Abstract
Many viruses, with distinct replication strategies, activate DNA-damage response pathways, including the lentivirus human immunodeficiency virus (HIV) and the DNA viruses Epstein–Barr virus (EBV), herpes simplex virus 1, adenovirus and SV40. DNA-damage response pathways involving DNA-dependent protein kinase, ataxia-telengiectasia mutated (ATM) and ‘ataxia-telengiectasia and Rad3-related’ (ATR) have all been implicated. This review focuses on the effects of HIV and EBV replication on DNA repair pathways. It has been suggested that activation of cellular DNA repair and recombination enzymes is beneficial for viral replication, as illustrated by the ability of suppressors of the ATM and ATR family to inhibit HIV replication. However, activation of DNA-damage response pathways can also promote apoptosis. Viruses can tailor the cellular response by suppressing downstream signalling from DNA-damage sensors, as exemplified by EBV. New small-molecule inhibitors of the DNA-damage response pathways could therefore be of value to treat viral infections.
Viruses are obligatory parasites that can replicate only within host cells. The genomes of viruses that infect human cells range in size from a few thousand nucleotides to several hundreds of kilobases and consist of single-stranded or double-stranded molecules of either DNA or RNA. All viruses require components from the host cell in order to replicate their genomes, in conjunction with virus-encoded enzymes. Recently, it has been observed that the replication of several types of viruses, including the DNA viruses Epstein–Barr virus (EBV), herpes simplex virus 1 (HSV-1), adenovirus and SV40, as well as the lentivirus human immunodeficiency virus (HIV), leads to activation of host DNA-damage response pathways (Refs 1, 2); this is achieved by a variety of mechanisms, some of which have been reviewed recently (Ref. 1). Here, we focus on the activation of DNA-damage response pathways by two viruses with distinct replication strategies: the small RNA virus HIV, which replicates through a pathway involving integration into the host genome; and the large DNA virus EBV, which replicates extra-chromosomally. It has been suggested that the DNA-damage response is required to facilitate the replication of some viruses and that viruses can tailor the DNA-damage response to promote the survival of cells; here we examine the evidence for these notions for HIV and EBV. Finally, the implications of these recent developments for future drug design are discussed.
The DNA-damage response
The DNA-damage response is tightly interlinked with the cell cycle checkpoints; together these signalling pathways act to prevent permanent genome damage. The components of three DNA-damage response pathways are detailed in Table 1. Sensors, which may interact directly with DNA, instigate the response to DNA damage; these then communicate the message to transducers, such as members of the phosphoinositide-3-kinase-related protein kinase (PI3K) family (Ref. 3). Transducers transmit the message to effectors, usually by direct phosphorylation of effectors, which can then modulate cellular processes.
Table 1. Overview of the components of the DNA damage responses to single-strand and double-strand breaksa.
| DNA damage | DNA repair pathway | Sensors | Transducers | Effectors | Cellular effects |
|---|---|---|---|---|---|
| SSBs | ATR | ATRIP Rad17-RFC complex |
ATR Chk1 BRCA1 |
H2AX p53 p21 Smc1 |
Cell cycle arrest DNA repair |
| DSBs | ATM | p53BP1 MRN complex |
ATM Chk1 Chk2 |
H2AX p53 p21 53BP1 Mdm2 Smc1 Cdc25A and C E2F1 |
Cell cycle arrest DNA repair Apoptosis |
| DNA-PK | Ku heterodimer | ATM DNA-PK Artemis |
H2AX p53 p21 Mdm2 MRN complex XRCC4 Ligase IV |
Cell cycle arrest DNA repair Apoptosis |
Abbreviations: ATM, ataxia-telengiectasia mutated; ATR, ataxia-telengiectasia and Rad3-related; ATRIP, ATR-interacting protein; BRCA1, breast cancer type 1 susceptibility protein; Cdc25, cell division cycle 25; Chk, checkpoint homologue; DNA-PK, DNA-dependent protein kinase; DSBs, double-strand DNA breaks; E2F1, E2F transcription factor 1; H2AX, H2A histone family, member X; Mdm2, double minute 2 gene product; MRN complex, a complex of the nuclease Mre11, the DNA-binding protein Rad50 and the Nijmegen breakage syndrome 1 gene product Nbs1; 53BP1, p53-binding protein 1; Rad17, a post-replication repair protein; RFC, replication factor complex; Smc1, structural maintenance of chromosomes 1; SSBs, single-strand DNA breaks; XRCC4, X-ray repair complementing defective repair in Chinese hamster cells 4.
The pathways of particular interest in this review are those associated with single-strand and double-strand DNA breaks (SSBs and DSBs, respectively), because these have been identified as being associated with the replication of several viruses (Refs 1, 4, 5, 6). Three members of the PI3K family – DNA-dependent protein kinase (DNA-PK), ataxia-telengiectasia mutated (ATM) and ‘ataxia-telengiectasia and Rad3-related’ (ATR) – are intimately involved in detecting and transducing DNA-damage response signals through three similar, yet different, signalling pathways (Ref. 3) (Table 1).
DNA repair through the PI3K family of kinases has been observed to have a fast and a slower repair phase: if the initial fast repair phase cannot eliminate the damage then the cell cycle is thought to be delayed while slower mechanisms are used (reviewed by Refs 7, 8, 9). All three PI3K family members are able to stimulate DNA repair through the phosphorylation of histone H2AX and the subsequent formation of foci containing DNA repair enzymes. The repair proteins commonly associated with the fast mode of DNA repair include DNA-PK, XRCC4 and DNA ligase IV, whereas the slower mode requires the MRN complex (a complex of the nuclease Mre11, the DNA-binding protein Rad50 and the Nijmegen breakage syndrome 1 gene product Nbs1), ATM, p53-binding protein 1 (53BP1), H2AX and Artemis (Ref. 10).
DNA-damage response to SSBs
SSBs are sensed through several complexes including the ATR-interacting protein (ATRIP) and a complex of Rad17 and replication factor complex (RFC). These proteins recruit transducers such as ‘breast cancer type 1 susceptibility protein’ (BRCA1) and the kinases Chk1 and ATR, which become phosphorylated. This leads to recruitment of the effectors, including the histone H2AX, the chromosome- and spindle-associated protein Smc1, the cyclin-dependent-kinase inhibitor p21 and the tumour suppressor protein p53, which act together to stimulate DNA repair and promote cell cycle arrest at the G2-M checkpoint (Refs 9, 11, 12).
DNA-damage response to DSBs
Signals from DSBs can be relayed through both ATM and DNA-PK. It is not known how these proteins preferentially bind to certain types of DNA ends but their mechanisms of activation and the recruitment of accessory proteins appear to be similar. Both require a sensor: for ATM this is either 53BP1 or the MRN complex; and for DNA-PK it is a heterodimer of the ATP-dependent DNA helicases Ku70 and Ku80 (Ref. 3). The pathways of signal transduction from the transducers are complex and seem to be intertwined; however, activation of different combinations of proteins appears to produce very different outcomes.
Unlike the other two PI3K family proteins, ATM can autophosphorylate on Ser1981 to cause the release of active monomers from an inactive homodimer complex (Ref. 13). Monomeric ATM is then recruited to damaged DNA by the MRN complex (Ref. 14), which is itself phosphorylated by ATM, and further phosphorylation of downstream targets ensues, such as p53 (Ser20 and Ser15), Chk2 (Thr68) and Smc1 (Ser966) (Refs 3, 7, 12). ATR can also phosphorylate p53 on Ser20 but additionally phosphorylates Nbs1 (Ser345), BRCA1 (Ser1423) and H2AX (Ser139), which leads preferentially to DNA repair. The phosphorylation of p53 on Ser20 causes accumulation of p53 protein by preventing its interaction with Mdm2 (the double minute 2 gene product), and consequently causes a G1-S cell cycle arrest through activation of p21 expression (Ref. 7). ATM also causes G1-S arrest through phosphorylation of Chk2, which then leads to Cdc25A being targeted for degradation, resulting in a build up of Cdk2-cyclin-E complexes (Ref. 3).
Another important outcome of ATM pathway activation is the induction of damage-induced apoptosis (reviewed extensively in Refs 10, 15). The transcription factor E2F1 is an important regulator during the S phase of the cell cycle (Ref. 16) and is normally labelled for degradation through ubiquitination of Ser31; this is prevented by phosphorylation of Ser31 by ATM (Ref. 17). As a consequence of the elevated abundance of E2F1 protein and activation of Chk2, expression of p73 is upregulated (Refs 18, 19), leading to the induction of apoptosis (Refs 19, 20).
In a similar mechanism to that of ATR and ATM, DNA-PK can phosphorylate p53. A single phosphorylation of Ser15 (by ATR or DNA-PK) can prevent p53 from binding to the transcription factor TFIID and consequently transactivating downstream genes; however, phosphorylation of Ser37 by DNA-PK, in addition to Ser15, can restore the transactivating activity but blocks Mdm2 binding (Refs 21, 22).
HIV replication and DNA-damage response pathways
Infection of permissive cells, such as lymphocytes and macrophages, with HIV leads to viral replication, and at least one cellular DNA-damage response pathway is activated during this process (see below). The question arises as to how this is achieved and what impact this has on the virus and host cell. Retroviral genomes undergo a complex replication strategy (Fig. 1); this subject has been reviewed recently (Ref. 23) and is not discussed in detail here. Following infection, the single-stranded positive-sense RNA genome, delivered to cells within retroviral virions, is converted to a double-stranded DNA form by reverse transcriptase, a retroviral enzyme carried within the virion. A second viral enzyme essential for replication is integrase. The function of this enzyme has been elucidated recently: integrase first cleaves nucleotides at the 3′ ends of the viral DNA, within the long terminal repeat (LTR), then simultaneously cleaves host DNA and ligates the 3′ ends of the viral genome and the 5′ ends of the host genome together (Ref. 24). The outcome of this reaction is to leave SSBs and/or short gaps at the ends of the viral genome at the integration site within the host genome. Unless these breaks and/or gaps are repaired, they would be converted into DSBs during host cell replication, with catastrophic consequences for the host genome.
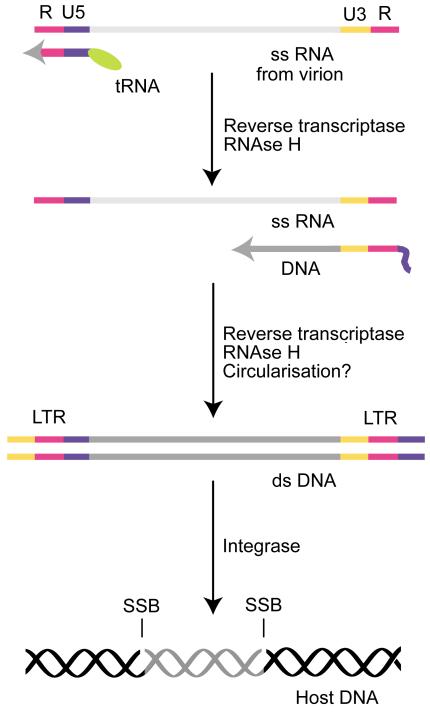
Figure 1. Overview of the transition of the retroviral genome from ss RNA to integrated ds DNA.
Retroviral virions contain a single-stranded (ss) RNA version of their genome. Upon infection of permissive cells, the ss RNA is converted to double-stranded (ds) DNA by the action of the virion enzyme reverse transcriptase. Reverse transcriptase employs a tRNA molecule bound near the U5 region of the ss RNA genome as an initial primer to synthesise a short region of single-stranded viral DNA, containing U5 and R regions. The DNA then dissociates from the genome and anneals to another copy of the R region, at the 3′ end of the genome. DNA synthesis proceeds along the genome to generate the negative strand. The positive strand is replicated from this template by a mechanism that uses DNA as its template and might involve circularised molecules. The RNA primer is destroyed by the RNAse H activity of reverse transcriptase. The double-stranded DNA genome is then recognised by a second virion enzyme, integrase, which cleaves two nucleotides from the 3′ end of each DNA strand at the long terminal repeat (LTR) and mediates their ligation into the host DNA. This process leaves the 5′ ends of the viral DNA unligated, resulting in single-strand breaks (SSBs).
Thus, during the HIV lifecycle there are two points at which the replicating genome could trigger a DNA-damage response: as unintegrated double-stranded viral DNA, or as integrated viral DNA containing DNA breaks and/or gaps. The use of viral mutants revealed that the ability to integrate into the genome is required to initiate the DNA-damage response, which suggests that the latter is the trigger (Ref. 4).
A further route by which HIV activates the DNA-damage response pathway occurs following the expression of the HIV gene vpr. Expression of this gene induces cell cycle arrest at the G2-M checkpoint (Ref. 25). The activation of this DNA-damage response pathway appears to occur in the absence of damaged DNA, with the underlying mechanism involving a physical interaction of Vpr protein with chromatin (Ref. 26).
Is activation of the DNA-damage response pathway an unfortunate consequence of HIV replication or part of a deliberate strategy to enlist the aid of cellular enzymes to accomplish an essential step in viral replication? This question has been addressed using specific inhibitors of individual components of the response pathways. Much of the work that has been undertaken has employed HIV-based viral vectors to transduce primary or established cell lines; some, but not all, conclusions have been verified studying HIV infection of primary cells. Transduction of retroviral vectors involves the infection of cells with vector-containing virions, replication of the vector RNA to a double-stranded DNA form and integration of the vector as a provirus. The efficiency of the process is measured by the stable expression of a marker gene. As detailed below, this body of research reveals that at least one cellular DNA-damage response pathway is essential to allow HIV replication in permissive cells. This suggests that HIV requires catalytic activities activated during the DNA-damage response to complete its replicative cycle.
Biochemical evidence for DNA-damage response activation by HIV
There is clear evidence that the ATM pathway is activated by HIV replication. ATM itself becomes phosphorylated on Ser1981 following infection of human T cells with HIV. Importantly, this does not occur with an integrase-deficient strain of HIV (Ref. 27). In addition, downstream signalling events such as phosphorylation of Chk2 at Thr68, p53 at Ser15 and Nbs1 at Ser345 are observed (Ref. 27). Biochemical evidence supporting the activation of other DNA repair pathways awaits further investigation.
There is also evidence that the ATR pathway is activated and promotes phosphorylation of Chk1 following expression of Vpr (Ref. 25). Involvement of the ATR pathway in Vpr-mediated cell cycle arrest has been shown using both a dominant negative form of ATR and knock-down of ATR expression using short interfering (Si) RNA (Ref. 25). A subsequent report revealed that nuclear foci containing phosphorylated H2AX are formed following Vpr expression and genetic evidence shows that two gene products on the ATR pathway, Rad1 and Hus1, are required to mediate the G2-M arrest (Ref. 28).
Genetic evidence for the involvement of DNA-damage response pathways by HIV
DNA-PK
It was initially demonstrated that pre-B cells from scid mice with DNA-PK defects had a dramatically reduced ability to transduce an HIV vector and also underwent apoptosis in response to HIV infection (Ref. 4). HIV integrase was rapidly identified as a key viral component of this process (Ref. 4). Further investigation into the residual ability of the scid cells to replicate viral vectors led to the discovery of additional contributions from the ATM and ATR pathways and implicated contributions from the xrcc4 and ligase IV genes (Refs 29, 30, 31). Thus, although DNA-PK clearly plays a key role in the transduction of an HIV vector, other genes can also substitute for it.
The evidence that DNA-PK activity is required for integration or post-integration repair of HIV genomes is compelling but not universally accepted. In support, packaged retroviral vectors encoding drug-resistance genes or reporter genes are unable to express the reporter genes or confer drug resistance to cells lacking DNA-PK (Ref. 4). The DNA-binding subunit of DNA-PK, Ku80, has also been implicated. Cells with reduced Ku80 expression show delayed HIV replication when compared with cells expressing normal levels of Ku80 (Ref. 32). Furthermore, ribozymes directed against Ku80 were able to prevent HIV infection of CD4+ human lymphocytes (Ref. 33). However, not all reports agree with the conclusion that DNA-PK plays a critical role in HIV replication. For example, Baekelandt and colleagues showed that while high titres of infection of viral vectors depended on the presence of the DNA-PK gene, at low titres no dependence was observed (Ref. 34). Additional evidence against an absolute requirement for DNA-PK function for HIV replication comes from the work of Ariumi and colleagues who used HIV vectors to measure expression in cells lacking DNA-PK expression. Since no reduction in HIV vector transduction was detected, they concluded that DNA-PK is not required for stable integration (Ref. 35). In future, the use of Si RNA may clarify this issue.
ATM
The role of ATM in HIV replication is also controversial. Daniel et al. originally showed that the ATM/ATR inhibitor caffeine inhibits retroviral integration in cells deficient in nonhomologous end joining (NHEJ), suggesting a role for ATM (Ref. 4). However, while some groups report no effect of ATM deficiency on the ability of HIV vectors to transduce cells, others have shown evidence for clear involvement. Three types of genetic experiment have been undertaken to address this question. Cells lacking a functional ATM gene, obtained either from subjects with the genetic disorder ataxia-telengiectasia or from ATM−/− mice have been employed, as have cells where ATM expression is knocked down using Si RNA. In addition, a series of caffeine-related inhibitors of the ATM/ATR family have been used to oblate the function of ATM and ATR in cells, although in these experiments it can be difficult to distinguish between the effects of ATM and ATR inhibition.
The case against an involvement of ATM in HIV integration is put by Ariumi and colleagues, who used Si RNA to knock-down expression of ATM but found no detrimental effect on the ability of HIV vectors to transduce cells. Although the Si RNA approach does not completely obliterate ATM expression, additional evidence comes from the obervation that no difference was observed in the efficiency of transducing HIV vectors into ATM−/− compared with wild-type mouse embryonic fibroblasts (Ref. 35). In addition, no inhibitory effect of caffeine was observed following HIV vector transduction of HeLa cells (Ref. 35). DeHart and colleagues reached the same conclusion, using a similar approach (Ref. 36). Although these two reports show that ATM is not absolutely required for integration of retroviral DNA, there are two caveats that impact on the question of whether ATM is required for HIV replication: the transduction of HIV vectors rather than infection with HIV virus was analysed and the cell types tested did not include the natural host, human T cells.
Evidence in favour of a role for ATM in HIV replication comes from studies on the effects of caffeine and related compounds on infection of human T cells (Ref. 37). The case in favour of an involvement of ATM in HIV replication was sealed by a report from Lau and colleagues who demonstrated that: (1) ATM−/− murine embryonic stem cells and primary T cells isolated from ataxia-telengiectasia patients displayed a reduced efficiency of transduction of a viral vector; and (2) that transduction of a human T-cell line was compromised by an ATM-specific inhibitor (KU-55933) (Ref. 27). This requirement does not appear to be unique to HIV as the transduction and integration of other retroviruses has also been shown to require ATM function (Refs 27, 38). Furthermore, the authors went on to show that replication of HIV in human T cells was suppressed by the inhibitor, as was replication of drug-resistant HIV isolates. It thus appears to be clear that although ATM is not absolutely required for successful integration of retroviral vectors in all cell types, it is required in human primary T cells, which are the natural target cells for HIV. This means that ATM is an excellent target against which to design novel drugs, such as KU-55933, to block HIV replication.
ATR
Strong evidence in favour of a role for ATR in HIV replication is provided by the demonstration that an expression vector that directs expression of a dominant negative form of ATR led to reduced integration of viral vector DNA (Ref. 31). However, experiments using Si RNA against ATR and the inhibitor caffeine could not detect a detrimental effect on the ability of HIV vectors to transduce cells (Refs 35, 36). In a similar manner to the ATM story, these reports suggest that ATR is not absolutely required for integration of retroviral DNA, but again HIV virus was not used and the cell types tested did not include the natural host, human T cells.
EBV and DNA-damage response pathways
Several large double-stranded DNA viruses also activate DNA-damage response pathways; EBV is discussed as an example here. EBV undergoes productive replication in both B cells and epithelial cells. Its genome is about 172 kb and exists in a double-stranded DNA linear form within virions. Following infection, the genome circularises through the association of the terminal repeat regions at each end of the genome. The virus is maintained as an extra-chromosomal episome in infected cells and can replicate once per cell cycle, using the host replication machinery, in combination with the action of a single EBV protein, EBNA1, on the viral origin of plasmid replication (OriP) (Ref. 39). However, an alternate replication strategy, using a second viral origin of replication (OriLyt), is used to amplify the viral genome during the productive replication or lytic cycle of EBV, where large copy numbers of the linear viral genome are produced (Ref. 40). Productive replication of EBV is initiated by the action of a single EBV gene, BZLF1; this encodes the protein Zta, which acts as both a transcription and a replication factor (Refs 41, 42). Zta induces the expression of other viral transactivators that in concert activate the expression of the remaining 80 or so viral genes. Zta also interacts with the OriLyt and acts as an origin-binding protein. EBV encodes seven essential EBV replication genes that are necessary for OriLyt-based replication in uninfected cells (Ref. 40). It is thought that the viral genome replicates from the circular episome, with the start of viral replication from a single-stranded nick in the DNA. A ‘rolling circle’ of multiple genome-length linear arrays of newly synthesised double-stranded genomes then emerge from the episome (Ref. 40) (Fig. 2). These are cut within the terminal repeats, by unknown enzymes, to yield individual linear genomes that are packaged into virions and released from the cell.
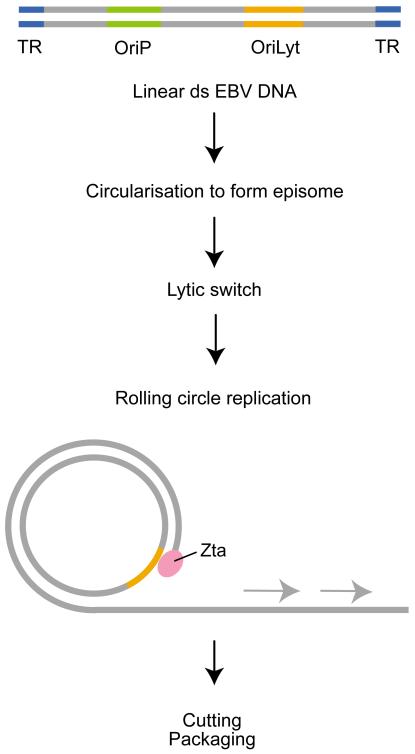
Figure 2. Overview of lytic replication of the Epstein–Barr virus genome.
Epstein–Barr virus (EBV) virions contain a double-stranded linear DNA genome. Upon infection of permissive cells, the double-stranded linear DNA is converted to circular double-stranded DNA by association of the terminal repeats (TRs). This episome form of the genome replicates once per cell division in latently infected cells (not shown), using the origin of replication OriP. Following the binding of the lytic switch transactivator Zta to the alternative replication origin OriLyt, a rolling-circle replication occurs, generating multiple copies of the linear genome. In the figure, new DNA synthesis is shown by the grey arrows. The new DNA is cut at each TR, releasing single genome-length units, which are packaged and released from the cell.
There appear to be two points during the life cycle of EBV that could activate a DNA-damage response: (1) during initial infection, the exposed ends of the viral linear DNA could resemble DSBs; and (2) during the lytic replication of the viral genome, the rolling-circle structure or the newly synthesised free linear genomes could appear as SSBs or DSBs. There is no evidence for or against the generation of a DNA-damage response during initial infection; a definitive answer awaits further investigation. However, it has been recently shown that components of the DNA-damage response pathway are activated during viral replication.
Biochemical evidence for DNA-damage response activation by EBV
Components of the ATM DNA-damage response pathway are activated rapidly following the induction of EBV lytic replication. Within 24 h of BZLF1 expression in cells containing EBV genomes, ATM was phosphorylated on Ser1981, Chk2 on Thr68, p53 on Ser15 and H2AX on Ser139 (Ref. 5). This strongly suggests activation of the ATM pathway, which is further corroborated by the colocalisation of ATM, Nbs1 and Mre11 at sites of viral replication (Ref. 5). However, signal transduction through the ATM pathway is blocked; despite activation of p53 by phosphorylation on Ser15, the protein appears to be compromised in its ability to activate downstream targets (Ref. 5). The EBV protein that interacts with OriLyt also physically interacts with p53 (Refs 5, 43, 44) and Kenney’s group have shown it to inhibit the transactivation ability of p53 (Ref. 43). Thus, it appears that an abortive DNA-damage response is generated during EBV infection. This raises the question as to whether the DNA-damage response is required for EBV replication, as it is for HIV replication. Indeed, the use of caffeine to inhibit ATM (and the other PI3K-related proteins) revealed that in conditions where caffeine inhibits the phosphorylation of ATM, Chk2 and Nbs1, it did not disrupt the amount of viral genome being produced in the cell (Ref. 5).
DNA-damage responses induced by replication of other viruses
As already mentioned, several other viruses have also been shown to activate DNA-damage response pathways during replication, and some appear to interfere with the outcome of those pathways. Like HIV, the DNA viruses HSV-1, polyoma virus and SV40 require the activation of DNA-damage response pathways to achieve efficient replication. Replication of the large DNA virus HSV-1 activates the ATM signal transduction pathway, causing phosphorylation of downstream components (Ref. 1). HSV-1 requires the response in order to form viral replication centres (Ref. 45). Thus, HSV-1 and EBV differ in the dependence of replication on activation of DNA-damage response pathways, despite belonging to the same viral family (Herpesviridae). Polyoma virus both induces and utilises the ATM DNA-damage response during its replication (Ref. 46), and a key viral protein for the replication of SV40 requires phosphorylation by ATM for function (Ref. 6). In addition, adenoviruses are prime examples of other viruses that, like EBV, modulate the DNA-damage response: viral genes from the E4 region of the adenovirus genome interfere with the MRN complex (Refs 47, 48).
Clinical implications/applications
In conclusion, infection with HIV activates at least two cellular DNA-damage response pathways and although these are not absolutely required for the transduction of genomes into cells or their subsequent integration, they are required for HIV infection and replication in its natural environment. Thus, it is interesting to speculate that HIV might have evolved mechanisms to deliberately activate these pathways in order to exploit downstream cellular enzymes that might facilitate viral replication. By contrast, replication of EBV during the viral lytic cycle activates a DNA-damage response pathway that does not appear to be required for efficient replication of the genome. Indeed, activation of the ATM pathway might result in apoptosis of the cell, interfering with replication. It is tempting to speculate that the interaction of Zta with p53 might act to modulate the function of p53 and, by halting the transduction of signals through the ATM pathway, prevent apoptosis.
The research reviewed here raises an important question of how the activation of the DNA-damage response pathways by viral replication might be exploited to generate antiviral drugs. A cellular gene that is required for viral replication presents a better target for drug design than viral genes do, since, unlike a viral gene, a cellular gene would not be subject to selective pressures to generate drug-resistant mutants. Progress in this direction has already been made for HIV, where it is clear that inhibition of integration prevents viral replication and promotes cellular apoptosis (Ref. 4). The ATM-specific inhibitor KU-55933 is able to inhibit HIV replication in primary T cells (Ref. 27), thus providing proof of principle. Furthermore, since KU-55933 is effective for isolates of HIV that are resistant to current drug regimes (Ref. 27), it may prove to be useful as an alternative line of therapy for HIV. For viruses such as EBV, where DNA-damage response pathways are activated but do not appear to be required for viral replication, a different approach is required. An obvious target for drug design is the point at which the response is stalled; for EBV, this appears to reside with p53. If p53 function could be restored in cells undergoing viral lytic replication, they might be directed to complete the ATM pathway and undergo apoptosis, so preventing replication. Thus, the Zta-p53 protein interaction might prove to be a suitable target against which to design inhibitory molecules.
Acknowledgements and funding
Research in the authors’ laboratory is funded by the Medical Research Council, the Leukaemia Research Fund and the Royal Society, UK. The authors thank the anonymous peer reviewers for their comments.
Contributor Information
Alison Sinclair, School of Life Sciences, University of Sussex, Brighton, BN1 9QG, UK.
Sarah Yarranton, School of Life Sciences, University of Sussex, Brighton, BN1 9QG, UK. Tel: +44 (0)1273 678 194; Fax: +44 1273 678 433; s.g.yarranton@sussex.ac.uk.
Celine Schelcher, School of Life Sciences, University of Sussex, Brighton, BN1 9QG, UK. Tel: +44 (0)1273 678 194; Fax +44 1273 678 433; c.schelcher@sussex.ac.uk.
References
- 1.Shirata N, et al. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J Biol Chem. 2005;280:30336–30341. doi: 10.1074/jbc.M500976200. [DOI] [PubMed] [Google Scholar]
- 2.Weitzman MD, et al. Interactions of viruses with the cellular DNA repair machinery. DNA Repair (Amst) 2004;3:1165–1173. doi: 10.1016/j.dnarep.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 4.Daniel R, Katz RA, Skalka AM. A role for DNA-PK in retroviral DNA integration. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 5.Kudoh A, et al. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J Biol Chem. 2005;280:8156–8163. doi: 10.1074/jbc.M411405200. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y, et al. Ataxia-telangiectasia-mutated (ATM) is a T-antigen kinase that controls SV40 viral replication in vivo. J Biol Chem. 2005;280:40195–40200. doi: 10.1074/jbc.C500400200. [DOI] [PubMed] [Google Scholar]
- 7.Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol. 2001;13:738–747. doi: 10.1016/s0955-0674(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 8.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 9.Zhou BB, Mattern MR, Khanna KK. Role of tumor suppressors in DNA damage response. Methods Mol Biol. 2003;223:39–50. doi: 10.1385/1-59259-329-1:39. [DOI] [PubMed] [Google Scholar]
- 10.Lobrich M, Jeggo PA. The two edges of the ATM sword: co-operation between repair and checkpoint functions. Radiother Oncol. 2005;76:112–118. doi: 10.1016/j.radonc.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Ball HL, Cortez D. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J Biol Chem. 2005;280:31390–31396. doi: 10.1074/jbc.M504961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyberg KA, et al. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 13.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 14.Paull TT, Lee JH. The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle. 2005;4:737–740. doi: 10.4161/cc.4.6.1715. [DOI] [PubMed] [Google Scholar]
- 15.Zgheib O, et al. ATM signaling and 53BP1. Radiother Oncol. 2005;76:119–122. doi: 10.1016/j.radonc.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 16.Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 17.Lin SC, et al. The proliferative and apoptotic activities of E2F1 in the mouse retina. Oncogene. 2001;20:7073–7084. doi: 10.1038/sj.onc.1204932. [DOI] [PubMed] [Google Scholar]
- 18.Irwin M, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 19.Stevens C, Smith L, La Thangue NB. Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol. 2003;5:401–409. doi: 10.1038/ncb974. [DOI] [PubMed] [Google Scholar]
- 20.Urist M, et al. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 2004;18:3041–3054. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pise-Masison CA, et al. Phosphorylation of p53: a novel pathway for p53 inactivation in human T-cell lymphotropic virus type 1-transformed cells. J Virol. 1998;72:6348–6355. doi: 10.1128/jvi.72.8.6348-6355.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 23.Nisole S, Saib A. Early steps of retrovirus replicative cycle. Retrovirology. 2004;1:9. doi: 10.1186/1742-4690-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skalka AM, Katz RA. Retroviral DNA integration and the DNA damage response. Cell Death Differ. 2005;12(Suppl 1):971–978. doi: 10.1038/sj.cdd.4401573. [DOI] [PubMed] [Google Scholar]
- 25.Roshal M, et al. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J Biol Chem. 2003;278:25879–25886. doi: 10.1074/jbc.M303948200. [DOI] [PubMed] [Google Scholar]
- 26.Lai M, et al. Activation of the ATR pathway by human immunodeficiency virus type 1 Vpr involves its direct binding to chromatin in vivo. J Virol. 2005;79:15443–15451. doi: 10.1128/JVI.79.24.15443-15451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau A, et al. Suppression of HIV-1 infection by a small molecule inhibitor of the ATM kinase. Nat Cell Biol. 2005;7:493–500. doi: 10.1038/ncb1250. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman ES, et al. Human immunodeficiency virus type 1 Vpr-mediated G2 arrest requires Rad17 and Hus1 and induces nuclear BRCA1 and gamma-H2AX focus formation. Mol Cell Biol. 2004;24:9286–9294. doi: 10.1128/MCB.24.21.9286-9294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daniel R, et al. Evidence that stable retroviral transduction and cell survival following DNA integration depend on components of the nonhomologous end joining repair pathway. J Virol. 2004;78:8573–8581. doi: 10.1128/JVI.78.16.8573-8581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daniel R, et al. Evidence that the retroviral DNA integration process triggers an ATR-dependent DNA damage response. Proc Natl Acad Sci U S A. 2003;100:4778–4783. doi: 10.1073/pnas.0730887100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daniel R, et al. Wortmannin potentiates integrase-mediated killing of lymphocytes and reduces the efficiency of stable transduction by retroviruses. Mol Cell Biol. 2001;21:1164–1172. doi: 10.1128/MCB.21.4.1164-1172.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeanson L, et al. Effect of Ku80 depletion on the preintegrative steps of HIV-1 replication in human cells. Virology. 2002;300:100–108. doi: 10.1006/viro.2002.1515. [DOI] [PubMed] [Google Scholar]
- 33.Waninger S, et al. Identification of cellular cofactors for human immunodeficiency virus replication via a ribozyme-based genomics approach. J Virol. 2004;78:12829–12837. doi: 10.1128/JVI.78.23.12829-12837.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baekelandt V, et al. DNA-Dependent protein kinase is not required for efficient lentivirus integration. J Virol. 2000;74:11278–11285. doi: 10.1128/jvi.74.23.11278-11285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ariumi Y, et al. DNA damage sensors ATM, ATR, DNA-PKcs, and PARP-1 are dispensable for human immunodeficiency virus type 1 integration. J Virol. 2005;79:2973–2978. doi: 10.1128/JVI.79.5.2973-2978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dehart JL, et al. The ataxia telangiectasia-mutated and Rad3-related protein is dispensable for retroviral integration. J Virol. 2005;79:1389–1396. doi: 10.1128/JVI.79.3.1389-1396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nunnari G, et al. Inhibition of HIV-1 replication by caffeine and caffeine-related methylxanthines. Virology. 2005;335:177–184. doi: 10.1016/j.virol.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Hasegawa M, et al. Resistance against Friend leukemia virus-induced leukemogenesis in DNA-dependent protein kinase (DNA-PK)-deficient scid mice associated with defective viral integration at the Spi-1 and Fli-1 site. Leuk Res. 2005;29:933–942. doi: 10.1016/j.leukres.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Sugden B. Origins of bidirectional replication of Epstein-Barr virus: models for understanding mammalian origins of DNA synthesis. J Cell Biochem. 2005;94:247–256. doi: 10.1002/jcb.20324. [DOI] [PubMed] [Google Scholar]
- 40.Tsurumi T, Fujita M, Kudoh A. Latent and lytic Epstein-Barr virus replication strategies. Rev Med Virol. 2005;15:3–15. doi: 10.1002/rmv.441. [DOI] [PubMed] [Google Scholar]
- 41.Sinclair AJ. bZIP proteins of human gammaherpesviruses. J Gen Virol. 2003;84:1941–1949. doi: 10.1099/vir.0.19112-0. [DOI] [PubMed] [Google Scholar]
- 42.Speck SH, Chatila T, Flemington E. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 1997;5:399–405. doi: 10.1016/S0966-842X(97)01129-3. [DOI] [PubMed] [Google Scholar]
- 43.Mauser A, et al. The Epstein-Barr virus immediate-early protein BZLF1 regulates p53 function through multiple mechanisms. J Virol. 2002;76:12503–12512. doi: 10.1128/JVI.76.24.12503-12512.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Q, Gutsch D, Kenney S. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol Cell Biol. 1994;14:1929–1938. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lilley CE, et al. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc Natl Acad Sci U S A. 2005;102:5844–5849. doi: 10.1073/pnas.0501916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dahl J, You J, Benjamin TL. Induction and utilization of an ATM signaling pathway by polyomavirus. J Virol. 2005;79:13007–13017. doi: 10.1128/JVI.79.20.13007-13017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans JD, Hearing P. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J Virol. 2005;79:6207–6215. doi: 10.1128/JVI.79.10.6207-6215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weitzman MD, Ornelles DA. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene. 2005;24:7686–7696. doi: 10.1038/sj.onc.1209063. [DOI] [PubMed] [Google Scholar]
Further reading, resources and contacts
- Review of ATM: Pandita TK. A multifaceted role for ATM in genome maintenance. Expert Rev Mol Med. 2003:1–21. doi: 10.1017/S1462399403006318. 2003. PubMed: 14987398.
- Review of HIV replication: Freed EO. HIV-1 replication. Somat Cell Mol Genet. 2001;26:13–33. doi: 10.1023/a:1021070512287. PubMed: 12465460.
- Review of EBV replication: Tsurumi T, Fujita M, Kudoh A. Latent and lytic Epstein-Barr virus replication strategies. Rev Med Virol. 2005;15:3–15. doi: 10.1002/rmv.441. PubMed: 15386591.