Abstract
Plants form symbiotic associations with endophytic bacteria within tissues of leaves, stems, and roots. It is unclear whether or how plants obtain nitrogen from these endophytic bacteria. Here we present evidence showing nitrogen flow from endophytic bacteria to plants in a process that appears to involve oxidative degradation of bacteria. In our experiments we employed Agave tequilana and its seed-transmitted endophyte Bacillus tequilensis to elucidate organic nitrogen transfer from 15N-labeled bacteria to plants. Bacillus tequilensis cells grown in a minimal medium with 15NH4Cl as the nitrogen source were watered onto plants growing in sand. We traced incorporation of 15N into tryptophan, deoxynucleosides and pheophytin derived from chlorophyll a. Probes for hydrogen peroxide show its presence during degradation of bacteria in plant tissues, supporting involvement of reactive oxygen in the degradation process. In another experiment to assess nitrogen absorbed as a result of endophytic colonization of plants we demonstrated that endophytic bacteria potentially transfer more nitrogen to plants and stimulate greater biomass in plants than heat-killed bacteria that do not colonize plants but instead degrade in the soil. Findings presented here support the hypothesis that some plants under nutrient limitation may degrade and obtain nitrogen from endophytic microbes.
Like all living things, plants require nitrogen (N) throughout their development. The N incorporated as NO3− and NH4+ represents about 2% of total plant dry matter, and is a component of proteins, nucleic acids, cofactors, signalling molecules, storage and numerous plant secondary products. The availability of N to plant roots is often an important limiting factor for plant growth. Plants obtain N from nitrogen fixing bacteria and decomposition of dead tissues of both plants and animals by microorganisms1,2,3,4,5. Moreover, a tiny fraction (0.00024%) of planetary N is available to plants in soils, where plants compete with microbes to absorb N6. The atmospheric formation of nitrate by photo-oxidation through the effect of lightning also is minimal9,10. The limited bioavailability of N, and the need to enhance crop growth, have provoked a continual expansion in use of N fertilizer with from 12 to 104 additional teragrams used each year7,8. On average only 30–50% of the applied N is taken up by plants, with the remainder being lost to surface run-off, leaching of nitrates and ammonia volatilization11. The excessive use of fertilizers is a highly contaminating process9 and the energy required to produce N through the Haber-Bosch process requires at least 1% of the world annual energy supply12.
Historically, it was thought that plants derived all N nutrition from the inorganic forms of N, NO3− and NH4+. However, it is now known that the principal form of N entering soils that do not receive inorganic fertilizer is organic N derived from microbial breakdown of organic matter, including amino acids, di- and tri-peptides, DNA and proteins13,14,15. In addition, some plants can scavenge organic N from insects (carnivorous plants) or by entomopathogenic fungus-mediated N translocation from insects16,17. Further, some plants have been shown to consume and degrade bacteria and yeasts through endocytosis3. However, it is unclear whether endocytosis of microbes by plants is a purely defensive process or rather is a means to acquire nutrients3,4,5. Plants are naturally colonised by endophytic bacteria that inhabit internal spaces without deleterious effects on host plants1,2. Endophytic bacteria that spend some time in soil enter roots or shoots and may spread to all plant tissues, often transmitting in seeds, thus ensuring transfer to the next plant generation2. However, controversy exists regarding the significance of nitrogen-fixing endophytes in plants1,2. It is frequently argued that positive effects of bacteria on plant growth may be the result of auxins and other growth regulators rather than enhanced N acquisition due to microbial N fixation18,19,20,21. Also there is no clear evidence as to how N is transferred from endophytic diazotrophs to the host plant. It has been suggested that N transfer to the host is via amino acids, ammonia, small organic molecules or mineralization of dead bacterial cells22. Whether N is actually transferred from endophytic bacteria to plant host has proven difficult to resolve. The question of the mechanism of transfer of N from endophyte to host plant is an important gap in our knowledge. One possible mechanism for N transfer to host is that plants may scavenge organic nitrogen by oxidation and degradation of endophytic bacteria or their proteins using reactive oxygen species (ROS) to lyse cells and denature proteins. This mechanism has been termed ‘oxidative nitrogen scavenging'4,23. The experiments reported here were done to evaluate whether 15N incorporated into bacterial endophyte biomolecules such as proteins and nucleic acids could be traced into plant molecules; and whether the transfer process involves evidence of oxidative degradation of microbes.
Results
15N2 gas assimilation into seedlings
Shoots of seedlings grown in 15N2-enriched air showed higher δ 15N vs air content (54.93 ± 5.65 δ 15N vs (‰); mean ± standard error of mean) than shoots from seedlings grown in non-enriched air (3.07 ± 1.12 δ 15N vs (‰)). This large difference in the 15N/14N ratios between the two treatments shows that N was fixed in the seedling tissues.
Bacillus tequilensis endophyte
For use in our tests to evaluate 15N transfer/translocation from bacterial endophyte to plant, we isolated an endophyte from seeds, seedlings and young plantlets (bulbillos) of Agave tequilana Weber, and identified it as endospore-forming Bacillus tequilensis (accession number KF792125)24 (Fig. 1) using sequence data. Bacillus tequilensis was present in both roots (Fig. 1C) and shoots of the host plant.
Figure 1. Bacillus tequilensis: an endophyte of Agave tequilana.
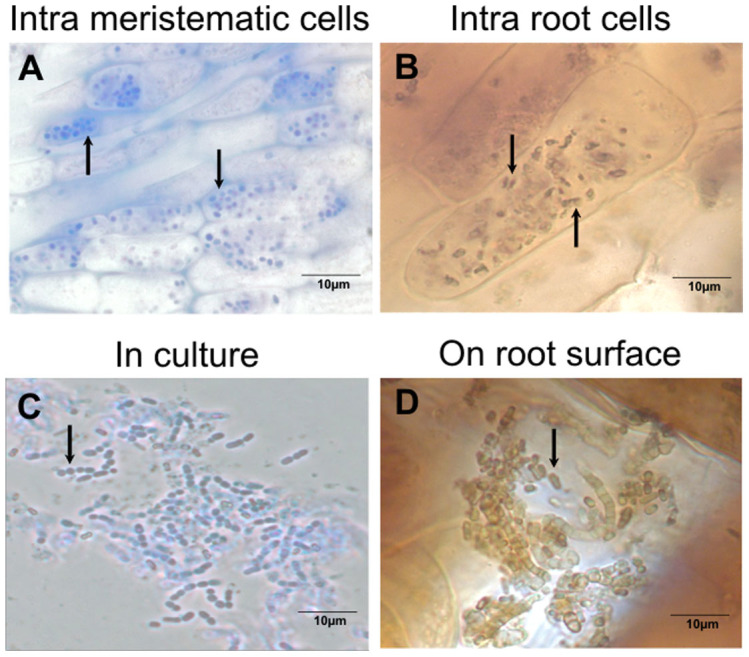
Intracellular bacteria in meristematic root cells (A) and oxidation within root cortical cell (B), detail of cell morphology of the isolate grown on TSA medium (C), and bacteria on root surface showing H2O2 concentrations (brown) around bacterial cells (D).
Visualization of bacteria in plant tissues
In seedling infection experiments using B. tequilensis we observed intracellular colonization by the bacterium into meristems and root epidermal cells of seedlings (Fig. 1A and 1B) and young plantlets. We further observed oxidation of bacterial cells within and on the surface of root epidermal cells (Fig. 1B and 1D)25.
Experiment 1
15N tracking experiments
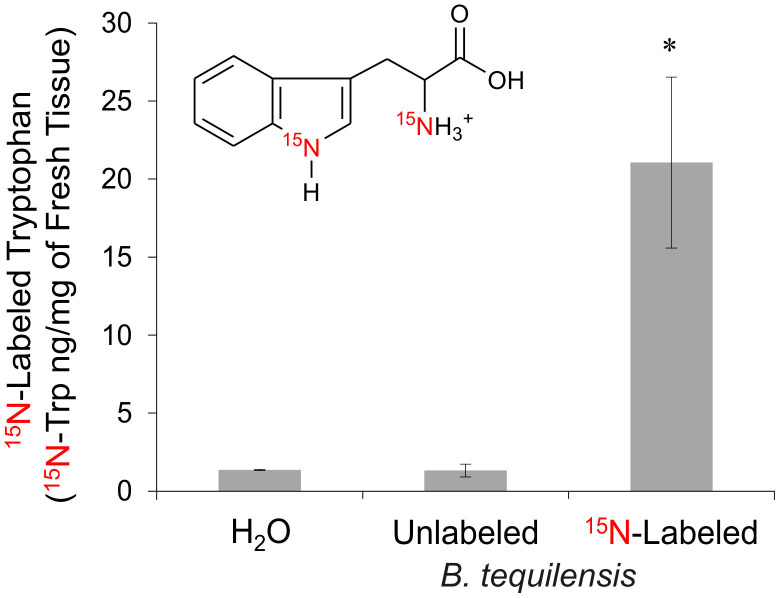
A significant difference (p<0.05) was observed in 15N-labeled tryptophan (15N-Trp) concentration between the controls (H2O and unlabeled B. tequilensis, 14N-Bteq) and plants inoculated with 15N-labeled B. tequilensis (15N-Bteq). 15N-labeled Trp in the 15N-Bteq treated plants was about 16-fold higher (21.05 ± 5.47 ng/mg) than 14N-Bteq treated (1.32 ± 0.40 ng/mg) and H2O treated plants (1.36 ng/mg) (Fig. 2, and Supplementary Fig. S5). Further quantification of 15N-Trp demonstrated that plants supplemented with 15NH4Cl had higher levels than plants supplemented with H2O or 14NH4Cl (Supplementary Fig. S1). We detected tryptophan by high-performance liquid chromatography coupled to mass spectrometry in tandem (HPLC-MS/MS), although this amino acid was not quantified (Supplementary Fig. S4)26,27,28.
Figure 2. Quantification of 15N-Trp in foliar tissue of A. tequilana using HPLC-MS/MS.
The sample groups analyzed were H2O treated, unlabeled B. tequilensis (14N-Bteq) and 15N-labeled B. tequilensis (15N-Bteq). Nitrogen content was calculated as the quantity of 15N-Trp (ng/mg). Data are the mean values ± standard error of the mean from three independent experiments. (*)15N-labeled B. tequilensis data are significantly different when compared with the H2O and unlabeled B. tequilensis groups (p<0.05). For multiple comparisons, t-test was applied.
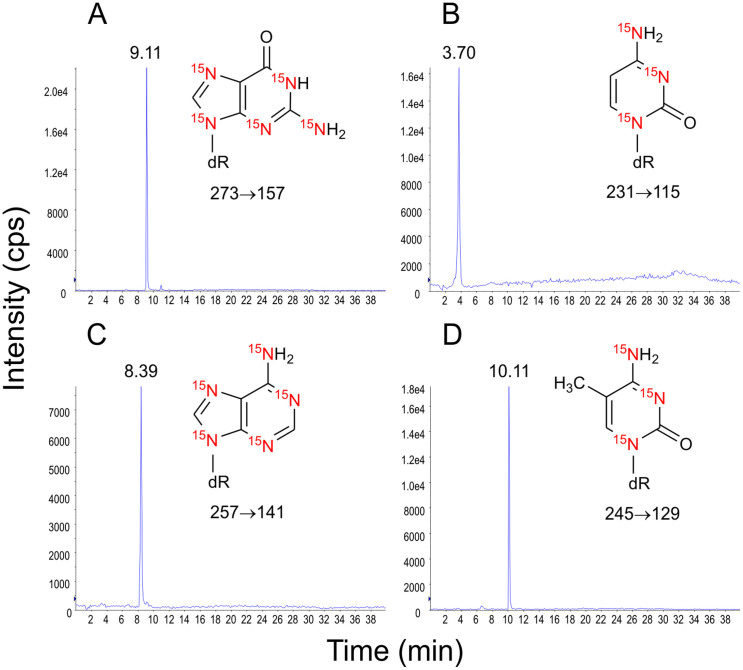
Nitrogen incorporation into tissues of A. tequilana was also confirmed by HPLC-MS/MS detection of 15N-labeled nucleosides (Fig. 3). Additionally, the same 15N-labeled nucleosides were detected from A. tequilana supplemented with 15N-labeled NH4Cl (Supplementary Fig. S9). Detection of 2′-deoxynucleosides methylated as 5-methyl-2′-deoxycytidine (15N3- MedC) and N′-methyl-2′-deoxyadenosine (15N5-MedA) was also observed in plants of A. tequilana supplemented with 15N-labeled B. tequilensis or 15N-labeled NH4Cl (Supplementary Fig. S10 and 11).
Figure 3. HPLC-MS/MS analysis of 15N-labeled 2′-deoxynucleosides from A. tequilana supplemented with 15N-labeled B. tequilensis.
2′-Deoxynucleosides were detected by the loss of the 2-deoxyribose moiety: 15N5-dG, m/z 273→157 (A), 15N3-dC, m/z 231→115 (B), 15N5-dA, m/z 257→141 (C) and 15N2-dT, m/z 245→129 (D).
Pheophytin analysis
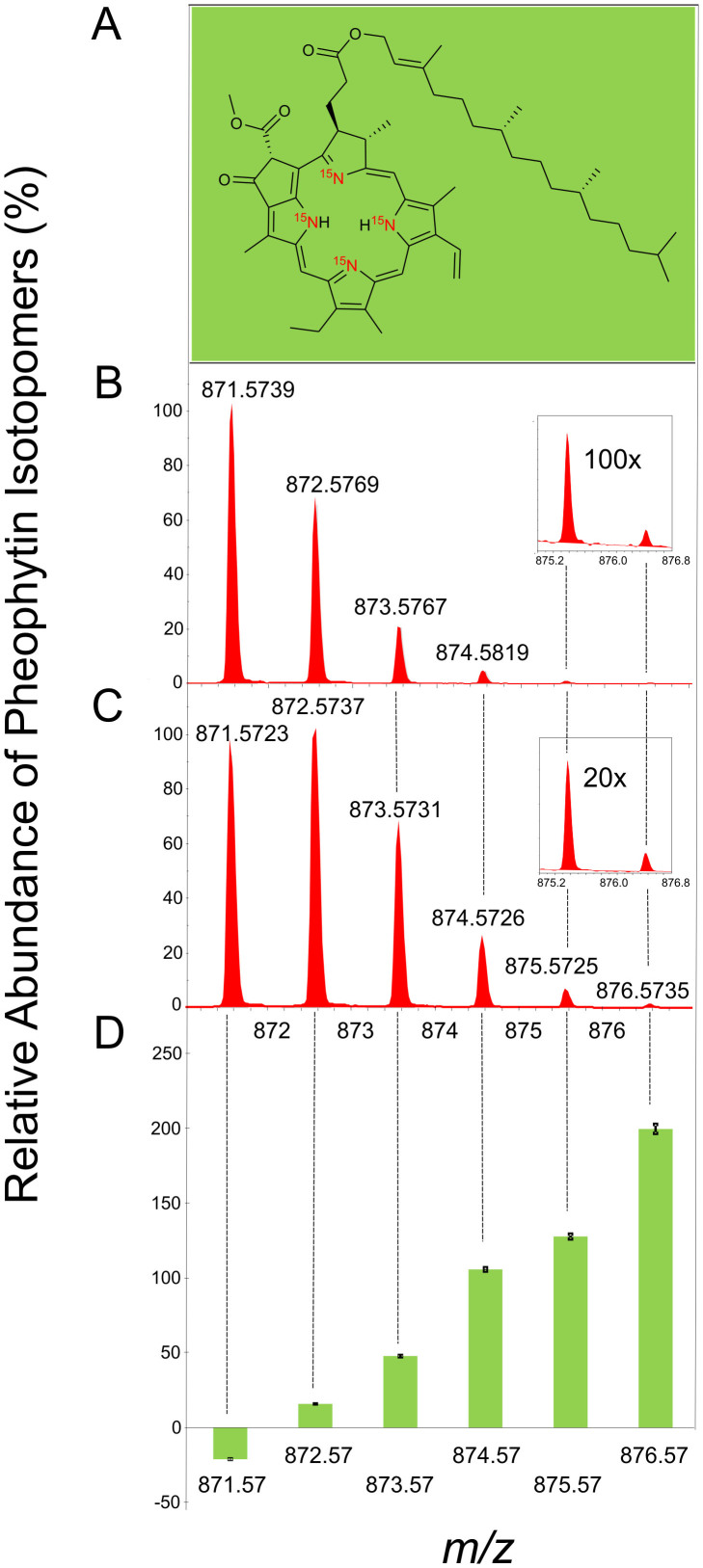
Analysis of unlabeled pheophytin a (Fig. 4B)26,27 showed a distribution of isotope peaks in agreement with the theoretical values of this molecular formula (C55H74N4O5, [M+H]+ = 871.5731) containing a base peak with m/z 871.5739 (Fig. 4B). Incubation of A. tequilana with the 15N-labeled B. tequilensis resulted in incorporation of 15N into the isotopomers of pheophytin. The relative abundance of the isotopomers m/z 872.57, 873.57, 874.57, 875.57, 876.57 increase by 16, 48, 106, 128 and 200% respectively (p<0.08; p<0.02; p<0.05; p<0.01 and p<0.003, respectively for m/z 871.57, 872.57, 873.57, 874.57 and 875.57, comparing 14N and 15N-labeled B. tequilensis groups, Fig. 4D and inset 4B and 4C). The 15N uptake to form the isotopomer m/z 876.57 pheophytin was notably greater, than that of unlabeled leaves, attesting to the incorporation of 15N atoms into the tetrapyrrole ring of the pheophytin molecule.
Figure 4. Relative abundance of pheophytin isotopomers from A. tequilensis.
(A) Chemical structure; (B) Percentage increase of pheophytin isotopomers calculated from plants supplemented with unlabeled B. tequilensis (C) and with 15N-labeled B. tequilensis (D). The data points shown are the mean values ± standard error of the mean from three independent experiments. p<0.08; p<0.02; p<0.05; p<0.01 and p<0.003, respectively for m/z 871.57, 872.57, 873.57, 874.57 and 875.57, comparing 14N and 15N-labeled B. tequilensis groups.
Experiment 2
Biomass increases in plants
In this experiment we treated plants with live or heat-killed 15N-labeled Bacillus (121°C, 10 min); and with MMN solution (see methods), non-endophytic E. coli (not labeled) and water as controls. Biomass measurements after two months of treatment showed that the greatest biomass increase was seen in plants treated with living B. tequilensis, where mean biomass increase was 1.63 ± 0.28 g (mean ± standard deviation). Plants treated with heat-killed B. tequilensis or mineral nutrient solution (MMN) showed biomass increases of 0.83 ± 0.16 g for heat-killed B. tequilensis; and 0.52 ± 0.34 g for MMN. Plants treated with non-endophytic Escherichia coli showed a biomass increase of 0.71 ± 0.14 g. Plants treated only with water showed the least biomass increase (0.15 ± 0.05 g).
Comparison of 15N absorption from living verses dead B. tequilensis
To evaluate the extent to which 15N was moving into plant tissues after the death of bacteria in soil or through endophytic colonization, we included in the experiment living and heat-killed B. tequilensis that had been labeled with 15N. On analysis we found that incorporation of 15N into nucleosides, 2′-deoxycytidine (15N3-dC) and 2′-deoxyadenosine (15N5-dA) was significantly greater (p<0.05) in plants treated with live Bacillus (a.u.: 15N3-dC 1.13 × 102 ± 13.93; 15N5-dA 4.50 × 101 ± 9.05) than plants treated with the heat-killed Bacillus (a.u.: 15N3-dC 3.40 × 101 ± 26.51; 15N5-dA 1.10 × 101 ± 6.83)28,29,30,31,32.
Discussion
The assimilation of 15N2 gas into tissues of Agave seedlings is indicative of the presence of nitrogen-fixing microorganisms within seedling tissues. From seedlings, seeds and bulbillos, we consistently isolated Bacillus tequilensis; however, plants may also have contained other non-cultured bacteria that could have been responsible for N fixation in plant tissues. We do not consider B. tequilensis to be responsible for N2 fixation in plant tissues; however, because of its ease of culturing and its endophytic nature, we employed B. tequilensis in experiments to evaluate the hypothesis that nitrogenous nutrients may flow from bacterial endophyte populations to plants.
We labeled proteins and nucleic acids of B. tequilensis with 15N; then inoculated plants with suspensions of bacteria. Using these bacteria we conducted two experiments. In the first experiment we watered plants weekly with a 4 mL suspension of 80•106 CFU mL−1 of 15N-labeled Bacillus for a six-month period. Our results show that plants incubated with the bacterial endophyte came to contain N that was originally contained within the bacterium. Detection of 15N-labeled tryptophan and 2′-deoxynucleosides suggests that 15N-labeled bacterial endophytes enter into or otherwise associate with plant tissues. Because both bacteria and plants possess tryptophan and 2′-deoxynucleosides, with this data alone we cannot evaluate transfer of nitrogen from bacterium to plant. However, we were able to detect the 15N in pheophytin a, a molecule derived from plant chlorophylls. Because chlorophyll is unique to the plant cells, the presence of the 15N label there is a definitive confirmation of transference of N from bacterium to plant tissues.
A second experiment was conducted to evaluate whether soil absorption from dead bacteria could account for some 15N movement into plant tissues, we developed an experiment in which we treated plants with live or heat-killed 15N-labeled bacteria (121°C, 10 min); and with MMN solution, non-endophytic E. coli (not labeled) and water as controls. Plants were watered weekly for a two-month period using 4 mL of bacterial suspension at a concentration of 80•106 CFU mL−1or MMN solution. In this experiment we found significantly more incorporation of 15N into 2′-deoxynucleosides in plants treated with live Bacillus than in plants treated with heat-killed Bacillus. We further evaluated the growth of these plants compared with those treated with water, E. coli or mineral nutrient solution. The amount of total acquired biomass was almost double in plants treated with living Bacillus than in plants treated with heat-killed Bacillus and E. coli; and more than 3.3 times that seen in plants treated with mineral solution. Enhanced growth of plants, and incorporation of the 15N label into plants treated with living 15N-labeled bacteria are results that are consistent with a scenario where N is transferred to plants from bacteria within tissues of plants rather than being absorbed primarily from dead bacteria in the soil. The fact that non-endophytic E. coli-treated plants demonstrated a biomass increase half that of living endophyte cultures suggests that efficient movement of N from bacteria to plants is a function of living, plant-colonizing, endophytic bacteria. The results of our experiments suggest that some nitrogen may come from decomposition of microbes in the soil. Decomposition of microbes in the soil could explain biomass increases in plants treated with heat-killed B. tequilensis and non-endophytic E. coli.
Microscopic observations of plant roots treated with the living Bacillus and stained using a reactive oxygen probe showed that degrading bacterial cells were associated with reactive oxygen and were often internalized into plant roots. Our observations are similar to those of Paungfoo-Lonhienne et al.3, where microbes were shown to enter into root cells of tomato and Arabidopsis where they were degraded. The involvement of reactive oxygen in procurement of organic nutrients from bacteria is consistent with ‘oxidative nitrogen scavenging', where reactive oxygen and proteases may be involved in nutrient extraction from bacteria4,33. We did not visualize bacteria or their degradation in shoot tissues; however, this is likely due to failure of the aqueous stains to penetrate into the shoot, rather than their absence from shoots.
In a recent study that evaluated the contribution of organic N in wheat from direct microorganism consumption against nitrate, L-alanine and L-tetraalanine absorption, it was found that plants absorbed nutrients through endocytosis of soil microbes, but the N obtained through this process was up to two orders of magnitude slower than other forms of organic and inorganic N in the soil5. Our study examining nutrient transfer from B. tequilensis to Agave suggests a process where endophytic bacterial degradation may supply N for plant growth; however, this mechanism may not be rapid and it could depend on occurrence of N deficiency or other nutrient depletion in soils. Our initial experiment to evaluate movement of 15N into plant molecules lasted six months. We do not know the minimum amount of time or precise plant growth conditions needed to see movement of N from bacterial endophytes to host plants. We propose that efficiency of this mode of nutrition depends on the particular host and microbe association. Agave tequilana is a desert plant adapted to growth under low N soil conditions and B. tequilensis is a native endophytic microbe of that plant. Internal colonization of plant tissues may increase the probability of transference of nutrients from endophytes to host plants. In this respect endophytic microbes that fix N may represent a nutritional resource that may be tapped into when soil N is limiting. Equally important to plants could be N derived from soil microbes that grow and obtain nutrients in the soil then colonize growing plants where they may be degraded. Whether N derived from microbes is a significant source of N for plants is a question that requires additional investigation. While our experiments on N transfer from endophyte to host in Agave are not exhaustive, they do provide further evidence that plants may obtain N through degradation of symbiotic microbes3,4,5,33.
Methods
Plant materials
For experiments we used one-yr-old seeds and asexual plantlets (bulbillos) derived from plants of Agave tequilana Weber that were originally collected on an A. tequilana plantation near Atotonilco el Alto, Jalisco, Mexico at coordinates 20°34′22.71″N, 102°32′00.85, 1900masl.
15N2 gas assimilation experiment with seedlings
An experiment was conducted to evaluate whether endophytic microbes fix nitrogen within intact plant tissues. In this experiment seeds were surface disinfected in 3% sodium hypochlorite for 20 min with constant agitation to remove external bacteria, then rinsed three times using sterile water. Five seeds were plated onto 0.7% agarose media in each of eight Petri dishes. Four Petri plates were placed in a 1-liter gas chamber in which the air was enriched with 33 mL of 15N2 gas. The other four Petri dishes were placed in a chamber in which the air was not enriched with 15N2. Both chambers were placed under fluorescent lighting with alternating light/dark periods (10 hr/14 hr) at laboratory ambient temperature for 21 days. After incubation shoots were excised from roots, washed to remove any superficial bacteria, then dried for 14 hr in an oven at 60°C. All shoots from a plate were combined to ensure sufficient material for analysis. For mass-spectroscopic 14N/15N ratio analysis, we sent 0.9–1.0 g of dried shoot material to the Stable Isotope/Soil Biology Laboratory at the Odum School of Ecology at the University of Georgia, Athens, USA.
Isolation of bacteria from A. tequilana seeds
Seeds were surface sterilized to remove epiphytic microbes. In this process seeds were immersed in a 3% hypochlorite solution for twenty minutes with constant agitation. The seeds were then rinsed with sterile distilled water and placed in 85% ethyl alcohol for ten minutes, followed by three rinses with sterile distilled water. To confirm the disinfection process, aliquots of the sterile water used in the final rinse were plated in tryptic soy agar (TSA; Difco, Sparks, MD, USA) and incubated at 28°C for 15 days, after which plates were examined for the presence of microorganism growth. To isolate bacteria, we used two procedures: 1) 30 sterilized seeds were placed onto 15 cm diameter Petri plates containing potato dextrose agar (PDA, Difco, Sparks, MD); and 2) Ten seeds were ground with 5 mL of aqueous solution (0.9% NaCl) using a sterile mortar and pestle and the seed extract was plated on TSA plates with different volumes (50 to 350 microliters per plate). All plates were incubated at 28°C for seven days until cream-colored colonies appeared.
Identification of bacterium by 16S rRNA and sequencing
For bacterial identification, 980 base pairs of the 16S rDNA region were sequenced24. Sequences were compared to sequences available in the NCBI GenBank database to identify the closest matches. We submitted a representative sequence to NCBI (GenBank accession number KF792125).
Experiment 1
Plant growth conditions
To reactivate bulbillos of A. tequilana collected from the field, plants were grown in sterile soil in a growth chamber at 27°C with 50% humidity under 14 h light and 10 h dark photoperiods and lamp intensities of 80 Watts/m2 for three weeks. Then plantlets were rinsed with sterile water to eliminate organic soil on roots and transferred immediately to N-free sand (30 g per glass bottle) for another three weeks maintained under the same conditions.
Visualization of bacterial oxidation in A. tequilana seedlings and bulbillos
To visualize bacterial oxidation in plant tissues, we stained seedlings and bulbillos with DAB/horseradish peroxidase for a 6–12 h period. We then excised seedling roots and shoots and placed them on a slide containing 0.1% aqueous toluidine blue. The DAB/peroxidase probe was prepared with a 5 mL solution of 100 mM potassium phosphate buffer, pH 6.9, 2.5 mM diaminobenzidine tetrachloride and 5-purpurogallin units/mL of horseradish peroxidase (Type VI, Sigma, St. Louis, MO, USA)25,33. Slides were examined using bright field microscopy on a Zeiss Axioskope® with a Spot Insight™ 4 megapixel digital camera.
Bacterial growth and [15N]-labeling
Bacillus tequilensis was grown and maintained in TSA. To evaluate transferral of organic N from bacteria to plants under limited nutrient conditions, we labeled B. tequilensis using 14NH4Cl or 15NH4Cl as the sole N sources in minimal medium M9 and incubated for 18 h at 200 rpm. All bacteria cultured with 15N contained nucleic acids labeled with isotopic N (Supplementary Fig. S8). 15N-Labeling of Bacillus tequilensis was confirmed by reversed phase HPLC-MS analysis of DNA bases (Supplementary Fig. S8). The HPLC-MS results showed complete 15N-labeling of nitrogen atoms of purine and pyrimidine bases through detection of 15N-labeled guanine at m/z 157 (Supplementary Fig. S8B), 15N-labeled cytosine at m/z 115 (Supplementary Fig. S8C), 15N-labeled adenine at m/z 157 (Supplementary Fig. S8D) and 15N-labeled thymine at m/z 129 (Supplementary Fig. S8E). Agave plantlets growing in sterile sand (SiO2) were inoculated with 15N-labeled B. tequilensis (15N-Bteq) or unlabeled B. tequilensis (14N-Bteq). Four mL of bacteria adjusted to 1 at OD600 (equivalent to 80•106 CFU mL−1) were administered to plants every week for six months. Controls included plants treated with H2O, 14N and 15N MMN solution.
Plant treatments and inoculation with bacteria
The plants used in this study were treated with 4 mL of: 1) water, 2) unlabeled MMN medium containing NH4Cl (14NH4Cl), 3) labeled MMN medium containing 15NH4Cl (15NH4Cl), 4) unlabeled bacteria suspension (14N-Bteq) or 5.) labeled bacteria suspension (15N-Bteq). NH4Cl medium or 15NH4Cl medium was prepared with 0.5 g/L 14NH4Cl or 15NH4Cl and 0.05 g/L CaCl2, 0.025 g/L NaCl, 0.05 g/L KH2PO4, 0.15 g/L MgSO4.7H2O, 1 mg/L FeCl3.6H2O, 5 g/L glucose monohydrate and 10 mL of trace elements solution. Trace element solution contained 3.728 g/L KCl, 1.546 g/L H3BO3, 0.845 g/L MnSO4.H2O, 0.05 g/L ZnSO4.7H2O, 0.0125 g/L CuSO4 and 0.05 g/L (NH4)6Mo7O24.4H2O. Bacillus tequilensis cells were grown for 18 h at 37°C in minimal medium with 14N and 15N as NH4Cl, pelleted by centrifugation (5,000 × gravity, 20 min, 4°C), and washed three times in sterilized glucose solution (0.08%). Four-mL of labeled or unlabeled bacterial cells (see above section ‘Bacterial growth and 15N labeling') were adjusted to 1 at OD600 (equivalent to 80•106 CFU mL−1) and used to inoculate each plant of A. tequilana maintained in sterile sand (30 g/dry wt). Additional treatments included plants watered with a mineral solution (50% MMN) supplemented with isotopic or non-isotopic NH4Cl; and plants watered with sterilized distilled water. The plants were watered once a week with 4 mL of each treatment during a six-month period. All plants were approximately 10–12 cm in height with 1 or 2 open leaves. Plants were grown in glass bottles in a growth chamber at 27°C day/night temperature, 14 h photoperiod.
Analysis of plant for 15N incorporation
The central leaf ‘cogollo' and new leaves of B. tequilensis-inoculated plants were taken and the presence of 15N-labeled tryptophan (15N-Trp) was quantified by High-Performance Liquid Chromatography coupled to Mass Spectrometry in tandem (HPLC-MS/MS). To reduce the chances that the 15N-labeled tryptophan was from the living bacterium rather than the host plant, the extraction was carried out seven days after inoculation with 15N-labeled bacteria. The detection and quantification of 15N-Trp in A. tequilana was performed by HPLC-MS/MS, in the Selected Reaction Monitoring (SRM) mode (m/z 207→189) (Supplementary Fig. S2–S4). The concentration of 15 N-Trp was determined in the samples through a calibration curve with standard solution of 15 N-Trp and melatonin D3-labeled (Mel-D3), as internal standard (Supplementary Fig. S5–S7)28.
Protein extraction from A. tequilana
See Supplementary Note 1.
HPLC-MS/MS analysis of tryptophan
See Supplementary Note 2.
Extraction of pheophytin from A. tequilana
To confirm that N transferred from microbes to plant tissue, an analysis in high-resolution mass spectrometry was carried out to evaluate the relative abundance of isotopomers of pheophytin (Fig. 4). Pheophytin is a magnesium-free derivative of chlorophyll, which has advantages over chlorophylls for isotopic analysis because of its stability, simpler mass spectra and better ionization characteristics. Pheophytin was extracted following the method of Perkins and Roberts26,27, with some modifications. Briefly, 0.5 g fresh leaves were frozen in liquid N2 and ground to a fine powder using mortar and pestle. For pheophytin extraction equal volumes of 85% acetone in 1 mL water, and ethyl ether were added and centrifuged at 10, 000 rpm for 10 min. The ether phase was collected and the procedure was repeated two times. Chlorophyll was converted to pheophytin by adding 10 µL diluted 6 M HCl to the ether extract. The excess of HCl was removed by adding 500 µL of water and centrifuging at 10, 000 rpm for 10 min, three times. The ether phase was collected, dried and prepared for HPLC/MS analysis by adding methanol to obtain a final sample concentration of 1 mg/mL.
HPLC/MS analysis of pheophytin
Pheophytin samples were analyzed using a MicroTOFQ-II mass spectrometer (Bruker) coupled to a Shimadzu HPLC system (Tokyo, Japan) with two pumps LC-20AD, automatic injector SIL-20A, column oven CTO-20A, UV detector SPD-20A and controller CBM-10A. A column Phenomenex Luna 5 µm (PFP2 150 × 2 mm, 100A particle size) was used and chromatography was performed with a flux of 200 µL/min using acetonitrile:H2O (+ 0.1% formic acid) as mobile phase in a gradient of 0 to 5 min 60% of acetonitrile, from 5 to 30 min 60 to 100% of acetonitrile. The column oven was kept in 40°C, UV detector was recorded at 400 and 600 nm. The mass spectrophotometer was operating in electrospray positive mode, with a nebulization and drying gas at 4 Bar and 8 L/min, respectively. Capillar voltage was set to 4500 V and drying temperature in 200°C. Collision cell and quadrupole energy were set to 20 eV and 10 eV, respectively. The molecular formula of pheophytin a is C55H74N4O5 and the base peak was detected at [M+H]+ = 872.5731.
Extraction of DNA from A. tequilana
DNA extraction from A. tequilana leaves was made after six months of treatments as described above. Freshly collected leaves (1 g) were ground to powder in liquid nitrogen and DNA was extracted with 1 mL of 50 mM extraction buffer (pH 8.0) containing 2% CTAB (p/v), 1.4 M NaCl, 100 mM Tris-HCl, 20 mM EDTA and 0.2% (v/v) β-mercaptoethanol. The solution was kept at 60°C, 800 × r.p.m for 30 minutes. Following, eppendorf tubes were left at room temperature and 500 µl of chloroform: isoamyl alcohol (24:1 v/v, −20°C) was added. The solution was maintained under agitation (10 minutes, 300 × r.p.m), centrifuged (12, 000 × r.p.m, 10 min, room temperature) and the organic phase discarded. RNAse A was added (in Tris 50 mM, pH 8.0, final concentration: 0.1 mg/mL) and the solution was incubated at 37°C for 30 minutes and agitated at 300 × r.p.m. Following this, isopropyl alcohol (−20°C) was added and the mixture was gently shaken and centrifuged (20 minute, 13,000 × r.p.m). This procedure was repeated twice. The mixture was washed with 60% isopropyl alcohol (−20°C) twice, centrifuged (5 min, 3,000 × r.p.m) and supernatant discarded. The DNA pellet was washed twice with 500 µL of 70% ethanol (−20°C), centrifuged, dried on speed vac and re-suspended in 0.1 mM desferroxamine. DNA concentration was measured spectrophotometrically at 260 nm.
Enzymatic hydrolysis of DNA from A. tequilana
See Supplementary Note 3.
HPLC-MS/MS analysis of DNA from A. tequilana
HPLC-MS/MS analysis was performed using an Agilent HPLC (1200 series, Agilent Waldbronn, Germany) coupled to a linear ion trap mass spectrometer (4000 QTRAP mass spectrometer, Applied Biosystems, Foster City, CA, USA) with electrospray ionization source. The column oven and auto sampler temperatures were set at 25°C and 4°C, respectively. For the separation, a reversed phase column was used [(C18(2)-HST Luna, Phenomenex, 100 mm × 2.0 mm, 2.5 μm particle size)]. Flow rate was set at 0.2 mL/min. Gradient elution was carried out with 0.1% formic acid (A) and acetonitrile:0.1% formic acid (8:2, v/v) (B). The separation was conducted with 0 to 40% B during the first 15 minutes, 40% B for 10 minutes, 40% to 0% B for 1 minute and 0% B for 35 minutes. MS/MS spectrometry analysis was performed in positive ionization mode and using SRM mode for each 2′-deoxynucleoside that corresponded to the loss of 2-deoxyribose moiety: 2′-deoxyguanosine (15N5-dG, m/z 273→157), 2′-deoxycytidine (15N3-dC, m/z 231→115), 2′-deoxyadenosine (15N5-dA, m/z 257→141), 2′-deoxythymidine (15N2-dT, m/z 245→129), 5-methyl-2′-deoxycytidine (15N3- MedC, m/z 245→129) and N′-methyl-2′-deoxyadenosine (15N5-MedA, m/z 271→155) (Fig. 3 and Supplementary Fig. S9 to 11)29,30,31,32. Mass spectrometry analyses were performed with the following parameters: collision excitation potential, 10 V; collision activated dissociation gas flow, medium; pause time, 5 ms; dwell time, 300 ms; curtain gas, 12 psi; ion source, 5500 V; temperature, 650°C; gas 1 and gas 2, 40 psi; declustering potential, 31 V and entrance potential, 10 V.
Analysis of 15N3-dC and 15N5-dA extracted from DNA of plants treated for two months with live or heat-killed 15N-labeled bacteria was performed as described above.
HPLC-MS analysis of DNA from bacteria
See Supplementary Note 4.
Experiment 2
Plant mass accumulation study
To evaluate whether soil absorption from dead bacteria could account for some 15N movement into plant tissues, we developed an experiment in which we treated plants (six replicates per treatment) over a two month period with live or heat-killed 15N-labeled B. tequilensis (heated to 121°C, 10 min), and with 50% MMN solution, non-endophytic E. coli (not labeled) and water as controls. Bacteria were applied as described in Experiment 1. To assess increase of 15N in leaves of Agave plants treated with 15N-labeled bacteria, deoxynucleosides were extracted from leaves and 2′-deoxycytidine and 2′-deoxyadenosine were measured. Measurements of plant mass with water-washed roots were made before and after two months of treatments. Biomass increase is the change in whole plant wet weight as a result of the treatments.
Statistical analyses
Statistical tests were performed with Origin (version 8.0) and GraphPad PRISM (version 5.0) programs. Significant differences were determined by t-test and one-way ANOVA (applying Dunnett's post-test) with the level of significance set at P < 0.05 for 15N-Trp quantification, detection of nucleosides (15N3-dC and 15N5-dA) and biomass increase, respectively.
Author Contributions
M.J.B.-G., P.D.M. & J.F.W. developed the concept of the experiments and analyses. M.J.B.-G., F.M.P., K.R.P. and M.S.T. implemented experiments. F.M.P., K.R.P., M.H.G.M., L.F.Y. & M.J.K. conducted biochemical analyses. M.S.T. isolated and identified bacteria. All authors contributed to data interpretation and writing of the manuscript.
Supplementary Material
SUPPLEMENTARY INFORMATION Nitrogen acquisition in Agave tequilana from degradation of endophytic bacteria
Acknowledgments
The authors acknowledge the research funding institutions FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo; No. 2012/12663-1, 2011/10048-5 and 2009/51850-9), CNPq (Conselho Nacional para o Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), PRONEX/FINEP (Programa de Apoio aos Núcleos de Excelência), PRPUSP (Pro-Reitoria de Pesquisa da Universidade de São Paulo), Instituto do Milênio-Redoxoma (No. 420011/2005-6), INCT Redoxoma (FAPESP/CNPq/CAPES; No. 573530/2008-4), NAP Redoxoma (PRPUSP; No. 2011.1.9352.1.8), CEPID Redoxoma (FAPESP; No. 2013/07937-8), John Simon Guggenheim Memorial Foundation (P.D.M. Fellowship), the John E. and Christina C. Craighead Foundation, USDA-NIFA Multistate Project W3147, the New Jersey Agricultural Experiment Station, National Council of Science and Technology of Mexico and the Program of Estancias Sabaticas y Posdoctorales para la Consolidacion de Grupos de Investigacion (fellowship number 186241), Proyectos de Desarrollo Cientifico para atender Problemas Nacionales (CONACYT 212875) and Project 207400 of Bilateral Cooperation México-Brazil funded by CONACYT and CNPq (Brazil, No. 490440/2013-4). We also acknowledge Cristobal Fonseca-Sepulveda for conducting nitrogen fixation tests on B. tequilensis, and Joan W. Bennett for reviewing an early draft of this manuscript.
References
- James E. K. Nitrogen fixation in endophytic and associative symbiosis. Field Crop Res. 65, 197–209 (2000). [Google Scholar]
- Reinhold-Hurek B. & Hurek T. Living inside plants: bacterial endophytes. Curr. Opin. Plant Biol. 4, 435–443 (2011). [DOI] [PubMed] [Google Scholar]
- Paungfoo-Lonhienne C. et al. Turning the Table: Plants Consume Microbes as a Source of Nutrients. PLoS ONE 5, e11915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. F. et al. A proposed mechanism for nitrogen acquisition by grass seedlings through oxidation of symbiotic bacteria. Symbiosis 57, 161–171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill P. W., Marsden K. A. & Jones D. L. How significant to plant N nutrition is the direct consumption of soil microbes by roots? New Phytol. 199, 948–955 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J. P. & Hart S. C. Competition for nitrogen between plants and soil micro-organisms. Trends Ecol. Evol. 12, 139–143 (1997). [DOI] [PubMed] [Google Scholar]
- Lawlor D. W., Lemaire G. & Gastal F. [Nitrogen, plant growth and crop yield]. Plant Nitrogen. [Lea, P. J. & Morot-Gaudry, J. F. (eds.)] [343–367](Springer-Verlag, Berlin 2001). [Google Scholar]
- Cleveland C. et al. Global patterns of terrestrial biological nitrogen (N) fixation in natural ecosystems. Global Biogeochem. Cycles 13, 623–645 (1999). [Google Scholar]
- Herridge D. F., Peoples M. B. & Boddey R. M. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311, 1–18 (2008). [Google Scholar]
- Mulvaney L. R., Khan S. A. & Elisworth T. R. Synthetic nitrogen fertilizers deplete soil nitrogen: A global dilema for sustainable cereal production. J. Environ. Qual. 38, 2295–2314 (2009). [DOI] [PubMed] [Google Scholar]
- Garnett T., Conn V. & Kaiser B. N. Root based approaches to improving nitrogen use efficiency in plants. Plant Cell Environ. 32, 1272–3040 (2009). [DOI] [PubMed] [Google Scholar]
- Smith B. E. Nitrogenase reveals its inner secrets. Science 297, 1654–1700 (2002). [DOI] [PubMed] [Google Scholar]
- Paungfoo-Lonhienne C. et al. Plants can use protein as a nitrogen source without assistance from other organisms. Proc. Natl. Acad. Sci. USA 105, 4524–4529 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paungfoo-Lonhienne C. et al. DNA is taken up by root hairs and pollen, and stimulates root and pollen tube growth. Plant Physiol. 153, 799–805 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell M. et al. Oligopeptides Represent a Preferred Source of Organic N Uptake: A Global Phenomenon? Ecosystems 16, 133–145 (2013). [Google Scholar]
- Adlassnig W. et al. Endocytotic uptake of nutrients in carnivorous plants. Plant J. 71, 303–313 (2012). [DOI] [PubMed] [Google Scholar]
- Behie S. W., Zelisko P. M. & Bidochka M. J. Endophytic insect-parasitic fungi translocate nitrogen directly from insects to plants. Science 336, 1576–1577 (2012). [DOI] [PubMed] [Google Scholar]
- Sturz A. V., Christie B. R. & Nowak J. Bacterial endophytes: potential role in developing sustainable systems of crop production. Crit. Rev. Plant Sci. 19, 1–30 (2000). [Google Scholar]
- Anand R. & Chanway C. P. Detection of GFP-labeled Paenibacillus polymyxa in auto-flourescing pine seedling tissues. Biol. Fertil. Soils 49, 111–118 (2013). [Google Scholar]
- Gyaneshwar P., James E. K., Reddy P. M. & Ladha J. K. Herbaspirillum colonization increase growth and nitrogen accumulation in aluminium tolerant rice varieties. New Phytol. 154,131–145 (2002). [Google Scholar]
- Patten C. L. & Glick B. R. Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microb. 42, 207–22 (1996). [DOI] [PubMed] [Google Scholar]
- Baset Mia M. A. & Shamsuddin Z. H. Nitrogen fixation and transportation by rhizobacteria: a scenario of rice and banana. Int. J. Botany. 6, 235–242 (2010). [Google Scholar]
- White J. F., Johnson H., Torres M. S. & Irizarry I. Nutritional endosymbiotic systems in plants: bacteria function like ‘quasi-organelles' to convert atmospheric nitrogen into plant nutrients. J. Plant Pathol. Microb. 3, e104 (2012). [Google Scholar]
- Baker G. C., Smith J. J. & Cowan D. A. Review and re-analysis of domain-specific 16S primers. J. Microb. Methods 55, 541–555 (2003). [DOI] [PubMed] [Google Scholar]
- Munkres K. D. Histochemical detection of superoxide radicals and hydrogen peroxide by Age-1 mutants of Neurospora. Fungal Genet. Newsletter 37, 24–25 (1990). [Google Scholar]
- Kahn M. L. et al. A mass spectroscopy method for measuring 15N incorporation into pheophytin. Analyt. Biochem. 307, 219–225 (2002). [DOI] [PubMed] [Google Scholar]
- Perkins H. J. & Roberts D. W. A. Purification of chlorophylls, pheophytins and pheophorbides for specific activity determinations. Biochim. Biophys. Acta 58, 486–498 (1962). [DOI] [PubMed] [Google Scholar]
- Almeida E. A. et al. Synthesis of internal labeled standards of melatonin and its metabolite N1-acetyl-N2-formyl-5-methoxykynurenin for their quantification using an on-line liquid chromatography-electrospray tandem mass spectrometry system. J. Pineal Res 36, 64–71 (2004). [DOI] [PubMed] [Google Scholar]
- Hua Y. et al. Comparison of negative and positive ion electrospray tandem mass spectrometry for the liquid chromatography tandem mass spectrometry analysis of oxidized deoxynucleosides. J. Am. Soc. Mass Spec. 12, 80–87 (2001). [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F. & Ashapkin V. V. DNA methylation in higher plants: past, present and future. Biochim. Biophys. Acta, Gene Regul. Mech. 1809, 360–368 (2011). [DOI] [PubMed] [Google Scholar]
- Barrientos Y. E.,Wrobel K. W., Torres A. L., Corona F. G. & Wrobel K. Application of reversed-phase high-performance liquid chromatography with fluorimetric detection for simultaneous assessment of global DNA and total RNA methylation in Lepidium sativum: effect of plant exposure to Cd(II) and Se(IV). Anal. Bioanal. Chem. 405, 2397–2404 (2013). [DOI] [PubMed] [Google Scholar]
- Charles M. P. et al. N-6-methyideoxyadenosine, a nucleoside commonly found in prokaryotes, induces C2C12 myogenic differentiation. Biochem. Biophys. Res. Commun. 314, 2476–482 (2004). [DOI] [PubMed] [Google Scholar]
- White J. F. et al. Hydrogen peroxide staining to visualize bacterial infections of seedling root cells. Microsc. Res. Techniq. 10.1002/jemt.22375 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY INFORMATION Nitrogen acquisition in Agave tequilana from degradation of endophytic bacteria