Abstract
microRNA (miRNA)-dependent regulation of gene expression is increasingly linked to development and progression of melanoma. In this study we evaluated the functions of miR-340 in human melanoma cells. Here, we show that miR-340 inhibits the tumorigenic phenotype of melanoma cells. We also found that miR-340 regulates RAS-RAF-Mitogen Activated Protein Kinase (MAPK) signaling by modulating the expression of multiple components of this pathway. Given the importance of MAPK signaling in melanoma, these results provide further insight into the pathogenesis of melanoma.
Keywords: microRNA, miR-340, melanoma, tumor suppressor, proliferation, invasion, RAS, RAF, MAPK
Introduction
microRNAs are a class of evolutionarily conserved small (22–25 nucleotides), non-coding RNAs that post-transcriptionally regulate nearly every process within the cell [1–4]. Mature miRNAs incorporated into the RNA-induced silencing complex (RISC) regulate target mRNAs via complementary base pairing in the 3′UTR as well as in the 5′UTR and protein coding regions. Binding of a miRNA-containing RISC to its target induces translational repression and/or mRNA degradation, thereby regulating target protein expression. Predictive algorithms suggest each miRNA regulates many targets, estimating approximately 60% of all human mRNAs are subject to miRNA-dependent regulation [5, 6]. Increasing evidence demonstrates miRNAs are critically involved in cancer biology, playing both tumor suppressor and oncogenic roles [7–9]. Although a significant body of evidence points to the importance of miRNA-dependent regulation in biology, the function of each individual miRNA remains to be elucidated, particularly as it pertains to the development and progression of human cancers.
Metastatic melanoma is a melanocyte-derived cancer with few successful treatment options leading to a poor prognosis that is underscored by a five-year survival rate of approximately 15% [10, 11]. Activating mutations in BRAF and NRAS are the first and second most common mutations in melanomas, respectively, suggesting the importance of aberrant MAPK signaling in melanoma [10, 12–15]. Specifically, BRAF missense mutations occur in 50–70% of melanomas and all are located within the kinase domain of the protein [12, 14, 15]. In a majority of BRAF melanoma mutations present, a single substitution occurs at codon 600 where the substitution of a T for an A causes a change from glutamic acid to valine in the sequence leading to its constitutive activation [15]. Mutations in NRAS occur in 15–30% of all melanomas and are often found at codon 61 (NRASQ61R) causing the protein to be in the activated conformation and therefore induces a loss of its GTPase activity [13, 15]. Mutations also occur in MAP3K5 in 24% of melanomas leading to increased survival [15]. Additionally, recent research highlighted a novel melanoma MAPK pathway mutation in RAC1P29S, a Rho family GTPase that has important roles in the regulation of adhesion, migration and invasion [15].
Our report demonstrates the pleiotropic effects of one miRNA, miR-340, functions to suppress tumorigenic properties in melanoma cells by regulating multiple components of the RAS-RAF-MAPK pathway, including the previously mentioned targets.
Materials and Methods
Cell culture
Normal human epidermal melanocytes (NHEMs) were grown in M254 (Life Technologies, Grand Island, NY) and HMGS supplement (Life Technologies). Human melanoma cells 451 Lu, WM35 and WM115 were a kind gift of Dr. Meenhard Herlyn, Wistar Institute, Philadelphia, PA. 451 Lu were grown in MEM basal media (Life Technologies). SK-MEL-2 (ATCC, Manassas, VA), WM35 and WM 115 melanoma cells were grown in EMEM basal media (ATCC). SK-MEL-28 (kind gift of Dr. Yiqun Shellman, University of Colorado Denver, Denver, CO) were grown in RPMI medium (Cellgro, Manassas, VA). Mel 928 (kind gift of Dr. Paul Robbins, Center for Cancer Research, National Cancer Institute, Bethesda, MD), Mel Ju (kind gift of Dr. Anja Katrin Bosserhoff, University of Regensburg, Regensberg, Germany) and Hs294T (ATCC) melanoma cells were grown in DMEM (Cellgro). All basal media was supplemented with 4.5g/L glucose, L-glutamine, 1mM sodium pyruvate, 1mM Non-essential amino acids supplemented with 10% (% v/v) heat-inactivated FBS. All cells were grown at 37°C in a humidified atmosphere of 5% CO2.
Expression vectors and cell culture experiments
miRZip scrambled hairpin RNA control and miRZip 340 microRNA inhibitor constructs (System Biosciences Inc., Mountain View, CA) were used to inhibit hsa-miR-340 and PCMV-MIR340 and PCMV-MIR control microRNA over expression vectors (Origene, Rockville, MD) were used to over express hsa-miR-340. Plasmid DNA was transfected with Lipofectamine 2000 (Life Technologies), 24 hours later the transfection mixture was replaced with fresh media and after 48 hours post-transfection, cells were collected for RNA, protein or cell-based assays and analysis. For the phosphorylated Erk1/2 experiments, 48 hours post-transfection, the MEK1 inhibitor PD98059 (Invivogen, San Diego, CA) in DMSO was applied to cells at a 50mM final concentration to cells for a 3 hour period, after which cells were collected for downstream applications.
RNA Isolation, real time quantitative PCR and droplet digital PCR
Total RNA was isolated using TRI Reagent (Molecular Research Center, Inc., Cincinnati, OH) according to manufacturer’s instructions and utilizing a Phase-Lock Gel Tube (5 Prime, Gaithersburg, MD). RNA quality was assessed on a Synergy H1 multi-mode microplate reader and Gen5 2.0 software (BioTek, Winooski, VT) and the Advantage RT-for-PCR cDNA Synthesis kit (Clontech, Mountain View, CA) was used to make cDNA according to the manufacturer’s protocol. TaqMan probes for BRAF, MITF, NRAS and 18s rRNA were used to evaluate transcript expression (Life Technologies), per the manufacturer’s suggested protocol. TaqMan Gene Expression Master Mix (Life Technologies) or iTaq Universal Probes Master Mix (Bio-Rad, Hercules, CA) was used in the qRT-PCR reactions, per the manufacturer’s suggested parameters. Additionally, to evaluate the effect of miR-340 inhibition on potential RAS-RAF-MAPK components, a custom Prime PCR array plate was used with the iTaq Universal SYBR Green Master Mix along with the Reference Genes H96 Array Plate to determine the appropriate endogenous control, in this case TATA-binding protein (TBP) (Bio-Rad). For the above quantitative RT-PCR reactions, a StepOnePlus qRT-PCR machine with StepOne 2.2.2 software (Life Technologies) was used to determine differences in transcript expression based on the ΔΔCT method. To quantify hsa-miR-340 RNAs, droplet digital PCR (ddPCR) was performed on the QX100 ddPCR system (Bio-Rad) using TaqMan FAM labeled probes for hsa-miR-340-5p and RNU6B, as an endogenous control (Life Technologies). 20μL reactions were assembled with cDNA, probes, ddPCR Supermix for Probes (no dUTP) and droplets were generated using the manufacturer’s suggested protocol with the QX100 Droplet Generator machine, droplet generator cartridges and gaskets and droplet generation oil for probes (Bio-Rad). Generated droplets were placed into a twin.tec PCR 96-well plate (Eppendorf, Hauppauge, NY). Plates were sealed using a PX1 PCR plate sealer (Bio-Rad) and pierceable foil heat seal and a PCR reaction under the following conditions was performed on a Mastercycler Gradient thermal cycler (Eppendorf): 5 minutes at 95°C, 30 seconds at 95°C, 1 minute at 60°C for 40 cycles with a ramp speed of 2°C/second, 98°C for 10 minutes and hold at 12°C overnight. FAM generated fluorescence in the droplets was evaluated using the QX100 Droplet Reader machine and QuantaSoft Software 1.3.2.0 (Bio-Rad). ddPCR data presented are representative of absolute copies of transcripts in reaction samples.
Cell-based Assays
Cell growth was evaluated using the CellTiter 96 AQueous One Solution (Promega, Madison, WI) according to the manufacturer’s instructions and evaluated on a Synergy H1 (BioTek) multi-mode microplate reader at 490nm using Gen5 2.0 software (BioTek). Cell motility/migration was evaluated by applying 4.9×105 cells to each well of Culture-Inserts (ibidi, LLC, Verona, WI). 24 hours later, inserts were removed, creating a 500μm cell free zone, at which point the cells were placed into an environmental chamber equipped with temperature and gaseous controls (InVivo Scientific, Valley Park, MO) creating constant humidity and a controlled temperature of 37°C with 5% CO2 mixed with 95% medical air (Airgas, Radnor, PA). The closure of the cell-free zone was evaluated every 6 hours automatically over multiple samples and z – planes to ensure capture of the correct plane of focus using time-lapse microscopy on an Eclipse Ti inverted microscope and NIS-Elements Ar 4.13.00 (Build 914) software (Nikon, Tokyo Japan) placed on a Vibraplane microscopy table (Kinetic Systems, Boston, MA) floated with Nitrogen gas (Airgas). Best focus photos were evaluated using NIS-Elements Ar 4.13.00 (Build 914) software (Nikon). Invasion was evaluated using Boyden invasion chambers (BD Biocoat Matrigel Invasion Chambers, BD Biosciences, San Jose, CA) by applying 2.5×104 cells/insert in 1% (v/v) heat-inactivated FBS media to the top portion of the chamber. The bottom portion of the chamber contained media with 10% (v/v) heat-inactivated FBS. After 24 hours, the amount of cells invaded was quantified by staining cells on the underside of the chamber with 5μM Calcein AM fluorescent dye (BD Biosciences) for 30 minutes at 37°C in a humidified atmosphere of 5% CO2, removing the cells with 0.25% Trypsin-2.21mM EDTA (Corning, Corning, NY) for 10 minutes at 37°C in a humidified atmosphere of 5% CO2 and evaluating on a Synergy H1 (BioTek) multi-mode microplate reader at 485/520nm (excitation/emission) using Gen5 2.0 software (BioTek).
Western Blotting Analysis and Antibodies
Total cellular lysates were prepared using radio immunoprecipitation assay (RIPA) buffer (Millipore, Billerica, MA) and protein concentration was evaluated using the DC Protein Assay (Bio-Rad) according the manufacturer’s protocol. Proteins were separated by SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes (Amersham GE Healthcare, Little Chalfont, Buckinghamshire, UK). After blocking with 3% milk, blots were probed with primary antibodies against phosphorylated Erk1/2 (#4370S Cell Signaling Technology, Danvers, MA), Erk1/2 (#4695 Cell Signaling Technology) and GAPDH (#NB300-328 Novus Biologicals, Littleton, CO) overnight, washed and incubated with HRP-conjugated secondary antibodies for 1 hour. Detection was performed using ECL Substrate (Perkin Elmer, Waltham, MA) and imaged on a Kodak Image Station 4000MM machine using Carestream MI SE 5.0.6.20 Software for image capture and analysis.
miRWalk and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
We used the miRWalk database to find predicted hsa-miR-340 targets (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/) [16]. The list of predicted miR-340 3′UTR targets was based on prediction overlap between at least 4 of 6 miRNA target prediction databases including DIANA-microT (version 3.0), miRanda (August 2010 release), miRDB (April 2009), miRWalk (March 2011), RNA22 (May 2008) and TargetScan (Version 5.1) [1, 5, 16–26]. The output list of predicted targets was imported into the KEGG pathway analysis KEGG Mapper tool (http://www.genome.jp/kegg/, Releases 65.1–69.0) to map the various pathways and cellular functions the miR-340 predicted targets are involved in [27].
Statistical analysis
Data represent at least 3 independent experiments expressed as means ± standard deviation (SD). A student’s t-test was used for statistical analysis with significance defined as a p value ≤ 0.05 or 0.01.
Results
miR-340 expression in melanoma cells
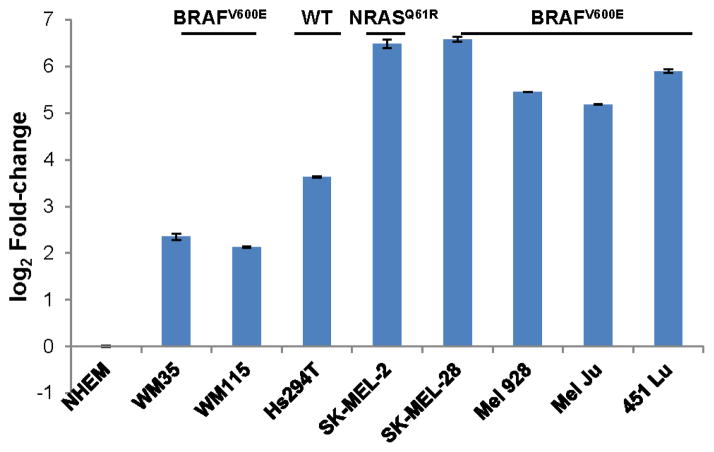
We previously noticed that miR-340 affects the proliferation of melanoma cells (Goswami et al personal communication). Additionally, several other research groups highlighted miR-340’s effects on proliferation in various cancers such as breast cancer, osteosarcoma, colorectal cancer and neuroblastoma [28–31]. It was also recently shown that expression of miR-340 correlates with an invasive phenotype in melanoma [32]. As a first step to better understand the involvement of miR-340 in melanoma pathogenesis, we evaluated the expression of miR-340 in a panel of melanoma cells, depicted in Figure 1. The panel of cell lines comprises NHEMs and melanoma cell lines representing early stage radial growth phase (WM35, BRAFV600E mutation) and vertical growth phase (WM115, BRAFV600E mutation) as well as metastatic melanoma cell lines with mutations in NRASQ61R (SK-MEL-2) and BRAFV600E (SK-MEL-28, Mel 928, Mel Ju, 451 Lu) or wild type for NRAS and BRAF (Hs294T) [12, 33–39]. Contrary to our expectations, miR-340 was significantly elevated in all of the melanoma cell lines, compared to NHEMs.
FIGURE 1. Expression of miR-340 in NHEMs and a panel melanoma cells.
Cells were collected and assayed for expression of miR-340, normalized to 18s rRNA and presented as log 2 fold-change in miR-340 expression ± SD, compared to NHEMs. In all cell lines, the expression of miR-340 was significantly over expressed compared to NHEMs (p value of ≤ 0.05).
miR-340 acts as a tumor suppressor in melanoma cells
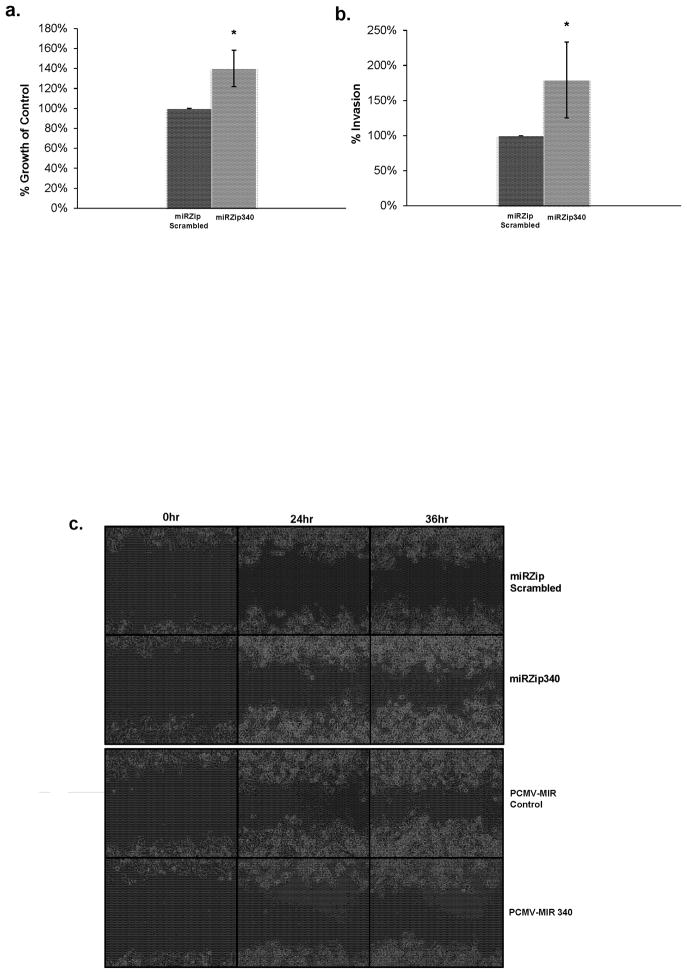
To analyze effects of miR-340 on tumorigenic properties of melanoma cells, we inhibited the function of miR-340 by ectopic expression of miR-340 inhibitors in 451 Lu melanoma cells, shown in Figure 2. Our results show a 40% increase in cell growth was found when miR-340 was inhibited (Figure 2a). Inhibiting miR-340 in melanoma cells also caused a 75% increase in invasion using Boyden invasion chambers (Figure 2b). Our data also demonstrate that overexpression as well as inhibition of miR-340 affects migration when a 500μm cell free zone is created using a cell culture insert (Figure 2c). It is understood that evaluating the effect of miR-340 overexpression and inhibition on motility/migration using the wound healing assay is the concurrent result of both motility and proliferation. However, the impact of proliferation is minimized in this case as the time frame evaluated is early (24hr and 36hr). Taken together, these results suggest miR-340 suppresses tumorigenic properties of melanoma cells.
FIGURE 2. miR-340 inhibits melanoma proliferation, invasion and migration.
451 Lu metastatic melanoma cells transfected with miRZip scrambled hairpin RNA control or miR-340 inhibiting miRZip 340 constructs. a. Cell proliferation comparing miRZip scrambled hairpin RNA control (left) and and miR-340 inhibiting miRZip 340 (right) transfected cells. 48 hours after transfection cells were re-plated for growth analysis, after an additional 48 hours absorbance was read on a plate reader, normalized to background and presented as percentages of control (scrambled hairpin RNA) ± SD. b. Invasion assay comparing miRZip scrambled hairpin RNA control (left) and miR-340 inhibiting miRZip 340 (right) transfected cells. 48 hours after transfection cells were plated in Boyden invasion chambers containing low/high serum concentrations on the top and bottom chambers, respectively. 24 hours later fluorescence was measured on a plate reader, normalized and presented as percentages of control (scrambled hairpin RNA) ± SD. c. Migration assay comparing miRZip scrambled hairpin RNA control (top) and miR-340 inhibiting miRZip340 (second) PCMV-MIR over expression control (third) and PCMV-MIR340 miR-340 over expressed (bottom) transfected cells. 48 hours after transfection cells were plated using cell culture inserts. 24 hours later, the cell culture inserts were removed and a 500μm cell free zone was created and cells were then allowed to migrate. Time-lapse microscopy of the cell free zone closure was automatically evaluated in the same region every 6 hours amongst the four treatment groups.
Inhibition of MAPK signaling pathway by miR-340
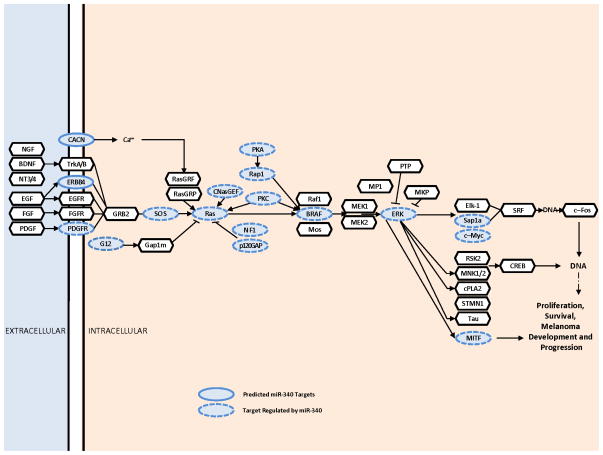
It is unlikely that miRNA function in melanoma can be explained by its effect(s) on a few targets. To understand the function of miR-340 in melanoma progression, it is critical to elucidate the multitude of targets miR-340 regulates. To do this, we first performed target prediction searches using the miRWalk miRNA target prediction database (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/) [16]. The miRWalk database allows the simultaneous query of up to 10 miRNA putative target databases. In our miR-340 predicted target search, we queried six databases including: DIANA-microT, miRanda, miRDB, miRWalk, RNA22 and TargetScan [1, 5, 16–26]. A list of possible miR-340 targets was generated and putative targets of interest were defined as a predicted overlap of at least 4 of 6 databases. Next, the KEGG Mapper tool (http://www.genome.jp/kegg/) was used to generate an array of pathways and functions the predicted targets were associated with [27]. From the mapping results, we found the MAPK signaling network contained a high proportion of predicted miR-340 targets (Figure 3). As demonstrated in Figure 3, 17 of 46 targets in the classical MAPK pathway were predicted to be regulated by miR-340.
FIGURE 3. miR-340 targets in the MAPK Pathway.
Predicted miR-340 targets are oval and outlined in a solid line and targets found to be predicted and regulated by miR-340 are oval and outlined in a dashed line. MAPK components not predicted to be targeted by miR-340 are depicted as a solid outlined hexagon. miR-340 target predictions are based on prediction overlap between at least 4 of 6 miRNA target prediction databases including DIANAmT, miRanda, miRDB, miRWalk, RNA22, and TargetScan.
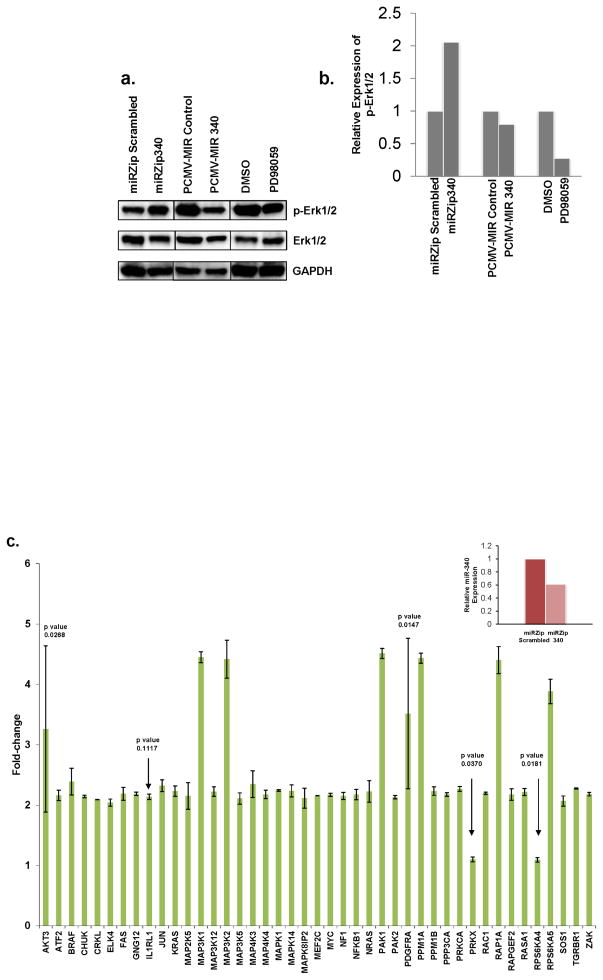
Given the importance of RAS-RAF-MAPK signaling in melanoma, we analyzed whether miR-340 had a functional impact on the activity of the MAPK pathway. As a hallmark of the activity of MAPK signaling pathway, we evaluated the expression of phosphorylated Erk1/2 upon the alteration of miR-340 function. Figure 4a and b show over expression of miR-340 reduced the levels of phosphorylated Erk1/2, similar to the effect of the MEK1 inhibitor PD98059 in 451 Lu melanoma cells. Although over expression of miR-340 did not affect the expression of phosphorylated Erk1/2 to the same degrees as PD98059, this result could be explained by further overexpression of miR-340 in this model is not physiologically relevant as miR-340 is already overexpressed in human melanoma cells (Figure 1). Conversely, down-regulation of miR-340 function resulted in a significant increase in phosphorylated Erk1/2 levels in melanoma cells (Figure 4a). These data suggest that miR-340 is important for the regulation and activity of MAPK signaling pathway. The effect of miR-340 inhibition resulting in the increased expression of phosphorylated Erk1/2 is not unique to 451 Lu melanoma cells as demonstrated by Supplemental Figure 1a and b.
FIGURE 4. miR-340 effects on the MAPK pathway.
a. Western blot analysis comparing phosphorylated Erk1/2, Erk1/2 and GAPDH expression in 451 Lu cells transfected or treated with the following; Lane 1 miRZip scrambled hairpin RNA control, Lane 2 miR-340 inhibiting miRZip340, Lane 3 PCMV-MIR over expression control, Lane 4 PCMV-MIR340 miR-340 over expression, Lane 5 DMSO control, Lane 6 50mM MEK1 inhibitor PD98059. b. GAPDH normalized densitometry of phosphorylated Erk1/2 expression amongst treatment groups. Presented as relative expression of phosphorylated Erk1/2. c. Expression of 43 components in the MAPK pathway upon inhibition of miR-340 in 451 Lu melanoma cells. 48 hours after transfection cells were collected and assayed for mRNA expression by qRT-PCR, normalized to TATA-binding protein and presented as fold-change in transcript expression ± SD, relative to scrambled hairpin RNA control. Changes in transcript expression are significant (p ≤ 0.01), except where p value is noted in figure and not considered significant. Insert. miR-340 expression levels 48 hours after transfection in 451 Lu cells transfected with miRZip scrambled hairpin RNA control (left) or miR-340 inhibiting miRZip340 (right). Normalized to RNU6B and presented as percentages of control.
To further study the function of miR-340-dependent regulation on the RAS-RAF-MAPK pathway, we inhibited miR-340 and evaluated the resultant transcript expression in a panel of predicted miR-340 targets within the RAS-RAF-MAPK pathway (Figure 4c and insert). The Figure 4 insert depicts the inhibition of miR-340 in 451 Lu cells, compared to scrambled hairpin RNA control. Overall, inhibition of miR-340 significantly affected the expression of 39 (p ≤ 0.01) out of 45 total RAS-RAF-MAPK components evaluated (Figure 4c). The fold-change in expression among the panel of transcripts ranged from 2 to 4.5, compared to control. Affected targets include BRAF, KRAS, NRAS and MYC. These particular targets are key oncogenes in cancer in general and, as noted previously, BRAF and NRAS in particular are important oncogenes in melanoma. Other notable targets that were significantly affected include several MAPK, MAP2K, MAP3K and MAP4K proteins including MAP3K5 as well as RAC1 and NF1. These results, taken together, suggest miR-340 is a regulator of the RAS-RAF-MAPK pathway in melanoma cells through pleiotropic effects on multiple components of the cascade.
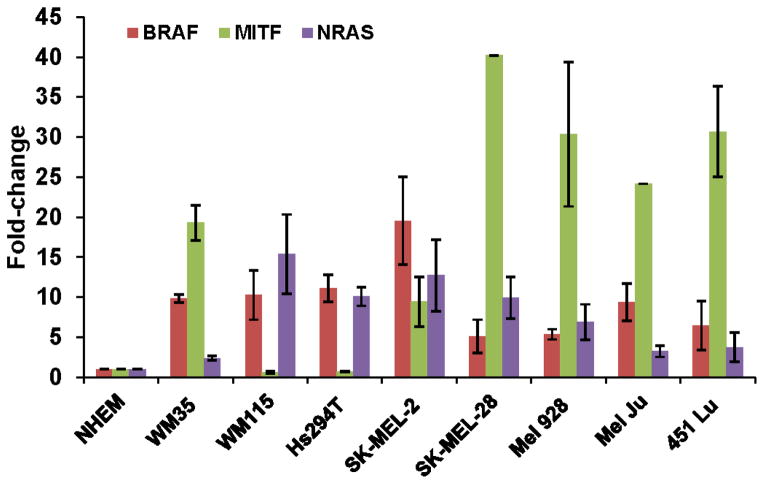
Expression of miR-340 targets in melanoma cells
To evaluate the function of miR-340 in the melanoma cell lines, we evaluated the expression of several miR-340 targets including BRAF, MITF and NRAS, shown in Figure 5. Expression of the targets did not inversely correlate with the expression of miR-340, as indicated by Pearson correlation coefficients of 0.31, 0.42 and 0.48 for correlations between miR-340 expression and BRAF, NRAS and MITF expression, respectively, amongst the cell lines. These data suggest that despite being over expressed in melanoma cells, the function of miR-340 is not up-regulated to significantly affect the expression of its targets. Additionally, as shown in Figure 4c, upon inhibiting the function of miR-340 the expression of BRAF and NRAS increased 2.35 and 2.20 fold, respectively, compared to control. Further supporting the hypothesis that miR-340 is a modulator of the MAPK pathway.
FIGURE 5. Expression patterns of miR-340 targets in a panel melanoma cells.
Expression of BRAF, MITF and NRAS in a panel of melanoma cell lines, compared to NHEMs. Cells were collected and assayed for expression of the aforementioned transcripts, normalized to 18s rRNA and presented as fold-change in miR-340 expression ± SD.
Discussion
As the importance of miRNAs in disease, especially cancer, is being elucidated it is critical to fully understand the functions of miRNAs related to the development and progression of disease. miRNAs are widely recognized for their ability to regulate mRNAs by modulating their stability and translation. However, the effect of a miRNA on each target is usually modest making it difficult to explain the pathophysiological effects of a miRNA through its regulation of a single target [40, 41]. This research is the first attempt to examine the pleiotropic regulation of the RAS-RAF-MAPK pathway by a single miRNA as it pertains to the development and progression of melanoma. In this report we demonstrate miR-340 functions as a tumor suppressor in melanoma cells most likely through down-regulation of the RAS-RAF-MAPK pathway by targeting multiple components of this pathway.
To establish miR-340 as a tumor suppressor in melanoma, we performed assays evaluating proliferation, invasion and migration. Inhibiting miR-340 resulted in a significant increase in proliferation, invasion and migration in melanoma cells. Our findings are in line with previous reports of miR-340 affecting proliferation, migration and invasion both in melanoma and other cancers [28–32].
A challenge toward understanding the biological relevance of miRNAs and their functions hinges on the identification of validated targets. Here, we attempt to characterize the function of miR-340 by connecting the dots from predicted targets to understanding the overall biological context of miR-340’s regulation. By mapping out the targets, we found that miR-340 has a high proportion of predicted targets in one particular cellular signaling cascade critical to the development and progression of melanoma, the RAS-RAF-MAPK pathway. We demonstrated that miR-340 regulates the RAS-RAF-MAPK pathway by showing significant effects on the expression of phosphorylated Erk1/2 upon over expression and inhibition of miR-340 as well as by showing a significant increase in transcript expression of a panel of 39 components in the RAS-RAF-MAPK pathway when miR-340 is inhibited. A majority of the targets evaluated demonstrated an increase in fold-change expression between 2 and 4.5, compared to control. To the best of our knowledge none of these targets were previously reported to be regulated by miR-340. Additionally, to our knowledge, 9 of the targets evaluated (CRKL, ELK4, MAP3K5, MAPK8IP2, PPM1A, PPM1B, PPP3CA, PRKX, RAPGEF2 and ZAK) were not previously described to be regulated by a miRNA prior to this report.
Although we evaluated miR-340 expression in melanoma and found it was over expressed, we also evaluated the expression of miR-340 targets and found there was no correlation between the expression of miR-340 in melanoma cell lines and the expression of a few miR-340 targets. These results suggest that possibly the function of miR-340 in suppressing the expression of its targets does not follow the pattern of its expression in melanoma cells. This is not surprising as the function of miRNAs are reported to depend on the abundance of targets (both coding and non-coding, i.e. pseudogenes) and the expression of RNA binding proteins that can affect miRNA-mRNA interactions [42]. Maybe this observation can serve as a cautionary tale when assessing the function of a miRNA based on the levels of its expression. Perhaps the analysis of miRNA function rather than expression would provide more relevant information on their role in cell biology and pathophysiology of diseases.
Although, our findings support previous research regarding tumor suppressor and oncogenic activities of miRNAs in cancer, our data also highlight the mechanism by which one miRNA acts as a tumor suppressor by exhibiting pleiotropic effects on multiple components of the RAS-RAF-MAPK signal transduction pathway in melanoma [7–9]. Future studies on the identification of additional miRNA regulatory networks in melanoma will provide greater understanding of melanoma biology and will present further avenues for therapeutic interventions to improve the prognosis of metastatic melanoma.
Supplementary Material
miR-340 is a novel inhibitor of tumorigenic phenotype in melanoma cells
miR-340 inhibits RAS-RAF-MAPK signaling pathway
miR-340 targets multiple components of RAS-RAF-MAPK pathway
Acknowledgments
We would like to thank Edwarda Desouza and Emily Adochio for their technical assistance. We would also like to thank Drs. Meenhard Herlyn, Yiqun Shellman, Paul Robbins and Anja Katrin Bosserhoff for their generous gifts of reagents. This research is supported by NIEHS training grant ES007015 (to A.M.P) and NIAMS grant AR063361 (to V.S.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Kloosterman WP, Plasterk RH. Developmental cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Friedman RC, Farh KK, Burge CB, Bartel DP. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. PLoS biology. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medina PP, Slack FJ. Cell Cycle. 2008;7:2485–2492. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 8.Krutovskikh VA, Herceg Z. BioEssays : news and reviews in molecular, cellular and developmental biology. 2010;32:894–904. doi: 10.1002/bies.201000040. [DOI] [PubMed] [Google Scholar]
- 9.Rovira C, Guida MC, Cayota A. IUBMB life. 2010;62:859–868. doi: 10.1002/iub.399. [DOI] [PubMed] [Google Scholar]
- 10.Miller AJ, Mihm MC., Jr The New England journal of medicine. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 11.Chin L, Garraway LA, Fisher DE. Genes & development. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 12.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 13.Fedorenko IV, Gibney GT, Smalley KS. Oncogene. 2013;32:3009–3018. doi: 10.1038/onc.2012.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray-Schopfer VC, da Rocha Dias S, Marais R. Cancer metastasis reviews. 2005;24:165–183. doi: 10.1007/s10555-005-5865-1. [DOI] [PubMed] [Google Scholar]
- 15.Solus JF, Kraft S. Adv Anat Pathol. 2013;20:217–226. doi: 10.1097/PAP.0b013e3182976c94. [DOI] [PubMed] [Google Scholar]
- 16.Dweep H, Sticht C, Pandey P, Gretz N. Journal of biomedical informatics. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Betel D, Koppal A, Agius P, Sander C, Leslie C. Genome biology. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betel D, Wilson M, Gabow A, Marks DS, Sander C. Nucleic acids research. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Nature structural & molecular biology. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Molecular cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis BP, Burge CB, Bartel DP. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 22.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. Nucleic acids research. 2007;35:D149–155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 24.Ritchie W, Flamant S, Rasko JE. Nature methods. 2009;6:397–398. doi: 10.1038/nmeth0609-397. [DOI] [PubMed] [Google Scholar]
- 25.Wang X. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, El Naqa IM. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 27.Kyoto Encyclopedia of Genes and Genomes (KEGG) release 65.0. 2013 [Google Scholar]
- 28.Sun Y, Zhao X, Zhou Y, Hu Y. Oncology reports. 2012;28:1346–1352. doi: 10.3892/or.2012.1958. [DOI] [PubMed] [Google Scholar]
- 29.Surgucheva I, Chidambaram K, Willoughby DA, Surguchov A. Journal of ocular biology, diseases, and informatics. 2010;3:41–52. doi: 10.1007/s12177-010-9054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu ZS, Wu Q, Wang CQ, Wang XN, Huang J, Zhao JJ, Mao SS, Zhang GH, Xu XC, Zhang N. Cancer. 2011;117:2842–2852. doi: 10.1002/cncr.25860. [DOI] [PubMed] [Google Scholar]
- 31.Zhou X, Wei M, Wang W. Biochemical and biophysical research communications. 2013;437:653–658. doi: 10.1016/j.bbrc.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 32.Bell RE, Khaled M, Netanely D, Schubert S, Golan T, Buxbaum A, Janas MM, Postolsky B, Goldberg MS, Shamir R, Levy C. The Journal of investigative dermatology. 2014;134:441–451. doi: 10.1038/jid.2013.340. [DOI] [PubMed] [Google Scholar]
- 33.Aziz SA, Davies M, Pick E, Zito C, Jilaveanu L, Camp RL, Rimm DL, Kluger Y, Kluger HM. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:3029–3036. doi: 10.1158/1078-0432.CCR-08-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies MA, Stemke-Hale K, Lin E, Tellez C, Deng W, Gopal YN, Woodman SE, Calderone TC, Ju Z, Lazar AJ, Prieto VG, Aldape K, Mills GB, Gershenwald JE. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:7538–7546. doi: 10.1158/1078-0432.CCR-09-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haass NK, Sproesser K, Nguyen TK, Contractor R, Medina CA, Nathanson KL, Herlyn M, Smalley KS. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:230–239. doi: 10.1158/1078-0432.CCR-07-1440. [DOI] [PubMed] [Google Scholar]
- 36.Herlyn D, Iliopoulos D, Jensen PJ, Parmiter A, Baird J, Hotta H, Adachi K, Ross AH, Jambrosic J, Koprowski H, et al. Cancer research. 1990;50:2296–2302. [PubMed] [Google Scholar]
- 37.Hinselwood DC, Abrahamsen TW, Ekstrom PO. Electrophoresis. 2005;26:2553–2561. doi: 10.1002/elps.200410427. [DOI] [PubMed] [Google Scholar]
- 38.Maddodi N, Huang W, Havighurst T, Kim K, Longley BJ, Setaluri V. The Journal of investigative dermatology. 2010;130:1657–1667. doi: 10.1038/jid.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massoumi R, Kuphal S, Hellerbrand C, Haas B, Wild P, Spruss T, Pfeifer A, Fassler R, Bosserhoff AK. The Journal of experimental medicine. 2009;206:221–232. doi: 10.1084/jem.20082044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dorn GW., 2nd Cell Cycle. 2013;12:707–708. doi: 10.4161/cc.23858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciafre SA, Galardi S. RNA Biol. 2013;10:935–942. doi: 10.4161/rna.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.