Abstract
Stro-1 has proved an efficacious marker for enrichment of skeletal stem and progenitor cells although isolated populations remain heterogeneous, exhibiting variable colony-forming efficiency and osteogenic differentiation potential. The emerging findings that skeletal stem cells originate from adventitial reticular cells have brought two further markers to the fore including CD146 and CD105 (both primarily endothelial and perivascular). This study has compared CD146-, CD105- and Stro-1 (individual and in combination)-enriched human bone marrow stromal cell subsets and assessed whether these endothelial/perivascular markers offer further selection over conventional Stro-1. Fluorescent cell sorting quantification showed that CD146 and CD105 both targeted smaller (2.22% ± 0.59% and 6.94% ± 1.34%, respectively) and potentially different human bone marrow stromal cell fractions compared to Stro-1 (16.29% ± 0.78%). CD146+, but not CD105+, cells exhibited similar alkaline phosphatase–positive colony-forming efficiency in vitro and collagen/proteoglycan deposition in vivo to Stro-1+ cells. Molecular analysis of a number of select osteogenic and potential osteo-predictive genes including ALP, CADM1, CLEC3B, DCN, LOXL4, OPN, POSTN and SATB2 showed Stro-1+ and CD146+ populations possessed similar expression profiles. A discrete human bone marrow stromal cell fraction (2.04% ± 0.41%) exhibited positive immuno-labelling for both Stro-1 and CD146. The data presented here show that CD146+ populations are comparable but not superior to Stro-1+ populations. However, this study demonstrates the critical need for new candidate markers with which to isolate homogeneous skeletal stem cell populations or skeletal stem cell populations which exhibit homogeneous in vitro/in vivo characteristics, for implementation within tissue engineering and regenerative medicine strategies.
Keywords: Skeletal stem cells, Stro-1, CD146, CD105, osteogenesis, pericyte and perivascular
Introduction
Skeletal stem cells (SSCs), originally described as colony-forming unit-fibroblasts (CFU-Fs),1–3 comprise a multipotent non-haematopoietic stem cell fraction within human bone marrow (HBM). SSCs offer an ideal cell source for bone tissue engineering strategies due to their self-renewal and multi-lineage differentiation capacity toward osteoblasts, chondrocytes and adipocytes.4–9 To date, SSC enrichment remains largely based on cell surface expression profile.10–15 Their expression profile appears complex, and to date, isolation of homogeneous SSCs remains to be demonstrated,16 since many markers thought of as SSC-specific are in fact expressed on the surface of fibroblastic cells. However, it is unclear whether a true homogeneous bone stem cell is required for successful in vivo tissue regeneration. Indeed, Tilley et al.17 have studied host derived-condensed heterogeneous bone marrow implanted into patients undergoing surgery which demonstrated bone tissue formation. Thus, homogeneity may not be a prerequisite for clinical advancement.
One surface marker that has shown robust enrichment for SSCs is the trypsin-resistant cell surface antigen 1 (Stro-1).18–21 Stro-1+ populations exhibit enhanced CFU-F capacity and elevated osteogenic differentiation both in vitro and in vivo in comparison to unsorted human bone marrow stromal cells (HBMSCs).22–25 However, Stro-1-enriched SSCs remain highly heterogeneous,26–29 reflected by their in vivo localisation and in vitro multipotency and behaviour, demonstrated by the work of MacArthur et al.30 on Stro-1 distribution and subpopulation behaviour in heterogeneous cell cultures of HBM.
Two alternative enrichment markers comprising melanoma cell adhesion molecule (MCAM; CD146) and Endoglin (CD105) have been identified on the cell surface of isolated populations exhibiting a significant fold increase in CFU-F capacity and in vitro osteogenic differentiation.31–33 CD146 and CD105 are both primarily endothelial cell markers, but have previously been shown to target perivascular cells. Both markers have attained significant interest on the premise that SSCs originate from a sub-endothelial or perivascular niche. There is emerging evidence within the musculoskeletal field that SSCs originate from adventitial reticular cells.34,35 However, it remains to be ascertained whether either of these markers are indeed SSC-specific and, at present, whether these markers offer improved SSC isolation compared to conventional Stro-1.
In this study, we have compared CD146 and CD105 with conventional Stro-1-enriched populations. Self-renewal and growth potential within resultant populations were assessed by colony-forming efficiency (CFE) assay and alkaline phosphatase (ALP) expression. Isolated populations were also examined under differentiation culture conditions and analysed by real-time polymerase chain reaction (rtPCR) analysis for differences in gene expression profiles. Analysis has focussed on traditional osteogenic genes and newer potentially osteo-predictive genes detailed by Larsen et al.36 In addition, in vivo growth and differentiation capacity were assessed following subcutaneous transplantation within MF1 nu/nu immunodeficient mice for 28 days.
This study demonstrates that CD146 and CD105 highlight a smaller fraction of HBMSCs compared with Stro-1, and we hypothesised that the two markers may be targeting a subset of Stro-1+ populations. Consequently, Stro-1, CD146 and CD105 were utilised individually and in combination (dual labelling) to isolate populations for comparative analysis. Dual labelling provided a tool to dissect and further interrogate the reported heterogeneous nature of Stro-1+ populations. CD146 has previously been shown to be co-expressed with Stro-1 on a subset of HBMSCs.37 Furthermore, the dual marker approach has previously defined a putative primitive SSC population using Stro-1 and CD106, clearly demonstrating an advantage over single markers.18 The aim of these studies was to determine whether CD146+ and/or CD105+ populations were comparable or different to Stro-1+ populations. Clarification through comparison of the current accepted SSC markers may eventually help elucidate a select surface expression profile which identifies either the true bone stem cell or minimally a SSC population exhibiting enhanced homogeneous in vitro/in vivo characteristics.
Methods and materials
All reagents were supplied by Sigma–Aldrich unless otherwise stated.
Ethics statement
These studies were conducted under ethical approval – LREC 194/99 (National Research Ethics Service – Southampton and South West Hampshire Research Ethics Committee) including Professional Indemnity and Clinical Trials Insurance. Written informed consent from each donor or their next of kin was obtained for use of samples in the research.
Bone marrow preparation and cell culture
Donated HBM was collected during routine total hip-replacement surgery from haematologically normal osteoarthritic and trauma patients at the Southampton General Hospital. Samples were suspended in modified Eagle’s medium–alpha (α-MEM) and centrifuged to remove fat. The cell pellet was suspended in 20 mL α-MEM and passed through a 40-µm sieve to remove bone fragments and blood clots. The cell suspension was then carefully loaded on top of Lymphoprep™ (Lonza) and centrifuged for 40 min at 2200 r/min and 18°C to remove red blood cells. A buffy layer above the Lymphoprep containing mononuclear cells was washed with phosphate buffered saline (PBS; Lonza) and suspended in basal culture media (α-MEM, 10% foetal calf serum (FCS), penicillin (100 U/mL) and streptomycin (0.1 mg/mL)).
Cell isolation – immunocytochemistry
Freshly prepared HBM cells were washed in PBS, suspended in blocking buffer (α-MEM, 10% human serum, 5% FCS and 10 mg/mL bovine serum albumin (BSA)) and incubated at 4°C for 15 min. Cells were then incubated with primary antibody solution (1:50), washed three times with isolation buffer (1% BSA, in PBS) and incubated with magnetic and/or fluorescent-conjugated secondary antibody solution (1:200). Antibody incubations were 60 min at 4°C under dark conditions diluted in isolation buffer.
Quantification
Immuno-labelled populations were quantified by flow cytometry (Guava easyCyte; Millipore) (Figure 2). Following immuno-labelling as described previously, 1 × 106 cells were fixed in 4% paraformaldehyde (PFA in dH2O/PBS), washed in PBS and suspended ready for analysis. Flow cytometry analysed 5000 events, and thresholds were set according to both negative and isotype controls. Data are presented as mean ± standard deviation (SD).
Figure 2.
Quantification of marker expression within HBMSC populations. Adult human bone marrow was fluorescently immuno-labelled with antibodies against Stro-1, CD146 and CD105 (a) individually and (b) in combination prior to quantification by flow cytometry. (a) x-axis depicts fluorescence intensity (logarithmic scale) and y-axis depicts cell count. (b) x- and y-axes depict fluorescence intensities (logarithmic scale) of each marker used for immuno-labelling. The red linear markers indicate the threshold for positive fluorescent labelling (assessed by negative controls). The green line in single marker populations indicates non-immuno-labelled control cells. Additional line depicts immuno-labelled populations.
HBMSC: human bone marrow stromal cell.
Magnetic-activated cell sorting (single-labelled cells)
Cell suspensions were divided into four populations comprising a control non-labelled fraction and Stro-1 (neat (mouse IgM) – ‘in-house’ Hybridoma), CD146 (1:50 (mouse IgG) – BD Biosciences (clone: 1MCAM)) and CD105 (1:50 (mouse IgG) – BD Biosciences (clone: 35/CD105)) immuno-labelled fractions. Following primary antibody adhesion and washing, cells were incubated with both magnetic (200 µL – anti-mouse IgM and IgG microbeads; Miltenyi Biotec) and fluorescent (1:200 – AlexaFluor 488–conjugated anti-mouse IgM and IgG and AlexaFluor 546–conjugated anti-mouse IgG; Invitrogen) secondary antibodies. Isolation was by magnetic cell sorting.
Fluorescent cell sorting (dual-labelled cells)
Cell suspensions were divided into four populations comprising a control non-labelled fraction and Stro-1/CD146 (neat Hybridoma/1:50 clone: 1MCAM), Stro-1/CD105 (neat Hybridoma/1:50 clone: 35/CD105) and CD146/CD105 (1:50 clone: 1/MCAM/1:50 (rabbit IgG) – Novus Biologicals) immuno-labelled fractions. Following incubation with primary antibody combinations and washing, cells were incubated with fluorescent (1:200, AlexaFluor 488–conjugated anti-mouse IgM and AlexaFluor 546–conjugated anti-mouse IgG (Stro-1/CD146 and Stro-1/CD105) and AlexaFluor 488–conjugated anti-mouse IgG and AlexaFluor PE/647–conjugated anti-rabbit IgG (CD146/CD105)) secondary antibodies. Isolation was by fluorescent cell sorting (FACSAria; Becton Dickinson) and FACS Diva software version 5.0.3.
Visualisation
Post isolation, a small proportion of each immuno-labelled population was seeded onto tissue culture plastic, allowed to adhere, fixed and imaged by fluorescence microscopy after 4′,6-diamidino-2-phenylindole (DAPI) counterstaining.
HBMSC populations were processed, immuno-labelled and sorted by magnetic-activated cell sorting (MACS) or fluorescent-activated cell sorting (FACS) as appropriate prior to in vitro expansion. Single-labelled populations were isolated by MACS due to the potential for bulk separation. However, MACS cannot be used to isolate dual-labelled populations as the system is not selective for A+/B+ from A+/B− and A−/B+ populations. Data presented for fraction percentages are an average from three to five individual patient-derived samples. Important to note here is that triple labelling was initially also investigated, but preliminary quantification by flow cytometry suggested triple-labelled cells constituted <1% HBMSCs and isolation by conventional FACS was unsuccessful in our hands.
Unselected cells were utilised as control populations for comparison and authentication of marker enrichment. However, it is important to note that unselected populations contained both target and non-target cells in combination. Therefore, comparison with immuno-selected populations should be interpreted on the premise of enrichment (removal of non-target cells), not comparison with a true negative control (negative fractions following target cell depletion). Indeed, the data presented here could be described as an assessment of non-stem cell–stem cell interaction and the effect on stem cell properties in culture, where immuno-selection offers increasing depletion of the non-stem cell fraction within HBM. Consequently, the aim of this study was not to achieve 100% purity (in the absence of specific markers), but rather use conventional isolation methods to enrich cell populations expressing Stro-1, CD146 and CD105 alone and in combination, representative of those equivalent populations previously published within the literature, and characterise for direct comparison.
CFE assay and ALP expression
Isolated cell samples were counted using a haemocytometer and seeded in tissue culture flasks with basal media at either 102 (P2 cultures – dual-labelled) or 103 (P0 cultures – single-labelled) cells/cm2 within T25-cm2 flasks. Cultures were PBS washed after 3 h and incubated at 37°C and 5% CO2 in a humidified atmosphere for 14 days without media change. Flasks were then fixed with 85% ethanol in dH2O. Fixed cultures were air dried and then incubated with Fast Violet B salt (2.5 µg/mL) and Naphthol AS-MX phosphate (40 µL/mL) in dH2O for 30–45 min at 37°C and 5% CO2 in a humidified atmosphere under dark conditions. Cultures were washed with dH2O and counterstained with haematoxylin for 5 min at room temperature.
MACS separation usually demonstrates approximately 70% purity, therefore non-labelled cells and potentially labelled non-mononuclear cells would have been present, both adding to the end cell count, but which may not have had the potential for colony formation. FACS separation demonstrated approximately 80%–85% purity. Seeding densities chosen were based on previous work within the group which initially investigated a range of densities including 0.5 × 101, 1 × 101, 1 × 102 and 1 × 103 cells/cm2. A seeding density of 103 cells/cm2 for MACS-separated P0 cultures was found to generate sufficient numbers of colonies for accurate quantification. A lower seeding density of 102 cells/cm2 for FACS-separated P2 cultures was chosen as higher densities resulted in confluent monolayer growth, possibly due to emergence of a clonogenic phenotype during in vitro expansion. Higher seeding densities for assessment of clonogenic capacity, compared to other published studies, were used to accommodate for incorporation of non-mononuclear cells within the initial cell count of MACS-separated populations. ALP expression was quantified as a relatively simple and routine indicator, but not predictor, of osteogenic differentiation potential. Colonies comprising ≥50 cells in distinct clusters and/or ≥50% ALP+ cells were counted. Single and dual CFE data were collected from four patient samples. The number of cells isolated and collected following FACS was too low to quantify reliably, and therefore, seeding densities could not be ascertained. All cells were culture expanded (P0); however, limited cells were cultured as colonies rather than monolayers. Colonies were subsequently passaged and reseeded (P1). Once monolayers were established and cell numbers were sufficient for quantification, flasks were seeded for colony growth analysis (P2 – CFE assay).
Differentiation culture
Isolated cell populations were cultured to approximately 80% confluency in media, trypsinised and seeded into four individual culture flasks. Flasks were incubated in basal (α-MEM, 10% FCS) or differentiation media (α-MEM, 10% FCS, 10 nM dexamethasone and 100 µM ascorbate-2-phosphate) for 10 and 21 days at 37°C and 5%CO2 in a humidified atmosphere. Cultures received twice weekly media changes.
Single-labelled populations were placed under basal and differentiation media conditions at P1. Dual-labelled populations required additional in vitro expansion and therefore were cultured to P2 before basal and differentiation conditions were applied.
Quantitative rtPCR
RNA isolation
At days 10 and 21 of culture, sorted populations were treated with Collagenase IV (1 mg/mL α-MEM) for 1 h at 37°C to digest extensive matrix deposition before PBS washing and incubation on ice in 1 mL Trizol® reagent for 2 min. Cell scrapers were used to detach any remaining cells from the culture flasks and resultant Trizol suspensions were stored at −80°C. Samples were incubated at room temperature with 200 µL chloroform for 2–3 min prior to centrifugation at 13,000 r/min for 15 min (4°C). Phase separation resulted in protein at the bottom, DNA in the middle and RNA at the top. Top aqueous phase was added to 500 µL isopropanol and incubated at room temperature for 10 min before further centrifugation at 12,000 r/min for 10 min (4°C). Precipitated RNA pellets were washed in 75% ethanol before dissolution in ultra-pure H2O. RNA concentration was evaluated using a NanoDrop 1000 spectrophotometer (Thermo Scientific).
Complementary DNA synthesis
RNA was subjected to DNAse treatment using a Zymo RNA clean-up kit (Cambridge Bioscience). A volume of 2 µg of RNA per sample was reverse transcribed to form complementary DNA (cDNA) using a SuperScript kit (Invitrogen) and stored at −20°C. Manufacturer protocols were employed for both kits.
rtPCR
cDNA was analysed using a 7500 Real-Time PCR System (Applied Bioscience) and accompanying software, in a reaction volume of 24 µL per gene (forward and reverse primers 5 µL each (Sigma–Aldrich or DNA Technology, Denmark), SYBR Green 12.5 µL and ultra-pure H2O 6.5 µL). cDNA samples were analysed for expression of a panel of traditional osteogenic genes including ALP and osteopontin (OPN) and novel osteo-predictive genes including Cell Adhesion Molecule 1 (CADM1),38 C-type lectin domain family 3, member B (CLEC3B; DNA Technology), Decorin (DCN; DNA Technology), Lysyl oxidase–like protein 4 (LOXL4; DNA Technology), Periostin (POSTN; DNA Technology) and Special AT-rich sequence Binding protein 2 (SATB2).36,39–41 Housekeeping (β-actin) and negative control genes Cathepsin C (CTSC; DNA Technology) and Myxovirus Resistance 1 (MX1; DNA Technology) were also assessed (Figure 1). CTSC and MX1 were chosen due their upregulation within non-bone-forming HBMSC-telomerase reverse transcriptase (HBMSC-TERT) cell populations.36 However, an important note to make is that a recent study has reported MX1 expression in an osteo lineage-restricted HBMSC subset.42 Consequently, MX1 data were not included here. Relative expression levels were normalised to the housekeeping gene β-actin. The comparative CT method was employed to quantify expression levels, where basal unsorted samples were assigned a value of 1 and all other groups were determined relative to this group. Duplicates were performed for all groups across all patient samples (three patients for dual populations and four patients for single populations).
Figure 1.
Gene primers for rtPCR. β-actin was used as the housekeeping gene for sample standardisation. Traditional osteogenic marker genes included ALP and OPN. Potential osteo-predictive genes included CADM1, CLEC3B, DCN, LOXL4, POSTN and SATB2. Inflammatory marker CTSC was used as a negative control.
rtPCR: real-time polymerase chain reaction.
Gene expression profiles of isolated populations were investigated to compare and assess the validity of isolations representing distinct fractions of HBMSCs capable of osteogenic differentiation in vitro. ALP and OPN were selected as standard markers of osteogenic differentiation.43,45 CADM1 is an osteo-progenitor surface marker.38 Mutated or knocked out SATB2 has been shown to correlate with skeletal defects in vivo,40,41 and reduction in the SATB2 protein leads to lower osteogenic gene expression.46 CLEC3B, DCN, LOXL4 and POSTN were chosen following their identification as potential osteo-predictive genes within bone-forming HBMSC-TERT cells by Larsen et al.,36 while immune regulatory gene CTSC was selected as a control following identified upregulation within non-bone-forming HBMSC-TERT cells.
In vivo differentiation
Diffusion chambers were constructed with Durapore™ membrane filters (hydrophilic polyvinylidene fluoride (PVDF) membrane; Merck Millipore) glued either side of a Plexiglas® ring (10 mm × 2 mm; diameter × thickness; Merck Millipore) and filled with 5% hydroxyapatite-loaded poly-lactic acid (HA-PLA) granules (molecular weight 56 kDa, porous supercritical CO2 synthesised foamed scaffold; gift from Dr Lisa White and Prof. Kevin M. Shakesheff). Confluent cell cultures were trypsinised (1% in PBS), suspended in basal media and seeded at 5 × 105 cells per diffusion chamber47 prior to dorsal subcutaneous implantation within male MF1 nu/nu immunodeficient mice (Harlan) midway between the front and hind legs either side of the spine. Chambers were incubated for 28 days (four chambers per mouse) without osteoinduction. Upon harvest, chambers were fixed in 4% PFA for 24 h and incubated in 20 mL rapid demineralisation buffer (4 M formic acid and 0.5 M sodium formate in dH2O, pH 2.15) at 4°C for 3 days to ensure smooth sectioning. Once demineralised, samples were removed from the Plexiglas ring and methanol dehydrated (50%, 70% and 90% in H2O, and 2 × 100%; 45–60 min each) before transfer to Histo-Clear (National Diagnostics). Samples were then incubated in paraffin wax at 60°C for 1 h before being embedded in wax blocks using an automated Shandon Citadel 2000 ready for histological assessment. Demineralisation of all samples was confirmed using Faxitron analysis.
PLA granules were used as a biocompatible scaffold to support cell growth within diffusion chambers. A volume of 5% HA was incorporated for osteoconductivity, optimised to a level whereby scaffolds could be foamed and later cut to size: higher concentrations generated scaffolds that could not be easily cut to size and shape. Implanted cell numbers reflected in vitro proliferation within a single passage. Single-labelled populations were isolated from four patient samples, while dual-labelled populations were isolated from three patient samples. Single-labelled populations and control unselected cells were each seeded in four separate chambers and all implanted subcutaneously within one mouse. A second mouse had corresponding dual-labelled populations and control unselected cells implanted within four separate chambers. This was repeated for each patient sample. Chambers were incubated in vivo for a period of 28 days as previous work has shown this time period to be sufficient for both cartilage and bone formation and was cost-effective.48 Harvested chambers were demineralised before histological analysis to improve sectioning as non-demineralised samples were found to tear sections. The diffusion chamber delivery system was employed to assess in vivo growth and differentiation capacity of isolated cell populations absent confounding host tissue invasion and complicating vasculature, as both may mask results originating from the isolated populations. The diffusion chamber does, however, allow movement of liquids, gases and nutrients across the permeable PVDF Durapore membranes which would allow for potential exogenous osteoinduction.
Histology
Paraffin-embedded diffusion chambers were sectioned at a thickness of 7 µm using a Microm HM330 D-6900 microtome (Heidelberg Instruments). Eight cross sections at 40–50 section intervals were collected on Superfrost glass slides (Thermo Scientific), representing approximately 50% of the total chamber. All sections were subsequently stained with Alcian Blue (0.5% w/v in 3% acetic acid)/Sirius Red (1% w/v in saturated picric acid) (A/S) for proteoglycan and collagen deposition, to indicate growth and matrix production in vivo.
Alcian Blue/Sirius Red
Sectioned samples were de-waxed in Histo-Clear and rehydrated through a series of reverse methanol washes. Slides were then incubated through a series of solutions at room temperature including haematoxylin (10 min), acid/alcohol dip, Alcian Blue (10 min), molybdophosphoric acid (3% w/v in dH2O, 10–20 min) and Sirius Red (45–60 min). After each incubation step, the slides were rinsed with H2O. Once stained, slides were methanol/Histo-Clear dehydrated and mounted with DPX.
Quantification
A/S-stained sections were photographed using an Olympus dotSlide microscope and accompanying OlyVIA software. Collagen and proteoglycan deposition within each section were quantified using an overlaid 150-µm square grid. The number of squares containing red collagen, blue proteoglycan, or both were counted and represented as a percentage of the total number of squares covering all tissues. An average was calculated across each set of eight sections and from all patients for comparison between isolated populations. Digital image analysis was not pursued due to inconsistencies in strength of stain presenting difficulties for software programs to analyse robustly. Spectrophotometric quantification of retrieved stain was not sensitive enough to assess histological differences.
Statistical analysis
All data sets were tested for Gaussian distribution, determined by a Kolmogorov–Smirnov test. A Mann–Whitney test was performed to assess significance between individual unpaired non-parametric data sets. An unpaired t-test (Welch corrected) was performed to assess significance between individual unpaired parametric data sets. Significance was denoted by *p ≤ 0.05, **p ≤ 0.01 and ***p ≤ 0.001.
Results
Identification and quantification of marker expression
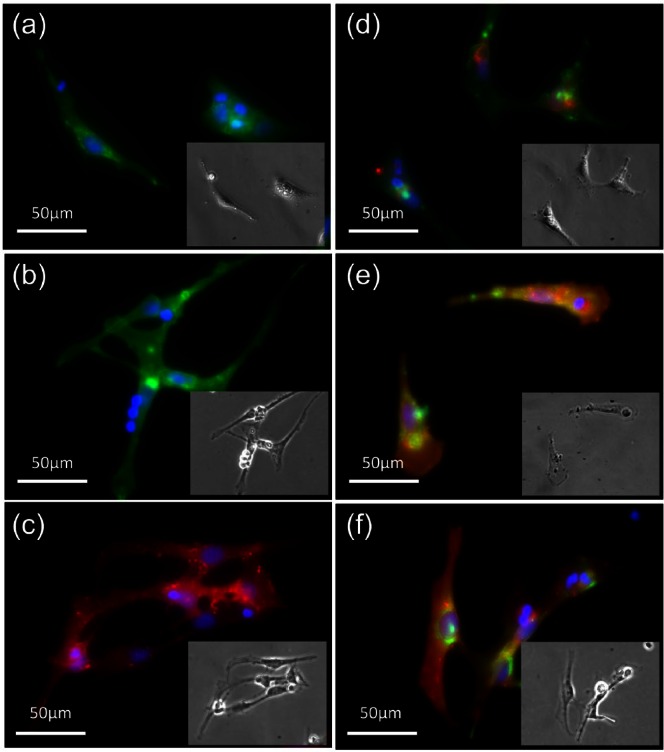
Following routine sieving and Lymphoprep separation of HBM samples, HBMSCs were immuno-labelled with Stro-1, CD146 and CD105 antibodies, alone and in combination. FACS quantification of immuno-labelled HBMSC populations before in vitro expansion showed 16.29% ± 1.73% expressed Stro-1, 2.22% ± 1.32% expressed CD146 and 6.94% ± 3.01% expressed CD105 (Figure 2(a)). Dual-labelled populations constituted a smaller percentage of the whole HBMSC fraction with 2.04% ± 0.71% expressing Stro-1/CD146, 3.05% ± 1.05% expressing Stro-1/CD105 and 0.68% ± 0.53% expressing CD146/CD105 (Figure 2(b)). Separated cells were left to adhere to tissue culture plastic before marker expression was visualised by fluorescence microscopy (Figure 3). HBMSCs exhibited expression of all three markers alone (Figure 3(a)–(c)) and in combination (Figure 3(d)–(f)). Populations sorted by MACS were quantified following isolation and recovered Stro-1 cells constituted 12.52% ± 1.29% of the total number loaded, while CD146 and CD105 cells constituted 3.75% ± 0.84% and 5.06% ± 2.70%, respectively.
Figure 3.
Fluorescence imaging of cell surface markers. HBMSCs were immuno-labelled with antibodies against (a) Stro-1, (b) CD146, (c) CD105, (d) Stro-1/CD146, (e) Stro-1/CD105 and (f) CD146/CD105. Single-labelled populations were tagged with both magnetic antibodies for MACS separation and fluorescent antibodies for post-separation visualisation. Dual-labelled cells were fluorescently tagged for FACS separation. Post isolation, cells were seeded onto tissue culture plastic, allowed to adhere, fixed and imaged following DAPI counterstaining. Picture inserts show corresponding bright-field images.
HBMSC: human bone marrow stromal cell; MACS: magnetic-activated cell sorting; FACS: fluorescent-activated cell sorting; DAPI: 4′,6-diamidino-2-phenylindole.
Isolated population purity following MACS separation was typically 70%, and following FACS separation, around 80%–85%. Ultimately, 100% pure population selection would require marker validation and application of single-cell sorting techniques such as a microfluidic approach.15 Higher isolation purity could be achieved by running the positively selected cell fraction through FACS for a second time. Previous experiments have shown purity to increase ≤90%. However, due to issues with cell viability following second isolation runs, only a single FACS separation was employed. The central aim was enrichment of populations using the markers mentioned for comparison and assessment of whether CD146 or CD105 provided superior selection over Stro-1. Pivotal would then be robust culture methodology for the maintenance of cultures, as in vitro expansion could lead to heterogeneous characteristic changes within isolated populations prior to any experimental investigation.
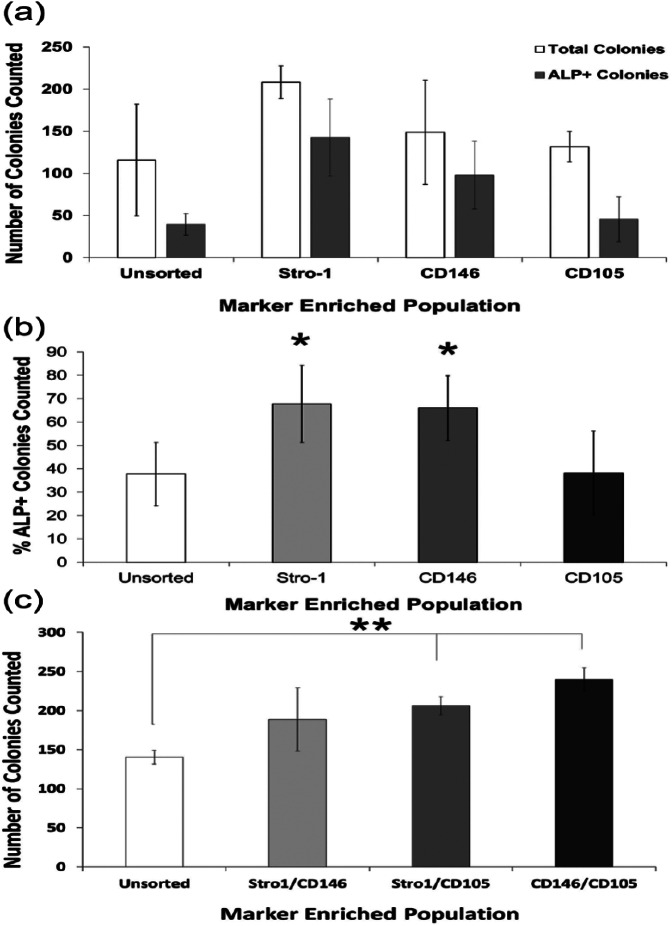
CFE assessment of marker-enriched populations
Following isolation, populations were assessed for clonal capacity by CFE assay. No significant differences were observed in total colony numbers between unsorted and single-labelled populations in the patients examined (Figure 4(a)). Stro-1- and CD146-enriched populations were comparable displaying significant enrichment for ALP+ colonies (1.79-fold Stro-1 ‘vs’ 1.74-fold CD146; p ≤ 0.05) over unsorted control samples (Figure 4(b)). Stro-1-/CD105- and CD146-/CD105-enriched populations also displayed significant enrichment in CFE (1.46- and 1.70-fold, respectively) compared to unsorted control samples (Figure 4(c)). CD105- and Stro-1-/CD146-enriched populations did not show significant enrichment. Isolated dual populations from all four samples did not exhibit measurable ALP expression.
Figure 4.
CFE assessment of isolated populations. (a and b) Following MACS isolation, single-labelled populations were seeded at a limiting dilution of 103 cells/cm2 within T25-cm2 flasks and cultured for 14 days. (c) Dual-labelled populations isolated by FACS were culture expanded to P2 prior to seeding at a limiting dilution of 102 cells/cm2 within T25-cm2 flasks and cultured for 14 days. Colony number and ALP expression were quantified and expressed as percentage ALP+ colonies. Dual-labelled populations, however, did not exhibit measurable levels of ALP expression. Consequently, dual-labelled population data are expressed as colony number only. Error bars are SD (*p ≤ 0.05).
CFE: colony-forming efficiency; MACS: magnetic-activated cell sorting; FACS: fluorescent-activated cell sorting; SD: standard deviation.
Interestingly, following MACS isolation of single-labelled populations, negative fractions also demonstrated ALP+ colony formation. Negative fractions following Stro-1 isolation generated 54% ± 13% ALP+ colonies (average 50/91 colonies), while CD146 and CD105 negative fractions generated 45% ± 19% (average 22/49 colonies) and 25% ± 3% ALP+ colonies (average 15/60 colonies). Negative fractions following FACS isolation were not collected.
Although CD146- and Stro-1-enriched populations exhibited comparable ALP+ CFE capacity, total colony counts were not significantly different between all isolations, suggesting the emergence of a clonogenic subpopulation. However, 14-day culture reduces this likelihood and may therefore be due to high variability between patients and low sample numbers. Although negative fractions following MACS isolation of single-labelled populations revealed a capacity for ALP+ colony formation at lower levels, this can be explained by differential marker expression between populations; Stro-1 negative fractions can express either or both CD146 and CD105 as can CD146 and CD105 negative fractions.
Gene expression analysis
Both single- and dual-labelled populations were cultured for 10 and 21 days in basal and differentiation media and analysed using rtPCR to assess gene expression.
Single-labelled populations
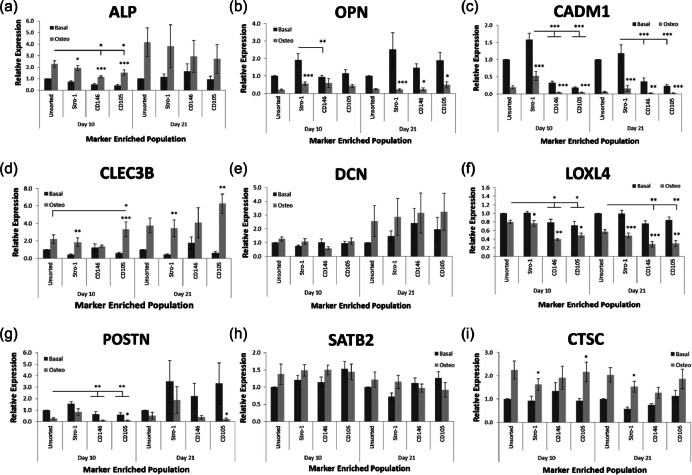
An elevation in ALP and reduction in LOXL4 expression were observed in all populations following 10 days of osteoinduction (Figure 5(a) and (f), respectively). Reduction in LOXL4 expression continued to 21 days of osteoinduction (Figure 5(f)). Elevated ALP expression was not observed after 21 days of osteoinduction (Figure 5(a)). CD146- and CD105-enriched populations exhibited decreased ALP, LOXL4 and POSTN expression compared to unsorted and Stro-1-enriched populations after 10 days with and without osteoinduction (Figure 5(a), (f) and (g), respectively). However, decreased ALP expression was only observed within osteogenic cultures (Figure 5(a)). Trends in ALP and POSTN expression between populations were not conserved after 21 days of osteoinduction (Figure 5(a) and (g), respectively). Only CD105-enriched populations exhibited decreased POSTN expression in response to osteoinduction (Figure 5(g)). Stro-1- and CD105-enriched populations exhibited increased CLEC3B expression in response to osteoinduction (Figure 5(d)). All isolated populations showed decreased OPN expression in response to osteoinduction after 21 days, although only Stro-1+ populations exhibited this same decrease after 10 days (Figure 5(b)). CADM1 exhibited decreased expression in both CD146- and CD105-enriched populations compared with Stro-1-enriched populations in both basal and osteogenic cultures after 10 days, but only in basal cultures after 21 days (Figure 5(c)). Osteoinduction resulted in decreased CADM1 expression within all populations. Under all conditions examined, no significant observations were noted between populations over time with and without osteoinduction in DCN and SATB2 expression (Figure 5(e) and (h)). Immune regulatory control gene CTSC displayed no significant differences between populations over time, although osteogenic cultures did exhibit elevated expression compared to basal cultures in Stro-1- and CD105-enriched isolations (Figure 5(i)). Data were derived from four patient samples.
Figure 5.
Gene expression analysis within single-labelled populations. Cell populations were isolated via MACS from HBMSCs according to Stro-1, CD146 and CD105 expression. Unsorted HBMSCs were used as a control. Samples were cultured for 10 and 21 days in either basal or osteogenic media before rtPCR analysis for (a) ALP, (b) OPN, (c) CADM1, (d) CLEC3B, (e) DCN, (f) LOXL4, (g) POSTN and (h) SATB2 expression. (i) The immune regulatory gene, CTSC, was used as a negative control. Error bars are SD (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001).
MACS: magnetic-activated cell sorting; HBMSC: human bone marrow stromal cell; rtPCR: real-time polymerase chain reaction; SD: standard deviation.
Dual-labelled populations
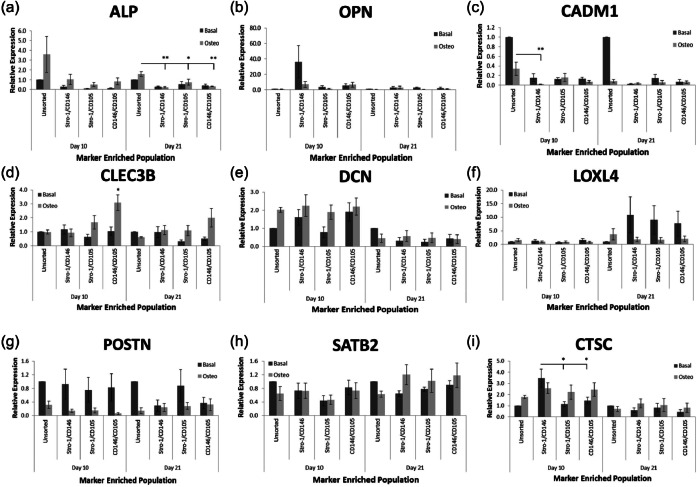
Dual-labelled populations demonstrated a delayed response to osteoinduction with increased ALP expression visible at 21 days (Figure 6(a)). Stro-1-/CD146-enriched populations exhibited reduced CADM1 expression following 10 days of osteoinduction compared to unsorted populations (Figure 6(c)). Furthermore, only CD146-/CD105-enriched populations exhibited elevated CLEC3B expression in response to osteoinduction for 10 days (Figure 6(d)). No reproducible significant differences were observed between dual-labelled populations with and without osteoinduction in OPN, DCN, LOXL4, POSTN and SATB2 expression (Figure 6(b), (e)–(h), respectively). No significant differences in expression were observed in immune regulatory gene CTSC between populations with and without osteoinduction at day 21 (Figure 6(i)). However, higher CTSC expression in Stro-1-/CD146-enriched populations compared to Stro-1-/CD105- and CD146-/CD105-enriched populations after 10 days without osteoinduction was observed (Figure 6(i)). Data were derived from three patient samples.
Figure 6.
Gene expression analysis within dual-labelled populations. Cell populations were isolated via FACS from HBMSCs according to Stro-1/CD146, Stro-1/CD105 and CD146/CD105 expression. Unsorted HBMSCs were used as a control. Samples were cultured for 10 and 21 days in either basal or osteogenic media before rtPCR analysis for (a) ALP, (b) OPN, (c) CADM1, (d) CLEC3B, (e) DCN, (f) LOXL4, (g) POSTN and (h) SATB2 expression. (i) The immune regulatory gene, CTSC, was used as a negative control. Error bars are SD (*p ≤ 0.05, **p ≤ 0.01).
MACS: magnetic-activated cell sorting; HBMSC: human bone marrow stromal cell; rtPCR: real-time polymerase chain reaction; SD: standard deviation.
Histological assessment of in vivo function
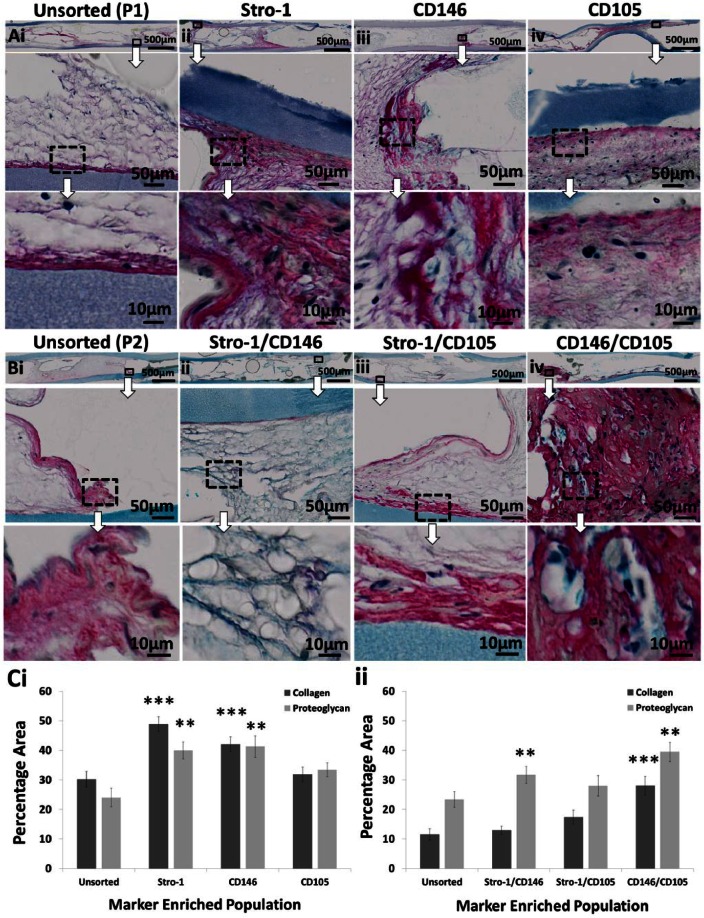
Both single- and dual-labelled populations were cultured in vivo within sealed diffusion chambers implanted subcutaneously in MF1 nu/nu mice. Immunoselected populations generated new disorganised collagen matrix and proteoglycan deposition (Figure 7). Quantification via cross-sectional image analysis demonstrated a significant (p ≤ 0.01) increase in both collagen matrix production and proteoglycan deposition within Stro-1-, CD146- and CD146-/CD105-enriched populations compared to control unsorted populations (Figure 7(C)). Stro-1-/CD146-enriched populations only exhibited enhanced proteoglycan deposition compared to unsorted populations (Figure 7(C)ii). Immunoselected CD105- and Stro-1-/CD105-enriched populations were not significantly different to control unsorted populations in their capacity for collagen synthesis and proteoglycan deposition (Figure 7(C)).
Figure 7.
Histological assessment of single- and dual-labelled population viability/function in vivo. (Ai and Bi) Unsorted, (Aii to Aiv) single- and (Bii to Biv) dual-labelled populations were expanded in vitro and seeded as a cell suspension into diffusion chambers at 5 × 105 cells per chamber without osteoinduction. Chambers were implanted subcutaneously within MF1 nu/nu mice and incubated for 28 days. Following harvest, chambers were sectioned and stained with Alcian Blue/Sirius Red. Collagen and proteoglycan were identified and quantified by image analysis and represented as a percentage of the total sample (C). Overview images of the entire implant are shown on top (scale bar 500 µm). Large images underneath show enlarged regions (scale bar 50 µm). Highly magnified images are shown at the bottom (scale bar 10 µm). Error bars are SD (**p ≤ 0.01, ***p ≤ 0.001).
SD: standard deviation.
Discussion
This study provides evidence that distinct cell populations can be isolated from HBM based on their expression of surface markers Stro-1, CD146 and CD105 alone and in combination.21,33,49 Stro-1 expression is considered a robust marker for the presence of functional SSCs within a given population, and alternative markers CD146 and CD105 enable interrogation of Stro-1-related heterogeneity. Marker-enriched populations exhibited proliferation and differentiation potential characteristic of SSCs compared to unsorted HBMSCs.52 Furthermore, this study indicates that these populations were functional in vivo, capable of generating new collagen matrix and proteoglycan deposition in the absence of host tissue and exogenous osteoinduction.
Heterogeneity within conventional Stro-1-enriched populations highlights the need for new markers to identify and target SSCs.16,28 Accumulating data for a perivascular origin of SSCs has highlighted two enrichment markers, namely, CD146 and CD105.31–33 Interestingly, an alternative approach has identified HOP26, CD49a and SB-10 (non-perivascular specific) as potential SSC markers based on their expression within populations exhibiting enriched CFU-F capacity.53
FACS quantification demonstrated the potential of CD146 and CD105, both alone and in combination, to target a smaller population within HBM than Stro-1 alone. However, not all CD146- and CD105-enriched cells co-expressed Stro-1, showing potential heterogeneity within these enriched populations. Indeed, only a fraction of CD146-enriched cells shared CD105 expression, and vice versa.54
Enrichment was assessed by CFE assay and ALP expression, both accepted markers of skeletal progenitor cells.2,55,56 However, ALP expression has not yet been proven as a predictor, rather a simple indicator of osteogenic potential.57 Although significant fold enrichment of ALP+ colonies was observed within isolated populations, all secondary CFE recorded here was of a significantly lower magnitude than previously published data (6%–10%, compared with 30%–40% recorded by Lo Surdo and Bauer58 at P3). This may be accounted for by the fact that all HBMs used within this study were derived from donors over the age of 65 years. Indeed, D’Ippolitio et al.59 demonstrated reduced ALP+ CFU-F capacity over increasing donor age. However, further clarification is required as other reports indicate that ALP+ CFU-F capacity was maintained across donor age, while Oreffo and colleagues61 found the number and percentage of ALP+ CFU-F showed a significant decrease in osteoporotic patients as compared with controls and osteoarthritic patients, indicating altered differentiation potential. Alternatively, CFE deficit may possibly be due to differences in protocol; a PBS wash 3 h after initial seeding provides inherent selection for the fast adherent cell fraction, thereby making CFE assessment across studies difficult.
In the population of patients examined, Stro-1-/CD146-enriched cell populations did not display significantly increased CFE which contrasts with findings that CFU-F capable Stro-1-/CD146-enriched osteo-precursors can be isolated from periodontal ligament.49 A number of reasons may account for this observation, including prolonged in vitro expansion, population impurity, CFE ‘versus’ CFU-F assay, patient variability and different protocols. Where Stro-1-/CD105-enriched populations showed increased CFE, CD105-enriched populations did not. The CD105-enriched population is itself heterogeneous, and the CD105+/Stro-1− fraction may elicit a negative effect on overall differentiation potential. Significantly increased CFE within CD146/CD105-enriched populations supports their published use as SSC markers.
Our studies indicate Stro-1- and CD146-enriched populations are capable of increased ALP+ colony formation, while Stro-1-/CD105- and CD146-/CD105-enriched populations are capable of increased colony formation. The absence of ALP expression within all dual-labelled populations isolated by FACS may be attributable to additional in vitro culture expansion compared to single-labelled populations, which is known to affect cell proliferation and differentiation potentials with the emergence of a dominating fibroblastic phenotype.62–65
CD146- and CD105-enriched populations appeared to comprise lower proportions of osteo-responsive cells compared to both unsorted and Stro-1-enriched populations due to their significant (p ≤ 0.05) decrease in ALP expression. Interestingly, Stro-1-enriched populations exhibited greater propensity for osteogenic differentiation suggested by increased expression of both early and late osteogenic markers following osteoinduction, POSTN and LOXL4, respectively. Stro-1-/CD146- and CD146-/CD105-enriched populations may comprise more osteo-responsive cells due to a reduction in CADM1 expression (early marker) and increase in CLEC3B expression (late marker). Taken together, the lack of more distinct gene expression profiles for OPN, DCN, LOXL4, POSTN and SATB2 between dual-labelled populations may again be explained by the emergence of a fibroblastic morphology,66–68 potentially through isolation impurity.69 Pro-inflammatory CTSC gene expression may explain low-level osteogenic gene expression as inflammation has previously been shown to down-regulate osteogenic differentiation.36,70,71
Osteochondral differentiation capacity is a prerequisite of SSCs, and as such, isolated populations were assessed in vivo using a MF1 nu/nu mouse model.72–75 Stro-1-, CD146- and CD146-/CD105-enriched populations exhibited enhanced collagen matrix production and proteoglycan deposition (precursors to bone). Stro-1 and CD146 markers can both isolate HBMSC subsets with similar in vitro and in vivo characteristics, although enriched populations remain heterogeneous.76 Stro-1-/CD146-enriched populations did not exhibit significantly increased collagen deposition. However, significantly increased proteoglycan deposition was observed indicating a potential increase in cartilaginous differentiation and possible pre-osteoid formation. Given a longer in vivo period, this proteoglycan-rich matrix could develop toward immature osteoid and subsequently to mature mineralised bone, given the correct spatiotemporal developmental signals. The relationship between Stro-1 and CD146 therefore requires further investigation.
CD105-enriched populations, in this study, did not show elevated collagen synthesis and proteoglycan deposition which correlated with their low ALP+ CFE. The closed nature of the diffusion chamber rather than an open system was pursued to eliminate haematopoietic marrow formation, as vascular infiltration compounds simple preliminary readouts. However, this may have hindered more advanced cartilage and bone formation often observed within a 28-day timescale. Additional studies would be required to elucidate this further since the pericytes lining marrow sinusoids are thought to be the SSC origin.34 It is possible that chambers contained sub-optimal cell numbers, limiting cartilage and bone formation.
Conclusion
This study has provided insight into the composition of heterogeneous HBMSC subsets using alternative and additional markers to Stro-1 including CD146 and CD105. Stro-1 is a robust SSC marker; however, isolation using only Stro-1 expression generates heterogeneous populations. Here, we demonstrate Stro-1-enriched populations exhibited both enhanced ALP+ CFE and differentiation capacity, expected of potential SSCs (the diminutive true bone stem cell fraction within conventional heterogeneous mesenchymal stem cell populations), and that CD146-enrichment demonstrated equivalent enhancement, indicating a perivascular SSC origin. Interestingly, CD146-enriched populations constituted a significantly narrower fraction (2.22%) of HBMSCs compared to Stro-1 (16.29%); however, heterogeneity within these populations remains unclear. Although CD146 provides an alternative to Stro-1, neither CD146 nor CD105 provided superior SSC isolation. Therefore, Stro-1 remains an acceptable SSC marker; however, critical to note is the continued need for specific SSC markers which can isolate pure populations for bone biology investigation in vitro and clinical translation in vivo.
Acknowledgments
The authors would like to thank Mr Douglas Dunlop and colleagues (Southampton General Hospital) for provision of patient bone marrow samples and Dr Janos Kanczler for assistance with the in vivo studies. The authors also thank Prof. Moustapha Kassem, Prof. Basem Abdallah and Ms Bianca Jørgensen for provision of and technical help with osteo-predictive gene primer sequences. Finally, the authors thank Dr Lisa White and Professor Kevin Shakesheff at the Wolfson Centre for Stem cells, Tissue Engineering and Modelling, Nottingham, for provision of HA-PLA scaffold.
Footnotes
Declaration of conflicting interests: No competing financial interests exist.
Funding: The research presented here was funded by the BBSRC (BB/G010579/1).
References
- 1. Caplan AI. Mesenchymal stem cells. J Orthop Res 1991; 9(5): 641–650. [DOI] [PubMed] [Google Scholar]
- 2. Owen M, Friedenstein AJ. Stromal stem cells: marrow-derived osteogenic precursors. Ciba Found Symp 1988; 136: 42–60. [DOI] [PubMed] [Google Scholar]
- 3. Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts and assays. Cell Stem Cell 2008; 2(4): 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parekkadan B, Milwid JM. Mesenchymal stem cells as therapeutics. Annu Rev Biomed Eng 2010; 12: 87–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Panetta NJ, Gupta DM, Quarto N, et al. Mesenchymal cells for skeletal tissue engineering. Panminerva Med 2009; 51(1): 25–41. [PubMed] [Google Scholar]
- 6. Kwan MD, Slater BJ, Wan DC, et al. Cell-based therapies for skeletal regenerative medicine. Hum Mol Genet 2008; 17(R1): R93–R98. [DOI] [PubMed] [Google Scholar]
- 7. Bork S, Horn P, Castoldi M, et al. Adipogenic differentiation of human mesenchymal stromal cells is down-regulated by microRNA-369-5p and up-regulated by microRNA-371. J Cell Physiol 2011; 226(9): 2226–2234. [DOI] [PubMed] [Google Scholar]
- 8. Hudson JE, Mills RJ, Frith JE, et al. A defined medium and substrate for expansion of human mesenchymal stromal cell progenitors that enriches for osteo- and chondrogenic precursors. Stem Cells Dev 2011; 20(1): 77–87. [DOI] [PubMed] [Google Scholar]
- 9. Dawson JI, Kanczler J, Tare RS, et al. Concise review: bridging the gap: bone regeneration using skeletal stem cell-based strategies – where are we now? Stem Cells 2014; 32(1): 35–44. [DOI] [PubMed] [Google Scholar]
- 10. Niehage C, Steenblock C, Pursche T, et al. The cell surface proteome of human mesenchymal stromal cells. PLoS One 2011; 6(5): e20399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harichandan A, Buhring HJ. Prospective isolation of human MSC. Best Pract Res Clin Haematol 2011; 24(1): 25–36. [DOI] [PubMed] [Google Scholar]
- 12. Barry FP, Murphy JM. Mesenchymal stem cells: clinical applications and biological characterization. Int J Biochem Cell Biol 2004; 36(4): 568–584. [DOI] [PubMed] [Google Scholar]
- 13. Bianco P, Kuznetsov SA, Riminucci M, et al. Postnatal skeletal stem cells. Methods Enzymol 2006; 419: 117–148. [DOI] [PubMed] [Google Scholar]
- 14. Lecourt S, Marolleau JP, Fromigue O, et al. Characterization of distinct mesenchymal-like cell populations from human skeletal muscle in situ and in vitro. Exp Cell Res 2010; 316(15):2513–2526. [DOI] [PubMed] [Google Scholar]
- 15. Gothard D, Tare RS, Mitchell PD, et al. In search of the skeletal stem cell: isolation and separation strategies at the macro/micro scale for skeletal regeneration. Lab Chip 2011; 11(7): 1206–1220. [DOI] [PubMed] [Google Scholar]
- 16. Russell KC, Phinney DG, Lacey MR, et al. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells 2010; 28(4): 788–798. [DOI] [PubMed] [Google Scholar]
- 17. Tilley S, Bolland BJ, Partridge K, et al. Taking tissue-engineering principles into theatre: augmentation of impacted allograft with bone marrow stromal cells. Regen Med 2006; 1(5): 685–692. [DOI] [PubMed] [Google Scholar]
- 18. Gronthos S, Zannettino AC, Hay SJ, et al. Molecular and cellular characterisation of highly purified stromal stem cells derived from human bone marrow. J Cell Sci 2003; 116(9): 1827–1835. [DOI] [PubMed] [Google Scholar]
- 19. Simmons PJ, Torok-Storb B. Identification of stromal cell precursors in human bone marrow by a novel monoclonal antibody, STRO-1. Blood 1991; 78(1): 55–62. [PubMed] [Google Scholar]
- 20. Tare RS, Khan F, Tourniaire G, et al. A microarray approach to the identification of polyurethanes for the isolation of human skeletal progenitor cells and augmentation of skeletal cell growth. Biomaterials 2009; 30(6): 1045–1055. [DOI] [PubMed] [Google Scholar]
- 21. Zannettino AC, Paton S, Kortesidis A, et al. Human multipotential mesenchymal/stromal stem cells are derived from a discrete subpopulation of STRO-1bright/CD34/CD45-/Glycophorin-A-bone marrow cells. Haematologica 2007; 92(12): 1707–1708. [DOI] [PubMed] [Google Scholar]
- 22. Gronthos S, Graves SE, Ohta S, et al. The STRO-1+ fraction of adult human bone marrow contains the osteogenic precursors. Blood 1994; 84(12): 4164–4173. [PubMed] [Google Scholar]
- 23. Gronthos S, Simmons PJ. The growth factor requirements of STRO-1-positive human bone marrow stromal precursors under serum-deprived conditions in vitro. Blood 1995; 85(4): 929–940. [PubMed] [Google Scholar]
- 24. Yang XB, Webb D, Blaker J, et al. Evaluation of human bone marrow stromal cell growth on biodegradable polymer/bioglass composites. Biochem Biophys Res Commun 2006; 342(4): 1098–1107. [DOI] [PubMed] [Google Scholar]
- 25. Yang X, Walboomers XF, van den Beucken JJ, et al. Hard tissue formation of STRO-1-selected rat dental pulp stem cells in vivo. Tissue Eng Part A 2009; 15(2): 367–375. [DOI] [PubMed] [Google Scholar]
- 26. De Bari C, Dell’Accio F, Karystinou A, et al. A biomarker-based mathematical model to predict bone-forming potency of human synovial and periosteal mesenchymal stem cells. Arthritis Rheum 2008; 58(1): 240–250. [DOI] [PubMed] [Google Scholar]
- 27. Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair – current views. Stem Cells 2007; 25(11): 2896–2902. [DOI] [PubMed] [Google Scholar]
- 28. Pevsner-Fischer M, Levin S, Zipori D. The origins of mesenchymal stromal cell heterogeneity. Stem Cell Rev 2011; 7(3): 560–568. [DOI] [PubMed] [Google Scholar]
- 29. Ho AD, Wagner W, Franke W. Heterogeneity of mesenchymal stromal cell preparations. Cytotherapy 2008; 10(4): 320–330. [DOI] [PubMed] [Google Scholar]
- 30. MacArthur BD, Tare RS, Please CP, et al. A non-invasive method for in situ quantification of subpopulation behaviour in mixed cell culture. J R Soc Interface 2006; 3(6): 63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sorrentino A, Ferracin M, Castelli G, et al. Isolation and characterization of CD146+ multipotent mesenchymal stromal cells. Exp Hematol 2008; 36(8): 1035–1046. [DOI] [PubMed] [Google Scholar]
- 32. Aslan H, Zilberman Y, Kandel L, et al. Osteogenic differentiation of noncultured immunoisolated bone marrow-derived CD105+ cells. Stem Cells 2006; 24(7): 1728–1737. [DOI] [PubMed] [Google Scholar]
- 33. Jarocha D, Lukasiewicz E, Majka M. Advantage of mesenchymal stem cells (MSC) expansion directly from purified bone marrow CD105+ and CD271+ cells. Folia Histochem Cytobiol 2008; 46(3): 307–314. [DOI] [PubMed] [Google Scholar]
- 34. Sacchetti B, Funari A, Michienzi S, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell 2007; 131(2): 324–336. [DOI] [PubMed] [Google Scholar]
- 35. Bianco P, Robey PG, Saggio I, et al. Mesenchymal stem cells in human bone marrow (skeletal stem cells): a critical discussion of their nature, identity, and significance in incurable skeletal disease. Hum Gene Ther 2010; 21(9): 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Larsen KH, Frederiksen CM, Burns JS, et al. Identifying a molecular phenotype for bone marrow stromal cells with in vivo bone-forming capacity. J Bone Miner Res 2010; 25(4): 796–808. [DOI] [PubMed] [Google Scholar]
- 37. Shi S, Gronthos S. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res 2003; 18(4): 696–704. [DOI] [PubMed] [Google Scholar]
- 38. Mentink A, Hulsman M, Groen N, et al. Predicting the therapeutic efficacy of MSC in bone tissue engineering using the molecular marker CADM1. Biomaterials 2013; 34(19): 4592–4601. [DOI] [PubMed] [Google Scholar]
- 39. Burns JS, Rasmussen PL, Larsen KH, et al. Parameters in three-dimensional osteospheroids of telomerized human mesenchymal (stromal) stem cells grown on osteoconductive scaffolds that predict in vivo bone-forming potential. Tissue Eng Part A 2010; 16(7): 2331–2342. [DOI] [PubMed] [Google Scholar]
- 40. Leoyklang P, Suphapeetiporn K, Siriwan P, et al. Heterozygous nonsense mutation SATB2 associated with cleft palate, osteoporosis, and cognitive defects. Hum Mutat 2007; 28(7): 732–738. [DOI] [PubMed] [Google Scholar]
- 41. Hassan MQ, Gordon JA, Beloti MM, et al. A network connecting Runx2, SATB2, and the miR-23a-27a-24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A 2010; 107(46): 19879–19884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park D, Spencer JA, Koh BI, et al. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell 2012; 10(3): 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu H, Toh WS, Lu K, et al. A subpopulation of mesenchymal stromal cells with high osteogenic potential. J Cell Mol Med 2009; 13(8B): 2436–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gothard D, Roberts SJ, Shakesheff KM, et al. Engineering embryonic stem-cell aggregation allows an enhanced osteogenic differentiation in vitro. Tissue Eng Part C Methods 2010; 16(4): 583–595. [DOI] [PubMed] [Google Scholar]
- 45. Song SJ, Jeon O, Yang HS, et al. Effects of culture conditions on osteogenic differentiation in human mesenchymal stem cells. J Microbiol Biotechnol 2007; 17(7): 1113–1119. [PubMed] [Google Scholar]
- 46. Deng Y, Wu S, Zhou H, et al. Effects of a miR-31, Runx2 and Satb2 regulatory loop on the osteogenic differentiation of bone marrow mesenchymal stem cells. Stem Cells Dev 2013; 22(16): 2278–2286. [DOI] [PubMed] [Google Scholar]
- 47. Gundle R, Beresford JN. The isolation and culture of cells from explants of human trabecular bone. Calcif Tissue Int 1995; 56(Suppl. 1): S8–S10.7719993 [Google Scholar]
- 48. Scott MA, Levi B, Askarinam A, et al. Brief review of models of ectopic bone formation. Stem Cells Dev 2012; 21(5): 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu J, Wang W, Kapila Y, et al. Multiple differentiation capacity of STRO-1+ CD146+ PDL mesenchymal progenitor cells. Stem Cells Dev 2009; 18(3): 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ning H, Lin G, Lue TF, et al. Mesenchymal stem cell marker Stro-1 is a 75kd endothelial antigen. Biochem Biophys Res Commun 2011; 413(2): 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lin G, Liu G, Banie L, et al. Tissue distribution of mesenchymal stem cell marker Stro-1. Stem Cells Dev 2011; 20(10): 1747–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Delorme B, Ringe J, Gallay N, et al. Specific plasma membrane protein phenotype of culture-amplified and native human bone marrow mesenchymal stem cells. Blood 2008; 111(5): 2631–2635. [DOI] [PubMed] [Google Scholar]
- 53. Stewart K, Monk P, Walsh S, et al. STRO-1, HOP-26 (CD63), CD49a and SB-10 (CD166) as markers of primitive human marrow stromal cells and their more differentiated progeny: a comparative investigation in vitro. Cell Tissue Res 2003; 313(3): 281–290. [DOI] [PubMed] [Google Scholar]
- 54. MacArthur BD, Tare RS, Murawski K, et al. Identification of candidate regulators of multipotency in human skeletal progenitor cells. Biochem Biophys Res Commun 2008; 377(1): 68–72. [DOI] [PubMed] [Google Scholar]
- 55. Delorme B, Charbord P. Culture and characterization of human bone marrow mesenchymal stem cells. Methods Mol Med 2007; 140: 67–81. [DOI] [PubMed] [Google Scholar]
- 56. Mostafa NZ, Fitzsimmons R, Major PW, et al. Osteogenic differentiation of human mesenchymal stem cells cultured with dexamethasone, vitamin D3, basic fibroblast growth factor, and bone morphogenetic protein-2. Connect Tissue Res 2011; 53(2): 117–131. [DOI] [PubMed] [Google Scholar]
- 57. Friedenstein AJ, Latzinik NW, Grosheva AG, et al. Marrow microenvironment transfer by heterotopic transplantation of freshly isolated and cultured cells in porous sponges. Exp Hematol 1982; 10(2): 217–227. [PubMed] [Google Scholar]
- 58. Lo Surdo J, Bauer SR. Quantitative approaches to detect donor and passage differences in adipogenic potential and clonogenicity in human bone marrow-derived mesenchymal stem cells. Tissue Eng Part C Methods 2012; 18(11): 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. D’Ippolitio G, Schiller PC, Ricordi C, et al. Age-related osteogenic potential of mesenchymal stromal stem cells from human vertebral bone marrow. J Bone Miner Res 1999; 14(7): 1115–1122. [DOI] [PubMed] [Google Scholar]
- 60. Stenderup K, Jutesen J, Eriksen EF, et al. Number and proliferative capacity of osteogenic stem cells are maintained during aging and in patients with osteoporosis. J Bone Miner Res 2001; 16(6): 1120–1129. [DOI] [PubMed] [Google Scholar]
- 61. Oreffo RO, Bord S, Triffitt JT. Skeletal progenitor cells and ageing human populations. Clin Sci (Lond) 1998; 94(5): 549–555. [DOI] [PubMed] [Google Scholar]
- 62. Huang G, Zheng Q, Sun J, et al. Stabilization of cellular properties and differentiation multipotential of human mesenchymal stem cells transduced with hTERT gene in a long-term culture. J Cell Biochem 2008; 103(4): 1256–1269. [DOI] [PubMed] [Google Scholar]
- 63. Lindner U, Kramer J, Behrends J, et al. Improved proliferation and differentiation capacity of human mesenchymal stromal cells cultured with basement-membrane extracellular matrix proteins. Cytotherapy 2010; 12(8): 992–1005. [DOI] [PubMed] [Google Scholar]
- 64. Javazon EH, Beggs KJ, Flake AW. Mesenchymal stem cells: paradoxes of passaging. Exp Hematol 2004; 32(5): 414–425. [DOI] [PubMed] [Google Scholar]
- 65. Fong CY, Subramanian A, Biswas A, et al. Derivation efficiency, cell proliferation, freeze-thaw survival, stem-cell properties and differentiation of human Wharton’s jelly stem cells. Reprod Biomed Online 2010; 21(3): 391–401. [DOI] [PubMed] [Google Scholar]
- 66. Baxter MA, Wynn RF, Jowitt SN, et al. Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion. Stem Cells 2004; 22(5): 675–682. [DOI] [PubMed] [Google Scholar]
- 67. Bonab MM, Alimoghaddam K, Talebian F, et al. Aging of mesenchymal stem cell in vitro. BMC Cell Biol 2006; 7: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wagner W, Horn P, Castoldi M, et al. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One 2008; 3(5): e2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Halfon S, Abramov N, Grinblat B, et al. Markers distinguishing mesenchymal stem cells from fibroblasts are downregulated with passaging. Stem Cells Dev 2011; 20(1): 53–66. [DOI] [PubMed] [Google Scholar]
- 70. Mann GN, Jacobs TW, Buchinsky FJ, et al. Interferon-gamma causes loss of bone volume in vivo and fails to ameliorate cyclosporin A-induced osteopenia. Endocrinology 1994; 135(3): 1077–1083. [DOI] [PubMed] [Google Scholar]
- 71. Theill LE, Boyle WJ, Penninger JM. RANK-L and RANK: T cells, bone loss, and mammalian evolution. Annu Rev Immunol 2002; 20: 795–823. [DOI] [PubMed] [Google Scholar]
- 72. Stiehler M, Seib FP, Rauh J, et al. Cancellous bone allograft seeded with human mesenchymal stromal cells: a potential good manufacturing practice-grade tool for the regeneration of bone defects. Cytotherapy 2010; 12(5): 658–668. [DOI] [PubMed] [Google Scholar]
- 73. Zou XH, Cai HX, Yin Z, et al. A novel strategy incorporated the power of mesenchymal stem cells to allografts for segmental bone tissue engineering. Cell Transplant 2009; 18(4): 433–441. [DOI] [PubMed] [Google Scholar]
- 74. Goodman SB. Allograft alternatives: bone substitutes and beyond. Orthopedics 2010; 33(9): 661. [DOI] [PubMed] [Google Scholar]
- 75. Lang P, Schumm M, Taylor G, et al. Clinical scale isolation of highly purified peripheral CD34+ progenitors for autologous and allogeneic transplantation in children. Bone Marrow Transplant 1999; 24(6): 583–589. [DOI] [PubMed] [Google Scholar]
- 76. James AW, Zara JN, Corselli M, et al. Use of human perivascular stem cells for bone regeneration. J Vis Exp 2012; 25(63): e2952. [DOI] [PMC free article] [PubMed] [Google Scholar]