and innovative therapies to cure advanced melanomas are needed. The underlying molecular biology of melanomas is complex and involves interactions between networks of genes, signaling pathways, and gene-regulatory mechanisms, and a better understanding of these underlying molecular mechanisms is essential for translational research. In addition, the histopathologic interpretation of cutaneous melanoma remains one of the most frustrating and difficult diagnostic areas in dermatopathology, and histopathologists would benefit from sensitive and specific diagnostic biomarkers. A number of protein-coding genes [2] have been identified as potential diagnostic and prognostic biomarkers [3-9], several of which exhibit distinct expression profiles between the spectrum of malignant melanomas and their benign forms [2]. In addition, non-protein-coding RNAs (microRNAs and long non-coding RNAs (lncRNAs))are emerging as early prognostic markers and therapeutic targets in a variety of diseases, and microRNAs (miRNAs) is particular have gained increasing attention due to their potential roles in tumorigenesis [10-14], not least in melanoma [15-19]. miRNAs are thought to influence cancer development by regulating transcription and translation of tumor suppressor genes and oncogenes [20-26]. Several genome-wide expression studies have implicated a number of miRNAs and lncRNAs that are potentially important regulators of melanoma development [9, 19, 27, 28].
Melanocytes are skin cells that originate from neural crest cells and have the ability to produce the pigment melanin [1]. Melanocyte differentiation occurs via a series of steps, ultimately resulting in lineage specification of melanoblasts and transportation of mature melanosomes to keratinocytes [29, 30]. Melanocytes are characterized by the expression of melanocyte-specific proteins, including tyrosinase, tyrosinase-related protein 1 and 2, melanosomal matrix proteins (Pmel17, MART-1), and microphthalmia transcription factor (MITF) [1]. Genes such as MITF, [31] PAX3, SOX10 [32-34], members of the Wnt and Notch signaling pathways [35-37], KIT, and cyclins [38] all play an important role in the development and regulation of melanocytes.
Melanogenesis is a stepwise metamorphic process in which normal melanocytes in the epidermis gradually transform into the vertical growth phase characteristic of malignant melanomas [39]. Several factors influence the transformation of melanocytes into melanomas, such as UV exposure [40], melanocyte integrity [41], melanocyte homeostatic mechanisms [42], and neural crest invasion and differentiation [43, 44]. In addition to the many protein-coding genes that regulate cancer development, many non-coding genes have also been shown to play important roles in cancer prognosis, diagnosis, and therapy. These include the small RNAs, in particular miRNAs and lncRNAs. Due to wide spread increase and mortality of melanoma globally, it is important to discuss ways and means for the insight mechanism of transformation of melanocytes into melanoma. On the basis of basic information, many methods have been proposed for the prognosis and treatment of melanoma. In this review, we have discussed the possible role of miRNAs in the pathology, diagnosis, and treatment of melanoma. Although role of lncRNAs in melanoma is not fully established, still lncRNAs were given due consideration for their involvement in melanoma.
miRNAs in melanocytes and melanoma biology
miRNAs are small, non-coding RNAs that play a physiological role in the post-transcriptional fine-tuning of the expression of up to 60% of mammalian protein-coding genes [45, 46]. The aberrant expression and function of miRNAs has been linked to the development and progression of many human diseases, including various cancers [10-14], not least melanoma [15-19]. As a result of systematic experimental screens for miRNAs involved in the development and progression of melanoma, several groups have identified miR-211 as the miRNA most differentially expressed between normal melanocytes and non-pigmented melanoma cell lines and primary melanomas from patients [47-52]. Ectopic expression of miR-211 in melanoma cell lines results in significant inhibition of growth and invasion compared to parental cells, suggesting that miR-211 normally functions as a tumor suppressor in melanocytes. This hypothesis is supported by the finding that miR-211 is encoded by a region in the sixth intron of TRPM1 (transient receptor potential cation channel subfamily M member 1), a candidate suppressor of melanoma metastasis [47, 48]. Moreover, we have also reported that the expression of TRPM1 and miR-211 are controlled by MITF, a master regulator of melanocyte development and function. It is therefore possible that the tumor suppressor activities of MITF and/or TRPM1may be mediated, at least in part, by miR-211. Recently, several miR-211 target genes have been identified, including Runt-related transcription factor 2 (RUNX2), insulin-like growth factor 2 receptor (IGF2R), TGF-beta receptor 2 (TGFBR2), the POU domain-containing transcription factor BRN2, and nuclear factor of activated T cells 5 (NFAT5) [48, 53].
miR-211 may also directly regulate melanocyte pigmentation and invasion, since it is highly expressed in melanocytes and pigmented melanomas but not in non-pigmented melanomas (Mazar et al, personal communication). Melanomas with greatly reduced miR-211 expression are highly invasive [47, 54, 55]. Conversely, melanoma cells that highly express miR-211 have reduced invasive potential [52], independent of expression of melastatin that was able to block formation of tumor nodules [56]. Together, these findings provide strong evidence that miR-211 plays a critical role in melanoma invasiveness and progression.
In an attempt to explain the mechanistic basis for these findings, a melanoma-specific metastasis gene network was scrutinized for overlaps between metastatic genes and miR-211 target genes [54]. Six genes overlapped: IGF2R, NFAT5, TGFBR2, FBXW7, ANGPT1, IGFBPS and VHL. Functional validation showed that knockdown of ‘central node genes’ had the same effect on melanoma cell invasion as up-regulation of miR-211. Of these, TGFB had previously been linked to melanoma progression via promotion of tissue and blood vessel invasion. More recently, Bell et al. [52] tried to establish which relationships between transcription factors and miRNAs were important for melanoma proliferation and invasion using gene expression profiling of normal and melanoma cells. Several miRNAs known to regulate proliferation and invasion were identified, including miR-211. A new miR-211 target, NUAK1, was also identified, which was shown to play a role in melanoma cell adhesion with downregulation of miR-211upregulatingNUAK1 and promoting adhesion, and vice versa.
miR-196a has also been shown to act as a tumor suppressor miRNA in melanoma [57]. Using a high-throughput miRNA expression profiling approach in cell lines and tissue samples, miR-196a expression was found to be significantly reduced in malignant lesions. Over-expression of miR-196a significantly reduced the invasive capacity of melanoma cells, and HOX-C8, cadherin-11, calponin-1, and osteopontin were identified asmiR-196a targets. These authors then went on to show that miR-196a downregulation led to upregulation of HOX-B7 and consequent stimulation of basic fibroblast growth factor (bFGF) signaling, with resulting ETS-1 transcription factor and bone morphogenetic protein 4 (BMP-4) expression, which is known play a role in melanoma progression [58]. Using similar methodology, Chen et al. showed that miR-193b also acts as a tumor suppressor via cyclin D1, and it was the most downregulated of 31 differentially expressed miRNAs in malignant tissue samples.
It has been shown that miRNA regulatory effect on their targets is directly correlated to mRNA decay [59, 60]. mRNA decay rates in animal cells changes rapidly and with half-lives varying from minutes to days [61]. For example, for mRNAs stability in mouse embryonic stem cells the median is around 7 h, whereas some genes, including Foxa2, Hes5 and Trib1, have halflives under an hour [62]. In a recent study, Larsson et al, [59] reported that the short-lived transcripts are more difficult to perturb using microRNAs. Therefore, it is important to take this (mRNA stability/decay rate) in to account when designing a miRNA perturbation study.
The role of miRNAs in the immune response
Hypoxia influences the microenvironment of solid tumors, including in melanoma. One effect of hypoxia in melanoma is that it is thought to facilitate escape from immune control and promote cancer via downregulation of antigens and proteins that are necessary for an effective immune response [134, 135].
Hypoxia is known to stimulate expression of several miRNAs. miR-210 is regulatedduringHIF1-α-dependent hypoxia in non-small cell lung cancer (IGR-Heu) and melanoma cell lines (NA-8) [124]. Reduced expression of miR-210 in melanoma cells facilitates cell lysis by antigen-specific cytotoxic T lymphocytes (CTLs). At a gene regulatory level,PTPN1, HOXA1, and TP53I11 are target genes of miR-210 and are thought to mediate the immunosuppressive response and have been shown to be involved in immune regulation and tumor initiation [125-129]. Another microRNA, miR-34a/c, has been reported to regulate innate immune responses in melanoma cells [109]. miR-34a/c control the expression of ULBP2,which is a ligand for natural killer cell immunoreceptor (NKG2D). NKG2D usually detects early tumors, eliminates cytotoxic lymphocytes, and provides an innate barrier to tumor development; removal of ligand protects malignant cells from NKG2D-mediated immune surveillance [130, 131]and overexpression of miR-34 significantly downregulates ULBP2 expression. In another study, ectopic expression of miR-30b and miR-30 enhanced melanoma metastasis by creating an immunosuppressive environment [113] via GALNT7 and increased synthesis of immunosuppressive molecules such asIL-10, and consequent reduced immune cell activation and recruitment. GALNT7 is a glycosylating protein, and changes in glycosylation patterns are believed to be associated with tumor progression [133]. Similarly, reduced GALANT7 expression levels alter O-glycans which leads to tumor growth and survival and interaction with immune cells [132].
The role of miRNAs in melanoma cell cycle and cell proliferation
Uncontrolled cellular proliferation is a hallmark of cancer. The cell cycle is principally controlled and regulated by cyclin-dependent kinases (CDKs) and the E2F transcription factor [136, 137]. Other proteins, such asc-myc, p27, and PTEN, activate CDKs and in turn regulate the cell cycle [138-140]. One can postulate that miRNAs that regulate cell proliferation might directly target these cell cycle regulators. The NF-kB pathway is also known to be associated with proliferation in melanoma [141], and it is also well-documented that cyclin D1,cyclinD3, and CDK4 play an important role in tumor progression [63-65, 142].
let-7b is an miRNA known to target cell cycle regulators. Acting as a tumor suppressor, increased let-7b expression results in significantly decreased proliferation [18] via reduced gene and protein expression of CDK4, cyclin D1, and cyclin D3[49]. miR-193b targets cyclin D1 (CCND1) and is significantly downregulated in melanocytic nevi [66]. Increased expression of miR-193b in melanoma cell lines (Malme-3M cells) significantly reduces CCND1 gene and protein expression and inhibits proliferation of melanoma cells.
Elevated expression of miR-145 in canine and human melanoma cell lines significantly reduces proliferation [143] via targeting of c-MYC [143] or Erbb3 [144]. It has been also recently been shown that miR-206 is significantly downregulated in melanoma cells and reduces cell growth and migration in many melanoma cell lines by targeting cell cycle proteins including CDK4, cyclin D1, and cyclin C [70].
Some microRNAs indirectly control the cell cycle via p27 [76], a tumor suppressor protein that binds to, and inhibits the function of, the cyclin D1-CDK4 complex and in doing so acts as a key regulator of the G1-S cell cycle transition; indeed, reduced expression of p27 is one of the main causes of uncontrolled cell-cycle progression into S phase [145]. Two studies have shown that miR-221/222 directly target p27 and is involved in cell cycle regulation [15, 17]; increased proliferation was observed in melanoma cells overexpressing miR-221/222, with the opposite true both in vitro and in vivo [15, 17]. Furthermore, the same studies show that miR-221/222 target the oncogenic tyrosine kinase receptor c-kit [74, 75]. Garofalo et al. [71] showed that miR-221/222 targets the PTEN and TIMP3 tumor suppressors [72, 73], induces TRAILmediated resistance, and enhances migration via activation of the AKT/PI3K signaling pathway and metallopeptidases.
Other miRNAs, includingmiR-205, miR-149, miR-18b, miR-21, miR-203, and miR-26a, also regulate cell cycle proteins but in a cyclin-independent manner. Dar et al. [77] showed that in primary melanomas, the expression of miR-205 was significantly reduced and inversely correlated with melanoma progression. Overexpression of miR-205 significantly reduced melanoma cell proliferation and colony formation in vitro and tumor growth in vivo, and induced a senescent phenotype [77] and apoptosis via the E2F1 transcription factor [146]. miR-205 targets the E2F1 and E2F5 transcription factors known to play a critical role in malignant melanoma [78, 79]. Jiang et al. [83] studied 106 primary melanomas and metastases and showed that inhibition of miR-21 was significantly associated with increased apoptosis and growth of human cutaneous melanoma, via inhibition of PTEN, Akt phosphorylation, Bax upregulation, and Bcl-2 inhibition. Stazger et al. [84] similarly determined that miR-21 is upregulated in primary melanomas and melanoma cell lines, and miR-21 knockdown in melanoma cell lines induced apoptosis, but not proliferation. miRNA-203 plays a crucial role in cell cycle arrest, and has been shown to be expressed at reduced levels inhuman and canine malignant melanoma cells and inhibits cell cycle arrest and the senescence phenotype. miR-203 might induced cell cycle arrest and senescence via an E2F3-dependent mechanism [85], kinesin superfamily protein 5b (kif5b), or the MITF/Rab27a pathway, which is one of the main pathways active in melanoma cells [67]. miR-195 was shown to be downregulated in primary melanomas and to target WEE1 [89]. Ectopic expression of miR-195 in melanoma cell lines reduced WEE1 expression and stress-induced G2-M cell cycle arrest and increased proliferation. A few other studies have demonstrated a role for miR-786-3p, miR-214, miR-15b, miR-155,and miR-126 in cellular proliferation;miR-786-3p has been shown to regulate proliferation and apoptosis by targeting the eukaryotic translation initiation factor 4 (eIF4E) in melanoma cells [90], overexpression ofmiR-214 targets the adhesion receptor ITGA3 and reduces cell death[91],and downregulation of miR-15breduced proliferation and increased apoptosis [93].
Two miRNAs have been shown to regulate the cell cycle by targeting p53. miR-18b suppresses proliferation and overexpression leads to suppression of the proto-oncogene MDM2 [98], activation of the p53 pathway, and reduced survival of melanoma cells [97]. miR-149 has been shown to effect cell cycle via a p53-mediated mechanism; p53 regulates miR-149, which in turn targets glycogen synthase kinase-3α (GSK3α) [100], increased expression ofMcl-1, and ultimately resistance to apoptosis [99]. Greenberg et al. [80] sought to identify those miRNAs that regulate the aggressive phenotype of melanoma using a comparative high-throughput miRNA profiling approach; two isogenic human melanoma cell lines were molecularly and phenotypically profiled and shown to display major differences in their capacity for proliferation, invasion, and tube formation.
miR-9 has been shown to be downregulated in metastatic melanomas compared to primary melanomas [81], and overexpression of miR-9 in melanoma cells resulted in significantly decreased proliferation and migration via an NF-kappaB1-dependent mechanism. Levati et al. demonstrated that ectopic expression of miR-155 significantly inhibits proliferation and induces apoptosis in four melanoma cell lines [101]. miR-126 is also thought to play a vital tumor suppressor role in human melanoma; Felli et al. [94] showed that miR-126 & 126* expression were downregulated during melanoma progression, and two metalloproteases, metalloprotease domain 9 (ADAM9) and metalloprotease 7 (MMP7), were identified as direct targets of miR-126 & 126*, both of which are thought to be involved in melanoma progression [95, 96].
The role of miRNAs in melanoma cell invasion
Many factors are known to drive melanoma cell migration and invasion, including FSCN1, BSG, β3-integrin, MARKS, GALANT7, c-MET, and NFkB [104-106, 108, 110, 111]; a number of miRNAs are thought to target these proteins and pathways. miR-145is thought to downregulate fascin actin-bundling protein 1 (FSCN1),a known regulator of cell migration [103, 104]. Segura et al. [28]investigated the role of miR-182 in melanoma cell invasion and showed that miRNA genes are located in genomic regions that harbor frequent gains and losses in primary melanomas [3, 28]. miR-182 was differentially expressed in melanoma cell lines compared to benign melanocytes, and FOXO3 and MITF were shown to be direct targets of miR-182 [28] and contribute in the migratory and invasive potential of the SK-MEL melanoma cell line.
In a mouse model of liver metastasis, immunocompromised mice received intra-splenic injections of A375 melanoma cells and subsequently intra peritoneal injections of chemically modified anti-miR-182 or negative control anti-miRNA [112]. Treatment of mice with anti-miR-182 resulted in significantly fewer liver metastases compared to controls. Similar findings were observed when mice were pre-treated with three doses of anti-miR-182 oligonucleotides followed by three weeks of miRNA-182. Suppression of miR-182 gene expression in liver tissues was verified by real-time PCR and was accompanied by upregulation of the miR-182 targets ADCY6 and FOXO3. Moreover, mRNA expression profiles of anti-miR-182-treated tumors differed from those of controls, supporting the notion that anti-miR-182 has a transcriptional impact on gene expression. Differentially expressed genes included genes involved in cell adhesion, migration, and apoptosis; upregulated genes after anti-miR-182 treatment included NFASC, CASP2, NCAM1, and CLDN17. CASP2, already identified as a miR-182 target [28], is a member of the caspase family of pro-apoptotic genes. Overall, the treatment of mice was well tolerated with no gross abnormalities and only slight derangement of liver function. The authors concluded that miR-182 targeting might be a promising therapeutic strategy for metastatic melanoma.
Gaziel-Sovran et al. [113] demonstrated that the invasive capability of two melanoma cell lines (113/6-4L and 131/4-5B1) was mediated by expression of miR-30b and miR-30d, which regulated invasion but not proliferation. The miR-30b/30d cluster is located near to c-MYC (8q24.21), an archetypal oncogene. Global transcriptomic analysis showed differential expression of 784 genes, with 58 genes downregulated in both cell lines. The most highly downregulated targets were GALNT1, GALNT7, and SEMA3A. Microarray analysis further revealed that ectopic expression of miR-30dupregulated the expression of immune modulators, such as IL-10, via repression of GALNT7 by miR-30d.
It has been shown that let-7b targets basigin (BSG), an invasion-associated protein [106]. Reduced let-7b expression in melanoma cells leads to increased metastases due to enhanced expression of BSG and consequently enhanced expression of extracellular matrix metalloproteinases (MMPs). Overexpression of let-7b resulted in reduced BSG and MMP-9 protein expression and reduced distant metastases [147]. let-7a targets ITGB3, which is known to increase the invasiveness of melanoma cells [105], and increased ITGB3 expression due to depletion of let-7ahas been shown to increase the invasive potential of melanoma cells[148]. Schwab et al. [107] showed that reduced expression of miR-200 family members promotes invasion and metastasis by adapting the cell to different local microenvironments via two interconvertible modes of invasion: elongated mesenchymal-type and rounded amoeboid-like. These morphological changes were phenocopied by reducing expression of the miR-200c target, MARCKS, which is involved in the formation of cell protrusions [108].
The expression of c-MET is associated with cell invasion and inhibition of apoptosis; it is also a prognostic factor in clinical studies [149, 150]. miR-199 and miR-34b/c have been shown to target MET and decrease its mRNA and protein expression, and inhibition of miR-199 and miR-34b/c increase expression of MET and increased cell adhesion and migration [151]. miR-137 has also been shown to target c-Metand YB1, with functional studies indicating that miR-137 suppresses melanoma cell invasion and vice versa [152].
Role of miRNAs in apoptosis
Many studies have highlighted the role of microRNAs in apoptosis, including miR-26a, miR-155, miR-205, miR-768-3p, miR-21, miR-15b, miR-126, the miR-506-514 cluster and miR-149. Reuland et al. [117] showed that miR-26a was downregulated in melanoma cell lines compared to primary melanocytes, and overexpression of miR-26a caused significant and rapid cell death and repressed expression of SODD [118], which protects melanoma cells from apoptosis. miR-155 was also shown to be down regulated in melanoma cells compared to normal melanocytes [101, 116], and ectopic expression of miR-155 significantly inhibited proliferation and induced apoptosis by targeting SKI, a transcriptional coregulator; downregulation of SKI in vitro inhibited melanoma cell growth [102]. miR-205 expression was reduced in melanoma cells compared to nevi [77], with further analysis showing that miR-205 targets and reduces expression of E2F1, thereby decreasing proliferation and inducing apoptosis or, as shown in [78], via its capacity to activate p73 family members in advanced malignant melanomas.
Role of miRNAs in melanoma epigenetics
Epigenetics refers to the biological process by which changes in phenotype or gene expression occur without changes to the DNA sequence. Cellular epigenetic events occur at greater frequency than mutations, may be sustained during the life of the cell, and can be transmitted to progeny [153].
Our group has conducted the most comprehensive characterization of the role of epigenetics in melanoma development. Using next generation sequencing (NGS), were ported on a global analysis of methylation events occurring in melanoma cell lines and melanocytes [119]. The CpG islands in the upstream regulatory regions of many coding and non-coding RNA genes, such as TERC, were hypermethylated, while repeated elements, such as LINE-2 and LTR, showed widespread hypomethylation in advanced stage melanoma cell lines; these results are currently being validated in human patient samples. We demonstrated that miR-375 is epigenetically regulated in melanoma cells and patient samples [115]; CpG island methylation regulated expression of miR-375 in WM1552C melanoma cells after treatment with the demethylating agents, 5-aza-2-deoxycytidine and 4-phenyl-butyrate (4-PBA). Methylation of miR-375 CpG islands was stage dependent and was significantly greater in stage II and III melanoma cell lines compared to stage I melanoma lines or benign melanocytes. In a third study, regulation of miR-34b by CpG island methylation was investigated, and increased methylation was apparent in stage III and IV melanoma cells compared to stage I and II melanoma cells, melanocytes, and keratinocytes [120].
miR-182 has been shown to be overexpressed in human melanoma cells after epigenetic modulation, and CpG islands upstream of mature miR-182 were hypermethylated in melanoma cells [121]. Similar work was carried out by Lodygin et al, [154] which demonstrated that expression of miR-34a, target of the tumor suppressor gene p53, was highly reduced in various cancers due to aberrant CpG methylation of miR-34a promoter. Apart of other cancers, CpG methylation of miR-34a promoter was detected in melanoma cell lines (19/44; 43.2%) and primary melanoma (20/32 samples; 62.5%). Expression of miR-137 was much lower in uveal melanoma than in uveal melanocytes [122], with ectopic expression of miR-137 reducing cell growth and significantly reducing expression of MITF, c-Met, and CDK6. From an epigenetic perspective, miR-137 expression was significantly increased with treatment with 5-aza-2-deoxycytidine and trichostatin A, indicating a potential role in epigenetic regulation. It has also been shown that miR-29 downregulates the DNA methyltransferases, DNMT3A and DNMT3B, which have been reported to be essential for methylation of the promoter region of tumor-related genes [123]. High expression of miR-29c was inversely correlated to both DNMT3A and DNMT3B protein expression in melanomas, and the authors concluded that both miR-29c and DNMT3B play an important role in melanoma progression and may be useful as epigenetic biomarkers. Another microRNA miR-31 was observed to be down regulated in melanoma tumor and cell lines. The authors of the study put forward three possible reasons for the reduced expression of miR-31. One reason was the genomic loss in a subset of samples, second reason was the epigenetic silencing by DNA methylation and third reason was EZH2-mediated histone methylation [155]. Dar et al, [97] targeted the MDM2-p53 pathway to investigate the biological role of miR-18b in melanoma. They demonstrated that expression of miR-18b was substantially reduced in melanoma specimens and cell lines due to hypermethylation and was reinduced in melanoma cell lines after 5-AZA-deoxycytidine treatment by 1.5 to 5.3-fold.
miRNAs as biomarkers for the diagnosis and prognosis of patients with melanoma
There is promise that measuring the expression of circulating miRNAs could be used to diagnose and determine the prognosis of a number of cancers [156-159]. Similarly, in melanoma patients, circulating miRNAs in the serum have been proposed as potential biomarkers. Recently, Shiiyama et al. [160] examined the expression profiles of 2000 miRNAs in the serum of melanoma patients and identified six microRNAs (miR-9, miR-145, miR-150, miR-155, miR-203, and miR-205) that were differentially expressed in metastatic melanoma patients compared to normal individuals. Similarly, in another study, five miRNAs, miR-150, miR-15b, miR-339, miR-199a-5p, and miR-424 were highly and differentially expressed in the serum of melanoma patients with a high risk of recurrence. The diagnostic accuracy of identification of primary melanomas using serum miRNA measurements has also been tested by screening 900 human miRNAs in 24 blood samples from melanoma patients and 20 samples from normal individuals [161]; 51 miRNAs showed dysregulated expression in the serum of melanoma patients [162]. One study focused on miR-221, and found significantly upregulated expression of miR-221 in malignant melanoma patients compared to healthy individuals [163]. miRNA transcriptomic analysis of tissue biopsies of nevi, thick primary melanomas, and metastatic melanomas were analyzed using the Illumina NGS platform, and again miR-211 was identified as having reduced expression in melanomas vs. nevi [164].
miRNA biomarker profiling has also been attempted in tissue samples of melanomas resected from patients for diagnosis purposes, including from formalin-fixed paraffin-embedded (FFPE) samples commonly used in diagnostic pathology laboratories. These studies identified a number of miRNAs that may be suitable diagnostic biomarkers for melanoma [165-167].
The utility of miRNAs as prognostic biomarkers in melanoma has also been investigated in a number of studies. In a pioneering study, the metastases in patients with uveal melanoma were investigated through microarray analysis and classified into low and high metastatic risk groups. The most significant expression was observed for let-7b and miR-199a and may be utilized as biomarker for metastatic risk in uveal melanoma [168]. Stazger et al. [93] studied 128 primary melanomas and identified differential expression of three miRNAs, miR-15b, miR-210, and miR-34a, the first two being upregulated and the latter downregulated in melanomas compared with melanocytic nevi; only miR-15b expression was associated with the poor recurrence-free survival. Xu et al. [49] revealed that out of 20 miRNAs that exhibited differential expression in primary or metastatic melanomas in comparison to benign nevi, only two miRNAs, miR-203 and miR-205, were significantly upregulated in malignant lesions and could be used as a biomarker for prognostic or diagnostic purposes.
Two recently published reports have focused on NGS for the identification of miRNAs with potential use as diagnostic biomarkers in patients with melanoma. In the first study of 698 known miRNAs, seven miRNAs (miR-203, miR-204-5p, miR-205-5p, miR-211-5p, miR-23b-3p, miR-26a-5p, and miR-26b-5p) showed decreased expression in melanomas vs. nevi [164]. In the second study[169], four differentially expressed miRNAs (hsa-miR-146, hsa-miR-27, hsa-miR-877, and hsa-miR-186) were detected in metastatic melanomas and primary cutaneous melanoma samples [169]. The miRNAs that have been investigated as biomarkers are shown in Table 2.
Table 2. The miRNAs involved in prognosis, diagnosis, and therapeutics.
| Type of biomarker | Name | Expression | Year | References | |
|---|---|---|---|---|---|
| Prognostic anddiagnosticmiRNAs | Circulating miRNAs |
miR-9, miR-145, miR-150 miR-155, miR-203, miR-205 |
Upregulation | 2013 | [160] |
|
miR-150, miR-15b, miR-339 miR-199a-5p and miR-424 |
Upregulation | 2012 | [161] | ||
| Identification of 51 miRNAs | Dysregulation | 2010 | [162] | ||
| miR-221 | Upregulation | 2011 | [163] | ||
| miRNAs in solid tumors | Identification of 84 miRNAs | 2009 | [165] | ||
| 2009 | [166] | ||||
| Identification of 70 miRNAs | Dysregulation | 2009 | [167] | ||
|
let-7b miR-199 |
Downregulation Upregulation |
2008 | [168] | ||
| miR-203, miR-204-5p, miR-205-5p, miR-211-5p, miR-23b-3p, miR-26a-5p and miR-26b-5p | Downregulation | 2013 | [164] | ||
|
miR-146, miR-27 miR-877 miR-186 |
2014 | [169] | |||
|
miR-203 miR-205 |
Upregulation Upregulation |
2012 | [49] | ||
|
miR-15b miR-210 miR-34a |
Upregulation Upregulation Downregulation |
2010 | [93] | ||
| Therapeutic miRNAs | Identification of 15 miRNAs | Upregulation | 2013 | [170] | |
| miR-26a | Downregulation | 2013 | [117] |
miRNAs as therapeutic agents
Recently, the focus of miRNAs in melanoma in translational research has shifted towards their use as therapeutic agents. Reuland et al. [117] identified miR-26a as significantly downregulated in human melanoma cells and established that the silence of death domain (SODD) may act as a novel target site for miR26a that mediates melanoma cell death. miR-26a may therefore serve as potential therapeutic molecule in the treatment of melanoma.
Wagenseller et al. [170] studied global miRNA expression profiles in melanoma tissues using microarrays in patients treated with anticancer drug and angiogenesis inhibitors, temsirolimus and bevacizumab [171, 172]; there was significant upregulation of 15 miRNAs in treated vs. non-treated tissues, 12 of which having tumor suppressor function via the targeting of 15 oncogenes. These miRNAs are therefore attractive candidates for further investigation as therapeutic agents.
The role of lncRNAs in melanoma
lncRNAs are very similar to mRNAs and are mainly regulated by RNA polymerase II. Like mRNAs, they are spliced, capped, and polyadenylated, but they do not have protein-coding capacity. The first functional lncRNA, XIST, was discovered in the early 1990s [173-175]; until then, lncRNAs were generally regarded as transcriptional noise or junk sequences. Recent studies have illustrated several important functions of lncRNAs in many biological processes, including regulation of gene expression, dosage compensation, genomic imprinting, nuclear organization and compartmentalization, and nuclear to cytoplasmic trafficking [176, 177], and more functions are likely to be discovered in the future.
In contrast to miRNAs, there is a paucity of literature discussing the role of specific lncRNAs in melanomas. In the seminal lncRNA melanoma study by Khaitan et al. [178], 77 lncRNAs were identified as significantly dysregulated in the melanoma cell line WM1552C and in primary melanomas from patients. A more detailed study was carried out on SPRY4-IT1, an intronic lncRNA of the SPRY4 gene, and knockdown of SPRY4-IT1 in the melanoma cell lines A375 and WM1552C resulted in significant cell death, invasion, and induction of apoptosis. Dysregulation of SPRY4-IT1 may therefore have an important role in melanomagenesis, be used as an early biomarker, and be a key regulator for melanoma pathogenesis in humans. This study formed the basis for investigation of various other lncRNAs in melanoma [179, 180] and stimulated interest in the role of SPRY4-IT1 in other cancers [181]. Follow-up studies have detailed the mechanistic role of SPRY4-IT1 in melanoma cells and shown that ectopic expression of SPRY4-IT1 in normal human melanocytes forms multinuclear, multidendrite structures [182] .
In another study, the lncRNA HOTAIR was shown to be overexpressed in metastatic tumor in lymph nodes compared to matched primary melanomas [180]. Knockdown of HOTAIR suppressed metastases and resulted in a three-fold reduction in the invasiveness of the melanoma cell line A375. A wound healing assay indicated better healing in cells transfected with control constructs than those transfected with HOTAIR in A375 cells [180]. Suppression of matrix degradation using HOTAIR knockdown upregulated the MMPs required for metastasis [183], which was confirmed by elevated gelatinase activity.
Wu et al. [184] showed that aberrant expression and increased binding of the lncRNA Llme23 to polypyrimidine tract-binding protein-associated splicing factor (PSF) was associated with mouse and human tumors through repression of proto-oncogene Rab23. Llme23 knockdown suppressed the malignant properties of the human melanoma cell line YUSAC and repressed expression of the proto-oncogene Rab23 [185].
In one study, 39 differentially regulated lncRNAs were identified by Flockhart et al. [179] in BRAF V600E melanomas cells, and the BANCR lncRNA was found to be overexpressed and associated with malignant melanoma. BANCR knockdown reduced melanoma cell migration by upregulating the chemokine CXCL11, a mediator of cell migration. This elegant study was an important discovery of an oncogene-regulated lncRNA transcript of potential clinical relevance in cancer.
Pasmant et al. identified single nucleotide polymorphisms (SNPs) on chromosome 9p21 that were associated with melanoma and other diseases through the allelic expression of the lncRNA ANRIL [186] . Expression studies have confirmed the coregulation of p15/CDKN2B, p16/CDKN2A, p14/ARF, and ANRIL. ANRIL in involved in regulation of CDKN2A/B expression through a cis-acting mechanism and its implication in proliferation and senescence. Modulation in ANRIL expression mediated susceptibility to several important human diseases, including melanoma. Similarly, genetic aberrations of the GAS5 locus have been found in several tumors, including melanoma, breast, and prostate cancers [187]. A list of all the lncRNAs currently implicated in melanoma is shown in Table 3.
Table 3. List of lncRNAs currently implicated in melanoma.
| Name | Cell utilized | Expression | Type of lncRNA | Role of lncRNA | Year of reporting | References |
|---|---|---|---|---|---|---|
| SPRY4-IT1 | WM1552C | Upregulated | Antisense long non-coding RNA | Regulation of cells viability apoptosis and melanoma cell motility | 2011, 2014 | [178] [182] |
| BANCR | Upregulated | lincRNA | Migratory capacity of melanoma cells by regulating CXC11 | 2012 | [179] | |
| HOTAIR | lincRNA | Motility, invasion, and metastasic potential of metastatic melanoma | 2013 | [180] | ||
| LlME23 | YUSAC melanoma cell line | Upregulated | lincRNA | Regulate process of melanoma through binding of PSF | 2013 | [184] |
| TUG1 | lincRNA | --- | 2013 | [188](Article withdrawn) | ||
| ANRIL | Human blood cells | Upregulated | Antisense long non-coding RNA | Risk of melanoma due to SNPs in 9P21 region | 2011 | [186] |
| GAS5 | ----- | ----- | Malignant melanomas due tospecial break points at 1p36 and at several sites throughout 1p22-q21 | 2000 | [187] |
Conclusions
Although many drugs have been introduced for the treatment of melanoma, clinical outcomes for metastatic disease remain poor and resistance to therapy is common. An increased understanding of the molecular and cellular biology of non-coding RNAs shows promise for the diagnosis, prognosis, and treatment of patients with melanoma. Although there is a lot of data relating to miRNAs in melanoma, more work is required to develop sensitive and specific molecular tests. To achieve this goal, there needs to be focused attention on the early diagnosis of melanoma recurrence, which is devastating for the patient. While established imaging techniques, such as MRI, PET, and CT are being used to detect early stage cancer, they lack the sensitivity required for very early tumor detection [189], in contrast to molecular-based techniques.
lncRNAs might also be used as biomarkers for the early diagnosis of melanomas. An optimal approach might be by using a combination of miRNAs and lncRNAs for the diagnosis and treatment of melanoma. The combinatorial approach will focus on protein genes that are generally oncogenes and are involved in melanoma progression and are generally targeted by miRNAs as well as role on lncRNAs which are beginning to become major molecules in cancer detection and treatment especially melanoma. Our group has performed extensive research on the early detection of melanoma using both miRNAs and lncRNAs, and the combination of these approaches might offer a solution for achieving this goal in the near future.
Figure 1. Schematic representation of the miRNAs involved in proliferation, invasion, apoptosis, and the immune response.
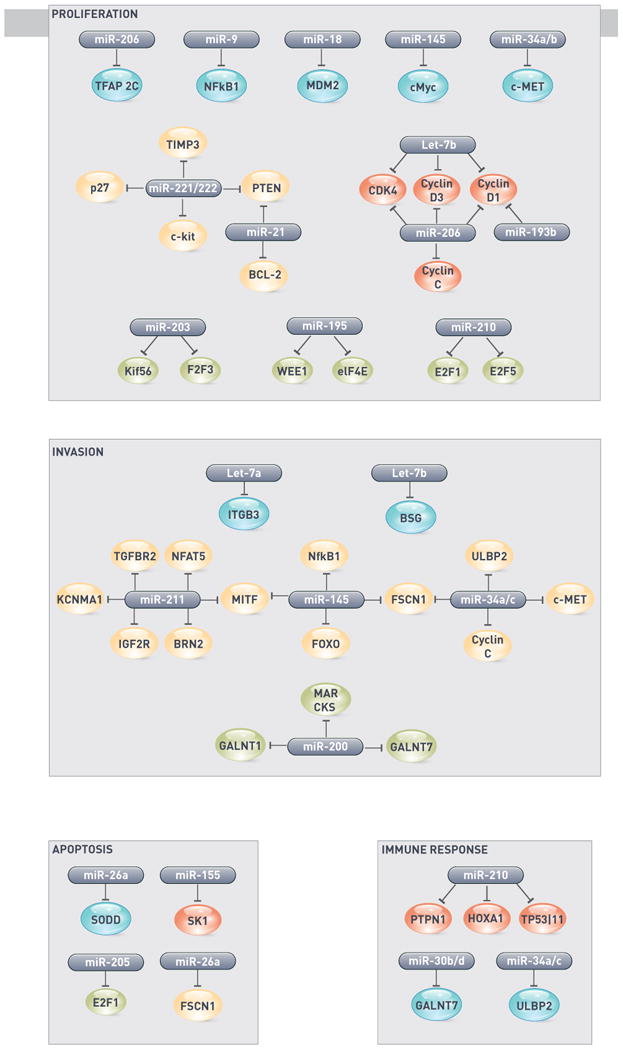
Table 1. The miRNAs involved in melanomagenesis.
| Name of miRNA | Expression of miRNAs | Year of reporting | Reference(s) indicating specific role of miRNAs in melanoma | Target | Reference(s) indicating specific role of targeted protein in melanoma | |
|---|---|---|---|---|---|---|
| miRNAs involved in proliferation and cell cycle | let-7b | Down | 2008 | [18] | Cyclin D1 Cyclin D3 CDK4 |
[63] [64] [65] |
| miR-193b | Down | 2010 | [66] | Cyclin D1 | [63] | |
| miR-145 | Down | 2014 | [67] | c-MYC | ||
| MITF | [68, 69] | |||||
| miR-206 | Down | 2014 | [70] | CDK4 Cyclin D1 Cyclin C |
[63] | |
| miR-221/222 | Up | 2009 2009 2008 |
[71][15][17] | PTEN TIMP3 c-KIT p27 c-KIT p27 |
[72] [73] [74, 75] [76] [74, 75] [76] |
|
| miR-205 | Down | 2011 | [77] | E2F1 E2F5 |
[78] [79] |
|
| miR-34a/c | Down | 2011 | [80] | c-MET | ||
| miR-9 | Down | 2012 | [81] | NFkB1 | [82] | |
| miR-21 | Up | 2012 2012 |
[83][84] | PTEN BCL-2 |
[72] | |
| miR-203 | Down | 2012 | [85] | F2F3 Kif5b |
[86][87] | |
| miR-195 | Down | 2013 | [88] | WEE1 | [89] | |
| miR-786-3p | 2013 | [90] | eIF4E | |||
| miR-214 | 2011 | [91] | TFAP2C | [92] | ||
| miR-15b | 2010 | [93] | ||||
| miR-126 | Down | 2013 | [94] | ADAM9 MMP7 |
[95] [96] |
|
| miR-18b | 2013 | [97] | MDM2 | [98] | ||
| miR-149 | 2011 | [99] | GSK3α | [100] | ||
| miR-155 | 2011 | [101] | SK1 | [102] | ||
| miRNAs involved in invasion | miR-145 | Down | 2013 | [103] | FSCN1 | [104] |
| let-7a | Down | Integrin β3 | [105] | |||
| let-7b | Down | BSG | [106] | |||
| miR-200 family | Down | 2010 | [107] | MARCKS | [108] | |
| miR-34a/b/c | Down | 2011, 2012 | [80, 109] | c-MET ULBP2 |
[110] | |
| miR-199 | Down | c-MET | [110] | |||
| miR-9 | Down | NfkB1 | [111] | |||
| miR-182 | Up | 2009, 2011 | [28, 112] | MITF FOXO3 |
||
| miR-30b/d | Up | 2011 | [113] | GALNT1 GALNT7 |
[114] | |
| miR-375 | Down | 2011 | [115] | |||
| miR-211 | Down | 2010 | [47, 52-54] | BRN2, KCNMA1, NFAT5, TGFBR2 | ||
| miR-214 | Up | 2011 | [91] | TFAP2C ITGA3 |
[92] | |
| miR-137 | MITF | |||||
| miR-148 | MITF | |||||
| miR-182 | MITF | |||||
| miRNAs involved in apoptosis | miR-15b | Up | 2010 | [93] | ||
| miR-155 | Down | 2009, 2009 | [101, 116] | SK1 | [102] | |
| miR-211 | 2010 | [47] | ||||
| miR-26a | Down | 2013 | [117] | SODD | [118] | |
| miR-768-3p | [90] | |||||
| miR-21 | ||||||
| miRNAs involved in epigenetics | Global effect | 2013 | [119] | |||
| miR-375 | 2011 | [115] | ||||
| miR-34b | 2011 | [120] | ||||
| miR-182 | 2014 | [121] | ||||
| miR-34a | ||||||
| miR-137 | 2011 | [122] | ||||
| miR-29c | 2011 | [123] | DNMT3A DNMT3B |
|||
| miR-31 | ||||||
| miR-18b | ||||||
| miRNAs involved in the immune response | miR-210 | 2012 | [124] | PTPN1 P53I11 HOXA1 |
[125, 126] [127, 128] [129] |
|
| miR-34a/c | Down | 2012 | [109] | ULBP2 | [130, 131] | |
| miR-30b/d | Up | 2011 | [113] | GALNT7 | [132, 133] |
Highlights.
RNA biology research demonstrates noncoding RNA connection to melanoma development
Integrative approach of miRNA and lncRNA is necessary to understand melanoma genesis
ncRNAs extend our understanding beyond confines of protein-coding genes in melanomas
Acknowledgments
This work was supported by National Institutes of Health grants NCI 5P30CA030199, CA165184, and CA172847 to RJP. We also thank Ms. Debbie McFadden (Sanford-Burnham Medical Research Institute) for formatting the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maddodi N, Setaluri V. Photochem Photobiol. 2008;84:528–536. doi: 10.1111/j.1751-1097.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 2.Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, Wu T, Niinobe M, Yoshikawa K, Hannigan GE, Halaban R. Cancer Res. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 5.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco F, Grignani F, Nervi C. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Grady WM, Parkin RK, Mitchell PS, Lee JH, Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz AM, Kroh EM, Allen A, Fritz BR, Markowitz SD, Tewari M. Oncogene. 2008 doi: 10.1038/onc.2008.10. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann U, Hasemeier B, Christgen M, Muller M, Romermann D, Langer F, Kreipe H. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 9.Mueller DW, Rehli M, Bosserhoff AK. J Invest Dermatol. 2009 doi: 10.1038/jid.2008.452. [DOI] [PubMed] [Google Scholar]
- 10.Shenouda SK, Alahari SK. Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 11.Aqeilan RI, Calin GA, Croce CM. Cell Death Differ. 17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 12.Lynam-Lennon N, Maher SG, Reynolds JV. Biol Rev Camb Philos Soc. 2009;84:55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- 13.Schmittgen TD. J Cell Mol Med. 2008;12:1811–1819. doi: 10.1111/j.1582-4934.2008.00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen A, Luo M, Yuan G, Yu J, Deng T, Zhang L, Zhou Y, Mitchelson K, Cheng J. Biotechnol Lett. 2008;30:2045–2052. doi: 10.1007/s10529-008-9800-8. [DOI] [PubMed] [Google Scholar]
- 15.Igoucheva O, Alexeev V. Biochem Biophys Res Commun. 2009;379:790–794. doi: 10.1016/j.bbrc.2008.12.152. [DOI] [PubMed] [Google Scholar]
- 16.Baugh LR, Demodena J, Sternberg PW. Science. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- 17.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini M, Colombo MP, Peschle C, Care A. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 18.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 19.Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, Erickson PF, Shellman YG, Robinson WA. Cancer Res. 2008;68:1362–1368. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- 20.Chen CZ. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 21.Dalmay T, Edwards DR. Oncogene. 2006;25:6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- 22.Esquela-Kerscher A, Slack FJ. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 23.Perera RJ, Ray A. BioDrugs. 2007;21:97–104. doi: 10.2165/00063030-200721020-00004. [DOI] [PubMed] [Google Scholar]
- 24.Kent OA, Mendell JT. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 25.Zhang B, Pan X, Cobb GP, Anderson TA. Dev Biol. 2006 doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 26.Hammond SM. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Felicetti F, Errico MC, Segnalini P, Mattia G, Care A. Expert Rev Anticancer Ther. 2008;8:1759–1765. doi: 10.1586/14737140.8.11.1759. [DOI] [PubMed] [Google Scholar]
- 28.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Rahilly R, Muller F. J Anat. 2007;211:335–351. doi: 10.1111/j.1469-7580.2007.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ernfors P. Experimental cell research. 2010;316:1397–1407. doi: 10.1016/j.yexcr.2010.02.042. [DOI] [PubMed] [Google Scholar]
- 31.Yajima I, Kumasaka MY, Thang ND, Goto Y, Takeda K, Iida M, Ohgami N, Tamura H, Yamanoshita O, Kawamoto Y, Furukawa K, Kato M. J Skin Cancer. 2011;(2011):730170. doi: 10.1155/2011/730170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wegner M. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2005;18:74–85. [Google Scholar]
- 33.Verastegui C, Bille K, Ortonne JP, Ballotti R. The Journal of biological chemistry. 2000;275:30757–30760. doi: 10.1074/jbc.C000445200. [DOI] [PubMed] [Google Scholar]
- 34.Medic S, Ziman M. PloS one. 2010;5:e9977. doi: 10.1371/journal.pone.0009977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weeraratna AT, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, Trent JM. Cancer cell. 2002;1:279–288. doi: 10.1016/s1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 36.Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, Brown EJ, Capobianco AJ, Herlyn M, Liu ZJ. J Clin Invest. 2005;115:3166–3176. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zabierowski SE, Baubet V, Himes B, Li L, Fukunaga-Kalabis M, Patel S, McDaid R, Guerra M, Gimotty P, Dahmane N, Herlyn M. Stem Cells. 2011;29:1752–1762. doi: 10.1002/stem.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grichnik JM, Burch JA, Burchette J, Shea CR. The Journal of investigative dermatology. 1998;111:233–238. doi: 10.1046/j.1523-1747.1998.00272.x. [DOI] [PubMed] [Google Scholar]
- 39.Bevona C, Goggins W, Quinn T, Fullerton J, Tsao H. Archives of dermatology. 2003;139:1620–1624. doi: 10.1001/archderm.139.12.1620. discussion 1624. [DOI] [PubMed] [Google Scholar]
- 40.Rigel DS. Journal ofrthe American Academy of Dermatology. 2008;58:S129–132. doi: 10.1016/j.jaad.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 41.Westerhof W. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2006;19:183–193. [Google Scholar]
- 42.Haass NK, Smalley KS, Li L, Herlyn M. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2005;18:150–159. doi: 10.1111/j.1600-0749.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- 43.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasemeier-Kulesa JC, Teddy JM, Postovit LM, Seftor EA, Seftor RE, Hendrix MJ, Kulesa PM. Dev Dyn. 2008;237:2657–2666. doi: 10.1002/dvdy.21613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedman RC, Farh KK, Burge CB, Bartel DP. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mazar J, DeYoung K, Khaitan D, Meister E, Almodovar A, Goydos J, Ray A, Perera RJ. PloS one. 2010;5:e13779. doi: 10.1371/journal.pone.0013779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levy C, Khaled M, Iliopoulos D, Janas MM, Schubert S, Pinner S, Chen PH, Li S, Fletcher AL, Yokoyama S, Scott KL, Garraway LA, Song JS, Granter SR, Turley SJ, Fisher DE, Novina CD. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Y, Brenn T, Brown ER, Doherty V, Melton DW. British journal of cancer. 2012;106:553–561. doi: 10.1038/bjc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakurai E, Maesawa C, Shibazaki M, Yasuhira S, Oikawa H, Sato M, Tsunoda K, Ishikawa Y, Watanabe A, Takahashi K, Akasaka T, Masuda T. Int J Oncol. 2011;39:665–672. doi: 10.3892/ijo.2011.1084. [DOI] [PubMed] [Google Scholar]
- 51.Jukic DM, Rao UN, Kelly L, Skaf JS, Drogowski LM, Kirkwood JM, Panelli MC. J Transl Med. 2010;8:27. doi: 10.1186/1479-5876-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bell RE, Khaled M, Netanely D, Schubert S, Golan T, Buxbaum A, Janas MM, Postolsky B, Goldberg MS, Shamir R, Levy C. The Journal of investigative dermatology. 2014;134:441–451. doi: 10.1038/jid.2013.340. [DOI] [PubMed] [Google Scholar]
- 53.Boyle GM, Woods SL, Bonazzi VF, Stark MS, Hacker E, Aoude LG, Dutton-Regester K, Cook AL, Sturm RA, Hayward NK. Pigment Cell Melanoma Res. 2011;24:525–537. doi: 10.1111/j.1755-148X.2011.00849.x. [DOI] [PubMed] [Google Scholar]
- 54.Levy C, Khaled M, Iliopoulos D, Janas MM, Schubert S, Pinner S, Chen PH, Li S, Fletcher AL, Yokoyama S, Scott KL, Garraway LA, Song JS, Granter SR, Turley SJ, Fisher DE, Novina CD. Mol Cell. 2010;40:841–849. doi: 10.1016/j.molcel.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Margue C, Philippidou D, Reinsbach SE, Schmitt M, Behrmann I, Kreis S. PloS one. 2013;8:e73473. doi: 10.1371/journal.pone.0073473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu N, Lapcevich RK, Underhill CB, Han Z, Gao F, Swartz G, Plum SM, Zhang L, Green SJ. Cancer research. 2001;61(3):1022–1028. [PubMed] [Google Scholar]
- 57.Mueller DW, Bosserhoff AK. International journal of cancer. Journal international ducancer. 2011;129:1064–1074. doi: 10.1002/ijc.25768. [DOI] [PubMed] [Google Scholar]
- 58.Braig S, Mueller DW, Rothhammer T, Bosserhoff AK. Cell Mol Life Sci. 2010;67:3535–3548. doi: 10.1007/s00018-010-0394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larsson E, Sander C, Marks D. Molecular systems biology. 2010;6:433. doi: 10.1038/msb.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Molecular systems biology. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ross J. Microbiological reviews. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS. DNA research : an international journal for rapid publication of reports on genes and genomes. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sauter ER, Yeo UC, von Stemm A, Zhu W, Litwin S, Tichansky DS, Pistritto G, Nesbit M, Pinkel D, Herlyn M, Bastian BC. Cancer research. 2002;62:3200–3206. [PubMed] [Google Scholar]
- 64.Malumbres M, Barbacid M. Trends in biochemical sciences. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 65.Sotillo R, Garcia JF, Ortega S, Martin J, Dubus P, Barbacid M, Malumbres M. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13312–13317. doi: 10.1073/pnas.241338598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J, Feilotter HE, Pare GC, Zhang X, Pemberton JG, Garady C, Lai D, Yang X, Tron VA. The American journal of pathology. 2010;176:2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noguchi S, Kumazaki M, Yasui Y, Mori T, Yamada N, Akao Y. The Journal of investigative dermatology. 2014;134:461–469. doi: 10.1038/jid.2013.310. [DOI] [PubMed] [Google Scholar]
- 68.Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, Schadendorf D, Dummer R. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 69.Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Genes & development. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Georgantas RW, 3rd, Streicher K, Luo X, Greenlees L, Zhu W, Liu Z, Brohawn P, Morehouse C, Higgs BW, Richman L, Jallal B, Yao Y, Ranade K. Pigment cell & melanoma research. 2014;27:275–286. doi: 10.1111/pcmr.12200. [DOI] [PubMed] [Google Scholar]
- 71.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM. Cancer cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Di Cristofano A, Pandolfi PP. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 73.Ahonen M, Baker AH, Kahari VM. Cancer research. 1998;58:2310–2315. [PubMed] [Google Scholar]
- 74.Alexeev V, Yoon K. J Invest Dermatol. 2006;126:1102–1110. doi: 10.1038/sj.jid.5700125. [DOI] [PubMed] [Google Scholar]
- 75.Montone KT, van Belle P, Elenitsas R, Elder DE. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 1997;10:939–944. [PubMed] [Google Scholar]
- 76.Li B, Jia N, Kapur R, Chun KT. Blood. 2006;107:4291–4299. doi: 10.1182/blood-2005-08-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. The Journal of biological chemistry. 2011;286:16606–16614. doi: 10.1074/jbc.M111.227611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nelson MA, Reynolds SH, Rao UN, Goulet AC, Feng Y, Beas A, Honchak B, Averill J, Lowry DT, Senft JR, Jefferson AM, Johnson RC, Sargent LM. Cancer biology & therapy. 2006;5:407–412. doi: 10.4161/cbt.5.4.2512. [DOI] [PubMed] [Google Scholar]
- 79.Umemura S, Shirane M, Takekoshi S, Kusakabe T, Itoh J, Egashira N, Tokuda Y, Mori K, Osamura YR. British journal of cancer. 2009;100:764–771. doi: 10.1038/sj.bjc.6604900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Greenberg E, Hershkovitz L, Itzhaki O, Hajdu S, Nemlich Y, Ortenberg R, Gefen N, Edry L, Modai S, Keisari Y, Besser MJ, Schachter J, Shomron N, Markel G. PloS one. 2011;6:e18936. doi: 10.1371/journal.pone.0018936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu S, Kumar SM, Lu H, Liu A, Yang R, Pushparajan A, Guo W, Xu X. The Journal of pathology. 2012;226:61–72. doi: 10.1002/path.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karst AM, Gao K, Nelson CC, Li G. International journal of cancer. Journal international du cancer. 2009;124:494–501. doi: 10.1002/ijc.23973. [DOI] [PubMed] [Google Scholar]
- 83.Jiang L, Lv X, Li J, Li J, Li X, Li W, Li Y. Acta histochemica. 2012;114:582–588. doi: 10.1016/j.acthis.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 84.Satzger I, Mattern A, Kuettler U, Weinspach D, Niebuhr M, Kapp A, Gutzmer R. Experimental dermatology. 2012;21:509–514. doi: 10.1111/j.1600-0625.2012.01510.x. [DOI] [PubMed] [Google Scholar]
- 85.Noguchi S, Mori T, Otsuka Y, Yamada N, Yasui Y, Iwasaki J, Kumazaki M, Maruo K, Akao Y. The Journal of biological chemistry. 2012;287:11769–11777. doi: 10.1074/jbc.M111.325027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hurst CD, Tomlinson DC, Williams SV, Platt FM, Knowles MA. Oncogene. 2008;27:2716–2727. doi: 10.1038/sj.onc.1210934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ju YS, Lee WC, Shin JY, Lee S, Bleazard T, Won JK, Kim YT, Kim JI, Kang JH, Seo JS. Genome research. 2012;22:436–445. doi: 10.1101/gr.133645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bhattacharya A, Schmitz U, Wolkenhauer O, Schonherr M, Raatz Y, Kunz M. Oncogene. 2013;32:3175–3183. doi: 10.1038/onc.2012.324. [DOI] [PubMed] [Google Scholar]
- 89.Watanabe N, Broome M, Hunter T. The EMBO journal. 1995;14:1878–1891. doi: 10.1002/j.1460-2075.1995.tb07180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jiang CC, Croft A, Tseng HY, Guo ST, Jin L, Hersey P, Zhang XD. Oncogene. 2013 doi: 10.1038/onc.2013.237. [DOI] [PubMed] [Google Scholar]
- 91.Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De Pitta C, Pinatel E, Stadler MB, Provero P, Bernengo MG, Osman I, Taverna D. The EMBO journal. 2011;30:1990–2007. doi: 10.1038/emboj.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Berger AJ, Davis DW, Tellez C, Prieto VG, Gershenwald JE, Johnson MM, Rimm DL, Bar-Eli M. Cancer research. 2005;65:11185–11192. doi: 10.1158/0008-5472.CAN-05-2300. [DOI] [PubMed] [Google Scholar]
- 93.Satzger I, Mattern A, Kuettler U, Weinspach D, Voelker B, Kapp A, Gutzmer R. International journal of cancer. Journal international du cancer. 2010;126:2553–2562. doi: 10.1002/ijc.24960. [DOI] [PubMed] [Google Scholar]
- 94.Felli N, Felicetti F, Lustri AM, Errico MC, Bottero L, Cannistraci A, De Feo A, Petrini M, Pedini F, Biffoni M, Alvino E, Negrini M, Ferracin M, Mattia G, Care A. PloS one. 2013;8:e56824. doi: 10.1371/journal.pone.0056824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kivisaari AK, Kallajoki M, Ala-aho R, McGrath JA, Bauer JW, Konigova R, Medvecz M, Beckert W, Grenman R, Kahari VM. The British journal of dermatology. 2010;163:726–735. doi: 10.1111/j.1365-2133.2010.09924.x. [DOI] [PubMed] [Google Scholar]
- 96.Ohashi S, Natsuizaka M, Nakagawa H. Cancer biology & therapy. 2011;11:184–187. doi: 10.4161/cbt.11.2.14140. [DOI] [PubMed] [Google Scholar]
- 97.Dar AA, Majid S, Rittsteuer C, de Semir D, Bezrookove V, Tong S, Nosrati M, Sagebiel R, Miller JR, 3rd, Kashani-Sabet M. Journal of the National Cancer Institute. 2013;105:433–442. doi: 10.1093/jnci/djt003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Z, Li M, Wang H, Agrawal S, Zhang R. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11636–11641. doi: 10.1073/pnas.1934692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jin L, Hu WL, Jiang CC, Wang JX, Han CC, Chu P, Zhang LJ, Thorne RF, Wilmott J, Scolyer RA, Hersey P, Zhang XD, Wu M. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15840–15845. doi: 10.1073/pnas.1019312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dhawan P, Singh AB, Ellis DL, Richmond A. Cancer research. 2002;62:7335–7342. [PubMed] [Google Scholar]
- 101.Levati L, Pagani E, Romani S, Castiglia D, Piccinni E, Covaciu C, Caporaso P, Bondanza S, Antonetti FR, Bonmassar E, Martelli F, Alvino E, DAtri S. Pigment cell & melanoma research. 2011;24:538–550. doi: 10.1111/j.1755-148X.2011.00857.x. [DOI] [PubMed] [Google Scholar]
- 102.Tonelli F, Lim KG, Loveridge C, Long J, Pitson SM, Tigyi G, Bittman R, Pyne S, Pyne NJ. Cellular signalling. 2010;22:1536–1542. doi: 10.1016/j.cellsig.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dynoodt P, Speeckaert R, De Wever O, Chevolet I, Brochez L, Lambert J, Van Gele M. International journal of oncology. 2013;42:1443–1451. doi: 10.3892/ijo.2013.1823. [DOI] [PubMed] [Google Scholar]
- 104.Shonukan O, Bagayogo I, McCrea P, Chao M, Hempstead B. Oncogene. 2003;22:3616–3623. doi: 10.1038/sj.onc.1206561. [DOI] [PubMed] [Google Scholar]
- 105.Seftor RE, Seftor EA, Gehlsen KR, Stetler-Stevenson WG, Brown PD, Ruoslahti E, Hendrix MJ. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:1557–1561. doi: 10.1073/pnas.89.5.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Muramatsu T, Miyauchi T. Histology and histopathology. 2003;18:981–987. doi: 10.14670/HH-18.981. [DOI] [PubMed] [Google Scholar]
- 107.Elson-Schwab I, Lorentzen A, Marshall CJ. PloS one. 2010;5 doi: 10.1371/journal.pone.0013176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Estrada-Bernal A, Gatlin JC, Sunpaweravong S, Pfenninger KH. Journal of cell science. 2009;122:2300–2310. doi: 10.1242/jcs.047860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Heinemann A, Zhao F, Pechlivanis S, Eberle J, Steinle A, Diederichs S, Schadendorf D, Paschen A. Cancer research. 2012;72:460–471. doi: 10.1158/0008-5472.CAN-11-1977. [DOI] [PubMed] [Google Scholar]
- 110.Chattopadhyay C, Ellerhorst JA, Ekmekcioglu S, Greene VR, Davies MA, Grimm EA. International journal of cancer Journal international du cancer. 2012;131:E56–65. doi: 10.1002/ijc.26487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boukerche H, ZZ Su, Emdad L, Sarkar D, Fisher PB. Cancer research. 2007;67:1812–1822. doi: 10.1158/0008-5472.CAN-06-3875. [DOI] [PubMed] [Google Scholar]
- 112.Huynh C, Segura MF, Gaziel-Sovran A, Menendez S, Darvishian F, Chiriboga L, Levin B, Meruelo D, Osman I, Zavadil J, Marcusson EG, Hernando E. Oncogene. 2011;30:1481–1488. doi: 10.1038/onc.2010.523. [DOI] [PubMed] [Google Scholar]
- 113.Gaziel-Sovran A, Segura MF, Di Micco R, Collins MK, Hanniford D, Vega-Saenz de Miera E, Rakus JF, Dankert JF, Shang S, Kerbel RS, Bhardwaj N, Shao Y, Darvishian F, Zavadil J, Erlebacher A, Mahal LK, Osman I, Hernando E. Cancer cell. 2011;20:104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bhavanandan VP. Glycobiology. 1991;1:493–503. doi: 10.1093/glycob/1.5.493. [DOI] [PubMed] [Google Scholar]
- 115.Mazar J, DeBlasio D, Govindarajan SS, Zhang S, Perera RJ. FEBS Lett. 2011;585:2467–2476. doi: 10.1016/j.febslet.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 116.Levati L, Alvino E, Pagani E, Arcelli D, Caporaso P, Bondanza S, Di Leva G, Ferracin M, Volinia S, Bonmassar E, Croce CM, DAtri S. Int J Oncol. 2009;35:393–400. [PubMed] [Google Scholar]
- 117.Reuland SN, Smith SM, Bemis LT, Goldstein NB, Almeida AR, Partyka KA, Marquez VE, Zhang Q, Norris DA, Shellman YG. The Journal of investigative dermatology. 2013;133:1286–1293. doi: 10.1038/jid.2012.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tao H, Hu Q, Fang J, Liu A, Liu S, Zhang L, Hu Y. Journal of Huazhong University of Science and Technology. Medical sciences = Hua zhong ke ji da xue xue bao. Yi xue Ying De wen ban = Huazhong keji daxue xuebao. Yixue Yingdewen ban. 2007;27:326–329. doi: 10.1007/s11596-007-0328-2. [DOI] [PubMed] [Google Scholar]
- 119.Li JL, Mazar J, Zhong C, Faulkner GJ, Govindarajan SS, Zhang Z, Dinger ME, Meredith G, Adams C, Zhang S, Mattick JS, Ray A, Perera RJ. Scientific reports. 2013;3:2962. doi: 10.1038/srep02962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mazar J, Khaitan D, DeBlasio D, Zhong C, Govindarajan SS, Kopanathi S, Zhang S, Ray A, Perera RJ. PloS one. 2011;6:e24922. doi: 10.1371/journal.pone.0024922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu S, Howell PM, Riker AI. Annals of surgical oncology. 2013;20:1745–1752. doi: 10.1245/s10434-012-2467-3. [DOI] [PubMed] [Google Scholar]
- 122.Chen X, Wang J, Shen H, Lu J, Li C, Hu DN, Dong XD, Yan D, Tu L. Investigative ophthalmology & visual science. 2011;52:1193–1199. doi: 10.1167/iovs.10-5272. [DOI] [PubMed] [Google Scholar]
- 123.Nguyen T, Kuo C, Nicholl MB, Sim MS, Turner RR, Morton DL, Hoon DS. Epigenetics : official journal of the DNA Methylation Society. 2011;6:388–394. doi: 10.4161/epi.6.3.14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Noman MZ, Buart S, Romero P, Ketari S, Janji B, Mari B, Mami-Chouaib F, Chouaib S. Cancer research. 2012;72:4629–4641. doi: 10.1158/0008-5472.CAN-12-1383. [DOI] [PubMed] [Google Scholar]
- 125.Lu X, Malumbres R, Shields B, Jiang X, Sarosiek KA, Natkunam Y, Tiganis T, Lossos IS. Blood. 2008;112:4098–4108. doi: 10.1182/blood-2008-03-148726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Medgyesi D, Hobeika E, Biesen R, Kollert F, Taddeo A, Voll RE, Hiepe F, Reth M. The Journal of experimental medicine. 2014;211:427–440. doi: 10.1084/jem.20131196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liang XQ, Cao EH, Zhang Y, Qin JF. FEBS letters. 2004;569:94–98. doi: 10.1016/j.febslet.2004.05.057. [DOI] [PubMed] [Google Scholar]
- 128.Liu XM, Xiong XF, Song Y, Tang RJ, Liang XQ, Cao EH. Journal of gastroenterology. 2009;44:460–469. doi: 10.1007/s00535-009-0030-1. [DOI] [PubMed] [Google Scholar]
- 129.Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le QT, Giaccia AJ. Mol Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Groh V, Wu J, Yee C, Spies T. Nature. 2002;419:734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 131.Fernandez-Messina L, Ashiru O, Boutet P, Aguera-Gonzalez S, Skepper JN, Reyburn HT, Vales-Gomez M. The Journal of biological chemistry. 2010;285:8543–8551. doi: 10.1074/jbc.M109.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brockhausen I. EMBO reports. 2006;7:599–604. doi: 10.1038/sj.embor.7400705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dennis JW, Granovsky M, Warren CE. Biochimica et biophysica acta. 1999;1473:21–34. doi: 10.1016/s0304-4165(99)00167-1. [DOI] [PubMed] [Google Scholar]
- 134.Palazon A, Aragones J, Morales-Kastresana A, de Landazuri MO, Melero I. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:1207–1213. doi: 10.1158/1078-0432.CCR-11-1591. [DOI] [PubMed] [Google Scholar]
- 135.Zinkernagel AS, Johnson RS, Nizet V. Journal of molecular medicine. 2007;85:1339–1346. doi: 10.1007/s00109-007-0282-2. [DOI] [PubMed] [Google Scholar]
- 136.Suryadinata R, Sadowski M, Sarcevic B. Bioscience reports. 2010;30:243–255. doi: 10.1042/BSR20090171. [DOI] [PubMed] [Google Scholar]
- 137.Morris L, Allen KE, La Thangue NB. Nature cell biology. 2000;2:232–239. doi: 10.1038/35008660. [DOI] [PubMed] [Google Scholar]
- 138.Mateyak MK, Obaya AJ, Sedivy JM. Molecular and cellular biology. 1999;19:4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Walter KA, Hossain MA, Luddy C, Goel N, Reznik TE, Laterra J. Molecular and cellular biology. 2002;22:2703–2715. doi: 10.1128/MCB.22.8.2703-2715.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mamillapalli R, Gavrilova N, Mihaylova VT, Tsvetkov LM, Wu H, Zhang H, Sun H. Current biology : CB. 2001;11:263–267. doi: 10.1016/s0960-9822(01)00065-3. [DOI] [PubMed] [Google Scholar]
- 141.Madonna G, Ullman CD, Gentilcore G, Palmieri G, Ascierto PA. Journal of translational medicine. 2012;10:53. doi: 10.1186/1479-5876-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Alonso SR, Ortiz P, Pollan M, Perez-Gomez B, Sanchez L, Acuna MJ, Pajares R, Martinez-Tello FJ, Hortelano CM, Piris MA, Rodriguez-Peralto JL. The American journal of pathology. 2004;164:193–203. doi: 10.1016/s0002-9440(10)63110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Noguchi S, Mori T, Hoshino Y, Yamada N, Nakagawa T, Sasaki N, Akao Y, Maruo K. The Journal of veterinary medical science / the Japanese Society of Veterinary Science. 2012;74:1–8. doi: 10.1292/jvms.11-0264. [DOI] [PubMed] [Google Scholar]
- 144.Zhang K, Wong P, Duan J, Jacobs B, Borden EC, Bedogni B. Pigment cell & melanoma research. 2013;26:408–414. doi: 10.1111/pcmr.12089. [DOI] [PubMed] [Google Scholar]
- 145.Bashir T, Pagano M. Cell cycle. 2004;3:7:e27–e29. [PubMed] [Google Scholar]
- 146.Alla V, Kowtharapu BS, Engelmann D, Emmrich S, Schmitz U, Steder M, Putzer BM. Cell cycle. 2012;11:3067–3078. doi: 10.4161/cc.21476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fu TY, Chang CC, Lin CT, Lai CH, Peng SY, Ko YJ, Tang PC. Exp Cell Res. 2011;317:445–451. doi: 10.1016/j.yexcr.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 148.Muller DW, Bosserhoff AK. Oncogene. 2008;27:6698–6706. doi: 10.1038/onc.2008.282. [DOI] [PubMed] [Google Scholar]
- 149.Cruz J, Reis-Filho JS, Silva P, Lopes JM. Oncology. 2003;65:72–82. doi: 10.1159/000071207. [DOI] [PubMed] [Google Scholar]
- 150.Otsuka T, Takayama H, Sharp R, Celli G, LaRochelle WJ, Bottaro DP, Ellmore N, Vieira W, Owens JW, Anver M, Merlino G. Cancer research. 1998;58:5157–5167. [PubMed] [Google Scholar]
- 151.Migliore C, Petrelli A, Ghiso E, Corso S, Capparuccia L, Eramo A, Comoglio PM, Giordano S. Cancer Res. 2008;68:10128–10136. doi: 10.1158/0008-5472.CAN-08-2148. [DOI] [PubMed] [Google Scholar]
- 152.Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, Holland-Cunz S, Sinnberg T, Schittek B, Schadendorf D, Diederichs S, Eichmuller SB. The Journal of investigative dermatology. 2013;133:768–775. doi: 10.1038/jid.2012.357. [DOI] [PubMed] [Google Scholar]
- 153.Kyrgidis A, Tzellos TG, Triaridis S. Journal of carcinogenesis. 2010;9:3. doi: 10.4103/1477-3163.62141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 155.Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, Fullen DR, Johnson TM, Giordano TJ, Palanisamy N, Chinnaiyan AM. Oncotarget. 2012;3:1011–1025. doi: 10.18632/oncotarget.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhao H, Shen J, Medico L, Wang D, Ambrosone CB, Liu S. PloS one. 2010;5:e13735. doi: 10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sultmann H. International journal of cancer. Journal international du cancer. 2011;128:608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 158.Redova M, Sana J, Slaby O. Future oncology. 2013;9:387–402. doi: 10.2217/fon.12.192. [DOI] [PubMed] [Google Scholar]
- 159.Wei C, Henderson H, Spradley C, Li L, Kim IK, Kumar S, Hong N, Arroliga AC, Gupta S. PloS one. 2013;8:e64396. doi: 10.1371/journal.pone.0064396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shiiyama R, Fukushima S, Jinnin M, Yamashita J, Miyashita A, Nakahara S, Kogi A, Aoi J, Masuguchi S, Inoue Y, Ihn H. Melanoma research. 2013 doi: 10.1097/CMR.0b013e328363e485. [DOI] [PubMed] [Google Scholar]
- 161.Friedman EB, Shang S, de Miera EV, Fog JU, Teilum MW, Ma MW, Berman RS, Shapiro RL, Pavlick AC, Hernando E, Baker A, Shao Y, Osman I. Journal of translational medicine. 2012;10:155. doi: 10.1186/1479-5876-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Leidinger P, Keller A, Borries A, Reichrath J, Rass K, Jager SU, Lenhof HP, Meese E. BMC Cancer. 2010;10:262. doi: 10.1186/1471-2407-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kanemaru H, Fukushima S, Yamashita J, Honda N, Oyama R, Kakimoto A, Masuguchi S, Ishihara T, Inoue Y, Jinnin M, Ihn H. Journal of dermatological science. 2011;61:187–193. doi: 10.1016/j.jdermsci.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 164.Kozubek J, Ma Z, Fleming E, Duggan T, Wu R, Shin DG, Dadras SS. PloS one. 2013;8:e72699. doi: 10.1371/journal.pone.0072699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Glud M, Klausen M, Gniadecki R, Rossing M, Hastrup N, Nielsen FC, Drzewiecki KT. J Invest Dermatol. 2009;129:1219–1224. doi: 10.1038/jid.2008.347. [DOI] [PubMed] [Google Scholar]
- 166.Liu A, Tetzlaff MT, Vanbelle P, Elder D, Feldman M, Tobias JW, Sepulveda AR, Xu X. Int J Clin Exp Pathol. 2009;2:519–527. [PMC free article] [PubMed] [Google Scholar]
- 167.Ma Z, Lui WO, Fire A, Dadras SS. J Mol Diagn. 2009;11:420–429. doi: 10.2353/jmoldx.2009.090041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Worley LA, Long MD, Onken MD, Harbour JW. Melanoma Res. 2008;18:184–190. doi: 10.1097/CMR.0b013e3282feeac6. [DOI] [PubMed] [Google Scholar]
- 169.Qi M, Huang X, Zhou L, Zhang J. International journal of molecular medicine. 2014 doi: 10.3892/ijmm.2014.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wagenseller AG, Shada A, DAuria KM, Murphy C, Sun D, Molhoek KR, Papin JA, Dutta A, Slingluff CL., Jr Journal of translational medicine. 2013;11:218. doi: 10.1186/1479-5876-11-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Guba M, BP von, Steinbauer M, Koehl G, Flegel S, Hornung M, Bruns CJ, Zuelke C, Farkas S, Anthuber M, Jauch KW, Geissler EK. Nat Med. 2002;8:128–135. doi: 10.1038/nm0202-128. [DOI] [PubMed] [Google Scholar]
- 172.Molhoek KR, Griesemann H, Shu J, Gershenwald JE, Brautigan DL, Slingluff CLJ. Cancer research. 2008;68:4392–4397. doi: 10.1158/0008-5472.CAN-07-5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 174.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 175.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 176.Wang KC, Chang HY. Molecular cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nagano T, Fraser P. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 178.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. Cancer research. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 179.Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, Khavari PA. Genome research. 2012;22:1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tang L, Zhang W, Su B, Yu B. Biomed Res Int. 2013;2013:251098. doi: 10.1155/2013/251098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zou Y, Jiang Z, Yu X, Sun M, Zhang Y, Zuo Q, Zhou J, Yang N, Han P, Ge Z, De W, Sun L. PloS one. 2013;8:e79598. doi: 10.1371/journal.pone.0079598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mazar J, Zhao W, Khalil AM, Lee B, Shelley J, Govindarajan SS, Yamamoto G, Ratnam M, Aftab MN, Collins S, Finck BN, Han X, Mattick JS, Dinger ME, Perera RJ. Oncotarget. 2014 doi: 10.18632/oncotarget.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Boissier S, Ferreras M, Peyruchaud O, Magnetto S, Ebetino FH, Colombel M, Delmas P, Delaisse JM, Clezardin P. Cancer research. 2000;60:2949–2954. [PubMed] [Google Scholar]
- 184.Wu CF, Tan GH, Ma CC, Li L. Journal of genetics and genomics = Yi chuan xue bao. 2013;40:179–188. doi: 10.1016/j.jgg.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 185.Chi S, Xie G, Liu H, Chen K, Zhang X, Li C, Xie J. Cellular signalling. 2012;24:1222–1228. doi: 10.1016/j.cellsig.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pasmant E, Sabbagh A, Vidaud M, Bieche I. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 187.Smedley D, Sidhar S, Birdsall S, Bennett D, Herlyn M, Cooper C, Shipley J. Genes Chromosomes Cancer. 2000;28:121–125. doi: 10.1002/(sici)1098-2264(200005)28:1<121::aid-gcc14>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 188.Liu Y, Yang S, Zhang X. Biochemical and biophysical research communications. 2013 [Google Scholar]
- 189.Schwimmer J, Essner R, Patel A, Jahan SA, Shepherd JE, Park K, Phelps ME, Czernin J, Gambhir SS. The quarterly journal of nuclear medicine : official publication of the Italian Association of Nuclear Medicine. 2000;44:153–167. [PubMed] [Google Scholar]


