Abstract
Lipoprotein lipase (LPL) has been highly conserved through vertebrate evolution, making it challenging to generate useful antibodies. Some polyclonal antibodies against LPL have turned out to be nonspecific, and the available monoclonal antibodies (Mab) against LPL, all of which bind to LPL’s carboxyl terminus, have drawbacks for some purposes. We report a new LPL-specific monoclonal antibody, Mab 4-1a, which binds to the amino terminus of LPL (residues 5–25). Mab 4-1a binds human and bovine LPL avidly; it does not inhibit LPL catalytic activity nor does it interfere with the binding of LPL to heparin. Mab 4-1a does not bind to human hepatic lipase. Mab 4-1a binds to GPIHBP1-bound LPL and does not interfere with the ability of the LPL–GPIHBP1 complex to bind triglyceride-rich lipoproteins. Mab 4-1a will be a useful reagent for both biochemists and clinical laboratories.
Lipoprotein lipase (LPL) is a crucial enzyme for the hydrolysis of triglycerides in plasma lipoproteins [1–3]. LPL is synthesized by adipocytes and myocytes and secreted into the interstitial spaces. The LPL is then picked up by GPIHBP1 (a glycosylphosphatidylinositol-anchored protein of capillary endothelial cells) and shuttled to the luminal face of capillaries. In the absence of GPIHBP1, LPL remains in the interstitial spaces around adipocytes and myocytes and never reaches its site of action within the capillary lumen [4]. A recent study by Gin and coworkers [5] suggested that the GPIHBP1–LPL complex may be crucial for the binding of triglyceride-rich lipoproteins (TRLs) to endothelial cells [5]. TRLs bound to the LPL–GPIHBP1 complex on the cell surface but not to GPIHBP1 alone [5]. GPIHBP1 and LPL are essential for the lipolytic processing of TRLs. A deficiency of either protein results in severe hypertriglyceridemia (chylomicronemia) [6, 7] and impairs the delivery of lipid nutrients to parenchymal cells [8, 9].
LPL is a key player in human plasma triglyceride metabolism, but studies of LPL biochemistry and function have been hampered by a paucity of antibody reagents. LPL is highly conserved in vertebrates, making it challenging to generate antibodies [10]. Widely used polyclonal antibodies against LPL have proven to be nonspecific [11]. Two mouse monoclonal antibodies (Mab) against bovine LPL, 5D2 and 5F9 [12–14], have been widely used. Both bind to the carboxyl-terminal portion of bovine LPL and cross-react with human LPL (hLPL) [13]. Mab 5D2 has been useful for measurements of LPL mass [12, 15], but it is not suitable for some studies because it blocks the catalytic activity of LPL [12, 14]. Mab 5F9 binds to denatured human LPL but only weakly to native LPL [13].
Here, we report a new mouse monoclonal antibody against hLPL, 4-1a. Mab 4-1a binds to the amino terminus of LPL, does not inhibit catalytic activity, and binds avidly to GPIHBP1-bound LPL.
MATERIAL AND METHODS
Lipase purification
Human lipoprotein lipase (hLPL) for the immunization of mice was purified from post-heparin human plasma [16]. The hLPL used to characterize Mab 4-1a was produced in suspension cultures of Chinese hamster ovary (CHO) cells and partially purified by heparin-Sepharose chromatography. The concentration of hLPL was measured with a sandwich ELISA with Mabs 5F9 and 5D2 [13]. Mouse lipoprotein lipase (mLPL) was produced in suspension cultures of stably transfected CHO-Lec1 cells and purified by ceramic hydroxyapatite, heparin–Sepharose, and Superdex 200 chromatography. The concentration of mLPL was measured with an ELISA [17]. Chicken LPL (cLPL) was purified from chicken adipose tissue [18], and the concentration of cLPL was measured with an ELISA [19]. Bovine LPL (bLPL) was purified from fresh milk [20] by heparin-Sepharose, CHT hydroxyapatite, and Superdex 200 chromatography. LPL catalytic activity was determined with a [3H]triolein substrate [21]. Human hepatic lipase (hHL) was prepared from CHO-K1 cells that had been transiently transfected with a hHL expression vector, pγk5-hHL, provided by Dr. Shau-Feng Chang (Heinrich-Pette-Institut, Hamburg, Germany). hHL was purified by heparin–Sepharose chromatography, and hHL mass was measured with an ELISA [22].
Monoclonal antibody production
Mice were immunized with hLPL, and hybridomas were selected after fusing splenocytes with myeloma cell line P3X [16, 23]. The cells were plated on 96-well plates with mouse peritoneal macrophages. Ten days later, aliquots of the medium were tested for hLPL antibodies with an ELISA. 96-well plates were coated with hLPL (5 ng/well), and samples of the conditioned medium (100 μl) were added to the wells and incubated overnight. Mab binding was detected with an anti-mouse IgG coupled to horseradish peroxidase. One hybridoma, 4-1a, produced an antibody that bound hLPL; it was cloned twice by limiting dilution and grown in serum-free media (Gibco PFHM-II) in CELLine Two-Compartment Bioreactors (Wilsom Wolf). The isotype of Mab 4-1a was IgG2a (Pierce Rapid Isotyping Kit). Mab 4-1a was purified on protein G–Sepharose columns (GE Healthcare); gel filtration revealed a single IgG peak.
Characterization of Mab 4-1a
Binding of Mab 4-1a to purified preparations of LPL and HL were assessed by western blotting. To localize the epitope for Mab 4-1a, CHO cells were transiently transfected with expression vectors for V5-tagged wild-type and mutant versions of hLPL and mLPL. Mutant LPLs were created by site-directed mutagenesis using the QuickChange Lightning Site-Directed Mutagenesis kit (Stratagene). After 48 h, western blots were performed on cell extracts (or conditioned medium samples) with Mab 4-1a and either Mab 5D2 or a V5 Mab. Antibody binding was detected with an Odyssey infrared scanner (Li-Cor).
Binding of Mab 4-1a to hLPL was also assessed with an ELISA. 96-well plates were coated with Mab 4-1a (1–3 μg/well), and the plates were blocked with Superblock (Pierce) for 2 h at room temperature. Next, either native LPL or heat-denatured hLPL (LPL placed at 45°C for 60 min) was added to the wells and incubated overnight. After washing the plates with PBS/0.05%Tween 20, biotinylated Mab 5D2 [13], or biotinylated Mab 4-1a was added and incubated for 5 h at room temperature. After washing the wells, NeutrAvidin–horseradish peroxidase (ThermoScientific) was added and incubated for 30 min at 20°C. Color was developed with O-phenylenediamine dihydrochloride.
To test the ability of Mab 4-1a to bind GPIHBP1-bound LPL on the surface of cells, CHO cells were transfected with empty vector, an S-protein–tagged GPIHBP1 expression vector, or a mutant GPIHBP1 expression vector containing a C65S mutation (which abolishes the ability of GPIHBP1 to bind LPL). One day later, the cells were washed and incubated with hLPL at 4°C. After 2 h, the cells were washed and incubated with Mab 4-1a conjugated to Alexa-488 (Life Technologies) for 1 h. The cells were then fixed with 3% paraformaldehyde. GPIHBP1 was detected with a rabbit anti-S-protein antibody (Abcam) followed by an Alexa-568–conjugated donkey anti-rabbit IgG (Life Technologies). To determine if Mab 4-1a interferes with the binding of TRLs to the LPL–GPIHBP1 complex on the surface of cells, CHL cells expressing S-protein–tagged GPIHBP1 were plated on coverslips in 24-well plates. Cells were then incubated at 4°C for 2 h with V5-tagged hLPL. After washing, the cells were incubated with Mab 5D2, Mab 4-1a, or a V5-specific Mab (Life Technologies) for 1 h. After washing, the cells were then incubated with DiI-labeled TRLs (d < 1.006 g/ml lipoproteins from Gpihbp1−/− mice) for 1 h [5], fixed in 3% paraformaldehyde, and blocked in 10% donkey serum. GPIHBP1 was detected with a rabbit anti-S-protein antibody (Abcam) followed by an Alexa-488–conjugated donkey anti-rabbit IgG (Life Technologies), and LPL-Mab complexes were detected with an Alexa-647 conjugated donkey anti-mouse IgG (Life Technologies). Images were obtained with an Axiovert 200 MOT microscope (Zeiss) with 63×/1.25 oil immersion objective and processed with AxioVision 4.2 software (Zeiss).
To test the utility of Mab 4-1a to detect LPL in immunohistochemistry studies, skeletal muscle from a wild-type mouse and a hLPL transgenic mouse [24] were collected and snap-frozen in OCT. Frozen sections were prepared for immunohistochemistry with the Mouse on Mouse (M.O.M.) immunodetection kit (Vector Laboratories). Briefly, methanol/acetone fixed sections (8–10-μm) were incubated with a rat monoclonal antibody against mouse GPIHBP1 (11A12) and Mab 4-1a (5 μg/ml each), followed by an incubation with an Alexa-568 conjugated donkey anti-rat IgG (1:200) and the M.O.M. biotinylated anti-mouse IgG reagent (1:200). The sections were then incubated with Alexa-488–conjugated avidin. The sections were then post-fixed with 4% paraformaldehyde and mounted on coverslips with DAPI antifade (Life Technologies).
To measure the Kd of hLPL and mLPL for Mab 4-1a, microtiter plates were coated with Mab 4-1a or mouse IgG (300 ng/well) for 48 h at 4°C. The plates were blocked with 5% bovine serum albumin, 0.05% Tween 20 in phosphate buffered saline for 4 h. hLPL or mLPL (0–400 ng/well) were added to wells and incubated overnight at 4°C. The plates were washed 9 times with PBS/0.05% Tween 20 and biotinylated Mab 5D2 (for human LPL) or biotinylated goat anti-mouse LPL (for mouse LPL) were added to wells for an overnight incubation. After washing wells extensively, NeutrAvidin Horseradish Peroxidase Conjugate (Thermo Scientific) was added to wells and incubated for 30 min at 20°C. After washing the wells and adding of o-phenylenediamine substrate, the OD490 nm was read after a 20-min incubation in the dark. The bound LPL mass was determined from standard curves for hLPL and mLPL. To correct for non-specific binding, the values obtained for wells coated with control mouse IgG were subtracted from the values from wells coated with Mab 4-1a. The Kd was calculated using the least-squared fitting program in GraphPad Prism.
RESULTS
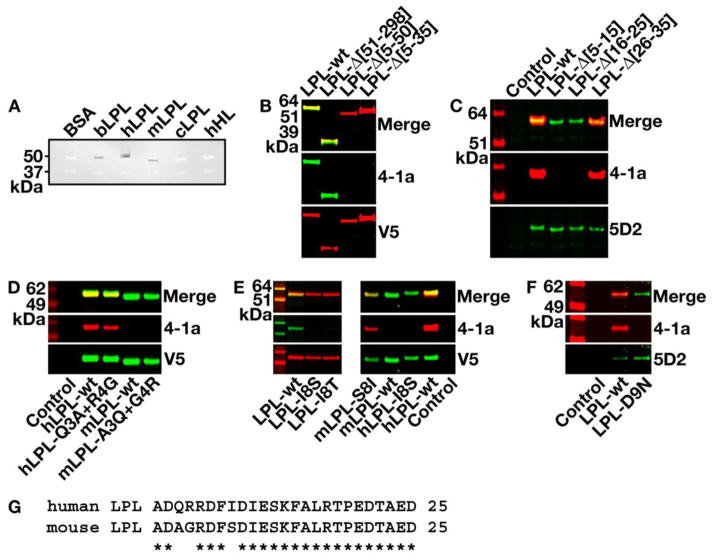
The reactivity of Mab 4-1a with purified lipase preparations was assessed with western blots. Mab 4-1a bound avidly to hLPL and bLPL but weakly to mLPL and cLPL (Fig. 1A). There was no binding of Mab 4-1a to hHL (Fig. 1A). The epitope of Mab 4-1a was localized by assessing the binding of Mab 4-1a to wild-type and mutant versions of hLPL and mLPL. Mab 4-1a bound avidly to wild-type hLPL and a hLPL mutant lacking residues 51–298 (hLPL-Δ[51–298]), but Mab 4-1a did not bind to hLPL-Δ[5–35] or hLPL-Δ[5–50] (Fig. 1B). Further studies showed that Mab 4-1a binds to hLPL-Δ[26–35] but not to hLPL-Δ[5–15] or hLPL-Δ[16–25] (Fig. 1C), demonstrating that Mab 4-1a binds within the first 25 amino acids of hLPL. Because Mab 4-1a binds weakly to mLPL, we compared the amino-terminal sequences of mLPL and hLPL and found that they differed at three amino acid residues (residues 3, 4, and 8 of hLPL; Fig. 1G). Changing amino acid residues 3 and 4 in a V5-tagged hLPL construct to the amino acids found in mLPL had minimal effects on Mab 4-1a binding to hLPL (Fig. 1D). Similarly, changing amino acid residues 3 and 4 in a V5-tagged version of mLPL to the residues found in hLPL had little effect on Mab 4-1a binding (Fig. 1E). However, changing Ile-8 in hLPL to Ser (the residue found in mLPL) or to Thr abolished Mab 4-1a binding. Also, Mab 4-1a bound avidly to a mLPL mutant containing Ile rather than Ser at residue 8 (Fig. 1E). In Fig. 1D and Fig. 1E, Mab 4-1a binding to wild-type mLPL in CHO cell extracts was extremely low—seemingly lower than the level of Mab 4-1a binding to highly purified mLPL in Fig. 1A. The reason for this difference is unclear but could relate to the large amount of mLPL (350 ng) loaded on the gel in Fig. 1A.
Fig. 1.
Characterization of Mab 4-1a binding to LPL. A, Mab 4-1a binding to purified lipases in a western blot assay (350 ng/lane, as determined with an ELISA specific for each lipase). To each sample, we added Laemmli buffer that had been spiked with internal molecular weight standards (37 and 50 kDa) labeled with a fluorophor absorbing at 700 nm (white; serving as a loading control). The presence of the 4-1a antibody was detected with an IRDye800-labeled donkey anti-mouse IgG (black). Signals were detected with a Li-Cor infrared scanner. BSA, bovine serum albumin; bLPL, bovine LPL purified from milk; hLPL, human LPL purified from hLPL-transfected CHO cells; mLPL, mouse LPL purified from CHO-Lec1 cells; cLPL, chicken LPL from chicken adipose tissue; hHL, human hepatic lipase. B, Binding of Mab 4-1a to protein extracts prepared from CHO cells that had been transfected with a wild-type hLPL expression vector or a mutant hLPL vector containing deletions of amino acids 51–298, 5–35, or 5–50. Western blots were performed with Mab 4-1a (green) or with an anti-V5 antibody (red). C, Binding of Mab 4-1a (red) or 5D2 (green) to wild-type hLPL or mutant hLPLs containing deletions of amino acids 5–15, 16–25, or 26–35. D, Binding of Mab 4-1a (red) or a V5 Mab (green) to wild-type hLPL and mutant hLPLs containing Q3A and R4G mutations, and to wild-type mLPL and mutant mLPLs containing A3Q and G4R mutations. E, On the left, binding of Mab 4-1a (green) or the V5 Mab (red) to culture supernatants from CHO cells transfected with either a wild-type hLPL expression vector or mutant hLPL constructs containing I8S or I8T mutations. On the right, binding of Mab 4-1a (red) and the V5 Mab (green) to culture supernatants of cells that had been transfected with mLPL-S8I, wild-type mLPL, hLPL-I8S, and wild-type hLPL. The Control lane shows nontransfected CHO cells. F, Binding of Mab 4-1a (red) and Mab 5D2 (green) to culture supernatants from cells that had been transfected with a wild-type hLPL vector or a mutant hLPL vector with a D9N mutation. G, Amino acid sequence alignment for the first 25 amino acids of mature human and mouse LPL. Conserved residues are indicated by an asterisk.
A common LPL polymorphism, found in ~3% of the population, changes Asp-9 to Asn (the “D9N polymorphism”) [25–27]. We suspected that the D9N polymorphism might interfere with Mab 4-1a binding. Indeed, Mab 4-1a did not bind to hLPL-D9N (Fig. 1F).
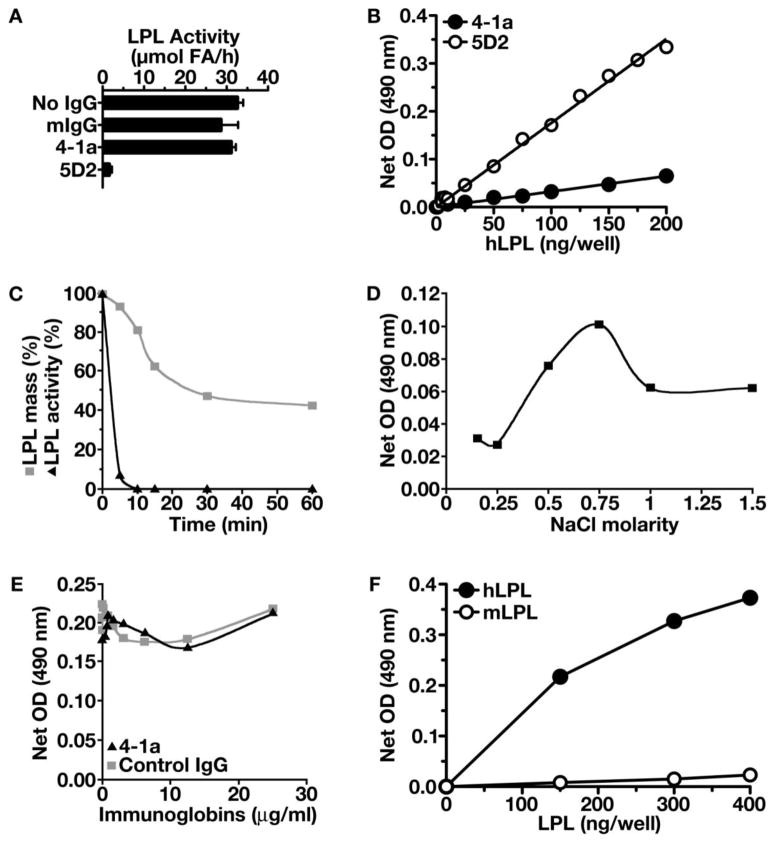
Mab 4-1a did not inhibit the catalytic activity of hLPL. When 30 μg of Mab 4-1a was added to 0.25 μg of hLPL, LPL catalytic activity was not reduced (Fig. 2A). In contrast, adding Mab 5D2 reduced catalytic activity by ~95% (Fig. 2A). When Mab 4-1a was immobilized on 96-well plates, it captured purified hLPL and was able to detect the homodimer, as judged by a sandwich ELISA (Fig. 2B). When the hLPL was subjected to heat-denaturation (by placing the LPL at 45°C), the amount of hLPL binding to immobilized Mab 4-1a fell 6% by 5 min and ~50% by 30 min (Fig. 2C). As expected, catalytic activity was markedly inhibited by heat-denaturation. After 5 min, hLPL catalytic activity fell by 93% (Fig. 2C). These data suggest that Mab 4-1a binds more avidly to catalytically active homodimers than to the heat-denatured catalytically inactive LPL monomers. The binding of hLPL to immobilized Mab 4-1a was influenced by the concentration of NaCl. As the NaCl concentration increased from 0.15 M to 0.75 M, the binding of Mab 4-1a increased threefold (Fig. 2D). Mab 4-1a did not impair the binding of hLPL to heparin-coated 96-well plates (Fig. 2E). The Kd for the equilibrium binding of Mab 4-1a for hLPL was determined to be 8.5 × 10−9 (Fig. 2F). Binding of mLPL under the same conditions, was extremely low and just above non-specific binding (Fig. 2F), consistent with the Western blot observations (Fig. 1).
Fig. 2.
Properties of Mab 4-1a. A, Assessing effects of Mabs 4-1a and 5D2 on the catalytic activity of hLPL. 30 μg of Mab 4-1a, Mab 5D2, or mouse IgG (mIgG, Sigma) was added to 0.25 μg of hLPL, and LPL catalytic activity was determined for 15 min with a [3H]triolein substrate [21]. The results are the average of 4 independent measurements. B, Testing the ability of Mab 4-1a to detect hLPL homodimers. Plates were coated with Mab 4-1a or Mab 5D2 (1 μg/well) and incubated overnight with hLPL (0–200 ng/well). The captured LPL was detected with biotinylated Mab 4-1a (filled circles) or biotinylated Mab 5D2 (open circles). C, Testing the ability of native and heat-denatured LPL to bind to Mab 4-1a (3 μg/well) immobilized on 96-well plates. Bound hLPL (50 ng/well) was detected with biotinylated Mab 5D2 and NeutrAvidin–horseradish peroxidase. Points show means of triplicates; variability between wells averaged 4.2%. Simultaneously, LPL catalytic activity was measured. Heat exposure inhibited LPL activity. Points depict duplicate measurements of LPL catalytic activity; duplicates varied by <9%. The specific activity of the substrate was 67,571 cpm/μmol fatty acid; the catalytic activity at t = 0 was 41.6 μmol/h/ml. D, Effect of NaCl molarity on the binding of hLPL to Mab 4-1a that was immobilized on 96-well plates. Each well was coated with Mab 4-1a (1 μg/well). 220 ng of hLPL were added to wells in different concentrations of NaCl (0.15–1.5 M). hLPL binding to the immobilized Mab 4-1a was detected with Mab 5D2 as described earlier. E, Assessing the effect of Mab 4-1a on the binding of hLPL to heparin-coated 96-well plates. Each well was coated with heparin (25 μg/well). hLPL (1.1 μg) was pre-incubated with increasing amounts of Mab 4-1a or a control mouse IgG (Sigma) for 30 min and then added to the heparin-coated wells and incubated overnight at 4°C. After washing the plate with PBS/Tween 0.05%, hLPL bound to the immobilized heparin was detected with Mab 5D2. F, Determination of the affinity of Mab 4-1a for hLPL. Plates were coated with 300 ng/well of Mab 4-1a or control mouse IgG for 48 h at 4°C. After blocking the wells, hLPL or mLPL was added to wells and incubated overnight. hLPL was detected with biotinylated 5D2; mLPL was detected with a biotinylated goat anti-mouse LPL. The Kd of Mab 4-1a for hLPL and mLPL was calculated as described in the Materials and Methods.
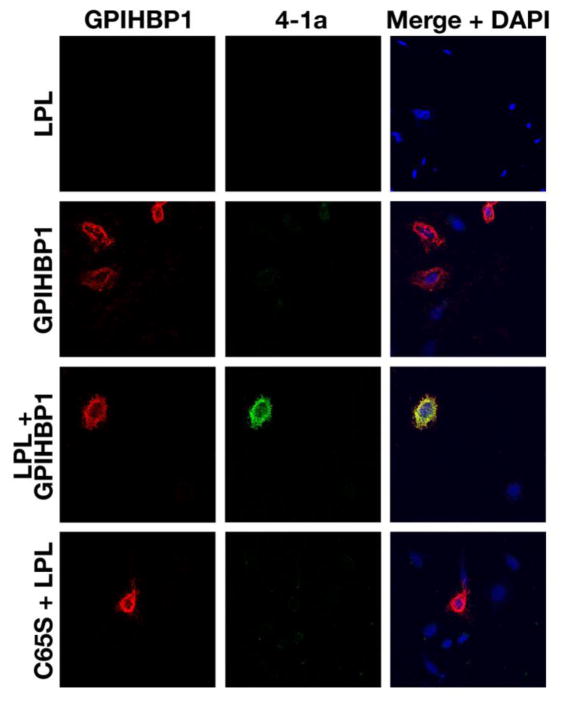
To investigate the ability of Mab 4-1a to bind to GPIHBP1-bound LPL, CHO cells expressing an S-protein–tagged GPIHBP1 were incubated with V5-tagged hLPL for 2 h. After washing, the cells were incubated with Mab 4-1a for 1 h and then fixed. Mab 4-1a bound avidly to GPIHBP1-bound LPL on the surface of cells, as judged by the binding of a fluorescently labeled antibody against mouse IgG (Fig. 3). There was no binding of Mab 4-1a to cells when hLPL was added to cells expressing a mutant GPIHBP1 (C65S) that lacked the ability to bind LPL [28].
Fig. 3.
Assessing the ability of Mab 4-1a to bind to GPIHBP1-bound LPL on the surface of CHO cells. CHO cells were transfected with empty vector (top row), or an S-protein–tagged GPIHBP1 expression vector (middle two rows), or a mutant GPIHBP1 expression vector (C65S) that cannot bind LPL (bottom row). One day later, the cells were washed and incubated with hLPL at 4°C. 2 h later, the cells were washed and incubated with Mab 4-1a conjugated to Alexa-488 for 1 h. Washed cells were fixed and then incubated with a rabbit anti-S-protein tag antibody followed by an Alexa-568-labeled donkey anti-rabbit antibody (red) to detect GPIHBP1. DNA was stained with DAPI (blue).
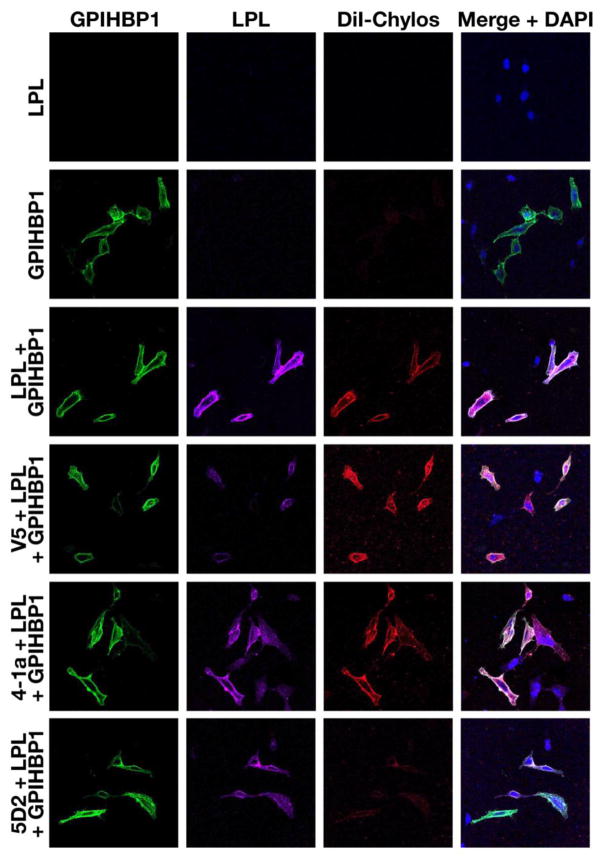
Recent studies suggested that the LPL–GPIHBP1 complex is important for the binding of TRLs to cells [5]. TRLs did not bind to nontransfected cells or to GPIHBP1-transfected CHL cells; however, when the GPIHBP1-transfected cells were pre-incubated with hLPL (thereby creating LPL–GPIHBP1 complexes on the cell surface), TRLs bound avidly [5]. Mab 5D2 has been reported to interfere with the binding of TRLs to the LPL–GPIHBP1 complex [29]. Here, we used fluorescence microscopy to compare the abilities of Mab 5D2 and Mab 4-1a to block binding of DiI-labeled TRLs to GPIHBP1-expressing cells that had been pre-incubated with hLPL. The results were unequivocal: Mab 4-1a had no effect on the binding of TRLs to the GPIHBP1–LPL complex on the cell surface, whereas Mab 5D2 blocked the binding of TRLs to the GPIHBP1–LPL complex (Fig. 4).
Fig. 4.
Mab 4-1a does not interfere with the binding of DiI-labeled TRLs to LPL–GPIHBP1 complexes on the surface of cells. CHL cells were transfected with empty vector or a mouse GPIHBP1 expression vector. After 24 h, the cells were washed and incubated with V5-tagged hLPL, followed by incubations with Mab 4-1a, Mab 5D2, or a V5-specific Mab, and then by an incubation with DiI-labeled TRLs (red). After washing the cells, the cells were fixed. GPIHBP1 (green) was detected with a rabbit anti-S-protein antibody followed by an Alexa-488-labeled donkey anti-rabbit antibody. The binding of Mab 4-1a, Mab 5D2, or the V5-Mab was detected with an Alexa-647-labeled donkey anti-mouse IgG (purple). DNA was stained with DAPI (blue).
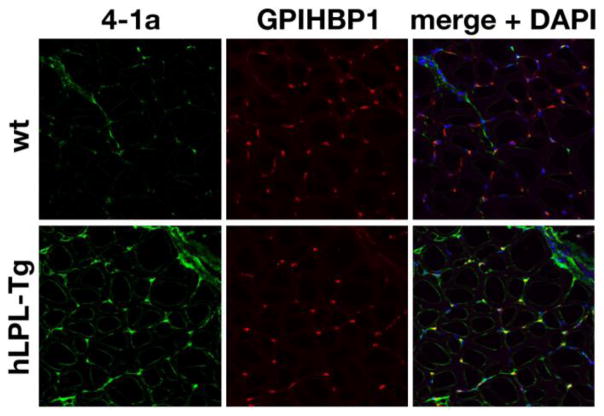
To determine whether antibody 4-1a could be a useful tool for immunohistochemistry studies, we assessed the ability of Mab 4-1a to detect hLPL in tissue sections of transgenic mice that express hLPL in skeletal muscle. Mab 4-1a bound avidly to capillaries in tissue sections from the hLPL transgenic mice (Fig. 5), which was not surprising since the vast majority of LPL in tissues is normally bound to GPIHBP1, which is expressed exclusively in endothelial cells [4, 30]. The binding of Mab 4-1a to hLPL colocalized with the binding of Mab 11A12 to GPIHBP1 (Fig. 5). In the skeletal muscle of the nontransgenic mouse, antibody 11A12 binding to capillaries was intense but there was little or no specific binding of Mab 4-1a (Fig. 5).
Fig. 5.
Mab 4-1a binds to hLPL in immunohistochemistry studies. Confocal microscopy images showing the binding of a mouse GPIHBP1–specific rat monoclonal antibody (Mab 11A12) and Mab 4-1a to skeletal muscle sections from a wild-type mouse (wt) and a mouse expressing a muscle-specific hLPL transgene (hLPL-Tg). GPIHBP1 (red) was detected with Mab 11A12 followed by an Alexa568-labeled donkey anti-rat IgG. The binding of Mab 4-1a was detected with the M.O.M. biotinylated anti-mouse IgG reagent (Vector Laboratories) followed by Fluo-avidin (green). DNA was stained with DAPI (blue).
DISCUSSION
Nearly 60 years ago, LPL was identified as a heparin-releasable enzyme that hydrolyzes triglycerides in TRLs [1]. More than 25 years ago, the cDNA for LPL was cloned [31], leading to increased understanding of LPL structure, physiology, and genetics, but it is still fair to say that progress in the field has been hampered by a paucity of useful antibody reagents. Widely used polyclonal antibodies against LPL have been shown to bind nonspecifically to other proteins [11], and there are drawbacks to currently available monoclonal antibodies, all of which bind within the carboxyl terminus of LPL. For example, Mab 5D2 blocks catalytic activity [14], limiting its utility for certain biochemical applications, and it binds poorly and inconsistently to hLPL when it is bound to soluble GPIHBP1. Here, we report the development and characterization of a new monoclonal antibody against hLPL, Mab 4-1a, which binds to the amino terminus of hLPL. Mab 4-1a binds avidly to LPL when it is bound by GPIHBP1 on the cell surface, and, unlike Mab 5D2, does not inhibit LPL catalytic activity.
Studies with mutant versions of hLPL showed that the epitope for Mab 4-1a involves residues 5–25. Because Mab 4-1a was generated in mice, we reasoned that its epitope would involve amino acid residues that differed in mLPL and hLPL sequences. Indeed, this was the case. Mab 4-1a was not capable of binding hLPL when Ile-8 was changed to Ser (the residue found in mLPL). Also, while Mab 4-1a binds very poorly to wild-type mLPL, it bound avidly to a mutant mLPL in which Ser-8 was changed to Ile (the residue found in hLPL). Residues 3 and 4 also differed in the human and mouse sequences, but changing those residues had minimal effects on Mab 4-1a binding. Interestingly, Mab 4-1a binding to hLPL was also abolished by the D9N polymorphism, which has an allele frequency of ~3% in human populations [32, 33]. Although the D9N polymorphism is relatively uncommon, it remains an important consideration for biochemical and immunocytochemical studies involving Mab 4-1a. When planning experiments with Mab 4-1a, it is important to avoid the use of hLPL containing the D9N substitution.
The development of Mab 4-1a is a welcome development for the LPL field. It binds native LPL in immunoassays and yields extremely robust, background-free signals on western blots. Also, it binds strongly to GPIHBP1-bound LPL and does not interfere with LPL catalytic activity.
Highlights.
We report a new LPL-specific monoclonal antibody, Mab 4-1a, which binds to the amino terminus of LPL (residues 5–25).
Mab 4-1a binds human and bovine LPL avidly.
Mab 4-1a does not inhibit LPL catalytic activity nor does it interfere with the binding of LPL to heparin.
Mab 4-1a binds to GPIHBP1-bound LPL.
Mab 4-1a does not interfere with the ability of the LPL–GPIHBP1 complex to bind triglyceride-rich lipoproteins.
Acknowledgments
We thank Dr. John Brunzell (University of Washington) for Mabs 5D2 and 5F9. This work was supported by grants from the NIH (HL090553 and HL087228), a Leducq Transatlantic Network grant (12CVD04), and the American Heart Association, Western States Affiliate.
Footnotes
The authors have no financial interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Korn ED. Clearing factor, a heparin-activated lipoprotein lipase. I. Isolation and characterization of the enzyme from normal rat heart. J Biol Chem. 1955;215:1–14. [PubMed] [Google Scholar]
- 2.Kompiang IP, Bensadoun A, Yang MW. Effect of an anti-lipoprotein lipase serum on plasma triglyceride removal. J Lipid Res. 1976;17:498–505. [PubMed] [Google Scholar]
- 3.Olivecrona T, Hultin M, Bergo M, Olivecrona G. Lipoprotein lipase: regulation and role in lipoprotein metabolism. Proc Nutr Soc. 1997;56:723–729. doi: 10.1079/pns19970072. [DOI] [PubMed] [Google Scholar]
- 4.Beigneux AP, Davies B, Gin P, Weinstein MM, Farber E, Qiao X, Peale P, Bunting S, Walzem RL, Wong JS, Blaner WS, Ding ZM, Melford K, Wongsiriroj N, Shu X, de Sauvage F, Ryan RO, Fong LG, Bensadoun A, Young SG. Glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 2007;5:279–291. doi: 10.1016/j.cmet.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gin P, Beigneux AP, Voss C, Davies BS, Beckstead JA, Ryan RO, Bensadoun A, Fong LG, Young SG. Binding preferences for GPIHBP1, a glycosylphosphatidylinositol-anchored protein of capillary endothelial cells. Arterioscler Thromb Vasc Biol. 2011;31:176–182. doi: 10.1161/ATVBAHA.110.214718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunzell JD, Deeb SS. Familial lipoprotein lipase deficiency, apo C-II deficiency, and hepatic lipase deficiency. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Disease. Vol. 2. McGraw-Hill; New York: 2001. pp. 2789–2816. [Google Scholar]
- 7.Young SG, Davies BS, Voss CV, Gin P, Weinstein MM, Tontonoz P, Reue K, Bensadoun A, Fong LG, Beigneux AP. GPIHBP1, an endothelial cell transporter for lipoprotein lipase. J Lipid Res. 2011;52:1869–1884. doi: 10.1194/jlr.R018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinstein MM, Goulbourne CN, Davies BS, Tu Y, Barnes RH, 2nd, Watkins SM, Davis R, Reue K, Tontonoz P, Beigneux AP, Fong LG, Young SG. Reciprocal metabolic perturbations in the adipose tissue and liver of GPIHBP1-deficient mice. Arterioscler Thromb Vasc Biol. 2012;32:230–235. doi: 10.1161/ATVBAHA.111.241406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginzinger DG, Clee SM, Dallongeville J, Lewis ME, Henderson HE, Bauje E, Rogers QR, Jensen DR, Eckel RH, Dyer R, Innis S, Jones B, Fruchart JC, Hayden MR. Lipid and lipoprotein analysis of cats with lipoprotein lipase deficiency. Eur J Clin Invest. 1999;29:17–26. doi: 10.1046/j.1365-2362.1999.00435.x. [DOI] [PubMed] [Google Scholar]
- 10.Doolittle MH, Ben-Zeev O. Immunodetection of lipoprotein lipase: antibody production, immunoprecipitation, and western blotting techniques. Methods Mol Biol. 1999;109:215–237. doi: 10.1385/1-59259-581-2:215. [DOI] [PubMed] [Google Scholar]
- 11.Casanovas A, Carrascal M, Abian J, Lopez-Tejero MD, Llobera M. Application of proteomic tools to detect the nonspecificity of a polyclonal antibody against lipoprotein lipase. J Proteome Res. 2008;7:4173–4177. doi: 10.1021/pr800131n. [DOI] [PubMed] [Google Scholar]
- 12.Chang SF, Reich B, Brunzell JD, Will H. Detailed characterization of the binding site of the lipoprotein lipase-specific monoclonal antibody 5D2. J Lipid Res. 1998;39:2350–2359. [PubMed] [Google Scholar]
- 13.Peterson J, Fujimoto WY, Brunzell JD. Human lipoprotein lipase: relationship of activity, heparin affinity, and conformation as studied with monoclonal antibodies. J Lipid Res. 1992;33:1165–1170. [PubMed] [Google Scholar]
- 14.Liu MS, Ma Y, Hayden MR, Brunzell JD. Mapping of the epitope on lipoprotein lipase recognized by a monoclonal antibody (5D2) which inhibits lipase activity. Biochim Biophys Acta. 1992;1128:113–115. doi: 10.1016/0005-2760(92)90264-v. [DOI] [PubMed] [Google Scholar]
- 15.Auwerx JH, Babirak SP, Fujimoto WY, Iverius PH, Brunzell JD. Defective enzyme protein in lipoprotein lipase deficiency. Eur J Clin Invest. 1989;19:433–437. doi: 10.1111/j.1365-2362.1989.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 16.Cheng CF, Bensadoun A, Bersot T, Hsu JST, Melford KH. Purification and characterization of human lipoprotein lipase and hepatic triglyceride lipase. Reactivity with monoclonal antibodies to hepatic triglyceride lipase. J Biol Chem. 1985;260:10720–10727. [PubMed] [Google Scholar]
- 17.Weinstein MM, Yin L, Beigneux AP, Davies BS, Gin P, Estrada K, Melford K, Bishop JR, Esko JD, Dallinga-Thie GM, Fong LG, Bensadoun A, Young SG. Abnormal patterns of lipoprotein lipase release into the plasma in GPIHBP1-deficient mice. J Biol Chem. 2008;283:34511–34518. doi: 10.1074/jbc.M806067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bensadoun A, Hsu J, Hughes B. Large scale lipoprotein lipase purification from adipose tissue. Methods Mol Biol. 1998;109:145–150. doi: 10.1385/1-59259-581-2:145. [DOI] [PubMed] [Google Scholar]
- 19.Cisar LA, Hoogewerf AJ, Cupp M, Rapport CA, Bensadoun A. Secretion and degradation of lipoprotein lipase in cultured adipocytes. Binding of lipoprotein lipase to membrane heparan sulfate proteoglycans is necessary for degradation. J Biol Chem. 1989;264:1767–1774. [PubMed] [Google Scholar]
- 20.Socorro L, Jackson RL. Monoclonal antibodies to bovine milk lipoprotein lipase. Evidence for proteolytic degradation of the native enzyme. J Biol Chem. 1985;260:6324–6328. [PubMed] [Google Scholar]
- 21.Hocquette JF, Graulet B, Olivecrona T. Lipoprotein lipase activity and mRNA levels in bovine tissues, Comparative biochemistry and physiology. Part B. Biochemistry & molecular biology. 1998;121:201–212. doi: 10.1016/s0305-0491(98)10090-1. [DOI] [PubMed] [Google Scholar]
- 22.Bensadoun A. Sandwich immunoassay for measurement of human hepatic lipase. Methods Enzymol. 1996;263:333–338. doi: 10.1016/s0076-6879(96)63025-0. [DOI] [PubMed] [Google Scholar]
- 23.Kwan SP, Yelton DE, Scharff MD. Production of monoclonal antibodies. In: Seltlow JK, Hollander A, editors. Genetic Engineering. Vol. 2. Plenum Press; New York: 1980. pp. 31–45. [Google Scholar]
- 24.Levak-Frank S, Weinstock PH, Hayek T, Verdery R, Hofmann W, Ramakrishnan R, Sattler W, Breslow JL, Zechner R. Induced mutant mice expressing lipoprotein lipase exclusively in muscle have subnormal triglycerides yet reduced high density lipoprotein cholesterol levels in plasma. J Biol Chem. 1997;272:17182–17190. doi: 10.1074/jbc.272.27.17182. [DOI] [PubMed] [Google Scholar]
- 25.van Hoek M, Dallinga-Thie GM, Steyerberg EW, Sijbrands EJ. Diagnostic value of post-heparin lipase testing in detecting common genetic variants in the LPL and LIPC genes. Eur J Hum Genet. 2009;17:1386–1393. doi: 10.1038/ejhg.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sagoo GS, Tatt I, Salanti G, Butterworth AS, Sarwar N, van Maarle M, Jukema JW, Wiman B, Kastelein JJ, Bennet AM, de Faire U, Danesh J, Higgins JP. Seven lipoprotein lipase gene polymorphisms, lipid fractions, and coronary disease: a HuGE association review and meta-analysis. Am J Epidemiol. 2008;168:1233–1246. doi: 10.1093/aje/kwn235. [DOI] [PubMed] [Google Scholar]
- 27.Henderson HE, Kastelein JJ, Zwinderman AH, Gagne E, Jukema JW, Reymer PW, Groenemeyer BE, Lie KI, Bruschke AV, Hayden MR, Jansen H. Lipoprotein lipase activity is decreased in a large cohort of patients with coronary artery disease and is associated with changes in lipids and lipoproteins. J Lipid Res. 1999;40:735–743. [PubMed] [Google Scholar]
- 28.Beigneux AP, Gin P, Davies BSJ, Weinstein MM, Bensadoun A, Fong LG, Young SG. Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. J Biol Chem. 2009;284:30240–30247. doi: 10.1074/jbc.M109.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goulbourne C, Gin P, Tatar A, Nobumori C, Hoenger A, Jiang H, Grovenor C, Adeyo O, Esko J, Goldberg I, Reue K, Tontonoz P, Bensadoun A, Beigneux A, Young S, Fong L. The GPIHBP1–LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metab. 2014 doi: 10.1016/j.cmet.2014.01.017. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies BSJ, Beigneux AP, Barnes RH, II, Tu Y, Gin P, Weinstein MM, Nobumori C, Nyrén R, Goldberg IJ, Olivecrona G, Bensadoun A, Young SG, Fong LG. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 2010;12:42–52. doi: 10.1016/j.cmet.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wion KL, Kirchgessner TG, Lusis AJ, Schotz MC, Lawn RM. Human lipoprotein lipase complementary DNA sequence. Science. 1987;235:1638–1641. doi: 10.1126/science.3823907. [DOI] [PubMed] [Google Scholar]
- 32.Razzaghi H, Aston CE, Hamman RF, Kamboh MI. Genetic screening of the lipoprotein lipase gene for mutations associated with high triglyceride/low HDL-cholesterol levels. Hum Genet. 2000;107:257–267. doi: 10.1007/s004390000367. [DOI] [PubMed] [Google Scholar]
- 33.McGladdery SH, Pimstone SN, Clee SM, Bowden JF, Hayden MR, Frohlich JJ. Common mutations in the lipoprotein lipase gene (LPL): effects on HDL-cholesterol levels in a Chinese Canadian population. Atherosclerosis. 2001;156:401–407. doi: 10.1016/s0021-9150(00)00670-5. [DOI] [PubMed] [Google Scholar]