Abstract
Expression of Melanoma AntiGen Encoding (MAGE) genes, particularly MAGE-A3, has been correlated with aggressive clinical course, the acquisition of resistance to chemotherapy and poor clinical outcomes of melanoma and other malignancies. MAGE proteins bind to KAP1, a gene repressor and ubiquitin E3 ligase which also binds KRAB domain containing zinc finger transcription factors (KZNFs), and MAGE expression may affect KZNF mediated gene regulation. To investigate mechanisms for these effects, we tested the hypothesis that differences in KRAB domain composition affect KZNF poly-ubiquitination and determine whether MAGE expression increases, decreases, or has no effect on KZNFs mediated gene repression. Using an integrated reporter gene responsive to repression by KRAB domain fusion proteins, we found that MAGE-A3 relieved KZNF mediated repression and induced KZNF poly-ubiquitination and degradation in association with expression of the A+B box KRAB domain. In contrast, MAGE-A3 enhanced KAP1 mediated repression of KZNFs expressing A or A+b box KRAB domains but caused no increase in poly-ubiquitination or degradation. MAGE-A3 has no significant impact on KZNFs with KRAB domains containing the Scan box motif. These data support our hypothesis by showing that the effects of MAGE-A3 on gene repression depend on the type of KZNF KRAB domain involved.
Introduction
The MAGE antigens are proteins that were first discovered because they elicited cytotoxic T cell and humoral responses in patients with malignant melanoma (Knuth et al., 1989). The genes were called MAGE genes for the acronym “Melanoma AntiGen Encoding gene” (Basarab et al., 1999; van der Bruggen et al., 1991). The original MAGE antigens were found to be normally expressed only in male gametes and became the first proteins identified in what is now a much larger group of antigens that are expressed in malignant tumors, placenta and testes, and that are known as Cancer Testes (CT) antigens (Simpson et al., 2005). The original MAGE genes included MAGE-A, B, and C, which are encoded on the X chromosome and have been called CT-X MAGE proteins. A number of autosomal gene families, named MAGE D through MAGE L, have more recently been found to be homologous with the CT-X MAGE genes but appear to be more widely expressed and include some genes which are expressed in all normal tissues (Chomez et al., 2001; Pold et al., 1999). The CT-X genes are now also known as Class I MAGE genes and the non-X encoded genes are called Class II MAGE genes. All MAGE genes are characterized by a MAGE homology domain (MHD) a region encoded by a single exon, showing as much as 98% homology within MAGE families. In this report we will concentrate on Class I MAGE proteins which, because of their tendency for expression in many cancers and hematopoetic malignancies and their very limited expression in normal adult tissues, have been used as tumor specific targets for immunotherapy of melanoma and other malignancies (Brossart, 2002; Coulie et al., 2002; Godelaine et al., 2003; Lonchay et al., 2004; Park et al., 1999; Park et al., 2002; Simpson et al., 2005; Sun et al., 2002; Zhang et al., 2003). The functions of most MAGE proteins remain unknown but several studies have shown correlations between Class I MAGE expression and tumor development, aggressive clinical course, or resistance to chemotherapeutic agents (Bertram et al., 1998; Duan et al., 2003; Gure et al., 2005; Hoek et al., 2004; Park et al., 2002; Simpson et al., 2005). However, it has not yet been conclusively determined whether Class I MAGE gene expression is a functionally irrelevant by-product of cellular transformation or could actually contribute to the development of malignancies (Simpson et al., 2005).
The high degree of homology between Class I MAGE family proteins and the fact they are often co-expressed, suggests that many perform common or complementary functions (Chomez et al., 2001; Lucas et al., 1998; Simpson et al., 2005). For instance, MAGE-A3 and MAGE-A6 differ mainly in un-translated regions and show 98% identity at the nucleotide level in their coding regions. Similarly, the murine mMage-b family, composed of mMage-b1, b2, and b3, are 98–99% homologous at the nucleotide levels and 97–100% identical at the amino acid level. Due to these factors and due to a lack of antibodies that can differentiate between nearly identical sub-family members, most studies of MAGE gene expression rely on detection of mRNA, usually by reverse transcription followed by the polymerase chain reaction (RT-PCR) (Chomez et al., 2001; Lucas et al., 1998; Simpson et al., 2005). Our experiments in this report involve MAGE-A3, which is the most common of the Class I MAGE genes expressed in malignancies and is in many ways characteristic of the Class I MAGE genes.
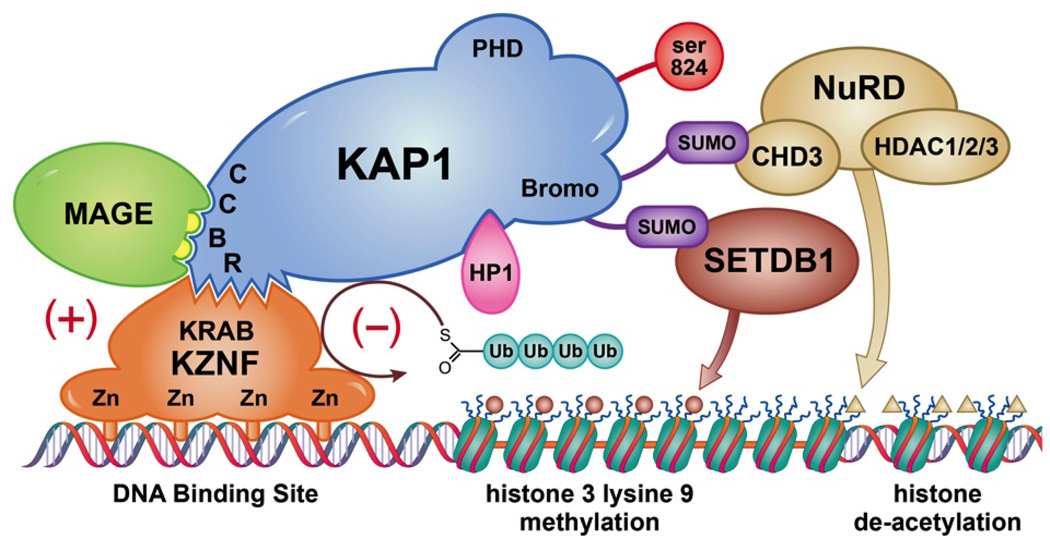
We and others have previously established that MAGE expression suppresses p53 and enhances the cellular response to DNA double strand breaks (Bhatia et al., 2013; Monte et al., 2006; Yang et al., 2007). We also discovered and others confirmed that a common function of the MHD is binding to KAP1, a universally expressed nuclear scaffolding protein and ubiquitin E3 ligase, also known as TRIM28, Tif1b, and Krip125 (Doyle et al., 2010; Yang et al., 2007). KAP1 causes chromatin condensation in several contexts, including a generalized condensation in response to DNA double strand breaks and more localized chromatin compaction causing repression of specific genes mediated by Kruppel-associated box (KRAB) domain containing zinc finger transcription factors (KZNFs)(Friedman et al., 1996) (please see Figure 1). KZNFs are the largest group of transcription factors in vertebrates and they direct KAP1 to specific chromatin sites recognized by their zinc finger motifs. KAP1 recruits SetDb1, histone de-acetylases, and heterochromatin protein 1 (HP1), inducing localized chromatin compaction and gene repression characterized by histone de-acetylation and histone-3-lysine-9- trimethylation (H3me3K9) an epigenetic signature also seen in senescence associated heterochromatin foci (SAHF)(Cammas et al., 2000; Friedman et al., 1996; Groner et al., 2010). Functionally, MAGE-KAP1 co-binding may: i. increase repression of specific genes, ii. have no effect on gene repression, or iii. increase KAP1 mediated ubiquitination of KZNFs with decreased target gene binding and de-repression of specific genes(Xiao et al., 2011). These variable effects mean that MAGE proteins can act as master transcription factors affecting cascades of gene activity.
Figure 1. KAP1 MAGE cartoon.
The KAP1 RING Finger-B Box-Coiled Coil (RBCC) domain binds to the KRAB domains of KZNFs, which target KAP1 to specific DNA sequences through their zinc finger DNA binding motifs. KAP1 mediates localized compaction of chromatin necessary for repression of gene transcription, and is associated with chromatin modifications including histone de-acetylation, histone 3 trimethylation on K9, and HP1 binding to both DNA and histones. All Class I MAGE proteins contain two leucines (yellow semi-circles) which enable binding to the same KAP1 RBCC region as binds KZNFs. In some cases MAGE expression enhances KAP1 E3 ubiquitin ligase activity, resulting in KZNF ubiquitination and degradation, thereby de-repressing the genes targeted by that KZNF. In other cases MAGE expression increases KAP1 binding causing increased gene repression. Modified from (Xiao et al., 2011).
To determine the mechanism(s) for the variable effects of MAGE proteins on gene repression and to identify MAGE effects on KZNFs, we used a comprehensive series of fusion proteins containing different KRAB domains fused to the Gal4-DNA binding domain (DBD), with a cell line containing an integrated KRAB-DBD responsive reporter gene, to test the hypothesis that differences in the composition of KZNF KRAB domains determine whether expression of MAGE proteins increases, decreases, or has no effect on KZNF mediated gene repression. Our work shows that the structure of KRAB domains is the main determinant of the outcome of MAGE mediated biological effects on KZNFS.
Materials and Methods
Tissue culture and transfections
HEK293T were purchased from American Type Culture Collection and were cultured in DMEM with 10% fetal bovine serum and 1% antibiotics (penicillin, streptomycin). CHO-5xGAL4-UAS-TK-Luc-2p cells, the kind gift of Dr. Raul Urrutia, were cultured in F12 with 10% fetal bovine serum and 100ug/ml hygromycin antibiotic(Bhatia et al., 2011; Urrutia, 2003). For transfections with MAGE-A3 plasmid, we used calcium phosphate (Invitrogen) and Lipofectamine 2000 (Invitrogen), the latter according to the manufacturer’s recommendations.
Expression Vectors and Strategy
FLAG-tagged wild type MAGE-A3 was expressed using p3xFLAG-CMV-2 plasmid (Sigma). We used the pSG424 vector for the construction and expression of GAL4-DBD fusion proteins in mammalian cells. The plasmid contains the SV40 ori/early promoter region fused to the coding sequencing for GAL4-DBD (AA 1–147), followed immediately by a polylinker and translation stop codons. Different KZNF KRAB domains were amplified and linked with GAL4 (1–147) at the 3’ end, to generate fused GAL4-DBD-KRAB constructs. Detailed information is show in Tables 1 and 2.
Table 1. Construction of GAL4-KRAB Fusion Constructs.
The clone ID of each KZNF clone from Thermo Fisher Scientific is shown. The corresponding GenBank accession number is also displayed.
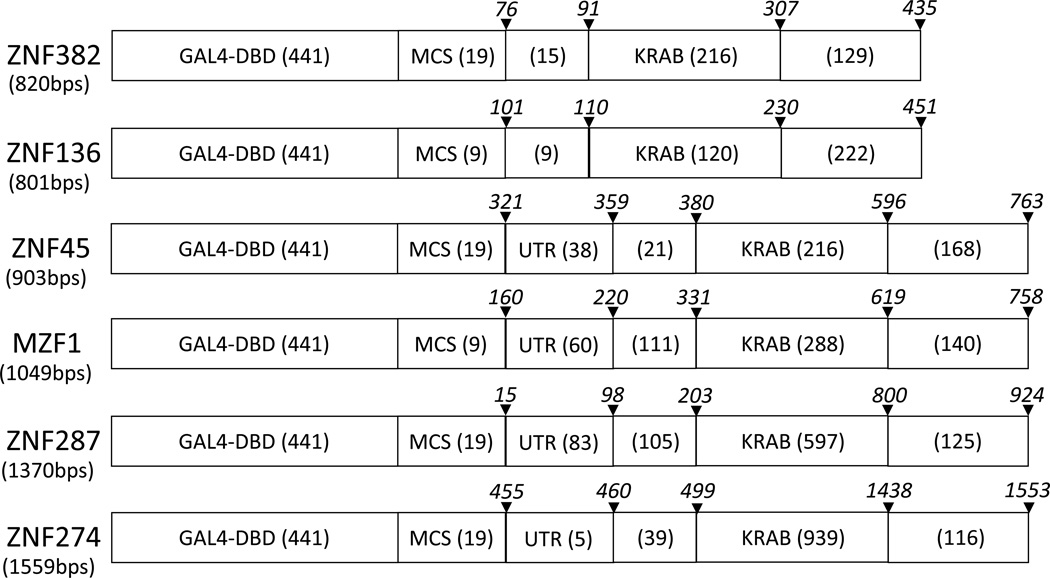
Table 2. Schematic Structure of GAL4-KRAB Hybrid Fusion Constructs.
The number of nucleotides in each region is shown in parentheses. The size of the multiple cloning site (MCS) in the resulting fusion construct is shown. The region and lengths of the KZNF cDNAs are also shown. UTR; untranslated region, KRAB; KRAB domain including A, B, b and SCAN domains. The italicized numbers in each region are base numbers that represent the location of the corresponding region and are adopted from the location numbering system of Genbank.
 |
Construction of Plasmids for GAL4-DBD-KRAB Fusion Protein Expression
Mammalian Gene Collection (MGC)-verified full length human KZNF cDNA clones were purchased from Thermo Fisher Scientific (Waltham, MA). PCR was performed to amplify ZNF cDNA using HotStar HiFidelity PCR kit (Qiagen, Valencia, CA). Primers were designed to amplify the region that includes KRAB and SCAN domains but not Zn finger motifs. Primers were also designed to include recognition sequences of the restriction enzymes as adaptors and three additional nucleotides to help binding of the restriction enzymes. The NCBI’s Primer-BLAST design tool was used to design primers and to calculate primer melting temperatures. After PCR, amplicons were digested with corresponding restriction enzymes to generate ligation-ready DNA fragments. For backbones, the pSG424 vector, which includes GAL4 DBD upstream of multiple cloning sites, was cut with the same restriction enzymes. Inserts were then ligated with the pSG424 backbone using Quick Ligation kit (New England BioLabs, Ipswich, MA). For transformation, 2µl of ligation reaction and One Shot MAX Efficiency DH5α-T1 competent cells (Invitrogen, Carlsbad, CA) were used according to the manufacturer’s protocol. Colonies were analyzed by restriction analysis and positive clones were sequenced (UW-Madison, Biotechnology Center) to verify correct incorporation and sequences of inserts.
Expression of the GAL4-DBD-KRAB Fusion Proteins Was Verified by Immunoblotting
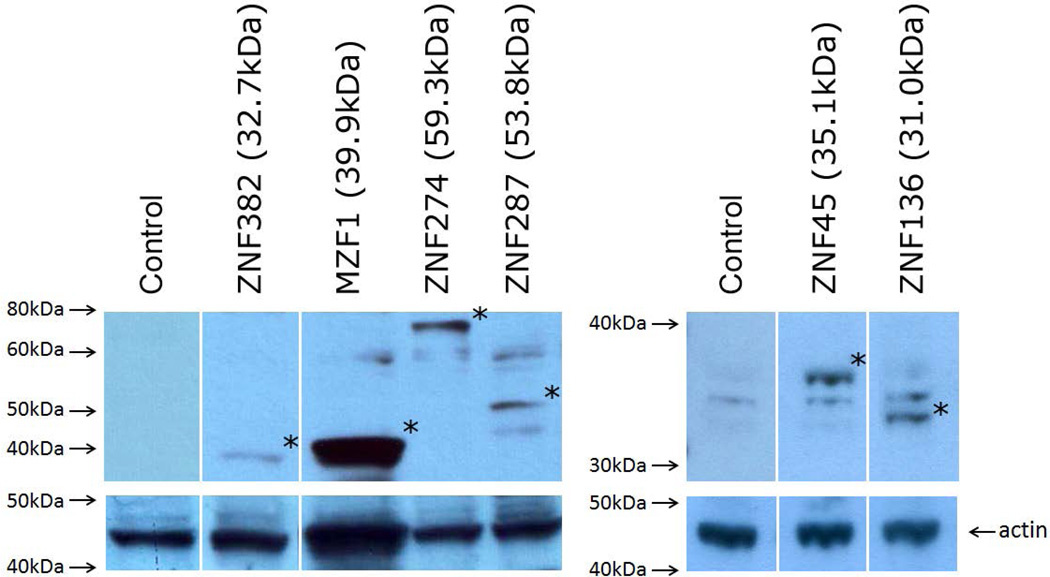
Briefly, HEK293T cells in 6-well plate were transiently transfected with either X-Fect (Clonetech, Mountain View, CA) or Lipofectamine 2000 (Invitrogen) and 5µg of ZNF expression vectors. Transfected cells were cultured for 48hrs before harvest. Cells were lysed in RIPA buffer (25mM Tris-HCl, pH 7.6, 150mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) supplemented with protease and phosphatase inhibitors. After clarification of the lysate by centrifugation, protein concentration was measured with Bio-Rad DC protein assay kit (Bio-Rad, Hercules, CA). The Mini-PROTEAN gel system (Bio-Rad) was used for gel electrophoresis and Western blotting. Antibody against the GAL4-DNA binding domain (US Biological, Swampscott, MA) was used as primary antibody to detect GAL4-ZNF fusion protein expression. Horseradish peroxidase (HRP)-linked anti-rabbit IgG was used as secondary antibody (Cell Signaling (Danvers, MA). For the detection of HRP on immunoblots, SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) was utilized (Figure 2).
Figure 2. Expression of KRAB Fusion Proteins.
The coding regions of all constructs were verified by forward and backward sequencing. The expression of diverse GAL4-KRAB fusion proteins (marked with asterisks) was verified by immunoblotting with anti-DBD antibody. Predicted sizes of the protein backbones are shown. Note that the ZNF287 runs at a higher than predicted molecular weight, perhaps due to glycosylation or other post translational modification. The membrane was stripped and re-probed with anti-actin antibody as a loading control.
Luciferase Reporter Assay
The transcriptional regulatory activity of KRAB domains was determined using the 5xGAL4-UAS-TK-Luc-luciferase reporter system, as described previously(Gebelein et al., 1998; Gebelein and Urrutia, 2001; Urrutia, 2003; Xiao et al., 2011). Briefly, CHO cells containing a stably integrated 5xGAL4-UAS-TK-Luc luciferase reporter gene were maintained under hygromycin selection and co-transfected with various 5xGAL4-DBD-KRAB constructs and reporter plasmid using LipofectAMINE™ (Life Technologies, Inc.). As a control for basal transcriptional activity, the CHO cells were co-transfected with the pcDNA3.1/HisC vectors. At 24 h after transfection, luciferase assays were performed using a Turner 20/20 luminometer and the dual luciferase assay system according to manufacturer's suggestions (Promega). The Rous sarcoma virus-Renilla reporter was added to each transfection as an indicator of transfection efficiency. The relative luciferase activity was expressed as a ratio between the luciferase activity in 5xGAL4-DBD transfected cells vs. the luciferase activity in cells transfected with various 5xGAL4-DBD-KRAB constructs. All studies were performed in triplicate in at least three independent experiments with similar results.
In Vivo Ubiquitination Assay
8 µg of wild-type or mutant MAGE-A3 expression plasmids, or empty vector control, were introduced into HEK293T cells at 70% confluency using the CaPO4 method (Clotech). 8 µg GAL4-KRAB-pSG424, 4 µg KAP1, 4 µg of pcDNA 3.1+ vector harboring HA tagged ubiquitin were co-transfected. After 20 h, the cells were treated with 25 µM of MG132 (C2211, Sigma) for 5 h, lysed in RIPA buffer, and subjected to immunoprecipitation. A 500 µg aliquot of total protein from the transfected HEK293T cells in 250 µl lysis solution was mixed with 10 µl of the anti-GAL4-DBD or anti-Flag affinity matrix (US Biological, Sigma-Aldrich) pre-blocked with 2% bovine serum albumin (BSA), and incubated overnight with gentle rotation at 4°C. The affinity matrix was washed with HIPA buffer three times, collected by centrifugation, and the precipitated proteins were denatured in sample buffer containing 0.1M dithiothreitol (DTT), subjected to 7% SDS-PAGE and transferred to a PVDF membrane. The transferred proteins were then incubated with anti-GAL4-DBD (G1018, Polyclonal, US Biological) or Anti-Flag (M2 F1804, Monoclonal, Sigma-Aldrich), or anti-HA (monoclonal, Cell signaling) antibodies. Anti-rabbit or mouse IgG, HRP-linked secondary antibodies (Cell signaling) were used for the western blotting procedure.
Plotting and Statistics
Plotting and statistics were performed using Sigma Plot 12 (Systat Software, San Jose, CA). The Mann-Whitney Rank Sum test was used to test for differences in repression with P<0.05.
Results
MAGE-KAP1 Mediated Gene Regulation Depends on KRAB Domain Structure
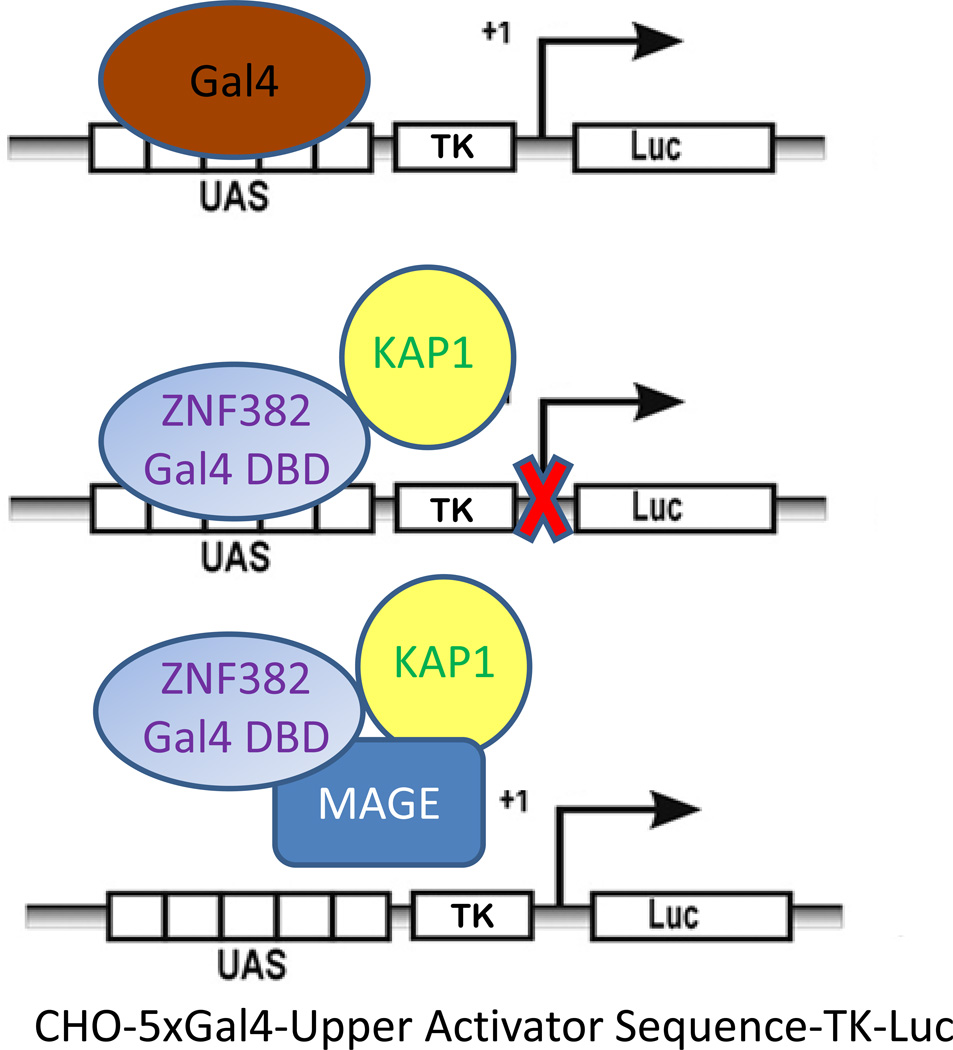
To determine the effects of MAGE expression with different KZNF KRAB domains we used the CHO-5xGAL4-UAS-TK-Luc cell line, a Chinese hamster ovary cell line that contains an integrated fusion gene composed of an optimized binding site (UAS) for the GAL4-DNA binding domain (GAL4-DBD) upstream of a constitutively active luciferase reporter gene(Gebelein et al., 1998; Gebelein and Urrutia, 2001; Urrutia, 2003; Xiao et al., 2011)(Figure 3). Plasmid based ectopic expression of the GAL4-DBD alone has no effect on luciferase production. However, ectopic expression of a fusion protein in which GAL4-DBD replaces the zinc finger motifs of a KZNF can recruit KAP1 to the promoter of the luciferase reporter gene, causing chromatin compaction and repression of luciferase expression. CHO-5xGAL4-UAS-TK-Luc-2p has limited expression of KAP1 so the degree of recruitment of KAP1 can be determined by measurement of luciferase activity. In our previous work we showed that the GAL4-DBD fused with the KRAB domain of ZNF382, a KZNF, recruits KAP1 to the CHO-5xGAL4-UAS-TK-Luc-2p fusion gene causing repression of the luciferase reporter gene. In addition, ectopic MAGE expression relieved that repression, causing increased expression of luciferase (Xiao et al., 2011). We also found that MAGE proteins have differential effects on different genes, decreasing expression of some and increasing expression of others.
Figure 3. CHO-5xGAL4-UAS-TK-Luc-2p Gene Repression Assay.
The CHO-5xGal4-UAS-TK-Luc-2p cell line has limited expression of KAP1 and contains an integrated fusion gene with an optimized recognition site (UAS) for the Gal4-DNA binding domain (Gal4-DBD). The UAS is upstream of a constitutively active promoter (TK) activating a luciferase (Luc) reporter gene. Ectopic expression of a KRAB domain-Gal4-DBD fusion protein can recruit KAP1 to the TK promoter region and cause repression of the luciferase reporter gene. This model indicates how MAGE may influence binding between KZNFs and KAP1. For instance, MAGE could block binding between KAP1 and a KZNF-Gal4-DBD fusion protein either by steric hindrance or by increasing ubiquitination and degradation of the fusion protein. Alternatively, MAGE could stabilize KAP1 and ZNF interaction, leading to increased gene repression. DBD=Gal4 DNA binding Domain. UAS=Upstream Activation Sequence. TK= constitutively active promoter. Luc = luciferase reporter gene.
To test the hypothesis that the variable effects of MAGE on KZNF regulated gene expression are determined by the structure of the KRAB domain of the individual KZNF, we made of series of GAL4-DBD fusion genes with different KRAB domains (Tables 1 and 2) and tested their affects on repression of the the luciferase reporter gene (Fig 4).
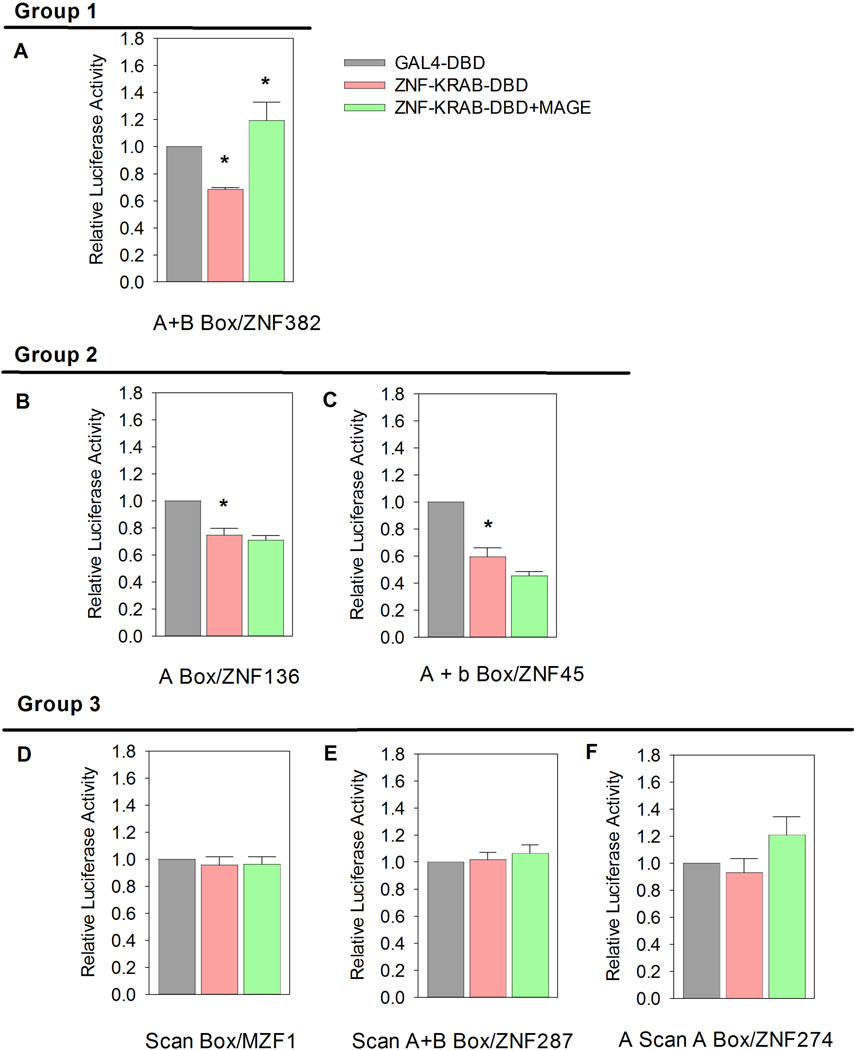
Figure 4. MAGE Effects on KAP1 Mediated Gene Repression Depend on KRAB Domain Structure.
CHO cells with a stably integrated 5xGAL4-UAS-TK-Luc reporter gene were maintained under hygromycin selection and co-transfected with various 5xGAL4-DBD- KRAB fusion constructs and reporter plasmid using LipofectAMINE™. The relative luciferase activity is expressed as a ratio between the activity of 5xGAL4-DBD with no KRAB domain vs. various 5xGAL4-DBD-KRAB fusion proteins. Results are shown normalized to reporter gene expression in the presence of 5xGAL4-DBD without KRAB domain. Luciferase activities were also measured in CHO-5xGAL4-UAS-TK-Luc-2p cells transfected with or without vector expressing full length MAGE-A3. Maximum luciferase repression occurs with the fusion protein containing the ZNF382 KRAB domain, which consists of A+B boxes (* P<0.05). Addition of a MAGE-A3 expression plasmid relieves ZNF382-KRAB related repression and increases luciferase activity (* P<0.05). Like the ZNF382-KRAB fusion protein, ZNF136 (A box) and ZNF45 (A+b boxes) fusion proteins cause significant repression (* P<0.05). However, neither shows significant rescue by MAGE-A3. No significant change in luciferase activity was observed with MZF1 (Scan), ZNF287 (Scan A+B boxes) and ZNF274 (A Scan A boxes) KRAB domain fusion proteins, with or without addition of MAGE-A3.
ZNF382 is a KZNF with an A and B Box KRAB domain that has been shown to be a tumor suppressor (Cheng et al., 2010). As we have previously reported and now confirm, ectopic expression of ZNF382 KRAB-Gal4-DBD fusion protein recruits KAP1 to the reporter gene and suppresses luciferase expression (P<0.05). Co-expression of MAGE proteins rescues reporter gene activity from ZNF382 KRAB domain mediated repression (P<0.05).
ZNF136 is a KZNF containing a KRAB domain composed only of an A box. We now report that like the ZNF382 KRAB domain, the ZNF136 KRAB domain causes repression in the CHO-5xGAL4-UAS-TK-Luc system (P<0.05). However, unlike repression caused by the ZNF382 KRAB domain, repression by ZNF136-KRAB is not rescued by MAGE-A3 expression.
ZNF45 is a KZNF containing a KRAB domain consisting of an A and b box. ZNF 45-KRAB repressed luciferase activity (P<0.05) but repression was not rescued by MAGE expression. In fact, there was a trend towards increased repression in the presence of MAGE but the trend did not reach statistical significance at the p≤ 0.05 level.
We also tested the MZF1, ZNF274 and ZNF287, all of which are KZNFs with KRAB domains containing a Scan motif. All of these fusion proteins showed no effect on luciferase expression, whether or not MAGE was present. These KRAB domains contained A and/or B box motifs, like those that repressed luciferase in the ZNFs 382, 136, and 45, but the presence of a Scan motif appears dominant in preventing regulation of genes in this system.
MAGE-A3 Enhances Ubiquitination of ZNF382 but not Other KZNFS
We and others have shown that MAGE expression enhances KAP1 ubiquitin ligase activity and specifically increases ubiquitination and degradation of p53 (Bhatia et al., 2013; Monte et al., 2006; Xiao et al., 2011). We have also shown that MAGE-C2 increases KAP1 mediated ubiquitination and degradation of ZNF382. To determine whether MAGE induced ubiquitination is a general phenomenon we investigated KZNF ubiquitination in the presence of MAGE-A3. As we expected from previous studies with MAGE-C2, we found that MAGE-A3 expression enhanced ZNF382 ubiquitination and degradation in an in vivo ubiquitination reaction (Figure 5). Because KZNF binding to KAP1 is necessary for repression of specific genes, these results show that MAGE-A3 may prevent significant repression of a KZNF target gene by increasing ubiquitination and degradation of KZNFs that direct KAP1 to specific chromatin sites. In contrast to KZNF ZNF382, ubiquitination was not seen in the presence or absence of MAGE with any of the other KZNFs we tested. For instance, no ubiquitination activity was detected with ZNF136 and ZNF45, KZNFs which increase repression of the luciferase reporter gene but are not de-repressed in the presence of MAGE-A3 (Figures 4 and 5). This finding is consistent with co-binding of MAGE-A3 to KAP1 with ZNF136 and ZNF45, perhaps with stabilization rather than degradation of the KAP1-KZNF complex by MAGE-A3. Likewise, no ubiquitination was found with MZF1, ZNF287 and ZNF274, all of which have KRAB domains containing the Scan box motif. This finding correlates with the finding that these KZNFs have no effect on repression in the luciferase reporter gene assay.
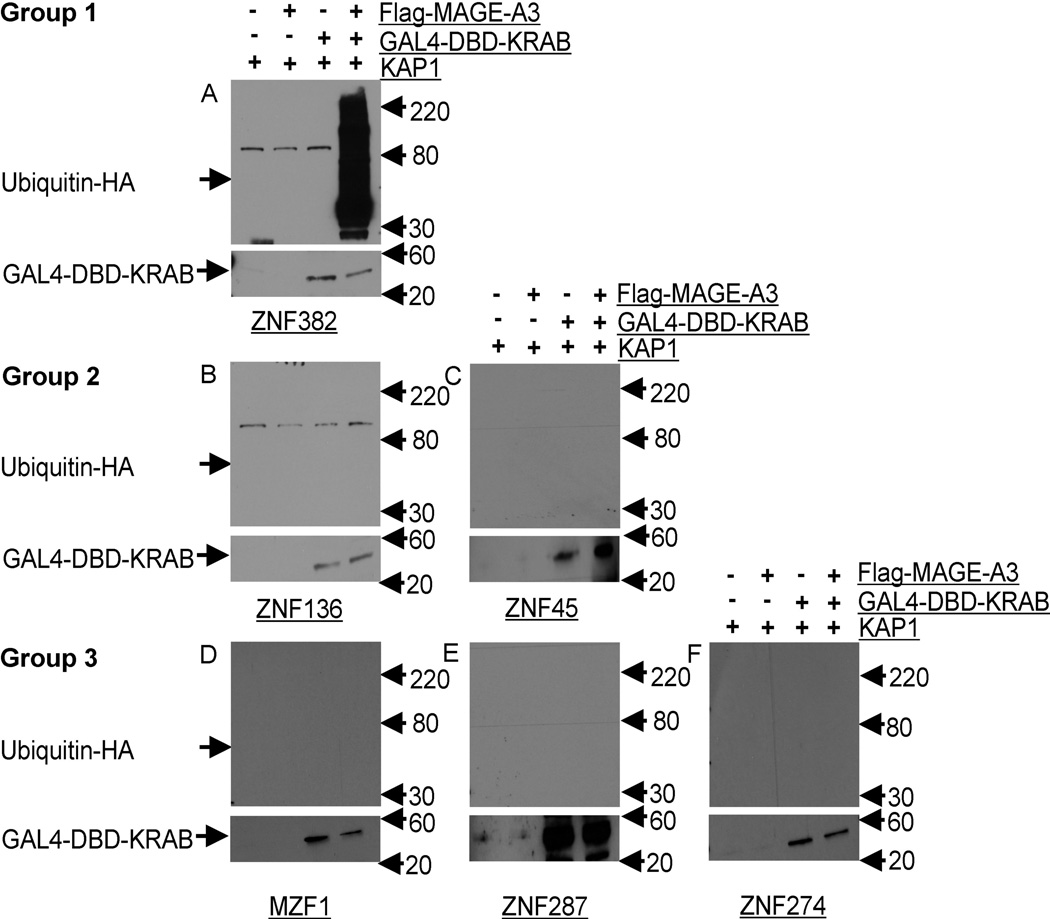
Figure 5. MAGE–KAP1 Binding Induces Ubiquitination Only With the ZNF382 KRAB domain.
HEK293T cells were transiently transfected with Flag tagged MAGE-A3 vector or empty vector, and KRAB domain GAL4-DBD fusion proteins, KAP1, and HA tagged ubiquitin. Proteasome activity was inhibited by incubation for 5 hours in the presence of 25 µM MG132. Top blots: GAL4-DBD tagged proteins were immunoprecipitated with anti-GAL4-DBD and ubiquitination was detected with anti-HA. Lower blots: immunoprecipitation and immunoblotting confirmed expression of all GAL4-DBD fused KRAB domains. Protein ladder was labeled at the right side. High-molecular-weight ubiquitinated species were seen only in blots of cells transfected with both GAL4-DBD-ZNF382-KRAB domain fusion protein and MAGE-A3 (panel A). No ubiquitination was observed in blots of cells transfected with GAL4-DBD fused KRAB domains from other KZNFs, regardless of MAGE-A3 expression, panels B to F (B=ZNF136, C=ZNF45, D=MZF1, E=ZNF287 and F=ZNF274).
KAP1 Mediated Ubiquitination of ZNF382 is Selective; MAGE-A3 is not Ubiquitinated in MAGE-KZNF-KAP1 complexes
Lastly, to test whether MAGE itself can be ubiquitinated by the KAP1 ubiquitin E3 ligase, we investigated ubiquitination of MAGE-A3 and found no appreciable ubiquitination in presence of any of the individual KRAB domains we studied. Thus, no self-ubiquitination of MAGE-A3 is mediated by KZNFs and the KAP1 ubiquitin ligase (Figure 6).
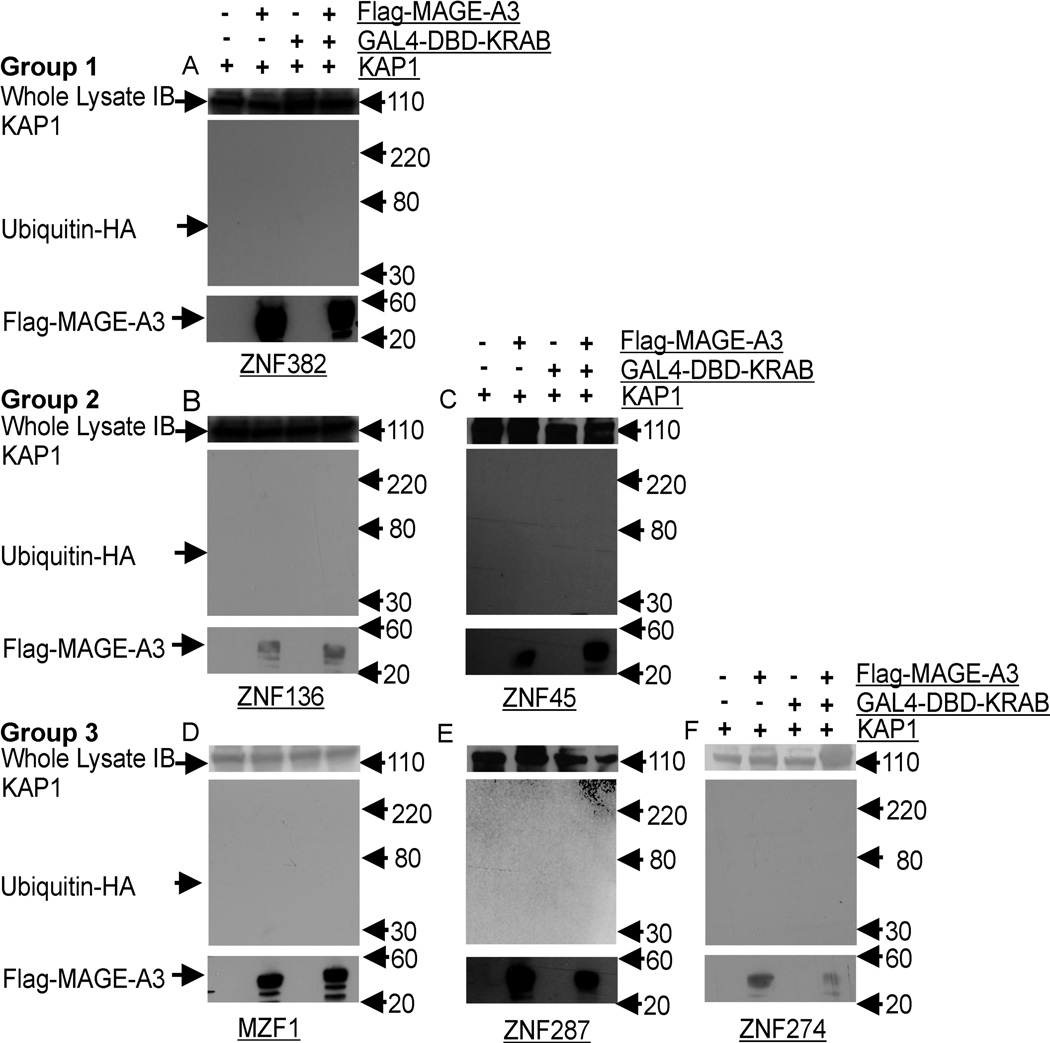
Figure 6. MAGE-A3 Is Not Ubiquitinated During KRAB-KAP1 Interactions.
HEK293T cells were transiently transfected with Flag tagged MAGE-A3 or empty vectors and KRAB domain GAL4-DBD constructs, KAP1, and HA tagged ubiquitin. Proteasome activity was inhibited by incubation for 5 hours in the presence of 25 µM MG132. Top panel, whole lysate immunoblot (IB) for KAP1; middle panel, Flag-MAGE-A3 tagged ubiquitinated proteins were immunoprecipitated with anti-Flag and detected with anti-HA; Lower panel: immunoprecipitation and immunoblotting confirmed expression of Flag-MAGE-A3. No ubiquitinated MAGE-A3 species were seen in blots of cells transfected with both KZNFs and MAGE-A3, panels A to F (A=ZNF382, B=ZNF136, C =ZNF45, D=MZF1, E=ZNF287 and F=ZNF274).
Discussion
Human Class I MAGE genes include the MAGE A, B, and C families. Class I MAGE genes are normally expressed mainly in developing germ cells and placenta. Class I MAGE genes differ from each other mainly in their un-translated regulatory regions, suggesting the proteins provide a common critical function that may be required at different times or sites during development of gametes and placenta. Promoter region hypomethylation, which may occur during epigenetic reprogramming in tumors, often results in the co-expression of multiple MAGE A and MAGE C proteins, especially MAGE-A3 and MAGE-C2, in a broad range of tumors including actinic keratoses, squamous cell carcinomas of skin, head, and neck, most breast carcinomas, some nevi and most advanced melanomas(Hoek et al., 2004; Jungbluth et al., 2005; Simpson et al., 2005). MAGE proteins have been increasingly recognized as tumor specific targets and MAGE expression has been correlated with aggressive clinical course, the acquisition of resistance to chemotherapy and poor clinical outcome, yet the dominant paradigm has been to view MAGE proteins as passive targets for immunotherapy and the functional contributions of MAGE to oncogenesis and the mechanisms involved have been relatively understudied.
Class I MAGE proteins are characterized by nearly identical MAGE homology domains (MHDs) containing two highly conserved leucines that we and others have shown are necessary for binding to KAP1 (Bhatia et al., 2013; Doyle et al., 2010). In particular, we have shown that they are necessary for the regulatory affects of MAGE-C2 on ZNF382 function and ubiquitination. Our current studies extend these results by showing that MAGE-A3 has similar regulatory effects on ZNF382, and show that these regulatory effects are specific since they are not seen with other KZNFs besides ZNF-328. Since MAGE-A3 and MAGE-C2 are members of different Class I MAGE sub families, these results show that diverse MAGE proteins can have identical effects, supporting the hypothesis that these proteins have common functions at the level of regulation of individual genes.
KZNFs are the largest group of transcription factors in vertebrates and function by directing the KAP1 complex to repress specific genes. KAP1 is a universally expressed 100kD nuclear scaffolding protein and ubiquitin E3 ligase also known as TF1B, RNF96, and TRIM28 (Friedman et al., 1996; Sherr and McCormick, 2002). KAP1 was first discovered as a transcriptional co-repressor required by all KZNFs. A current popular model predicts that zinc finger motifs of KZNFs bind to their target DNA sequences, recruiting KAP1, Setdb1, heterochromatin 1 (HP1), and histone de-acetylases (HDACs), leading to gene repression by formation of localized facultative heterochromatin regions in target gene promoter regions (Please see figure 1). In this model, KZNF KRAB domains bind to the RBCC domain of KAP1. It is not known whether the components of the KRAB domain itself help determine how the co-repressor complexes are assembled or bind to specific chromatin sites. Furthermore, it is not known what other factors may influence the ubiquitin E3 ligase activity of KAP1.
The KZNF family has been divided into four subfamilies on the basis of the primary structure of the KRAB amino-terminal repressor domain: those that contain an A box alone (A box), those with an A box combined with a B box (A+B box), those combining the A box with a divergent B box (A+b Box), and those containing a Scan domain (Urrutia, 2003) (Fig 7).
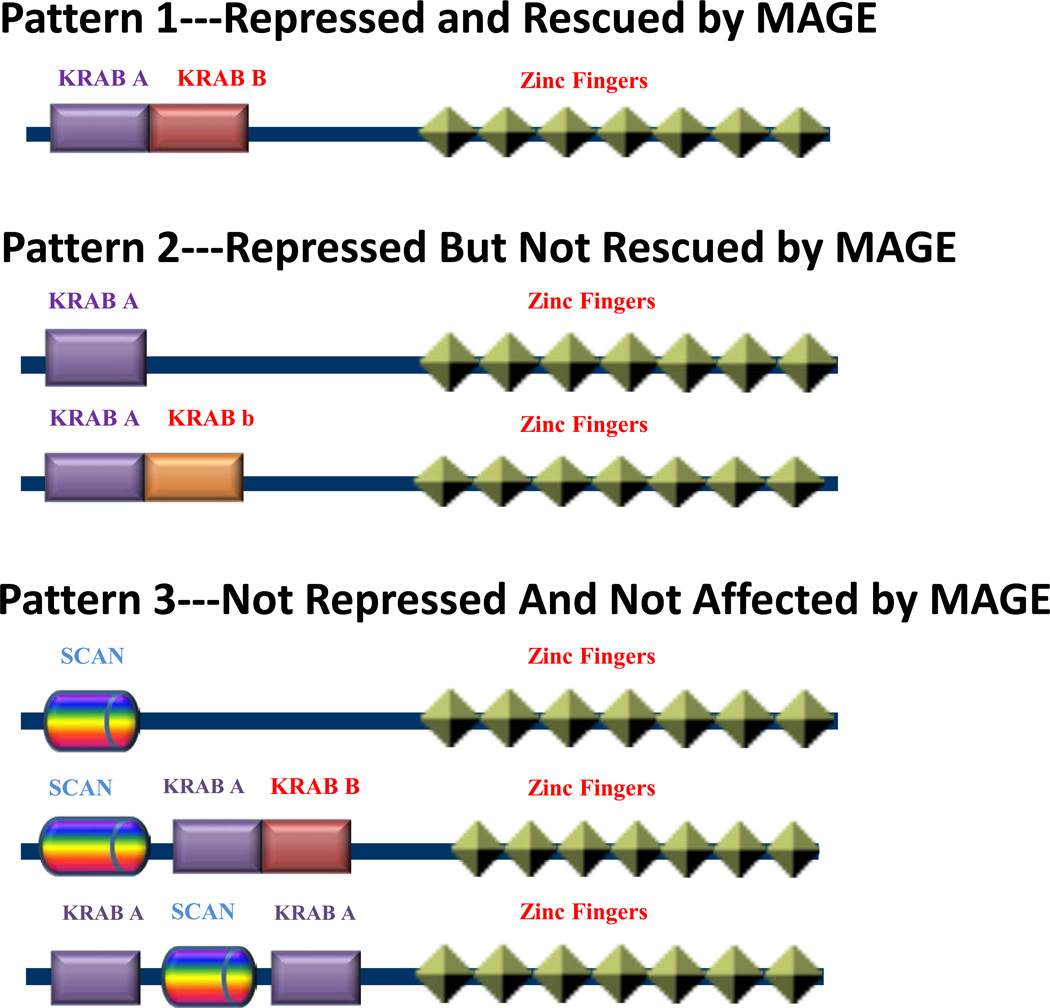
Figure 7. Schematic Models For Primary Structures of Typical KRAB–Domains.
KRAB domains can be divided into three groups depending on their interactions with MAGE and KAP1: Group 1 (KRAB A+B) is repressed by KRAB domains and de-repressed by MAGE; Group 2 (KRAB A, KRAB A+b) is repressed by KRAB domains but not rescued by MAGE; Group 3 (SCAN, SCAN+KRAB A+B, KRAB A+SCAN+KRAB A) is not repressed by KRAB domains and is not affected by MAGE.
Based upon our luciferase reporter gene analysis, all KZNFs without the Scan motif show significant reduction in luciferase activity, indicating the KAP1-KZNF complexes are efficiently assembled to form the co-repressor complexes necessary to induce and maintain transcriptional repression. However, effects are reversed by MAGE-A3 in ZNF382, which has a KRAB domain structure including the A+B boxes and shows a distinct pattern of repression without MAGE, and de-repression or rescue in the presence of MAGE-A3.
Similar to the effects of the ZNF382 KRAB A+B box motif, KZNFs with the A box alone (ZNF136) or the A+b box motifs (ZNF45) cause gene repression. However, in contrast to the ZNF382 A+B box motif, neither A+b nor A only types of KRAB domain show rescue with MAGE-A3. In fact, the ZNF45 A+b box motif KRAB domain shows a strong tendency to increase repression in the presence of MAGE-A3. This clearly shows MAGE-A3 can differentially modulate KZNFs depending on the KRAB domain involved. Ubiquitination only occurred with the ZNF382 A+B box motif, offering an attractive mechanism for MAGE mediated de-repression in which ubiquitination of the KZNF leads to degradation and loss of the KZNF necessary to anchor the KAP1 repressor complex at a particular site, with subsequent loss of heterochromatin and de-repression of specific genes.
Members of a defined subset of KRAB ZNFs contain a Scan domain, which has not been associated with transcriptional regulation but instead allows homo- and heter-dimerization with other Scan-containing KZNF proteins(Collins et al., 2001). Our luciferase data show that KZNFs containing a Scan motif, including those with Scan box only, Scan-A+B, or the A-Scan-A box motifs have no significant effect on luciferase activity, indicating the Scan motif is associated with weaker, if any, KAP1 mediated transcriptional repression in comparison with other KRAB domains. Additionally, MAGE-A3 has no significant influence on KZNFs containing Scan domains in terms of luciferase activity or ubiquitination. Since the ZNF274 and 287 KRAB domains contain A and B motifs, the Scan domain appears dominant and is associated with loss of repression by the A or B boxes.
In summary, we show here that: 1. Depending on the KRAB domain involved, MAGE expression has differential effects on KRAB-zinc finger transcription factor mediated KAP1 binding. 2. These binding patterns affect gene repression, and 3. MAGE-A3 expression can differentially affect ubiquitination of KZNF by KAP1 ubiquitin E3 ligase. Combined with our previous published work on MAGE-C2, these results allow generalization of our findings to most Class I MAGE proteins (Xiao et al., 2011). These results allow us to suggest a functional classification of KRAB domains (Fig7) to complement the structural classification previously proposed (Urrutia, 2003). The functional divisions would include those KRAB domains that repress transcription and are rescued from repression in the presence of MAGE, those KRAB domains that repress and are un-responsive to MAGE, and the scan domain KRAB regions that do not repress and are not responsive to MAGE.
Acknowledgments
We thank Dr. Vladimir Spiegelman (University of Wisconsin School of Medicine and Public Health, Madison) for providing us the HA tagged ubiquitin plasmid. We also thank Dr. Vijay Setaluri, Dr. Nihal Ahmed, and Dr. Hasan Mukhtar (University of Wisconsin School of Medicine and Public Health, Madison) for useful discussions. This work was supported by USPHS Grant R21 AR062773 (BJL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Basarab T, Picard JK, Simpson E, Russell-Jones R. Melanoma antigenencoding gene expression in melanocytic naevi and cutaneous malignant melanomas. Br J Dermatol. 1999;140:106–108. doi: 10.1046/j.1365-2133.1999.02616.x. [DOI] [PubMed] [Google Scholar]
- Bertram J, Palfner K, Hiddemann W, Kneba M. Elevated expression of S100P, CAPL and MAGE 3 in doxorubicin-resistant cell lines: comparison of mRNA differential display reverse transcription-polymerase chain reaction and subtractive suppressive hybridization for the analysis of differential gene expression. Anticancer Drugs. 1998;9:311–317. doi: 10.1097/00001813-199804000-00004. [DOI] [PubMed] [Google Scholar]
- Bhatia N, Xiao TZ, Rosenthal KA, Siddiqui IA, Thiyagarajan S, Smart B, Meng Q, Zuleger CL, Mukhtar H, Kenney SC, et al. MAGE-C2 promotes growth and tumorigenicity of melanoma cells, phosphorylation of KAP1, and DNA damage repair. J Invest Dermatol. 2013;133:759–767. doi: 10.1038/jid.2012.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia N, Yang B, Xiao TZ, Peters N, Hoffmann MF, Longley BJ. Identification of novel small molecules that inhibit protein-protein interactions between MAGE and KAP-1. Arch Biochem Biophys. 2011;508:217–221. doi: 10.1016/j.abb.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brossart P. Dendritic cells in vaccination therapies of malignant diseases. Transfus Apher Sci. 2002;27:183–186. doi: 10.1016/s1473-0502(02)00041-1. [DOI] [PubMed] [Google Scholar]
- Cammas F, Mark M, Dolle P, Dierich A, Chambon P, Losson R. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development. 2000;127:2955–2963. doi: 10.1242/dev.127.13.2955. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Geng H, Cheng SH, Liang P, Bai Y, Li J, Srivastava G, Ng MH, Fukagawa T, Wu X, et al. KRAB zinc finger protein ZNF382 is a proapoptotic tumor suppressor that represses multiple oncogenes and is commonly silenced in multiple carcinomas. Cancer Res. 2010;70:6516–6526. doi: 10.1158/0008-5472.CAN-09-4566. [DOI] [PubMed] [Google Scholar]
- Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- Collins T, Stone JR, Williams AJ. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulie PG, Karanikas V, Lurquin C, Colau D, Connerotte T, Hanagiri T, Van Pel A, Lucas S, Godelaine D, Lonchay C, et al. Cytolytic T-cell responses of cancer patients vaccinated with a MAGE antigen. Immunol Rev. 2002;188:33–42. doi: 10.1034/j.1600-065x.2002.18804.x. [DOI] [PubMed] [Google Scholar]
- Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Duan Y, Lamendola DE, Yusuf RZ, Naeem R, Penson RT, Seiden MV. Overexpression of MAGE/GAGE genes in paclitaxel/doxorubicin-resistant human cancer cell lines. Clin Cancer Res. 2003;9:2778–2785. [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Gebelein B, Fernandez-Zapico M, Imoto M, Urrutia R. KRAB-independent suppression of neoplastic cell growth by the novel zinc finger transcription factor KS1. J Clin Invest. 1998;102:1911–1919. doi: 10.1172/JCI1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebelein B, Urrutia R. Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger-Kruppel-associated box protein. Mol Cell Biol. 2001;21:928–939. doi: 10.1128/MCB.21.3.928-939.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godelaine D, Carrasco J, Lucas S, Karanikas V, Schuler-Thurner B, Coulie PG, Schuler G, Boon T, Van Pel A. Polyclonal CTL responses observed in melanoma patients vaccinated with dendritic cells pulsed with a MAGE-3.A1 peptide. J Immunol. 2003;171:4893–4897. doi: 10.4049/jimmunol.171.9.4893. [DOI] [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, Bucher P, Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6:1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gure AO, Chua R, Williamson B, Gonen M, Ferrera CA, Gnjatic S, Ritter G, Simpson AJ, Chen YT, Old LJ, Altorki NK. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin Cancer Res. 2005;11:8055–8062. doi: 10.1158/1078-0432.CCR-05-1203. [DOI] [PubMed] [Google Scholar]
- Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- Jungbluth AA, Ely S, DiLiberto M, Niesvizky R, Williamson B, Frosina D, Chen YT, Bhardwaj N, Chen-Kiang S, Old LJ, Cho HJ. The cancer-testis antigens CT7 (MAGE-C1) and MAGE-A3/6 are commonly expressed in multiple myeloma and correlate with plasma-cell proliferation. Blood. 2005;106:167–174. doi: 10.1182/blood-2004-12-4931. [DOI] [PubMed] [Google Scholar]
- Knuth A, Wolfel T, Klehmann E, Boon T, Meyer zum Buschenfelde KH. Cytolytic T-cell clones against an autologous human melanoma: specificity study and definition of three antigens by immunoselection. Proc Natl Acad Sci U S A. 1989;86:2804–2808. doi: 10.1073/pnas.86.8.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonchay C, van der Bruggen P, Connerotte T, Hanagiri T, Coulie P, Colau D, Lucas S, Van Pel A, Thielemans K, van Baren N, Boon T. Correlation between tumor regression and T cell responses in melanoma patients vaccinated with a MAGE antigen. Proc Natl Acad Sci U S A. 2004;1012(Suppl):14631–14638. doi: 10.1073/pnas.0405743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas S, De Smet C, Arden KC, Viars CS, Lethe B, Lurquin C, Boon T. Identification of a new MAGE gene with tumor-specific expression by representational difference analysis. Cancer Res. 1998;58:743–752. [PubMed] [Google Scholar]
- Monte M, Simonatto M, Peche LY, Bublik DR, Gobessi S, Pierotti MA, Rodolfo M, Schneider C. MAGE-A tumor antigens target p53 transactivation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc Natl Acad Sci U S A. 2006;103:11160–11165. doi: 10.1073/pnas.0510834103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kim CJ, Lee JH, Shin SH, Chung GH, Jang YS. Effective immunotherapy of cancer by DNA vaccination. Mol Cells. 1999;9:384–391. [PubMed] [Google Scholar]
- Park JW, Kwon TK, Kim IH, Sohn SS, Kim YS, Kim CI, Bae OS, Lee KS, Lee KD, Lee CS, et al. A new strategy for the diagnosis of MAGE-expressing cancers. J Immunol Methods. 2002;266:79–86. doi: 10.1016/s0022-1759(02)00105-9. [DOI] [PubMed] [Google Scholar]
- Pold M, Zhou J, Chen GL, Hall JM, Vescio RA, Berenson JR. Identification of a new, unorthodox member of the MAGE gene family. Genomics. 1999;59:161–167. doi: 10.1006/geno.1999.5870. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- Sun X, Hodge LM, Jones HP, Tabor L, Simecka JW. Co-expression of granulocyte-macrophage colony-stimulating factor with antigen enhances humoral and tumor immunity after DNA vaccination. Vaccine. 2002;20:1466–1474. doi: 10.1016/s0264-410x(01)00476-5. [DOI] [PubMed] [Google Scholar]
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- Xiao TZ, Bhatia N, Urrutia R, Lomberk GA, Simpson A, Longley BJ. MAGE I transcription factors regulate KAP1 and KRAB domain zinc finger transcription factor mediated gene repression. PLoS One. 2011;6:e23747. doi: 10.1371/journal.pone.0023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, O'Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM, Gravekamp C, Setaluri V, Peters N, Hoffmann FM, et al. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res. 2007;67:9954–9962. doi: 10.1158/0008-5472.CAN-07-1478. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chaux P, Stroobant V, Eggermont AM, Corthals J, Maillere B, Thielemans K, Marchand M, Boon T, Van Der Bruggen P. A MAGE-3 peptide presented by HLA-DR1 to CD4+ T cells that were isolated from a melanoma patient vaccinated with a MAGE- 3 protein. J Immunol. 2003;171:219–225. doi: 10.4049/jimmunol.171.1.219. [DOI] [PubMed] [Google Scholar]