Abstract
S-glutathionylation (Pr–SSG) is a specific post-translational modification of cysteine residues by the addition of glutathione. S-Glutathionylated proteins induced by oxidative or nitrosative stress play an essential role in understanding the pathogenesis of the aging and age-related disorder, such as Alzheimer’s disease (AD). The purpose of this research is to develop a novel and ultrasensitive method to accurately and rapidly quantify the Pr–SSG by using capillary gel electrophoresis with laser-induced fluorescence detection (CGE-LIF). The derivatization method is based on the specific reduction of protein-bound S-glutathionylation with glutaredoxin (Grx) and labeling with thiol-reactive fluorescent dye (Dylight 488 maleimide). The experiments were performed by coupling the derivatization method with CGE-LIF to study electrophoretic profiling in in vitro oxidative stress model–S-glutathionylated bovine serum albumin (BSA-SSG), oxidant-induced human colon adenocarcinoma (HT-29) cells, brain tissues, and whole blood samples from an AD transgenic (Tg) mouse model. The results showed almost an eightfold increase in S-glutathionyl abundance when subjecting HT-29 cells in an oxidant environment, resulting in Pr–SSG at 232±10.64 (average ±SD; n=3) nmol/mg. In the AD–Tg mouse model, an initial quantitative measurement demonstrated the extent of protein S-glutathionylation in three brain regions (hippocampus, cerebellum, and cerebrum), ranging from 1 to 10 nmol/mg. Additionally, we described our developed method to potentially serve as a highly desirable diagnostic tool for monitoring S-glutathionylated protein profile in minuscule amount of whole blood. The whole blood samples for S-glutathionyl expression of 5-month-old AD–Tg mice are quantified as 16.3 μmol/L (=7.2 nmol/mg protein). Altogether, this is a fast, easy, and accurate method, reaching the lowest limit of Pr–SSG detection at 1.8 attomole (amol) level, reported to date.
Keywords: S–glutathionylation, AD–Tg mice, Pr–SSG, Grx, Thiol quantification, CGE-LIF
Introduction
Glutathione (GSH) exists in both free and protein-bound (S-glutathionylated) forms. Free form of glutathione exists primarily in its reduced form (GSH), which can be converted to the oxidized glutathione disulfide (GSSG) during oxidative stress. The S-glutathionylation of protein (Pr–SSG), representing an important post-translational modification, plays an essential role in studying the etiology of the aging disorder, such as Alzheimer’s disease (AD). The pathway to generate Pr–SSG involves thiol/disulfide exchange between GSSG and thiol residues at Cys residues on proteins [1]. GSSG, when elevated, can promote this thiol/disulfide exchange and cause a greater expression of Pr–SSG [2]. Currently, the S-glutathionylation is investigated to elucidate pathological changes in AD because some proteins are sensitive to cysteine oxidation initiated by increased oxidative stress in AD brain [1, 2]. Using a redox proteomic approach, several proteins have been identified as specific targets of S-glutathionylation in AD [1, 2]. For instance, glyceraldehyde 3 phosphate dehydrogenase (GAPDH), a well-known enzyme in intermediary metabolism, plays a crucial role in AD [2]. These oxidatively modified proteins in the AD brain demonstrate impaired protein function, which resulted in altered cell function, apoptosis, and the development of amyloid plaques [1, 2]. Also, studies show that S-glutathionylation has potential to serve for redox regulation mechanism [3]. Currently, a great number of methods have been developed so far to quantify the free (non-protein bound) glutathione. To study GSH, proteins existing as the most abundant component in biological samples are removed by either acidification or ultrafiltration to eliminate interference [3, 4]. Additionally, prior to the chromatographic or electrophoretic analysis of GSH, determination of total GSH requires the reduction of all disulfide, which can be achieved enzymatically by glutathione reductase (GR) [5] or chemically with various reducing compounds such as dithiothreitol (DTT) [6, 7].
All these methods to study GSH, except electrochemical detection, depend on derivatization of glutathione to improve the detection limit either by spectrophotometric or fluorescence detections [4]. Various labeling agents for GSH have been developed for possible derivatization at sites including carboxylic, amino, and thiol functional groups [6]. The most commonly used labeling agent is Ellman’s reagent, 5,5-dithiobis-(2-nitrobenzoic)-acid (DTNB). After labeling thiol groups, it gives the absorbance of the colored product 5-thionitrobenzoate at 412 nm [8, 9]. Also, fluorophore such as o-phthaldialdehyde is widely employed to form fluorescent adduct of GSH [10, 11]. However, it has its limited use due to its poor specificity and significant interference by amino acids and other thiols [12]. Alternatively, with thiol group trapped by blocking reagent such as iodoacetamide (IAA), the free amino group of alkylated GSH and GSSG can be converted to N-(2,4-dinitrophenyl) derivatives upon reaction with 2,4-dinitrofluorobenzene and measured calorimetrically [13–15].
After derivatization, high-performance liquid chromatography and capillary electrophoresis are the most used separation techniques to determine GSH as an indicator of oxidative stress level in biological systems. Both methods can be coupled with different detection systems, such as UV–vis, fluorimetric or electrochemical, or mass spectrometry detector. With these various derivatization and detection methods, the limits of detection (LOD) for the GSH detection lie in a wide range from 65 fmol [16, 17] to 10 nmol [6, 18].
As stated above, many researchers applied GSH/GSSG redox ratio for measuring oxidative stress level in biological samples. However, the GSH/GSSG ratio is quite dynamic because high enzyme activity of GR can reduce GSSG to GSH [19]. Also, GSSG can be exported to maintain intracellular redox status due to its high potential [20]. Unlike GSSG, the protein-bound form (Pr–SSG) appears more stable and was also considered as a useful biomarker for oxidative stress [1]. However, there are little-developed approaches to facilitate quantifying low abundant Pr–SSG in a miniscule amount of biological sample.
To assess the Pr–SSG level, the free form of S-glutathionyl was removed to eliminate the interference, and the protein pellet obtained from acid precipitation was solubilized first and subsequently measured. The mostly widely used method to analyze Pr–SSG is Ellman’s reagent (DTNB) assay [8]. Also, an ELISA assay for detection of actin S-glutathionylation in vitro was reported using anti-glutathione antibody [21]. Both of these two methods cannot be applied for studying extreme low abundance of samples due to their detection limits.
In addition, imaging analysis has been widely used in localizing and visualizing Pr–SSG. In situ detection of stained S-glutathionylated proteins in murine lungs was reported using glutaredoxin (Grx)-catalyzed cysteine derivatization and confocal laser scanning microscopy [22]. A recent mass-spectrometry-based proteomics study also used a similar three-step derivatization procedure including blocking free thiols, Grx-3-catalyzed reduction of S-glutathiol moieteies, and N-ethylmaleimide (NEM)–biotin labeling followed with avidin affinity purification [23]. Both imaging and proteomic studies allow the localization and identification of Pr–SSG, respectively. However, they are qualitative or semi-quantitative, thus the concentrations of Pr–SSG adducts cannot be determined [19].
Therefore, to address the needs to accurately and sensitively quantify protein disulfides, our group has developed an ultrasensitive method to simply and rapidly quantify the Pr–SSG by using capillary gel electrophoresis with laser-induced fluorescence detection (CGE-LIF). The Pr–SSG derivatization procedure comprises of (1) blocking the native thiols with alkylating agent, NEM; (2) the deglutathionylation of disulfides (–SSG) into sulfhydryl groups (–SH); and (3) fluorescence labeling of the newly formed –SH group with Dylight 488 maleimide (excitation at 493 nm, emission at 528 nm). The indirectly fluorophore labeled S-glutathionyls were then monitored using CGE-LIF. This method proved to be sufficient to analyze nanogram amounts of proteins and sensitive to detect attomole levels of S-glutathiols in less than 15 min.
Three applications of monitoring Pr–SSG from in vitro S-glutathionylated bovine serum albumin (BSA), menadione (2-methyl-1, 4-naphtoquinone; MQ) treated/untreated cells, and an AD transgenic mouse model have been performed in this study. First, to optimize the derivatization procedure and validate the CGE-LIF analytical system, in vitro S-glutathionylated BSA was utilized. It was generated from BSA subjective to 1,1′-azobis (N,N′-dimethylformamide) (diamide) oxidation.
Secondly, to illustrate the potential of this analytical method to study Pr–SSG in biological samples, we studied menadione (MQ) induced oxidative protein S-glutathionylation damage in human colon adenocarcinoma cells (HT-29) [14]. The results showed a significant increase in S-glutathionyl damage in MQ-treated cells, resulting in Pr–SSG at 232±10.64 (average ±SD; n=3) nmol/mg. Thirdly, to understand the early and progressive cellular changes in Alzheimer’s disease development and progression, we also initiated investigating S-glutathionylation of proteins in the 5-month-old Alzheimer’s disease transgenic mouse model (B6.Cg-Tg), which carries Swedish amyloid precursor protein mutation (APP) and exon 9 deletion of the PSEN1 gene [24]. AD-Tg mouse at 5 month old is a critical-age stage because it is right before the amyloid plaque appearance that may be associated with protein S-glutathionylation. Additionally, we also displayed the potential application of this method in studying electrophoretic patterns in various brain anatomical regions, such as cerebrum, cerebellum, and hippocampus, with their S-glutathionyl levels ranging from 1 to 10 nmol/mg. We also explored the analysis of Pr–SSG level in whole blood and detected 16.3 μM which is equal to 7.2 nmol/mg protein in this initial application.
All these taken together, the reported method of attomole detection limit will offer a great tool for future diagnostic investigation of S-glutathionylation in various settings such as low amount of biopsy samples and single cells. We envision that this new tool will provide a wide venue for scientists to examine S-glutathionylation related biological questions, especially when sample amounts are limited.
Through the demonstration of examining electrophoretic profiles in biological samples including cells and animal studies, we envision that this method can further be used to (1) study and potentially classify the S-glutathionylation electrophoretic patterns; (2) monitor longitudinal changes in Pr–SSG profiles; and (3) further potentially serve as a prescreening platform for the biomarkers in the AD progression.
Experimental section
Materials and reagents
Sodium dodecyl sulfate (SDS), 2-[4-(2-hydroxyethyl) piperazin-1-yl] ethanesulfonic acid (HEPES), sodium phosphate dibasic, potassium phosphate monobasic, tris (hydroxymethyl)-aminomethane, tricine, ethylenediaminetetraacetic acid (EDTA), BSA, methanol, sodium hydroxide, hydrochloric acid, neocuproine, dextran from Leuconostoc mesenteroides (average molecular weight (Mw), 64,000–76,000), trichloroacetic acid (TCA) were all from Sigma. Amicon ultra-4 (Mw cutoff of 3,000 Da) spin column was purchased from Millipore. Identification tag was from Hasco Tag Company. APP and PS1 primers were from IDT. Master Mix was purchased from Idaho Technology Inc. Recombinant glutaredoxin (Grx-3) (C14S, C65Y) from Escherichia coli was obtained from IMCO. Glutathione reductase (GR) was from Roche. Nicotinamide adenine dinucleotide phosphate (NADPH), reduced glutathione (GSH), and glutathione disulfide (GSSG) were all from Calbiochem. Diamide was from Research Organics Inc. DTNB was from Biodynth. DL–DTT was from Fluka. IAA and Triton X-100 were obtained from Alfa Aesar. Ultra Trol dynamic pre-coat LN was from Target Discovery. NEM, Dylight 488 maleimide, dimethyl sulfoxide, immobilized tris-2-carboxyethyl phosphate disulfide reducing gel (TCEP), bicinchoninic acid (BCA) protein assay kit were all purchased from Thermo Scientific. HT-29 cells, as generous gifts, came from Dr. Tak Aw’s lab at LSUHSC in Shreveport, LA.
In vitro S-glutathionylation of BSA
BSA solution 5 mg/ml was firstly obtained by dissolving BSA in 50 mM Tris/1 mM EDTA with 0.1% SDS at pH 7.5 and subsequently mixed with 5 mM DTT at 37 °C for 1 h [25]. Because native BSA has few free thiol groups, DTT was used to reduce exposed disulfides from denatured BSA after treating with 0.1% SDS [26]. DTT was then removed by using spin columns before subjecting BSA to 2.5 mM diamide at 37 °C for 30 min to generate S-glutathionylation in vitro [25, 26]. After desalting using spin columns to remove diamide, final concentration of in vitro S-glutathionylated BSA was 5 mg/ml.
In vitro S-glutathionylated BSA disulfide determination
The S-glutathionyl level in stress model–S-glutathionylated bovine serum albumin (BSA-SSG) was determined using modified Ellman’s assay [8, 27]. After neutralizing the BSA-SSG to pH 8, the samples were split into two sets (40 μl of 5 mg/ml S-glutathionylated BSA each). Set I was pretreated with 40 μl immobilized TCEP gel to reduce all disulfides into thiols at 37 °C for 30 min with occasional shaking. Hence, set I contains total thiols (total Pr-SH = reduced Pr-SH + native Pr-SH), whereas set II contains only native Pr-SH. Both sets were incubated with 70 μl phosphate buffer (1.5 mM KH2PO4, 6.36 mM K2HPO4, 1.57 mM EDTA) containing 3.95 mM DTNB at room temperature for 15 min. Total Pr-SH and native Pr-SH were subsequently determined spectrometrically at 412 nm from sets I and II, respectively [28]. Hence, the concentration of S-glutathionyls in BSA-SSG (nanomole glutathiols per milligram of BSA) was quantified and determined as a half of the thiols difference between set II and set I.
Cell culture and MQ treatment
The human colon epithelial cell line HT-29 were grown in McCoy’s medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 units/ml) and incubated in 5% CO2 at 37 °C [29]. For the MQ-mediated oxidative stress treatment, cells were treated with 200 μM MQ for 30 min in serum-free DMEM (Dulbecco’s modified Eagle’s medium) [14]. After gentle rinsing, scrapping cells into cold phosphate-buffered saline (PBS) with a scraper and then centrifugation at 1,000 rpm, cell pellets were harvested and immediately frozen in liquid nitrogen. For cell lysis, cells were sonicated in 250 μL 1× PBS with 2% SDS for 20 min at 4 °C [23]. The same procedures were applied to untreated HT-29 cells.
Transgenic mice and genotyping
AD-Tg mice [B6.Cg-Tg (APPswe, PSEN1dE9) 85Dbo/J, stock no. 005864, Bar Harbor, ME] were purchased from Jackson Laboratory. Real-time polymerase chain reaction and melting curve analysis (HRM) have been used for genotyping of the AD–Tg mice. The primer sets for APP transgenic gene are (forward 5′-GAC TGA CCA CTC GAC CAG GTT CTG-3′, reverse 5′-CTT GTA AGT TGG ATT CTC ATA TCC G-3′) [24]. Primer sets for the PS1 mutant gene are (forward 5′-AAT AGA GAA CGG CAG GAG CA-3′, reverse 5′-GCC ATG AGG GCA CTA ATC AT-3′). DNA was extracted by immersing mouse tail snip in 25 mM NaOH/0.2 mM EDTA at 98 °C for 1 h. The extracted DNA samples were mixed with 40 mM Tris–HCl (pH 5.5) and centrifuged at 4,000×g for 3 min. The sequence of APP gene was amplified by quantitative PCR and then analyzed by melting curve analysis using LC 32 Scanner (Idaho Technology, UT). The melting temperature of App gene is 86±0.6 °C.
Protein preparations from AD–Tg mice whole blood and brain
In this initial study, whole blood samples and brain tissue were isolated from one pair of 5-month AD–Tg/wild-type (WT) mice. The mice were anesthetized using sodium pentobarbital (60 mg/kg) through intraperitoneal injection and perfused transcardially with ice-cold saline. The brains were rapidly removed and dissected to harvest three brain regions including cerebrum, cerebellum, and hippocampus on ice. Samples were rinsed with ice-cold PBS and stored in −80 °C for further study. After homogenization of the brain tissue, proteins from each preparation was precipitated in 5% TCA at 4 °C for 15 min, and centrifuged at 10,000×g for 5 min. The protein pellets were kept and then dissolved in PBS solution with 5% SDS at pH 8 [15, 27]. All procedures for handling of mice were approved by the Institutional Animal Care and Use Committee.
S-glutathionylated protein derivatization and detection
This procedure comprises of blocking the native thiols using NEM, the deglutathionylation of disulfides into free sulfhydryl (–SH) group, followed with fluorescence labeling of the newly formed SH group by Dylight 488 maleimide (as shown in Fig. 1a). The derivatized samples were then separated using size-based CGE and detected using LIF. Deglutathionylation assay (depicted in Fig. 1b) based on the precise enzymatic recycling reaction using Grx-3 includes the following three steps [30, 31]: (1) specific reduction of Pr–SSG by Grx-3 to generate newly formed Pr-SH. (2) The tripeptide GSH from Grx-SSG intermediate is transferred to GSH to generate GSSG. (3) Glutathione reductase (GR) reduces GSSG with presence of NAPDH.
Fig. 1.
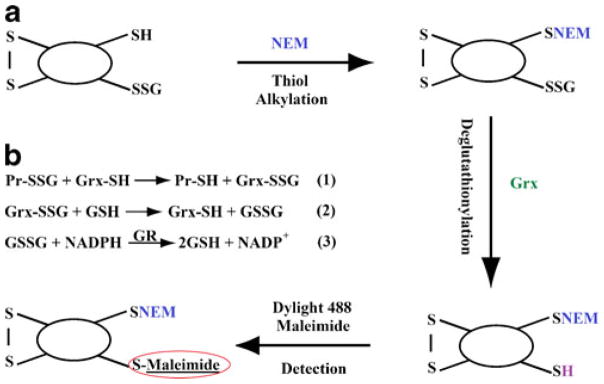
Mechanism of deglutathionylation and detection of Pr–SSG. a Schematic representation of the method to detect glutathionylated proteins by labeling with Dylight 488 maleimide. b Deglutathionylation assay is divided by three steps. (1) The Pr–SSG is reduced by Grx. (2) The Grx-SSG intermediate is transferred to GSH. (3) Glutathione reductase (GR) reduces GSSG with presence of NAPDH
Briefly, for alkylation of free thiol groups, 20 μl protein sample of each preparation (in vitro BSA-SSG, MQ induced HT-29 cells, brain tissues, and whole blood from AD mouse model) was incubated in 40 mM NEM and 25 mM HEPES, 0.2 mM EDTA, 0.01 mM neocuproine, and 2.5% SDS at pH 7.7 for 30 min at 4 °C [23, 25]. The excess NEM was removed by extensive wash (six times) with spin columns. The washing solvent was PBS at pH 7.4 and each wash involved centrifugation at 4,000×g for 20 min. For deglutathionylation, the blocked Pr–SSG sample was subsequently incubated with 2.5 μg/ml Grx-3, 4 U/ml GSSG reductase, 1 mM GSH, and 1 mM NADPH for 50 min at 37 °C [31]. The excess GSH and NADPH were removed by using spin columns again. Lastly, for the labeling of newly generated free thiols, 2 μl of 12.5 mM Dylight 488 maleimide was used and vortex-mixed overnight. The excess dye was later removed by spin columns with obtaining a final volume of the protein sample of 100 μl.
Validation of the developed method
Various controls were performed to validate the selectivity of S-glutathionyl labeling of the method. These controls included: (a) 1.25 μM Dylight 488 maleimide dye alone; (b) BSA-SSG without Grx-3 catalyzed deglutathionylation step; (c) BSA-SSG pretreated with 5 mM DTT to reduce the protein-bound disulfides; (d) native BSA without S-glutathionylation; and (e) MQ-treated HT-29 cells without Grx-3 reduction.
Additionally, we investigated (1) desalting efficiency of NEM and (2) NEM alkylating efficiency. Excess blocking reagent needs to be removed before deglutathionylation to avoid alkylation of freshly reduced disulfides, which could cause underestimation of Pr–SSG. On the other hand, incomplete NEM blocking of free thiols will result in false artifacts of Pr–SSG signals.
Protein concentration measurement
BCA assay kit was used to measure the protein concentration according to manufacturer’s protocol. The absorbance of the protein samples at 562 nm was determined by NanoDrop 2000/2000c Spectrophotometer.
CE instrument
CE with LIF has been considered as a highly efficient, admirably sensitive, and fast analytical separation technique for the analysis of low abundance of proteins. A commercial capillary electrophoresis instrument, Beckman Coulter P/ACE MDQ system (Fullerton, CA), was used for the analysis of protein S-glutathionylation. For excitation, the LIF detector used an argon-ion laser (488 nm line, 3 Mw) that was directed to a detector window in the capillary using a fiber optic [32]. A 520DF20 bandpass filter (~510–530 nm) was used select the Dylight 488 fluorescence before the photo-multiplier tube (PMT). The PMT output signals were sampled at 4 Hz. Separation was carried out using a 30.8-cm-long, 50-μm-ID, 361-μm-OD fused-silica capillary. UltraTrol LN, a class of linear polyacrylamide made of N-substituted acrylamide copolymers, was used for pre-coating the capillary walls to decrease the electroosmotic flow and inhibit protein binding to the capillary wall [33]. The sample was injected hydrodynamically at 11 kPa for 4 s into the capillary containing the sieving matrix buffer, which consisted of 15% dextran (64–76 kDa), 0.5% SDS, 20 mM Tris, and 20 mM Tricine. Separation was performed at −570 V/cm. The capillary was reconditioned between consecutive runs with sequential pressure-driven flushes (20 psi) of 0.5 M sodium hydroxide for 5 min, H2O for 3 min, the dynamic coating reagent UltraTrol LN for 2 min, the separation buffer for 2 min, and the sieving matrix buffer for 4 min. The separation buffer consisted of 20 mM Tris, 20 mM Tricine, and 0.5% SDS at pH 8.
Data analysis
Igor Pro software (Wavemetrics, Lake Oswego, OR) has been utilized to analyze data and electrophorograms, including migration time and peak area for each individual sample. To generate a calibration curve of S-glutathionyls, in vitro BSA-SSG was used for plotting S-glutathionyls amounts with respect to fluorescence peak area. Therefore, amounts of Pr–SSG from HT-29 cells and brain tissue and whole blood of AD–Tg/WT mice were calculated based on this curve.
Results and discussion
Grx-catalyzed deglutathionylation
Proteins were first heavily alkylated with NEM to block residual protein thiols and then subjected to deglutathionylation to generate newly formed free thiols. The ability to specifically identify and further reduce Pr–SSG is crucial for the selectivity of this derivatization method. Grx belongs to the thiol-disulfide oxidoreductase family [34]. Grx catalyzes the reversible reduction the Pr–SSG to free SH group through a monothiol mechanism relying solely on the N-terminal Cys 22 [30, 35, 36]. Cys 22 residue of Grx will become S-glutathionylated itself. The reduced form of Grx can then be regenerated using GSH coupled to GSSG reductase [36]. Since native Grx can recognize protein disulfides and reduce them also, it potentially produces false-positive artifacts [37]. So, we elected to utilize the mutant enzyme C14S, C65Y of gluaredoxin-3 (Grx-3), mainly due to its preference for the reduction of protein bound S-glutathiols [38]. The selectivity of this reductive treatment was also demonstrated in a Pr–SSG proteomic study [23]. It was noted by Mannervik that at least 0.5 mM GSH is required to ensure optimal activity of Grx [39]. We followed the optimal concentrations of all enzymes involved in deglutathionylation cycling [31].
Detection and validation of S-glutathionylation using CGE-LIF analysis
In the present work, we provide a novel approach for monitoring S-glutathionylated proteins based on the selective enzymatic recognition of Pr–SSG, fluorescent labeling, and quantification by CGE-LIF analysis. To validate the analytical system, we chose to analyze in vitro oxidized BSA.
As stated in the method section, various controls including (a) Dylight 488 maleimide dye alone; (b) BSA-SSG without Grx-3-catalyzed deglutathionylation step; (c) BSA-SSG pretreated with DTT to reduce the protein bound disulfides; (d) native BSA without S-glutathionylation were performed to validate the detection of S-glutathionylation using this method.
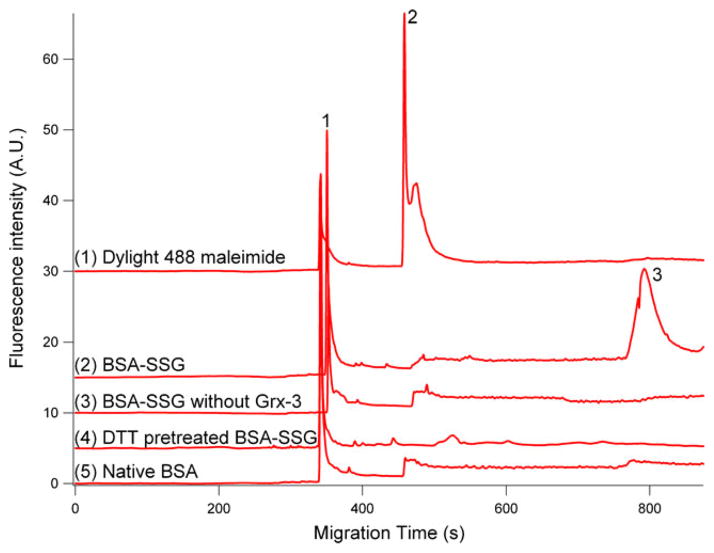
Firstly, we studied the CGE-LIF analysis of the fluorescent dye, Dylight 488 maleimide. This is to differentiate the peaks of dye itself from those of S-glutathionylated proteins and further identify how differently they distribute in terms of migration time. Trace 1 in Fig. 2 demonstrated that the Dylight 488 maleimide alone shows multiple peaks occurring between 347 and 573 s, whereas in vitro S-glutathionylated BSA (trace 2 in Fig. 2) exhibits a new additional peak appearing at 796 s indicating the detection of S-glutathiols. Altogether, it validated the reactivity of Dylight 488 maleimide toward the disulfide group in the treated BSA.
Fig. 2.
In vitro S-glutathionylated BSA (BSA-SSG) and validation of the method using controls. 1 Electropherograms of Dylight 488 maleimide; 2 BSA-SSG; 3 BSA-SSG without Grx reduction; 4 BSA-SSG pretreated with DTT; 5 native BSA. Hydrodynamic injection at pH 8 and 11 kPa for 4 s; separation in gel containing 15% dextran (64–76 kDa), 0.5% SDS, 20 mM Tris, and 20 mM Tricine at −570 V/cm. 1, 2 Indicate Dylight 488 maleimide. Top trace is offset in the Y axis for the clarity
Secondly, a control experiment was performed in the presence of all the derivatization steps except Grx deglutathionylation, and no peak at 796 s for the BSA-SSG was observed (Trace 3 in Fig. 2). Grx-mediated deglutathionylation is essential to enzymatically reduce S-glutathionyls and further generate newly generated free thiols for consecutive labeling. Thus, the omission of the deglutathionylation step and no S-glutathionyl detection in this negative control further validates the requirement of the enzymatic cycling for the labeling reaction.
Furthermore, the selectivity of S-glutathionyls labeling was also exploited in native BSA and BSA-SSG pretreated with reducing agent, DTT. After the DTT treatment, the protein-bound S-glutathionyls was reduced, and hence, the consecutive labeling of S-glutathionyls was abolished. In native BSA, no S-glutathiols are available for labeling. As we expected, in both controls, no peak around 796 s was observed in both traces 4 and 5 of Fig. 2. All these observations further indicate the selectivity of this method.
Dylight 488 fluorophore-tagged Pr–SSG will lead to an increase in molecular weight. The addition of NEM (124 Da), reduction of S-glutathionyls (−307 Da), and maleimide labeling (800 Da) will result in an increase of Mw of 617 Da. Proteomic study disclosed that the majority of identified S-glutathionylated proteins have Mw ranging from 25 to 70 kDa [23]. Therefore, the Mw shift of 617 Da is insignificant compared with the S-glutathionylated protein size itself, and this derivatization procedure has little effect on the protein mobility in the CGE separation.
Evaluation of NEM removal
After alkylating the free thiols, the NEM was removed using Amicon ultra-4 (Mw cutoff of 3,000 Da) spin columns. If the removal were not complete, the remaining NEM will compete with Dylight 488 maleimide by reacting with freshly reduced thiols from S-glutathionyls. This would cause the decreased or no signals for the detection of S-glutathionylated proteins. Therefore, we investigated whether the levels of NEM removal were associated with the numbers of ultracentrifugation wash. Electronic Supplementary Material Fig. S1 displays a peak at approximately 796 s representing BSA-SSG after six washes with desalting column, whereas no peak detected for S-glutathionyl after only three times wash. This observation indicates that, by doubling the desalting wash times, the excess NEM was completely removed, which eliminates its interference of binding reduced S-glutathionyls.
Assessment of blocking efficiency
The derivatization procedure only works when only S-glutathionylated cysteine residue will be derivatized and fluorescently labeled. If the free thiols were not completely blocked with NEM, then the remaining nonblocked cysteines would be labeled with Dylight 488 maleimide and produce a false artifact in the electropherogram. Hence, we studied the blocking efficiency using various concentrations (10, 30, and 40 mM) of two blocking reagents, NEM and IAA without deglutathionylation step. As seen in Electronic Supplementary Material Fig. S2, 40 mM of NEM was considered as the optimal concentration to completely block all the free thiols in BSA-SSG.
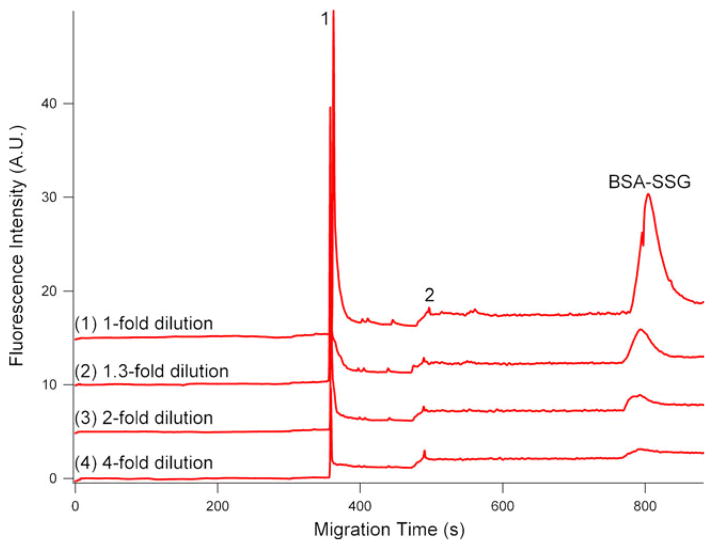
Calibration curve, assay reproducibility, and accuracy
The concentration of S-glutathionyl groups in S-glutathionylated BSA was determined by Ellman’s colorimetric assay, which was expressed as 92.41 nmol S-glutathionyl per 1 mg BSA protein. Therefore, the concentrations of a serial dilution (1-, 1.3-, 2-, and 4-fold dilutions) of BSA-SSG were calculated ranging from 92.41 to 23.10 nmol/mg protein. In Ellman’s assay, immobilized TCEP was utilized due to following reasons: (1) It allows recovering reduced BSA in high yield without dialyzing or desalting steps; (2) It eliminates the possibility of remaining TCEP reacting with Ellman’s reagent, DTNB; (3) It does not interfere with consecutive thiols-reactive reagent, i.e., maleimide cross-linkers. The calibration curve was generated from separately injecting a serial dilution of S-glutathionylated BSA in triplicate onto CE (Fig. 3). The integrated fluorescence peak area and corresponding S-glutathiol (in moles) are indicated as y and x in the calibration curve, respectively [40].
Fig. 3.
Electropherograms of a series of dilution (1-, 1.3-, 2-, and 4-fold) of BSA-SSG labeled with Dylight 488 maleimide. 1, 2: Dylight 488 maleimide. All the experimental conditions are the same as in Fig. 2. Top trace is offset in the Y axis for the clarity
| (1) |
LOD and limits of quantitation (LOQ) are defined as the amount for which the S/N is 3 and 10, respectively. In this CGE-LIF study, the LOD and LOQ of S-glutathionyl groups assessed by Eq. 1 were 1.8 and 6 amol, respectively, which are the lowest LOD and LOQ reported to date.
The reproducibility of the method was determined by performing three replicate injections of the in vitro S-glutathionylated BSA solution. The relative standard deviation (RSD) values for migration time and peak area were 2.0% and 0.5%, respectively, suggesting good reproducibility in the measurement of S-glutathionylation abundance in one sample. The precision of the method was determined by replicate injection of the test model—in vitro S-glutathionylated BSA on three consecutive days. The RSD values of migration time and peak area were 5.5% and 0.7%, respectively, for the day-to-day precision assay. The accuracy was determined by performing a standard spiking method, where known amounts of S-glutathionylated BSA (BSA-SSG) standard was mixed with previous analyzed BSA-SSG and further detected by CGE-LIF method in triplicate. The concentration of BSA-SSG was subsequently quantified by the calibration curve. The percentage of the spiked BSA-SSG analyte recovered by the assay indicates the accuracy of the method was 93.5%.
S-glutathionylation determination in MQ-mediated HT-29 cells
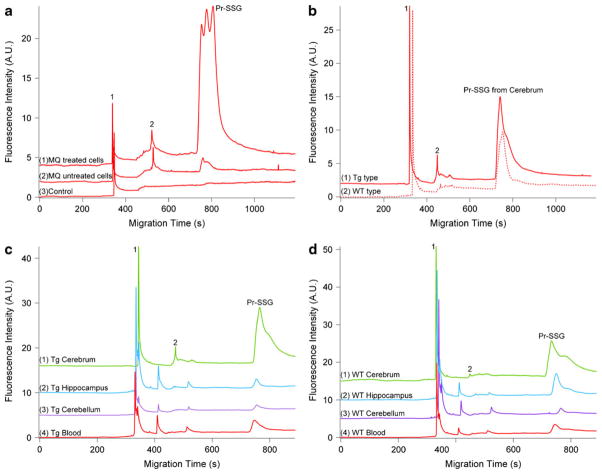
To illustrate the potential of this novel analytical method, we directly applied it to the analysis of MQ-induced S-glutathionylation. Fig. 4a (traces 1 and 2) demonstrates wide peaks (738–1,103 s), indicating a broad range of proteins susceptible to S-glutathionylation in this oxidative stress cell model. A negative control carried by eliminating the Grx-catalyzed deglutathionylation (shown in trace 3 of Fig. 4a) also highlighted the specificity of this derivatization method and the necessity of this reduction step.
Fig. 4.
Quantitative electrophoretic profiling of MQ-mediated glutathionylation in HT-29 cells and Pr–SSG of brain tissues and blood in AD–Tg/WT mice by using CGE-LIF. a Electropherograms of Pr–SSG from HT-29 cells labeled with Dylight 488 maleimide. Trace 1 demonstrates Pr–SSG from MQ-treated HT-29 cells. Traces 2 and 3 indicate MQ untreated HT-29 cell and MQ-treated HT-29 cells without deglutathionylation, respectively. b Electropherograms of Pr–SSG in cerebrum of AD–Tg/WT mice at 5 months. c Electropherograms of Pr–SSG from three anatomical positions (cerebrum, cerebellum, and hippocampus) in brain tissue and blood sample of AD–Tg mouse. d Electropherograms of Pr–SSG from three anatomic brain regions and blood of AD-WT mouse. 1, 2: Dylight 488 maleimide. All the experimental conditions are the same as in Fig. 2. Top trace is offset in the Y axis for the clarity
Based on the Eq. 1 and the measured peak area for the S-glutathionylated peaks, the total S-glutathiols content in the injected volume for MQ-treated and untreated HT-29 cells were calculated as (326±15) fmol (average ±SD; n=3) and (41±3.4) fmol, (average ±SD; n=3), respectively. For the CE analysis, only 1.1 nL volumes of samples were hydrodynamically injected, which leads to 1.41 and 1.40 ng of total proteins for the MQ-treated and untreated cells, respectively. Therefore, the treatment with MQ resulted in eightfold increase in protein-bound S-glutathiol contents (232±10.6 nmol/mg protein) compared with untreated cells (29.3±2.5 nmol/mg protein). MQ, a redox cycling quinine, can form the toxic species, the semi-quinone (SQ) radical, through the one-electron metabolism catalyzed by NADPH–ubiquinone oxidoreductase (ubQO). Also, MQ can be replenished by reaction between SQ and O2 with concomitant free radical generation of superoxide anion (O2−) [15]. Our results are in close agreement with other reported values for Pr–SSG in this MQ-mediated oxidative stress model [15]. In addition, the RSD was 4–7% indicative of reproducibility of LIF detection of Dylight 488-labeled S-glutathionyls in multiple CE injections.
Monitoring S-glutathionylated proteins in AD–Tg mice brain
Another direct biological application of this CGE-LIF method was demonstrated in characterizing the electrophoretic profiles in nanogram amounts of whole blood and brain tissues including hippocampus, cerebrum, and cerebellum from 5-month-old AD–Tg mice. The fluorescence of Dylight 488 maleimide-labeled S-glutathiols was detected in a migration window between 725 and 927 s in both Tg and WT mice (Fig. 4c, d). The quantitative amounts of Pr–SSG in different biological samples were calculated and listed in Table 1. To further improve separation of protein mixture based on size, an additional experiment for gel condition test was performed by detection of Pr–SSG in cerebrum of AD–Tg/WT mice at 5 month old using gel matrix containing 10% dextran (Mw, 64–76 KDa), 0.5% SDS, 20 mM Tris, and 20 mM Tricine. The only difference between original (15% dextran) and this new experiment (10% dextran) is gel percentage, where the same Mw dextran was used. The result is shown in Electronic Supplementary Material Fig. S3, which indicates gel percentages affect the Pr–SSG separation efficiency. To obtain further optimum resolution, gel concentration and 2D-CE will be investigated in future study.
Table 1.
S-glutathionyl contents in different brain tissues and blood of AD–Tg mice model
| Sample sources | AD–Tg mice | WT mice |
|---|---|---|
| Cerebrum | 10.38±4.51 nmol/mg | 11.99±4.29 nmol/mg |
| Cerebellum | 3.31±0.96 nmol/mg | 1.06±0.13 nmol/mg |
| Hippocampus | 1.36±0.11 nmol/mg | 3.37±0.58 nmol/mg |
| Whole blood | 16.3±2.25 μM | 27.4±2.96 μM |
The B6.Cg mice model is associated with early onset of AD so that it was selected for study of neurodegenerative alternation, S-glutathionylation, with the AD progression. Mice genotyping was accomplished with melting curve analysis, and the results are shown in Electronic Supplementary Material Fig. S4. Since the appearance of amyloid pathology and cognitive impairment are generated in transgenic mice at 6 and 7 month of age, it is intriguing for us to study 5-month-old mice pair and explore whether S-glutathionylation formation precedes the appearance of amyloid plaque. We demonstrated the feasibility of applying this newly developed CE-LIF method to detect and quantify S-glutathionylated proteins in whole blood and specific brain regions including hippocampus, cerebrum, and cerebellum from a 5-month-old AD mice pair (data shown in Table 1). Although no statistical evaluation regarding genotyping can be undertaken in this initial animal study, this method proves to be a valuable tool for future investigation into characterizing S-glutathionylation abundance and electrophoretic profile with AD progression. To further understand the cause of early-onset AD, a large-scale animal study including AD–Tg/WT mice pair at different age stages, such as 1, 5, and 11 month old, indicative of long before, immediately before, and long after amyloid plaque accumulation, is necessary for examining whether S-glutathionylation formation is an early event in the AD progression. Additionally, we envision that this method can potentially serve as a platform for studying disease etiology and serve as prescreening of AD if different S-glutathionylation patterns indeed occur with progression.
The quantitative data in Table 1 listed S-glutathionylated protein levels in specific brain regions. Previous studies reported that the levels of reduced GSH in 4-month-old mouse brain varied in different anatomical positions, with cortex > cerebellum > hippocampus [41, 42]. However, in another rat study, no brain region associated difference in S-glutathiol expression was observed [43]. The discrepancy might be due to the use of different species. Currently, our data is not conclusive to discuss whether S-glutathionylation level varies with brain regions with AD progression. However, this new method has explicitly displayed the potential to lead a complete study to answer the biological question, such as whether regional vulnerability of the AD brain tissue is reflected in differences in S-glutathionylated protein pattern.
The results demonstrated the extent of protein S-glutathionylation in whole blood, averaging 16.3 μM which is equal to 7.15 nmol/mg protein. The level of Pr–SSG in human blood was reported as 138 uM [1]. The discrepancy between the two values might be due to the differences of species [1]. Blood protein-bound glutathione concentration may reflect glutathione status in other less readily accessible tissues, such as brain biopsy, thus that of blood measurement has been considered as an index for of whole body oxidative stress status and serves as an important biomarker diagnostically [19]. Altogether, with Mw pattern distribution characterized from the CGE separation, we envision that the method can be further extended to investigate the S-glutathionyls pattern classification in blood samples from the evolvement of AD disease progression using chemometric tools such as principal component analysis. If difference in electrophoretic profiles exists, this method could potentially become a pre-screening tool for early detection of AD.
Our method of coupling Grx-catalyzed derivatization and labeling of protein-bound S-glutathiols to CGE-LIF detection has exhibited several great merits, including low LOD, fast analysis time, and potential for Mw pattern classification. It reached the limit of S-glutathiol detection of 1.8 amol, which is 109-fold less than the conventional Ellman’s assay. It facilitates the glutathione homeostasis study in minuscule biological samples, such as single cells or biopsy tissues. The CGE separation lasted less than 15 min, significantly faster than traditional SDS-PAGE analysis.
Conclusion
In conclusion, we have demonstrated here a Grx-catalyzed derivatization and labeling approach coupled with CGE-LIF analysis to accurately monitor the Pr–SSG levels in cells and tissues. This method allows detection of 1.8 amol amount of S-glutathionylated protein in nanogram amount of proteins. It reached the lowest LOD reported to date. This method provides a venue to better understand the regulation of Pr–SSG and to evaluate its potential role in oxidative stress-related disease, such as AD in future large-scale studies.
Acknowledgments
This study was supported by the National Institutes of Health and the National Center for Research Resources Grant P20RR016456 and NIH grant # DK44510. Special thanks go to Dr. Edgar Arriaga’s group for providing their house-written Wide Peak analysis software in our study. We also thank Dr. Spaulding for his great assistance on animal care.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00216-011-5311-x) contains supplementary material, which is available to authorized users.
Contributor Information
Cheng Zhang, Department of Biomedical Engineering, Louisiana Tech University, 818 Nelson Avenue, Ruston, LA 71272, USA.
Cynthia Rodriguez, Department of Molecular and Cellular Physiology, Louisiana State University Health Sciences Center, Shreveport, LA 71130-3932, USA.
Magdalena L. Circu, Department of Molecular and Cellular Physiology, Louisiana State University Health Sciences Center, Shreveport, LA 71130-3932, USA
Tak Yee Aw, Department of Molecular and Cellular Physiology, Louisiana State University Health Sciences Center, Shreveport, LA 71130-3932, USA.
June Feng, Email: junefeng@latech.edu, Department of Biomedical Engineering, Louisiana Tech University, 818 Nelson Avenue, Ruston, LA 71272, USA.
References
- 1.Kleinman WA, Komninou D, Leutzinger Y, Colosimo S, Cox J, Lang CA, Richie JP., Jr Protein glutathiolation in human blood. Biochem Pharmacol. 2003;65(5):741–746. doi: 10.1016/s0006-2952(02)01560-5. [DOI] [PubMed] [Google Scholar]
- 2.Cotgreave IA, Gerdes RG. Recent trends in glutathione biochemistry–glutathione-protein interactions: a molecular link between oxidative stress and cell proliferation? Biochem Biophys Res Commun. 1998;242(1):1–9. doi: 10.1006/bbrc.1997.7812. [DOI] [PubMed] [Google Scholar]
- 3.Muscari C, Pappagallo M, Ferrari D, Giordano E, Capanni C, Caldarera CM, Guarnieri C. Simultaneous detection of reduced and oxidized glutathione in tissues and mitochondria by capillary electrophoresis. J Chromatogr B Biomed Sci Appl. 1998;707 (1–2):301–307. doi: 10.1016/s0378-4347(97)00595-1. [DOI] [PubMed] [Google Scholar]
- 4.Monostori P, Wittmann G, Karg E, Turi S. Determination of glutathione and glutathione disulfide in biological samples: an in-depth review. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(28):3331–3346. doi: 10.1016/j.jchromb.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 5.Ercal N, Yang P, Aykin N. Determination of biological thiols by high-performance liquid chromatography following derivatization by ThioGlo maleimide reagents. J Chromatogr B Biomed Sci Appl. 2001;753(2):287–292. doi: 10.1016/s0378-4347(00)00560-0. [DOI] [PubMed] [Google Scholar]
- 6.Camera E, Picardo M. Analytical methods to investigate glutathione and related compounds in biological and pathological processes. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;781 (1–2):181–206. doi: 10.1016/s1570-0232(02)00618-9. [DOI] [PubMed] [Google Scholar]
- 7.Yang CS, Chou ST, Liu L, Tsai PJ, Kuo JS. Effect of ageing on human plasma glutathione concentrations as determined by high-performance liquid chromatography with fluorimetric detection. J Chromatogr B Biomed Appl. 1995;674(1):23–30. doi: 10.1016/0378-4347(95)00287-8. [DOI] [PubMed] [Google Scholar]
- 8.Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82(1):70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 9.Ellman GL, Lysko H. Disulfide and sulfhydryl compounds in TCA extracts of human blood and plasma. J Lab Clin Med. 1967;70 (3):518–527. [PubMed] [Google Scholar]
- 10.Molnar-Perl I. Derivatization and chromatographic behavior of the o-phthaldialdehyde amino acid derivatives obtained with various SH-group-containing additives. J Chromatogr A. 2001;913(1–2):283–302. doi: 10.1016/s0021-9673(00)01200-0. [DOI] [PubMed] [Google Scholar]
- 11.Yan CC, Huxtable RJ. Fluorimetric determination of monobromobimane and o-phthalaldehyde adducts of gamma-glutamylcysteine and glutathione: application to assay of gamma-glutamylcysteinyl synthetase activity and glutathione concentration in liver. J Chromatogr B Biomed Appl. 1995;672 (2):217–224. doi: 10.1016/0378-4347(95)00226-9. [DOI] [PubMed] [Google Scholar]
- 12.Hissin PJ, Hilf R. A fluorometric method for determination of oxidized and reduced glutathione in tissues. Anal Biochem. 1976;74 (1):214–226. doi: 10.1016/0003-2697(76)90326-2. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida T. Determination of reduced and oxidized glutathione in erythrocytes by high-performance liquid chromatography with ultraviolet absorbance detection. J Chromatogr B Biomed Appl. 1996;678(2):157–164. doi: 10.1016/0378-4347(95)00489-0. [DOI] [PubMed] [Google Scholar]
- 14.Circu ML, Moyer MP, Harrison L, Aw TY. Contribution of glutathione status to oxidant-induced mitochondrial DNA damage in colonic epithelial cells. Free Radic Biol Med. 2009;47(8):1190–1198. doi: 10.1016/j.freeradbiomed.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Circu ML, Rodriguez C, Maloney R, Moyer MP, Aw TY. Contribution of mitochondrial GSH transport to matrix GSH status and colonic epithelial cell apoptosis. Free Radic Biol Med. 2008;44(5):768–778. doi: 10.1016/j.freeradbiomed.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones DP, Carlson JL, Samiec PS, Sternberg P, Jr, Mody VC, Jr, Reed RL, Brown LA. Glutathione measurement in human plasma. Evaluation of sample collection, storage and derivatization conditions for analysis of dansyl derivatives by HPLC. Clin Chim Acta. 1998;275(2):175–184. doi: 10.1016/s0009-8981(98)00089-8. [DOI] [PubMed] [Google Scholar]
- 17.Cereser C, Guichard J, Drai J, Bannier E, Garcia I, Boget S, Parvaz P, Revol A. Quantitation of reduced and total glutathione at the femtomole level by high-performance liquid chromatography with fluorescence detection: application to red blood cells and cultured fibroblasts. J Chromatogr B Biomed Sci Appl. 2001;752(1):123–132. doi: 10.1016/s0378-4347(00)00534-x. [DOI] [PubMed] [Google Scholar]
- 18.Russell J, McKeown JA, Hensman C, Smith WE, Reglinski J. HPLC determination of biologically active thiols using pre-column derivatisation with 5,5′-dithio-(bis-2-nitrobenzoic acid) J Pharm Biomed Anal. 1997;15(11):1757–1763. doi: 10.1016/s0731-7085(96)02019-5. [DOI] [PubMed] [Google Scholar]
- 19.Pastore A, Federici G, Bertini E, Piemonte F. Analysis of glutathione: implication in redox and detoxification. Clin Chim Acta. 2003;333(1):19–39. doi: 10.1016/s0009-8981(03)00200-6. [DOI] [PubMed] [Google Scholar]
- 20.Sies H, Graf P. Hepatic thiol and glutathione efflux under the influence of vasopressin, phenylephrine and adrenaline. Biochem J. 1985;226(2):545–549. doi: 10.1042/bj2260545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson M, Lundberg M. Glutathionylation of beta-actin via a cysteinyl sulfenic acid intermediary. BMC Biochem. 2007;8:26. doi: 10.1186/1471-2091-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aesif SW, Janssen-Heininger YMW, Reynaert NL. Protocols for the detection of S-glutathionylated and S-nitrisylated proteins in situ. Thiol Redox Transituins cell Signal. 2010;474:289–296. doi: 10.1016/S0076-6879(10)74017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lind C, Gerdes R, Hamnell Y, Schuppe-Koistinen I, von Lowenhielm HB, Holmgren A, Cotgreave IA. Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch Biochem Biophys. 2002;406(2):229–240. doi: 10.1016/s0003-9861(02)00468-x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q, Zhang X, Chen J, Miao Y, Sun A. Role of caspase-3 in tau truncation at D421 is restricted in transgenic mouse models for taupathies. J Neurochem. 2009;109:476–484. doi: 10.1111/j.1471-4159.2009.05959.x. [DOI] [PubMed] [Google Scholar]
- 25.Reynaert NL, Ckless K, Guala AS, Wouters EFM, Vliet Avd, Janssen-Heininger YMW. In site detection of S-glutathionylated proteins following glutaredoxin-1 catalyzed cysteine derivatization. Biochimica et Biophysica Acta. 2006;1760:380–387. doi: 10.1016/j.bbagen.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Cheng G, Ikeda Y, Iuchi Y, Fujii J. Detection of S-glutathionylated proteins by glutathione S-transferase overlay. Arch Biochem Biophys. 2005;435(1):42–49. doi: 10.1016/j.abb.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 27.Patsoukis N, Georgiou CD. Determination of the thiol redox state of organisms: new oxidative stress indicators. Anal Bioanal Chem. 2004;378(7):1783–1792. doi: 10.1007/s00216-004-2525-1. [DOI] [PubMed] [Google Scholar]
- 28.Klatt P, Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur J Biochem. 2000;267(16):4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x. [DOI] [PubMed] [Google Scholar]
- 29.Circu ML, Stringer S, Rhoads CA, Moyer MP, Aw TY. The role of GSH efflux in staurosporine-induced apoptosis in colonic epithelial cells. Biochem Pharmacol. 2009;77(1):76–85. doi: 10.1016/j.bcp.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynaert NL, Ckless K, Guala AS, Wouters EF, van der Vliet A, Janssen-Heininger YM. In situ detection of S-glutathionylated proteins following glutaredoxin-1 catalyzed cysteine derivatization. Biochim Biophys Acta. 2006;1760(3):380–387. doi: 10.1016/j.bbagen.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Hamnell-Pamment Y, Lind C, Palmberg C, Bergman T, Cotgreave IA. Determination of site-specificity of S-glutathionylated cellular proteins. Biochem Biophys Res Commun. 2005;332(2):362–369. doi: 10.1016/j.bbrc.2005.04.130. [DOI] [PubMed] [Google Scholar]
- 32.Feng J, Arriaga EA. Quantification of carbonylated proteins in rat skeletal muscle mitochondria using capillary sieving electrophoresis with laser-induced fluorescence detection. Electrophoresis. 2008;29(2):475–482. doi: 10.1002/elps.200700262. [DOI] [PubMed] [Google Scholar]
- 33.Feng J, Navratil M, Thompson LV, Arriaga EA. Principal component analysis reveals age-related and muscle-type-related differences in protein carbonyl profiles of muscle mitochondria. J Gerontol A Biol Sci Med Sci. 2008;63(12):1277–1288. doi: 10.1093/gerona/63.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shelton MD, Chock PB, Mieyal JJ. Glutaredoxin: role in reversible protein S-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid Redox Signal. 2005;7(3–4):348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 35.Gravina SA, Mieyal JJ. Thioltransferase is a specific glutathionyl mixed disulfide oxidoreductase. Biochemistry. 1993;32 (13):3368–3376. doi: 10.1021/bi00064a021. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Jao S, Nanduri S, Starke DW, Mieyal JJ, Qin J. Reactivity of the human thioltransferase (glutaredoxin) C7S, C25S, C78S, C82S mutant and NMR solution structure of its glutathionyl mixed disulfide intermediate reflect catalytic specificity. Biochemistry. 1998;37(49):17145–17156. doi: 10.1021/bi9806504. [DOI] [PubMed] [Google Scholar]
- 37.Holmgren A, Aslund F. Glutaredoxin. Methods Enzymol. 1995;252:283–292. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- 38.Nordstrand K, slund F, Holmgren A, Otting G, Berndt KD. NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism. J Mol Biol. 1999;286(2):541–552. doi: 10.1006/jmbi.1998.2444. [DOI] [PubMed] [Google Scholar]
- 39.Mannervik B, Axelsson K. Role of cytoplasmic thioltransferase in cellular regulation by thiol-disulphide interchange. Biochem J. 1980;190(1):125–130. doi: 10.1042/bj1900125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chassaing C, Gonin J, Wilcox CS, Wainer IW. Determination of reduced and oxidized homocysteine and related thiols in plasma by thiol-specific pre-column derivatization and capillary electrophoresis with laser-induced fluorescence detection. J Chromatogr B Biomed Sci Appl. 1999;735(2):219–227. doi: 10.1016/s0378-4347(99)00425-9. [DOI] [PubMed] [Google Scholar]
- 41.Kang Y, Viswanath V, Jha N, Qiao X, Mo JQ, Andersen JK. Brain gamma-glutamyl cysteine synthetase (GCS) mRNA expression patterns correlate with regional-specific enzyme activities and glutathione levels. J Neurosci Res. 1999;58(3):436–441. doi: 10.1002/(SICI)1097-4547(19991101)58:3<436::AID-JNR9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 42.Abbott LC, Nejad HH, Bottje WG, Hassan AS. Glutathione levels in specific brain regions of genetically epileptic (tg/tg) mice. Brain Res Bull. 1990;25(4):629–631. doi: 10.1016/0361-9230(90)90124-i. [DOI] [PubMed] [Google Scholar]
- 43.Zhu Y, Carvey PM, Ling Z. Age-related changes in glutathione and glutathione-related enzymes in rat brain. Brain Res. 2006;1090(1):35–44. doi: 10.1016/j.brainres.2006.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]