Abstract
Fibrosis of the subsynovial connective tissue (SSCT) is a predominant feature of carpal tunnel syndrome (CTS). While the nature of CTS has been extensively studied, little is known about the etiology of this disease. We investigated SSCT tissue from patients with CTS and control subjects using fibrosis arrays and cell culture analysis. Two-fold changes in fibrotic gene expression were found in multiple genes from patient SSCT using fibrosis arrays. This data was confirmed via qRT-PCR on a subset of genes; collagen I (Col1), collagen III (Col3), connective tissue growth factor (CTGF), transforming growth factor β (TGF-β) and SMAD3 (p<0.05), which significantly corroborate the fold changes found in the fibrosis arrays. To further explore the nature of SSCT fibrosis, cells were isolated from patient and control tissue. Col1, Col3, TGF-β, and SMAD3 were highly expressed in patient SSCT fibroblasts as compared to control (p<0.05). Further, fibrotic genes expression was decreased by inhibiting TGF-β receptor I (TβRI) activity (p<0.05). TGF-β second messenger SMAD activity was significantly activated in SSCT fibroblasts from patients and this activation was abrogated by inhibiting TβRI signaling (p<0.05). These findings suggest that blocking TGF-β signaling may be an important therapeutic approach to treating the underlying fibrosis of SSCT in CTS patients.
Keywords: Carpal Tunnel Syndrome, Subsynovial Connective Tissue, Fibrosis, TGF-β
INTRODUCTION
Carpal tunnel syndrome (CTS) is a compression neuropathy of the median nerve that has been associated histologically with a non-inflammatory fibrosis and thickening of the subsynovial connective tissue (SSCT)1–6. Therapeutic and preventative options remain limited, as the initiating events of this non-inflammatory fibrosis are poorly understood7,8.
Previous studies have identified the increased expression of fibrotic factors such as transforming growth factor - beta (TGF-β), connective tissue growth factor, (CTGF), type 1 collagen (Col1) and type 3 collagen (Col3) in the SSCT from CTS patients6. TGF-β is a well characterized pleiotropic growth factor that is involved in fibrosis in many tissue and organ systems9–11.
The goal of this study was to identify mechanisms involved in development and maintenance of SSCT fibrosis in CTS, with the aim of identifying targets for specific anti-fibrotic treatments, in particular TGF-β fibrotic gene expression and regulation was explored. In order to achieve this goal, we examined the SSCT from CTS patients and individuals with no history of CTS using tissue fibrosis arrays and cell culture models. Given that TGF-β is known to play a central role in many fibrotic disorders12,13, and the increases of TGF-β responsive genes in CTS SSCT we targeted TGF-β signaling at the receptor level using the TβRI inhibitor, SD208, to evaluate TGF-β activity and fibrotic gene expression after inhibition.
METHODS
The study was approved by our IRB. Patients were diagnosed with CTS by clinical examination, history, and neurophysiological testing, including electromyography and nerve conduction studies. Human SSCT biopsy tissue from CTS patients (n=5) undergoing carpal tunnel release were harvested as previously reported6. Age matched SSCT from fresh cadavers (n=5) with no history of CTS, harvested within 12 hours postmortem, were used as control tissue. All patient and control tissue was from patients ranging in age from 25–64 years. The SSCT specimen from each patient was divided into two segments, one for cell culture and the other for tissue analysis. The tissue segment was flash frozen and stored at −80C until RNA isolation, as described below. The remaining tissue was used to generate cell cultures, also as described below.
SSCT Tissue Fibrosis Array
Harvested tissue was minced on ice and immersed in 1 mL of Trizol reagent (Invitrogen Life Technologies, Grand Island, NY). Total RNA was isolated from SSCT using Trizol reagent. RNA was quantitated using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Equal amounts of total RNA from tissue were used to synthesize cDNA, using the iScriptTM cDNA Synthesis Kit (Bio-Rad). Gene expression analysis was completed using Human Fibrosis PCR arrays (SA Biosciences –PAHS-120Z, Frederick, MD) according to manufacturer’s protocol. Quantitative real-time PCR using 2× SYBR Green Master Mix was performed at 95C for 10 min, followed by 40 cycles of 95C for 15 sec and 60C for 1 min. Gene expression was normalized to GAPDH as determined by SA Biosciences RT2 Profiler™ PCR Array Data Analysis software (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). Fold changes were calculated by log 2 scale between patient and control tissue.
SSCT Cell Culture
After biopsy, the SSCT was minced in standard 37C culture in Dulbecoco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 1% antibiotic/antimycotic. Cells were then allowed to grow in DMEM, 10% FCS and 1% antibiotic/antimycotic with feeding every third day. Cell cultures were treated either with 5 ng/mL TGF-β1 (R&D Systems, Minneapolis, MN), 50 nM TβRI specific inhibitor, SD208 (Tocris, Minneapolis, MN), or combination of 5 ng/mL TGF-β1 and 50 nM SD208, as per experimental design. The cell cultures were analyzed by real-time PCR for gene expression and for SMAD activity using a luciferase reporter assay.
Quantitative Real-Time PCR
Total RNA was isolated from SSCT and cell culture using Trizol reagent (Invitrogen Life Technologies, Grand Island, NY). RNA was quantitated using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Equal amounts of total RNA from tissue and cell culture were used to synthesize cDNA, using the iScriptTM cDNA Synthesis Kit (Bio-Rad). Real-time PCR was performed in triplicate as previously described14 and values were normalized using β-tubulin as a control. Primers were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/) and were purchased from Integrated DNA Technologies (Coralville, IA). Results were presented normalized to either control or to vehicle conditions.
SMAD Luciferase Reporter Assay
SMAD reporter assays were completed by transiently transfecting cells with a SMAD binding element (SBE) reporter construct, CAGA12-MLP-Luc into SSCT cells as previously described15. Cells were plated in 6 well plates at approximately 50% confluence. The following day cells were transfected with 250 ng of the promoter reporter construct or the empty vector pcDNA4 using fugene-6 transfection reagent (Roche, NH as specified by the manufacturer). SMAD activity was assessed in cells as per experimental design. Briefly, cell extracts were harvested 24 h after transfection using 100 µl of passive lysis buffer. Cell lysates (20 µl) were evaluated for luciferase activity using the Dual-Luciferase reporter assay kit (Promega). Relative luciferase units of activity were reported after subtraction from values for basal levels of expression found in vehicle treated control cells.
Statistical Analysis
Statistical analysis for SA Biosciences RT2 Profiler™ PCR Array significance was determined by the change in gene expression and was evaluated by unpaired Student t-test for each gene. The level of statistical significance is set at p <0.05. Statistical analysis for all cell culture was determined by unpaired 2-tailed Student's t-test at p< 0.05 using Microsoft Excel software. Results are reported as mean ± SE.
RESULTS
SSCT Tissue Fibrosis Analysis
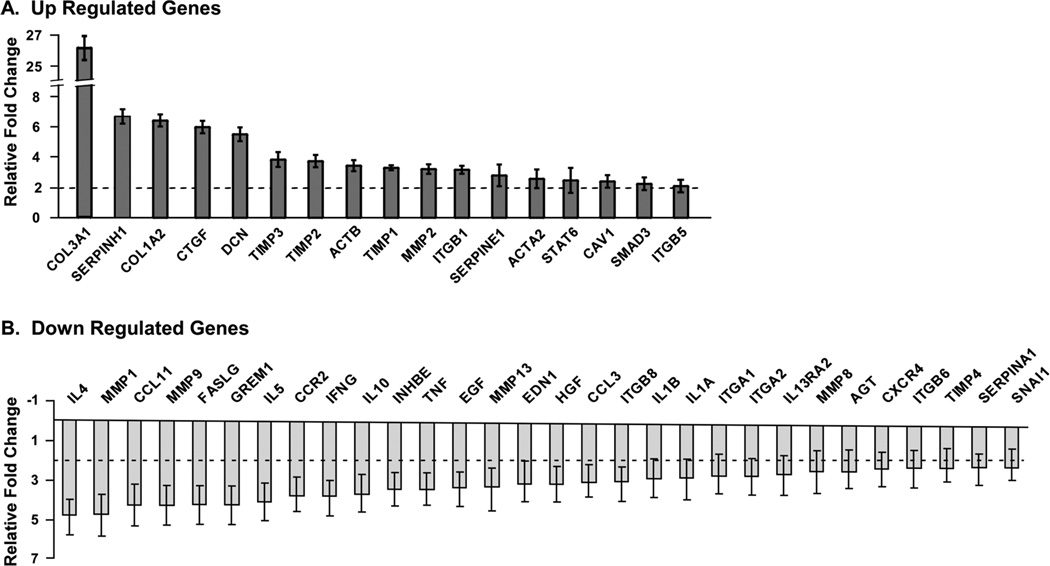
Fibrosis array comparison of CTS patient SSCT with control SSCT resulted in 18 genes regulated at least 2 fold greater than control tissue. In addition 30 genes were down regulated at least 2 fold in patient tissue compared to control tissue. (Figure 1) Genes that were up regulated included extracellular matrix proteins or receptors such as collagen type IIIA1 (Col3), collagen type IA2(Col1), decorin (DCN), smooth muscle actin (ACTA2), beta-actin (ACTB), caveolin (CAV1) and fibronectin receptor integrin beta 1 (ITGB1); signaling second messengers involved in regulating fibrosis including connective tissue growth factor (CTGF), SMAD3 and STAT6 and increases in extracellular matrix regulators such as tissue inhibitors of metalloproteases (TIMPs), matrix metalloproteases (MMPs), serpine peptidase inhibitors (SERPINE1H and SERPINE1) and integrin beta 5 (ITGB5). Genes that were down regulated included interleukins (IL) and MMPs. A full list of genes and fold changes are provided in supplemental data Table 1.
Figure 1.
Fibrosis tissue array relative fold change in genes from patient SSCT compared to control tissue A) increased gene expression and B) decreased gene expression relative to control tissue. Results are reported as mean ± standard error, n=5.
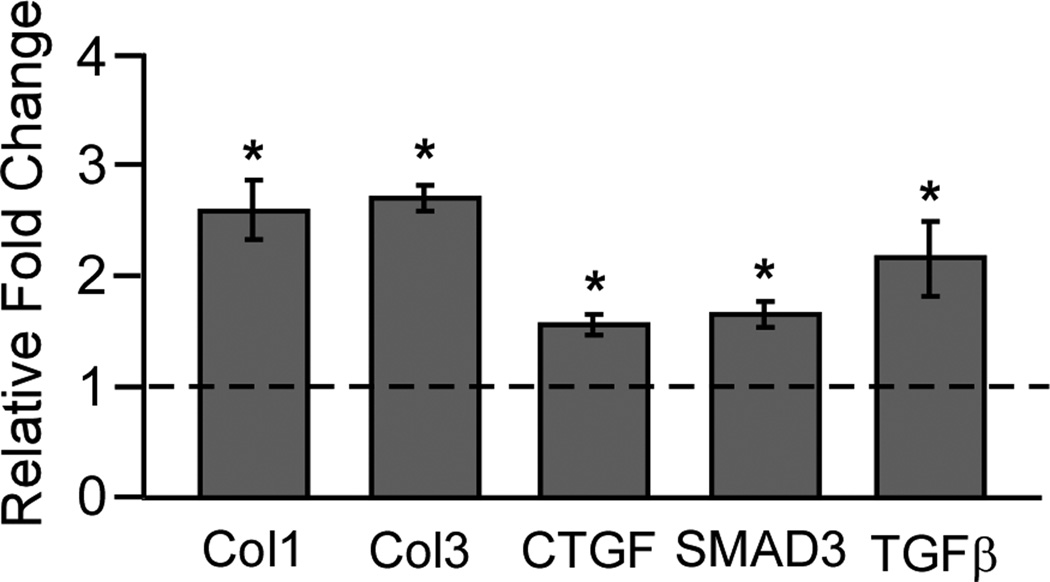
Previous work has shown increases in TGF-β, TGF-β receptors, CTGF, Col1 and Col3 in the SSCT of CTS patients6,16,17. To confirm the tissue microarray data, TGF-β pathway and collagen genes were analyzed by quantitative RT-PCR. Genes were selected based on 2 fold or greater expression differences and/or TGF-β pathway genes known to be important markers of SSCT fibrosis including Col1, Col3, TGF-β, CTGF and SMAD3. Tissue gene expression found in fibrosis array was confirmed using unique primers. (Table 1) The results shown in Figure 2 indicate that genes that had two-fold or greater changes in the microarray Col1, Col3, CTGF, and SMAD3 were significantly up regulated as compared to control tissue. In addition, TGF-β1 which had a 1.6 fold increase in the microarray and is found highly expressed in CTS SSCT was significant in patient SSCT compared to control tissue.
Table 1.
Primers Sets Used in Real-Time PCR
| Gene | Forward | Reverse |
|---|---|---|
| CTGF | TCCCACCCAATTCAAAACAT | TGCTCCTAAAGCCACACCTT |
| COL1A2 | TCCAAAGGAGAGAGCGGTAA | CAGATCCAGCTTCCCCATTA |
| COL3A1 | CCAGGAGCTAACGGTCTCAG | CAGGGTTTCCATCTCTTCCA |
| TGFB1 | GTGGAAACCCACAACGAAAT | CGGAGCTCTGATGTGTTGAA |
| SMAD3 | GGGCTCCCTCATGTCATCTA | TTGAAGGCGAACTCACACAG |
| TUBULIN | GAGTGCATCTCCATCCACGTT | TAGAGCTCCCAGCAGGCATT |
Figure 2.
Confirmation of tissue fibrosis array gene expression was analyzed by quantitative RT-PCR. Results are normalized to control tissue, mean ± SE, n=5.* denotes significance at P<0.05.
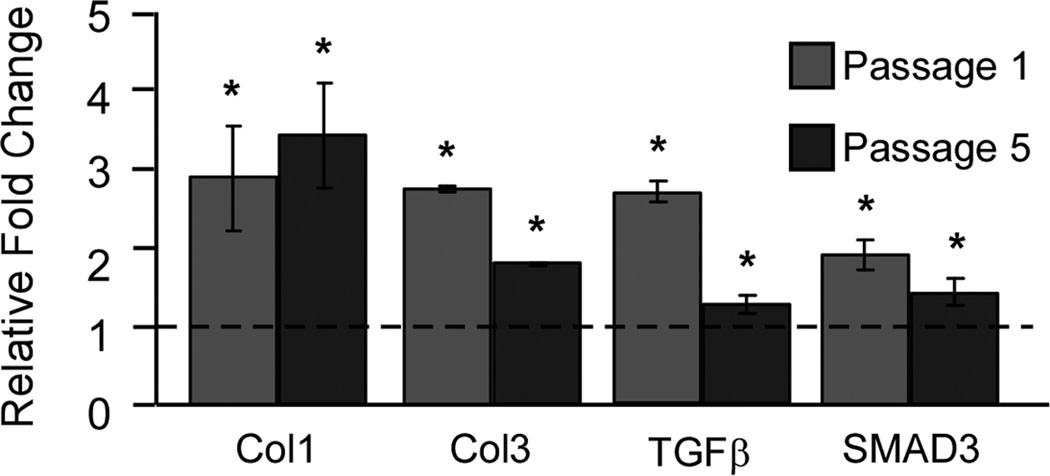
In order to further explore the underlying fibrosis in CTS, SSCT cell culture analysis was completed. Gene expression of Col1, Col3, TGF-β1 and SMAD 3 remained significantly elevated in normal cells as compared to control SSCT. Cells serially passaged five times retained this significantly increased expression. (Figure 3)
Figure 3.
Cultured SSCT fibroblasts maintain fibrotic gene signature using quantitative RT-PCR analysis of Col1, Col3, TGF-β, and SMAD3 in cells isolated from CTS patients. Cells maintain fibrotic gene expression through serial passages. Results are normalized to control cells, mean ± SE, n=3.* denotes significance at P<0.05.
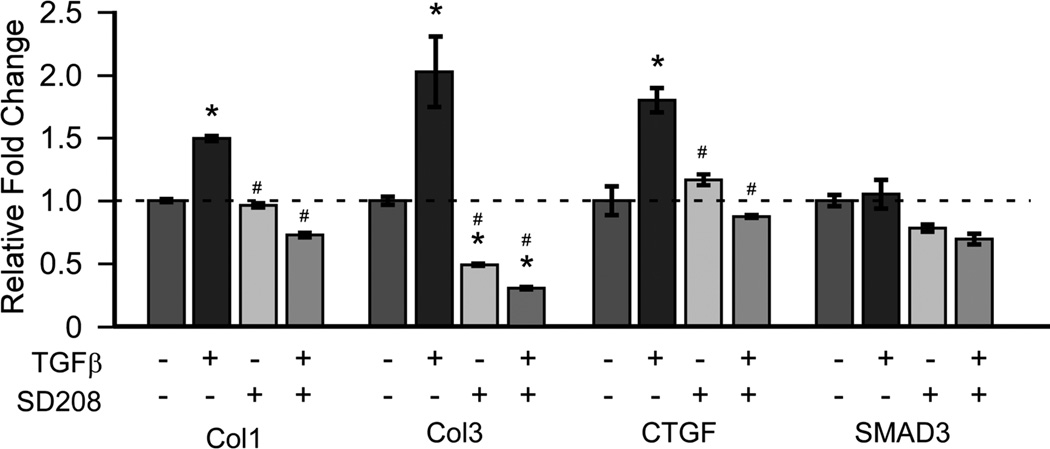
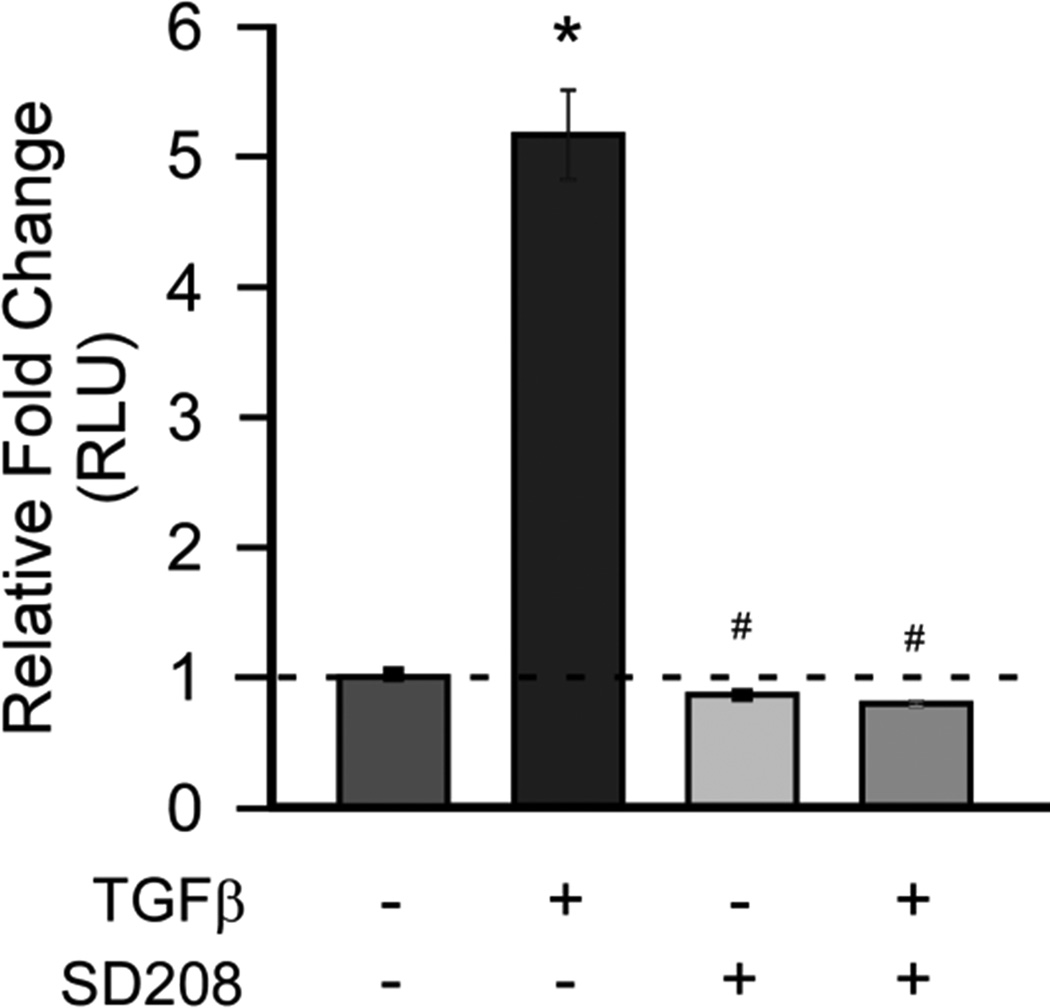
To evaluate TGF-β mediated gene expression and inhibition cultures were treated with TGF-β1, TβRI inhibitor (SD208) or combined TGF-β1 and SD208. Blocking TβRI activity resulted in significant decreases in the expression of Col1, Col3, and CTGF after 24 hours. (Figure 4) To assess the activation of canonical TGF-β signaling a SMAD reporter assay was used to determine responsiveness to TGF-β and to the TβRI inhibitor (SD208) in CTS patient SSCT fibroblast cell cultures. TGF-β1 treatment resulted in significantly increased SMAD reporter activity, while this activity was completely blocked by the TβRI inhibitor after 24 hours of treatment. (Figure 5)
Figure 4.
Inhibition of TβRI in SSCT patient cells down regulates fibrotic gene expression. Quantitative RT-PCR analysis of Col1, Col3, CTGF and SMAD3 in SSCT cells isolated from CTS patients and were treated for 24 hours ± TGF-β (5ng/mL) and ± SD208 (50 nM). The mean ± SE are depicted, n=3.* denotes significance from vehicle, # denotes significance from TGF-β treatment at P<0.05.
Figure 5.
Patient SSCT fibroblasts have increased responsiveness to TGF-β which is significantly reduced by SD208 (TβRI inhibitor). Relative fold changes in relative SMAD luciferase units (RLU) are reported. Cells were treated for 24 hours ± TGF-β (5ng/mL) and ± SD208 (50 nM). The mean ± SE are depicted, n=3.* denotes significance from vehicle, # denotes significance from TGF-β treatment at P<0.05.
DISCUSSION
Non-inflammatory synovial fibrosis is commonly observed in idiopathic CTS1–4,6,18. Since fibrosis appears to play an important role in the pathology of this disease we sought to characterize the fibrosis in CTS and to identify potential therapeutic targets.
In this study we explored the nature of fibrosis in SSCT from five patients. Tissue fibrosis arrays were used to identify candidate genes involved in this fibrosis. We found increased expression of fibrotic genes, TGF-β second messengers (SMAD3, CTGF), and in downstream endpoints such as collagens, TIMPs as well as overall decreases in MMPs. We also observed decreases in inflammatory markers such as interleukins and interferon gamma, which correlate with reports on the non-inflammatory nature of CTS2,4, but these changes were not significant.
TGF-β is secreted in a latent form and can be bound by latency binding proteins, which allow for the sequestering and localization in extracellular matrix. However beyond storage of TGF-β, extracellular matrix also provides for the targeted presentation of TGF-β to cells and for the activation of TGF-β, thereby making the cross-talk between matrix deposition and TGF-β availability and activation an important mechanisms in fibrosis and for the expression of TGF-β responsive genes. Indeed prolonged TGF-β signaling is a characteristic of fibrosis in many tissues19.
TGF-β signaling is propagated through serine/threonine receptors of the activin receptor-like kinase receptor family20. TGF-β binds to TGF-β receptor II (TβRII), which recruits and cross phosphorylates TGF-β receptor I (TβRI). Once activated TβRI phosphorylates receptor-SMAD proteins, canonical modulators of TGF-β signaling21. SMADs form a trimer of two receptor SMADs and common mediator SMAD (co-SMAD), that translocates to the nucleus and acts as a transcription factor initiating gene expression. Common targets of TGF-β signaling include extracellular matrix proteins such as collagen, fibronectin, proteoglycan and tissue regulators such as MMPs and TIMPs22–24.
Previous work has shown that TGF-β expression levels are high in SSCT6. Serum levels of TGF-β have been found at least as high as 22 ng/mL25 While TGF-β is known to be highly bound by latency proteins, activation rates have been shown to be 0.5 ng/mL per hour26. Given the rate of TGF-β1 activation, the increased availability of TGF-β1 in the SSCT and extended culture conditions we believe 5ng/mL represents a conservative estimate of the active TGF-β1 that would be found in vivo and therefore represents a reasonable dose in cell culture. In addition it has been previously shown that TGF-β in fetal calf serum and fetal bovine serum is not active27. However given that TGF-β in the serum may be activated by cells in cell culture this is adjusted for by comparison with control treatments.
In addition, connective tissue growth factor (CTGF), also known as CCN2, strongly associates with both TGF-β signaling and fibrosis6,28–31. CTGF, despite its name, is a matricellular protein rather than a growth factor. CTGF acts by modulating responsiveness of growth factors and receptors and by proteolytic activity. Indeed overexpression of CTGF in animal models does not predictably result in fibrotic activity and may act more in a co-stimulatory manner32,33. CTGF expression is induced by TGF-β34,35. CTGF can also potentiate TGF-β signaling by enhancing the binding of TGF-β to TGF-β receptors34,36. In addition, CTGF in conjunction with TGF-β has been found to sustain fibrotic activity37.
In order to validate tissue fibrosis arrays and explore genes of interest, unique primers were designed for quantitative RT-PCR. In particular TGF-β1 expression was evaluated despite only showing a 1.6 fold increase in fibrosis arrays as protein levels of TGF-β1 are highly expressed in patient SSCT6. Previous work has shown that even small increases in TGF-β expression can have profound effects in the local environment and changes in receptors or second messengers such as SMAD3 can amplify the TGF-β associated responsiveness38. Given that TGF-β is a common mediator of fibrosis in tissue and organ systems19,39,40 and that significant increases are found in both TGF-β1 and TGF-β receptors in SSCT of CTS patients, we chose to focus on this pathway as a potential regulator and therapeutic target6,16,18.
At the cellular level we cultured fibroblasts isolated from the SSCT in both patients and control tissue and evaluated the expression of TGF-β, SMAD3, and collagens (Col1 and Col3). TGF-β remained significantly expressed in SSCT fibroblasts from patients as compared to control fibroblasts. CTGF was not significantly regulated in these cultures; however this may be due to the lack of exogenous TGF-β1 in these cultures as CTGF expression is regulated in large part via TGF-β activation6,28,30,41. It is interesting to note that SMAD3 total expression was increased in patient fibroblasts as well as tissues, which may confer an increased responsiveness to TGF-β signaling. Furthermore Col1 and Col3 were significantly increased in both tissue and fibroblast cultures. The accumulation of Col1 and Col3 that is seen in SSCT fibrosis may result from a TGF-β mediated increase in extracellular matrix and impaired degradation. Additionally, TGF-β promotes a positive feedback loop between deposition of matrix and activation of TGF-β42,43. Verrecchia et al. identified Col1A2, Col3A1 as well as other collagens and TIMP1 as TGF-β and SMAD3 target genes in fibroblasts.24 Indeed in the tissue fibrosis array TIMP1 levels are increased over three fold. Exploration and validation of extracellular matrix enzymes such as TIMPs and MMPs were not explored in this study, but are a focus on ongoing research.
Interestingly, SMAD expression also remains high in the SSCT cell culture compared to normal cells even without TGF-β1 induction. (Figure 3) Furthermore significantly increased expression Col1 and Col3, both of which are regulated by TGF-β1 signaling, were maintained in cell culture in patient SSCT fibroblasts compared to normal SSCT fibroblasts. This increased fibrotic expression was significantly maintained through five serial passages, indicating a potential long-term phenotypic change in patient SSCT fibroblasts.
To further evaluate the effect of TGF-β1 activity in patient SSCT fibroblasts we evaluated the effect of a TβRI inhibitor to block fibrotic gene expression. All cells were treated with TGF-β1 ± SD208, a TβRI inhibitor. For all genes analyzed, Col1, Col3, and CTGF the TβRI inhibitor significantly reduced expression in patient SSCT fibroblasts compared to normal SSCT fibroblasts. Interestingly, CTGF is induced by TGF-β as well as by mechanical stimulation and this induction is mediated at least in part through the activation of SMAD3 signaling41,44,45, indicating again, the link between CTGF, TGF-β and the underlying fibrosis in CTS patients. SMAD3 expression was not significantly regulated in culture by addition of TGF-β or by inhibiting TβRI activity. However, SMAD action results from activation of the SMAD protein by phosphorylation and translocation to the nucleus rather than total SMAD levels. Therefore changes in total SMAD amounts after 24 hours of treatment may not be the mechanism of SMAD action; rather SMAD3 activation and activity are a more relevant measure.
SMAD activity was analyzed in SSCT fibroblasts from CTS patients to further examine TGF-β signaling. Patient SSCT fibroblasts were treated with TGF-β1 and a significant increase in SMAD activity was evident. In addition we looked to specifically block TGF-β signaling using the TβRI inhibitor, SD20813,40. Upon treatment, SMAD activation was reduced to control levels, indicating that TβRI blocks canonical SMAD3 signaling in patient SSCT fibroblasts.
The net accumulation of collagen in fibrotic tissue comes as a result of misregulation between deposition and degradation of extracellular matrix. In this case Col1 and Col3 expression is significantly enhanced in both patient tissue and fibroblast derived from this tissue. This expression corresponds to the previously reported increased protein expression of Col1 and Col3 in patient SSCT6. When TGF-β1 signaling is blocked by inhibiting TβRI, expression levels of Col1 and Col3 are decreased indicating that TGF-β1 could be an important regulator of extracellular matrix deposition in patient SSCT fibrosis.
Limitations of this study include a relatively small sample size. The tissue fibrosis arrays provided indications of fold regulation of multiple genes in the SSCT of CTS patients, however likely due to the idiopathic nature of CTS and resulting variation and sample size no significance was determined. Fibrosis arrays served as an indicator of potentially regulated genes in SSCT fibrosis. In contrast when quantitative RT-PCR was completed on the same tissue samples significance was determined for Col1, Col3, CTGF, SMAD3, and TGF-β. In addition, the use of cadaver tissue as normal control tissue may not be ideal; however given the nature of surgical requirement for tissue acquisition it would not be ethical, in our opinion, to take such tissue from people who do not have CTS. The strengths of this study include the significant gene expression changes in this cohort, both in the tissue and fibroblasts derived from this tissue
Targeting the TGF-β pathway has impacts on life threatening diseases ranging from cancer, induce pulmonary fibrosis and diabetic related fibrosis to life-long quality of life diseases such as scleroderma and carpal tunnel syndrome. Multiple drugs have been developed to target TGF-β pathway including drugs that target ligands, receptors, and second messenger SMAD pathways20,46. Many of these drugs have been delivered systemically with some success, however concerns over systemic treatments make local delivery of antifibrotic agents appealing20. In addition studies are now being initiated to look at short-term or combinatorial treatments that may have substantial therapeutic benefits for treating TGF-β related pathologies such as fibrosis. Indeed topical treatment of scleroderma has shown some benefit in reducing fibrosis47, indicating that both systemic and local delivery of TGF-β modulators may be therapeutic options to treat CTS. It remains of great importance to modulate this fibrotic signal without interfering with the beneficial systemic actions of TGF-β.
In summary, in this study we explored the nature of the fibrotic expression found in the CTS SSCT and cells of patients with CTS. We noted increased fibrotic gene expression, including but not limited to increases in Col1, Col3, CTGF and SMAD3. These genes were confirmed using quantitative RT-PCR and were found to be significantly up regulated in patient tissue as compared to control tissue. Fibroblasts were isolated from SSCT from both patient and cadaver tissue. We confirmed significantly increased gene expression for Col1 and 3, TGF-β and SMAD3 in CTS fibroblasts maintained over time. Further, we found that the TβRI inhibitor, SD208, significantly reduced fibrotic gene expression and canonical TGF-β signaling. These studies are the first to examine blocking TGF-β signaling in CTS and suggest that therapies targeted for the TGF-β signaling pathway may ultimately have utility for the prevention and/or treatment of CTS.
Supplementary Material
Acknowledgments
Supported by grants from NIH/NIAMS (AR49823); NIH/NIAMS F32 (AR063596); and NIH/NIAMS T32 (AR056950).
REFERENCES
- 1.Lluch AL. Thickening of the synovium of the digital flexor tendons: cause or consequence of the carpal tunnel syndrome? J Hand Surg Br. 1992;17:209–212. doi: 10.1016/0266-7681(92)90091-f. [DOI] [PubMed] [Google Scholar]
- 2.Donato G, Galasso O, Valentino P, et al. Pathological findings in subsynovial connective tissue in idiopathic carpal tunnel syndrome. Clin Neuropathol. 2009;28:129–135. doi: 10.5414/npp28129. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs PC, Nathan PA, Myers LD. Synovial histology in carpal tunnel syndrome. J Hand Surg Am. 1991;16:753–758. doi: 10.1016/0363-5023(91)90208-s. [DOI] [PubMed] [Google Scholar]
- 4.Schuind F, Ventura M, Pasteels JL. Idiopathic carpal tunnel syndrome: histologic study of flexor tendon synovium. J Hand Surg Am. 1990;15:497–503. doi: 10.1016/0363-5023(90)90070-8. [DOI] [PubMed] [Google Scholar]
- 5.Ettema AM, Zhao C, An KN, Amadio PC. Comparative anatomy of the subsynovial connective tissue in the carpal tunnel of the rat, rabbit, dog, baboon, and human. Hand (N Y) 2006;1:78–84. doi: 10.1007/s11552-006-9009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chikenji T, Gingery A, Zhao C, et al. Transforming growth factor-beta (TGF-beta) expression is increased in the subsynovial connective tissues of patients with idiopathic carpal tunnel syndrome. J Orthop Res. 2014;32:116–122. doi: 10.1002/jor.22485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keith MW, Masear V, Chung KC, et al. American Academy of Orthopaedic Surgeons Clinical Practice Guideline on diagnosis of carpal tunnel syndrome. J Bone Joint Surg Am. 2009;91:2478–2479. doi: 10.2106/JBJS.I.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keith MW, Masear V, Chung KC, et al. American Academy of Orthopaedic Surgeons clinical practice guideline on the treatment of carpal tunnel syndrome. J Bone Joint Surg Am. 2010;92:218–219. doi: 10.2106/JBJS.I.00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prud'homme GJ. Pathobiology of transforming growth factor beta in cancer, fibrosis and immunologic disease, and therapeutic considerations. Lab Invest. 2007;87:1077–1091. doi: 10.1038/labinvest.3700669. [DOI] [PubMed] [Google Scholar]
- 10.Warburton D. Developmental responses to lung injury: repair or fibrosis. Fibrogenesis Tissue Repair. 2012;5(Suppl 1):S2. doi: 10.1186/1755-1536-5-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauldie J, Bonniaud P, Sime P, et al. TGF-beta, Smad3 and the process of progressive fibrosis. Biochem Soc Trans. 2007;35:661–664. doi: 10.1042/BST0350661. [DOI] [PubMed] [Google Scholar]
- 12.Hawinkels LJ, Ten Dijke P. Exploring anti-TGF-beta therapies in cancer and fibrosis. Growth Factors. 2011;29:140–152. doi: 10.3109/08977194.2011.595411. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Shi-wen X, Eastwood M, et al. Contribution of activin receptor-like kinase 5 (transforming growth factor beta receptor type I) signaling to the fibrotic phenotype of scleroderma fibroblasts. Arthritis Rheum. 2006;54:1309–1316. doi: 10.1002/art.21725. [DOI] [PubMed] [Google Scholar]
- 14.Hawse JR, Cicek M, Grygo SB, et al. TIEG1/KLF10 Modulates Runx2 Expression and Activity in Osteoblasts. PLoS One. 2011;6:e19429. doi: 10.1371/journal.pone.0019429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnsen SA, Subramaniam M, Monroe DG, et al. Modulation of transforming growth factor beta (TGFbeta)/Smad transcriptional responses through targeted degradation of TGFbeta-inducible early gene-1 by human seven in absentia homologue. J Biol Chem. 2002;277:30754–30759. doi: 10.1074/jbc.M204812200. [DOI] [PubMed] [Google Scholar]
- 16.Ettema AM, Amadio PC, Zhao C, et al. A histological and immunohistochemical study of the subsynovial connective tissue in idiopathic carpal tunnel syndrome. J Bone Joint Surg Am. 2004;86-A:1458–1466. doi: 10.2106/00004623-200407000-00014. [DOI] [PubMed] [Google Scholar]
- 17.Kim JK, Koh YD, Kim JS, et al. Oxidative stress in subsynovial connective tissue of idiopathic carpal tunnel syndrome. J Orthop Res. 2010;28:1463–1468. doi: 10.1002/jor.21163. [DOI] [PubMed] [Google Scholar]
- 18.Sun YL, Moriya T, Zhao C, et al. Subsynovial connective tissue is sensitive to surgical interventions in a rabbit model of carpal tunnel syndrome. J Orthop Res. 2012;30:649–654. doi: 10.1002/jor.21565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samarakoon R, Overstreet JM, Higgins PJ. TGF-beta signaling in tissue fibrosis: redox controls, target genes and therapeutic opportunities. Cell Signal. 2012;25:264–268. doi: 10.1016/j.cellsig.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nature reviews. Drug discovery. 2012;11:790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millan FA, Denhez F, Kondaiah P, Akhurst RJ. Embryonic gene expression patterns of TGF beta 1, beta 2 and beta 3 suggest different developmental functions in vivo. Development. 1991;111:131–143. doi: 10.1242/dev.111.1.131. [DOI] [PubMed] [Google Scholar]
- 22.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 23.Verrecchia F, Mauviel A. Transforming growth factor-beta signaling through the Smad pathway: role in extracellular matrix gene expression and regulation. J Invest Dermatol. 2002;118:211–215. doi: 10.1046/j.1523-1747.2002.01641.x. [DOI] [PubMed] [Google Scholar]
- 24.Verrecchia F, Chu ML, Mauviel A. Identification of novel TGF-beta /Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J Biol Chem. 2001;276:17058–17062. doi: 10.1074/jbc.M100754200. [DOI] [PubMed] [Google Scholar]
- 25.Pscherer S, Freude T, Forst T, et al. Anti-diabetic treatment regulates pro-fibrotic TGF-beta serum levels in type 2 diabetics. Diabetology & metabolic syndrome. 2013;5:48. doi: 10.1186/1758-5996-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albro MB, Cigan AD, Nims RJ, et al. Shearing of synovial fluid activates latent TGF-beta. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2012;20:1374–1382. doi: 10.1016/j.joca.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khosla S, Oursler MJ, Schroeder MJ, Eberhardt NL. Transforming growth factor-beta 1 induces growth inhibition of a human medullary thyroid carcinoma cell line despite an increase in steady state c-myc messenger ribonucleic acid levels. Endocrinology. 1994;135:1887–1893. doi: 10.1210/endo.135.5.7956909. [DOI] [PubMed] [Google Scholar]
- 28.Leask A. Signaling in fibrosis: targeting the TGF beta, endothelin-1 and CCN2 axis in scleroderma. Front Biosci (Elite Ed) 2009;1:115–122. doi: 10.2741/E12. [DOI] [PubMed] [Google Scholar]
- 29.Abraham D. Connective tissue growth factor: growth factor, matricellular organizer, fibrotic biomarker or molecular target for anti-fibrotic therapy in SSc? Rheumatology (Oxford) 2008;47(Suppl 5):v8–v9. doi: 10.1093/rheumatology/ken278. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Liu H, Meyer C, et al. TGF-beta mediated connective tissue growth factor (CTGF) expression in hepatic stellate cells requires Stat3 activation. J Biol Chem. 2013 doi: 10.1074/jbc.M113.478685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh J, Zhao C, Zobitz ME, et al. Morphological changes of collagen fibrils in the subsynovial connective tissue in carpal tunnel syndrome. J Bone Joint Surg Am. 2006;88:824–831. doi: 10.2106/JBJS.E.00377. [DOI] [PubMed] [Google Scholar]
- 32.Panek AN, Posch MG, Alenina N, et al. Connective tissue growth factor overexpression in cardiomyocytes promotes cardiac hypertrophy and protection against pressure overload. PLoS One. 2009;4:e6743. doi: 10.1371/journal.pone.0006743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong Z, Chen R, Alt DS, et al. Susceptibility to liver fibrosis in mice expressing a connective tissue growth factor transgene in hepatocytes. Hepatology. 2009;50:939–947. doi: 10.1002/hep.23102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abreu JG, Ketpura NI, Reversade B, De Robertis EM. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol. 2002;4:599–604. doi: 10.1038/ncb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parisi MS, Gazzerro E, Rydziel S, Canalis E. Expression and regulation of CCN genes in murine osteoblasts. Bone. 2006;38:671–677. doi: 10.1016/j.bone.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Ikawa Y, Ng PS, Endo K, et al. Neutralizing monoclonal antibody to human connective tissue growth factor ameliorates transforming growth factor-beta-induced mouse fibrosis. J Cell Physiol. 2008;216:680–687. doi: 10.1002/jcp.21449. [DOI] [PubMed] [Google Scholar]
- 37.Mori T, Kawara S, Shinozaki M, et al. Role and interaction of connective tissue growth factor with transforming growth factor-beta in persistent fibrosis: A mouse fibrosis model. J Cell Physiol. 1999;181:153–159. doi: 10.1002/(SICI)1097-4652(199910)181:1<153::AID-JCP16>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 38.Ihn H, Yamane K, Kubo M, Tamaki K. Blockade of endogenous transforming growth factor beta signaling prevents up-regulated collagen synthesis in scleroderma fibroblasts: association with increased expression of transforming growth factor beta receptors. Arthritis Rheum. 2001;44:474–480. doi: 10.1002/1529-0131(200102)44:2<474::AID-ANR67>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 39.Satish L, Gallo PH, Baratz ME, et al. Reversal of TGF-beta1 stimulation of alpha-smooth muscle actin and extracellular matrix components by cyclic AMP in Dupuytren's-derived fibroblasts. BMC Musculoskelet Disord. 2011;12:113. doi: 10.1186/1471-2474-12-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohnuma-Koyama A, Yoshida T, Tajima-Horiuchi H, et al. Didecyldimethylammonium chloride induces pulmonary fibrosis in association with TGF-beta signaling in mice. Exp Toxicol Pathol. 2013;65:1003–1009. doi: 10.1016/j.etp.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Grotendorst GR. Connective tissue growth factor: a mediator of TGF-beta action on fibroblasts. Cytokine Growth Factor Rev. 1997;8:171–179. doi: 10.1016/s1359-6101(97)00010-5. [DOI] [PubMed] [Google Scholar]
- 42.Hubmacher D, Apte SS. The biology of the extracellular matrix: novel insights. Curr Opin Rheumatol. 2013;25:65–70. doi: 10.1097/BOR.0b013e32835b137b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horiguchi M, Ota M, Rifkin DB. Matrix control of transforming growth factor-beta function. J Biochem. 2012;152:321–329. doi: 10.1093/jb/mvs089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grotendorst GR, Duncan MR. Individual domains of connective tissue growth factor regulate fibroblast proliferation and myofibroblast differentiation. Faseb J. 2005;19:729–738. doi: 10.1096/fj.04-3217com. [DOI] [PubMed] [Google Scholar]
- 45.Wang S, Denichilo M, Brubaker C, Hirschberg R. Connective tissue growth factor in tubulointerstitial injury of diabetic nephropathy. Kidney Int. 2001;60:96–105. doi: 10.1046/j.1523-1755.2001.00776.x. [DOI] [PubMed] [Google Scholar]
- 46.Nagaraj NS, Datta PK. Targeting the transforming growth factor-beta signaling pathway in human cancer. Expert opinion on investigational drugs. 2010;19:77–91. doi: 10.1517/13543780903382609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merkel PA, Silliman NP, Denton CP, et al. Validity, reliability, and feasibility of durometer measurements of scleroderma skin disease in a multicenter treatment trial. Arthritis Rheum. 2008;59:699–705. doi: 10.1002/art.23564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.