ABSTRACT
For centuries, cholera has been one of the most feared diseases. The causative agent Vibrio cholerae is a waterborne Gram-negative enteric pathogen eliciting a severe watery diarrheal disease. In October 2010, the seventh pandemic reached Haiti, a country that had not experienced cholera for more than a century. By using whole-genome sequence typing and mapping strategies of 116 serotype O1 strains from global sources, including 44 Haitian genomes, we present a detailed reconstructed evolutionary history of the seventh pandemic with a focus on the Haitian outbreak. We catalogued subtle genomic alterations at the nucleotide level in the genome core and architectural rearrangements from whole-genome map comparisons. Isolates closely related to the Haitian isolates caused several recent outbreaks in southern Asia. This study provides evidence for a single-source introduction of cholera from Nepal into Haiti followed by rapid, extensive, and continued clonal expansion. The phylogeographic patterns in both southern Asia and Haiti argue for the rapid dissemination of V. cholerae across the landscape necessitating real-time surveillance efforts to complement the whole-genome epidemiological analysis. As eradication efforts move forward, phylogeographic knowledge will be important for identifying persistent sources and monitoring success at regional levels. The results of molecular and epidemiological analyses of this outbreak suggest that an indigenous Haitian source of V. cholerae is unlikely and that an indigenous source has not contributed to the genomic evolution of this clade.
IMPORTANCE
In this genomic epidemiology study, we have applied high-resolution whole-genome-based sequence typing methodologies on a comprehensive set of genome sequences that have become available in the aftermath of the Haitian cholera epidemic. These sequence resources enabled us to reassess the degree of genomic heterogeneity within the Vibrio cholerae O1 serotype and to refine boundaries and evolutionary relationships. The established phylogenomic framework showed how outbreak isolates fit into the global phylogeographic patterns compared to a comprehensive globally and temporally diverse strain collection and provides strong molecular evidence that points to a nonindigenous source of the 2010 Haitian cholera outbreak and refines epidemiological standards used in outbreak investigations for outbreak inclusion/exclusion following the concept of genomic epidemiology. The generated phylogenomic data have major public health relevance in translating sequence-based information to assist in future diagnostic, epidemiological, surveillance, and forensic studies of cholera.
INTRODUCTION
Comparative whole-genome sequence studies have helped to delineate the origin, phylogeographic spread, and detailed pathogenome evolution of past and present pandemic waves of cholera (1, 2). In October 2010, for the first time in nearly a century, Haiti was devastated by a cholera epidemic that garnered worldwide attention. Both the public and research communities debated the source (2). The outbreak began in the village of Meille, just south of Mirebalais (3, 4) in the Centre Department (5) of Haiti (Fig. 1). The camp for the United Nations Stabilization Mission in Haiti (MINUSTAH) was suspected of introducing Vibrio cholerae into the watershed located in Meille by discharging sewage into a tributary of the Artibonite River (3, 4). Given that Haiti had been free of cholera for more than a century, that Nepalese soldiers had arrived in the camp in the preceding days, and that V. cholerae is endemic in Nepal, the introduced strain was suspected to be Nepalese in origin (3, 4). Molecular genotyping added further support for this hypothesis (6, 7). The genomic analysis of a Haitian outbreak strain indicated that the epidemic was caused by bacteria introduced into Haiti as a result of human activity with the current seventh epidemic southern Asian type V. cholerae isolate (8). Analyses based on larger and more comprehensive genomic and epidemiological data sets allowed more comprehensive assessment of genomic diversity and evolutionary dynamics during and after the Haitian epidemic (9, 10). The results of these molecular and epidemiological investigations supported this initial hypothesis and pointed to a likely Nepalese origin of the Haitian cholera pandemic that followed contamination of the Artibonite River (3, 4, 7, 11). After the presentation of the first case on 16 October, the outbreak quickly spread around Meille and downstream to the Artibonite delta by 19 October (3, 4). In the lower Artibonite areas, 3,020 cholera cases were reported in less than 48 h, sparking massive panic and flight of people out of the affected areas and in turn, spread of the disease to other areas (3, 4). A less severe outbreak occurred in Port-au-Prince after patients arrived from Mirebalais, and even as the outbreak spread to the southern part of the country, incidences and mortality was significantly less in the Port-au-Prince metropolitan area where health care and clean water were more readily accessible (12).
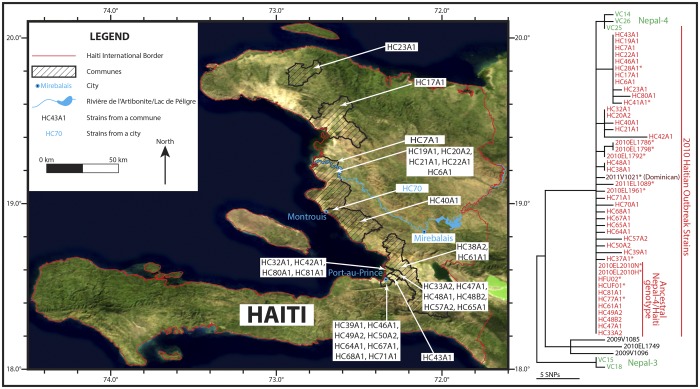
FIG 1 .
Geographic and phylogenetic locations of Haitian strains. One of two equally parsimonious trees showing relationships among Haitian strains is shown and is based upon 25 parsimony informative SNPs out of 71 total. Only one of the 25 SNPs is homoplastic, leading to a consistency index of 0.9615. The map is adapted from reference 35. Strain locations not depicted in the geographical map are indicated by an asterisk.
The 2010 Haitian epidemic provides a unique outbreak scenario and opportunity to apply genomic analyses to study the evolution and epidemiological spread of an outbreak following a single-source introduction in a naive environment without the confounding effects of other circulating clones of the V. cholerae O1 serotype. To study the genomic plasticity and subtle polymorphic changes, we analyzed a comprehensive panel of 116 genome sequences available in GenBank that encompasses (i) contemporary outbreak isolates, (ii) recent and legacy reference strains to place the current outbreaks into a historical context (1), and (iii) isolates known to be phylogenetically related to the Haitian outbreak strains (Fig. 1; see Table S1 in the supplemental material) (7, 9, 13). The use of whole-genome single-nucleotide polymorphism (SNP) typing (WGST) of the conserved genomic backbone and rigorous polymorphism discovery in the genome architecture of these 116 V. cholerae O1 isolates allowed the tracking of V. cholerae pathogenome evolution.
RESULTS
Discovery of polymorphisms.
To investigate the phenotypic variations on the level of individual polymorphisms, we applied a high-throughput bioinformatics pipeline for SNP discovery and validation, taking into account the coverage and quality of the consensus genome underlying sequence reads. To establish this high-resolution phylogenomic framework, all genomes were subjected to SNP discovery and subsequent manual in silico validation. Next-generation sequencing reads and assembled contigs were mapped to the completed and high-quality V. cholerae strain N16961 genome sequence to detect high-quality SNPs. This analysis yielded a panel of 670 high-quality SNPs in the V. cholerae O1 core genome and was specifically developed to achieve high-resolution evolutionary insights within the epidemic clade (see Table S1 in the supplemental material). Identified SNPs are distributed stochastically on chromosome I (500 SNPs) and II (170 SNPs), without any indication of mutational hot spots (see Fig. S1 and S2 in the supplemental material).
Phylogenetic analysis.
We used the identified SNPs and metadata associated with each isolate to create a global phylogeny of the O1 serotype and to calibrate a molecular clock (see Table S2 in the supplemental material). A phylogenetic reconstruction of the evolutionary relationships among 116 V. cholerae strains was performed using maximum parsimony analysis to understand the dynamics of the Haitian outbreak and its phylogenetic position within the seventh epidemic relatives (Fig. 2). Of the 670 core genome SNPs analyzed, 330 were parsimony informative. The limited homoplasy in these data with only 22 homoplastic SNPs and a consistency index of 0.9651 is an indication of a highly accurate tree of a clonal organism without intraclade lateral gene transfer events (14, 15). A single tree is presented (Fig. 2), though there were 20 equally parsimonious hypotheses for the evolution of these strains. The phylogenetic ambiguity was restricted to only a few areas of the tree, which can be seen in the consensus tree shown in the supplemental material (see Fig. S3 in the supplemental material).
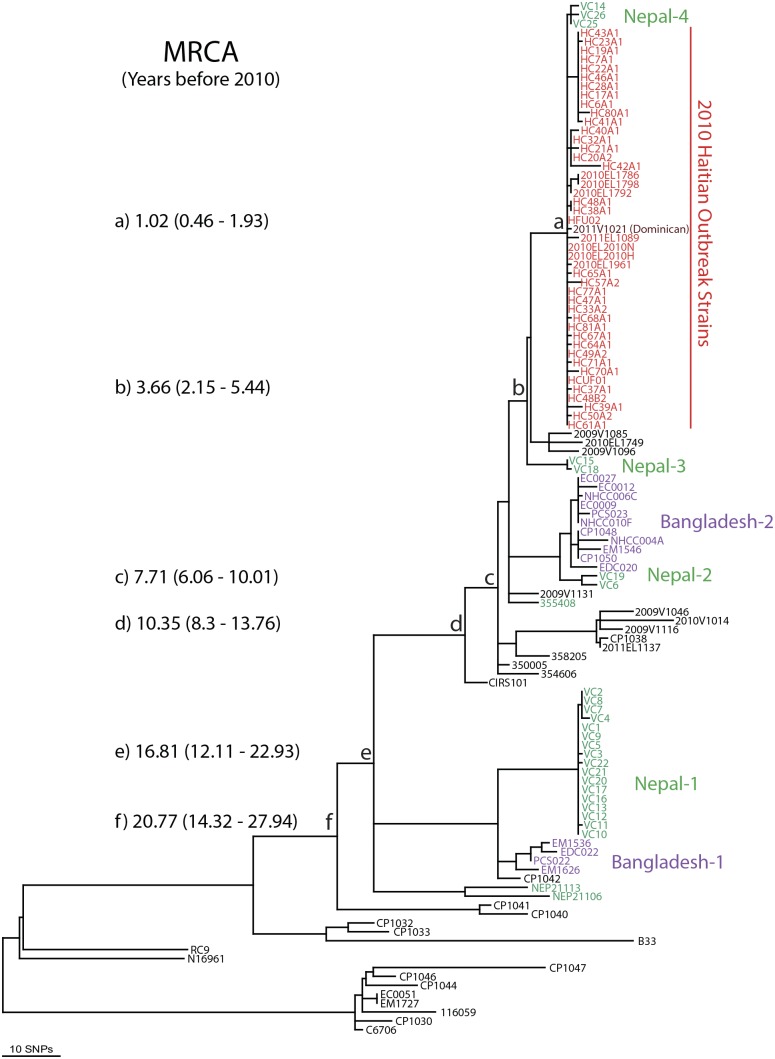
FIG 2 .
Maximum parsimony tree of global V. cholerae O1 genomes. Comparisons of 116 genomes yielded 670 total SNPs of which 330 were parsimony informative. The tree shown is one of 20 equally parsimonious trees with a consistency index of 0.9651. Trees were recovered using a heuristic search in PAUP 4.0b10. The outgroup used for rooting was identified as basal following a larger analysis that included diverse non-O1 strain genomes (see Table S1 in the supplemental material). The branches (a to f) are arbitrarily labeled for discursive purposes; note the BEAST estimates for most recent common ancestors (MRCA) including the 95% confidence interval.
All Haitian isolates and three Nepalese isolates representing the Nepal-4 subclade were contained in a monophyletic clade supported by 6 synapomorphic SNPs (Fig. 1 and Fig. 2, branch a). We note here that the investigated Nepalese isolates (including Nepal-4) were collected before the Nepalese peacekeepers were deployed to Haiti, thus ruling out an export of this genotypic profile from Haiti into Nepal (7). A single isolate from the neighboring Dominican Republic (2011V-1021) falls within the Haitian clade, indicating a likely spread of the epidemic clone from Haiti into the neighboring country (Fig. 1 and Fig. 2; see Table S1 in the supplemental material) (13). We identified 34 SNPs among the 45 Haitian strains; in most cases (n = 27), these were strain specific. The 7 remaining parsimony informative SNPs provide evidence for phylogenetic substructure within these outbreak samples. Four Haitian subclades were identified, with the largest containing 11 isolates and supported by 2 SNPs. Within this subclade, there was no additional subpopulation structure, as polymorphisms within this group were strain specific and limited to only one genome. This limited phylogenetic structuring is expected in an outbreak situation where mutations are more likely to be found in single genomes and before variant lineages go extinct. Phylogeographic patterns are consistent with epidemiological records of the source and accounts of the spread of cholera around Haiti. The first cases occurred in Mirebalais, Haiti; however, we have no samples from that immediate region. Interestingly, the basal Haitian genotype can be found only in samples from the arrondissements surrounding Port-au-Prince, suggesting that patients fled the Mirebalais region to the capital city with higher sanitation standards and health care facilities. Samples collected from patients in areas far downstream in the Artibonite River watershed and its delta showed evidence of genetic differentiation from the source genotype. Isolates within each of the smaller subclades were scattered around the country, indicating rapid movement of infected persons out of the lower Artibonite River watershed. The molecular data are consistent with epidemiological accounts of the dispersal of rice agricultural workers from the Artibonite delta (Fig. 1) (3, 4).
The non-Haitian genomes from contemporary V. cholerae isolates mostly fell into well-supported monophyletic clades consistent with current disease outbreaks. The numerous isolates in the Bangladesh-1/Nepal-1, Bangladesh-2/Nepal-2, and Haitian/Nepal-4 clades were notable. A smaller number of recent isolates were also found in well-supported clades, with some examples from disparate geographic locations (e.g., 2009/2010 from Zimbabwe) (see Table S1 in the supplemental material), providing other examples of dissemination outside southern Asia.
This 2010/2011 Haitian/Nepal-4 clade is nested inside another well-supported (supported by 3 synapomorphic SNPs and found in all 20 trees) monophyletic clade that also contains the two 2010 Nepal-3 isolates and three 2009/2010 Sri Lankan/Indian/Cameroonian isolates (Fig. 2, branch b). Phylogenetically, the closest strains originate from Sri Lanka, India, and Cameroon and were identified in a PulseNet study of 380 V. cholerae isolates for their pulsed-field gel electrophoresis (PFGE) similarity to the Haitian outbreak isolates (16). There were seven other isolates with identical PFGE patterns (13), but all are connected within a deeper monophyletic clade using WGST (Fig. 2, branch c). The PulseNet database represents a broad survey from 27 countries, so these 11 isolates are representatives of a much larger set of V. cholerae isolates. Their dispersed origins clearly show that this particular clade (Fig. 2, branch c) has been disseminated to geographically distant regions. It is notable that representatives of recent Bangladesh and Nepalese outbreaks are closely related. The Bangladesh/Nepal-1 and -2 clades contain isolates from 2010, and both have very limited diversity. While the two countries’ isolates are phylogenetically distinct, they are also more closely related than strains from the same locations isolated in different years, suggesting very rapid dissemination. These data also allow inferences concerning the ecological niches of V. cholerae. Human and perhaps environmental movement of the pathogen must be occurring very quickly within days, weeks, or months as opposed to years or decades. It is interesting that several major clones are coexisting and circulating across this southern Asian region.
BEAST analysis.
The vertex component analysis (VCA) Bayesian approach to phylogenetic analysis allowed us to estimate the coalescence times to the most recent common ancestors (MRCA) for the different clades. The MRCA dates for clades a to f are shown in Fig. 2 along with their 95% confidence interval (95% CI). As evident from the topology shown in Fig. 2, a small number of recently (decades not centuries) derived clones seem to be dominating the disease occurrence. The overall rate of SNP generation in this phylogeny was 2.50 per year per genome, which is slightly lower than the 3.3 value reported by Mutreja et al. (1). All recent Nepalese and Bangladesh strains share a MRCA within the last two decades (17.4 years, branch e). The Haitian/Nepal-4 clade isolates shared an MRCA of only 1 year (95% CI, 0.46 to 1.85 years) before their isolation in 2010. This coalescence would certainly be in southern Asia, as no cases of cholera were reported in Haiti during this time period. This is again evidence for the rapid and extremely long distance dissemination of the cholera pathogen prior to an outbreak scenario. We also noted that the members of the individual phylogenetic clades (e.g., including Nepal-1 to -3) are dispersed across significant distances in southern Asia and in other countries, with the exception of the Haitian/Nepal-4 clade. The three closely related Nepal-4 strains were differentiated from the Haitian isolates by a single synapomorphic SNP, but they were still contained in the same monophyletic clade as all of the Haitian isolates. Most of the isolates in the Haitian/Nepal-4 clade are the sole representatives of their lineages, radiating out from a recent common ancestor. The monophyly and limited population structure of the Haitian/Nepal-4 clade strongly argue for a genetic bottleneck (single introduction into Haiti) followed by a large population expansion.
Genomic heterogeneity in the integrative conjugative element and ToxR-binding repeats.
To assess the genome-wide plasticity in the Haiti/Nepal-4 clade, we further analyzed differences in the overall genome architectures by comparing whole-genome maps from representative isolates of the four Nepalese subgroups to Haitian outbreak isolates and strains from concurrent, contemporary, and historic disease occurrences (see Fig. S4 and Table S1 in the supplemental material). The clustering topology showed that all tested clinical and environmental O1 serotype strains cluster in a single group apart from the non-O1/O139 strains and confirmed similarities of the Haitian/Nepal-4 clade (Fig. S4). Within the Haitian epidemic, only very limited genomic heterogeneity was observed among the strains, indicating their clonal nature. Identified polymorphic loci are the toxin-linked cryptic (TLC) element, mannose-sensitive hemagglutinin (MSHA) pilus, tRNA regions, wbeT, rfb, and the integrative conjugative element ICE/SXT element (10, 17). The SXT/R391 type element found in the Haitian epidemic strains introduces multidrug resistance against chloramphenicol (floR), streptomycin (strBA), sulfamethoxazole (sul2), and trimethoprim (dfrA1) and differs by only five SNPs from ICEVchInd5, first described in an epidemic V. cholerae O1 El Tor strain from India (17, 18) (Fig. S5). Comparison of the gene inventory by Katz et al. (10) and our whole-map comparisons of Nepalese genomes identified an ∼19,929-kb indel within the ICE encompassing 13 genes (Vch1786_I0089 [mutL] to Vch1786_I0102 [traI]; Fig. S5). This length variation can serve as a structural marker to further differentiate the Haitian outbreak strains from Nepal-4. This finding is indicative of a likely loss of this segment after divergence from the common ancestor of the Haitian/Nepal-4 cluster and argues against a direct evolutionary relationship of the Nepal-4 group to the Haitian epidemic strains, while related Nepalese lineages 2 (VC6) and 3 (VC18) carry an intact SXT element (Fig. S5). Similarities in the number of the 7-mer repeat (TTTTGAT) required for binding of the global transcriptional regulator ToxR promoting cholera toxin (CtxA/B) production support the close relatedness of Haitian to Nepal-4 isolates (19). The Haitian/Nepal-4 cluster features five repeat copies, while four copies are found in Nepal-2 (VC6) and -3 (VC18). Such an increased number of ToxR-binding repeats has been associated with a more severe form of cholera (20).
DISCUSSION
In the current postgenomics era, outbreak investigations have transitioned from sequencing single archetypical outbreak strains to examining large numbers of isolates for source determination and understanding outbreak dynamics and evolution at a population level. The 2010 Haitian cholera epidemic is one of the best-studied and largest infectious disease outbreaks with thorough associated epidemiological data, with more than 100 complete clinical genomes and nearly as many reference isolates sequenced in the aftermath of the epidemic (1, 5, 8, 9). High-resolution analytical approaches catalogued subtle polymorphisms and genotypic (and phenotypic) heterogeneity among clinical strains derived from the Haitian epidemic to resolve the relatively genetically monomorphic O1 serotype (5, 7–9). In this study, we placed the genomic and epidemiological patterns of the Haitian outbreak into a phylogeographic context to better understand the spread and persistence of the pathogen. As expected, genetic and phylogenetic discriminatory power was lacking, as the narrow time window in which isolates were collected allowed for few polymorphisms and thus hindered our ability to differentiate isolates and place them in a hierarchical model of evolution. Nonetheless, our data confirmed a Nepali source of the outbreak (7) and provided additional evidence against the involvement of non-O1 strains and molecular evidence for patterns of geographic spread within Haiti. Such phylogeographic information is essential for linking the outbreak to its source, tracking and controlling its spread and transmission, and for risk assessment delineated from the carried virulence genotypes (21).
The phylogeographic patterns demonstrate the ability of this pathogen to move quickly across the landscape on a global (mediated solely by human carriers) and local (mediated by rivers and human carriers) scale. Closely related strains collected in different years can often be found in different countries, demonstrating the necessity of a robust contemporary global reference database with genomic and epidemiological metadata for source attribution (21). Our analyses suggest that the MRCA to the Haitian/Nepal-4 clade existed in Nepal, but the genotype did not change before introduction into Haiti, causing this ancestral genotype to appear in Haiti. This finding is consistent with the work of Hendriksen et al. (7), but not Katz et al. (10), who placed the root along the branch leading to the Nepal-4 clade. The existence of the ancestral genotype in the Port-au-Prince area is suggestive of the environmental source of the Haitian outbreak; however, epidemiological evidence has established the village of Meille just south of Mirabalais as the Haitian source (12). Rather, the presence of this ancestral genotype in Port-au-Prince is indicative of the destination and speed of the first cholera patients fleeing the Mirabalais region. The general lack of hierarchical phylogenetic structuring is expected in an outbreak scenario where multiple variants can be found before they ultimately go extinct. The fact that these variants were found throughout the country and even in neighboring Dominican Republic is a testament to the speed and extent of patient dispersal. This pattern of human mediated dispersal was not limited to the onset of the outbreak, as phylogeographic patterns show a lack of structuring even within the two largest Haitian subclades. The mobility of the human population may continue to complicate eradication efforts in the long term. The lack of additional genomic regions specific to the Haitian isolates and the lack of conflicting phylogenetic signal (homoplasy) in both global and Haitian/Nepal-4 phylogenies are indicative that horizontal gene transfer probably plays no role in the evolution of this group of V. cholerae. Indeed, Katz et al. (10) showed that Haitian isolates are poorly transformable even in a laboratory, casting doubt on the possibility that non-O1 V. cholerae may have contributed to the evolution of this outbreak. This phylogenomic framework with high phylogenetic accuracy and resolution, coupled with the growing genome database, lays the foundation for rapid assessment of future outbreaks. These whole-genome-based analyses have been remarkable in confirming epidemiological observations (1, 5, 7–9, 12) regarding the Nepali source of the outbreak and the flight of persons throughout Haiti. Our ability to reconstruct the source, spread, and evolution of this outbreak from molecular data provides important confidence in the value of these types of investigations for outbreaks where epidemiological data are sparse.
MATERIALS AND METHODS
Whole-genome polymorphism discovery. (i) Whole-genome mapping.
Whole-genome maps of representative Nepal-4 (VC14 and VC26), Nepal-3 (VC18), and Nepal-2 (VC6) strains were generated in support of the comparative genome analysis of the draft assemblies of the Nepalese isolates. Optical mapping facilitated the discovery of structural polymorphisms in the genome architectures compared to Haitian O1 outbreak strains (maps HC20A2 and HC46A1) (see Table S1, Fig. S4, and Fig. S5 in the supplemental material). Maps were prepared by OpGen, Gaithersburg, MD, USA. Briefly, following gentle lysis and dilution, high-molecular-mass genomic DNA molecules were spread and immobilized on derivatized glass slides and digested with NheI. The DNA digestions were stained with YOYO-1 fluorescent dye and photographed using a fluorescence microscope interfaced with a digital camera. Automated image analysis software located, sized, and assembled the fragments from multiple scans into whole-chromosome whole-genome maps.
(ii) SNP discovery.
The genomes of all strains listed in Table S1 in the supplemental material were subjected to SNP discovery. SNPs were identified both from discovery in assembled contigs and/or read-based discovery using strain N16961 as a reference. The Nepalese sequence reads deposited in the SRA archive were assembled to allow for contig-based discovery. The SNP discovery and verification pipelines are implemented in the Ergatis framework. Ergatis (3) is an open-source, web tool that is used to create, run, and monitor reusable computational analysis pipelines (http://ergatis.sourceforge.net). Ergatis is built on Workflow (4), an extensible markup language (XML)-based pipelining system. Ergatis allows the development of distributed pipelines to be run across a computational grid easily and efficiently. Software to support conversion of output formats has been developed within the Ergatis framework. Genomic regions with identities of more than 98% on chromosomes I and II were excluded, and regions that were not conserved among strains were also excluded from further analysis. The resulting SNP data set is then converted to standard variant output format (variant call format [VCF]) for further phylogenomic analysis.
(iii) Contig-based SNP discovery.
The contig-based discovery module uses (i) nucmer, delta-filter and show-snps distributed with the Mummer package for reference-based SNP discovery with a minimum alignment threshold of >90%; and (ii) kSNP, a k-mer algorithm analysis using suffix arrays without the need of prior sequence alignment (22).
(iv) Read-based SNP discovery.
The read-based discovery module uses (i) the Java-based NGS variant calling tool SolSNP (http://solsnp.sourceforge.net/) that deploys Kolmogorov-Smirnov statistic and data filtering to call variants on high-coverage aligned genomes (7) and (ii) Burrows-Wheeler Aligner (BWA) for efficient gapped alignment algorithm for short-read alignments (<200 bp; bwa-short) and long-read alignments (bwa-sw) (23, 24). Alignments are sorted and manipulated with Picard, a Java-based set of utilities (http://picard.sourceforge.net), and SAMtools have been used for SNP discovery (25). After BWA alignment and sorting with Picard tools, positions with a minimum base quality and mapping quality of 20 were extracted and called only when 9 out of 10 bases showed the variant base with a minimum of 10× coverage.
(v) SNP verification and curation.
Predictions from the contig-based or read-based discovery modules in this study and published SNP panels for V. cholerae were merged into a single SNP data set (7, 9). To build a robust phylogeny, SNP regions that colocalized with known repeats and/or mobile elements, such as phages or transposons, which evolve at different rates, were excluded from further analysis, a strategy previously used by our group for the typing of bacterial pathogens (26, 27). By default, 41 bp surrounding each predicted SNP position were extracted from the reference genome N16961 and searched against all query genomes using a comprehensive all versus all BLASTN analysis (28, 29) to exclude false-positive calls. BLAST alignments were then parsed and analyzed for (i) alignment quality (full-length/partial) to correct for misalignments (e.g., repetitive regions), (ii) prevalence of this sequence in the analyzed query genomes (insufficient genome coverage and indels), (iii) number of resulting hits in a query genome and excluded for ambiguous and/or noncorresponding paralogous hits (unjoined contigs and genome duplications), and (iv) manual inspection.
(vi) Assessment of SNP quality and coverage.
The bioinformatics pipeline takes into account genome coverage and base quality for each query genome for which underlying sequence reads are available. Reads used in the assembly were aligned to the assembled genome and coverage and average quality calculated for each base in the query genomes (see Fig. S6 and S7 in the supplemental material). For Illumina assemblies using Velvet, reads were aligned to the assembly using BWA, and quality and coverage statistics were gathered after processing the alignment through mpileup (23, 24). For 454 or hybrid assemblies generated with the Celera assembler, sets of reads were taken from the gatekeeper store produced during the assembly and processed as described for Illumina-assembled genomes. The resulting quality and coverage scores were used to assess the relative reliability of individual SNP predictions with queries and to correct for false-positive results by applying a filter with cutoff settings for coverage of <50× and quality of <25.
(vii) Phylogenetics.
Maximum parsimony was used to generate the phylogenetic hypothesis in Fig. 1 and S2 in the supplemental material and the consensus tree from 80 equally parsimonious trees in Fig. S1. We used the Phylogenetic Analysis Using Parsimony (PAUP 4.0b10) software (30) to perform a heuristic search. Additional distantly related genomes of the O1 serotype were used to establish that the eight genome groups (e.g., strain 116059) at the base of the tree were closely related but are phylogenetically positioned outside the genomes of interest (Table S1, outgroup). These groups were then used as outgroups to root the tree and infer the ancestral state of SNPs in the ingroup.
(viii) BEAST.
Divergence date estimates were obtained using the Bayesian Markov chain Monte Carlo framework implemented in the BEAST 1.6.2 software package (31) using the general time reversible (GTR) model of nucleotide evolution. The molecular clock was calibrated using the uncorrelated lognormal clock model (32), which allows rates to vary on branches, and the Bayesian Skyline coalescent prior (33), which minimizes model assumptions about demographic history. Three independent chains were run for 40 million generations with sampling every 4,000 generations allowing 4 million generations for burn-in. Proper mixing of chains and effective sample size (ESS) were assessed using Tracer 1.5 (34). Chains were then combined achieving an ESS greater than 200 for all parameters. The nodes at the bases of the Nepal-4 and Haitian/Nepal-4 clades were specified in the model to ensure that these estimates were properly sampled. The maximum clade credibility tree (see Fig. S1 in the supplemental material) was constructed using Tree Annotator, which is part of the BEAST software package. Nodes recovered with a posterior probability of >0.5 are annotated with median node heights (years before 2010).
Nucleotide sequence accession numbers.
GenBank and/or SRA accessions and metadata of the V. cholerae genomes analyzed in this study are listed in Table S1 in the supplemental material. Whole-genome sequences (WGS) of globally and temporally diverse collection of 116 V. cholerae O1 strains, including 44 strains from Haiti, were downloaded as annotated consensus draft assemblies and/or raw reads from GenBank and/or the Sequence Read Archive (SRA) repositories. Deposited sequence reads for Nepalese strains VC6, VC14, VC18, and VC26 were downloaded from SRA and assembled to facilitate whole-genome map comparisons (7).
SUPPLEMENTAL MATERIAL
SNP discovery on V. cholerae O1 chromosome I. Circles from the outer circle to the inner circle are as follows: identified SNP positions referenced to N16961 positions on the 4,033,464-Mbp chromosome I (circle 1), GC skew using a 5-kb window size (circle 2), and predicted open reading frames (ORFs) of strain N16961 encoded on the plus strand (circle 3) and minus strand (circle 4). The SNP backbone of V. cholerae O1 chromosome comprising 500 SNPs is shown: 107 synonymous SNPs (green), 323 nonsynonymous SNPs (17 stop mutations) (red), 65 intergenic SNPs (gray), and 9 multiallelic SNPs (orange). Individual SNP patterns for the 116 query genomes from the outer circle to the inner circle show the reference base sequence (circle 5) and the query sequence base (circles 6 to 121) whenever it differs from the reference. SNPs are depicted according to their phylogenetic relatedness revealed by the SNP-derived topology in Fig. 2. SNP deserts and genomic regions (≥5 kb) with no SNP prediction are highlighted in pink and colocalize with the complement of V. cholerae genomic island (Vibrio pathogenicity island [VPI], Vibrio seventh pandemic island [VSP], cholera toxin prophage [CTX], toxin-linked cryptic element [TLC], and genomic island [GI] [black]) that have been excluded from SNP discovery in the core genome. Predicted SNPs with a consistency index (CI) of <1 indicative of homoplasy are highlighted in purple in the outermost circle. Download
SNP discovery on V. cholerae O1 chromosome II. Circles from the outer circle to the inner circle are as follows: identified SNP positions referenced to N16961 positions on the 1,072,315-Mbp chromosome I (circle 1), GC skew using a 5-kb window size (circle 2), and predicted ORFs of strain N16961 encoded on the plus strand (circle 3) and minus strand (circle 4). The SNP backbone of V. cholerae O1 chromosome II comprising 170 SNPs is shown as follows; 33 synonymous SNPs (green), 121 nonsynonymous SNPs (red) (5 stop mutations), 16 intergenic SNPs (gray), and 3 multiallelic SNPs (orange). Individual SNP patterns for the 116 query genomes from the outer circle to the inner circle show the reference base sequence (circle 5) and the query sequence base (circles 6 to 121) whenever it differs from the reference. SNPs are depicted according to their phylogenetic relatedness revealed by the SNP-derived topology in Fig. 2. SNP deserts and genomic regions (≥5 kb) with no SNP prediction are highlighted in pink and colocalize with the genomic islands (GI) (labeled in black) that have been excluded from the core genome SNP discovery. Predicted SNPs with a consistency index (CI) of <1 indicative of homoplasy are highlighted in purple in the outermost circle. Download
Consensus tree. A strict consensus tree of all 80 equally parsimonious trees. One of these trees is shown in Fig. 2. That most of the hierarchical topology seen in Fig. 2 is retained in the consensus tree is indicative of the lack of conflicting phylogenetic signal and the accuracy of clades found in Fig. 2. For clonally inherited genomic DNA, a single character can accurately define a branch. Bootstrapping (36) is a commonly used metric for assessing the robustness of a tree; however, it is an inappropriate metric for such data sets, as it will underestimate the support for short, but real, branches (14, 36, 37). Measures of character state conflict are more appropriate for indicating the accuracy of trees generated with such data (14, 37). The homoplasy (or consistency) index and strict consensus trees provide both a measure of conflicting phylogenetic signal as well as the portions of the tree that are affected by homoplasy and may not truly reflect the evolution of the organism. Download
Whole-genome map similarity cluster analysis. The data are comprised of experimental and in silico maps of both chromosomes of V. cholerae O1 and non-O1/O139. Maps are compared by using the unweighted-pair group method with arithmetic means (UPGMA). The analysis includes maps of two representative Nepal-4 strains VC14 and VC26, which have been identified as close phylogenetic relatives of the Haitian outbreak strains featuring many of the genetic hallmarks found in the outbreak clade. Download
Length polymorphism in the integrative conjugative element. (A) Map comparisons of the prototypical ICEVchInd5 element at the prfC locus (97,952 bp; GenBank accession no. GQ463142) (18) compared to the ICE prevalent in the prototypical Haitian outbreak strain 2010EL-1786 (97,915 bp; GenBank accession no. JN648379) (17) and Nepalese group 2 to 4 strains. (B) Nepal-4 strains are distinguished by the absence of a 19.929-bp segment within the ICE encompassing 13 genes (Vch1786_I0089 [mutL] to Vch1786_I0102 [traI]). The deletion occurs next to a transposase mutator family gene (Vch1786_I0088). Restriction sites of NheI (GCTAGC) used for optical mapping and identified palindromic sequences are marked. Download
Sequence read coverage for predicted SNP positions. Computation of underlying sequence read coverage on (A) chromosome I and (B) chromosome II for each of the predicted SNP positions in the analyzed V. cholerae serotype O1 genomes. Download
Sequence read quality for predicted SNP positions. Computation of underlying sequence read base quality on (A) chromosome I and (B) chromosome II for each of the predicted SNP positions in the analyzed V. cholerae serotype O1 genomes. Download
Strain history and metadata of strains analyzed in this study. The geographical origin of the strains are shown in column A according to the color scheme used in Fig. 2
SNP discovery in V. cholerae O1. This table lists 670 intra- and intergenic identified SNPs detected in the 116 V. cholerae O1 serotype strains analyzed in this study. The SNP positions are referenced to the respective chromosome and position in strain N16961. The table contains information for each SNP position in the reference genome distinguishing intergenic, synonymous, and nonsynonymous SNPs and annotations. The data table lists number of discovered alleles for every strain at each assayed position (bi- or multiallelic) and flags potential low or high character homoplasies and branch-specific SNPs (see Fig. S1 and S2 in the supplemental material). The data set was used to compute the number of SNPs per gene, SNP density, and genome-wide SNP distribution to facilitate the identification of potential mutational hot spots.
ACKNOWLEDGMENTS
This study received support from the South Texas Center of Emerging Infectious Diseases (STCEID) and Department of Biology at the University of Texas at San Antonio and computational support from the Computational System Biology Core funded by the National Institutes of Health under contractG12MD007591. F.S. is supported in part by the STCEID.
M.E., T.P., J.R., and P.S.K. interpreted the findings and wrote the manuscript. All authors have read and approved the manuscript.
Footnotes
Citation Eppinger M, Pearson T, Koenig SSK, Pearson O, Hicks N, Agrawal S, Sanjar F, Galens K, Daugherty S, Crabtree J, Hendriksen RS, Price LB, Upadhyay BP, Shakya G, Fraser CM, Ravel J, Keim PS. 2014. Genomic epidemiology of the Haitian cholera outbreak: a single introduction followed by rapid, extensive, and continued spread characterized the onset of the epidemic. mBio 5(6):e01721-14. doi:10.1128/mBio.01721-14.
REFERENCES
- 1. Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JL, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465. 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grad YH, Waldor MK. 2013. Deciphering the origins and tracking the evolution of cholera epidemics with whole-genome-based molecular epidemiology. mBio 4(5):e00670-13. 10.1128/mBio.00670-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Frerichs RR, Boncy J, Barrais R, Keim PS, Piarroux R. 2012. Source attribution of 2010 cholera epidemic in Haiti. Proc. Natl. Acad. Sci. U. S. A. 109:E3208 (Author reply, 109:E3209.) 10.1073/pnas.1211512109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frerichs RR, Keim PS, Barrais R, Piarroux R. 2012. Nepalese origin of cholera epidemic in Haiti. Clin. Microbiol. Infect. 18:E158–E163. 10.1111/j.1469-0691.2012.03841.x. [DOI] [PubMed] [Google Scholar]
- 5. Llanes R, Somarriba L, Pedroso P, Mariscal E, Fuster C, Zayas Y. 2013. Did the cholera epidemic in Haiti really start in the Artibonite Department? J. Infect. Dev. Ctries. 7:753–755. 10.3855/jidc.3311. [DOI] [PubMed] [Google Scholar]
- 6. Orata FD, Keim PS, Boucher Y. 2014. The 2010 cholera outbreak in Haiti: how science solved a controversy. PLoS Pathog. 10(1):e1003967. 10.1371/journal.ppat.1003967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hendriksen RS, Price LB, Schupp JM, Gillece JD, Kaas RS, Engelthaler DM, Bortolaia V, Pearson T, Waters AE, Upadhyay BP, Shrestha SD, Adhikari S, Shakya G, Keim PS, Aarestrup FM. 2011. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. mBio 2(4):e00157-11. 10.1128/mBio.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. 2011. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364:33–42. 10.1056/NEJMvcm0904262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. 2012. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc. Natl. Acad. Sci. U. S. A. 109:E2010–E2017. 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, Turnsek MA, Guo Y, Wang S, Paxinos EE, Orata F, Gladney LM, Stroika S, Folster JP, Rowe L, Freeman MM, Knox N, Frace M, Boncy J, Graham M, Hammer BK, Boucher Y, Bashir A, Hanage WP, Van Domselaar G, Tarr CL. 2013. Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. mBio 4(4):e00398-13. 10.1128/mBio.00398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lantagne D, Balakrish Nair G, Lanata CF, Cravioto A. 2014. The cholera outbreak in Haiti: where and how did it begin? Curr. Top. Microbiol. Immunol. 379:145–164. 10.1007/82_2013_331. [DOI] [PubMed] [Google Scholar]
- 12. Piarroux R, Barrais R, Faucher B, Haus R, Piarroux M, Gaudart J, Magloire R, Raoult D. 2011. Understanding the cholera epidemic, Haiti. Emerg. Infect. Dis. 17:1161–1168. 10.3201/eid1707.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reimer AR, Van Domselaar G, Stroika S, Walker M, Kent H, Tarr C, Talkington D, Rowe L, Olsen-Rasmussen M, Frace M, Sammons S, Dahourou GA, Boncy J, Smith AM, Mabon P, Petkau A, Graham M, Gilmour MW, Gerner-Smidt P, V. cholerae Outbreak Genomics Task Force 2011. Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa. Emerg. Infect. Dis. 17:2113–2121. 10.3201/eid1711.110794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pearson T, Okinaka RT, Foster JT, Keim P. 2009. Phylogenetic understanding of clonal populations in an era of whole genome sequencing. Infect. Genet. Evol. 9:1010–1019. 10.1016/j.meegid.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 15. Pearson T, Hornstra HM, Sahl JW, Schaack S, Schupp JM, Beckstrom-Sternberg SM, O’Neill MW, Priestley RA, Champion MD, Beckstrom-Sternberg JS, Kersh GJ, Samuel JE, Massung RF, Keim P. 2013. When outgroups fail; phylogenomics of rooting the emerging pathogen, Coxiella burnetii. Syst. Biol. 62:752–762. 10.1093/sysbio/syt038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Talkington D, Bopp C, Tarr C, Parsons MB, Dahourou G, Freeman M, Joyce K, Turnsek M, Garrett N, Humphrys M, Gomez G, Stroika S, Boncy J, Ochieng B, Oundo J, Klena J, Smith A, Keddy K, Gerner-Smidt P. 2011. Characterization of toxigenic Vibrio cholerae from Haiti, 2010-2011. Emerg. Infect. Dis. 17:2122–2129. 10.3201/eid1711.110805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sjölund-Karlsson M, Reimer A, Folster JP, Walker M, Dahourou GA, Batra DG, Martin I, Joyce K, Parsons MB, Boncy J, Whichard JM, Gilmour MW. 2011. Drug-resistance mechanisms in Vibrio cholerae O1 outbreak strain, Haiti, 2010. Emerg. Infect. Dis. 17:2151–2154. 10.3201/eid1711.110720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Déry C, Burrus V, Waldor MK. 2009. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5:e1000786. 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pfau JD, Taylor RK. 1996. Genetic footprint on the ToxR-binding site in the promoter for cholera toxin. Mol. Microbiol. 20:213–222. 10.1111/j.1365-2958.1996.tb02502.x. [DOI] [PubMed] [Google Scholar]
- 20. Nair GB, Qadri F, Holmgren J, Svennerholm AM, Safa A, Bhuiyan NA, Ahmad QS, Faruque SM, Faruque AS, Takeda Y, Sack DA. 2006. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J. Clin. Microbiol. 44:4211–4213. 10.1128/JCM.01304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aarestrup FM, Brown EW, Detter C, Gerner-Smidt P, Gilmour MW, Harmsen D, Hendriksen RS, Hewson R, Heymann DL, Johansson K, Ijaz K, Keim PS, Koopmans M, Kroneman A, Lo Fo Wong D, Lund O, Palm D, Sawanpanyalert P, Sobel J, Schlundt J. 2012. Integrating genome-based informatics to modernize global disease monitoring, information sharing, and response. Emerg. Infect. Dis. 18(11):e1. 10.3201/eid1808.120277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gardner S, Slezak T. 2010. Scalable SNP analyses of 100+ bacterial or viral genomes. J. Forensic Res. 1:107. 10.4172/2157-7415.1000107. [DOI] [Google Scholar]
- 23. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43:491–498. 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, Feldkamp M, Kusecek B, Vogler AJ, Li Y, Cui Y, Thomson NR, Jombart T, Leblois R, Lichtner P, Rahalison L, Petersen JM, Balloux F, Keim P, Wirth T, Ravel J, Yang R, Carniel E, Achtman M. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 42:1140–1143. 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. 2011. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc. Natl. Acad. Sci. U. S. A. 108:20142–20147. 10.1073/pnas.1107176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karlin S, Altschul SF. 1990. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl. Acad. Sci. U. S. A. 87:2264–2268. 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30. Swofford D. 1998. Phylogenetic analysis using parsimony. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 31. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Drummond AJ, Rambaut A, Shapiro B, Pybus OG. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22:1185–1192. 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 34. Rambaut A, Drummond A. 2007. Tracer v1.4: MCMC trace analyses tool. [Google Scholar]
- 35. Stoeckli R, Vermote E, Saleous N, Simmon R, Herring D. 2005. The Blue Marble Next Generation - a true color earth dataset including seasonal dynamics from MODIS. NASA Earth Observatory. http://earthobservatory.nasa.gov/Features/BlueMarble/bmng.pdf.
- 36. Felsenstein D, D’Amico DJ, Hirsch MS, Neumeyer DA, Cederberg DM, de Miranda P, Schooley RT. 1985. Treatment of cytomegalovirus retinitis with 9-[2-hydroxy-1-(hydroxymethyl)ethoxymethyl]guanine. Ann. Intern. Med. 103:377–380. 10.7326/0003-4819-103-3-377. [DOI] [PubMed] [Google Scholar]
- 37. Archie J. 1996. Measures of homoplasy, p 153–188 In Sanderson M, Huord L. (ed), Homoplasy: the recurrence of similarity in evolution. Academic Press, San Diego, CA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SNP discovery on V. cholerae O1 chromosome I. Circles from the outer circle to the inner circle are as follows: identified SNP positions referenced to N16961 positions on the 4,033,464-Mbp chromosome I (circle 1), GC skew using a 5-kb window size (circle 2), and predicted open reading frames (ORFs) of strain N16961 encoded on the plus strand (circle 3) and minus strand (circle 4). The SNP backbone of V. cholerae O1 chromosome comprising 500 SNPs is shown: 107 synonymous SNPs (green), 323 nonsynonymous SNPs (17 stop mutations) (red), 65 intergenic SNPs (gray), and 9 multiallelic SNPs (orange). Individual SNP patterns for the 116 query genomes from the outer circle to the inner circle show the reference base sequence (circle 5) and the query sequence base (circles 6 to 121) whenever it differs from the reference. SNPs are depicted according to their phylogenetic relatedness revealed by the SNP-derived topology in Fig. 2. SNP deserts and genomic regions (≥5 kb) with no SNP prediction are highlighted in pink and colocalize with the complement of V. cholerae genomic island (Vibrio pathogenicity island [VPI], Vibrio seventh pandemic island [VSP], cholera toxin prophage [CTX], toxin-linked cryptic element [TLC], and genomic island [GI] [black]) that have been excluded from SNP discovery in the core genome. Predicted SNPs with a consistency index (CI) of <1 indicative of homoplasy are highlighted in purple in the outermost circle. Download
SNP discovery on V. cholerae O1 chromosome II. Circles from the outer circle to the inner circle are as follows: identified SNP positions referenced to N16961 positions on the 1,072,315-Mbp chromosome I (circle 1), GC skew using a 5-kb window size (circle 2), and predicted ORFs of strain N16961 encoded on the plus strand (circle 3) and minus strand (circle 4). The SNP backbone of V. cholerae O1 chromosome II comprising 170 SNPs is shown as follows; 33 synonymous SNPs (green), 121 nonsynonymous SNPs (red) (5 stop mutations), 16 intergenic SNPs (gray), and 3 multiallelic SNPs (orange). Individual SNP patterns for the 116 query genomes from the outer circle to the inner circle show the reference base sequence (circle 5) and the query sequence base (circles 6 to 121) whenever it differs from the reference. SNPs are depicted according to their phylogenetic relatedness revealed by the SNP-derived topology in Fig. 2. SNP deserts and genomic regions (≥5 kb) with no SNP prediction are highlighted in pink and colocalize with the genomic islands (GI) (labeled in black) that have been excluded from the core genome SNP discovery. Predicted SNPs with a consistency index (CI) of <1 indicative of homoplasy are highlighted in purple in the outermost circle. Download
Consensus tree. A strict consensus tree of all 80 equally parsimonious trees. One of these trees is shown in Fig. 2. That most of the hierarchical topology seen in Fig. 2 is retained in the consensus tree is indicative of the lack of conflicting phylogenetic signal and the accuracy of clades found in Fig. 2. For clonally inherited genomic DNA, a single character can accurately define a branch. Bootstrapping (36) is a commonly used metric for assessing the robustness of a tree; however, it is an inappropriate metric for such data sets, as it will underestimate the support for short, but real, branches (14, 36, 37). Measures of character state conflict are more appropriate for indicating the accuracy of trees generated with such data (14, 37). The homoplasy (or consistency) index and strict consensus trees provide both a measure of conflicting phylogenetic signal as well as the portions of the tree that are affected by homoplasy and may not truly reflect the evolution of the organism. Download
Whole-genome map similarity cluster analysis. The data are comprised of experimental and in silico maps of both chromosomes of V. cholerae O1 and non-O1/O139. Maps are compared by using the unweighted-pair group method with arithmetic means (UPGMA). The analysis includes maps of two representative Nepal-4 strains VC14 and VC26, which have been identified as close phylogenetic relatives of the Haitian outbreak strains featuring many of the genetic hallmarks found in the outbreak clade. Download
Length polymorphism in the integrative conjugative element. (A) Map comparisons of the prototypical ICEVchInd5 element at the prfC locus (97,952 bp; GenBank accession no. GQ463142) (18) compared to the ICE prevalent in the prototypical Haitian outbreak strain 2010EL-1786 (97,915 bp; GenBank accession no. JN648379) (17) and Nepalese group 2 to 4 strains. (B) Nepal-4 strains are distinguished by the absence of a 19.929-bp segment within the ICE encompassing 13 genes (Vch1786_I0089 [mutL] to Vch1786_I0102 [traI]). The deletion occurs next to a transposase mutator family gene (Vch1786_I0088). Restriction sites of NheI (GCTAGC) used for optical mapping and identified palindromic sequences are marked. Download
Sequence read coverage for predicted SNP positions. Computation of underlying sequence read coverage on (A) chromosome I and (B) chromosome II for each of the predicted SNP positions in the analyzed V. cholerae serotype O1 genomes. Download
Sequence read quality for predicted SNP positions. Computation of underlying sequence read base quality on (A) chromosome I and (B) chromosome II for each of the predicted SNP positions in the analyzed V. cholerae serotype O1 genomes. Download
Strain history and metadata of strains analyzed in this study. The geographical origin of the strains are shown in column A according to the color scheme used in Fig. 2
SNP discovery in V. cholerae O1. This table lists 670 intra- and intergenic identified SNPs detected in the 116 V. cholerae O1 serotype strains analyzed in this study. The SNP positions are referenced to the respective chromosome and position in strain N16961. The table contains information for each SNP position in the reference genome distinguishing intergenic, synonymous, and nonsynonymous SNPs and annotations. The data table lists number of discovered alleles for every strain at each assayed position (bi- or multiallelic) and flags potential low or high character homoplasies and branch-specific SNPs (see Fig. S1 and S2 in the supplemental material). The data set was used to compute the number of SNPs per gene, SNP density, and genome-wide SNP distribution to facilitate the identification of potential mutational hot spots.