Abstract
Aptamers are a class of small nucleic acid ligands that are composed of RNA or single-stranded DNA oligonucleotides and have high specificity and affinity for their targets. Similar to antibodies, aptamers interact with their targets by recognizing a specific three-dimensional structure and are thus termed “chemical antibodies.” In contrast to protein antibodies, aptamers offer unique chemical and biological characteristics based on their oligonucleotide properties. Hence, they are more suitable for the development of novel clinical applications. Aptamer technology has been widely investigated in various biomedical fields for biomarker discovery, in vitro diagnosis, in vivo imaging, and targeted therapy. This review will discuss the potential applications of aptamer technology as a new tool for targeted cancer therapy with emphasis on the development of aptamers that are able to specifically target cell surface biomarkers. Additionally, we will describe several approaches for the use of aptamers in targeted therapeutics, including aptamer-drug conjugation, aptamer-nanoparticle conjugation, aptamer-mediated targeted gene therapy, aptamer-mediated immunotherapy, and aptamer-mediated biotherapy.
Keywords: cell surface biomarker, nanomedicine, oligonucleotide aptamer, SELEX, targeted cancer therapy
Introduction
The terms “aptamer” and “SELEX” were introduced by two independent groups in 1990.1,2 The term “aptamer” refers to small nucleic acid ligands that exhibit specific therapeutic functions and an unambiguous binding affinity for their targets. Conversely, Systematic Evolution of Ligands by EXponential enrichment (SELEX) technology is the method used for aptamer development. Although using small molecule nucleic acids as therapeutics has been explored for decades, development of SELEX and aptamer technology revolutionized this field.
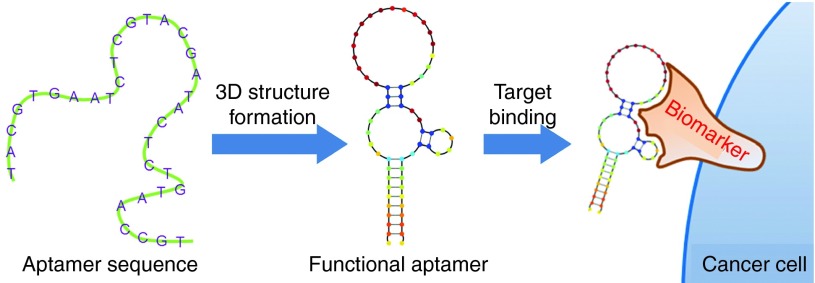
The most important property of an aptamer, from the Latin aptus (to fit), is its high target selectivity. These short, chemically synthesized, single-stranded (ss) RNA or DNA oligonucleotides fold into specific three-dimensional (3D) structures with dissociation constants usually in the pico- to nano-molar range.3 Moreover, in contrast to other nucleic acid molecular probes, aptamers interact with and bind to their targets through structural recognition (Figure 1), a process similar to that of an antigen-antibody reaction. Thus, aptamers are also referred to as “chemical antibodies.”
Figure 1.
Schematic diagram of aptamer binding to its target.
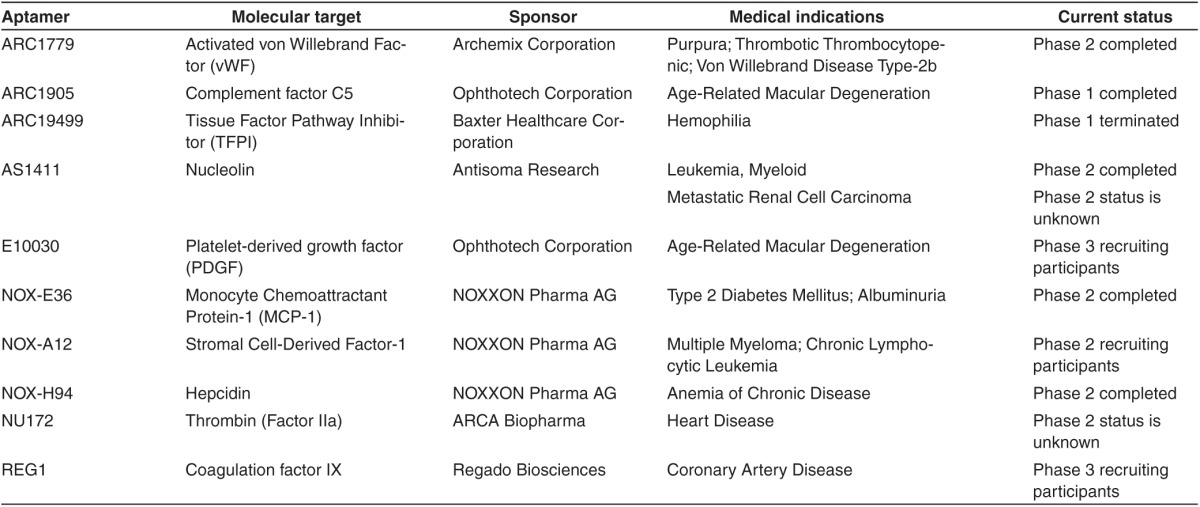
Due to their small size and oligonucleotide properties, aptamers offer several advantages over protein antibodies in both their extensive clinical applicability and a less challenging industrial synthesis process. Specifically, (i) aptamers can penetrate tissues faster and more efficiently due to their significantly lower molecular weight (8–25 kDa aptamers versus ~150 kDa of antibodies). Therefore, aptamers penetrate tissues barriers and reach their target sites in vivo more efficiently than the larger-sized protein antibodies. (ii) Aptamers are virtually nonimmunogenic in vivo. In principal, as aptamers are oligonucleotides they should not be recognized by the immune system. In practice, a recent clinical study showed that aptamers did not stimulate an immune response in vivo,4,5 as compared to protein antibodies that are highly immunogenic, especially following repeat injections. (iii) Aptamers are thermally stable. Based on the intrinsic property of oligonucleotides, even after a 95 °C denaturation, aptamers can refold into their correct 3D conformations once cooled to room temperature. In comparison, protein-based antibodies permanently lose their activity at high temperatures. More importantly, a well-established synthesis protocol and chemical modification technology lead to (iv) rapid, large-scale aptamer synthesis and modification capacity that includes a variety of functional moieties; (v) low structural variation during chemical synthesis; and (vi) have lower production costs. Moreover, aptamers specifically recognize a wide range of targets, such as ions, drugs, toxins, peptides, proteins, viruses, bacteria, cells, and even tissues.6,7,8,9,10,11,12 In the clinic, aptamer-based therapeutics are gaining momentum. For example, Macugen, a modified RNA aptamer, specifically targets vascular endothelial growth factor. It has been approved by the US Food and Drug Administration (FDA)13 for the treatment of wet age-related macular degeneration and is under evaluation for other conditions.14 In the cancer setting, AS1411 targets nucleolin, a protein over-expressed in a variety of tumors. It is currently being evaluated as a potential treatment option in solid tumors and acute myeloid leukemia.15 An updated list of therapeutic aptamers undergoing clinical trials is included in ref. 16 and Table 1. Taken together, these clinical studies highlight many possible uses that aptamers may have in a variety of biomedical fields, including therapeutics.17
Table 1. A list of therapeutic aptamers undergoing clinical trials.
Since aptamer technology was first introduced, the RNA-based sequence library has been widely used for SELEX. Based on the existing evidence, it is believed that the presence of a 2′-OH group and non-Watson-Crick base pairing allows RNA aptamer oligonucleotides to fold into more diverse 3D structures than ssDNA molecules. Consequently, using the more flexible RNA sequences simplifies the development of high-affinity and -specificity aptamers. Despite their advantages, RNA sequences are very sensitive to nucleases present in biological environments and can be rapidly degraded.18 To increase nuclease resistance of RNA-based aptamers, several chemical modifications have been investigated. Evidence shows that 2′-OH group and phosphodiester linkages of RNA sequences are the sites of nuclease hydrolysis. Subsequently, substitutions of the 2′-OH functional group by 2′-fluoro, 2′-amino, or 2′-O-methoxy motifs, and/or changes to the phosphodiester backbone with boranophosphate or phosphorothioate are the most common modifications aimed at increasing nuclease resistance.19 More recently, Wu et al. developed a novel chemical modification method to increase siRNA stability, in which phosphorodithioate and 2′-O-Methyl were simultaneously substituted in the same nucleotide.20 This modification method significantly enhanced siRNA stability and represents a potential new direction for utilization of RNA-based therapies in complex biological systems. Other effective modifications recently reported utilize the locked nucleic acid technology16,21 or generate “mirror” RNA sequence structures, termed spiegelmers.22 These modifications result in structural changes to the RNA sequences, which cannot be digested by nucleases.
In addition to RNA aptamers, ssDNA-based aptamers have also been developed. Due to their lack of 2′-OH groups, DNA molecules are naturally resistant to 2′-endonucleases and are stable in biological environments. Recently, our group developed a biostable DNA-based aptamer specific for CD30, a protein biomarker that is over-expressed in Hodgkin and anaplastic large cell lymphomas. Functional analysis demonstrated that this ssDNA-based aptamer exhibited high CD30 binding affinity as low as 2 nmol/l and was stable in human serum for up to 8 hours. Conversely, an RNA-based CD30 aptamer was digested within 10 minutes under similar conditions.23
In summary, unique chemical features and biological functions have made aptamers a very attractive tool in biomedical research over the past two decades. Currently, there are over 4,000 published articles referenced in the PubMed database that include the term “aptamer.” Research areas that include aptamer technology cover bioassays, drug development, cell detection, tissue staining, in vitro and in vivo imaging, nanotechnology, and targeted therapy. As chemical antibodies, aptamers represent an excellent alternative to replace or supplement protein antibodies, which have been extensively used in the clinic.
Aptamers Specifically Targeting Cell Surface Biomarkers
Using SELEX technology to develop aptamers for cell surface biomarkers
SELEX, the methodology used to develop aptamers specific for a target of interest, is based on a repetitive amplification and enrichment process. The SELEX process follows several steps: first, a random ssDNA oligonucleotide library is chemically synthesized to contain between 1014–1015 unique random sequences flanked by conserved primer binding sites. This step utilizes the following universal scheme: 5′-sense primer sequence-(random sequence)-antisense primer sequence-3′, where the primer sequence ranges from 18 to 22 bases and the random sequence contains 20–40 nucleic acids. The general procedure consists of labeling the 5′-sense primer with a fluorochrome reporter for monitoring aptamer selection, while the 3′-antisense primer is labeled with an affinity molecule, such as biotin, that is used to separate single-stranded oligonucleotides generated in each amplification round. This random ssDNA library can be used directly to select an initial pool of DNA aptamers. Conversely, generation of RNA aptamers requires two extra steps. Specifically, a pool of random ssDNA oligonucleotides is generated, T7 RNA polymerase promoter sequence is added to the 5′-sense primer, and the DNA is then used as a template for T7 RNA polymerase-based transcription in the 5′ to 3′ direction. During the second SELEX step, the oligonucleotide library is heated and rapidly cooled to promote the formation of 3D structures. The library is then mixed with the target of interest for specific binding enrichment. In the third step, the unbound sequences are discarded through the use of membranes, columns, magnetic beads, and capillary electrophoresis.6,24,25 In the fourth step, the enriched sequences are amplified in vitro by either PCR (DNA aptamers) or RT-PCR (RNA aptamers) to generate a new sequence library for the next round of SELEX. The amplified sequence library may go through further negative-target selection, which eliminates the nonspecific sequences generated by binding of nontarget moieties. Lastly, aptamer selection goes through 4–20 rounds of amplification and enrichment. The exact number of required amplification and selection steps depends on the aptamer target being a purified protein or a living cell, and on the evolution of the aptamer sequence library, as that established by gel electrophoresis, flow cytometry (for target binding), classical cloning or sequencing methods, or by high throughput Next-Generation Sequencing (NGS). In recent years, the traditional SELEX method had also been modified to include the capillary electrophoresis (CE) SELEX, toggle selection, photo-SELEX, bead-based selection, X-Aptamers, and Slow Off-rate Modified Aptamers (SOMAmers) in order to maximize affinity and specificity, to improve the speed of selection and success rate, and to provide additional properties to the selected aptamers.26,27,28,29,30,31
Similar to protein antibody development, purified recombinant proteins or peptides expressed in prokaryotic or eukaryotic systems can be used as targets for aptamers selected by the SELEX method. However, because of the posttranslational modifications, especially in the case of highly glycosylated proteins, purified proteins or peptides often cannot fold into the correct 3D structure that is formed under physiologic conditions.32 Consequently, the newly synthesized aptamers may not be able to selectively recognize and interact with their corresponding targets, which would result in failure of the biomedical application. As this is a common problem, it is very important to choose biomarkers in their native conformation for aptamers selection. Taking this issue into an account, a modified SELEX technology that uses whole living cells, Cell-based SELEX (or Cell-SELEX), was recently established.33 To develop cell-specific aptamers, the Cell-SELEX method uses whole living cells that express surface biomarkers of interest. However, the presence of many different cell surface molecules in addition to the target biomarker(s) results in the synthesis of many unrelated/unwanted aptamers. Therefore, in addition to all the SELEX steps described above, Cell-SELEX technology also utilizes control cells that do not express the target biomarker(s) during the counter-selection step.33
Well-characterized biomarkers that are endogenously expressed at high levels, such as the ErbB superfamily, MUC1, EpCAM, and CD30, offer the best potential for cell-based aptamer development. Subsequently, cell lines that have high endogenous expression of cell-specific or cancer type-specific biomarker(s) are commonly used for Cell-SELEX. However, if such cell lines are unavailable, a biomarker of interest could be over-expressed in a particular cell line via gene transfection and the parental cells used for counter-selection. Using this approach, aptamers targeting the cancer stem cell (CSC) biomarker CD133 have been recently developed.34 In this study, CD133 cDNA was transfected into HEK293T cells that were then used for aptamer enrichment, with the parental HEK293T cells serving as a negative control. Similarly, an aptamer specific for the human receptor tyrosine kinase was recently developed.35
Despite the advantages offered by the Cell-SELEX system, this method provides low aptamer enrichment efficiency because many off-target surface biomarkers/molecules are coexpressed on the cells of interest. To overcome this obstacle, our lab introduced a modified SELEX method that combines the cell-based SELEX with purified protein-based SELEX techniques. This hybrid (or cross-over) SELEX had been used to develop Tenascin-C-specific RNA aptamers.36 In our lab, by employing the hybrid-SELEX approach, we developed a DNA aptamer specific for CD30-positive lymphoma tumor cells.23 As shown in Figure 2, the synthesized ssDNA sequence library was initially selected through the cell-based SELEX with CD30-expressing cells, followed by further enrichment with the purified CD30 protein-based SELEX. The current thought is that aptamers developed through this hybrid-SELEX process will be more selective in recognizing and binding to their target biomarker(s). In addition to our hybrid-SELEX approach, other modified Cell-SELEX technologies have been developed, such as internalized Cell-SELEX, designed to select functional aptamers that could be internalized by human cells,37,38,39,40,41,42,43 and FACS-SELEX, that is used to eliminate dead cells that nonspecifically bind nucleic acids and affect subsequent aptamer selection results.44,45
Figure 2.
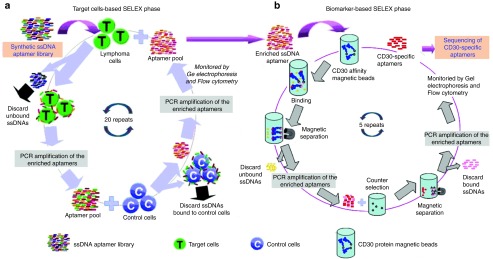
Schematic diagram of our hybrid-SELEX method for selection of CD30-specific ssDNA aptamer. In our experiment, the hybrid-SELEX process is divided into (a) the cell-based SELEX selection and (b) CD30 protein-based SELEX enrichment. First, CD30-expressing lymphoma cells are used for positive selection and CD30-negative Jurkat cells are used in negative counter-selection. After 20 rounds of selection, the enriched aptamer pool is incubated with CD30 protein immobilized on magnetic beads for five additional rounds of enrichment. SELEX, Systematic Evolution of Ligands by EXponential enrichment.
One of the most important applications for the Cell-SELEX technology lies in biomarkers discovery, a new and exciting area of personalized cancer research. Using the Cell-SELEX method, cell-specific aptamers can be selected without prior knowledge of biomarkers present at the cell surface and is achieved through the altered target cell-mediated enrichment and off-target cell-mediated elimination. Newly synthesized aptamers can be further characterized and biomarkers identified by combining affinity purification and mass spectrometry analysis.46,47,48 Through this approach, protein tyrosine kinase 7 (PTK7)11,49 and immunoglobin heavy mu chain47,48 were shown to be highly expressed in solid tumors and on lymphoma cells.
Aptamers specific for cell surface biomarkers
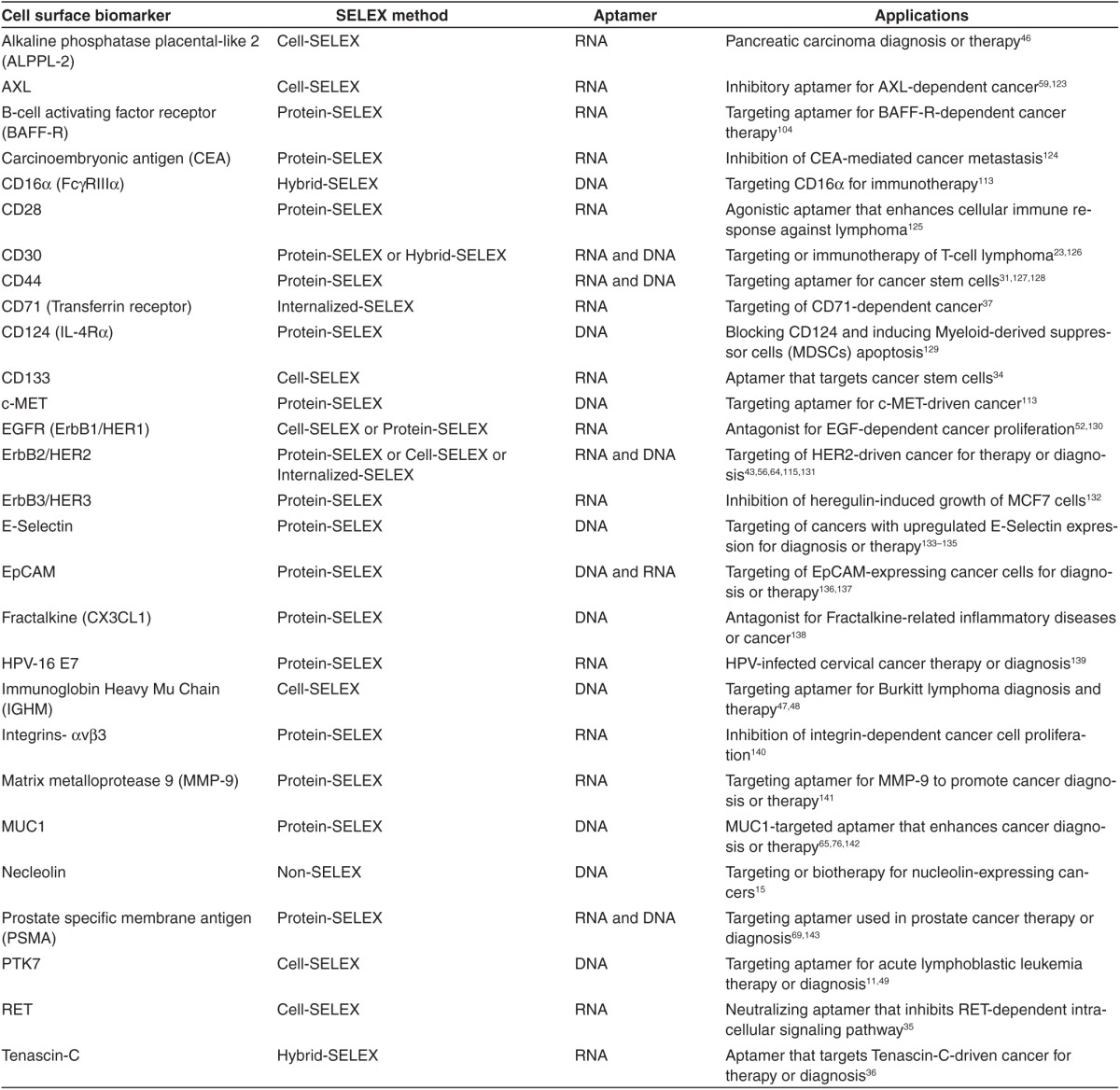
Cell surface biomarkers are functionally important molecules involved in many biological processes, such as signal transduction, cell adhesion and migration, cell–cell interactions, and communication between the intra- and extra-cellular environments. An abnormal expression of cell surface biomarkers is often related to tumorigenesis.50 Clinically, it is estimated that about 60% of cancer-targeting drugs, including therapeutic antibodies and small molecule inhibitors, target cell surface biomarkers,51 making them attractive for disease treatment. In the last decade, many aptamers targeting cell surface biomarkers have been developed through the advancement of both the protein- and/or cell-based SELEX technologies (see Table 2 for detailed list). These aptamers have been extensively studied for diagnosis and/or treatment of hematological malignancies,7,23,49 lung,52,53,54 liver,55 breast,56,57 ovarian,58 brain,59,60 colorectal,61 and pancreatic cancers,46 as well as for identification and characterization of CSCs.34,62
Table 2. Aptamers specifically targeting cell surface biomarkers used in cancer therapy.
Aptamer-Mediated Targeted Therapies
Traditional cancer treatment approaches, such as chemotherapy, radiotherapy, photodynamic therapy, and photothermal therapy can cause serious side effects in patients due to their associated nonspecific toxicity. To minimize these side effects, a concept of personalized, targeted therapy has been gaining momentum. One of the main clinical approaches for targeted cancer therapy employs antibody-based drugs. Although antibody-mediated therapy is highly specific and results in fewer side effects, potential immunogenicity and high cost of production may limit its clinical applications. To overcome these obstacles, oligonucleotide aptamer-based targeted therapeutics and specific drug delivery systems have recently been explored. These studies revealed numerous advantages offered by the aptamer technology over protein-based antibody therapies, with some of these described in the section below.
Aptamer-drug conjugates
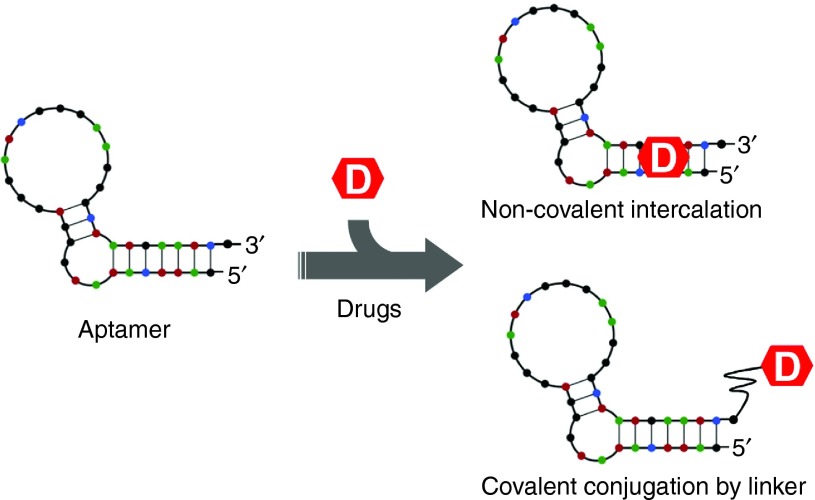
Aptamer-drug conjugation (ApDC) is a very simple yet effective model of noncovalently or covalently conjugating aptamer sequences directly with therapeutic agents (Figure 3). For example, aptamer-conjugated Doxorubicin (Dox), a chemotherapeutic agent extensively used in the treatment of various cancers, has recently been shown to have enhanced therapeutic efficacy over Dox alone. Mechanistically, Dox cytotoxicity is caused by its intercalation into the nucleic acid structure at the preferred paired CG or GC sites with subsequent inhibition of cancer cell proliferation. Taking advantage of its propensity for intercalation, Dox can be noncovalently conjugated to oligonucleotide aptamers containing CG/GC sequences through a simple incubation step. A recent report by Subramanian et al. describes the effectiveness of aptamer-Dox conjugates in the treatment of retinoblastoma.63 In their study, a 2′-fluoro modified RNA aptamer EpDT3 (specific for EpCAM, a CSC marker), was noncovalently conjugated with Dox. After binding to EpCAM molecules expressed at the cancer cell surface, the EpDT3-Dox conjugates were preferentially internalized by the cancer and not by the healthy cells, greatly enhancing therapeutic efficacy and reducing treatment-associated side effects. Several other studies also utilized aptamer-Dox conjugates for cancer therapy, such as HER2 aptamer-Dox conjugates targeting breast cancer,64 MUC1 aptamer-Dox conjugates targeting lung cancer,65 and PSMA aptamer-Dox conjugates targeting prostate cancer.66 Despite their obvious advantages, several concerns related to the use of aptamer-Dox conjugate have been raised. These include (i) instability of the aptamer-drug conjugate due to the reversible nature of noncovalent conjugation process; (ii) short circulating half-life of aptamer-drug conjugates in vivo due to their low molecular weight; and (iii) poor drug payload capacity due to a very simple structure of aptamers. These three disadvantages and technological approaches to improve them are described in greater detail below.
Figure 3.
Schematic diagram of noncovalent or covalent aptamer-drug conjugation.
To enhance the stability of drug loading, Dox can be covalently conjugated to aptamer sequences via a functional linker moiety. For example, the DNA aptamer sgc8 possesses a strong affinity for PTK7 kinase that is abundantly expressed on the surface of CCRF-CEM T-cell acute lymphoblastic leukemia cells. To enhance its stability, this aptamer was covalently conjugated with Dox through an acid-labile linker.67 Once the sgc8 aptamer-Dox conjugate was preferentially bound and internalized by the target cells, the acid-labile linker was easily cleaved in the acidic lysosomal environment, releasing Dox and effectively killing target cells.67 On the other side of the spectrum, covalent conjugation is the most commonly used method of aptamer-drug conjugation, especially for agents that cannot intercalate into the nucleic acid structure or whose intercalation would disrupt aptamer structure.68 Evidence suggests that these covalently conjugated aptamer-drug compounds are significantly more stable than the corresponding noncovalently conjugated intercalations.69
Conjugation of aptamers with high molecular weight polymers, such as polyethylene glycol (PEG), has been examined in order to increase aptamer molecular weight. Specifically, PEG has been widely used in drug modifications, including synthesis of Macugen aptamers. This modification, resulting in PEGylated aptamers, not only increased the aptamer molecular weight and prolonged its circulating half-life, but also enhanced its stability and decreased its toxic accumulation in nontarget tissues.70,71
Finally, in order to increase aptamer-drug payload capacity, an innovative model named aptamer-tethered DNA nanotrains (aptNTrs) was recently introduced by Zhu et al. to deliver Dox to cancer cells.72 In this study, structure of the sgc8 aptamer that targets PTK7 was modified by adding a DNA trigger probe on the 5′-end. Consequently, the modified aptamer acted as a locomotive for targeting, while two hairpin monomers containing Dox intercalation sites acted as boxcars to deliver the drug. After self-assembly, the newly synthesized sgc8 aptamer-NTrs displayed high drug payload capacity, with the drug/sgc8 aptamer-NTr molar ratio of 50:1. Importantly, sgc8 aptamer-NTrs-Dox conjugates were preferentially internalized by the target cells, thereby inhibiting tumor cell growth in vitro and in vivo.72
Another strategy for increasing the aptamer payload capacity involves the construction of polyvalent aptamers. Polyvalent aptamers exhibit an increased target affinity and are more rapidly internalized by their target cells. To demonstrate this, Boyacioglu et al. developed a new DNA aptamer they termed SZTI01 against PSMA.69 First, a dimeric aptamer complex (DAC) was created for specific delivery of Dox to PSMA-expressing cancer cells. Then, the SZTI01 aptamer was modified on the 3′-terminus with either a dA16 or dT16 single-stranded tail that contained CpG sites for loading Dox, and the two monomers were annealed in a 1:1 ratio to form the DAC structure. The results of the study showed that DACs have a high Dox payload capacity with the Dox/DAC molar ratio of about 4:1, and the DACs-Dox conjugates were stable under physiological conditions for up to 8 hours.69 In another study, a DNA aptamer targeting MUC1 was truncated and an aptamer containing three repeats of the active targeting region, termed L3, was synthesized. Although the Dox payload capacity was not specifically modified in the L3 aptamer, the L3-Dox conjugates showed a stronger affinity to target cells and lower cytotoxicity to off-target cells than the parental MUC1 aptamer.73 Finally, polyvalent aptamers can also be constructed through the rolling circle amplification (RCA) technology. Using the RCA method and the sgc8 aptamer sequence as a circular template, a polyvalent sgc8 aptamer, termed Poly-Aptamer-Drug, was synthesized.74 It was determined that the Dox payload capacity of the polyvalent sgc8 aptamer increased tenfold, as compared to the monovalent sgc8 aptamer. Moreover, because of their 40-fold greater binding affinity, the Poly-Aptamer-Drug conjugates were more effective than their monovalent counterparts in targeting and killing leukemia cells.74
Although Dox presents itself as a very attractive chemotherapeutic agent for use in aptamer conjugation, other drugs, such as Gemcitabine (Gem) and photosensitizers, can also be targeted to cancer cells through the aptamer technology. Gem is an FDA-approved deoxycytidine analog (dFdC) used for anticancer therapy. To deliver Gem specifically to pancreatic cancer cells, Ray et al. developed a novel aptamer-Gem polymer model. In this model, a single-stranded RNA polymer contained Gem that was enzymatically synthesized through a mutant T7 RNA polymerase-mediated transcription reaction and fused with a nuclease-resistant 2′-fluoro-modified RNA aptamer (E07) that selectively binds to EGFR on pancreatic cancer cells. The E07 aptamer structure was modified by introducing a 24-nucleotide sequence at the 3′ end and using it as an adaptor for Gem polymer binding. Following an annealing step, the Gem polymer complementary bound with the E07 aptamer and preferentially targeted the EGFR-expressing pancreatic cancer cells, inhibiting cell proliferation.75
Compared with the traditional chemotherapeutic agents, controlled conditional prodrug photosensitizers have also been extensively used for aptamer-mediated drug delivery. In this therapeutic approach, termed photodynamic therapy, or photodynamic therapy, photosensitizers are activated by light irradiation and induce production of intracellular reactive oxygen species, resulting in cytotoxicity. A study by Ferreira et al. describes the development of a DNA aptamer specific for MUC1 and covalently conjugated at the 5′ end with the photosensitizer chlorin e6.76 Upon light irradiation, MUC1-expressing epithelial cancer cells were preferentially killed with cytotoxicity about 500-fold higher than that of the control cells. Similar studies have reported using a necleolin aptamer (AS1411)-TMPyP4 for targeting breast cancer77 and the EGFR aptamer (R13)-TF70 for treatment of lung cancer.78
Finally, approaches to extend the scope of aptamer application have also been developed. Similar to bi-specific antibodies, bi-specific or even tri-specific aptamers can be constructed. A bi-specific aptamer for targeting different cells was recently described by Zhu et al. In their study, specific DNA aptamers sgc8 and sgd5a were conjugated through a dsDNA linker. Compared to each mono-aptamer, this bi-specific aptamer (named SD) could recognize its target cell simultaneously with equal specificity and affinity, while Dox intercalation into the dsDNA induced target cell cytotoxicity.79 In the same study, a Y-shape dsDNA linker was used to construct a tri-specific aptamer that also recognized its target cells with high specificity and affinity.79 Clinically, Min et al. proposed using a bi-specific aptamer for prostate cancer therapy. It is well established that prostate tumors may contain both PSMA-positive and -negative cell types. Thus, this study utilized two aptamers, a 2′-fluoro modified RNA aptamer targeting PSMA-expressing cells and a DUP-1 peptide aptamer specific to PSMA-negative cells, conjugated through streptavidin. Moreover, intercalating Dox into the PSMA aptamer of this bi-specific aptamer model could serve as a tool to target all prostate cancer cell types.80
Aptamer-nanoparticle therapeutics
Nanoparticles (NPs) are attractive vehicles to increase both the half-life and the drug payload capacity of aptamer-mediated drug delivery. In addition to their common features, such as biocompatibility for clinical applications, large surface for enhanced aptamer and drug loading, and uniform size and shape for excellent biodistribution, NPs have other individual physical and chemical properties defined by their materials. For example, copolymers and liposomes are biodegradable, while metal materials offer exceptional photothermal and magnetic performance. Thus, NPs are used extensively in drug delivery and controlled release systems, with several examples described below.
Copolymers and liposomes. Conjugation of aptamers with copolymers or liposomes offers excellent opportunities for targeted drug delivery, based on their enhanced biodegradability and biocompatibility. Pioneering work of Farokhzad et al. described the development of aptamer-NPs bioconjugates almost a decade ago.81,82 In their study, an anti-PMSA aptamer was conjugated with poly(lactic acid; PLA)-PEG or poly(lactic-co-gycolic-acid; PLGA)-PEG functional groups; in this model the anti-PSMA aptamer was used as a targeting molecule, PLA or PLGA help to encapsulate and control drug release, while PEG enhanced circulating half-life of the resultant bioconjugate. After optimization and Docetaxel (Dtxl) loading, these Dtxl-encapsulated functional aptamer-NPs significantly improved in vitro cellular toxicity by preferentially targeting LNCaP cells. More importantly, following a single intratumoral injection, Dtxl-encapsulated functional aptamer-NPs showed reduced systemic toxicity and a significant in vivo tumor burden reduction. In the years following these successful early studies, several publications described the development of novel aptamer-NPs bioconjugates. For example, due to its high level of expression, MUC1 protein is an important target for anticancer drug delivery in most adenocarcinomas. A functional MUC1 aptamer-NPs bioconjugate was created using an emulsion/evaporation method to include PLGA and paclitaxel (PTX). After PTX loading, the encapsulation efficacy was 83.66 ± 1.7% with a drug load of 4.26 ± 0.1%. A typical kinetic profile for sustained release was observed, with about 65% of the drug gradually released over the first 48 hours. Importantly, this PTX-loaded aptamer-NPs enhanced the in vitro drug delivery and cytotoxicity restricted to MUC1-expressing cancer cells.83
In another example, an AS1411 aptamer was used to target tumor cells. In this study, Aravind et al. developed a PLGA-lecithin-PEG and PTX-containing AS1411 aptamer-NPs bioconjugates for breast cancer therapy.84 After optimization, the NPs were about 85.5 nm in size, and exhibited high encapsulation efficiency (60.93 ± 3.4%) and superior sustained drug release, compared to the PLGA NPs. As expected, these NPs were effectively internalized by target cells and enhanced the in vitro cell killing effect.84 A study by Guo et al. utilized a similar approach for glioma therapy in vivo. Their results show that these novel conjugates exhibit a prolonged half-life in circulation, inhibit xenograft tumor growth, and prolong survival of animals bearing intracranial glioma.85 Finally, a recent study by Xing et al. described a functional AS1411 aptamer-liposome NPs for breast cancer therapy. These aptamer-liposome NPs exhibited enhanced tumor tissue penetration and improved antitumor efficacy in mice bearing MCF7 xenografts.86
In the study by Dhar et al., authors combined a modified RNA aptamer A10 specific for PSMA with a functionalized platinum [Pt(IV)]-based PLGA-PEG NP for prostate cancer therapy.87 The authors then investigated NP-associated pharmacokinetics, biodistribution, tolerability, and efficacy. The results show that functional NPs had high maximum tolerated dose, prolonged systemic blood circulation time, decreased kidney toxicity, and enhanced antitumor efficacy in vivo.87,88 Following a similar scenario, several recent studies utilized different aptamers and copolymers or liposome: TD05 aptamer-liposome NPs for Burkitt lymphoma89 and PSMA aptamer-PCL-PEG NPs for prostate cancer.90 All of these studies indicate that aptamer-guided functionalized copolymers or liposome NPs are an effective, universal, and safe platform for targeted drug delivery.
Metal nanomaterials. Excellent optical, electromagnetic, stability, and biocompatibility properties make metal nanomaterials useful tools in designing targeted drug delivery systems. Recently, our group reported a novel metal NP, Apt-HAuNS-Dox, which is comprised of a CD30-specific RNA aptamer (Apt), a functional hollow gold nanosphere (HAuNS), and conjugated Dox.91 Our results indicate that the formed Apt-HAuNS-Dox NPs, approximately 42 nm in diameter, have a drug payload efficiency of >90%, which equals approximately 30% w/w. Importantly, these NPs are ultrasensitive to pH changes, and release 80% of their Dox payload within 2 hours at pH 5.0. In vitro studies utilizing a mixture of various cell types revealed that the Apt-HAuNS-Dox NPs selectively kill lymphoma cells, but exhibit no cytotoxicity in other cell types present in the same culture (Figure 4).91
Figure 4.
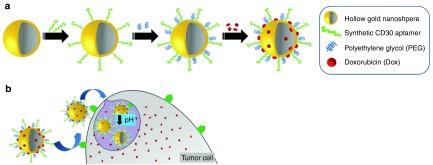
Formulation of the Apt-HAuNS-Dox nanoscale drug-delivery system and the mechanism of pH-dependent drugs release. (a) A schematic illustration of Apt-HAuNS-Dox synthesis. Aptamers and PEG were conjugated to the surface of HAuNS sequentially via covalent S-Au bonds, followed by loading with doxorubicin through a charge force. (b) Our hypothesis is that the Apt-HAuNS-Dox NPs selectively target lymphoma cells via the aptamer-mediated biomarker interaction, resulting in internalization and intracellular delivery into lysosomes. Due to their low pH sensitivity, lysosomal microenvironment triggers a rapid Dox release and initiates tumor cell apoptosis. PEG, polyethylene glycol; NPs, nanoparticles.
In another example of gold NP utilization, gold nanomaterials are incorporated into the photothermal therapy (PTT) strategy under near-infrared laser irradiation. A study by Wang et al. described conjugation of two specific aptamers, namely CSC1 and CSC13, to the surface of gold nanorods (AuNRs).92 Resulting conjugates were then used to target and kill both stem and nonstem cancer cells. Using this method, a 10-minute near-infrared laser irradiation decreased cell viability to 36% and 47%, respectively.92 In a follow up study, the same group developed a PTT/photodynamic therapy combination strategy based on the sgc8 aptamer tagged to both AuNRs and a photosensitizer. With this approach, the PTT/photodynamic therapy dual therapy resulted in a more effective therapeutic outcome than either therapeutic modality alone.93
Finally, super paramagnetic iron oxide nanoparticles (SPION) possess simultaneous drug delivery and in vivo imaging qualities. Using the 5TR1 aptamer against MUC1 as a targeting tool, Jalalian et al. developed an Epirubicin-5TR1 aptamer-SPION tertiary complex for imaging detection and treatment of murine colon carcinomas.94
Virus-like particles. Virus-like particles (VLPs) are naturally derived nanomaterials with multifunctional properties that can be used for drug delivery. A method for constructing VLP-aptamer conjugates was first reported by Tong et al. in 2009.95 Using an efficient oxidative coupling strategy, up to 60 copies of sgc8 aptamer were conjugated on the surface of each MS2 bacteriophage capsid. The functional sgc8 aptamer-VLPs conjugates exhibited a strong binding capacity to target cells, followed by internalization and degradation in lysosomes.95 More recently, Cohen and Bergkvist developed a functional VLP-AS1411 aptamer loaded with porphyrin photosensitizer that was used for breast cancer therapy. Their results showed that after irradiation, almost 100% of MCF7 cells underwent apoptosise, as compared to virtually no cell death observed in the off-target MCF-10A cells under the same treatment conditions.96
In conclusion, copolymer, liposome, metal, and virus-like NPs could be successfully used to enhance biodistribution, stability, and targeting affinity of aptamers. In addition, nanomaterials such as hydrogels, silica, quantum dots (QD), and single-walled carbon nanotubes are also promising vehicles for aptamer-mediated delivery of targeted therapeutics. However, several applications of these nanomaterials have been recently reviewed,97,98,99 and will not be discussed further in the present report.
Aptamer-mediated gene therapy
Small interfering RNA (siRNA) and microRNA (miRNA) molecules are powerful gene silencing tools that represent a new class of gene-mediated therapeutics, and their effectiveness as cancer gene therapy has been extensively evaluated. However, their use in clinical applications has been limited due to their lack of cell/tissue specificity during in vivo delivery. Therefore, combining aptamers that provide high targeting specificity with siRNA/miRNA technology can achieve selective gene targeting with high efficiency.
Similar to conjugation of chemotherapeutic agents, siRNA and miRNA can be covalently conjugated with aptamers to form aptamer-siRNA or aptamer-miRNA chimeras. McNamara et al. first developed an aptamer-siRNA chimera in which either Plk1 (polo-like kinase 1) siRNA or Bcl2 (B-cell lymphoma-2) siRNA were covalently conjugated with a modified A10 RNA aptamer against PSMA.100 Their results showed that this simple conjugation did not affect biological functions of either the aptamer or the siRNA. Moreover, aptamer-siRNA chimeras could specifically bind to the PSMA-expressing cells, thereby silencing Plk1 or Bcl2 gene expression and significantly inhibiting in vivo tumor growth.100 In a similar approach, Thiel et al. used a RNA aptamer against the rat HER2 that was conjugated with a Bcl2 siRNA. Consequently, these HER2 aptamer-Bcl2 siRNA chimeras specifically targeted HER2-expressing cancer cells and downregulated Bcl2 gene expression. Furthermore, these chimeras also enhanced cisplatin efficacy, thus providing a novel combinational strategy for cancer therapy.43 In fact, in addition to cancer therapy, utilization of this simple aptamer-siRNA chimera model is very effective and has been applied in other areas of biomedical research. For example, HIV infections101 are another serious public health problem. In an elegant study reported by Zhou et al., an inhibitory RNA aptamer against gp120 was covalently conjugated with a tat/rev siRNA. Their results showed that this dual inhibitory function of an anti-gp120 aptamer-siRNA chimera significantly inhibited HIV replication and host-to-host spread.102,103
In addition to covalent conjugation, noncovalent conjugation of aptamers and siRNA is also an effective method. Recently, Zhou et al. developed a series of BAFF-R aptamer-STAT3 siRNA chimeras through covalent and noncovalent conjugation methods. In their study, an inhibitory 2′-fluoro modified RNA aptamer against BAFF-R, a protein that is over-expressed in B-cell malignancies, was developed to deliver STAT3 siRNA. Two BAFF-R aptamer-STAT3 siRNA chimeras were conjugated covalently through transcription, in which the sense and antisense strands were swapped. The other two chimeras were conjugated through a RNA stick, in which the aptamer-stick and siRNA-stick were chemically synthesized and annealed. Results showed that all chimera combinations (i.e., aptamer-siRNA and aptamer-stick-siRNA) preferentially delivered STAT3 siRNA into BAFF-R-expressing cells with similar efficacy, subsequently downregulating STAT3 gene expression. Combined with the inhibitory function of a BAFF-R aptamer, this dual-function chimera represents an effective alternative for treatment of B-cell malignancies.104
Drawing from other branches of aptamer technology, multivalent aptamer-siRNA chimeras have been recently developed. A study by Wullner et al. describes a bivalent aptamer-siRNA chimera for prostate cancer therapy.81 In their design, two RNA aptamers directed against PSMA were joined together using a linker of the eukaryotic elongation factor 2 (EEF2) siRNA. Compared with a corresponding monovalent chimera, this bivalent PSMA aptamer-EEF2 siRNA chimera showed a greatly enhanced in vitro cytotoxicity, prolonged half-life, and superior tumor inhibition in vivo.105 In another example, a multivalent PSMA aptamer-siRNA chimera was noncovalently conjugated through a biotin-streptavidin connector and showed satisfactory specificity and effectiveness against PSMA-positive cells.106
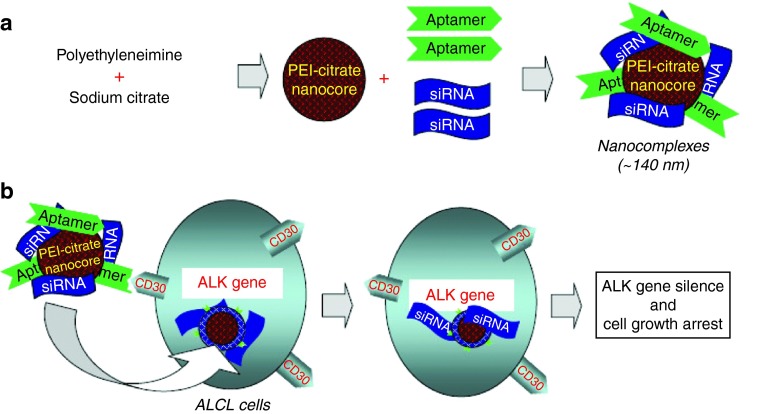
Although aptamer-siRNA chimeras could specifically deliver siRNA to their target cells, there are some challenges associated with systemic delivery, such as poor siRNA payload, short biological half-life, and undesirable biodistribution. One potential solution to these problems lies in nanotechnology. For example, our group developed a functional siRNA nanocomplex comprised of a CD30 aptamer and an ALK-targeted siRNA within nano-sized poly-(ethylenimine) (PEI) polymer carriers (Figure 5).107 PEI is a well-characterized cation carrier that exhibits high cell transfection efficiency, strong buffering capacity, and ability to release functional nucleic acids from endosomes into the cytoplasm by inducing osmotic endosomal rupture. Our study showed that functional siRNA/aptamer nanocomplexes preferentially bound to and were internalized by the CD30-expressing lymphoma cells, resulting in downregulation of cellular ALK gene expression, inhibition of cell proliferation, and apoptosis.107 To further improve this approach, Bagalkot et al. developed a two-step process in which the resultant chimeras were comprised of a siRNA, aptamer, PEI, and QDs-PMAT nanocore. In doing so, the thiol-reactive terminal group-modified siRNA molecules were first adsorbed electrostatically onto the PEI-QDs-PMAT nanocore, and then the PSMA aptamer with a thiol group was conjugated with a siRNA moiety to form an aptamer-siRNA chimera on the NPs surface. Using PSMA-expressing cells as a model, these multifunctional QDs-chimera NPs could be specifically internalized, thereby silencing expression of their target genes.108
Figure 5.
Development of a tumor cell type-selective and cancer gene-specific nanocomplex for ALCL cells. (a) A nano-sized carrier core structure was initially formed via aggregation of polyethyleneimine (PEI) and cross linking with sodium citrate (PEI-citrate nanocore). The synthetic RNA-based CD30 aptamers and ALK siRNA were then incorporated onto the PEI-citrate nanocore to form the nanocomplex. (b) When the functional RNA nanocomplex is added to cultures, the aptamer component will selectively target CD30-positive ALCL cells. Aptamer-mediated cell binding will facilitate intracellular delivery of the nanocomplex. The siRNA component will subsequently silence the cellular ALK gene, resulting in the growth arrest of ALCL cells. ALCL, anaplastic large cell lymphoma.
Similar to siRNA, miRNA can also be selectively targeted by specific aptamers to desired tissues. In a study by Dai et al., researchers developed a MUC1 aptamer-miR29b chimera to target ovarian carcinoma cells. In their study, a palindromic miRNA-29b sequence was synthetically joined at the 3′-end with a MUC1 aptamer. Results indicated that the Chi-29b chimera was preferentially internalized by MUC1-expressing OVCAR-3 cells, resulting in downregulation of DNA methyltransferase expression (DNMTs, specific targets of miR29b) and restoration of PTEN expression, and induction of apoptosis.109 In a follow up study, the same group determined that an intraperitoneal injection of the Chi-29b chimera significantly inhibited in vivo tumor growth by modulating the DNMTs-PTEN signaling pathway. More importantly, the Chi-29b chimeras also inhibited growth of therapy-resistant ovarian cancer cells, indicating that these chimeras may offer novel avenues for the treatment of relapsed ovarian carcinomas.110
Aptamer-mediated immunotherapy
A relatively novel concept in cancer therapy, immunotherapy has recently been getting a lot of attention due to its low potential side effects and high specificity. Of note, antibody-based therapies represent the most studied of immunotherapy approaches. The major therapeutic mechanisms of antibodies are antibody-dependent cell-mediated cytotoxicity (ADCC), complement-mediated cytotoxicity, and enhanced phagocytosis or opsonization through the Fc functional region. As chemical antibodies, oligonucleotide aptamers can imitate protein antibodies to execute immunotherapy functions. In a preliminary study, Bruno et al. developed a DNA aptamer-Fc conjugate to act as artificial antibodies.111 Although the study clearly showed that the aptamer-Fc conjugate could be recognized and internalized by macrophages, no further evaluation of its therapeutic antitumor efficacy was performed.111 In a more recent study, Stecker et al. conjugated a MUC1 aptamer with the C1q molecule through a biotin-streptavidin connector and evaluated the chimera's killing effect using the MCF7 breast cancer cell line. Treatment with this chimera induced significant membrane attack complex (MAC) formation on the MCF7 cell surface, leading to cell death and confirming therapeutic effectiveness of the aptamer-C1q conjugate.112
To improve recruitment of the immune cells towards a tumor, Boltz et al. developed a bi-specific c-MET-CD16α aptamer. Their design combined a c-MET aptamer for specific targeting of cancer cells with a CD16α aptamer that promoted recruitment of the CD16α-expressing natural killer cells. After optimizing the length of aptamers and DNA linkers, the two aptamers were conjugated. The resultant bi-specific aptamer promoted recruitment of the natural killer cells and induced killing of the target cells.113 In another study, Xiong et al. utilized a DNA aptamer specific for K562 leukemia cells that was modified by adding a PEG linker and a diacyllipid tail at the 5′-terminal. In this approach, the PEG linker protected the correct 3D conformation of the aptamer on the cell surface, while the diacyl lipid tail facilitated and enhanced incorporation of the aptamer into the cell membrane. Consequently, this multivalent aptamer increased K562 killing by the natural killer cells by 50%,114 indicating that this model may be further developed for adoptive immunotherapy.
Aptamer-mediated target cell biotherapy
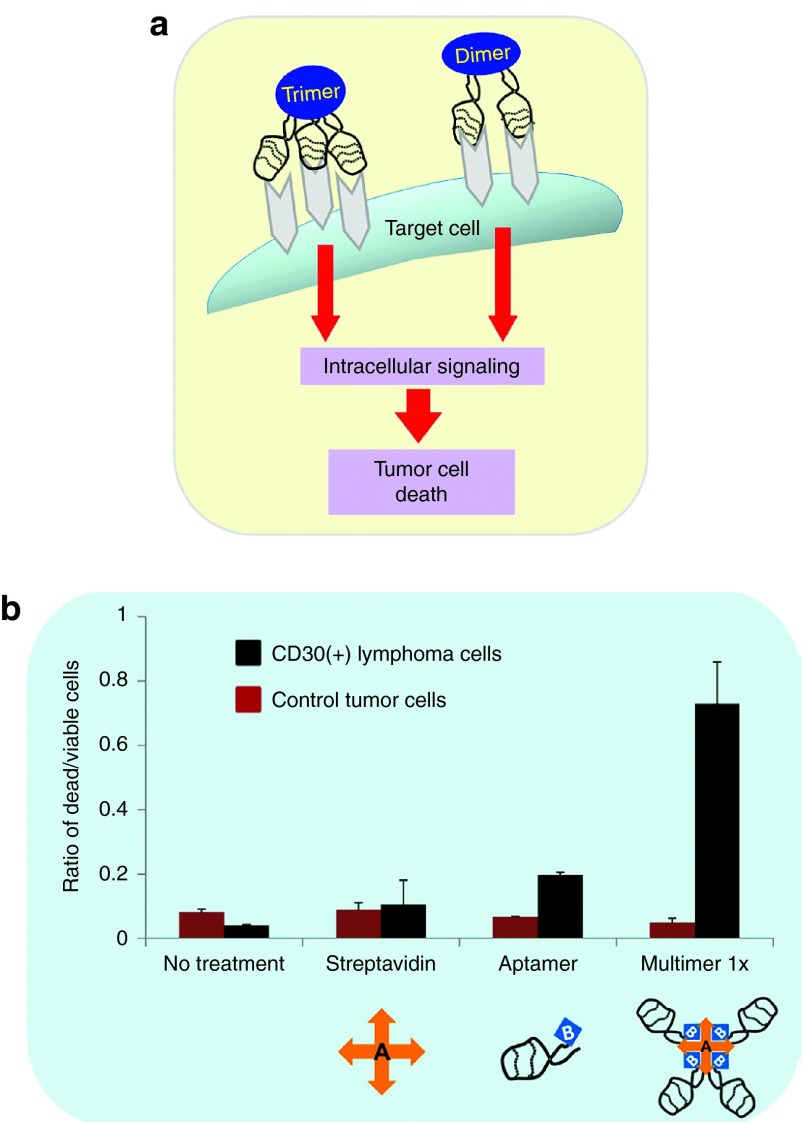
Many cell surface receptors and biomarkers play a role in specific cellular functions, such as signaling transduction pathways. Therefore, their interaction with aptamers could confer agonistic or antagonistic effects on their specific biologic functions, resulting in cancer cell death. In a therapeutic setting, monovalent aptamers, especially those targeted to cell surface biomarkers, are usually unable to activate the downstream signaling pathways. Conversely, multivalent aptamers induce receptor multimerization, thus triggering downstream signaling. Recently, Mahlknecht et al. reported development of a trimeric HER2 aptamer for biotherapy in a human gastric cancer model. This trimeric HER2 aptamer significantly inhibited in vitro and in vivo proliferation of tumor cells, as compared to the monomeric HER2 aptamer.115 When compared against a HER2 monoclonal antibody, the trimeric HER2 aptamer increased the antitumor efficacy by twofold. The molecular mechanism involved internalization and cytoplasmic translocation of the HER2 receptor induced by the aptamer binding, followed by their degradation in the lysosomes.115 In a similar approach, our group developed a highly stable DNA aptamer specific for CD30. We conjugated a biotinylated CD30 aptamer with a streptavidin connector to form multivalent aptamers (Figure 6).23 Our results show that the addition of multivalent aptamers into cell culture induced CD30 oligomerization and subsequently triggered lymphoma apoptosis, up to sixfold higher than that observed in the control group.23 Together, these results demonstrate the feasibility of using aptamers as a biotherapeutics and warrant their further evaluation in the clinical arena.
Figure 6.
Aptamer-based biotherapy. (a) Schema showing receptor oligomerization-inducing downstream signaling. CD30-associated signaling is activated by its ligand through trimerization of the receptor, leading to varied outcomes that range from apoptosis to proliferation. (b) CD30-positive and -negative cells were incubated without any treatment or in the presence of control streptavidin, monomeric aptamer, and multimeric aptamer. Following 72-hour incubation, the multivalent CD30 aptamer induced cell death in the CD30-positive lymphoma cells, but had no effect on the CD30-negative control cells. Ratio of the dead/live cells was calculated by costaining the cells with Hoechst 33342 (live cells) and propidium iodide (dead cells).
Conclusion
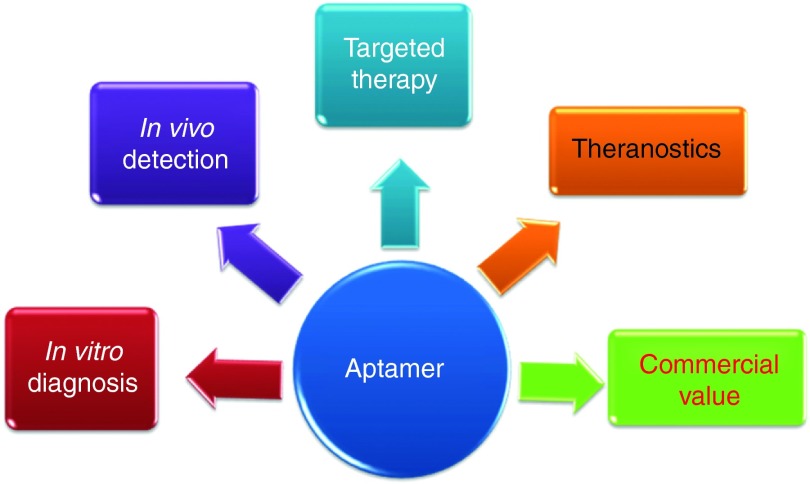
Antibody-based targeted therapeutics provide high target specificity and affinity. However, their potential for immunogenicity is of a great concern, as is their high production cost, both of which have limited their clinical applicability. As discussed in this review, when compared to protein antibodies, oligonucleotide aptamers offer many advantages, including simple chemical synthesis, virtual nonimmunogenicity, smaller size, faster tissue penetration, ease of modification with different functional moieties, low cost of production, and high biological stability. Therefore, aptamers have become a promising new class of molecular ligands that could replace or supplement protein antibodies. In summary, aptamer technology has a strong market value and may be applied in various biomedical fields, including in vitro cancer cell detection, in vivo tumor imaging, and targeted cancer therapy (Figure 7).
Figure 7.
Summary of various aptamer applications.
Although aptamer technology has a great potential in the biomedical field, several technical challenges remain and must be addressed. These include: (i) how can aptamers be rapidly adapted for specific targets by decreasing false-positive/-negative selection? Primarily dependent on the natural properties of targets of interest, such as proteins versus cells or tissues, the process of aptamer selection is usually time-consuming, and the success rate is sometimes low. To improve the speed and success rate, novel methods for aptamer selection have been recently described. They include bead-based selection, that can select aptamers as rapidly as a single round of selection,27,28 and the SOMAmer, which improves the aptamer production success rate from less than 30% to over 50%.29,30 More recently, a study by Cho et al. devised a Quantitative Parallel Aptamer Selection System (QPASS) method, which integrates microfluidic selection, NGS, and in situ-synthesized aptamer arrays. This approach allows for the simultaneous measurement of affinity and specificity for thousands of candidate aptamers in parallel.116 In addition to QPASS, evolving modifications to the Cell-SELEX approach are beginning to address difficulties with successful removal of the influence stemming from the presence of dead cells, slow enrichment aptamers recognizing targets of interest, and contamination with unwanted aptamer sequences. As described above, utilization of the above-mentioned FACS-mediated SELEX44,45 and hybrid-SELEX23 offers novel approaches that address these technical challenges.
(ii) How can we select cancer-relevant targets for aptamer development and clinical applications? Tumorigenesis is a dynamic process that includes multiple constantly changing factors. Therefore, a one-size-fits-all cancer-specific biomarker is unlikely to ever be identified. Yet, it has been established that certain biomarkers present in healthy tissues are highly expressed in cancer cells. Moreover, certain biomarkers are associated with particular cancer cell types making them to be considered as useful targets for development of targeted cancer therapy. However, while use of cancer cells to identify biomarkers and to develop therapeutic agents is a reasonable approach, cultured cells, especially immortalized cell lines, greatly differ from tumor tissues in vivo. To overcome these limitations and to select more reliable cancer-relevant biomarkers for aptamer development, several innovative SELEX methods have been recently described. Of particular interest are the tissue-based SELEX117 and the in vivo-SELEX,118 which offer target selection under more relevant pathologic conditions. This cell/tissue-specific biomarker selection can also be utilized for development of noncancer related therapies, as shown for aptamers targeting the adipose tissue in obesity119 and for aptamers designed to penetrate the blood-brain barrier in order to combat brain diseases.120 Hence, we believe that the careful selection of cancer-associated biomarkers and cell/tissue type-specific biomarkers will expand the scopes of aptamer applicability and improve the feasibility of clinical applications.
(iii) What methods could improve aptamer biostability in vivo? Unmodified RNA-based aptamers are very susceptible to the nuclease-mediated degradation in vivo. Although many chemical modifications aimed at increasing biostability of the RNA aptamers have been developed, including 2′-modifications, 3′-modifications, phosphodiester backbone modifications,19,20 and utilizations of novel nucleic acids (locked nucleic acid and Spiegelmers),16,21,22 their effectiveness is still limited. When it was first described, PEGylation was a very attractive strategy for prolonging aptamer circulation half-life and enhancing their biostability. However, a recent report showed that the in vivo use of PEGylated aptamers induced production of anti-PEG antibodies,121 emphasizing the need for the development of alternative approaches.
(iv) How can aptamer technology be modified to achieve a more effective drug delivery? Many drug delivery systems described in this review are tested in vitro or in animal models. Yet, as with any compound that is translated from the bench to the bedside, aptamer-drug conjugates may behave differently in a human patient than they do in laboratory animals. Therefore, aptamer-drug conjugation remains an important challenge that must be considered. Specifically, various coupling approaches lead to different pharmacokinetics, biodistribution, and tolerability in vivo, which in turn greatly affect treatment effectiveness. In the same vein, we must consider the effectiveness of aptamer-mediated target gene therapy. Gene therapy, including siRNA and miRNA aimed at silencing specific genes, is considered the next generation therapeutic approach. However, silencing a single pathogenic gene may not be a viable therapeutic option because tumorigenesis is a process regulated by multiple genes and signaling pathways. Therefore, combining targeted therapeutics with gene therapy may represent the most effective strategy. Such combinational therapy approaches can greatly improve the therapeutic efficacy while reducing the required dosages of both drugs and small molecule RNAs,122 and, more importantly, may offer new alternatives to combat chemotherapy-resistant cancers.110
(v) The last important point to consider is whether aptamer-mediated biotherapies can become effective, FDA-approved medications. Following Macugen approval by the FDA, many aptamer-mediated biotherapies have been evaluated in clinical trials. Of particular interest is AS1411, an antitumor aptamer that has completed several Phase I clinical trials.15 Trial results are promising and offer useful insights into further modifications that could be applied to therapeutic aptamer development.
Taken together, although some technical challenges remain to be addressed, oligonucleotide aptamers have become an attractive and promising tool for targeted cancer therapy. As more clinical data are accumulated, we and others will be better equipped to optimize aptamer formulations, leading to the expansion of aptamer use in the clinic.
Acknowledgments
This project was supported in part by NIH grants R01CA151955 and R33CA173382 to Y.Z.
References
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- Eyetech Study Group Preclinical and phase 1A clinical evaluation of an anti-VEGF pegylated aptamer (EYE001) for the treatment of exudative age-related macular degeneration. Retina. 2002;22:143–152. doi: 10.1097/00006982-200204000-00002. [DOI] [PubMed] [Google Scholar]
- Eyetech Study Group Anti-vascular endothelial growth factor therapy for subfoveal choroidal neovascularization secondary to age-related macular degeneration: phase II study results. Ophthalmology. 2003;110:979–986. doi: 10.1016/S0161-6420(03)00085-X. [DOI] [PubMed] [Google Scholar]
- Parekh P, Tang Z, Turner PC, Moyer RW, Tan W. Aptamers recognizing glycosylated hemagglutinin expressed on the surface of vaccinia virus-infected cells. Anal Chem. 2010;82:8642–8649. doi: 10.1021/ac101801j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefah K, Tang ZW, Shangguan DH, Chen H, Lopez-Colon D, Li Y, et al. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia. 2009;23:235–244. doi: 10.1038/leu.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayrac AT, Sefah K, Parekh P, Bayrac C, Gulbakan B, Oktem HA, et al. In vitro Selection of DNA Aptamers to Glioblastoma Multiforme. ACS Chem Neurosci. 2011;2:175–181. doi: 10.1021/cn100114k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno JG, Kiel JL. In vitro selection of DNA aptamers to anthrax spores with electrochemiluminescence detection. Biosens Bioelectron. 1999;14:457–464. doi: 10.1016/s0956-5663(99)00028-7. [DOI] [PubMed] [Google Scholar]
- Kirby R, Cho EJ, Gehrke B, Bayer T, Park YS, Neikirk DP, et al. Aptamer-based sensor arrays for the detection and quantitation of proteins. Anal Chem. 2004;76:4066–4075. doi: 10.1021/ac049858n. [DOI] [PubMed] [Google Scholar]
- Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Parekh P, Turner P, Moyer RW, Tan W. Generating aptamers for recognition of virus-infected cells. Clin Chem. 2009;55:813–822. doi: 10.1373/clinchem.2008.113514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que-Gewirth NS, Sullenger BA. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007;14:283–291. doi: 10.1038/sj.gt.3302900. [DOI] [PubMed] [Google Scholar]
- Sundaram P, Kurniawan H, Byrne ME, Wower J. Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci. 2013;48:259–271. doi: 10.1016/j.ejps.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- Schmidt KS, Borkowski S, Kurreck J, Stephens AW, Bald R, Hecht M, et al. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004;32:5757–5765. doi: 10.1093/nar/gkh862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KM, Lee S, Ban C. Aptamers and their biological applications. Sensors (Basel) 2012;12:612–631. doi: 10.3390/s120100612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Keefe AD, Cload ST. SELEX with modified nucleotides. Curr Opin Chem Biol. 2008;12:448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- Wu SY, Yang X, Gharpure KM, Hatakeyama H, Egli M, McGuire MH, et al. 2'-OMe-phosphorodithioate-modified siRNAs show increased loading into the RISC complex and enhanced anti-tumour activity. Nat Commun. 2014;5:3459. doi: 10.1038/ncomms4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster C, Zydek M, Rothkegel M, Wu Z, Gallin C, Geßner R, et al. Properties of an LNA-modified ricin RNA aptamer. Biochem Biophys Res Commun. 2012;419:60–65. doi: 10.1016/j.bbrc.2012.01.127. [DOI] [PubMed] [Google Scholar]
- Eulberg D, Klussmann S. Spiegelmers: biostable aptamers. Chembiochem. 2003;4:979–983. doi: 10.1002/cbic.200300663. [DOI] [PubMed] [Google Scholar]
- Parekh P, Kamble S, Zhao N, Zeng Z, Portier BP, Zu Y. Immunotherapy of CD30-expressing lymphoma using a highly stable ssDNA aptamer. Biomaterials. 2013;34:8909–8917. doi: 10.1016/j.biomaterials.2013.07.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallikaratchy P, Stahelin RV, Cao Z, Cho W, Tan W. Selection of DNA ligands for protein kinase C-delta. Chem Commun (Camb) 2006;30:3229–3231. doi: 10.1039/b604778e. [DOI] [PubMed] [Google Scholar]
- Martin JA, Parekh P, Kim Y, Morey TE, Sefah K, Gravenstein N, et al. Selection of an aptamer antidote to the anticoagulant drug bivalirudin. PLoS ONE. 2013;8:e57341. doi: 10.1371/journal.pone.0057341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath SC. Methods developed for SELEX. Anal Bioanal Chem. 2007;387:171–182. doi: 10.1007/s00216-006-0826-2. [DOI] [PubMed] [Google Scholar]
- Yang X, Bassett SE, Li X, Luxon BA, Herzog NK, Shope RE, et al. Construction and selection of bead-bound combinatorial oligonucleoside phosphorothioate and phosphorodithioate aptamer libraries designed for rapid PCR-based sequencing. Nucleic Acids Res. 2002;30:e132. doi: 10.1093/nar/gnf132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li X, Prow TW, Reece LM, Bassett SE, Luxon BA, et al. Immunofluorescence assay and flow-cytometry selection of bead-bound aptamers. Nucleic Acids Res. 2003;31:e54. doi: 10.1093/nar/gng054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer S, Vaught JD, Bock C, Gold L, Katilius E, Keeney TR, et al. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay. PLoS ONE. 2011;6:e26332. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Elizondo-Riojas MA, Li X, Lokesh GL, Somasunderam A, Thiviyanathan V, et al. X-aptamers: a bead-based selection method for random incorporation of druglike moieties onto next-generation aptamers for enhanced binding. Biochemistry. 2012;51:8321–8323. doi: 10.1021/bi300471d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P, Sullenger BA, White RR. Further characterization of the target of a potential aptamer biomarker for pancreatic cancer: cyclophilin B and its posttranslational modifications. Nucleic Acid Ther. 2013;23:435–442. doi: 10.1089/nat.2013.0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefah K, Shangguan D, Xiong X, O'Donoghue MB, Tan W. Development of DNA aptamers using Cell-SELEX. Nat Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- Shigdar S, Qiao L, Zhou SF, Xiang D, Wang T, Li Y, et al. RNA aptamers targeting cancer stem cell marker CD133. Cancer Lett. 2013;330:84–95. doi: 10.1016/j.canlet.2012.11.032. [DOI] [PubMed] [Google Scholar]
- Cerchia L, Ducongé F, Pestourie C, Boulay J, Aissouni Y, Gombert K, et al. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol. 2005;3:e123. doi: 10.1371/journal.pbio.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hicke BJ, Marion C, Chang YF, Gould T, Lynott CK, Parma D, et al. Tenascin-C aptamers are generated using tumor cells and purified protein. J Biol Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- Wilner SE, Wengerter B, Maier K, de Lourdes Borba Magalhães M, Del Amo DS, Pai S, et al. An RNA alternative to human transferrin: a new tool for targeting human cells. Mol Ther Nucleic Acids. 2012;1:e21. doi: 10.1038/mtna.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan A, Levy M. Cell internalization SELEX: in vitro selection for molecules that internalize into cells. Methods Mol Biol. 2014;1103:241–265. doi: 10.1007/978-1-62703-730-3_18. [DOI] [PubMed] [Google Scholar]
- Gourronc FA, Rockey WM, Thiel WH, Giangrande PH, Klingelhutz AJ. Identification of RNA aptamers that internalize into HPV-16 E6/E7 transformed tonsillar epithelial cells. Virology. 2013;446:325–333. doi: 10.1016/j.virol.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Bobbin ML, Burnett JC, Rossi JJ. Current progress of RNA aptamer-based therapeutics. Front Genet. 2012;3:234. doi: 10.3389/fgene.2012.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel WH, Bair T, Peek AS, Liu X, Dassie J, Stockdale KR, et al. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS ONE. 2012;7:e43836. doi: 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camorani S, Esposito CL, Rienzo A, Catuogno S, Iaboni M, Condorelli G, et al. Inhibition of receptor signaling and of glioblastoma-derived tumor growth by a novel PDGFRß aptamer. Mol Ther. 2014;22:828–841. doi: 10.1038/mt.2013.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, et al. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012;40:6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer G, Ahmed MS, Dolf A, Endl E, Knolle PA, Famulok M. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat Protoc. 2010;5:1993–2004. doi: 10.1038/nprot.2010.163. [DOI] [PubMed] [Google Scholar]
- Avci-Adali M, Metzger M, Perle N, Ziemer G, Wendel HP. Pitfalls of cell-systematic evolution of ligands by exponential enrichment (SELEX): existing dead cells during in vitro selection anticipate the enrichment of specific aptamers. Oligonucleotides. 2010;20:317–323. doi: 10.1089/oli.2010.0253. [DOI] [PubMed] [Google Scholar]
- Dua P, Kang HS, Hong SM, Tsao MS, Kim S, Lee DK. Alkaline phosphatase ALPPL-2 is a novel pancreatic carcinoma-associated protein. Cancer Res. 2013;73:1934–1945. doi: 10.1158/0008-5472.CAN-12-3682. [DOI] [PubMed] [Google Scholar]
- Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt's lymphoma cells. Mol Cell Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, et al. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josic D, Clifton JG, Kovac S, Hixson DC. Membrane proteins as diagnostic biomarkers and targets for new therapies. Curr Opin Mol Ther. 2008;10:116–123. [PubMed] [Google Scholar]
- Yildirim MA, Goh KI, Cusick ME, Barabási AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- Esposito CL, Passaro D, Longobardo I, Condorelli G, Marotta P, Affuso A, et al. A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death. PLoS ONE. 2011;6:e24071. doi: 10.1371/journal.pone.0024071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, Medley CD, Sefah K, Shangguan D, Tang Z, Meng L, et al. Molecular recognition of small-cell lung cancer cells using aptamers. ChemMedChem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Xu L, Shi X, Tan W, Fang X, Shangguan D. Recognition of subtype non-small cell lung cancer by DNA aptamers selected from living cells. Analyst. 2009;134:1808–1814. doi: 10.1039/b904476k. [DOI] [PubMed] [Google Scholar]
- Wang FB, Rong Y, Fang M, Yuan JP, Peng CW, Liu SP, et al. Recognition and capture of metastatic hepatocellular carcinoma cells using aptamer-conjugated quantum dots and magnetic particles. Biomaterials. 2013;34:3816–3827. doi: 10.1016/j.biomaterials.2013.02.018. [DOI] [PubMed] [Google Scholar]
- Kim MY, Jeong S. In vitro selection of RNA aptamer and specific targeting of ErbB2 in breast cancer cells. Nucleic Acid Ther. 2011;21:173–178. doi: 10.1089/nat.2011.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Sefah K, Tang L, Zhao Z, Zhu G, Ye M, et al. A novel aptamer developed for breast cancer cell internalization. ChemMedChem. 2012;7:79–84. doi: 10.1002/cmdc.201100457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Simaeys D, López-Colón D, Sefah K, Sutphen R, Jimenez E, Tan W. Study of the molecular recognition of aptamers selected through ovarian cancer cell-SELEX. PLoS ONE. 2010;5:e13770. doi: 10.1371/journal.pone.0013770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchia L, Esposito CL, Jacobs AH, Tavitian B, de Franciscis V. Differential SELEX in human glioma cell lines. PLoS ONE. 2009;4:e7971. doi: 10.1371/journal.pone.0007971. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kang D, Wang J, Zhang W, Song Y, Li X, Zou Y, et al. Selection of DNA aptamers against glioblastoma cells with high affinity and specificity. PLoS ONE. 2012;7:e42731. doi: 10.1371/journal.pone.0042731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefah K, Meng L, Lopez-Colon D, Jimenez E, Liu C, Tan W. DNA aptamers as molecular probes for colorectal cancer study. PLoS ONE. 2010;5:e14269. doi: 10.1371/journal.pone.0014269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefah K, Bae KM, Phillips JA, Siemann DW, Su Z, McClellan S, et al. Cell-based selection provides novel molecular probes for cancer stem cells. Int J Cancer. 2013;132:2578–2588. doi: 10.1002/ijc.27936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian N, Raghunathan V, Kanwar JR, Kanwar RK, Elchuri SV, Khetan V, et al. Target-specific delivery of doxorubicin to retinoblastoma using epithelial cell adhesion molecule aptamer. Mol Vis. 2012;18:2783–2795. [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Duan JH, Song YM, Ma J, Wang FD, Lu X, et al. Novel HER2 aptamer selectively delivers cytotoxic drug to HER2-positive breast cancer cells in vitro. J Transl Med. 2012;10:148. doi: 10.1186/1479-5876-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Duan J, Zhan Q, Wang F, Lu X, Yang X-D. Novel MUC1 aptamer selectively delivers cytotoxic agent to cancer cells in vitro. PloS One. 2012;7:e31970. doi: 10.1371/journal.pone.0031970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagalkot V, Farokhzad OC, Langer R, Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed Engl. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- Huang YF, Shangguan D, Liu H, Phillips JA, Zhang X, Chen Y, et al. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. Chembiochem. 2009;10:862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinely NL, Plumb JA, Wheate NJ. DNA-based aptamer fails as a simultaneous cancer targeting agent and drug delivery vehicle for a phenanthroline-based platinum(II) complex. J Inorg Biochem. 2013;128:124–130. doi: 10.1016/j.jinorgbio.2013.07.021. [DOI] [PubMed] [Google Scholar]
- Boyacioglu O, Stuart CH, Kulik G, Gmeiner WH. Dimeric DNA Aptamer Complexes for High-capacity-targeted Drug Delivery Using pH-sensitive Covalent Linkages. Mol Ther Nucleic Acids. 2013;2:e107. doi: 10.1038/mtna.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Neoh KG, Kang ET, Choe WS, Su X. PEGylated anti-MUC1 aptamer-doxorubicin complex for targeted drug delivery to MCF7 breast cancer cells. Macromol Biosci. 2011;11:1331–1335. doi: 10.1002/mabi.201100173. [DOI] [PubMed] [Google Scholar]
- Taghdisi SM, Danesh NM, Sarreshtehdar Emrani A, Tabrizian K, Zandkarimi M, Ramezani M, et al. Targeted delivery of Epirubicin to cancer cells by PEGylated A10 aptamer. J Drug Target. 2013;21:739–744. doi: 10.3109/1061186X.2013.812095. [DOI] [PubMed] [Google Scholar]
- Zhu G, Zheng J, Song E, Donovan M, Zhang K, Liu C, et al. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc Natl Acad Sci USA. 2013;110:7998–8003. doi: 10.1073/pnas.1220817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Gee Neoh K, Kang ET, Choe WS, Su X. Designer tridentate mucin 1 aptamer for targeted drug delivery. J Pharm Sci. 2012;101:1672–1677. doi: 10.1002/jps.23101. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ali MM, Eckert MA, Kang DK, Chen YY, Sender LS, et al. A polyvalent aptamer system for targeted drug delivery. Biomaterials. 2013;34:9728–9735. doi: 10.1016/j.biomaterials.2013.08.079. [DOI] [PubMed] [Google Scholar]
- Ray P, Cheek MA, Sharaf ML, Li N, Ellington AD, Sullenger BA, et al. Aptamer-mediated delivery of chemotherapy to pancreatic cancer cells. Nucleic Acid Ther. 2012;22:295–305. doi: 10.1089/nat.2012.0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira CS, Cheung MC, Missailidis S, Bisland S, Gariépy J. Phototoxic aptamers selectively enter and kill epithelial cancer cells. Nucleic Acids Res. 2009;37:866–876. doi: 10.1093/nar/gkn967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh YA, Yang SJ, Wei MF, Shieh MJ. Aptamer-based tumor-targeted drug delivery for photodynamic therapy. ACS Nano. 2010;4:1433–1442. doi: 10.1021/nn901374b. [DOI] [PubMed] [Google Scholar]
- Liu Q, Xu L, Zhang X, Li N, Zheng J, Guan M, et al. Enhanced photodynamic efficiency of an aptamer-guided fullerene photosensitizer toward tumor cells. Chem Asian J. 2013;8:2370–2376. doi: 10.1002/asia.201300039. [DOI] [PubMed] [Google Scholar]
- Zhu G, Meng L, Ye M, Yang L, Sefah K, O'Donoghue MB, et al. Self-assembled aptamer-based drug carriers for bispecific cytotoxicity to cancer cells. Chem Asian J. 2012;7:1630–1636. doi: 10.1002/asia.201101060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K, Jo H, Song K, Cho M, Chun YS, Jon S, et al. Dual-aptamer-based delivery vehicle of doxorubicin to both PSMA (+) and PSMA (-) prostate cancers. Biomaterials. 2011;32:2124–2132. doi: 10.1016/j.biomaterials.2010.11.035. [DOI] [PubMed] [Google Scholar]
- Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res. 2004;64:7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci USA. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Hu Y, Duan J, Yuan W, Wang C, Xu H, et al. Novel aptamer-nanoparticle bioconjugates enhances delivery of anticancer drug to MUC1-positive cancer cells in vitro. PloS One. 6:e24077. doi: 10.1371/journal.pone.0024077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind A, Jeyamohan P, Nair R, Veeranarayanan S, Nagaoka Y, Yoshida Y, et al. AS1411 aptamer tagged PLGA-lecithin-PEG nanoparticles for tumor cell targeting and drug delivery. Biotechnol Bioeng. 2012;109:2920–2931. doi: 10.1002/bit.24558. [DOI] [PubMed] [Google Scholar]
- Guo J, Gao X, Su L, Xia H, Gu G, Pang Z, et al. Aptamer-functionalized PEG-PLGA nanoparticles for enhanced anti-glioma drug delivery. Biomaterials. 2011;32:8010–8020. doi: 10.1016/j.biomaterials.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Xing H, Tang L, Yang X, Hwang K, Wang W, Yin Q, et al. Selective Delivery of an Anticancer Drug with Aptamer-Functionalized Liposomes to Breast Cancer Cells in Vitro and in Vivo. J Mater Chem B Mater Biol Med. 2013;1:5288–5297. doi: 10.1039/C3TB20412J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc Natl Acad Sci USA. 2008;105:17356–17361. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC. Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo. Proc Natl Acad Sci USA. 2011;108:1850–1855. doi: 10.1073/pnas.1011379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sefah K, Liu H, Wang R, Tan W. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc Natl Acad Sci USA. 2010;107:5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Siddiqui IA, Nihal M, Pilla S, Rosenthal K, Mukhtar H, et al. Aptamer-conjugated and doxorubicin-loaded unimolecular micelles for targeted therapy of prostate cancer. Biomaterials. 2013;34:5244–5253. doi: 10.1016/j.biomaterials.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, You J, Zeng Z, Li C, Zu Y. An ultra pH-sensitive and aptamer-equipped nanoscale drug-delivery system for selective killing of tumor cells. Small. 2013;9:3477–3484. doi: 10.1002/smll.201202694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sefah K, Altman MB, Chen T, You M, Zhao Z, et al. Aptamer-conjugated nanorods for targeted photothermal therapy of prostate cancer stem cells. Chem Asian J. 2013;8:2417–2422. doi: 10.1002/asia.201300375. [DOI] [PubMed] [Google Scholar]
- Wang J, You M, Zhu G, Shukoor MI, Chen Z, Zhao Z, et al. Photosensitizer-gold nanorod composite for targeted multimodal therapy. Small. 2013;9:3678–3684. doi: 10.1002/smll.201202155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalalian SH, Taghdisi SM, Shahidi Hamedani N, Kalat SA, Lavaee P, Zandkarimi M, et al. Epirubicin loaded super paramagnetic iron oxide nanoparticle-aptamer bioconjugate for combined colon cancer therapy and imaging in vivo. Eur J Pharm Sci. 2013;50:191–197. doi: 10.1016/j.ejps.2013.06.015. [DOI] [PubMed] [Google Scholar]
- Tong GJ, Hsiao SC, Carrico ZM, Francis MB. Viral capsid DNA aptamer conjugates as multivalent cell-targeting vehicles. J Am Chem Soc. 2009;131:11174–11178. doi: 10.1021/ja903857f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen BA, Bergkvist M. Targeted in vitro photodynamic therapy via aptamer-labeled, porphyrin-loaded virus capsids. J Photochem Photobiol B, Biol. 2013;121:67–74. doi: 10.1016/j.jphotobiol.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Liu J, Liu H, Kang H, Donovan M, Zhu Z, Tan W. Aptamer-incorporated hydrogels for visual detection, controlled drug release, and targeted cancer therapy. Anal Bioanal Chem. 2012;402:187–194. doi: 10.1007/s00216-011-5414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhang X, Ye M, Jiang J, Yang R, Fu T, et al. Aptamer-conjugated nanomaterials and their applications. Adv Drug Deliv Rev. 2011;63:1361–1370. doi: 10.1016/j.addr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P, Viles KD, Soule EE, Woodruff RS. Application of aptamers for targeted therapeutics. Arch Immunol Ther Exp (Warsz) 2013;61:255–271. doi: 10.1007/s00005-013-0227-0. [DOI] [PubMed] [Google Scholar]
- McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- Neff CP, Zhou J, Remling L, Kuruvilla J, Zhang J, Li H, et al. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci Transl Med. 2011;3:66ra6. doi: 10.1126/scitranslmed.3001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R, et al. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009;37:3094–3109. doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Tiemann K, Chomchan P, Alluin J, Swiderski P, Burnett J, et al. Dual functional BAFF receptor aptamers inhibit ligand-induced proliferation and deliver siRNAs to NHL cells. Nucleic Acids Res. 2013;41:4266–4283. doi: 10.1093/nar/gkt125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullner U, Neef I, Eller A, Kleines M, Tur MK, Barth S. Cell-specific induction of apoptosis by rationally designed bivalent aptamer-siRNA transcripts silencing eukaryotic elongation factor 2. Curr Cancer Drug Targets. 2008;8:554–565. doi: 10.2174/156800908786241078. [DOI] [PubMed] [Google Scholar]
- Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Bagaria HG, Wong MS, Zu Y. A nanocomplex that is both tumor cell-selective and cancer gene-specific for anaplastic large cell lymphoma. J Nanobiotechnology. 2011;9:2. doi: 10.1186/1477-3155-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagalkot V, Gao X. siRNA-aptamer chimeras on nanoparticles: preserving targeting functionality for effective gene silencing. ACS Nano. 2011;5:8131–8139. doi: 10.1021/nn202772p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai F, Zhang Y, Zhu X, Shan N, Chen Y. Anticancer role of MUC1 aptamer-miR-29b chimera in epithelial ovarian carcinoma cells through regulation of PTEN methylation. Target Oncol. 2012;7:217–225. doi: 10.1007/s11523-012-0236-7. [DOI] [PubMed] [Google Scholar]
- Dai F, Zhang Y, Zhu X, Shan N, Chen Y. The anti-chemoresistant effect and mechanism of MUC1 aptamer-miR-29b chimera in ovarian cancer. Gynecol Oncol. 2013;131:451–459. doi: 10.1016/j.ygyno.2013.07.112. [DOI] [PubMed] [Google Scholar]
- Bruno JG, Carrillo MP, Crowell R. Preliminary development of DNA aptamer-Fc conjugate opsonins. J Biomed Mater Res A. 2009;90:1152–1161. doi: 10.1002/jbm.a.32182. [DOI] [PubMed] [Google Scholar]
- Stecker JR, Savage AA, Bruno JG, García DM, Koke JR. Dynamics and visualization of MCF7 adenocarcinoma cell death by aptamer-C1q-mediated membrane attack. Nucleic Acid Ther. 2012;22:275–282. doi: 10.1089/nat.2012.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boltz A, Piater B, Toleikis L, Guenther R, Kolmar H, Hock B. Bi-specific aptamers mediating tumor cell lysis. J Biol Chem. 2011;286:21896–21905. doi: 10.1074/jbc.M111.238261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Liu H, Zhao Z, Altman MB, Lopez-Colon D, Yang CJ, et al. DNA aptamer-mediated cell targeting. Angew Chem Int Ed Engl. 2013;52:1472–1476. doi: 10.1002/anie.201207063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlknecht G, Maron R, Mancini M, Schechter B, Sela M, Yarden Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc Natl Acad Sci USA. 2013;110:8170–8175. doi: 10.1073/pnas.1302594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M, Soo Oh S, Nie J, Stewart R, Eisenstein M, Chambers J, et al. Quantitative selection and parallel characterization of aptamers. Proc Natl Acad Sci USA. 2013;110:18460–18465. doi: 10.1073/pnas.1315866110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xu H, Ding H, Huang Y, Cao X, Yang G, et al. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J Pathol. 2009;218:327–336. doi: 10.1002/path.2543. [DOI] [PubMed] [Google Scholar]
- Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA, et al. In vivo selection of tumor-targeting RNA motifs. Nat Chem Biol. 2010;6:22–24. doi: 10.1038/nchembio.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liu H, Sefah K, Liu B, Pu Y, Van Simaeys D, et al. Selection of aptamers specific for adipose tissue. PloS One. 2012;7:e37789. doi: 10.1371/journal.pone.0037789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Chen YH, Lennox KA, Behlke MA, Davidson BL. In vivo SELEX for Identification of Brain-penetrating Aptamers. Mol Ther Nucleic Acids. 2013;2:e67. doi: 10.1038/mtna.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifer MG, Williams LD, Sobczyk MA, Michaels SJ, Sherman MR. Selectivity of binding of PEGs and PEG-like oligomers to anti-PEG antibodies induced by methoxyPEG-proteins. Mol Immunol. 2014;57:236–246. doi: 10.1016/j.molimm.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Kim E, Jung Y, Choi H, Yang J, Suh JS, Huh YM, et al. Prostate cancer cell death produced by the co-delivery of Bcl-xL shRNA and doxorubicin using an aptamer-conjugated polyplex. Biomaterials. 2010;31:4592–4599. doi: 10.1016/j.biomaterials.2010.02.030. [DOI] [PubMed] [Google Scholar]
- Cerchia L, Esposito CL, Camorani S, Rienzo A, Stasio L, Insabato L, et al. Targeting Axl with an high-affinity inhibitory aptamer. Mol Ther. 2012;20:2291–2303. doi: 10.1038/mt.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Han SR, Kim NY, Lee SH, Jeong JS, Lee SW. An RNA aptamer that binds carcinoembryonic antigen inhibits hepatic metastasis of colon cancer cells in mice. Gastroenterology. 2012;143:155–65.e8. doi: 10.1053/j.gastro.2012.03.039. [DOI] [PubMed] [Google Scholar]
- Pastor F, Soldevilla MM, Villanueva H, Kolonias D, Inoges S, de Cerio AL, et al. CD28 aptamers as powerful immune response modulators. Mol Ther Nucleic Acids. 2013;2:e98. doi: 10.1038/mtna.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Oguro A, Ohtsu T, Nakamura Y. RNA aptamers selected against the receptor activator of NF-kappaB acquire general affinity to proteins of the tumor necrosis factor receptor family. Nucleic Acids Res. 2004;32:6120–6128. doi: 10.1093/nar/gkh949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ababneh N, Alshaer W, Allozi O, Mahafzah A, El-Khateeb M, Hillaireau H, et al. In vitro selection of modified RNA aptamers against CD44 cancer stem cell marker. Nucleic Acid Ther. 2013;23:401–407. doi: 10.1089/nat.2013.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasunderam A, Thiviyanathan V, Tanaka T, Li X, Neerathilingam M, Lokesh GL, et al. Combinatorial selection of DNA thioaptamers targeted to the HA binding domain of human CD44. Biochemistry. 2010;49:9106–9112. doi: 10.1021/bi1009503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth F, De La Fuente AC, Vella JL, Zoso A, Inverardi L, Serafini P. Aptamer-mediated blockade of IL4Ra triggers apoptosis of MDSCs and limits tumor progression. Cancer Res. 2012;72:1373–1383. doi: 10.1158/0008-5472.CAN-11-2772. [DOI] [PubMed] [Google Scholar]
- Li N, Nguyen HH, Byrom M, Ellington AD. Inhibition of cell proliferation by an anti-EGFR aptamer. PLoS ONE. 2011;6:e20299. doi: 10.1371/journal.pone.0020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastjerdi K, Tabar GH, Dehghani H, Haghparast A. Generation of an enriched pool of DNA aptamers for an HER2-overexpressing cell line selected by Cell SELEX. Biotechnol Appl Biochem. 2011;58:226–230. doi: 10.1002/bab.36. [DOI] [PubMed] [Google Scholar]
- Chen CH, Chernis GA, Hoang VQ, Landgraf R. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc Natl Acad Sci USA. 2003;100:9226–9231. doi: 10.1073/pnas.1332660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann AP, Somasunderam A, Nieves-Alicea R, Li X, Hu A, Sood AK, et al. Identification of thioaptamer ligand against E-selectin: potential application for inflamed vasculature targeting. PLoS ONE. 2010;5:e13050. doi: 10.1371/journal.pone.0013050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann AP, Tanaka T, Somasunderam A, Liu X, Gorenstein DG, Ferrari M. E-selectin-targeted porous silicon particle for nanoparticle delivery to the bone marrow. Adv Mater Weinheim. 2011;23:H278–H282. doi: 10.1002/adma.201101541. [DOI] [PubMed] [Google Scholar]
- Mann AP, Bhavane RC, Somasunderam A, Liz Montalvo-Ortiz B, Ghaghada KB, Volk D, et al. Thioaptamer conjugated liposomes for tumor vasculature targeting. Oncotarget. 2011;2:298–304. doi: 10.18632/oncotarget.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Zhu Z, An Y, Zhang W, Zhang H, Liu D, et al. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal Chem. 2013;85:4141–4149. doi: 10.1021/ac400366b. [DOI] [PubMed] [Google Scholar]
- Shigdar S, Lin J, Yu Y, Pastuovic M, Wei M, Duan W. RNA aptamer against a cancer stem cell marker epithelial cell adhesion molecule. Cancer Sci. 2011;102:991–998. doi: 10.1111/j.1349-7006.2011.01897.x. [DOI] [PubMed] [Google Scholar]
- Waybrant B, Pearce TR, Wang P, Sreevatsan S, Kokkoli E. Development and characterization of an aptamer binding ligand of fractalkine using domain targeted SELEX. Chem Commun (Camb) 2012;48:10043–10045. doi: 10.1039/c2cc34217k. [DOI] [PubMed] [Google Scholar]
- Toscano-Garibay JD, Benítez-Hess ML, Alvarez-Salas LM. Isolation and characterization of an RNA aptamer for the HPV-16 E7 oncoprotein. Arch Med Res. 2011;42:88–96. doi: 10.1016/j.arcmed.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Mi J, Zhang X, Giangrande PH, McNamara JO, 2nd, Nimjee SM, Sarraf-Yazdi S, et al. Targeted inhibition of alphavbeta3 integrin with an RNA aptamer impairs endothelial cell growth and survival. Biochem Biophys Res Commun. 2005;338:956–963. doi: 10.1016/j.bbrc.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Da Rocha Gomes S, Miguel J, Azéma L, Eimer S, Ries C, Dausse E, et al. (99m)Tc-MAG3-aptamer for imaging human tumors associated with high level of matrix metalloprotease-9. Bioconjug Chem. 2012;23:2192–2200. doi: 10.1021/bc300146c. [DOI] [PubMed] [Google Scholar]
- Ferreira CS, Matthews CS, Missailidis S. DNA aptamers that bind to MUC1 tumour marker: design and characterization of MUC1-binding single-stranded DNA aptamers. Tumour Biol. 2006;27:289–301. doi: 10.1159/000096085. [DOI] [PubMed] [Google Scholar]
- Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62:4029–4033. [PubMed] [Google Scholar]