Abstract

Changing ocean health and the potential impact on marine mammal health are gaining global attention. Direct health assessments of wild marine mammals, however, is inherently difficult. Breath analysis metabolomics is a very attractive assessment tool due to its noninvasive nature, but it is analytically challenging. It has never been attempted in cetaceans for comprehensive metabolite profiling. We have developed a method to reproducibly sample breath from small cetaceans, specifically Atlantic bottlenose dolphins (Tursiops truncatus). We describe the analysis workflow to profile exhaled breath metabolites and provide here a first library of volatile and nonvolatile compounds in cetacean exhaled breath. The described analytical methodology enabled us to document baseline compounds in exhaled breath of healthy animals and to study changes in metabolic content of dolphin breath with regard to a variety of factors. The method of breath analysis may provide a very valuable tool in future wildlife conservation efforts as well as deepen our understanding of marine mammals biology and physiology.
Cetacean species are long-lived, feed high in the food chain, and have a blubber layer that acts as an energy store as well as can store chemical and toxins that may be concentrated up through the food chain and environment. Due to these traits, their health is often reflective of their ecosystem.1 Some cetacean populations, including Alaska’s Cook Inlet stock of beluga whales, Southern Resident killer whales, and North Atlantic right whales, have become depleted over the years. Understanding baseline health and causes of disease in cetaceans may help to reverse occurring negative health trends. Health assessments of dolphin populations are especially important during unusual mortality events or other die offs occurring due to known or suspected emerging infectious diseases, contaminations, harmful algal bloom-associated biotoxins, or sudden changes in prey.2 Unfortunately, close and routine monitoring of cetaceans, especially large cetaceans, in their natural habitat can be fundamentally challenging. Robust health assessments often require the collection of biological samples that are difficult to acquire, such as skin biopsies or blood samples. It would be of great benefit for conservation efforts, as well as for monitoring the health status of managed cetacean populations, to develop a minimally invasive and diagnostically useful test to assess the health of evasive marine mammal species. For humans, breath analysis has been explored as a noninvasive alternative to traditional point-of-care testing. Prior studies in humans have sought to identify and link relevant biomarker compounds in the complex mixture of exhaled metabolites to specific health states, thus providing a noninvasive “window” into the physiological state of the organism.3,4 The main challenges associated with breath analysis are great variability and low abundance of exhaled metabolites. Consequently, to a large extent, the advances in the breath analysis field are contingent on advances in analytical chemistry.
Certain anatomic features of cetaceans may make breath analysis methods particularly suitable. In terrestrial mammals including humans, the nasal cavity and the mouth meet at the pharynx at the back of the nose and mouth. The pharynx is part of the digestive system as well as the respiratory system because it carries both food and air. The two systems share a common pathway allowing for incidental contamination of the breath chemicals by odors from the digestive system. Cetaceans, however, have separated digestive and pulmonary systems (the larynx extends up into the nasal cavity rather than opening into the throat). Further, the cetacean trachea is short, with a large diameter to allow small cetaceans like Tursiops to be explosive breathers.5 These animals exchange 70–90% of total lung capacity in 0.3 s,6 with peak air flow of 24 L per second, depending on the animal’s body size.6,7 This breathing behavior also leads to rapid gas exchange, which makes cetaceans ideal candidates for breath metabolomic studies. Furthermore, dolphins’ lungs contain significantly more alveoli than human lungs do. While most mammals have only one capillary layer, dolphin lungs are made up of two layers of capillaries, thereby increasing the efficiency of gas exchange. This double layer of capillaries means the surface area of the lungs is greatly increased and gas exchange can occur more quickly, potentially resulting in a higher content of metabolites that partition from the blood into the exhaled breath compared to terrestrial mammals. All of the above factors may lead to enhanced excretion of various compounds that can be potentially elucidated for diagnostic purposes or facilitate biomarker discovery. Breath analysis has been previously indicated to be potentially useful in physiological studies of bottlenose dolphins and California sea lions (Zalophus californianus), including measurement of oxygen (O2) consumption and carbon dioxide (CO2) production during dives.8 While these studies measured several fractions of gaseous components, they did not have the technology required to measure the comprehensive metabolite components present in breath.
Human breath is known to contain trace amounts of volatile organic compounds (VOCs) such as acetone, methanol, ethanol, aldehydes, and alkanes, typically in parts-per-billion (ppb) or parts-per-trillion (ppt) molar fraction ranges.4 Nonvolatile compounds such as lipids and proteins/peptides, as well as virus particles or entire cells such as bacteria or epithelial cells, may be also present in breath aerosol droplets carried in the exhaled breath stream. Because these aerosols are likely generated from the liquid lining of the lung, it is thought that these compounds represent a surrogate profile of the bloodstream. Hundreds of different endogenous and exogenous chemicals have been identified in the exhaled breath of humans.3 Diet, activity, environmental air/water quality, and health status can all leave a “mark” in exhaled breath. Thus, breath analysis provides the opportunity to simultaneously view physiological interactions between an organism and its environment.
Currently, very little knowledge exists on the baseline breath metabolite composition of marine mammals; however, several studies have shown that cetacean breath potentially has great diagnostic value. Previous breath collection devices relied simply on placing a surface such as glass beaker, nylon gauze, or plastic sheet in the stream of blow and collecting a small fraction of the exhalate. Even such a simple approach allowed for detection of DNA,9 hormones,10 and various bacteria11,12 in the cetacean blow. Detection of volatiles has also been reported for whale breath using sorbent.13 These methods cannot accommodate the collection of a higher volume of exhalate required to detect and quantify various low abundance trace metabolites. In the present work, we develop analytical methodology to efficiently trap cetacean exhaled breath and to lay the groundwork for application of breath analysis for marine mammal health diagnostics and conservation efforts. Integrating veterinary information with information gathered using the new technology developed here will further aid in assessing an animal’s health status. Furthermore, the potential of discovery of breath-based metabolite biomarkers in cetaceans may provide valuable insight for relevant indicators of health in human breath and help to better understand their metabolic origin. Here, we present a method and application that we believe will have an impact in not only analytical chemistry but also other disciplines including marine biology, ecology, and physiology.
Experimental Section
Sampling Device
We have developed an exhaled breath condensate (EBC) collection device specifically adapted for marine mammal anatomy and physiology (Figure 1a). The dimensions of the device were as follows: the length of the condenser tube, 37 in. (94 cm); the inner diameter of the condenser tube, 1 in. (2.54 cm); the outer diameter, 1–1/4 in. (3.18 cm); the length of the insulated housing, 36 in. (91.5 cm), the length of the ice bath section inside the housing, 32 in. (81.3 cm). The collection tube was made of borosilicate glass. The material used for the outer casing was plastic (PVC), and the intake manifold parts were made of PTFE. A standard soft rubber surgical mask was placed on the manifold and was the only part of the device that came into contact with the animal (each animal had an individual mask; the masks were degassed for several days in a vacuum oven at 50 °C prior to use). For EBC sample collection, a glass tube is placed inside the plastic casing and the space between the glass tube and casing is filled with crushed dry ice (Figure 1a). During exhalation, the expired breath passes through the chilled tube, while during inhalation a one way valve closes and allows incoming ambient air to bypass the chilled tube. This design is intended to prevent exposure to the animals from extreme temperature differences in inhaled air temperatures. After collection, the device is disassembled and condensed breath is removed from the tube using a plunger and placed in a vial using a scoopula (if frozen) or pipette (if thawed). In order to limit cross-contamination, the device was thoroughly cleaned in between collections. Each individual part that was in contact with the EBC sample or animal besides the glass tube was thoroughly rinsed with DI water, submerged in a 70% ethanol/water solution bath and soaked for ∼10 min, and then allowed to air-dry until complete solvent evaporation. The glass tube was also rinsed with DI water followed by a rinse with 70% ethanol and then thoroughly wiped with Kimwipe tissues before the next use. Brand new borosilicate vials with stainless steel silicon septa caps were used for sample storage. When possible, additional sample thawing and/or transfer between sample collection and analysis were avoided (e.g., the following headspace sampling or sample lyophilization occurred directly in the sample vial). Sea water and air condensate blanks were collected to account for possible environmental contaminants presence (Figures A3 and A4 in the Supporting Information). For the water blank, an aliquot of seawater in the general vicinity of the collection area was placed in the vial. For an air blank, the cleaned collection tube was chilled with the dry ice and exposed to open air in the general vicinity of the collection area for approximately 5 min. The condensate in the form of powdery snow was then scraped from the surface and placed in the vial. The blanks samples were sealed and stored at −80 °C until analysis similarly to the EBC samples.
Figure 1.
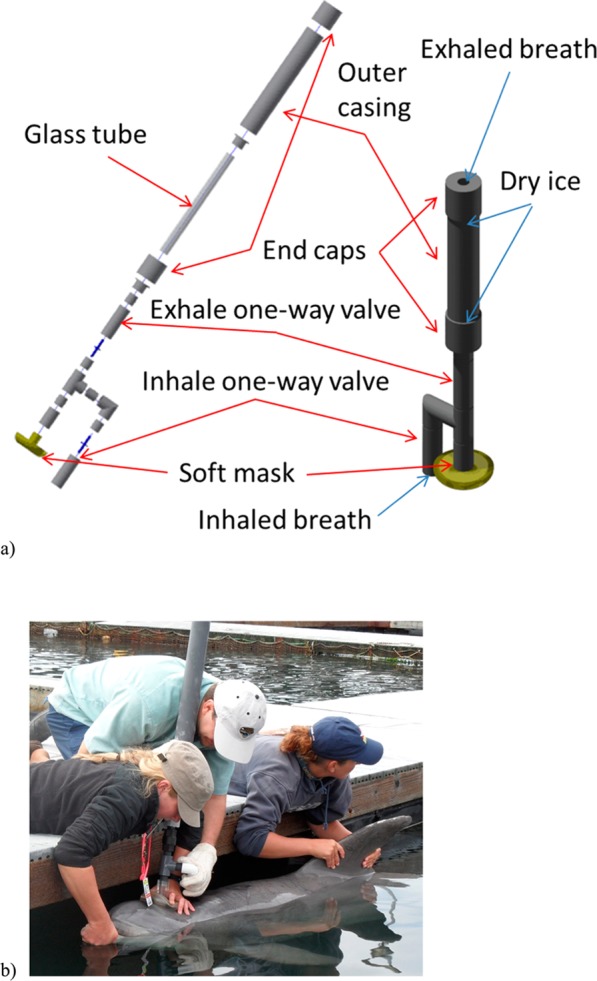
(a) Schematic view of the breath sampling device. Left: the exploded view of the device assembly. Right: the assembled device (not to scale). The space between the glass tube and outer casing is filled with cooling material (dry ice pellets), and the tube is locked in place with end-caps. For breath collection, the soft mask on the bottom of the device is placed around a dolphin’s blowhole. When the animal inhales, the inhale one-way valve opens, while the exhale valve closes and air is routed from the intake; when the animal exhales, the inhale valve closes, the exhale valve opens, and exhaled air is expelled through the chilled glass tube out of the exhaust. (b) Sampling breath from a trained dolphin.
EBC Sample Collection from Managed Animals under Human Care
An EBC sample collection with managed animals was carried out with the U.S. Navy Marine Mammal Program in San Diego, CA. Prior to the studies, several samples were collected and their chemical content was evaluated using GC/MS in order to establish further study feasibility and sample quality. In addition to preliminary feasibility studies, both a small-scale study (May 2011) and larger-scale study (December 2011) were conducted. For the small study, a total of 9 EBC samples was collected from 3 animals (3 from nonfasting, 6 from fasting animals: 2 males/1 female). For the large scale study, 46 EBC samples were collected from 13 animals: 6 males, 7 females. We collected samples from these animals on 5 consecutive days, both fasting and nonfasting. In total, 19 samples were collected from fasting animals (6 from males, 13 from females) and 27 from nonfasting animals (12 from males, 15 from females). For nonfasting animals, the samples were collected on average 3 h after a feeding of 1.4 kg of capelin, therefore likely catching the end of the postprandial phase. Some of the samples were collected from animals with identified health conditions.
For the collection, animals breathed normally while positioned by trainers at the water’s surface (Figure 1b). Some of the trained animals were asked to beach themselves onto a padded mat and had the sample collected outside of water. The collection device was positioned over the animal’s blowhole and held for the duration of the sampling (5 min or 10–20 breaths, depending on the animal). All but one dolphin breathed normally during the breath collection procedure. In the latter case, the dolphin had a pre-existing respiratory condition causing more rapid and shallow breaths. Immediately after collection, samples were placed on dry ice and later stored in a −80 °C freezer. The collected samples were further routed for mass spectrometry (MS) analysis as described below.
An additional set of experiments was conducted to establish the effect of rebreathing valves on metabolic content. We have determined that removing the valves and holding the device directly above the animal’s blow hole during exhalation did not result in any changes of metabolic content, although the collected volumes were typically smaller. The collection mode without valves was employed for the EBC collection from a wild population (described in the following section).
EBC Sample Collection from Wild Dolphins
The EBC collection device was utilized for collecting samples from the long-term resident wild dolphin population in Sarasota Bay, FL as part of a bottlenose dolphin health assessment effort conducted by the Chicago Zoological Society’s Sarasota Dolphin Research Program under National Marine Fisheries Service Scientific Research Permit No. 15543. The device was taken on a boat during the dolphin capture and release exercises, and the EBC samples were collected in a course of several days as a part of large suite of samples and measurements comprising a comprehensive panel of health assessment tests.2 Upon capture, each animal was transferred onto the padded and shaded deck of a specially designed veterinary examination boat for ∼40–50 min. The EBC samples were collected within that time frame when instructed by the project leader and veterinarians. The federal permit under which the studies were carried out did not allow for any objects touching the blowhole of a dolphin, as this might elicit a negative response from wild animals. Therefore, the EBC collection device without a rebreathing valve was utilized (the one way valve assembly was removed and a soft gas mask was placed directly on the tube inlet). The EBC collection device was then held immediately above the animal’s blowhole without contact. Ten breaths were collected for each animal (when possible) in about ∼2 to 10 min time frame. Since in this collection mode some of the breath may escape, the collected volumes were typically smaller compared to those for the managed population animals and varied in the range of ∼100–400 μL depending on the animal’s size and behavior during collection. During examinations and sampling, veterinary staff members were constantly assessing the animal’s condition and making decisions regarding whether some tests would be performed. As a result, some of the animals were released without collecting EBC samples. In a few cases, animals exhibited hyperventilation with small, shallow breaths. For these animals, smaller amounts of EBC were procured. In addition, several animals were moved from the deck back into the water to complete sampling. The EBC collection was allowed to be carried out directly in the water with the animal being held with its blowhole above the water by team members and the collection device was held above the blowhole.
In total, 21 samples were collected:12 samples in May 2011 and 9 samples in May 2012. Of these, 2 animals produced very small samples due to hyperventilation. Both males (13) and females (8) ranging in age from 2 to 41 years were tested. Individual health status varied as well, with some animals exhibiting symptoms of infection (e.g., brownish-colored discharge accumulated around the blowhole, breath malodors). Upon collection, all of the samples were immediately placed in dry ice and later stored in a −80 °C freezer for chemical analysis.
Gas Chromatography/Mass Spectrometry Analysis of Volatile Fraction
The volatile organic compound (VOC) chemical content of the samples collected from dolphins was studied using gas chromatography/mass spectrometry (GC/MS). For the analysis, a Varian 3800 GC with a 4000 Ion Trap MS (Varian, Walnut Creek, CA) with an electron ionization (EI) source and a VF-5 ms 5% phenol/95% PDMS GC column (Varian, Walnut Creek, CA) instrument was used. An aliquot of thawed sample was placed in a vial to be tested for VOC content in the headspace. The volatiles were sampled from the headspace using carboxen/polydimethylsiloxane (CAR/PDMS) d.f. 75 μm partially cross-linked (black hub) solid phase microextraction (SPME) fibers (Supelco, St. Louis, MO) to preconcentrate the compounds prior to measurement. An optimal sample volume that allows for good S/N while requiring minimal amount of sample was determined by performing experiments with a series of aliquots, ranging from 300 to 50 μL of sample. The minimum required volume was found to be 100 μL, while an optimal volume was found to be 500 μL. When available, 500 μL of each sample was used for analysis. For VOC extraction enhancement, 0.5 mL of saturated NaCl solution was added to the EBC.
For sample extraction, a borosilicate vial sample containing an EBC sample was agitated at 90 °C and the volatile chemicals contained in the EBC were collected on the SPME fiber. After sampling, the SPME was inserted into the heated GC inlet and the adsorbed/absorbed chemicals were desorbed and injected into the GC column. The GC column oven cycle was optimized to allow optimal peak separation for benchmark human EBC samples. The protocol was then adjusted for the dolphin breath samples based on the obtained results. The scanned m/z range was 35–600 Th. The ion source and detector were switched off for the first 4 min of the GC protocol to avoid detector saturation during elution of the carbon dioxide peak, since a large amount of carbon dioxide was trapped in the sample due to the high content in dolphin breath, as well as possible contamination from the dry ice collection tube chilling material. The obtained GC/MS data were then analyzed offline.
The chemicals in the sample were identified on the basis of electron ionization (EI) MS fragmentation data. Compound identification was carried out as follows; the GC peak of interest was selected, and the corresponding mass spectrum was matched against existing database entries. Mass Spectral Search Software v. 2.0 with NIST 2005 and Wiley 2009 MS libraries was used. If the match score exceeded 80%, the match was presumed to be correct. Otherwise, the list of the potential compounds was reviewed and the most likely tentative candidate(s) was selected on the basis of fragmentation patterns. If the match score for the best match was below 20%, the compound was presumed unidentified and not included into the list of detected metabolites. An example of fragmentation pattern matching is shown (Figure A1b) in the Supporting Information.
Additional GC/MS experiments were conducted using chemical derivatization of the sample at the Genome Center of University of California, Davis. A 675 μL aliquot of the sample was lyophilized and derivatized with methoxylamine hydrochloride (MeOx) solution in pyridine and N-methyl-N-trimethylsily trifluoroacetamide (MSTFA). The analyte was then directly injected into a 6890 GC (Agilent Technologies, Santa Clara, CA) equipped with a cryo-cooled injection system inlet (CIS4, GERSTEL, Inc.) interfaced to the Pegasus IV time-of-flight mass spectrometer (Leco, St. Joseph, MI) operated in the 1D reflectron mode. Chromatographic separation was performed on an Rtx-5SilMS column with a 10 m integrated guard column [95% dimethyl/5% diphenyl polysiloxane film; 30 m × 0.25 mm (inside diameter) × 0.25 μm d.f. (Restek, Bellefonte, PA)]. The GC oven temperature program was as follows: initial temperature of 45 °C with a 2 min hold followed by a 20 °C/min ramp up to 300 °C with a 2 min hold followed by a 20 °C/min ramp up to 330 °C with a 0.5 min hold. As expected, less volatile compounds such as carbonic acids were detected in this method (Table A1 in the Supporting Information).
Liquid Chromatography/Mass Spectrometry Analysis of Nonvolatile Fraction
For liquid chromatography/mass spectrometry (LC/MS) analysis, the 0.1 mL aliquot of sample (upon availability) was lyophilized and then redissolved in 100 μL of 90% acetonitrile in water. Five μL of resuspended sample was then injected for analysis. For the in-depth MS/MS analysis, 1 mL of total volume was pooled from multiple animals’ samples. The CUDA (12-[[(cyclohexylamino)carbonyl]amino]dodecanoic acid) in methanol/toluene, 9:1 v/v internal standard, was used for quality control and to assess reproducibility. Chromatography was performed on an Agilent 1200 Series high-performance liquid chromatography (HPLC) system (Agilent Technologies, Santa Clara, CA). Samples were housed in an autosampler maintained at 4 °C, and material was separated on a Kinetex 2.6 μm (HILIC) 100 Å HPLC column (150 mm × 2.10 mm) (Phenomenex, Torrance, CA), held at 40 °C during analysis. Mobile phase A consisted of water. Mobile phase B was 90% acetonitrile in water. Ammonium acetate and acetic acid were added to each to obtain a pH of 5. Starting mobile phase composition was 100% B; over 15 min, the flow of mobile phase B was decreased to 45% and replaced by mobile phase A. The flow rate was held constant at 0.35 mL/min over this time. LC eluents were analyzed with an Agilent 6530 accurate-mass Q-TOF mass spectrometer (Santa Clara, CA) equipped with an Agilent Jet Stream ESI source in positive and negative ion modes. The mass range was set to 60–1700 Thomson (m/z); scan rate was 4 spectra/second, and sample analysis time was 21 min per sample. Fragmentor voltage was 120 V; sheath gas flow was 11 L/min, and sheath gas temp was 350 °C. An additional experiment was conducted with the reverse phase Waters Acquity CSH C18 column 1.7 μm, UHPLC (2.1 × 100 mm) (Milford, MA USA) (column for broader metabolite coverage). For this mode, the CUDA (12-[[(cyclohexylamino)carbonyl]amino]dodecanoic acid) in methanol/toluene, 9:1 v/v internal standard, was also added for quality control and to assess reproducibility. The samples were separated on the column held at 65 °C during analysis. Mobile phase A consisted of 60% acetonitrile in water. Mobile phase B was 10% acetonitrile in isopropanol. Formic acid and ammonium formate were added to make the final concentration of each mobile phase 10 mM for both formic acid and ammonium formate. Mobile phase composition at time (minutes) 0 was 15% B, at time 4 was 30% B, at time 5 was 48% B, at time 22 was 82% B, and at time 23 was 99% B and held for 1 min before the column was re-equilibrated. Flow rate was 0.6 mL/min. The entire sample was dried down and then resuspended in 100 μL of 9:1 methanol/toluene. Three μL of sample was then injected. Samples were analyzed by MS and identified by MS/MS spectra using an Agilent 6530 accurate mass LC-QTOF in positive ionization mode. The mass range was 60–1700 Thomson (m/z); the scan rate was 2 spectra/second, and sample analysis time was 30 min per sample. Fragmentor voltage was set at 120 V; sheath gas flow was 11 L/min, and sheath gas temperature was 350 °C. For both analyses modes, compound annotations were performed by comparing sample MSMS spectra to NIST and Metlin libraries, using the software MSpepSearch and NISTMS.
Chemical identification was carried out automatically by MS/MS fragmentation of the parent ion from analytes in the LC eluent with the following matching of MS/MS patterns to METLIN and RESPECT mass spectral libraries with a 3 mTh window. For some of the selected compounds of interest for which MS/MS data were not obtained, tentative identification based on exact mass was carried out using the METLIN library. Automatic software matching may result in incorrect identification for some metabolites leading to repeating entries for identified compounds at different retention times. In order to establish the correct structures in each case, confirmation of identified compounds through use of chemical standards is necessary. Further tentative identification was carried out by manually selecting the best match. The manually identified compounds are listed in Table A13 in the Supporting Information.
Statistical Analysis
The LC/MS raw data files were first processed with the “Find By Molecular” feature in Agilent’s Mass Hunter Qualitative Analysis B.05.00SP1 software in order to deconvolve each peak. The deconvolved chromatograms were then exported to .cef data format, and the peaks were aligned using Mass Profiler Professional 12.1 software. The alignment window was set at 0.5 min. The files were then put to recursive analysis using the “Find by Formula” feature of the Mass Hunter Qualitative Analysis B.05.00SP1 software. In the compiled peak tables, the minimum intensity value of each individual peak over all the samples was found, and each of those values was divided by three. This was done in order to reduce the impact of zero values in PCA and PLS. Captive and wild dolphin data were treated as a single data set for deconvolution and alignment. All of the peaks that were present in any of the blanks were removed from the table. In order to remove any spurious peaks, a “70% filter” was applied: peaks which were present in less than 70% of the samples in one group and more than 20% in another were removed. Furthermore, peaks that were present in less than 20% of samples in both groups were also removed.
After preprocessing, the obtained peak tables data were analyzed using partial least square discriminant analysis (PLSDA).14 Partial least-squares (PLS) regression is a multivariate latent-variable method that relates one dependent variable or predictand (y) to a set of independent variables, or predictors, X. Partial least-squares discriminant analysis (PLS-DA) is the application of PLS to the classification of problems in which y is a vector that codifies the class of each sample (male/female, fasting/nonfasting, or managed/wild animals). The class label of an unknown sample is decided from the y-value predicted by the PLS model. Ideally, the predicted y should be close to the coded class values, such as organized in the range rather than stochastically assigned. In practice, it is a real number and different approaches can be used to convert the predicted y into a class label.
Results
We collected and analyzed exhaled breath from managed (San Diego Bay, CA USA) and wild (Sarasota Bay, FL USA) bottlenose dolphin (Tursiops truncatus) populations, as described in the Experimental Section. The design of the condenser was found to allow for minimal restriction to exhaled breath thus allowing animals to breathe comfortably while collecting a sufficient amount of exhalate. The breath collection process was found to be very efficient for dolphins, and the target sample volume of ∼0.5 mL could be collected from trained animals in under 5 min (approximately 10 full breaths from an animal). Prior to further large-scale sample collection, the chemical content of the samples collected from trained animals was analyzed using GC/MS to confirm that the collection method allows capturing metabolites in exhaled breath. Breath samples from three animals (one female, two males) were tested. In addition, sample collection with and without the rebreathing valve was conducted on several animals to verify that the valve presence/absence does not affect the metabolomics content of the samples. The sampler performance and the sampling method were evaluated/validated by comparing the samples obtained from the same animal back-to-back and on different days, as well as for different animals. Samples with abnormally small volumes (less than ∼0.2 mL for 10 full breaths) coupled with abnormal animal behavior during collection were verified for metabolic content. GC/MS analysis was not used for quantitative measurements but only to document the compounds in the volatile fraction. For the nonvolatile fraction, the internal standard was employed and data scaled as described above to allow for semiquantitative analysis. In the protocol development study, we have determined that an excellent run-to-run reproducibility was achieved for samples from the same animal, but sample collection conditions have had pronounced effects. Examples of gas chromatography/mass spectrometry (GC/MS) and liquid chromatography/mass spectrometry (LC/MS) data, as well as a list of identified compounds, are provided in the Supporting Information (Figures A1 and A2 and Tables A1–A13); note that chemical identification assignments are putative with varying degree of certainty.
Our results indicate that, similarly to human breath, dolphin breath is a very complex mixture of various compounds, many of which are present in trace amounts. A large number of detected compounds have been reported to be present in human breath, especially volatile compounds such as various aliphatic and aromatic alcohols, hydrocarbons, and carbonyls.3 Some of the prominent compounds found in the volatile fraction of dolphin breath are small amines, such as 1-pentanamine, bis (2-hydroxypropyl) amine, and 5-(hydroxymethyl)-2-pyrrolidinone, many of which have not been reported in humans.3 The latter compounds are presumed to be responsible for the characteristic “fishy” smell of dolphin breath. Compounds detectable in nonvolatile fractions of exhaled breath condensate of bottlenose dolphins included amino acids, peptides, lipids such as steroids, phospholipids, prostaglandins, carbohydrates, and small molecules such as carbonic acids, amines, and pharmaceuticals. Due to the “explosive” breathing of cetaceans, we presume that we are able to capture many nonvolatile compounds originating from the aerosolized droplets from the deep lung alveolar space.
In addition to a large number of compounds that are obviously of biogenic origin, some compounds may be attributed to external sources. Examples of such compounds are phthalates (plasticizers); 2,4-diisocyanato-1-methyl-benzene (environmental contaminant); 2,6-bis(1,1-dimethylethyl)-2,5-cyclohexadiene-1,4-dione (known water contaminant); and some of the long-chain alcohols (potentially a water-soluble fraction of urban runoff). A list of volatile compounds identified in seawater is given in Table A3 and in ambient air condensate, in Table A4 in the Supporting Information. The proposed chemical identities are tentative, and in upcoming years, we anticipate to confirm the identities of key metabolites of interest in the database through the use of chemical standards.
As in humans,4 we observed variations in the chemical content of dolphin EBC. This may be partially attributed to the effect of the breath sampling procedure itself on chemical content, which is well documented in human studies.15 However, we also expect normal biogenic variation among animals. An example of changing GC patterns for different animals is shown in Figure A1 in the Supporting Information. To further explore links between breath chemical composition and an animal’s life history and health status, we utilized established bioinformatics approaches. Animal breath metabolome patterns were analyzed as described in the “Online Methods”. Partial least square discriminant analysis (PLS-DA)14 was applied to the data for fasting versus nonfasting animals, as well as males versus females. We have determined which compounds have had the highest contribution to discrimination analysis between the two states indicating potential metabolites of interest. In addition, the total metabolome could be represented and compared using principal component analysis (PCA). An example of PCA plot for the LC/MS data is shown for the managed dolphin population (Figure 2). In order to know how to classify different groups, PLS-DA was applied and Figure 3 shows the PLS-DA score plot of male versus female animals in San Diego Bay. The PCA plot of fasting versus nonfasting animals is also shown (Figure 4). Finally, Figure 5 shows the PCA plot for the two populations: wild animals in Sarasota Bay and managed animals in San Diego Bay.
Figure 2.
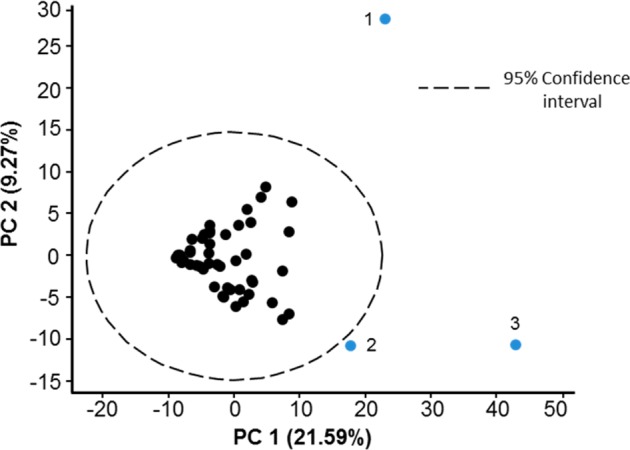
Principal component analysis (PCA) of LC/MS samples. Abnormal breathing behavioral or health issues were identified for each of the three dolphins outside of the cluster. All other dolphins were known to be healthy at the time of sample collection.
Figure 3.
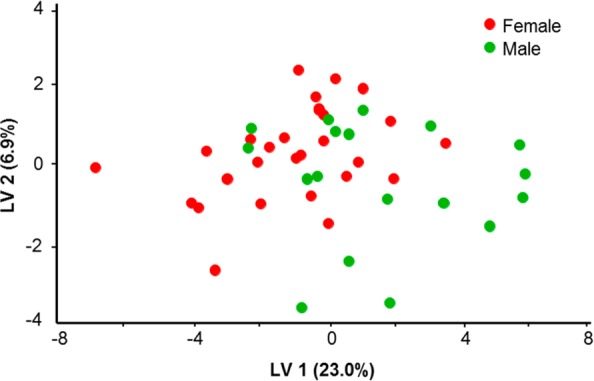
Partial least square discriminant analysis (PLS-DA) of LC/MS samples for male and female dolphins. No apparent clustering is observed on the basis of sex alone.
Figure 4.
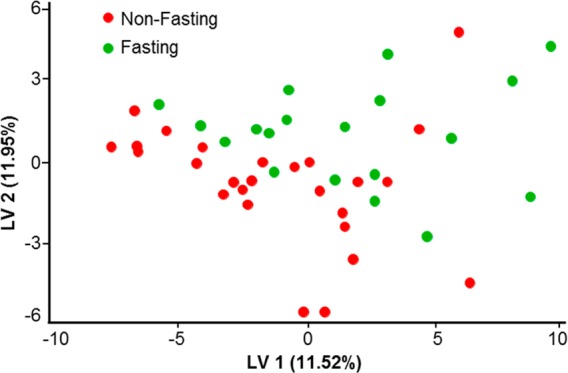
Partial least square discriminant analysis (PLS-DA) of LC/MS samples for fasting and nonfasting animals. Weak clustering is observed for the fasting status.
Figure 5.
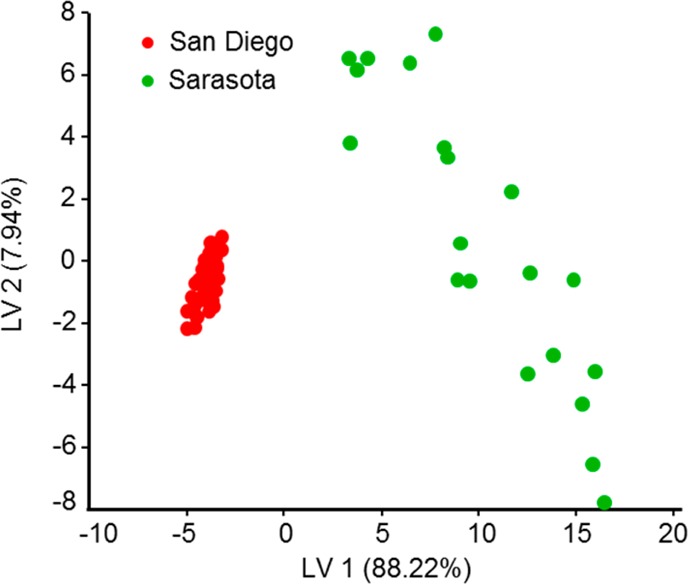
Partial least square (PLS) of LC/MS samples for wild (Sarasota Bay) and managed (San Diego Bay) populations. Apparent clustering is observed; animals in the wild population are significantly more dissimilar to each other compared to the animals in the managed population.
The metabolome comparison was carried out for the LC/MS data, as the chemical composition of the nonvolatile fraction of the sample was found to be more consistent with better day-to-day reproducibility (data not shown) than for the volatile fraction. The lower reproducibility for the volatile fraction is presumed to result from greater effects of the sampling conditions (such as ambient temperature, humidity, and wind speed) on the amount of trapped chemicals during sample collection. This variability in human studies has been also observed, and it has been previously suggested to confine EBC measurements to nonvolatile compounds16 due to this phenomenon.
Discussion
As discussed in the Results section, our data indicate that dolphin breath contains a large variety of low-abundance metabolites, many of which are common with those found in human breath.17 In humans, changes in the presence or abundance of some of these compounds have been considered as indicative of health status, e.g., linked to malignancy.18 In addition to biogenic compounds, potential environmental contaminants were also present. Such contaminants could be introduced into the EBC sample through sample handling; for example, sample contact with plastic surfaces may lead to introduction of phthalates. Thus, we took great efforts to minimize sample contact with any surface other than clean glass and conducted sample collection in a way to minimize cross-contamination, as described above. Since phthalates are easily released into the environment, an alternative introduction route into the samples may be through contamination of the animals’ habitat. As San Diego Bay and Sarasota Bay, Florida, are heavily urbanized environments, large amounts of chemical and biological waste are inevitably present in runoff waters, likely leading to contamination. As would be expected, several compounds known to be industrial environmental pollutants were found both in the water as well as the EBC (Table A3 in the Supporting Information). This is not surprising, as dolphins spend their entire lives in water and are exposed to contaminants over long durations of time. San Diego is home to several industries, a commercial airport, and high levels of commercial and recreational boat traffic. Sarasota Bay is also a highly urbanized coastal area with a high level of recreational boating. External contaminants may be absorbed through the skin as well as inhaled or ingested by dolphins, metabolized, and subsequently exhaled.
Figure 2 shows the PCA score plot for the managed population based on metabolites detected in the nonvolatile fraction of the EBC. As can be seen, several outliers are present. Interestingly, each outlier coincides with a specific health issue, including a bone lesion (animal 3) and pneumonia (upon retrospective analysis, animal 2). Animal 1, on the other hand, exhibited abnormal behavior and shallow breathing during the sample collection and was unlikely to be exhaling alveolar air. For this dolphin, a very small amount of sample was procured. Also, the sample was found to be diluted by water as it contained only a small amount of biogenic metabolites. Consequently, the animal was expected to be an outlier. The animals within the main cluster were all healthy. A future larger scale study may help to establish whether specific health status conditions (or sample collection and treatment differences) would result in distinct differentiation of animals with and without health or other issues. Upon removal of the outlier animals, no discernible clustering can be observed for the baseline healthy animals. The scatter indicates small variations in chemical composition among animals. In addition to individual differences between animals, variations in the metabolic content of exhaled breath are likely to occur due to a multitude of factors associated with a normal life cycle, including contaminants from the environment, diet, age, health status, and environment as well and many others. These factors will likely lead to fluctuations in the chemical signature of dolphin breath.19 Furthermore, changes in sampling conditions, such as ambient temperature and wind speed, may have also introduced bias into our sampling in determining concentrations of breath metabolites, especially for volatile compounds. Follow up studies using chemical standards are necessary to precisely measure abundances and changes in abundances of key metabolites of interest in EBC.
The PLS-DA score plot of males versus females (Figure 3) shows that breath metabolite differences between sexes are not immediately apparent. This suggests that either sex-specific chemicals such as hormones were present in insignificantly different amounts during the time of sampling or their relative contribution to the total metabolome is small enough to minimize group differences based on gender alone. This observation is in contrast with human studies where noticeable differences in breath composition between males and females were reported,20,21 although those studies did not comprehensively map the breath metabolome. At the same time, the PLS-DA score plot of fasting versus nonfasting animals (Figure 4) does show a weak separation. This implies that some of the differences in metabolism due to fasting status (such as glucose or lipid levels) may affect the composition of exhaled breath in dolphins. In the present study, the sample collection was carried out near the end of the postprandial phase.22 It is possible that collecting samples immediately at postprandial state may lead to better discrimination between fed and unfed animals. Metabolic differences in fasting and nonfasting status were previously investigated for humans, and subject-specific metabolite changes (specifically in lipids and amino acids) were consistently reflected in various biological fluids including breath.23 We expect that such metabolic changes would occur in dolphins as well. The most relevant biomarker discriminating fasting and nonfasting groups (base peak m/z 1144.345 Th/retention time 21.8415 min) was tentatively identified as (6Z,9Z,12Z,15Z,18Z)-3-oxotetracosapenta-6,9,12,15,18-enoyl-CoA. This compound has been putatively functionally linked to the HADHA gene, which provides instructions for making part of the mitochondrial trifunctional protein enzyme complex required to metabolize long-chain fatty acids, the major source of energy used by the heart and muscles during periods of fasting.24,25 Another compound that was found to be important for discrimination of both fasting and nonfasting female animals (base peak m/z 975.7548 Th/retention time 1.65733 min) was tentatively identified as a triglyceride.
Comparison of breath metabolite profiles for the managed (San Diego Bay, CA) and wild (Sarasota Bay, FL) dolphin populations, shown in Figure 5, indicates that apparent differences exist between these two groups. Although both populations of animals are of the same species and both originated in the Gulf of Mexico, the groups are clearly distinguishable from each other. When comparing the managed and wild populations, it is apparent that wild animals are more dissimilar to each other compared to managed population animals. A variety of factors such as geographic spread of the wild individuals may drive the difference between the two populations. However, the most likely explanation is that the managed population is maintained in a stable environment. The dolphins sampled from the managed population are fed a consistent diet composed of a predominant mixture of commercially caught capelin, herring, mackerel, and squid and are supplemented with vitamins specially formulated for marine mammals. The managed dolphins in this study were not exhibiting obvious reproductive behavior, had known health status, were housed in sea pens in the same general area each day, and received daily health checks and medical care when needed. In contrast, wild populations are subject to shifts in environmental conditions, prey species availability, and may have unidentified health conditions.26 Within the well-defined, long-term range of the resident community, wild dolphins may be exposed to differing levels of environmental conditions including runoff, algae, and natural micro bacteria populations. The wild dolphin samples included a greater variety of life history categories (for example, mother and calf were present in wild population but not in the managed population), as well as other factors (for example, sampling in the wild population occurred during the height of the reproductive season).
In summary, our results indicate that breath metabolite analysis is a promising noninvasive method of monitoring cetacean health, both in managed populations and in the wild. Due to respiratory system anatomy and breathing behavior, this method appears to be even more amenable to cetaceans than humans. This finding potentially opens up possibilities of learning from dolphin-based studies to help advance human breath research.27 Our newly developed method is intended to provide useful diagnostic information via monitoring of EBC metabolome content which can be used by veterinary personnel and conservation managers in their decision making processes. In the present study, we demonstrate how the developed methodology can be used to establish a baseline of the total metabolic “fingerprint” of an animal as reflected in exhaled breath. We also elucidated the chemical basis of specific differences in dolphin populations. As we continue to expand the information on the metabolome, the understanding of the metabolite’s origin and comparison across species can ultimately enhance our knowledge of underlying metabolic processes.
We hope the reported analytical methodology will lay the groundwork for further developments in cetacean health monitoring via exhaled breath. In the future, it will be important to establish which chemicals (biomarkers) in dolphin breath can be reliably linked to certain health conditions or exposures. Longitudinal comparison to other routinely acquired diagnostic measures (blood, urine, fecal, and ultrasound tests) would provide an excellent basis to assess the potential of dolphin breath diagnostics for “personalized medicine” approaches to managed animal care. In turn, this may enable rapid health assessment by monitoring for the presence or absence of specific biomarker compounds during routine health assessment or surveillance of wild populations. Tracking breath biomarkers may provide diagnostic information about a specific animal and also can be helpful in tracking the effects of environmental stress in marine life populations and monitoring ecosystem health and recovery progress. Further work is needed to assess the total metabolic variance over medium- and long-term studies. These longitudinal observations could also be further correlated with the existing studies in human models. When benchmarked, the proposed methods may serve as a future “gold standard” for reliable and noninvasive health status monitoring of marine mammals.
Acknowledgments
This study was supported by the Office of Naval Research (ONR) grant #N-00014-13-1-0580 [CED], under an animal care and use protocol reviewed and approved by the Navy Marine Mammal Program Institutional Animal Care and Use Committee and the Navy Bureau of Medicine, and partially by Dolphin Quest [RSW], The Hartwell Foundation [CED] and National Center for Advancing Translational Sciences (NCATS) through grant #UL1 TR000002.
Supporting Information Available
Examples of GC data for EBC of dolphin; examples of LC data for EBC of dolphin; loadings plot for the PLSDA analysis; compounds in dolphin breath identified with GC/MS using chemical derivatization; compounds in dolphin breath identified with GC/MS using head space SPME extraction; list of volatile metabolites (GC/MS) detected in seawater; list of volatile metabolites (GC/MS) in air condensate; lists of nonvolatile metabolites (LC/MS) for managed and wild population animals using HILIC and RP columns in positive and negative ion modes; manually selected best tentative matches for nonvolatile metabolites (LC/MS). This material is available free of charge via the Internet at http://pubs.acs.org/.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Bossart G. D. Vet. Pathol. 2011, 48, 676–690. [DOI] [PubMed] [Google Scholar]
- Wells R.; Rhinehart H.; Hansen L.; Sweeney J.; Townsend F.; Stone R.; Casper D. R.; Scott M.; Hohn A.; Rowles T. EcoHealth 2004, 1, 246–254. [Google Scholar]
- de Lacy Costello B.; Amann A.; Al-Kateb H.; Flynn C.; Filipiak W.; Khalid T.; Osborne D.; Ratcliffe N. M. J. Breath Res. 2014, 8, 014001. [DOI] [PubMed] [Google Scholar]
- Amann A., Smith D., Eds. Volatile Biomarkers: Non-Invasive Diagnosis in Physiology and Medicine; Elsevier: Boston, 2013; p 563. [Google Scholar]
- Kooyman G. L.; Cornell L. H. Physiol. Zool. 1981, 54, 55–61. [Google Scholar]
- Ridgway S. H.; Scronce B. L.; Kanwisher J. Science 1969, 166, 1651–1654. [DOI] [PubMed] [Google Scholar]
- Irving L.; Scholander P. F.; Grinnell S. W. J. Cell. Comp. Physiol. 1941, 17, 145–168. [Google Scholar]
- Ponganis P. J.; Kooyman G. L.; Winter L. M.; Starke L. N. J. Comp. Physiol., B 1997, 167, 9–16. [DOI] [PubMed] [Google Scholar]
- Frere C. H.; Krzyszczyk E.; Patterson E. M.; Hunter S.; Ginsburg A.; Mann J. PLoS One 2010, 5, e12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg C. J.; Rogers T. L.; Shorter A.; Barton K.; Miller P. J. O.; Nowacek D. Mar. Mammal Sci. 2009, 25, 605–618. [Google Scholar]
- Schroeder J. P.; Raverty S.; Cameron C.; Zabek E.; Eshghi A., Bain D.; Wood B., Hanson B.; Rhodes L.. Investigation into the Microbial Culture and Molecular Screening of Exhaled Breaths of Endangered Southern Resident Killer Whales (SRKW) and Pathogen Screening of the Sea-Surface Microlayer (SML) in Puget Sound. In Proceedings of the 2009 Puget Sound Georgia Basin Ecosystem Conference, Seattle, Washington USA, February, 8–11, 2009. [Google Scholar]
- Acevedo-Whitehouse K.; Rocha-Gosselin A.; Gendron D. Anim. Conserv. 2010, 13, 217–225. [Google Scholar]
- Cumeras R.; Cheung W. H. K.; Davis C. E.; Gulland F.; Goley D. Metabolites 2014, 4, 790–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhuis J. A.; Hoefsloot H. C. J.; Smit S.; Vis D. J.; Smilde A. K.; van Velzen E. J. J.; van Duijnhoven J. P. M.; van Dorsten F. A. Metabolomics 2008, 4, 81–89. [Google Scholar]
- Risby T. H.; Solga S. F. Appl. Phys. B: Lasers Opt. 2006, 85, 421–426. [Google Scholar]
- Effros R. M.; Casaburi R.; Porszasz J.; Morales E. M.; Saraswat A.; Rehan V. J. Breath Res. 2012, 6, 048001–048002. [DOI] [PubMed] [Google Scholar]
- Phillips M. Anal. Biochem. 1997, 247, 272–278. [DOI] [PubMed] [Google Scholar]
- Amann A.; Corradi M.; Mazzone P.; Mutti A. Expert Rev. Mol. Diagn. 2011, 11, 207–217. [DOI] [PubMed] [Google Scholar]
- Phillips M.; Herrera J.; Krishnan S.; Zain M.; Greenberg J.; Cataneo R. N. J. Chromatogr., B 1999, 729, 75–88. [DOI] [PubMed] [Google Scholar]
- Lechner M.; Moser B.; Niederseer D.; Karlseder A.; Holzknecht B.; Fuchs M.; Colvin S.; Tilg H.; Rieder J. Respir. Physiol. Neurobiol. 2006, 154, 478–483. [DOI] [PubMed] [Google Scholar]
- Tsang K. W.; Ip S. K.; Leung R.; Tipoe G. L.; Chan S. L.; Shum I. H.; Ip M. S.; Yan C.; Fung P. C.; Chan-Yeung M.; Lam W. Lung 2001, 179, 83–91. [DOI] [PubMed] [Google Scholar]
- Venn-Watson S.; Carlin K.; Ridgway S. Gen. Comp. Endocrinol. 2011, 170, 193–199. [DOI] [PubMed] [Google Scholar]
- Krug S.; Kastenmueller G.; Stueckler F.; Rist M. J.; Skurk T.; Sailer M.; Raffler J.; Roemisch-Margl W.; Adamski J.; Prehn C.; Frank T.; Engel K.-H.; Hofmann T.; Luy B.; Zimmermann R.; Moritz F.; Schmitt-Kopplin P.; Krumsiek J.; Kremer W.; Huber F.; Oeh U.; Theis F. J.; Szymczak W.; Hauner H.; Suhre K.; Daniel H. FASEB J. 2012, 26, 2607–2619. [DOI] [PubMed] [Google Scholar]
- Kamijo T.; Aoyama T.; Komiyama A.; Hashimoto T. Biochem. Biophys. Res. Commun. 1994, 199, 818–825. [DOI] [PubMed] [Google Scholar]
- Kamijo T.; Aoyama T.; Miyazaki J.; Hashimoto T. J. Biol. Chem. 1993, 268, 26452–26460. [PubMed] [Google Scholar]
- Wells R.; McHugh K. A.; Douglas D. C.; Shippee S.; Berens McCabe E. J.; Barros N. B.; Phillips G. T. Front. Endocrinol. 2013, 4, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schivo M.; Aksenov A. A.; Yeates L. C.; Pasamontes A.; Davis C. E. Front. Endocrinol. 2013, 4, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


