Abstract
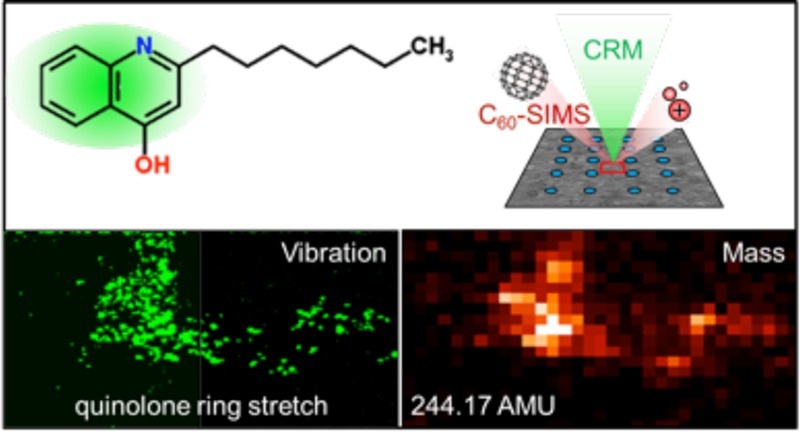
Secondary ion mass spectrometry (SIMS) and confocal Raman microscopy (CRM) are combined to analyze the chemical composition of cultured Pseudomonas aeruginosa biofilms, providing complementary chemical information for multiple analytes within the sample. Precise spatial correlation between SIMS and CRM images is achieved by applying a chemical microdroplet array to the sample surface which is used to navigate the sample, relocate regions of interest, and align image data. CRM is then employed to nondestructively detect broad molecular constituent classes—including proteins, carbohydrates, and, for the first time, quinolone signaling molecules—in Pseudomonas-derived biofilms. Subsequent SIMS imaging at the same location detects quinolone distributions in excellent agreement with the CRM, discerns multiple quinolone species which differ slightly in mass, resolves subtle differences in their distributions, and resolves ambiguous compound assignments from CRM by determining specific molecular identities via in situ tandem MS.
Biological systems are comprised of a vast, diverse assortment of chemical species, ranging in size and complexity from monatomic electrolytes up to massive biopolymers such as proteins, complex carbohydrates, and nucleic acids. Since function arises not just from what components are present in a system, but also how they are distributed spatially (and temporally), visualizing their distribution via chemical imaging is critical to gaining a comprehensive understanding of biological function. Although analyte labeling (e.g., with fluorescent dyes or radioisotopes) is one, well-established way to accomplish this, label-free imaging offers an alternative approach with several advantages: effective probes need not be developed for each analyte, nor must the system be perturbed by the introduction of exogenous compounds, and parallel imaging of multiple analytes is not limited by the number of simultaneously usable and/or detectable probes.
Mass spectrometry imaging (MSI)1−3 and confocal Raman microscopy (CRM)4−6 are two label-free molecular imaging techniques that operate on different fundamental principles. MSI detects and visualizes analyte distribution on the basis of molecular weight. This is commonly done by scanning a microprobe across the sample surface and ionizing constituents in a spatially registered fashion, which can then be analyzed, detected, and used to generate ion images or maps of relative abundance.1 A variety of MSI microprobes are available, each with unique characteristics and advantages; two of the most common are focused lasers for matrix-assisted laser desorption/ionization (MALDI),3 which provides a high upper mass range, and focused ion beams for secondary ion mass spectrometry (SIMS),7,8 which provides high spatial resolution, down to <100 nm in ideal cases.9 In contrast to mass-based detection by MSI, CRM visualizes chemical distributions based on the characteristic vibrational frequencies of different chemical functional groups. In the Raman scattering process, these vibrational frequencies shift scattered light away from the frequency of an incident laser beam by an amount that is characteristic of the functional groups present. CRM utilizes a standard confocal microscope; therefore, the lateral and axial spatial resolutions are defined by Δx = 0.61λ/NA and Δz = (2.2nλ)/(π(NA)2), respectively, where λ represents laser wavelength, NA is the numerical aperture of the microscope objective, and n is the refractive index of the medium.10,11 Image acquisition is performed by scanning the focal volume of the laser, which allows for nondestructive (and therefore potentially live sample) imaging in three dimensions at submicrometer-scale spatial resolution.6,12
Given the orthogonality of these techniques, combining MSI and CRM (or other vibrational imaging methods) for molecular imaging can be advantageous. More specifically, correlating mass and vibrational images by chemically imaging the same location using orthogonal detection modes imparts numerous benefits for biological studies, beyond their combined individual application. Molecules that do not ionize efficiently may produce a strong vibrational signature or vice versa; thus, by combining both methods, chemical coverage is expanded. One technique may be used to resolve subtly differing compounds that are indistinguishable by the other, for example structural isomers with different vibrational modes (e.g., CRM) or functionally similar molecules with slightly different masses (e.g., MSI). In cases where an analyte is mutually detectable by both CRM and MSI, observed distributions can be cross-validated;13 this is especially helpful in MSI experiments where artifacts can arise in ion images due to ion suppression14,15 or image processing.16
Several reports have illustrated the advantage of combining MSI with CRM or related vibrational imaging techniques for biological analysis, a subject we also reviewed in depth recently.17 Li et al.13 correlated CRM, SIMS, and laser desorption ionization (LDI) MSI to elucidate the subcellular localization of carbohydrates (cellulose and hemicellulose) in biofuel feedstock grass, allowing more definitive mass and vibrational assignments from mutually observed chemical features. Petit and co-workers18 demonstrated that synchrotron-Fourier transform-infrared (FT-IR) and -ultraviolet microspectroscopies and SIMS imaging could be combined for liver tissue analysis, with FT-IR used to visualize broad molecular classes (lipids, proteins, DNA, and sugars) and SIMS to resolve specific lipid species. More recently, Fagerer et al.19 combined single cell fluorescence and Raman imaging with MALDI mass spectrometry (MS) profiling of algae in order to visualize secondary metabolite production and the associated depletion of cellular adenosine triphosphate, impressively demonstrating how combining these techniques can yield a more comprehensive biological picture. Finally, a landmark-based histology registration strategy was developed for semiautomated alignment between MALDI-time-of-flight (TOF) and Raman imaging platforms by Bocklitz et al.20
Here, we present a method for correlating molecular images obtained from two label-free techniques, SIMS and CRM, and demonstrate how their complementarity can be exploited for enhanced molecular imaging of a biological sample. We applied the approach to investigate cultured bacterial biofilms of the opportunistic pathogen Pseudomonas aeruginosa. Unlike tissue sections, which are commonly used in imaging studies, bacterial biofilms do not present obvious anatomical features for image registry. Thus, we also developed a chemical microspot-based system for navigating and locating microscopic regions of interest (ROIs), a critical step in precisely correlating images acquired on a highly uniform surface using two different techniques and two separate instruments (at two institutions). The correlation of MSI and CRM data enabled us to broadly characterize the chemical composition of the biofilm microenvironment as well as specific constituent analytes including quinolones, a class of signaling molecules involved in Pseudomonas biofilm growth and maturation.
Experimental Section
A schematic representing the overall workflow of the sequential correlated imaging approach demonstrated in this work is shown in Figure 1, with procedural details described below.
Figure 1.
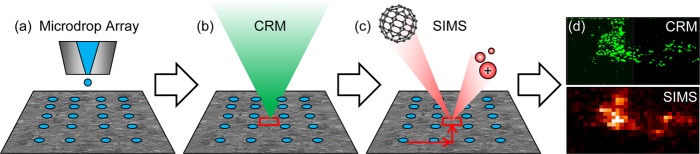
CRM/SIMS correlated imaging workflow. (a) A microdroplet array is applied to the dried biofilm. (b) CRM is performed to locate ROIs, and array coordinates are recorded. (c) The sample is transferred to the SIMS instrument, and the array is used to navigate back to the ROIs. (d) The CRM and SIMS data are correlated, using the array for alignment.
Materials and Chemicals
Silicon substrates were purchased from Silicon, Inc. (Boise, ID) as 4-in-diameter wafers of Si (100), then scored and broken into 2 × 2 cm2 tiles before use. Two quinolone standards, 2-heptyl-3-hydroxy-4(1H)-quinolone (Pseudomonas quinolone signal, PQS) and 2-heptyl-4-quinolone (HHQ), were purchased from Sigma-Aldrich (St Louis, MO) and dissolved in HPLC-grade methanol (Sigma-Aldrich), then deposited and air-dried on clean Si wafers for the SIMS and CRM measurements. The Ag nanoparticle solution (PELCO NanoXact, 50 nm, 0.02 mg/mL in 2 mM aqueous citrate) was purchased from Ted Pella, Inc. (Redding, CA) and diluted 1:1 with HPLC-grade methanol for inkjet printing.
Biofilm Preparation
Pseudomonas aeruginosa, “wild type” ATCC strain 15692 (ATTC, Manassas, VA) was used for all experiments. Cell suspensions were grown in fastidious anaerobe broth (FAB) culture medium with filter-sterilized glucose as a carbon source at 30 °C overnight. The FAB medium contained the following components: (NH4)2SO4 (2 g L–1), Na2HPO4·2H2O (6 g L–1), KH2PO4 (3 g L–1), NaCl (3 g L–1), MgCl2 (93 mg L–1), CaCl2 (11 mg L–1), and trace metals solution (1 mL L–1). The trace metals solution contained CaSO4·2H2O (200 mg L–1), FeSO4·7H2O (200 mg L–1), MnSO4·H2O (20 mg L–1), CuSO4·5H2O (20 mg L–1), ZnSO4·7H2O (20 mg L–1), CoSO4·7H2O (10 mg L–1), NaMoO4·H2O (10 mg L–1), and H3BO3 (5 mg L–1). The cell culture solution was deposited onto the silicon wafer tiles placed at the bottom of Petri dishes and additional growth culture medium added to a 50× dilution. The biofilms were allowed to grow under static conditions at 30 °C for 72 h. The culture mixture was then removed by pipet, and biofilms were permitted to air-dry fully in sterile conditions prior to microspotting and analysis.
Microdroplet Array Application
The Ag nanoparticle solution was printed onto dried biofilm surfaces with a ChIP-1000 Chemical Inkjet Printer (Shimadzu Corp., Kyoto, Japan). A single 100 pL droplet was dispensed at each position in a 500-μm pitch array across the entire biofilm surface. Visual monitoring during deposition and measurement via SIMS confirmed that droplets formed single 132 ± 9 μm (n = 6) diameter spots and did not spread on the biofilm surface. To use the array as a Cartesian coordinate grid, an origin was designated at one corner of the tile by inscribing a small unique feature into the biofilm with sharp tweezers.
CRM
Raman microscopy was performed on an Alpha 300R confocal Raman microscope (WITec GmbH, Ulm, Germany) with a 60×, NA = 1.0 coverslip-corrected water immersion objective (Nikon Corp., Tokyo, Japan) employing a frequency-doubled Nd:YAG laser (λ = 532 nm) delivered through a single-mode optical fiber, dichroic beam splitter, and focused onto the surface of the sample using a microscope objective operating in epi-illumination geometry. The backscattered radiation was transmitted through a 50-μm diameter multimode fiber to a UHTS 300 spectrometer with a 600 groove mm–1 diffraction grating and back-illuminated CCD camera cooled to −65 °C (Newton DU970 N–BV, Andor Technology Ltd., Belfast, UK). The incident laser power on the sample was adjusted to 10 mW. Raman spectra were recorded by accumulating 100 spectra at an integration time of 500 ms per spectrum. Images were acquired by collecting a full Raman spectrum at each image pixel (100 × 100 spectrum array per image) with a 100 ms integration time per pixel. Raman chemical images were generated by using a sum filter, which integrates the signal intensity over a defined wavenumber range that is representative of the molecular species of interest and subtracts the background as a linear baseline from the first to second border as defined by the sum filter. Data analysis on the Raman images was performed using WITec Project 2.1 software (WITec GmbH, Ulm, Germany), and Raman spectra were processed using Igor Pro 6.32A (Lake Oswego, OR).
MSI
MS experiments were performed on a customized hybrid MALDI/C60-SIMS Q-TOF mass spectrometer, described in detail elsewhere,21 operated in positive ion/C60-SIMS mode. This instrument is a modified version of the QSTAR XL (AB SCIEX, Framingham, MA) featuring a 20 kV DC C60 primary ion beam for SIMS, a translational sample stage that enables imaging experiments, tandem MS (MS/MS) capability via collision induced dissociation (CID), and high mass resolution (R > 10 000). C60+ was selected for the primary beam, operated with a 70 pA DC sample current and a ∼15-μm spot diameter. MS mode acquisition parameters were optimized for detection of quinolones and other small metabolites, collecting m/z 100–300 with ion guide Q1 transmission biased to the upper half of this range (25% at m/z 120, 75% at m/z 200). MS/MS was performed with 10 eV CID (collision induced dissociation) and Ar in the collision cell. MSI was performed in two modes: “step-mode” in which the probe is centered at each discrete pixel location for a specified accumulation time before stepping to the next position and “continuous raster mode” in which the probe is continuously moved across the sample in a horizontal line scan to acquire each row of the image. This allowed images to be acquired several-fold faster but limited spatial resolution and accumulation time. Step-mode acquisition was performed at 10 × 10 μm2 pixel/step size and 1 s/pixel, while rastering was performed at 20 × 20 μm2 pixel size and 0.25 s/pixel. In both cases, the ion dose was well beyond the traditional static limit (1 × 1012 primary ions cm–2 s–1); the step mode dose is estimated at ∼2.5 × 1014 primary ions cm–2 s–1, corresponding to an etch rate of ∼200 nm/s based on atomic force microscopy measurements in similar previous work,22 assuming similar sputter rates. Data were acquired with Analyst v1.2 and oMALDI Server v5.1 software (AB SCIEX), and images were converted to .img files at 20 bins/AMU for processing in BioMap (Novartis, Switzerland). The ion images shown here represent signal intensity (counts) in each pixel with a “thermal” false-color scale ranging from black (no signal) through red to white (high signal). Coating the biofilms with metal (1–2 nm of sputtered Au) was found to suppress rather than enhance biological ions, so biofilms were untreated prior to imaging, aside from the microspot application. Mass calibration was performed with indium/indium oxide clusters.
Scanning Electron Microscopy (SEM)
SEM was performed on an environmental scanning electron microscope (Philips XL30 ESEM-FEG, Philips/FEI Co., Hillsboro, OR) operating at 2 kV electron beam energy, 2.4 nm spot size, and 8.7 mm working distance. Biofilm samples were analyzed without surface modification (i.e., standard metal coating) in order to avoid generating artifact features on the biofilm surface.
Results and Discussion
Biofilm Profiling with CRM and C60-SIMS
Prior to the imaging experiments, a biofilm sample was profiled by CRM and C60-SIMS individually to determine the compositional coverage and overlap of the two techniques. A typical Raman spectrum acquired from the biofilm is shown in Figure 2a. Vibrational bands characteristic of known biofilm components can be assigned based on previous work,23 prominently including: (1) a band at 1005 cm–1 (band i shown in Figure 2a), assigned to symmetric ring vibrations in tryptophan and phenylalanine, and 1605 cm–1 (band iv), arising from C=C stretching vibrations in phenylalanine, both representative of proteins;24 and (2) bands at 1034 cm–1 (band ii) and 1163 cm–1 (band iii) in the carbohydrate region of the spectrum, attributed to C–O stretching vibrations with contributions from in-plane C–H deformations in phenylalanine (1034 cm–1) and C–C, and C–O asymmetric ring vibrations, characteristic of carbohydrate moieties.25,26 At some sample locations, a strong band at 1370 cm–1 (band v in Figure 2b) was also observed. This vibration, a common feature in quinolone-associated spectra,26,27 is tentatively assigned to a ring-stretching vibration in quinolones,27 a known P. aeruginosa secondary metabolite class28 detected in biofilms by MS in recent work from our laboratories,29 and from cultured colonies in the work of others,30 but not previously reported by Raman spectroscopy. Raman spectra acquired from ROIs marked by the 1370 cm–1 quinolone-associated peak also exhibited bands characteristic of ring vibrations at 1155 cm–1, arising from C–C and C–N asymmetric ring breathing vibrations,31 as well as CH deformation vibrations near 1460 cm–1,27,32,33 a vibration at 1557 cm–1 (band vi), and another at 1600 cm–1 (band vii), attributed to aromatic C=C stretching.33 Other significant vibrations arose from C=O stretching at 1650 cm–1,26,33 and CH bending and CH twisting, giving rise to a wide band centered near 1210 cm–1,34 and CH in-plane bending at 1256 cm–1.31,34 Figure 2b compares the Raman spectrum acquired from a quinolone-rich biofilm region overlaid with a spectrum acquired from a purified PQS standard. The similarity of the two spectra is striking, with the 1155, 1370, 1462, 1557, and 1600 cm–1 bands well-matched between the 1370 cm–1 ROI and the PQS standard. Raman profiling of a second quinolone standard (HHQ, Figure S1) produced a similar vibrational profile. Thus, the concordance of a number of peaks with known quinolone species is strong evidence for assigning these spectra to the quinolone class, although it is not possible to definitively distinguish between PQS and HHQ based on Raman imaging alone.
Figure 2.
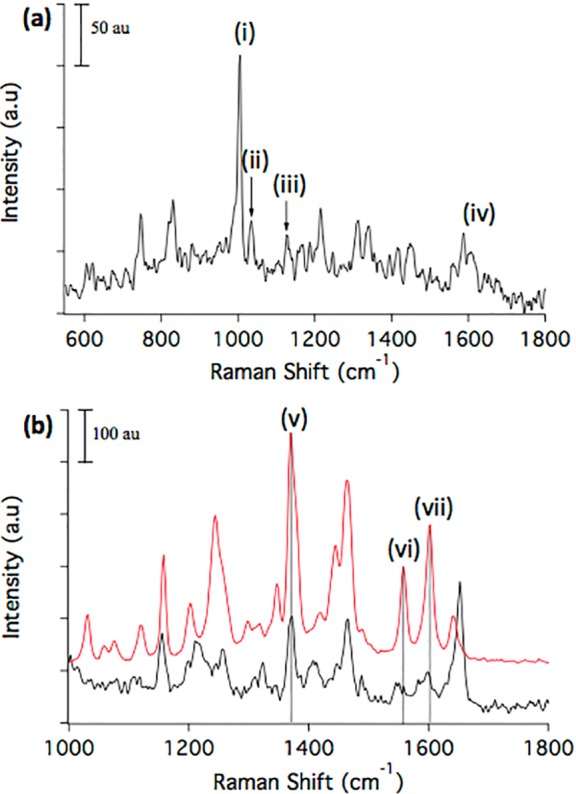
CRM spectral profiles of P. aeruginosa biofilm. (a) CRM profiling of a P. aeruginosa biofilm detects multiple biomolecular classes by characteristic vibrations, including (i) 1005 cm–1 from symmetric ring-breathing vibrations of phenylalanine and tryptophan (indicating proteins), (ii) 1034 cm–1 from C–O stretching of carbohydrate moieties with contributions from in-plane C–H deformations in phenylalanine, (iii) 1163 cm–1 from C–C and C–O asymmetric ring-breathing vibrations of carbohydrates, and (iv) 1605 cm–1 C=C stretching in phenylalanine. (b) Comparison of a quinolone-rich biofilm ROI (black trace) with a purified commercial PQS standard (red trace) reveals several matching vibrations, including (v) 1371 cm–1 quinolone ring stretch, (vi) C–C and C–N–C associated quinolone ring stretches, and (vii) 1603 cm–1 symmetric C=C stretching in the quinolone ring. Dotted vertical lines are added to facilitate comparison. All Raman spectra are baseline corrected using a fourth order polynomial function.
Quinolones are known secondary metabolites observed in several dozen unique species. They function as cell signals, virulence factors, and redox mediators, among other roles in P. aeruginosa,35 and, thus, are analytes of particular interest. Although most quinolones have not been functionally characterized, we recently observed cell-scale spatial distributions of quinolones on P. aeruginosa biofilms29 that may relate to their unique functions, and so we chose to focus on these analytes here.
To complement the CRM images, C60-SIMS was conducted in a semitargeted manner, i.e., the acquisition parameters were optimized for detection of quinolones and other small metabolites in the <300 AMU range. A detail of the P. aeruginosa biofilm MS profile is shown in Figure 3a; quinolones were consistently detected as MH+ ions, in agreement with previous reports,28,30 as well as with our own recent observations using MSI with other probe types (metal-assisted LDI and Au-SIMS, validated by CE–ESI MS/MS). Assignments were initially made by mass match with previous reports, and in situ MS/MS was also performed to confirm identities when signals were adequate, as shown for HHQ in Figure 3b and for other analytes in Table S1. A total of nine quinolones were detected and confirmed with MS/MS, including two isobaric quinolone pairs—PQS/HQNO and C9:1-PQS/C9:1-NQNO—yielding unique characteristic fragments. A mass list summary of these results is presented in Figure S2.
Figure 3.
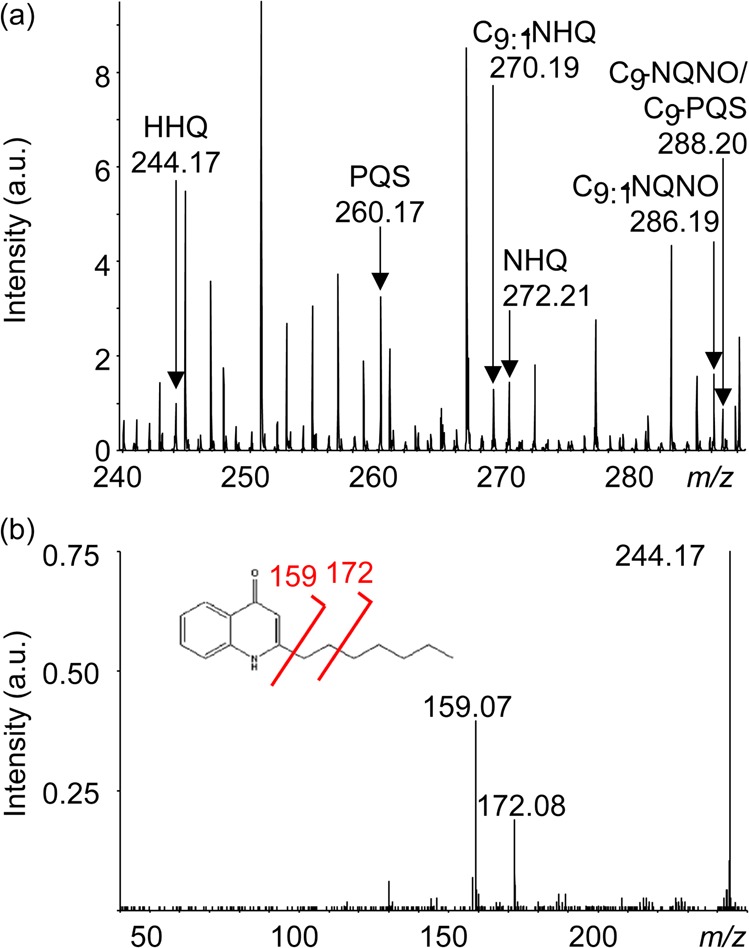
C60-SIMS and MS/MS of biofilm. (a) C60-SIMS direct analysis of an untreated biofilm surface yielded a profile that included multiple quinolones detected as MH+ pseudomolecular ions. (b) In situ MS/MS of putative quinolones supports the mass assignments, e.g., for HHQ at m/z 244.17 shown here, yielding characteristic fragments at m/z 159.07 and 172.08.
Correlated C60-SIMS/CRM Imaging
Following characterization, CRM and C60-SIMS were combined to investigate the biofilm surface via correlated imaging. Owing to the relatively large sample area (∼2 × 2 cm2) and limited imaging fields of view (<150 × 150 μm2 for CRM when working at 60× magnification), it is challenging to ensure precise sample navigation, reliable relocation of microscopic CRM ROIs for subsequent MSI, and proper alignment of imaged regions for precise spatial registry of the CRM and MSI data. To address these issues, we developed a fiducial array approach, wherein 100 pL droplets of an Ag nanoparticle solution were dispensed in a Cartesian grid (500-μm pitch) across the sample surface. Once dried, the nanoparticle spots were visible in optical, MS, and electron microscope images and could therefore be used to register spatial locations across these distinct imaging modes (the CRM microscope was operated in bright field mode for sample navigation and visualization of microspots). An origin was specified at one corner of the sample, and then features could be located to within a single “cell” of the grid using an (x,y) coordinate axis. The array also served as a visual indicator of sample alignment while remounting in a holder for MSI after CRM.
Results from a correlated imaging experiment are shown in Figure 4. CRM is performed first as it is nondestructive, thus providing a good way to survey the sample. A region with intense micrometer-scale quinolone features was found and imaged by CRM; the sample was then physically transferred to the C60-SIMS instrument where the ROI was relocated using the microspot array. MSI was performed over the entire grid cell, which included the CRM-imaged ROI. Figure 4a shows the CRM “composite quinolone” image aligned and superimposed with the SIMS ion image of quinolone HHQ (MH+ at m/z 244.17). The two images are in excellent agreement in spatial distribution and relative signal intensity within the feature, cross-validating the data and indicating that the observed ion and Raman scattering distributions are accurate, not artifactual (e.g., arising from ion suppression effects or similar vibrational modes in other biofilm constituents). The individual SIMS and CRM images are also shown separately in Figure 4b and c, respectively. The microspots around the ROI are visible in the optical image, Figure 4e, and are also detected as intense spots in several ion images, including m/z 250.81, Figure 4f, and in the total ion count, Figure S3. These marker ions were not identified, but they are likely adduct or inorganic cluster ions formed or enhanced by the presence of the Ag nanoparticles and/or the citrate buffer. Notably, ions associated with the microspots did not include silver clusters such as Ag+ or Ag2+, perhaps indicating that the silver was extensively associated with other compounds, or that the nanoparticles were not sufficiently fragmented by the C60+ primary ion beam. The spots are well-defined against the biofilm background, indicating that the nanoparticle solution dried into discrete 132 ± 9 μm (n = 6) diameter regions without diffusing into the adjacent sample.
Figure 4.
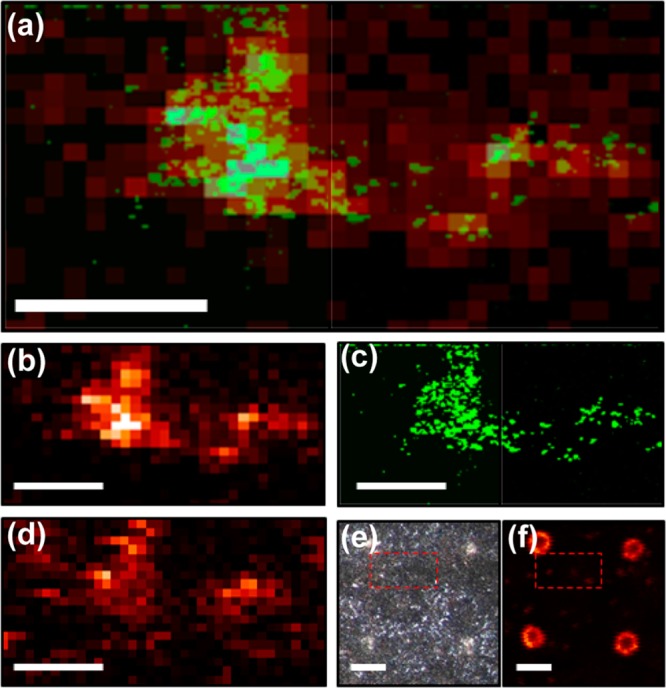
Correlated SIMS/CRM imaging provides additional information about signaling molecules in P. aeruginosa biofilm. (a) Superimposed CRM “composite quinolone” image (1350–1400 cm–1, quinolone ring stretch) and SIMS 2-heptyl-2-quinolone ion image (HHQ, MH+ at m/z 244.17) shows a similar molecular distribution in the selected ROI. The same images are shown individually for (b) SIMS and (c) CRM, where high spatial resolution enables visualization of micron-scale features within the ROI. (d) Another quinolone, 4-hydroxy-2-nonylquinolone-N-oxide (C9–NQNO, MH+ at m/z 288.20) appears to be colocalized, but the distribution is distinct from that of HHQ within the composite quinolone area. (e) Optical and (f) SIMS ion images (m/z 250.81) with a larger field of view show how the microspot array is visualized around the ROI, allowing precise navigation and image alignment. Red boxes specify the ROI of the CRM/SIMS detail. Scale bars = 100 μm in a–d and 200 μm in e and f.
The nanometer-scale spatial resolution provided by CRM here is complemented by the chemical specificity of the correlated SIMS data, which enabled detection of at least nine quinolones and additional related metabolites present in and around the ROI (images shown in Figure S3). This allowed unique distributions of specific quinolone species to be discerned, as observed with C9–NQNO (MH+ at m/z 288.20), shown in Figure 4d. Note that the distribution of this quinolone still falls within the composite quinolone distribution observed by CRM, based on the common frequency of the quinolone ring vibration, though the distributions of the two quinolones within that feature differed. We also observed an interesting trend between two quinolone subclasses in the SIMS data; the two detected 3-hydroxyquinolones (PQS at m/z 260.17 and C9–PQS at m/z 288.20) were similarly distributed in patches throughout the ROI, in contrast with the other (non-3-hydroxy) 4-quinolones, which were mostly concentrated in the ROI itself. Fragment ions characteristic of these classes, m/z 175/188 from 3-hydroxyquinolones and m/z 172/159 from 4-quinolones, reflected similar distributions (shown in Figure S4), further validating these mass assignments and the associated distributions. The significance of such a distribution is not apparent but suggests yet to be determined differences in the biological function of the quinolone classes.
SEM of the Correlated Imaging of a ROI
To investigate the physical nature of the quinolone “hotspots” that were observed by CRM and SIMS, SEM was performed following a combined imaging experiment. The resulting optical, CRM, SIMS, and SEM images are shown in Figure 5. The SEM image of this area reveals single cells exposed in patches on the biofilm, in contrast with a uniformly smooth biofilm surface elsewhere (shown in Figure S5). The cells are consistent in size with the quinolone features observed in the CRM image and also with the typical P. aeruginosa cell size (1–2 μm) and shape. In addition to detection of quinolones here, SIMS images show colocalization of phosphocholine (PC, M+m/z 184.08), a cell membrane phospholipid fragment. Detection of PC from the outer membranes of the exposed cells in this area is a likely explanation that would fit with the correlated SEM and CRM data, suggesting that quinolone “hotspots” may consist of perturbed biofilm regions where cells have been exposed.
Figure 5.
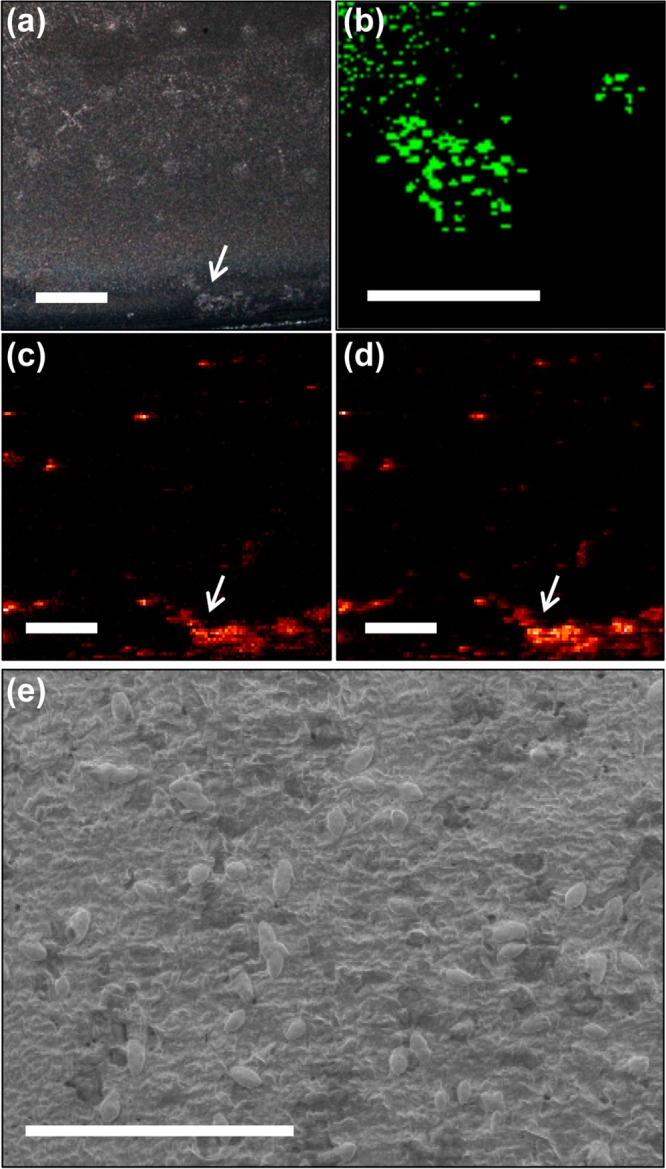
SEM of a quinolone “hot spot” revealing unique topography and cell features on a biofilm surface. (a) The optical image, (b) CRM image of a quinolone ring stretch (1350–1400 cm–1), (c) HHQ (m/z 244.17) ion image, and (d) PC (m/z 184.08) ion image suggest a chemical ROI at the edge of the biofilm; the CRM location is indicated with white arrows. (e) SEM of a region within the CRM field of view shows a rough patch and micrometer-sized single cells on the biofilm surface. Scale bars = 500 μm in a, c, and d; 100 μm in b; and 20 μm in e.
Conclusions
We report a method that combines two label-free molecular imaging techniques, CRM and MSI, and we demonstrate its utility with the analysis of metabolites on P. aeruginosa bacterial biofilms. A chemical microspot array printed on the sample allows precise navigation, relocation of analysis regions, and alignment of correlated image data. CRM enables nondestructive imaging with submicrometer spatial resolution of secreted quinolones and detection of multiple biomolecule classes and enables imaging with submicrometer spatial resolution of secreted quinolones. MSI with C60-SIMS allows mass-based discrimination of multiple specific quinolone species having subtly differing distributions, as well as confirmation of mass assignments with in situ MS/MS experiments. SEM of the imaged regions reveals that quinolone concentrations detected with SIMS and resolved by CRM correlate with single cells exposed on the biofilm surface; thus our CRM-MSI imaging approach may serve as an effective platform for in situ single cell metabolomics experiments in future work.
Ongoing efforts include incorporating the MALDI mode of the hybrid mass spectrometer used here in order to detect larger molecules, such as proteins and polysaccharides, in conjunction with high resolution CRM and SIMS imaging. We are also adapting sample preparation techniques to enable correlated 3D imaging with CRM and SIMS in order to explore the complex native 3D biofilm structure, as well as transitioning from bacterial monoculture to plant-microbe cocultures in order to study metabolic exchange at the biological interface. Finally, while image alignment was performed manually here, future improvements will include automatic image alignment using the novel microspot array approach developed for this study.
Acknowledgments
This work was funded by the Department of Energy Office of Biological and Environmental Research through grant DE SC0006642 (support of E.J.L. and R.N.M.) and by the National Institute of Allergies and Infectious Diseases under grant R01 AI113219. The authors are grateful for electron microscopy assistance from Scott Robinson and Cate Wallace in the Beckman Institute Imaging Technology Group, which is partially supported by the National Science Foundation grant DBI-9871103.
Supporting Information Available
Supplemental figures referenced in the text are contained in the Supporting Information document. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Rubakhin S.; Sweedler J. In Mass Spectrometry Imaging; Rubakhin S. S.; Sweedler J. V., Eds.; Humana Press: New York, 2010; pp 21–49. [Google Scholar]
- Setou M.Imaging Mass Spectrometry: Protocols for Mass Microscopy; Springer: Tokyo, 2010. [Google Scholar]
- Chughtai K.; Heeren R. M. A. Chem. Rev. 2010, 110, 3237–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieing T.Confocal Raman Microscopy; Springer: Berlin, 2011. [Google Scholar]
- Gierlinger N.; Schwanninger M. Plant Physiol. 2006, 140, 1246–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppels G. J.; Demul F. F. M.; Otto C.; Greve J.; Robertnicoud M.; Arndtjovin D. J.; Jovin T. M. Nature 1990, 347, 301–303. [DOI] [PubMed] [Google Scholar]
- Boxer S. G.; Kraft M. L.; Weber P. K. Annu. Rev. Biophys. 2009, 38, 53–74. [DOI] [PubMed] [Google Scholar]
- Lanni E. J.; Rubakhin S. S.; Sweedler J. V. J. Proteomics 2012, 75, 5036–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmer F.; Paul W.; Krehl M.; Niehuis E. Surf. Interface Anal. 2012, 45, 312–314. [Google Scholar]
- Everall N. J. Analyst 2010, 135, 2512–2522. [DOI] [PubMed] [Google Scholar]
- Juang C. B.; Finzi L.; Bustamante C. J. Rev. Sci. Instrum. 1988, 59, 2399–2408. [Google Scholar]
- Klein K.; Gigler A. M.; Aschenbrenne T.; Monetti R.; Bunk W.; Jamitzky F.; Morfill G.; Stark R. W.; Schlegel J. Biophys. J. 2012, 102, 360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Chu L.-Q.; Sweedler J. V.; Bohn P. W. Anal. Chem. 2010, 82, 2608–2611. [DOI] [PubMed] [Google Scholar]
- Jones E.; Lockyer N.; Kordys J.; Vickerman J. J. Am. Soc. Mass Spectrom. 2007, 18, 1559–1567. [DOI] [PubMed] [Google Scholar]
- Lay J. O.; Gidden J.; Liyanage R.; Emerson B.; Durham B. Lipid Technol. 2012, 24, 36–40. [Google Scholar]
- Deininger S.-O.; Cornett D.; Paape R.; Becker M.; Pineau C.; Rauser S.; Walch A.; Wolski E. Anal. Bioanal. Chem. 2011, 401, 167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masyuko R.; Lanni E. J.; Sweedler J. V.; Bohn P. W. Analyst 2013, 138, 1924–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit V. W.; Réfrégiers M.; Guettier C.; Jamme F.; Sebanayakam K.; Brunelle A.; Laprévote O.; Dumas P.; Le Naour F. Anal. Chem. 2010, 82, 3963–3968. [DOI] [PubMed] [Google Scholar]
- Fagerer S. R.; Schmid T.; Ibanez A. J.; Pabst M.; Steinhoff R.; Jefimovs K.; Urban P. L.; Zenobi R. Analyst 2013, 138, 6732–6736. [DOI] [PubMed] [Google Scholar]
- Bocklitz T. W.; Crecelius A. C.; Matthäus C.; Tarcea N.; von Eggeling F.; Schmitt M.; Schubert U. S.; Popp J. Anal. Chem. 2013, 85, 10829–10834. [DOI] [PubMed] [Google Scholar]
- Lanni E. J.; Dunham S. J. B.; Nemes P.; Rubakhin S. S.; Sweedler J. V. J. Am. Soc. Mass Spectrom. 2014, 10.1007/s13361-13014-10978-13369. [DOI] [PubMed] [Google Scholar]
- Robinson M. A.; Graham D. J.; Castner D. G. Anal. Chem. 2012, 84, 4880–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masyuko R. N.; Lanni E. J.; Driscoll C. M.; Shrout J. D.; Sweedler J. V.; Bohn P. W. Analyst 2014, 10.1039/c1034an00435c. [DOI] [PubMed] [Google Scholar]
- Puppels G. J.; Garritsen H. S.; Segers-Nolten G. M.; de Mul F. F.; Greve J. Biophys. J. 1991, 60, 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harz M.; Rosch P.; Peschke K. D.; Ronneberger O.; Burkhardt H.; Popp J. Analyst 2005, 130, 1543–1550. [DOI] [PubMed] [Google Scholar]
- Neugebauer U.; Schmid U.; Baumann K.; Ziebuhr W.; Kozitskaya S.; Deckert V.; Schmitt M.; Popp J. ChemPhysChem. 2007, 8, 124–137. [DOI] [PubMed] [Google Scholar]
- Neugebauer U.; Szeghalmi A.; Schmitt M.; Kiefer W.; Popp J.; Holzgrabe U. Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 2005, 61, 1505–1517. [DOI] [PubMed] [Google Scholar]
- Lépine F.; Milot S.; Déziel E.; He J.; Rahme L. G. J. Am. Soc. Mass Spectrom. 2004, 15, 862–869. [DOI] [PubMed] [Google Scholar]
- Lanni E. J.; Masyuko R. N.; Driscoll C. M.; Aerts J. T.; Shrout J. D.; Bohn P. W.; Sweedler J. V. Anal. Chem. 2014, 10.1021/ac5020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan V. V.; Liu W.-T.; Pogliano K.; Dorrestein P. C. Nat. Chem. Biol. 2012, 8, 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch P.; Schneider H.; Zimmermann U.; Kiefer W.; Popp J. Biopolymers 2004, 74, 151–156. [DOI] [PubMed] [Google Scholar]
- Bolboaca M.; Kiefer W.; Popp J. J. Raman Spectrosc. 2002, 33, 207–212. [Google Scholar]
- Frosch T.; Schmitt M.; Popp J. Anal. Bioanal. Chem. 2007, 387, 1749–1757. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Yu K.; Wang S. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2006, 65, 159–163. [DOI] [PubMed] [Google Scholar]
- Huse H.; Whiteley M. Chem. Rev. 2010, 111, 152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


