Abstract
The current antibody-mediated numeration assays of circulating tumor cells (CTCs) require multiple steps and are time-consuming. To overcome these technical limitations, a cancer cell-activatable aptamer-reporter was formulated by conjugating a biomarker-specific aptamer sequence with paired fluorochrome-quencher molecules. In contrast to the antibody probes, the intact aptamer-reporter was optically silent in the absence of cells of interest. However, when used in an assay, the aptamer selectively targeted cancer cells through interaction with a specific surface biomarker, which triggered internalization of the aptamer-reporter and, subsequently, into cell lysosomes. Rapid lysosomal degradation of the aptamer-reporter resulted in separation of the paired fluorochrome-quencher molecules. The released fluorochrome emitted bright fluorescent signals exclusively within the targeted cancer cells, with no background noise in the assay. Thus, the assays could be completed in a single step within minutes. By using this one-step assay, CTCs in whole blood and marrow aspirate samples of patients with lymphoma tumors were selectively highlighted and rapidly detected with no off-target signals from background blood cells. The development of the cancer cell-activatable aptamer-reporter system allows for the possibility of a simple and robust point-of-care test for CTC detection, which is currently unavailable.
Keywords: aptamer-reporter, cell-activatable, circulating tumor cell detection
Introduction
As a small molecule probe, aptamers are composed of short, single-stranded oligonucleotides (RNA or ssDNA) ranging from 30 to 60 bases.1,2,3,4 Similar to protein antibodies on the basis of their three-dimensional structures, aptamers specifically recognize and bind to their targets with high sensitivity and specificity.5,6,7,8,9 These targets can include small molecules, macromolecules, proteins, viruses, cells, and tissues.10,11,12,13,14 However, in contrast to antibodies, oligonucleotide aptamers are easily generated through chemical synthesis, and can be conveniently modified with a variety of functional molecules for different purposes. Therefore, aptamers have been widely studied for potential biomedical applications as a “chemical antibody”15,16,17 and, more importantly, for developing new clinical applications that cannot be performed with current protein antibody technology.18,19
Metastasis or dissemination of primary tumor is the major cause of mortality in cancer patients. Recent studies have demonstrated that circulating tumor cells (CTCs) in the bloodstream are key factors in the establishment of metastatic tumors.20,21,22 In addition, the number of CTCs present in whole blood is directly associated with cancer progression, recurrence, survival rate, and prognosis.23,24,25 Therefore, accurate detection of CTCs will provide critical information for the proper management of cancer patients. Antibody technology has been widely used for a variety of biomedical applications since it was first developed. The current CTC assays use antibodies as a specific probe to target tumor cells. These antibodies are preconjugated with fluorochrome as a reporter for imaging cell detection. However, the conjugated fluorochrome is constantly emitting a fluorescent signal, regardless of the status of whether the antibody is free or binding to cells, leading to a high level of background noise and potential off-target signals from normal blood cells. Therefore, the current antibody-mediated assays require multiple steps to isolate CTCs from normal blood cells and eliminate potential off-target signals. Repeated washes to remove excess free fluorochrome-conjugated antibody from the assays are also required. Notably, these multi-step assays are time-consuming and, more importantly, lead to loss of the blood sample and damage to CTCs, which adversely impacts assay accuracy. To overcome these technical obstacles in antibody-mediated assays, a new reporter system that can selectively highlight only tumor cells without generation of background noise is urgently needed.
Results
Design of the tumor cell-activatable aptamer-reporter
For point-of-care purposes, an “ideal” CTC assay should be (i) simple (one-step reaction), (ii) high throughput (complete in minutes), and (iii) accurate (no loss of blood sample and no damage of tumor cells through the assay process). However, no such assay could be developed using current antibody-mediated technology as described above. To this end, we designed a unique aptamer-reporter, which is composed of an oligonucleotide sequence specific for a tumor cell biomarker conjugated with a reporter of paired fluorochrome-quencher molecules. The underlining hypothesis is that the intact aptamer-reporter is optically silent in the absence of tumor cells (Figure 1a), but can be activated by tumor cells to emit fluorescence. As showed in Figure 1b, in assays containing patient blood the oligonucleotide sequence will selectively target CTCs through interaction with a specific surface biomarker, which triggers internalization of the aptamer-reporter through a natural cellular process and, subsequently, into cell lysosomes that are rich in different enzymes. This internalization mechanism has been successfully used for intracellular delivery of chemotherapeutic drugs for targeted therapy by a newly FDA-approved antibody-drug-conjugate, brentuximab.26 Rapid degradation of the aptamer-reporter by lysosomal nucleases results in separation of the paired fluorochrome-quencher molecules, the released fluorochrome becoming optically active and emitting bright fluorescent signals exclusively within the targeted tumor cells. Notably, because it is controlled by a specific aptamer-biomarker interaction and an intracellular activation switch, the aptamer-reporter will generate no background noise or off-target signals from normal blood cells and thus, the assay can be carried out in one-step, load-then-read (Figure 1c).
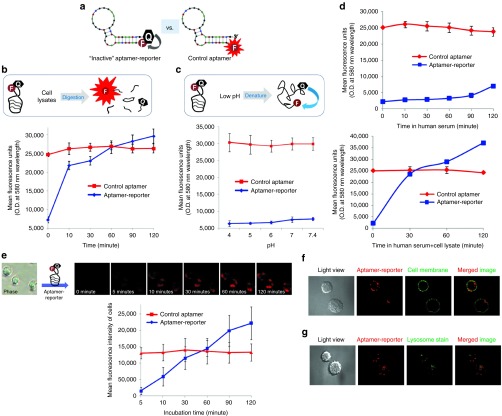
Figure 1.
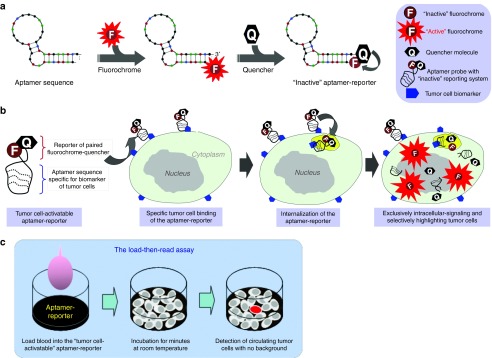
Schema of the tumor cell-activatable aptamer-reporter for one-step assay of circulating tumor cells (CTCs) in a whole blood sample. (a) A biomarker-specific and tumor cell-activatable aptamer-reporter, which is composed of two functional units: an oligonucleotide aptamer sequence specific for the biomarker of interest, and a tumor cell-activatable reporter composed of paired fluorochrome-quencher molecules at the 5′- and 3′-ends of the same sequence, respectively. Due to covalent chemical conjugation, the intact aptamer is able to sufficiently hold the quencher molecule close to the paired fluorochrome, rendering the fluorochrome optically “inactive” in the absence of tumor cells. (b) Tumor cell-triggered intracellular activation of the aptamer-reporter. In assays containing tumor cells, the aptamer-reporter will selectively target and specifically bind to surface biomarkers, resulting in internalization of the aptamer-reporter into the targeted tumor cells and subsequently into cell lysosomes via a natural cellular process. The degradation of the aptamer-reporter by lysosomal nucleases will lead to separation of the paired fluorochrome-quencher molecules and release of the fluorochrome, emitting bright fluorescent signals exclusively within the tumor cells. (c) Proposed one-step assay for rapid detection of CTCs. Simply load patient whole blood into the assay that contains the optically silent aptamer-reporter, and CTCs are selectively highlighted and then detected within minutes.
Tumor cell-activation and intracellular signaling of the aptamer-reporter
For the initial proof-of-concept studies, the aptamer-reporter was formulated by conjugating a 39-mer RNA sequence specific for the CD30 biomarker27,28,29 with a fluorochrome Cy3 compound and a BHQ2 at the 5′- and 3′-ends, respectively (Figure 1a and Figure 2a, left). Due to covalent chemical conjugation, aptamer sequence is able to hold fluorochrome and quencher molecules close to each other and, thus, the BHQ2 molecule will quench fluorescent signals emitted from the Cy3 compound on the same aptamer. A control aptamer was also synthesized by conjugating the same sequence with the fluorochrome Cy3 compound alone (Figure 2a, right).
Figure 2.
Tumor cell activation and intracellular signaling of the aptamer-reporter. (a) As a demonstration model, a tumor cell-activatable aptamer-reporter was formulated by conjugating a CD30 biomarker-specific aptamer sequence with the fluorochrome Cy3 compound and quencher molecule BHQ2 at the 5′- and 3′-ends, respectively (left). A control aptamer was also synthesized by conjugating the same aptamer sequence with fluorochrome Cy3 compound alone (right). (b) To test activation potential, the formulated aptamer-reporter was incubated with fresh lysates of Karpas 299 tumor cells as a cellular enzyme source. Resultant changes of fluorescent signals in reactions containing the aptamer-reporter or the control aptamer were kinetically quantified and compared in graph. (c) To rule out structure change effect, the aptamer-reporter and the control aptamer were treated in different pH conditions from pH 4.0 to pH 7.4, and changes in fluorescent signals were kinetically monitored and graphed. (d) To test biostability, the aptamer-reporter and control aptamer were incubated in human serum alone or in the presence of fresh cell lysates. The changes in fluorescence were kinetically monitored and graphed. (e) To study tumor cell-induced activation, the aptamer-reporter was incubated with fresh culture of Karpas 299 cells (a human anaplastic large cell lymphoma expressing surface CD30 biomarkers) at room temperature, and cells were directly examined by fluorescent microscope at different time points as indicated. Quantified fluorescent signals of cells were showed in graph. (f) To confirm intracellular signaling, cells were treated with the aptamer-reporter and cell membranes were then stained with Alexa 488. Fluorescent signals derived from the aptamer-reporters (red) and cell membranes (green) were detected by confocal microscope. (g) Similarly, cells treated with the aptamer-reporter and cell lysosomes were then stained with Lyso-ID. Co-location of fluorescent signals emitted from the aptamer-reporters (red) and stained lysosomes (green) were detected by confocal microscope.
First, to validate the optical-activatable property, the aptamer-reporter was incubated with fresh lysates of cultured Karpas 299 tumor cells of human ALCL at room temperature as a cellular enzyme source. Changes in fluorescence were kinetically quantified. Incubation with cell lysates activated the aptamer-reporter and triggered a rapid increase in florescent signals in a time-dependent fashion (Figure 2b). In contrast, cell lysates had no effect on the control aptamer which constantly emitted high levels of fluorescent signals. To rule out a potential effect due to conformation change, the aptamer-reporter was treated under different pH conditions, from pH 4.0 to physiological pH 7.4, at room temperature for 30 minutes (Figure 2c). Notably, cell lysosomes have a low pH environment (pH 4–5) which can cause denaturing of the aptamer sequence. Change in pH had no effect on fluorescent emission of the aptamer-reporter or the control aptamer. These findings indicated that the formed aptamer-reporter can be optically activated by cell lysates, but not affected by the low pH-induced structure change. To further strengthen these observations, the control and aptamer-reporter were also incubated in human serum alone or in serum containing fresh cell lysates under. Kinetic analysis of fluorescence revealed that the aptamer-reporter was stable up to 90 minutes in human serum with minimal background, and was rapidly activated by the addition of cell lysates (Figure 2d).
To study tumor cell-induced activation, the aptamer-reporter was simply added into a culture of Karpas 299 cells, which express high levels of the CD30 biomarker on the cell surfaces. Cells were then directly examined under fluorescent microscope. As shown in Figure 2e (upper panel), the aptamer-reporter was rapidly optically activated by the tumor cells and, thus, highlighted the tumor cells. The fluorescence was initially observed at 10 minutes postincubation and lasted up to 120 minutes, with no background noise. Quantified fluorescent signals of tumor cells at different time points were calculated and summarized in graph in Figure 2e.
Subsequently, to validate intracellular optical activation, cells were treated with the aptamer-reporter at room temperature for 30 minutes and cell membranes were then stained with the green fluorescent dye, Alexa 488. Cells were directly examined by a confocal fluorescent microscope (Figure 2f). Fluorescent signals derived from the aptamer-reporters (red) and prestained cell membranes (green) were recorded separately. Merged images confirmed that the aptamer-reporter emitted fluorescent signals exclusively confined within the targeted cells. Moreover, to detect intracellular distribution, the cells were treated with the aptamer-reporter and cell lysosomes were then stained with the fluorescent dye, Lyso-ID. Merging of confocal microscopic images revealed the colocation of fluorescent signals from the aptamer-reporter (red) and prestained lysosomes (green) within the same cells (Figure 2g). These findings confirmed that the aptamer-reporter could be optically activated and emit fluorescent signals exclusively within the targeted cells.
One-step assay of CTCs in whole blood sample
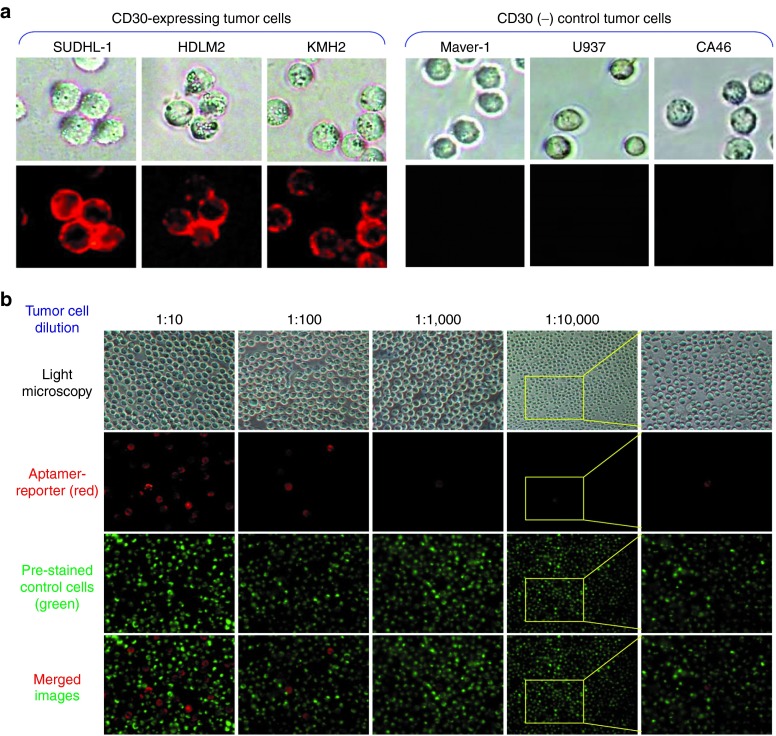
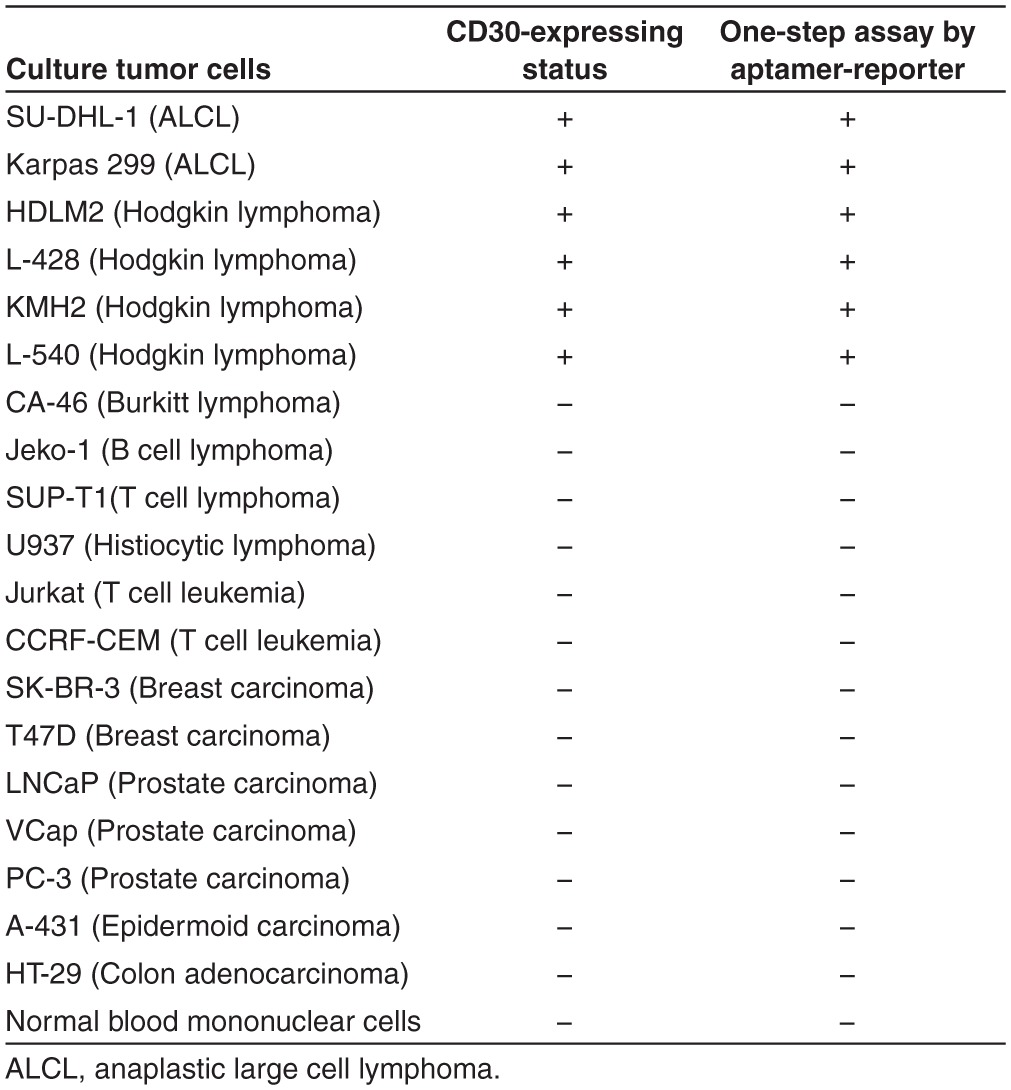
To validate assay specificity, multiple cultured cells were incubated with the aptamer-reporter at room temperature for 30 minutes, and directly examined under fluorescent microscope. As shown in Figure 3a, the aptamer-reporter was activated by the tumor cells that express the CD30 biomarker and, thus, highlighted the tumor cells of SUDHL-1, HDLM2, and KMH2 cells, but remained optically silent in control cultures of Maver-1, U937, and CA46 cells that do not express CD30, with no background noise. Additional studies with carcinoma tumor cells and normal peripheral cells are summarized in Table 1. To assess sensitivity, a cell mixture was made by diluting Karpas 299 tumor cells at ratios from 1/10 to 1/10,000 in U937 control cells, which were prestained with green fluorescence (Figure 3b). The cell mixtures were incubated with the aptamer-reporter, and fluorescent signals derived from the aptamer-reporters (red) and prestained control cells (green) were directly recorded. Merged images revealed that the aptamer-reporters were selectively activated and highlighted by CD30-expressing tumor cells, but did not react to control cells in the cell mixtures. Taken together, these findings indicate that the aptamer-reporter was both specific and sensitive for detection of tumor cells.
Figure 3.
Specific and selective highlighting of tumor cells by the aptamer-reporter. (a) To validate specificity, tumor cells with or without CD30-biomarker expression were used as indicated. The aptamer-reporter was selectively activated by and, thus, highlighted CD30-expressing tumor cells (left panels) and did not react to tumor cells that were negative for CD30 (right panels). Additional validation data with different tumor cells are listed in Table 1. (b) For a sensitivity test, a cell mixture was made by diluting CD30-expressing Karpas 299 tumor cells in CD30-negative U937 cells at ratios from 1:10 to 1:10,000. For identification purposes, U937 cells were pre-stained with green fluorescence. Cell mixtures were incubated with the aptamer-reporter and examined under a fluorescent microscope. The merged images revealed that the aptamer-reporter was sensitive and could selectively highlight tumor cells of interest (red) among many background off-target control cells (green).
Table 1. Specificity of the aptamer-reporter mediated one-step assay.
For preclinical evaluation, peripheral blood and marrow aspirate were collected from patients with ALCL tumors under a protocol approved by the IRB for the Houston Methodist Research Institute. Diagnosis of ALCL was made by tissue biopsy, and CD30 expression on lymphoma cells was confirmed by immunohistochemical staining of tumor tissues with anti-CD30 antibody (Figure 4a). To detect CTCs of ALCL, whole blood samples were added into a 12-well plate (40 million total blood cells/well), which was preloaded with the aptamer-reporter (5 nmol/l final concentration). After incubation for 30 minutes at room temperature, plates were directly examined under a microscope. Blood cells were well preserved for morphologic identification, including intact RBC, white blood cells, and possibly rare tumor cells that could not be distinguished (Figure 4b). However, the CTCs in whole blood samples were selectively highlighted by the aptamer-reporter with bright fluorescence and without background noise or off-target signals from normal blood (Figure 4b, right panels). Similarly, patient marrow aspirate was also added into the assays, and CTCs were selectively highlighted by the aptamer-reporter as detected by fluorescent microscopy (Figure 4c).
Figure 4.
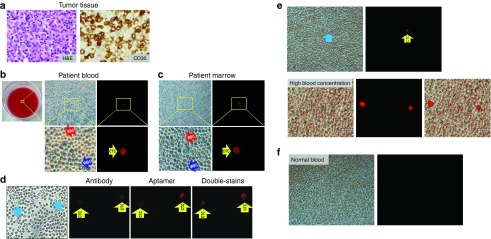
Specific detection of CTCs in patient samples by the aptamer-reporter-mediated load-then-read assay. (a) For clinical validation, diagnosis of an anaplastic large cell lymphoma (ALCL) tumor was made by histological examination (H&E stain) of biopsied tissues, and CD30 expression of tumor cells was confirmed by immunohistochemical stain with anti-CD30 antibody. (b) For CTC detection, whole blood samples were added into plate wells which were pre-loaded with the aptamer-reporter and incubated at room temperature for 30 minutes (left panel). Light microscopy revealed well preserved and intact red blood cells (RBC), white blood cells (WBC), and nucleated cells, but CTCs could not be distinguished by morphological examination alone (middle panels; upper: low magnification view, and lower: high magnification view). However, the CTCs in the whole blood sample were selectively highlighted by the aptamer-reporter and easily detected under fluorescent microscope due to a lack of background noise and off-target signals from normal blood cells (right panels; upper: low magnification view, and lower: high magnification view). (c) Similarly, the CTCs in the patient marrow aspirate sample were also selectively highlighted by the aptamer-reporter and detected under fluorescent microscope. (d) To confirm assay specificity, double-staining of marrow aspirate samples was conducted by initial probing with the FITC-conjugated anti-CD30 antibody, followed by treatment with the aptamer-reporter. Cellular fluorescent signals derived from antibody (green) and the aptamer-reporters (red) were recorded separately. Merged images revealed that CTCs were double-stained, confirming assay specificity. (e) CTCs were also detected in a blood sample from an additional patient with an ALCL tumor (upper panel). Furthermore, the assay was sensitive enough to detect fluorescent signals of the highlighted CTCs, which were overlapped by normal blood cells and not directly observed under light microscope due to the high concentration of cells (lower panel). (f) As a normal control, whole blood samples from healthy donors were also tested. No off-target signals or background fluorescence could be detected.
Currently, antibodies have been used as the “gold standard” probe for tumor cell detection in the clinical setting. Double-staining of CTCs were performed to confirm specificity of the aptamer-reporter. Patient marrow cells were initially probed by the FITC-conjugated anti-CD30 antibody. After removal of free antibody from the reaction, the cells were then treated with the aptamer-reporter and directly examined under a fluorescent microscope. Cell signals derived from antibody (green) and the aptamer-reporters (red) were recorded separately (Figure 4d). Merged images revealed that the CTCs in patient marrow were double-stained by both antibody and the aptamer-reporters, confirming specificity of the one-step assay. Notably, the aptamer and antibody did not compete for the same binding site(s) on the same tumor cells.23 Additional blood specimens from different patients with ALCL tumor were also analyzed, and CTCs were specifically highlighted and detected with a high signal-background noise ratio (Figure 4e, upper panel). To validate assay capacity, a high-concentration blood sample was loaded into a 12-well plate (120 million blood cells/well) to form multiple layers of cells (Figure 4e, lower panel). Interestingly, cellular fluorescent signals were easily detected, although the highlighted CTCs were overlapped by normal blood cells and could not be observed under a light microscope. As a normal control, whole blood samples from healthy donors were tested under the same assay and showed no off-target signals (Figure 4f).
Discussion
It is generally believed that there are only a scant number of detectable CTCs in patient blood, although the actual number ranges are not yet fully known.20,21,22 A recent study demonstrated that the average number of CTCs ranges from a mean of 79 to 196 cells per ml in whole blood specimens of patients with different types of cancers, which is much higher than the cut-off cell number (5 cells/ml) detected by current commercial tests.30 In addition, employing the Cell Search Profile Kit could detect more than 30-fold of the median CTC counts that are identified by the clinically used Cell Search Epithelial Kit in the same blood samples of patients with breast or lung cancers.31 These findings indicate the presence of much more CTCs in patient blood than previously thought, suggesting the possibility that, with a sensitive detection approach, the current standard numeration of CTCs could be achieved using smaller blood samples. Since our one-step assay will eliminate any potential loss of the blood sample and damage of tumor cells during the assay process, it will be able to detect the actual number of CTCs, and thus, require a much smaller blood sample. Notably, although the number of CTCs present in blood may vary among different types of tumors and different stages of disease, our assay scale can simply be justified by employing different-sized plates as clinically indicated with 96-, 24-, 12-, 6-, and single-well(s) to meet specific clinical needs. In addition, the load-then-read technology provides the possibility of performing high-throughput screening assays for rapid detection of CTCs. Moreover, good preservation of intact CTCs in this one-step assay will allow us to collect viable CTCs for further analysis of cellular protein and/or gene expression profiles at a single-cell level.
To eliminate background noise in whole-blood samples and enable intracellular highlighting of tumor cells, the one-step assay was designed by taking advantage of tumor cell internalization of the aptamer-reporter through a natural biological cell process, which is triggered by specific binding of aptamer sequences to surface CD30 receptors on tumor cells (Figure 1b). Interestingly, a similar internalization mechanism is employed for intracellular delivery of chemotherapeutic drug by an antibody-drug-conjugate, brentuximab, a newly FDA-approved, targeted therapeutic specific for CD30-expressing lymphomas.26 Thus, it is considerable that, in addition to the potential for diagnostic applications, aptamers can also be used as a chemical antibody for targeted therapy.32
In summary, our study introduced a unique tumor cell-activatable aptamer-reporter technology and demonstrated a simple, one-step assay for detection of CTCs in patient whole blood. Since this technology improves upon the existing antibody-driven methods, we believe it holds a high potential in early identification of patients who are at a higher risk for developing tumor metastases due to their elevated levels of systemic CTCs.
Materials and methods
Reagents and cells. In the proof of principal studies, a 39-mer RNA-based aptamer sequence, previously shown to specifically bind CD30-expressing tumor cells, was used.27,28,29 A novel tumor cell-activatable aptamer-reporter was then chemically synthesized (Bio-Synthesis, Lewisville, TX) by conjugating a fluorochrome Cy3 together with a Black Hole Quencher 2 (BHQ2) molecule at the opposing ends of the aptamer sequence as follows: Cy3-5′-rGrArUrUrCrArUrArUrGrGrGrUrGrGrGrArUrCrGrGrGrArArGrGrGrCrUrArUrGrArArUrCrG-3′-BHQ2. A control aptamer probe with the same sequence was conjugated with a fluorochrome Cy3 compound at the 5′ end. Alexa Fluor 488 conjugates for cell membrane staining were purchased from Invitrogen (Grand Island, NY) and Lyso-ID Green Detection Kit for lysosome staining was purchased from Enzo Life Sciences (Farmingdale, NY). The FITC-conjugated anti-human CD30 antibody was purchased from BD Biosciences (Franklin Lakes, NJ).
Cancer cells lines, including human anaplastic large cell lymphoma (ALCL) cells (Karpas 299 and SUDHL-1 cell lines from Mark Raffeld at NIH), Hodgkin lymphoma cells (HDLM2 and KMH2 cell lines from Barbara Savoldo, Baylor College of Medicine, Houston, TX), B cell lymphoma cells (Mino and Maver-1 cell lines from ATCC), and leukemia cells (U937 cell line from ATCC) were cultured in RPMI 1640 medium supplemented with 10% FBS, 100 u/ml penicillin, and 100 µg/ml streptomycin at 37 °C under the atmosphere of 5% CO2 and ≥95% humidity.
Activation of the aptamer-reporter. Fresh tumor cell lysates were prepared to confirm the cell-activatable property of the formed aptamer-reporter. Briefly, cultured Karpas 299 cells were harvested, washed twice, and suspended in PBS (5 × 105/ml). Cells were frozen at −80 °C, thawed at room temperature twice, and centrifuged at 12,000g for 20 minutes at 4 °C. Finally, 100 µl supernatants of cell lysates were collected and added into wells of a 96-well black wall plate, which was preloaded with the aptamer-reporter (5 nmol/l final concentration). After incubation at room temperature, fluorescence signals of the assays were kinetically quantified by a Synergy H4 microplate reader (BioTek, Winooski, VT) at different time points, as described. Each condition was tested in triplicate and experiments were repeated more than three times with similar results. The mean value of fluorescence was calculated and shown as the mean ± SD. Similarly, the control aptamer, containing the same sequence and conjugated with the fluorochrome Cy3 compound alone, was tested under the same treatment conditions and change in fluorescent signals of the control group was kinetically monitored as described above. To rule out the effects of low pH conditions, which is seen in cell lysosomes and might result in denature of the aptamer sequence 3D-structure, the aptamer-reporter (5 nmol/l final concentration) was added into PBS with pHs 4.0, 5.0, 6.0, 7.0, and 7.4 in a 96-well black wall plate. After incubation at room temperature for 30 minutes, fluorescence of the assays was quantified by a Synergy H4 microplate reader. Each condition was tested in triplicate and experiments were repeated more than three times with similar results. The mean value of fluorescence was calculated and shown as the mean ± SD. Finally, the aptamer-reporter (5 nmol/l final concentration) were incubated in 100 µl human serum (Atlanta Biological, Lawrenceville, GA, USA) in a 96-well black wall plate and changes in fluorescence were quantified as described above. In control group, 100 µl of cell lysate supernatants were also added into the human serum reaction and fluorescence was monitored. Each condition was tested in triplicate and experiments were repeated more than three times with similar results. The mean value of fluorescence was calculated and shown as the mean ± SD.
One-step assay and tumor cell detection. To validate the tumor cell-induced activation, 100 µl of cultured Karpas 299 cells (5 × 105/ml) were preseeded in a 96-well black wall plate in PBS, and 5 µl of the aptamer-reporter was then added to each well at 5 nmol/l final concentration. Without any additional preparation steps, the plates were directly examined under an Olympus IX81 fluorescent microscope and cell fluorescent signals were kinetically imaged using a TRITC filter under 547 nm of peak excitation wavelength and 572 nm of peak emission wavelength. For control purposes, cells were also treated with the control aptamer that was conjugated with fluorochrome Cy3 compound alone under the same condition. Fluorescent images were recorded using the same exposure time (300 microseconds) at different time points post treatment as described in Figures. Meanwhile, cell morphology was confirmed by light microscopy images. Each experiment with triple wells was repeated at least three times with similar results. For quantification, intensity of fluorescent signals from individual tumor cells was analyzed by Image J software (NIH, Bethesda, MD). A total of 90 cells from triplet wells (30 cells/well) at each time point were randomly gated and recorded. To get the background value of fluorescent intensity, 10 random noncellular locations with an average area of cell sizes were gated and calculated similarly. The mean fluorescent intensity of tumor cells was derived after background signals were deducted, calculated with standard deviation, and shown in graph with ± SD error bars.
Cell staining. For cell membrane staining, cultured Karpas 299 cells were treated with the aptamer-reporter at room temperature for 30 minutes. After washing once with PBS, the cells were suspended in PBS at room temperature, stained with Concanavalin A conjugated to Alexa Fluor 488 (1 mg/ml) for 2 minutes, fixed with 4% formaldehyde for 10 minutes, washed with PBS, and suspended in PBS/glycerin (weight/weight ratio of 1:1). Finally, 15 µl of cell solution was loaded onto glass slides and examined by a confocal microscope to determine intracellular localization of the aptamer-reporter (FITC filter for Alexa Fluor 488 dye, TRITC filter for Cy3-labeled aptamer-reporters). For lysosomal detection, Karpas 299 cells were treated with the aptamer-reporter as described above. After washing once with PBS, cells were stained with Lyso-ID green dye (1:1,000 dilution) at room temperature for 1 hour, fixed with 4% formaldehyde for 10 minutes, washed with PBS, and then suspended in PBS/glycerin. Finally, 15 µl of cell solution was loaded onto glass slides and examined by a confocal microscope as described above to determined possible colocalization of aptamer-reporters with the lysosomes.
Specificity and sensitivity validation. CD30-expressing cells (Karpas 299, SUDHL-1, HDLM2, and KMH2) and CD30-negative control cells (Maver-1, U937, and CA46) were tested to confirm specificity of the aptamer-reporter. The cells were incubated with the aptamer-reporter for 30 minutes at room temperature and the fluorescence was detected as described above. Additional cell lines (breast, prostate, and colon carcinomas) and peripheral blood collected from healthy donors were also tested and are listed in Table 1.
To test sensitivity of the aptamer-reporter, cell mixtures were prepared from CD30-expressing Karpas 299 cells and CD30-negative U937 control cells. For tracking purposes, U937 cells were stained with Concanavalin A conjugated to Alexa Fluor 488 and Karpas 299 cells were added in 1:10 to 1:10,000 ratios, as indicated. Subsequently, cell mixtures were added into 96-well black wall plates (2 × 105 cells/well) that were preloaded with the aptamer-reporting system (5 nmol/l final concentration). After 30 minutes incubation at room temperature, cells were examined by a fluorescent microscope (FITC filter for Alexa Fluor 488 to detect U937 cells, TRITC filter for Cy3 derived from aptamer-reporters). Merged images demonstrated CD30-selective cell staining by aptamer-reporters.
Detection of CTCs in patient whole blood and marrow aspirate samples in a single-step reaction. Under an approved IRB protocol, peripheral blood and bone marrow samples were collected from patients (n = 6) with stage IV ALCL tumors diagnosed by a histological evaluation of H&E-stained tumor tissue and immunohistochemical demonstration of CD30 expression on tumor cells. Ten blood and marrow samples collected from healthy donors were used as a control group. Following cell counting, whole blood and marrow aspirate specimens were added into wells of a 12-well plate that contained the aptamer-reporter (5 nmol/l final concentration) for a total of 40 million blood or marrow cells/well. After incubation at room temperature for 30 minutes, single-step labeled CTCs were directly detected by a fluorescent microscope. For a morphological evaluation, CTC images were also obtained with a light microscope within the same view fields.
To confirm specificity of the aptamer-reporter, marrow cells were double-stained with FITC-conjugated anti-human CD30 antibody and the aptamer-reporter. Briefly, bone marrow cells were diluted in 1 ml PBS and incubated with FITC-conjugated anti-human CD30 antibody (BD Biosciences, San Jose, CA) at room temperature for 30 minutes. To eliminate unbound antibody and minimize damage to CTCs, the reaction supernatant containing some red blood cells (RBCs) was gently removed by a pipette. The remaining cells were then suspended in 200 µl PBS and added into well plates that were preloaded with the aptamer-reporter at a 5 nmol/l final concentration. After 30 minutes incubation at room temperature, plates were directly examined by a fluorescent microscope and cell fluorescence (green: antibody, red: aptamer-reporter) recorded separately. Merged images showed that CTCs in patient marrow were double-stained, confirming specificity of the aptamer-reporter.
To test the effect of blood concentration, a total of 120 million blood cells from patients diagnosed with ALCL were added into 12-well plates to form multiple cell layers. After 30 minutes incubation with the aptamer-reporter, plates were examined by light and fluorescent microscopes to detect CTC fluorescence as described above. Merged images indicated that the assay was able to detect CTCs in high-concentration blood samples, even when CTCs overlapped with normal blood cells and could not be observed under a light microscope. To validate these results, whole blood samples from healthy donors were also tested in the single-step assay, which revealed no false-positive or off-target staining in the normal blood cells.
Acknowledgments
This project was supported in part by NIH grants R01CA151955 and R33CA173382 to Y.Z.
References
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Bunka DH, Stockley PG. Aptamers come of age - at last. Nat Rev Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- Gold L, Janjic N, Jarvis T, Schneider D, Walker JJ, Wilcox SK, et al. 2012Aptamers and the RNA world, past and present Cold Spring Harb Perspect Biol 4doi: 10.1101/cshperspect.a003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Held DM, Kissel JD, Patterson JT, Nickens DG, Burke DH. HIV-1 inactivation by nucleic acid aptamers. Front Biosci. 2006;11:89–112. doi: 10.2741/1782. [DOI] [PubMed] [Google Scholar]
- Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- Blank M, Blind M. Aptamers as tools for target validation. Curr Opin Chem Biol. 2005;9:336–342. doi: 10.1016/j.cbpa.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Kang M, Peterson R, Feigon J. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell. 2009;33:784–790. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, et al. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- Farokhzad OC, Karp JM, Langer R. Nanoparticle-aptamer bioconjugates for cancer targeting. Expert Opin Drug Deliv. 2006;3:311–324. doi: 10.1517/17425247.3.3.311. [DOI] [PubMed] [Google Scholar]
- McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- Zheng D, Seferos DS, Giljohann DA, Patel PC, Mirkin CA. Aptamer nano-flares for molecular detection in living cells. Nano Lett. 2009;9:3258–3261. doi: 10.1021/nl901517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlknecht G, Maron R, Mancini M, Schechter B, Sela M, Yarden Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc Natl Acad Sci USA. 2013;110:8170–8175. doi: 10.1073/pnas.1302594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, He X, Wang K, Wu X, Ye X, Guo Q, et al. Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration. Proc Natl Acad Sci USA. 2011;108:3900–3905. doi: 10.1073/pnas.1016197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- Mallikaratchy PR, Ruggiero A, Gardner JR, Kuryavyi V, Maguire WF, Heaney ML, et al. A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic Acids Res. 2011;39:2458–2469. doi: 10.1093/nar/gkq996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel KW, Giangrande PH. Therapeutic applications of DNA and RNA aptamers. Oligonucleotides. 2009;19:209–222. doi: 10.1089/oli.2009.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L, Xu J, Tan X, Liu Z, Xu L, Peng R. Dual-aptamer modification generates a unique interface for highly sensitive and specific electrochemical detection of tumor cells. ACS Appl Mater Interfaces. 2014;6:7309–7315. doi: 10.1021/am5006783. [DOI] [PubMed] [Google Scholar]
- Song Y, Zhu Z, An Y, Zhang W, Zhang H, Liu D, et al. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal Chem. 2013;85:4141–4149. doi: 10.1021/ac400366b. [DOI] [PubMed] [Google Scholar]
- Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- Liu MC, Shields PG, Warren RD, Cohen P, Wilkinson M, Ottaviano YL, et al. Circulating tumor cells: a useful predictor of treatment efficacy in metastatic breast cancer. J Clin Oncol. 2009;27:5153–5159. doi: 10.1200/JCO.2008.20.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethdorf S, Fritsche H, Müller V, Rau T, Schindlbeck C, Rack B, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A'Hern R, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–2918. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- Miller MC, Doyle GV, Terstappen LW. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J Oncol. 2010;2010:617421. doi: 10.1155/2010/617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senter PD, Sievers EL. The discovery and development of brentuximab vedotin for use in relapsed Hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nat Biotechnol. 2012;30:631–637. doi: 10.1038/nbt.2289. [DOI] [PubMed] [Google Scholar]
- Mori T, Oguro A, Ohtsu T, Nakamura Y. RNA aptamers selected against the receptor activator of NF-kappaB acquire general affinity to proteins of the tumor necrosis factor receptor family. Nucleic Acids Res. 2004;32:6120–6128. doi: 10.1093/nar/gkh949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhao N, Zeng Z, Feng Y, Tung CH, Chang CC, et al. Using an RNA aptamer probe for flow cytometry detection of CD30-expressing lymphoma cells. Lab Invest. 2009;89:1423–1432. doi: 10.1038/labinvest.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Zhang P, Zhao N, Sheehan AM, Tung CH, Chang CC, et al. Using oligonucleotide aptamer probes for immunostaining of formalin-fixed and paraffin-embedded tissues. Mod Pathol. 2010;23:1553–1558. doi: 10.1038/modpathol.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores LM, Kindelberger DW, Ligon AH, Capelletti M, Fiorentino M, Loda M, et al. Improving the yield of circulating tumour cells facilitates molecular characterisation and recognition of discordant HER2 amplification in breast cancer. Br J Cancer. 2010;102:1495–1502. doi: 10.1038/sj.bjc.6605676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EW, Shima DT, Calias P, Cunningham ET, Jr, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]