Abstract
Antisense oligonucleotides (ASOs) hold promise for gene-specific knockdown in diseases that involve RNA or protein gain-of-function. In the hereditary degenerative disease myotonic dystrophy type 1 (DM1), transcripts from the mutant allele contain an expanded CUG repeat1–3 and are retained in the nucleus4, 5. The mutant RNA exerts a toxic gain-of-function6, making it an appropriate target for therapeutic ASOs. However, despite improvements in ASO chemistry and design, systemic use of ASOs is limited because uptake in many tissues, including skeletal and cardiac muscle, is not sufficient to silence target mRNAs7, 8. Here we show that nuclear-retained transcripts containing expanded CUG (CUGexp) repeats are extraordinarily sensitive to antisense silencing. In a transgenic mouse model of DM1, systemic administration of ASOs caused a rapid knockdown of CUGexp RNA in skeletal muscle, correcting the physiological, histopathologic, and transcriptomic features of the disease. The effect was sustained for up to one year after treatment was discontinued. Systemically administered ASOs were also effective for muscle knockdown of Malat-1, a long noncoding RNA (lncRNA) that is retained in the nucleus9. These results provide a general strategy to correct RNA gain-of-function and modulate the expression of expanded repeats, lncRNAs, and other transcripts with prolonged nuclear residence.
Antisense silencing by the RNase H mechanism entails a three step process of oligonucleotide hybridization to its cognate RNA, cleavage of the target by RNase H1, and exonuclease degradation of the cleavage fragments. The relative efficiency of this mechanism in the nucleus and cytoplasm is uncertain. Observations that ASOs shuttle from cytoplasm to nucleus10 and that targeting introns11 or nuclear RNA12 can silence gene expression indicate that antisense is active in the nucleus. Activity in the cytoplasm, however, is less clear. While RNase H1 is not restricted to the nucleus13, recent studies indicate that the non-nuclear fraction is confined to mitochondria14. This suggests that ASO•RNase H cleavage is mainly a nuclear process, whose potency could be maximized by targeting transcripts with long nuclear residence.
To test this concept we used a transgenic mouse model of DM1. HSALR transgenic mice express CUGexp RNA at high levels in skeletal muscle. Whereas human DM1 is caused by an expanded CTG repeat in the 3′ UTR of DMPK3, in HSALR mice the expanded repeat is in the 3′ UTR of a human skeletal actin (hACTA1) transgene6. In both conditions the CUGexp transcripts are retained in nuclear foci, along with splicing factors in the Muscleblind-like (MBNL) family. Muscleblind sequestration leads to misregulated alternative splicing and other changes of the muscle transcriptome15–17. The RNA toxicity was mitigated in mice by CAG-repeat morpholino oligomers that compete with MBNL proteins for CUGexp binding, without activating RNase H. However, this approach required direct injection of a single muscle, followed by in vivo electroporation, a method to load muscle fibers with oligomers18. As an alternative, RNase H-active ASOs could produce widespread correction, provided that uptake of circulating ASOs was sufficient to induce target cleavage.
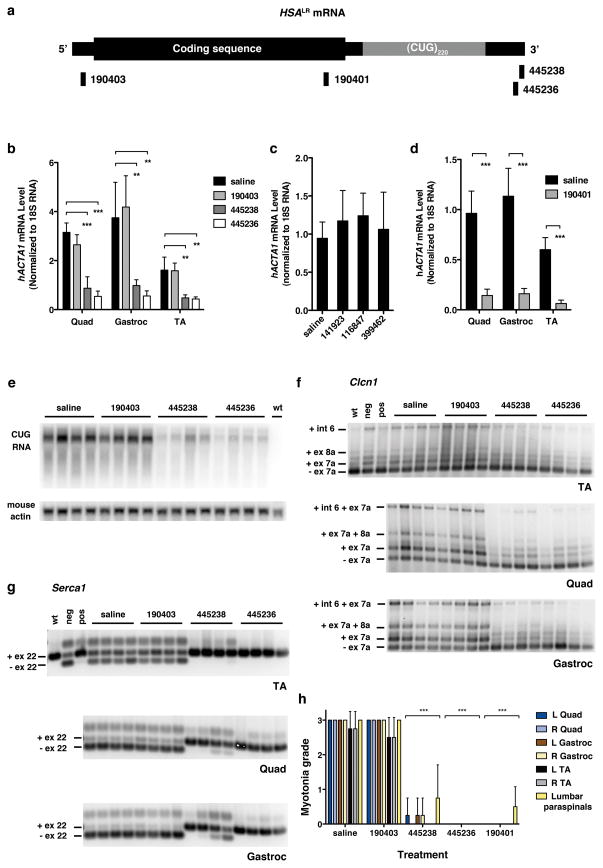
We identified ASOs showing (1) strong knockdown of hACTA1 in tissue culture; (2) good tolerability when systemically administered in wild-type mice; and (3) activity against hACTA1-CUGexp transcripts in vivo when electroporated into muscle (Supplementary Figs. 1–3). The ASOs had 2′-O-methoxyethyl (MOE) modifications at both ends to maximize biostability, and a central gap of 10 unmodified nucleotides to support RNase H activity (MOE gapmers; Supplementary Table 1). We tested three of the ASOs in HSALR transgenic mice by subcutaneous injection of 25 mg/kg twice weekly (Fig. 1a). After 4 weeks of administration (8 injections), ASO 445236 reduced the level of CUGexp RNA in hindlimb muscles by > 80% (Fig. 1b). Another ASO targeting 3′ of the repeat tract also showed strong CUGexp reduction, whereas an ASO targeting the 5′ UTR, or three oligonucleotides against other targets, had no effect (Fig. 1b–c).
Figure 1. Systemic administration of 2′-O-(2-methoxyethyl) (MOE) ASOs in the HSALR transgenic mouse model of DM1.
a, Location of ASO targeting sequences relative to hACTA1 coding region and expanded CUG repeat in the 3′ UTR. b, Quantitative real-time RT-PCR of hACTA1-CUGexp mRNA in quadriceps, gastrocnemius, and tibialis anterior (TA) muscle in HSALR mice treated with the indicated ASOs by subcutaneous injection of 25 mg/kg twice weekly for 4 weeks. Mice were anlayzed 1 week after the final dose (n = 4 per group). Shown are mean levels of transgene mRNA ± SD. ** P < 0.001, *** P < 0.0001 (1-way ANOVA). c, hACTA1-CUGexp transcript levels in quadriceps are not affected by ASOs targeting unrelated transcripts (141923, randomer; 116847, Pten; 399462, Malat-1; n = 4 per group; same dose as b). Error bars ± SD. d, Knockdown of hACTA1-CUGexp mRNA in muscle by ASO 190401 (n = 4 per group; same dose as b). Error bars ± SD. *** P ≤ 0.0005 (t-test). e, Northern analysis of RNA from quadriceps muscle. The level of CUGexp RNA was determined using a (CAG)9 oligonucleotide probe. Mouse actin serves as loading control. f, g, RT-PCR analysis of alternative splicing of Clcn1 (f) and Serca-1 (g) transcripts. For Clcn1, only the −ex7a isoform encodes a functional ion channel. −ex7a, exon 7a skipping; +ex7a, exon 7a inclusion; -ex22, exon 22 skipping; +ex22, exon 22 inclusion; wt, FVB/n wild-type; neg, negative control injected with GAC25 morpholino; pos, positive control injected with CAG25 morpholino. h, Blinded analysis of myotonia by electromyography, 1 week following final dose (n = 4 mice per group). Error bars ± SD. *** P < 0.0001 ASO- vs. saline-treated muscles (2-way ANOVA).
RNase H cleavage of mRNA is usually followed by rapid decay of cleavage fragments. However, stable cleavage fragments are observed occasionally19, and the CUGexp tract forms extensive hairpins20 and ribonucleoprotein complexes21 that could inhibit exonuclease activity. The failure of antisense targeting in the 5′ UTR also raised the possibility that cleavage downstream of the repeat tract was required for efficient silencing. We therefore tested an additional ASO, 190401, targeting the hACTA1 coding region, and found that it also was highly effective (Fig. 1d). Furthermore, northern blotting using a CAG-repeat probe showed no evidence for a stable CUGexp cleavage fragment (Fig. 1e), and in situ hybridization showed reduction of nuclear CUGexp foci (Supplementary Fig. 4). These results indicated that expanded CUG repeats are degraded following a cleavage event 5′ or 3′ of the repeat tract.
Reduction of CUGexp RNA would be expected to release sequestered MBNL1 protein and improve its splicing regulatory activity. Consistent with this prediction, alternative splicing of four MBNL1-dependent exons, Serca1 exon 22, Titin exon 362, Zasp exon 11, and Clcn1 chloride ion channel exon 7a, was normalized (Fig. 1f, g, Supplementary Figs. 5–6)15. The Clcn1 splicing defect causes loss of channel function, repetitive action potentials, and delayed muscle relaxation (myotonia)22, a cardinal feature of the disease. Blinded analysis showed that myotonic discharges in hindlimb muscles were eliminated by the active ASOs (Fig. 1h), indicating rescue of Clcn1 function.
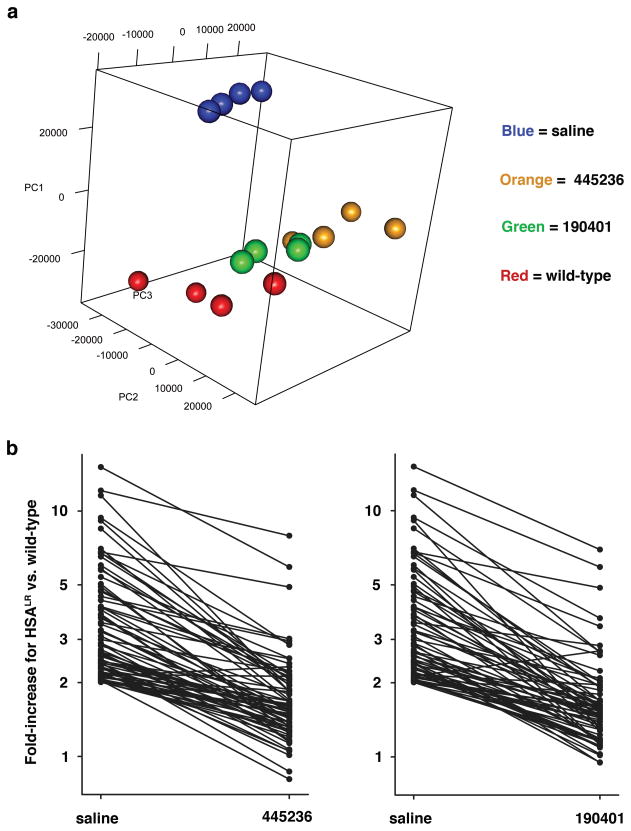
In addition to splicing defects, expression of CUGexp RNA or ablation of Mbnl1 causes extensive remodeling of the muscle transcriptome16, 17, 23. We used microarrays to examine transcriptomic effects of ASOs. Principle component analysis showed that gene expression in ASO-treated HSALR mice was shifted towards wild-type mice, indicating an overall trend for transcriptome normalization (Fig. 2a). Among transcripts that were up- or down-regulated in HSALR muscle, > 85% were normalized or partially corrected by ASOs, without evidence for off-target effects (Fig. 2b; Supplementary Fig 7; Supplementary Table 2). These results confirm that ASOs caused an overall improvement of the muscle transcriptome.
Figure 2. Transcriptomic effects of ASOs.
Microarray analysis of gene expression in quadriceps muscle (n = 4 mice per group). a, Principle component analysis shows segregation of HSALR (saline) away from wild-type mice in widely separated clusters. ASOs caused HSALR transgenic mice to cluster nearer to wild-type mice (25 mg/kg biweekly for 4 wks). b, Among transcripts upregulated in HSALR vs. wild-type mice (saline), > 85% showed complete or partial return to normal expression after treatment with ASOs.
ASO effects were evident within 2 weeks (Supplementary Fig. 8) and were dose-dependent. A 3-fold dose reduction (8.5 mg/kg biweekly for 4 weeks) caused partial myotonia and splicing correction, whereas a 10-fold dose reduction (2.5 mg/kg) caused partial myotonia correction in tibialis anterior but not in quadriceps (Supplementary Fig. 9a–c), the latter muscle having higher basal levels of CUGexp expression18. Serum chemistries showed no evidence for renal or liver toxicity (Supplementary Fig 10).
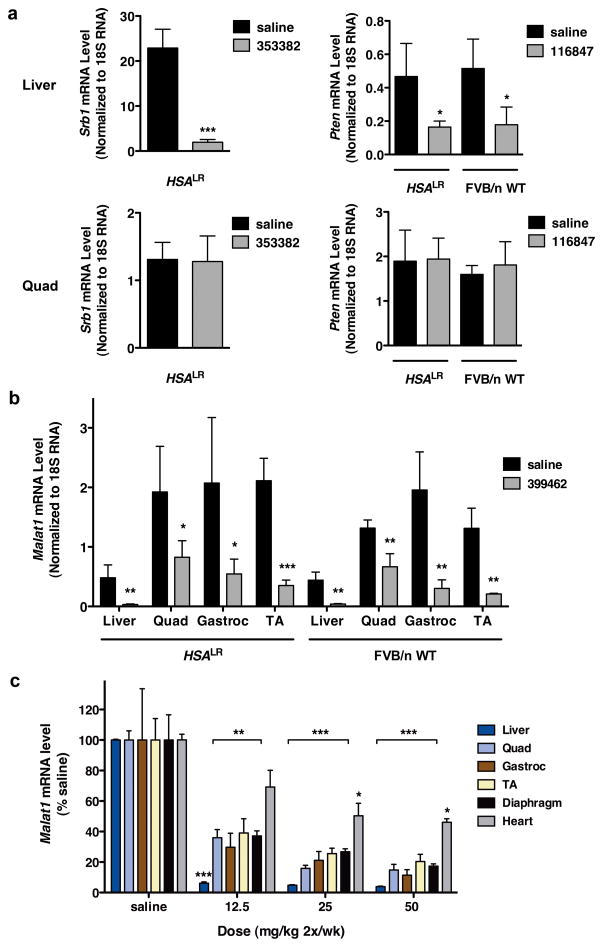
A uniform finding in previous studies of MOE gapmer ASOs was that systemic administration failed to cause significant target reduction in muscle, despite efficient knockdown in liver (n = 12 different mRNA targets; Supplementary Table 3), raising the possibility that muscle tissue in our model is unusually susceptible to antisense silencing. We examined the functional integrity of the muscle membrane, a physiological barrier to ASO uptake24, and found that muscle penetration of the extracellular dye, Evans Blue, was similar in HSALR and wild-type mice (Supplementary Fig. 11a). Direct analysis of muscle tissue indicated that ASO accumulation was no greater in HSALR mice than in wild-type controls (Supplementary Fig. 11b, c). The mRNA level for RNase H1 likewise was similar in HSALR and WT muscle (Supplementary Fig. 12). We tested ASOs targeting other muscle-expressed transcripts. ASOs for Pten phosphatase or Srb1 scavenger receptor showed efficient target knockdown in liver, but no appreciable knockdown in HSALR or WT muscle (Fig. 3a). Taken together with previous studies, our results argue for a specific sensitivity of hACTA1-CUGexp transcripts rather than a general enhancement of ASO activity in HSALR muscle.
Figure 3. Differential sensitivity of transcripts to ASO knockdown in skeletal muscle.
a, In HSALR or FVB/n wild-type mice, ASOs targeting Srb1 (353382) or Pten (116847) were effective for knockdown in liver but not in quadriceps muscle (qRT-PCR, ± SD; n = 4 per group). * P = 0.02; *** P < 0.0001 (t-test). b, HSALR and FVB/n wild-type mice were treated with ASO 399462 targeting Malat-1, a nuclear-retained lncRNA. Levels of Malat-1 transcript in the indicated tissues were determined by qRT-PCR (± SD). (n = 4 ASO, 3 saline) * P = 0.035; ** P < 0.007; and *** P = 0.001 for ASO vs. saline (t-test). c, Dose response of Malat-1 knockdown in BALB/c wild-type mice. BALB/c wild-type mice were treated with saline or ASO 399462 targeting Malat-1 at 12.5, 25, and 50 mg/kg twice per week for 3.5 weeks (7 total doses; n = 4 per group). Tissues were collected for RNA isolation two days after the final dose. Malat-1 transcript levels were determined by qRT-PCR (± SEM). * P < 0.01; ** P < 0.001; *** P < 0.0001 (2-way ANOVA).
A noteworthy metabolic feature of hACTA1-CUGexp and human DMPK-CUGexp mRNA is that processing and polyadenylation are normal yet the transcripts are retained in the nucleus5, 6. Recent studies have shown that RNase H1, the enzyme responsible for antisense knockdown, is localized to the nucleus and mitochondria14, suggesting that antisense cleavage of nuclear-encoded RNA occurs prior to nuclear export, and raising the possibility that nuclear-retained transcripts may exhibit enhanced sensitivity. To determine if other nuclear-retained transcripts show a similar effect we examined metastasis associated lung adenocarcinoma transcript 1 (Malat1), an endogenous nuclear lncRNA9. We identified MOE gapmer ASOs that produced strong Malat1 knockdown in cells, in an RNase H1-dependent manner (Supplementary Fig. 13). In WT and HSALR mice, subcutaneous administration of ASOs for four weeks caused > 80% Malat1 reduction in muscle (Fig. 3b, c), supporting the concept that nuclear-retained transcripts have enhanced sensitivity.
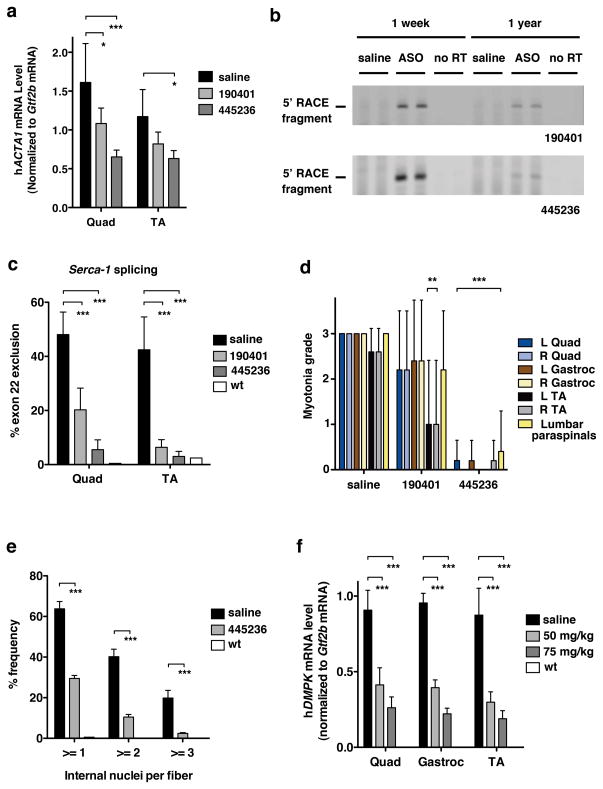
To determine the duration of ASO action in muscle we examined mice at 15 and 31 wks following ASO discontinuation, and found that hACTA1-CUGexp knockdown and splicing correction remained strong (not shown). One year after ASO injection was discontinued, target reduction by ASO 190401 had waned, but remained ~50% or more for ASO 445236 (Fig. 4a). Even at this late timepoint the appropriate cleavage products were detected by amplification of cDNA 5′ ends (5′ RACE), indicating persistent ASO-RNase H1 activity (Fig. 4b). Consistent with the extent of target reduction, there was partial return of myotonia and splicing defects for ASO 190401, whereas correction by ASO 445236 remained strong (Fig 4c, d; Supplementary Fig. 14a–e). Furthermore, the persistent knockdown of CUGexp RNA largely prevented the age-dependent myopathic changes in HSALR muscle, as evidenced by reduced frequency of central nuclei (Fig. 4e) and improved muscle fiber diameter (mainly, a prevention of fiber atrophy) (Supplementary Fig. 15). These findings indicate that ASO activity against hACTA1-CUGexp in muscle is remarkably durable and that long-term reduction of the toxic RNA can protect against structural changes in muscle fibers. Notably, the duration of Malat1 knockdown in muscle was also prolonged (>50% reduction at 31 weeks following discontinuation) and more persistent than in liver (Supplementary Fig. 16).
Figure 4. Duration of ASO activity and in vivo targeting of human DMPK.
(a-e) Two month-old HSALR mice received saline or indicated ASO by subcutaneous injection of 25 mg/kg twice weekly for 4 weeks (n = 5 each ASO, n = 6 saline). Tissues were isolated 1 year after the final dose. a, qRT-PCR analysis of HSALR transgene mRNA (± SD), normalized to the housekeeping gene Gtf2b mRNA. Results were similar when normalized to total RNA input. * P < 0.05; *** P < 0.0001 ASO- vs. saline-treated Quad or TA muscle (2-way ANOVA). b, 5′ rapid amplification of cDNA ends (RACE) was performed on muscle RNA obtained 1 week or 1 year after discontinuation of ASO 190401 (upper) or 445236 (lower). PCR products labeled “5′ RACE fragment” migrated at the expected position for ASO-RNase H cleavage products, and were confirmed by DNA sequencing. “No RT” = no reverse transcriptase. c, Quantitation of Serca-1 splicing (± SD). *** P < 0.0001 ASO- vs. saline-treated muscle (2-way ANOVA). “wt” = representative FVB/n wild-type. d, Myotonia graded by a blinded examiner (± SD). ** P < 0.001; *** P < 0.0001 ASO- vs. saline-treated muscle (2-way ANOVA). e, After prolonged knockdown of toxic RNA, the number of internal nuclei per muscle fiber (± SD) was determined by histologic analysis (age 14 months). (n = 4 ASO 445236; n = 3 saline). “WT” = untreated 3 month-old FVB/n wild-type control. *** P < 0.0001 ASO vs. saline-treated (2-way ANOVA). f, DM328XL mice received subcutaneous injections of saline or ASO 445569 targeting the 3′ UTR of hDMPK. The ASO dose was 50 or 75 mg/kg twice weekly for 4 weeks (n = 5 low dose; n = 4 high dose; n = 2 saline). Tissues were isolated 2 days after the final dose. Dose-dependent reduction of hDMPK (± SD) normalized to housekeeping gene Gtf2b mRNA. Note that hDMPK mRNA was undetectable in wild-type mice. “wt” = untreated wild-type littermates of DM328XL transgenic mice (n = 2).
Therapeutic application of this strategy to human DM1 will require transfer of the targeting sequence to hDMPK. We developed MOE gapmer ASOs that were active against hDMPK in cells. We examined in vivo activity after 4 weeks of twice weekly subcutaneous injection in transgenic mice that express hDMPK with 800 CUG repeats. The ASO produced significant knockdown of hDMPK-CUGexp transcripts in hindlimb muscle (Fig 4f; Supplementary Figs. 17, 18), supporting the feasibility of silencing the pathogenic DM1 RNA.
Despite physiological barriers to tissue uptake, our results indicate that systemic targeting of CUGexp RNA is feasible because small amounts of ASOs that enter muscle fibers can hybridize their target and productively engage RNase H1. Although the mechanisms for enhanced sensitivity of CUGexp RNA and Malat1 are not fully defined, our data suggest that residence time in the nucleus is an important determinant of transcript sensitivity. Features of the nuclear environment that may enhance antisense activity include nuclear localization of RNase H114 and auxiliary proteins that promote oligonucleotide hybridization25, and, in the case of CUGexp transcripts, spatial concentration of targets in a small volume4. A similar approach may be effective for other genetic disorders that have nuclear accumulation of repeat expansion RNA26, 27. Previous studies have used CAG-repeat ASOs that bind CUGexp RNA without activating RNase H, in an effort to block the protein interactions or modify the metabolism of the toxic RNA18, 28. While this approach was effective with local delivery, initial attempts at systemic delivery were less successful (Wheeler and Thornton, unpublished), which fits with the expectation that higher tissue concentrations of ASO are required to occupy CUGexp binding sites than to induce RNase H cleavage. Furthermore, the RNase H mechanism is attractive because it exploits the nuclear retention phenomenon to gain a therapeutic advantage, while posing less risk of off-target effects by avoiding a repetitive sequence. Recently, local delivery of RNase H-active CAG repeat ASOs induced partial CUGexp knockdown, but was accompanied by muscle damage29, again suggesting that direct targeting of the repeat tract may have pitfalls. Our results also suggest that ASOs are useful for in vivo functional characterization and therapeutic modulation of lncRNAs, a large and recently-recognized class of regulatory RNAs30.
Methods
Antisense oligonucleotides
ASOs were synthesized at Isis Pharmaceuticals as described previously31. All ASOs were MOE gapmer 20mers having (1) phosphorothioate as the intersubunit linkage; (2) 2′-O-(2-methoxyethyl) (MOE) modifications of 5 nucleotides at the 5′ and 3′ end; and (3) a central gap of 10 deoxynucleotides. The sequence of each ASO is listed in Supplementary Table 1. CAG25 and GAC25 morpholinos18 were purchased (Gene Tools, LLC, Philomath, OR).
Identification of active ASOs
Criteria for identifying active hACTA1-targeting ASOs were as follows: (1) selection of targeting sequence that were not conserved in mice, to avoid knockdown of endogenous skeletal actin; (2) > 50% hACTA1 knockdown when electroporated in HepG2 cells (Supplementary Fig. 1); and (3) absence of hepatotoxic or immunostimulatory effects in WT mice, when administered by subcutaneous injection of 50 mg/kg twice weekly for 4 weeks (Supplementary Fig. 2a–c). Of 11 candidate ASOs examined, 5 satisfied these criteria. For the ASO with highest activity in HepG2 cells, we also verified activity against hACTA1-CUGexp transcripts in vivo, by direct injection and electroporation of tibialis anterior muscle in HSALR mice (Supplementary Fig. 3). Four of the five ASOs were subsequently used for subcutaneous administration in HSALR mice. ASOs targeting Malat1 were identified by demonstration of > 50% target knockdown when electroporated in MHT mouse hepatoma cells, and absence of hepatotoxic or immunostimulatory effects in WT mice (dosing as above).
Cell transfection and gene analysis
HepG2 cells were electroporated in a 96 well plate format at 165V with ASOs in complete media containing MEM, NEAA, sodium pyruvate, and 10% FBS at room temperature. Cells were incubated overnight and lysed in RLT buffer (Qiagen). Total RNA was prepared using Qiagen RNeasy kit. Quantitative real time RT-PCR (qRT-PCR) was performed using the Qiagen QuantiTect Probe RT-PCR kit. Twenty μl qRT-PCR reactions were run in duplicate and normalized against total RNA, calculated using the Ribogreen assay (Invitrogen).
Experimental mice
Institutional Animal Care and Use Committees at the University of Rochester, Genzyme Corporation, and Isis Pharmaceuticals approved all animal experiments. HSALR mice in the line 20b were derived and maintained on the FVB/n background6. The (CTG)250 tract in this line is unstable, and has shortened to (CTG)220. DM328XL mice carry a 45 kb human genomic fragment that includes the mutant DMPK gene with 800 CTG repeats32,33. The DM328XL mice were hemizygous and display no histologic changes, myotonia, or splicing defects in skeletal muscle34, 35. FVB/n, BALB/c, C57Bl/10, and Mdx mice were obtained from Jackson Laboratories.
Muscle injection of ASOs
Tibialis anterior (TA) muscle was injected with 0.2, 0.4, or 0.8 nmole ASO in 20 μl saline, and the contralateral TA with 20 μl saline alone, followed by electroporation, as described previously36. Treatment assignments were randomized and injections blinded.
Subcutaneous injection of ASOs
All ASOs were dissolved in phosphate buffered saline (PBS). Twice-per-week doses of 2.5, 8.5, 12.5, 25, or 50 mg/kg were injected subcutaneously in the interscapular region for 3.5–4 weeks (7 or 8 total doses). Injection volumes ranged from 140–200 μl.
Real-time PCR Assay
Total RNA was purified from tibialis anterior (TA), gastrocnemius (Gastroc), or quadriceps (Quad) muscle using the RNeasy Lipid Tissue Mini Kit (Qiagen) according to the manufacturer’s instructions. qRT-PCR was used to determine mRNA levels for ACTA1, Srb1, Pten, Malat1, and RNase H1, with 18S rRNA as normalization control, on an Applied Biosystems 7500 Real-Time PCR System. General transcription factor 2b (Gtf2b) and total RNA (Ribogreen assay) served as normalization controls for human DMPK and mouse Dmpk.
Real-Time PCR Assay Primer-Probe Sequences
ACTA1 PPset # 1
Forward: 5′-GTAGCTACCCGCCCAGAAACT-3′
Reverse: 5′-CCAGGCCGGAGCCATT-3′
Probe: 5′-ACCACCGCCCTCGTGTGCG-3′
ACTA1 PPset # 2
Forward: 5′-GACGAGGCTCAGAGCAAGAGA-3′
Reverse: 5′-TGATGATGCCGTGCTCGATA-3′
Probe: 5′-CCTGACCCTGAAGTAC-3′
Srb1
Forward: 5′-TGACAACGACACCGTGTCCT-3′
Reverse: 5′-ATGCGACTTGTCAGGCTGG-3′
Probe: 5′-CGTGGAGAACCGCAGCCTCCATT-3′
Pten
Forward: 5′-ATGACAATCATGTTGCAGCAATTC-3′
Reverse: 5′-CGATGCAATAAATATGCACAAATCA-3′
Probe: 5′-CTGTAAAGCTGGAAAGGGACGGACTGGT-3′
Malat1
Forward: 5′-TGGGTTAGAGAAGGCGTGTACTG-3′
Reverse: 5′-TCAGCGGCAACTGGGAAA-3′
Probe: 5′-CGTTGGCACGACACCTTCAGGGACT-3′
RNase H1
Forward: 5′-ACTCAGGATTTGTGGGCAATG-3′
Reverse: 5′-CCTCAGACTGCTTCGCTCCTT-3′
Probe: 5′-AGAGGCCGACAGACTGGCACGG-3′
human DMPK
Forward: 5′-AGCCTGAGCCGGGAGATG-3′
Reverse: 5′-GCGTAGTTGACTGGCGAAGTT-3′
Probe: 5′-AGGCCATCCGCACGGACAACCX-3′
mouse Dmpk
Forward: 5′-GACATATGCCAAGATTGTGCACTAC-3′
Reverse: 5′-CACGAATGAGGTCCTGAGCTT-3′
Probe: 5′-AACACTTGTCGCTGCCGCTGGCX-3′
Ap2M1, sequences previously reported37
18s rRNA, proprietary sequences, (Applied Biosystems, Cat #4310893-E)
Gtf2b, proprietary sequences, (Applied Biosystems, Cat #4331182)
Northern analysis
Total RNA (6 μg) was separated on agarose/MOPS/formaldehyde gels, transferred to nylon membranes, and hybridized with (CAG)9 or mouse actin 32P-labeled oligonucleotide probes as described previously18.
Electromyography
EMG was performed under anesthesia by a blinded examiner as described previously18. Myotonic discharges were graded on a 4-point scale: 0, no myotonia; 1, occasional myotonic discharge in less than 50% of needle insertions; 2, myotonic discharge in greater than 50% of needle insertions; 3: myotonic discharge with nearly every insertion.
RT-PCR analysis of alternative splicing
RT-PCR was performed using the SuperScript III One-Step RT-PCR with Platinum Taq DNA Polymerase (Invitrogen) using gene-specific primers for cDNA synthesis and PCR amplification. The primers for Clcn1, Serca-1, Titin and ZASP were described previously15, 36. PCR products were separated on agarose gels, stained with SybrGreen I Nucleic Acid Gel Stain (Invitrogen), and imaged using a laser scanner (Fujifilm LAS-3000 Intelligent Dark Box or GE Healthcare Typhoon 9400). Band intensities were quantitated using ImageQuant software (GE Healthcare.)
Transcriptome analysis by microarray
RNA was isolated from quadriceps muscle of wild-type mice or HSALR transgenic mice treated with vehicle (saline), ASO 445236, or ASO 190401 (n = 4 per group, 25 mg/kg ASO twice weekly for 4 weeks). RNA integrity was verified (RIN values > 7.5 on Agilent Bioanalyzer). RNA was processed to cRNA and hybridized on microbeads using MouseRef-8 v2.0 Expression BeadChip Kits (Illumina, San Diego) according to the manufacturer’s recommendations. Image data were quantified using BeadStudio software (Illumina). Signal intensities were quantile normalized. We used row-specific offsets to avoid any values less than 2, prior to the normalization. Data from all probe sets with 6 or more nucleotides of CUG, UGC, or GCU repeats was suppressed to eliminate the possibility that expanded repeats in the hybridization mixture (CAG repeats in cRNA, originating from CUGexp RNA) could cross-hybridize with repeat sequences on probes. To eliminate genes whose expression was not readily quantified on the arrays, we suppressed probes that did not show a detection probability of P < 0.1 for all samples in the group that showed the higher mean expression level. Comparisons between groups were summarized and rank-ordered by fold-changes of mean expression level and t tests. The software package R 38 was used to perform principal components analysis (PCA)39, 40 on WT, ASO-treated, and saline-treated microarray samples. The principal components allowed the capture of the majority of the expression variation in each sample within 3 dimensions. We plotted the first three principal components of each sample. Array data have been submitted to the Gene Expression Omnibus, accession number GSE38962 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE38962).
Fluorescence in situ hybridization (FISH)
Localization of CUGexp RNA by FISH was performed using a 5′-Texas Red-labeled CAG repeat oligoribonucleotide probe on muscle cryosections from ASO- or saline-treated mice as described previously15. Images are maximum projections of deconvolved Z-plane stacks (9 images, 0.1 or 0.2 μM steps) captured under identical exposure and illumination conditions using a fluorescence microscope (Carl Zeiss Axioplan 2 or Nikon Eclipse E600), CCD digital camera (Hamamatsu ORCA R2 or Photometrics Cool Snap HQ), and Metamorph software (Molecular Devices). Optigrid structured illumination imaging system (Qioptiq) also was used to capture images of DM328XL muscle. Maximum gray level intensity was quantitated using Metamorph. Objectives: 100X Plan-APOCHROMAT 1.4 NA oil (Zeiss) or 63X Plan Apo 1.4 NA oil (Nikon).
Muscle fiber morphometry
To outline muscle fibers and label nuclei, 10 μM transverse cryosections of muscles from ASO- or saline-treated mice were fixed with 4% paraformaldehyde, pH 7.3, washed in PBS, and incubated in 20 μg/ml FITC-wheat germ agglutinin (WGA; Sigma) and 4,6 diamino-2 phenylindole dihydrochloride (DAPI; 1:20,000) in PBS for 1 hour at room temperature. Sections then were washed in PBS, mounted, and sealed. Images were captured using an Axioplan 2 fluorescence microscope (Zeiss), an ORCA R2 CCD digital camera (Hamamatsu Photonics), Metamorph software, and a 20X Plan-NEOFLUAR 0.5 NA objective (Zeiss). Using the calipers application in Metamorph, muscle fiber diameter, defined as the minimum “Feret’s diameter” (the mimimum distance of parallel tangents at opposing borders of the muscle fiber41) was determined. H&E images were captured using an Infinity2-1 1.4 megapixel color CCD digital camera (Lumenera), Infinity Analyze 5.0 software (Lumenera), and a 10X Plan-NEOFLUAR 0.3 NA objective (Zeiss).
5′ Rapid Amplification of cDNA Ends (RACE) analysis
5′ RACE was performed using the FirstChoice RLM-RACE Kit (Invitrogen). Briefly, 1 μg of total mRNA was ligated with 5′ RACE Adaptor (5′-GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA-3′), then reverse transcribed with a primer specific for the cleavage fragment (5′-TGAGAAGTCGCGTGCTGGAG-3′ for 190401, or 5′-TTTTTTTTACGCAGC-3′ for 445236). The synthesized cDNA was treated with RNase H, then amplified with 5′ RACE Outer Primer and 5′-TTGCGGTGGACGATGGAAGG-3′ (for 190401 fragment), or 5′-TGTGTAAAACGACGGCCAGTACGCAGCTTAACAGAATGAC-3′ (for 445236 fragment). The PCR products were analyzed on agarose gels stained with SYBR Green I (Invitrogen) and scanned with a laser fluorimager (Typhoon, GE Healthcare).
RNAse H1 siRNA experiments
Mouse hepatocellular SV40 large T-antigen carcinoma (MHT) cells were cultured in DMEM supplemented with 10% fetal calf serum, streptomycin (0.1 mg/ml), and penicillin (100 U/ml). siRNA treatments were performed using Opti-MEM containing 5 mg/ml Lipofectamine 2000 as previously described37. Briefly, MHT cells plated at 7500 cells per well and were incubated for either 24 or 48 hours with 75 nM of siRNA targeting RNaseH1 (5′-GCTTGGTGAGACGTGCTTATT-3′ and 5′-TAAGCACGTCTCACCAAGCTG-3′) or Ap2M1 (sequences reported previously37) in OPTI-MEM and Lipofectamine 2000. Twenty-four hours post-transfection, cells were treated with increasing doses of the Malat1-targeting ASO 399479 in DMEM-10% FBS. Twenty-four hours after addition of oligonucleotides, cells were lysed and RNA was isolated using RNAeasy and qRT-PCR was performed to determine the level of Malat1 mRNA.
Tissue drug level determination
Approximately 30–100 mg liver and muscle tissue were homogenized as described42. Capillary Gel Electrophoresis (CGE) methods were used to measure unlabeled drug concentrations in mouse tissues. The methods for the hACTA1 ASOs were slight modifications of previously published methods42, 43, which consist of a phenol-chloroform (liquid-liquid) extraction followed by a solid phase extraction. An internal standard (ASO 355868, a 27-mer 2′-O-methoxyethyl modified phosphorothioate oligonucleotide) was added prior to extraction. Tissue sample analyses were conducted using a Beckman MDQ capillary electrophoresis instrument (Beckman Coulter). Tissue sample concentrations were calculated using calibration curves, with a lower limit of quantitation (LLoQ) of approximately 1.14 μg/g.
Biochemical analysis/serum chemistry
Serum separated in serum separator tubes (BD cat # 365956) was used to determine aspartate transaminase (AST), alanine transaminase (ALT), blood urea nitrogen (BUN), and creatinine values using Olympus reagents and an Olympus AU400e analyzer (Melville, NY).
Evans blue dye uptake assay
Evans Blue Dye (EBD) was dissolved in phosphate-buffered saline at a concentration of 10 mg/mL and filter-sterilized. HSALR, FVB/n, Mdx, or C57Bl/10 mice were injected by intraperitoneal injection with 10 μL EBD solution per gram bodyweight. After a period of 24h, muscle tissues were harvested (quadriceps, gastrocnemius, tibialis anterior, diaphragm and heart). The mass of each muscle was determined prior to lysing each sample individually in a microfuge tube containing N,N-dimethylformamide and a 5 mm steel bead, which was subjected to 30 Hz shaking in a Qiagen TissueLyser II. Lysed muscle samples were heated at 55° C, centrifuged, and the absorbance of the supernatant was determined by spectrophotometric measurement at 636 nm. A standard curve of EBD in N,N-dimethylformamide allowed determination of EBD content in individual muscle samples.
Statistical analysis
Group data are presented as ± SD, except where ± SEM is indicated. Between-group comparison was performed by two-tailed Student’s t-test or ANOVA, as indicated. A P value of < 0.05 was considered significant.
Supplementary Material
Acknowledgments
From the Wellstone Muscular Dystrophy Cooperative Research Center and Center for RNA Biology at the University of Rochester, with support from NIH (AR049077, U54NS48843, AR/NS48143, K08NS064293, U01NS072323), the Saunders Family Neuromuscular Research Fund, Run America, and fellowship (M.N.) from the Muscular Dystrophy Association and Uehara Memorial Foundation. The authors thank Dr. Geneviève Gourdon for providing DM328XL mice, Dr. Mahyar Sabripour for assistance with principal components analysis, and Linda Richardson and Sarah Leistman for technical assistance.
Footnotes
Author contributions. All authors participated in the planning, design, and interpretation of experiments. T.M.W., A.J.L., S.K.P., A.R.M, and M.N. performed experiments. T.M.W. and C.A.T. wrote the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare competing financial interests.
S.K.P., A.R.M., C.F.B. are employees of Isis Pharmaceuticals, Inc, which develops antisense drugs. A.J.L., S.H.C., and B.M.W. are employees of Genzyme Corporation, which develops biological therapeutics. C.A.T. and T.M.W. received research support from Isis Pharmaceuticals. Isis Pharmaceuticals has applied for patents related to this work.
References
- 1.Harley HG, et al. Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature. 1992;355:545–546. doi: 10.1038/355545a0. [DOI] [PubMed] [Google Scholar]
- 2.Buxton J, et al. Detection of an unstable fragment of DNA specific to individuals with myotonic dystrophy. Nature. 1992;355:547–548. doi: 10.1038/355547a0. [DOI] [PubMed] [Google Scholar]
- 3.Brook JD, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 4.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci U S A. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mankodi A, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 7.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 8.Geary RS, et al. Pharmacokinetics of a tumor necrosis factor-alpha phosphorothioate 2′-O-(2-methoxyethyl) modified antisense oligonucleotide: comparison across species. Drug Metab Dispos. 2003;31:1419–1428. doi: 10.1124/dmd.31.11.1419. [DOI] [PubMed] [Google Scholar]
- 9.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorenz P, Misteli T, Baker BF, Bennett CF, Spector DL. Nucleocytoplasmic shuttling: a novel in vivo property of antisense phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 2000;28:582–592. doi: 10.1093/nar/28.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vickers TA, et al. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J Biol Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 12.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 13.Wu H, et al. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J Biol Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki Y, et al. An upstream open reading frame and the context of the two AUG codons affect the abundance of mitochondrial and nuclear RNase H1. Mol Cell Biol. 2010;30:5123–5134. doi: 10.1128/MCB.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin X, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 16.Osborne RJ, et al. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum Mol Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanadia RN, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler TM, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasselblatt P, Hockenjos B, Thoma C, Blum HE, Offensperger WB. Translation of stable hepadnaviral mRNA cleavage fragments induced by the action of phosphorothioate-modified antisense oligodeoxynucleotides. Nucleic Acids Res. 2005;33:114–125. doi: 10.1093/nar/gki155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Napierala M, Krzyzosiak WJ. CUG repeats present in myotonin kinase RNA form metastable “slippery” hairpins. J Biol Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- 21.Miller JW, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. Embo J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mankodi A, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 23.Du H, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alter J, et al. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- 25.Pontius BW, Berg P. Rapid assembly and disassembly of complementary DNA strands through an equilibrium intermediate state mediated by A1 hnRNP protein. J Biol Chem. 1992;267:13815–13818. [PubMed] [Google Scholar]
- 26.Li LB, Bonini NM. Roles of trinucleotide-repeat RNA in neurological disease and degeneration. Trends Neurosci. 2010;33:292–298. doi: 10.1016/j.tins.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulders SA, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci U S A. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JE, Bennett CF, Cooper TA. RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc Natl Acad Sci U S A. 2012;109:4221–4226. doi: 10.1073/pnas.1117019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Cheruvallath ZS, Kumar RK, Rentel C, Cole DL, Ravikumar VT. Solid phase synthesis of phosphorothioate oligonucleotides utilizing diethyldithiocarbonate disulfide (DDD) as an efficient sulfur transfer reagent. Nucleosides Nucleotides Nucleic Acids. 2003;22:461–468. doi: 10.1081/NCN-120022050. [DOI] [PubMed] [Google Scholar]
- 32.Seznec H, et al. Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum Mol Genet. 2000;9:1185–1194. doi: 10.1093/hmg/9.8.1185. [DOI] [PubMed] [Google Scholar]
- 33.Nakamori M, Gourdon G, Thornton CA. Stabilization of expanded (CTG)*(CAG) repeats by antisense oligonucleotides. Mol Ther. 2011;19:2222–2227. doi: 10.1038/mt.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seznec H, et al. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum Mol Genet. 2001;10:2717–2726. doi: 10.1093/hmg/10.23.2717. [DOI] [PubMed] [Google Scholar]
- 35.Gomes-Pereira M, et al. CTG trinucleotide repeat “big jumps”: large expansions, small mice. PLoS Genet. 2007;3:e52. doi: 10.1371/journal.pgen.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koller E, et al. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 39.Raychaudhuri S, Stuart JM, Altman RB. Principal components analysis to summarize microarray experiments: application to sporulation time series. Pac Symp Biocomput. 2000:455–466. doi: 10.1142/9789814447331_0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ringner M. What is principal component analysis? Nat Biotechnol. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 41.Briguet A, Courdier-Fruh I, Foster M, Meier T, Magyar JP. Histological parameters for the quantitative assessment of muscular dystrophy in the mdx-mouse. Neuromuscul Disord. 2004;14:675–682. doi: 10.1016/j.nmd.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Leeds JM, Graham MJ, Truong L, Cummins LL. Quantitation of phosphorothioate oligonucleotides in human plasma. Anal Biochem. 1996;235:36–43. doi: 10.1006/abio.1996.0088. [DOI] [PubMed] [Google Scholar]
- 43.Geary RS, Matson J, Levin AA. A nonradioisotope biomedical assay for intact oligonucleotide and its chain-shortened metabolites used for determination of exposure and elimination half-life of antisense drugs in tissue. Anal Biochem. 1999;274:241–248. doi: 10.1006/abio.1999.4290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.