ABSTRACT
There is an emerging paradigm that the human microbiome is central to many aspects of health and may have a role in preventing enteric infection. Entamoeba histolytica is a major cause of amebic diarrhea in developing countries. It colonizes the colon lumen in close proximity to the gut microbiota. Interestingly, not all individuals are equally susceptible to E. histolytica infection. Therefore, as the microbiota is highly variable within individuals, we sought to determine if a component of the microbiota could regulate susceptibility to infection. In studies utilizing a murine model, we demonstrated that colonization of the gut with the commensal Clostridia-related bacteria known as segmented filamentous bacteria (SFB) is protective during E. histolytica infection. SFB colonization in this model was associated with elevated cecal levels of interleukin 17A (IL-17A), dendritic cells, and neutrophils. Bone marrow-derived dendritic cells (BMDCs) from SFB-colonized mice had higher levels of IL-23 production in response to stimulation with trophozoites. Adoptive transfer of BMDCs from an SFB+ to an SFB− mouse was sufficient to provide protection against E. histolytica. IL-17A induction during BMDC transfer was necessary for this protection. This work demonstrates that intestinal colonization with a specific commensal bacterium can provide protection during amebiasis in a murine model. Most importantly, this work demonstrates that the microbiome can mediate protection against an enteric infection via extraintestinal effects on bone marrow-derived dendritic cells.
IMPORTANCE
Entamoeba histolytica is the causative agent of amebiasis, an infectious disease that contributes significantly to morbidity and mortality due to diarrhea in the developing world. We showed in a murine model that colonization with the commensal members of the Clostridia known as SFB provides protection against E. histolytica and that dendritic cells from SFB-colonized mice alone can recapitulate protection. Understanding interactions between enteropathogens, commensal intestinal bacteria, and the mucosal immune response, including dendritic cells, will help in the development of effective treatments for this disease and other infectious and inflammatory diseases. The demonstration of immune-mediated protection due to communication from the microbiome to the bone marrow represents an emerging field of study that will yield unique approaches to the development of these treatments.
INTRODUCTION
There are 58 million cases of childhood diarrhea (1, 2) each year, resulting in two million deaths annually (WHO). Diarrheal illness is a primary cause of mortality in children less than 5 years of age in developing countries and significantly contributes to malnutrition (3). Diarrhea in these populations is caused by many different enteropathogens, with Entamoeba histolytica contributing to morbidity and mortality. Forty-five percent of infants in our cohort in Dhaka are infected by 1 year of age, and 10.9% have E. histolytica diarrhea (3). E. histolytica was also shown to be the leading cause of unadjusted mortality from 12 to 24 months of age in a 7-site study of moderate to severe diarrhea in low-income countries (4). Amebiasis caused by E. histolytica varies greatly in presentation and can range from asymptomatic colonization to mild diarrhea to dysentery and finally to an invasive disease of the liver, lung, or brain (5).
Genetic variation between patients may explain some of the differences in disease manifestation but does not fully account for the wide variability in presentation (5, 6). Recent studies have suggested that the composition of the intestinal bacterial microbiota may influence the outcome of protozoan infections (7). Vaccination with a recombinant LecA fragment of the E. histolytica surface Gal/GalNAc lectin is protective during infection in a murine model of amebiasis (8). Blockade of interleukin 17A (IL-17A) abrogated this protection (8), suggesting that induction of the cytokine is protective. IL-17A has pleiotropic functions, including mucin and antimicrobial peptide induction and upregulation of IgA transport (9) across the intestinal epithelium. It can also support neutrophil infiltration in inflammatory-disease models (10–13). Neutrophils play a central role in cell-mediated clearance of parasites and may be important in clearance of E. histolytica (14). IgA was also implicated in protection against E. histolytica infection in a childhood cohort and thus may be involved in immunity against the parasite (15). Given that IL-17A and downstream mediators are important in immunity to E. histolytica, components of the intestinal microbiota that elicit exogenous IL-17A in the absence of intestinal pathology might prove to be protective during amebiasis.
Segmented filamentous bacteria (SFB), or “Candidatus Savagella,” are genetically and morphologically unique members of the Clostridia. They represent an uncultivable component of the mammalian intestinal microbiota, reported to colonize both humans and mice (16–18). It has been demonstrated that SFB colonization induces a potent Th17 helper response in the intestine that is dependent on intestinal dendritic cells (19, 20). SFB colonization is associated with higher intestinal levels of both IL-17A and the damage-associated molecular pattern molecule and antimicrobial peptide serum amyloid A (SAA) (20). SFB infection influences many models of intestinal and extraintestinal inflammatory disease, suggesting that it may have a systemic influence on the immune response (21–23). Indeed, a growing body of literature suggests that intestinal colonization with commensal microorganisms can have extraintestinal effects on dendritic-cell precursors that influence susceptibility to pathogens (24–26). It is also quite possible that mediators induced by the intestinal microbiota in the serum might influence the bone marrow in such a way as to prime DCs to provide protection against, or exacerbate, enteropathogen infections (27, 28).
Bone marrow dendritic cells (BMDCs) are known to be able to recapitulate effector functions of some in vivo antigen-presenting-cell populations, and lipopolysaccharide (LPS)-matured BMDCs have been effectively utilized in adoptive transfer experiments to examine the influence of dendritic cells in infection models (28–30). Thus, to begin to examine the influence of changes in the microbiota on resistance to amebic infection, we utilized a murine model and BMDCs to explore what influence colonization with SFB had on intestinal infection with E. histolytica. Here we demonstrate that colonization with SFB provided protection against E. histolytica colitis and that bone marrow dendritic cells (BMDCs) derived from SFB-colonized mice were able to recapitulate protection in mice that had not been colonized with the bacteria. Protection mediated by BMDCs from SFB+ mice was IL-17A dependent. These data suggest that intestinal colonization with a commensal bacterium can alter bone marrow in such a way as to provide protection against parasite infection.
RESULTS
Cohousing Charles River (SFB+) and Jackson mice (SFB−) protected Jackson CBA mice from E. histolytica infection.
To test if alteration if the intestinal microbiota could alter susceptibility to E. histolytica infection, mice from two different animal vendors, Charles River and Jackson Laboratories, were cohoused (31, 32). The SFB status of mice after 3 weeks of cohousing was measured via qPCR and Gram stains. The cohoused mice were then challenged with E. histolytica trophozoites (2 × 106) via intracecal injection. Seven days later, E. histolytica and SFB burden were measured by qPCR. Cohousing mice transferred SFB (Fig. 1A), and potentially many other bacteria, and provided protection against E. histolytica infection (Fig. 1B).
FIG 1 .
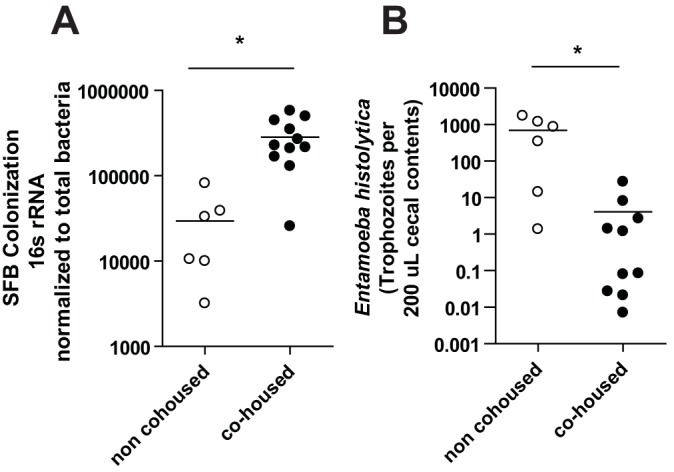
Cohousing mice from different vendors transfers SFB and confers protection against Entamoeba histolytica infection. qPCR was used to quantify relative expression compared to total bacteria and trophozoites of SFB (A) and Entamoeba histolytica (B), respectively, in cecal lysate from SFB− CBA/J mice that were cohoused with SFB+ C3H mice and noncohoused mice; mice were infected with 2 million E. histolytica trophozoites via cecal laparotomy. *, P < 0.05.
Segmented filamentous bacteria specifically protect against E. histolytica and induce increased IL-17A, IL-23, neutrophils, and dendritic cells in the intestine and SAA in the blood.
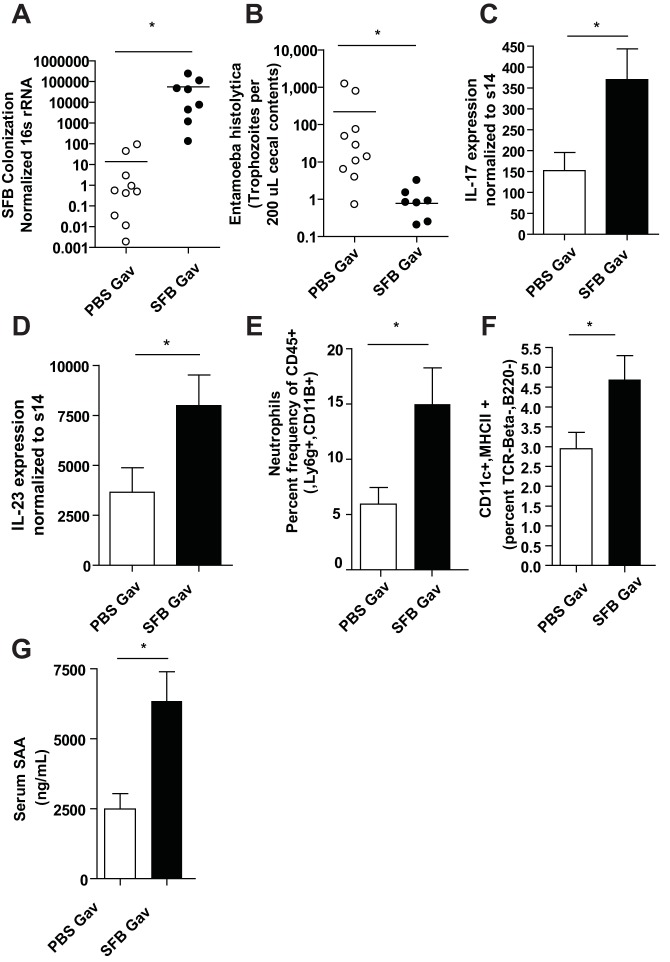
As cohousing of mice both transferred SFB and was protective against E. histolytica infection, we sought to determine if SFB specifically provided protection against the ameba. We directly colonized CBA/J mice from Jackson Laboratories with SFB by orogastric gavage with SFB-monoassociated feces resuspended in phosphate-buffered saline (PBS), provided by the Yakult Central Institute for Microbiological Studies, 1 week prior to E. histolytica infection. Mice became colonized with SFB following gavage (Fig. 2A) and were protected from infection with the ameba (Fig. 2B). Additionally, there was increased IL-17A and IL-23 expression before (Fig. S1) and after (Fig. 2C and D) E. histolytica infection. Neutrophils (Fig. 2E; also, see Fig. S1c in the supplemental material) and DCs (Fig. 2F; also, see Fig. S1d in the supplemental material) were also elevated in the intestine of SFB colonized mice after E. histolytica infection. We also observed increased SAA in serum in SFB-colonized mice after ameba infection (Fig. 2G).
FIG 2 .
SFB-colonized mice are protected from E. histolytica infection, have increased IL-17A, IL-23, neutrophils, and DCs in their intestines, and have more serum amyloid A (SAA) in their serum. Quantification via qPCR of SFB relative expression compared to total bacteria (A) and enumeration of trophozoites (B) was performed in cecal lysate from mice given gavages with PBS or SFB-monoassociated feces (Yakult) 7 days prior to intracecal infection with E. histolytica. IL-17A (C) and IL-23 (D) induction in ceca (lamina propria) was measured via qPCR, and neutrophil and DC infiltration was measured by flow cytometry and indicated markers (E and F). (G) Serum SAA was measured by ELISA. *, P < 0.05.
BMDCs derived from SFB-colonized mice have an increased capacity to produce IL-23 that is partially recapitulated with SAA treatment.
IL-23 is a key cytokine in the generation and maintenance of IL-17A-producing cells (33). SAA can directly induce IL-23 from both DCs and macrophages (20, 34) and is known to upregulate an epigenetic mediator, JMJD3 (33), that specifically increases IL-23 production in a macrophage cell line. Thus, as SFB increased frequency of intestinal DCs, IL-23, IL-17A expression, and circulating serum SAA, we examined the capacity of bone marrow DCs from SFB-infected mice to produce IL-23 in response to trophozoites. BMDCs were generated from SFB-free and SFB-infected mice and treated with trophozoites for 24 h. BMDCs and trophozoites were cocultured in RPMI with 10% fetal bovine serum (FBS) under aerobic conditions rather than in ameba culture medium under anaerobic conditions, as both DCs and trophozoites were viable after 24 h in these media; however, some cell death had occurred, as observed via trypan blue. Cytokines in the supernatants were then determined by enzyme-linked immunosorbent assay (ELISA) (IL-23; R&D Systems). BMDCs derived from mice that were colonized with SFB produced significantly more IL-23 in response to trophozoites (Fig. 3A). These data suggested that SFB altered bone marrow cells in such a way as to favor increased IL-23 production. We hypothesized that SAA induced in the serum by intestinal SFB colonization could influence BMDCs to produce increased IL-23. Therefore, we wanted to test if the presence of SAA during the differentiation of SFB-free BMDCs might alter them in such a way that they also would produce more IL-23. Thus, SAA (10 µg/µl) was added for the first 4 days of bone marrow DC culture. Addition of SAA to BMDC culture from SFB-free mice partially recapitulated the increased IL-23 seen in BMDCs derived from SFB+ mice (Fig. 3B). This suggested that SAA might be at least partially responsible for alteration of bone marrow DCs in this model.
FIG 3 .
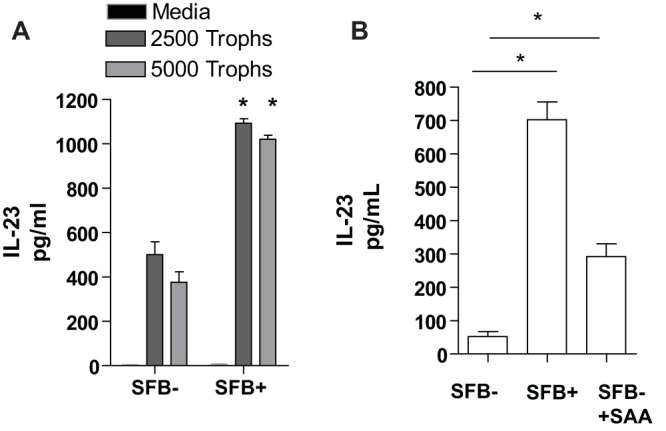
Bone marrow-derived DCs from mice colonized with SFB produce more IL-23 in response to E. histolytica trophozoites, and increased IL-23 production can be partially recapitulated in non-SFB-colonized mice with addition of SAA. Bone marrow cells were harvested from 4-week-old male CBA/J mice infected with or free of SFB, and cells were cultured in RPMI with 10% FBS, supplemented on days 0 and 3 with GM-CSF (10 ng/ml; Peprotech), and harvested on day 6. (A) A total of 2.5 × 105 cells were plated per well of a flat-bottom 96-well dish and treated with medium or trophozoites (Trophs) for 24 h. IL-23 in the supernatants was measured by ELISA. (B) BMDCs were cultured as described above with the addition of a third group with BMDCs from SFB-naive mice in which SAA was added to GM-CSF-containing medium for the first 4 days of culture only. Cells were washed and then plated with 2,500 trophozoites, and IL-23 was measured after 24 hours as described above. *, P < 0.05.
BMDCs from SFB+ but not SFB− mice migrated to the intestine.
To test if the increase in DCs in the gut of SFB+ mice was due to a direct effect on the bone marrow, BMDCs from SFB-colonized and SFB-free mice were adoptively transferred to SFB− mice. For in vivo transfers, day 6 BMDC from SFB− and SFB+ mice were matured with LPS (1 µg/ml), and 16 h later, cells were stained with carboxyfluorescein succinimidyl ester (CFSE; 10 nM), and 5 × 105 cells were administered via intraperitoneal injection. CFSE+ BMDC were detected in the lamina propria but only from SFB+ mice (Fig. 4). The ability of SFB+ BMDCs to home to the gut may help explain the increased numbers of DCs seen in the lamina propria of SFB-colonized mice during E. histolytica infection (Fig. 2E). These results were consistent with other studies that have shown that adoptive transfer of LPS-matured BMDCs results in DC trafficking to the intestine (28, 29, 35).
FIG 4 .
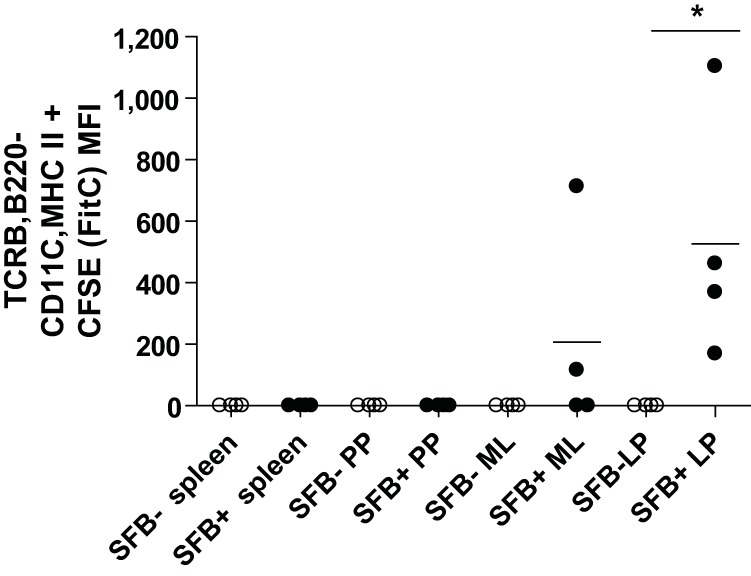
BMDCS from SFB-colonized mice migrated to the lamina propria of SFB-free mice upon adoptive transfer via the intraperitoneal route. Bone marrow cells (500,000) from SFB+ or SFB− mice were CFSE stained and adoptively transferred to 5-week-old CBA/J mice free of SFB. Flow cytometry for CFSE staining was performed on single cells isolated from spleens, Peyer’s patches (PP), mesenteric lymph nodes (MLN), and lamina propria (LP). *, P < 0.05.
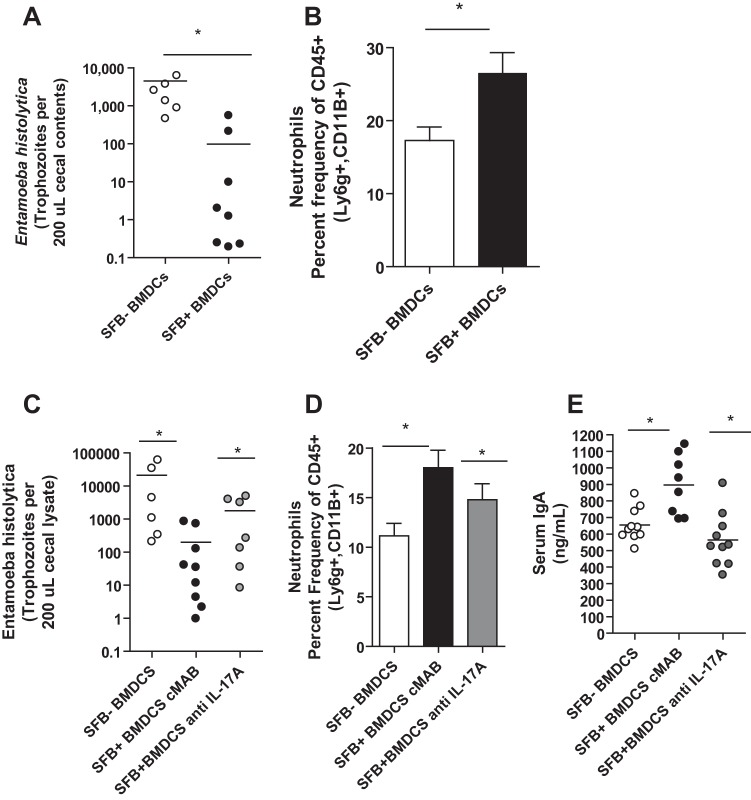
Adoptive transfer of BMDCs from SFB colonized mice was sufficient to provide protection against infection in an IL-17A-dependent manner.
We hypothesized that BMDCs from SFB-colonized mice may provide protection against amebiasis and that the mechanism of this protection might be via downstream induction of IL-17A. To explore this idea, we adoptively transferred BMDCs (5 × 105 cells) from SFB− or SFB+ mice to SFB− mice prior to E. histolytica infection. Indeed, we found that BMDCs from SFB+ mice were sufficient both to confer protection (Fig. 5A) and to recapitulate the increase in neutrophils (Fig. 5B) observed with SFB colonization alone. Blockade of IL-17A during transfer abrogated protection (Fig. 5C) and neutrophil influx (Fig. 5D) and led to a decrease in IgA induction (Fig. 5E). We concluded that IL-17A and downstream innate and perhaps adaptive immune responses underlie the protection observed during adoptive transfer of BMDCs from SFB+ mice.
FIG 5 .
BMDCs from mice colonized with SFB are sufficient to confer protection against E. histolytica colonization and recapitulate protective immune response, and blockade of IL-17A during adoptive transfer abrogates this protection. A total of 500,000 bone marrow cells from SFB+ or SFB− mice prepared as described in Materials and Methods were adoptively transferred to 5-week-old CBA/J mice free of SFB 1 week prior to infection with 2 million E. histolytica trophozoites via cecal laparotomy. E. histolytica (A) organisms in cecal lysate were quantified via qPCR. (B) Neutrophil infiltration in cecum by indicated markers. Four-week-old CBA/J mice were treated with 200 µg rat anti-mouse IL-17A MAb (clone M210; Amgen) or rat IgG2a isotype control (clone 2A3; BioXcell) 3 days before BMDC transfer (500,000 intraperitoneally from mice that received SFB gavage and mice that did not) and 7 days after and then infected with 2 million trophozoites that day. (C) E. histolytica in cecal lysate from mice was quantified via qPCR. (D) Neutrophil infiltration was measured via flow cytometry by indicated markers. (E) Total serum IgA was measured by ELISA. *, P < 0.05.
DISCUSSION
We have demonstrated that alteration of the microbiota via introduction of the commensal bacteria SFB impacts susceptibility to amebic infection in a murine model, likely via its impact on the mucosal immune system (36). There was an increase in IL-17A and IL-23 expression in the intestines of SFB-colonized mice prior to and following amebic infection and a relative increase in neutrophils after amebic infection. Our laboratory and others have shown that these effectors may provide protection against E. histolytica (14). We also observed increased CD11c+ MHCII+ cells in the intestines of mice, suggesting that dendritic cells induced by SFB may also help mediate protection against infection. This is not surprising given that Goto et al. recently demonstrated that major histocompatibility complex class II (MHC-II)-dependent presentation of SFB antigens by intestinal DCs is crucial for Th17 cell induction (37).
Interestingly, protection in our model appeared to involve not only the mucosal immune response in the gut but also the bone marrow. Resistance to amebic colitis was adoptively transferred to SFB-free mice with bone marrow-derived dendritic cells (BMDCs) from SFB-colonized mice. This suggested that SFB alteration of bone marrow cells might underlie protection against amebic colitis. BMDCs derived from SFB colonized mice produced more IL-23 in response to E. histolytica trophozoites than BMDCs from mice that were not colonized with the bacteria. However, this response was not specific to E. histolytica, as increased IL-23 production from SFB+ BMDCs was also seen with LPS and SAA treatment (data not shown). This suggests that SFB colonization of the intestine has conditioned bone marrow cells to be more responsive to stimulation with pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), such as SAA, or, alternatively, that there were shifts in the populations of dendritic-cell precursors present in the bone marrow prior to BMDC culture in SFB-colonized mice.
Therefore, in this model of SFB-induced alteration of the microbiome, it is not yet clear how SFB altered bone marrow cells. Future studies will be required to examine the possibility of shifts in dendritic-cell precursors or changes in responsiveness to DAMPS and PAMPS. In this context, intestinal inflammation or colonization with commensal organisms has been shown to alter populations of bone marrow progenitor cells (25, 26, 38). However, exactly how cross talk occurs between the intestine and bone marrow is not well understood. Recent work has shown that gut microbiota metabolism of dietary fiber can increase the concentration of circulating short-chain fatty acids (SCFAs) and that treatment of mice with the SCFA propionate led to alterations in bone marrow hematopoiesis (39). A similar mechanism might underlie protection in our model of E. histolytica/SFB coinfection. However, SFB-mediated alteration of the metabolome has not been explored. SFB do, however, have an intimate association with the gut intestinal mucosa, likely due to their limited metabolic capability (40, 41). These bacteria are thus known to alter host gene expression in the intestine (41).
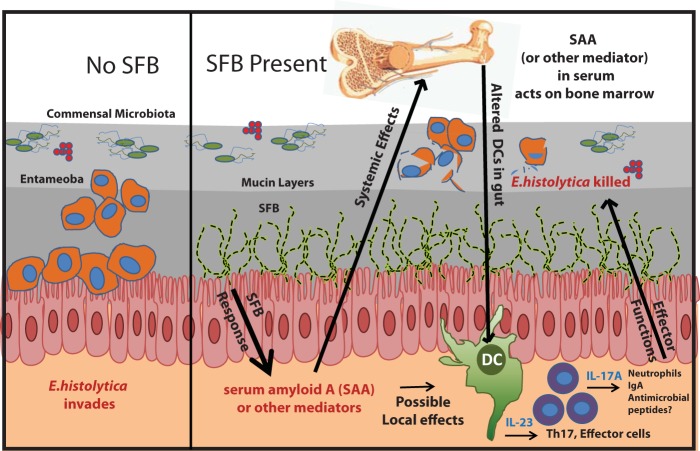
SAA has been shown to be upregulated in the intestines of mice colonized with SFB (20), and we have demonstrated that SAA is also increased in the serum of E. histolytica/SFB-infected mice. SAA is known to be produced by many cell types, including epithelial cells (3), and SFB tightly associate with the intestinal mucosa (4). SAA-dependent induction of Jmjd3-mediated epigenetic regulation of inflammatory cytokine gene expression has also been described (33), including IL-23, in SAA-stimulated macrophages. It is thus not hard to imagine that circulating SAA might act on bone marrow cells to alter IL-23 induction via epigenetic mechanisms (33). Indeed, we cultured BMDCs from SFB− mice in the presence of SAA for the first 4 days of culture and then removed the mediator, and the resulting differentiated BMDCs produced significantly more IL-23 than BMDCs from SFB− mice alone. However, this did not fully recapitulate the increase in IL-23 production seen in BMDCs from SFB-colonized mice. This study is not conclusive, however, and multiple mechanisms, perhaps including alteration of metabolic by-products, may underlie the increased IL-23 production seen in BMDCs from SFB-colonized mice. We propose a model in which intestinal colonization with SFB leads to induction of systemically circulating mediators, perhaps including SAA, that alter bone marrow dendritic-cell precursors so that they provide protection against E. histolytica via induction of downstream effectors, which may include IL-17A, neutrophils, and IgA (Fig. 6).
FIG 6 .
Model for SFB-mediated protection against E. histolytica colonization. SFB colonization of the intestine may induce soluble mediators, including SAA, which can have local effects as well as trigger systemic changes in bone marrow that support increased IL-23 production from dendritic-cell subsets and downstream IL-17A-mediated innate and adaptive immune responses. These effectors may then protect against intestinal E. histolytica infection.
In conclusion, we have demonstrated that colonization with a specific component of the intestinal microbiota, SFB, provided protection against infection with the protozoan pathogen E. histolytica and that BMDCs and downstream IL-17A induction recapitulated this protection. This work therefore suggests that alteration of the microbiome can mediate resistance to intestinal infection via extraintestinal effects on bone marrow cells. Future studies will be required to describe and understand the underlying mechanisms of these microbiome-induced changes in bone marrow dendritic-cell precursors.
MATERIALS AND METHODS
Mice.
Male 5-week-old CBA/J mice (Jackson Laboratories, Charles River) were housed in a specific-pathogen-free facility in micro-isolator cages and provided autoclaved food (Lab Diet 5010) and water ad libitum. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Virginia.
E. histolytica culture and intracecal injection.
Animal-passaged HM1:IMSS E. histolytica trophozoites were cultured from cecal contents of infected mice in complete trypsin-yeast-iron (TYI-33) medium supplemented with Diamond vitamin mix (JRH Biosciences), 100 U/ml of penicillin and streptomycin, and bovine serum (Sigma-Aldrich). Prior to injection, trophozoites were grown to log phase, and 2 × 106 parasites were suspended in 150 µl culture medium and injected intracecally (42).
SFB colonization.
Mice were colonized with SFB by cohousing for 3 weeks with SFB+ age-matched mice or directly colonized with SFB by orogastric gavage with SFB-monoassociated feces (a kind gift from the Yakult Central Institute for Microbiological Studies) 1 week prior to E. histolytica infection. SFB-monoassociated feces were independently confirmed to be associated with SFB only, with no presence of other bacteria or fungal contamination, by both Y. Umesaki and us via qPCR, Sanger sequencing, and culture (43).
Quantitative real-time RT-PCR.
SFB and E. histolytica colonization were measured by real-time PCR. For SFB colonization, qPCR with Sybr green was performed, and data were normalized to expression of a conserved Eubacteria 16S RNA gene (EUB). Primer sequences are as follows: EUB forward, 5′-ACTCCTACGGGAGGCAGCAGT-3′; EUB reverse, 5′-ATTACCGCGGCTGCTGGC-3′; SFB forward, 5′-GACGCTGAGGCATGAGAGCAT-3′; SFB reverse, 5′-GACGGCACGGATTGTTATTCA-3′. A 6-carboxyfluorescein (FAM)-labeled probe (44), a standard curve prepared from trophozoites, and quantitative PCR (qPCR) were utilized for E. histolytica quantification; the primer and probe sequences are as follows: Eh-f, AAC AGT AAT AGT TTC TTT GGT TAG TAA AA; Eh-r, CTT AGA ATG TCA TTT CTC AAT TCA T; Eh-YYT, ATT AGT ACA AAA TGG CCA ATT CAT TCA-dark quencher; IL-17A F, 5′-ACTACCTCAACCGTTCCACG-3′; IL-17A R, 5′-AGAATTCATGTGGTGGTCCAG-3′. Cytokines were measured via qPCR with Sybr green, and data were normalized to expression of S14: IL-23p19F, 5′-GACCCACAAGGACTCAAGGA-3′; IL-23p19 R, 5′-CATGGGGCTATCAGGGAGTA-3′; S14 F, 5′-TGGTGTCTGCCACATCTTTGCATC-3′; S14 R, 5′-AGTCACTCGGCAGATGGTTTCCTT-3′. Melting temperatures were 60°C for EUB, SFB, IL-17A, and IL-23 and for E. histolytica (44). Primers and probes were purchased from Integrated DNA Technologies, Coralville, IA, USA.
Serum SAA was measured by ELISA (ab157723; Abcam, Cambridge, England).
Bone marrow-derived-dendritic-cell (BMDC) culture.
Bone marrow cells were harvested from 4 weeks old male CBA/J mice from Jackson Laboratories that had been treated by gavage with PBS or SFB the week prior. Cells were cultured in RPMI with 10% FBS supplemented on days 0 and 3 with granulocyte-macrophage colony-stimulating factor (GM-CSF) (10 ng/ml; Peprotech) and harvested on day 6. For in vitro experiments, 2.5 × 105 cells were plated per well of a flat-bottom 96-well dish and treated with trophozoites for 24 h. Cytokines in the supernatants were determined by ELISA (IL-23; R&D Systems).
DC transfer.
For in vivo transfers, day 6 BMDCs were stimulated with LPS (1 µg/ml); 16 h later, cells were washed in PBS, and 5 × 105 cells were administered intraperitoneally. For CFSE experiments, cells were stained with CFSE in PBS for 10 min (10 nM) and then washed twice in PBS prior to adoptive transfer.
IL-17A blockade.
Four-week-old CBA/J mice were treated with 200 µg rat anti-mouse IL-17A monoclonal antibody (MAb) (clone M210; Amgen) or rat IgG2a isotype control (clone 2A3; BioXcell) at 3 days before and on the day of BMDC transfer (500,000 intraperitoneally from mice receiving SFB gavage or those not receiving SFB gavage). Mice were challenged intracecally with 2 × 106 trophozoites 7 days after BMDC transfer.
In vitro culture and flow cytometry of intestinal cells.
For intracellular staining, intestinal tissue was digested in Liberase TL (0.17 mg/ml Roche) and DNase (0.5 mg/ml; Sigma) for 45 min at 37°C and processed into a single-cell suspension. A total of 1 × 106 cells were incubated with CD16/CD32 MAb (BD Biosciences) to block Fc binding, followed by staining with CD11V-BV421, MHCII-FITC, TCR-beta-APC, B220-APC, CD45.2-APC, Ly6G-PE, Ly6C-Percp Cy 5.5, or CD11b-Pacific Blue and run on an LSR Fortessa cell analyzer (BD Biosciences). The data were analyzed with FlowJo (Tree Star Inc.).
Statistical analysis.
An analysis of variance (ANOVA) followed by the Tukey-Kramer test was used for analysis of differences among multiple groups. Student’s t test was used for comparisons between two groups. P values of less than 0.05 were considered significant. Results of representative experiments are shown; all experiments were replicated 2 to 4 times, with 4 to 12 individuals per group.
SUPPLEMENTAL MATERIAL
Representative flow cytometry and baseline IL-17A and IL-23 expression in the cecum following SFB gavage. Representative gating strategies utilized for neutrophils (A) and CD11c+ cells (B) are shown. IL-17A (C) and IL-23 (D) induction in ceca (lamina propria) was measured via qPCR 7 days after gavage with PBS (SFB−) or SFB (SFB+). SFB colonization was confirmed via qPCR and microscopy. Download
ACKNOWLEDGMENTS
This work was supported by NIH grant 5R01 AI026649-25 to W.P. We thank Yoshinori Umesaki, from the Yakult Central Institute for Microbial Research (Tokyo, Japan) for supplying the SFB-monocolonized feces. We also gratefully acknowledge the gift of anti-IL-17A monoclonal antibodies from Alison Budelsky at Amgen Incorporated (Seattle, WA) and Florent Ginhoux, Brian Kelsall, and Tom Braciale for helpful discussions.
S.B., W.P., and M.W.-K. contributed to the design, analysis and interpretation of the data. S.B., E.B., M.C., C.C., C.N., and Z.N. conducted experiments and critiqued the work. S.B. and W.P. wrote the manuscript.
Footnotes
Citation Burgess SL, Buonomo E, Carey M, Cowardin C, Naylor C, Noor Z, Wills-Karp M, Petri, Jr., WA. 2014. Bone marrow dendritic cells from mice with an altered microbiota provide interleukin 17A-dependent protection against Entamoeba histolytica colitis. mBio 5(6):e01817-14. doi:10.1128/mBio.01817-14.
REFERENCES
- 1. De La Sante OM. 2004. WHO/PAHO informal consultation on intestinal protozoal infections. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2. Pierce KK, Kirkpatrick BD. 2009. Update on human infections caused by intestinal protozoa. Curr. Opin. Gastroenterol. 25:12. [DOI] [PubMed] [Google Scholar]
- 3. Mondal D, Minak J, Alam M, Liu Y, Dai J, Korpe P, Liu L, Haque R, Petri WA. 2012. Contribution of enteric infection, altered intestinal barrier function, and maternal malnutrition to infant malnutrition in Bangladesh. Clin. Infect. Dis. 54:185–192. 10.1093/cid/cir807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O’Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 5. Verkerke HP, Petri WA, Marie CS. 2012. The dynamic interdependence of amebiasis, innate immunity, and undernutrition. Semin. Immunopathol. 34:771–785. 10.1007/s00281-012-0349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mondal D, Petri WA, Sack RB, Kirkpatrick BD, Haque R. 2006. Entamoeba histolytica-associated diarrheal illness is negatively associated with the growth of preschool children: evidence from a prospective study. Trans. R. Soc. Trop. Med. Hyg. 100:1032–1038. 10.1016/j.trstmh.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 7. Berrilli F, Di Cave D, Cavallero S, D’Amelio S. 2012. Interactions between parasites and microbial communities in the human gut. Front. Cell. Infect. Microbiol. 2:141. 10.3389/fcimb.2012.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo X, Barroso L, Lyerly DM, Petri WA, Houpt ER. 2011. CD4+ and CD8+ T cell- and IL-17-mediated protection against Entamoeba histolytica induced by a recombinant vaccine. Vaccine 29:772–777. 10.1016/j.vaccine.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cao AT, Yao S, Gong B, Elson CO, Cong Y. 2012. Th17 cells upregulate polymeric Ig receptor and intestinal IgA and contribute to intestinal homeostasis. J. Immunol. 189:4666–4673. 10.4049/jimmunol.1200955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blaschitz C, Raffatellu M. 2010. Th17 cytokines and the gut mucosal barrier. J. Clin. Immunol. 30:196–203. 10.1007/s10875-010-9368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fujisawa T, Velichko S, Thai P, Hung LY, Huang F, Wu R. 2009. Regulation of airway MUC5AC expression by IL-1beta and IL-17A; the NF-kappaB paradigm. J. Immunol. 183:6236–6243. 10.4049/jimmunol.0900614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mei J, Liu Y, Dai N, Hoffmann C, Hudock KM, Zhang P, Guttentag SH, Kolls JK, Oliver PM, Bushman FD, Worthen GS. 2012. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J. Clin. Invest. 122:974–986. 10.1172/JCI60588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maddur MS, Miossec P, Kaveri SV, Bayry JT. 2012. Th17 cells: biology, pathogenesis of autoimmune and inflammatory diseases, and therapeutic strategies. Am. J. Pathol. 181:8–18. 10.1016/j.ajpath.2012.03.044. [DOI] [PubMed] [Google Scholar]
- 14. Asgharpour A, Gilchrist C, Baba D, Hamano S, Houpt E. 2005. Resistance to intestinal Entamoeba histolytica infection is conferred by innate immunity and Gr-1+ cells. Infect. Immun. 73:4522–4529. 10.1128/IAI.73.8.4522-4529.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haque R, Ali IM, Sack RB, Farr BM, Ramakrishnan G, Petri WA. 2001. Amebiasis and mucosal IgA antibody against the Entamoeba histolytica adherence lectin in Bangladeshi children. J. Infect. Dis. 183:1787–1793. 10.1086/320740. [DOI] [PubMed] [Google Scholar]
- 16. Pamp SJ, Harrington ED, Quake SR, Relman DA, Blainey PC. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB). Genome Res. 22:1107–1119. 10.1101/gr.131482.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yin Y, Wang Y, Zhu L, Liu W, Liao N, Jiang M, Zhu B, Yu HD, Xiang C, Wang X. 2012. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 7:615–621. 10.1038/ismej.2012.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davis CP, Savage DC. 1974. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect Immun 10:948–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, Laufer TM, Ignatowicz L, Ivanov II. 2014. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40:594–607. 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498. 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. 2011. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. U. S. A. 108: 11548–11553. 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. 2010. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32:815–827. 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. 2011. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U. S. A. 108:4615–4622. 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khosravi A, Yáñez A, Price JG, Chow A, Merad M, Goodridge HS, Mazmanian SK. 2014. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 15:374–381. 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, Kambayashi T, Larosa DF, Renner ED, Orange JS, Bushman FD, Artis D. 2012. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 18:538–546. 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Griseri T, McKenzie BS, Schiering C, Powrie F. 2012. Dysregulated hematopoietic stem and progenitor cell activity promotes interleukin-23-driven chronic intestinal inflammation. Immunity 37:1116–1129. 10.1016/j.immuni.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ursell LK, Haiser HJ, Van Treuren W, Garg N, Reddivari L, Vanamala J, Dorrestein PC, Turnbaugh PJ, Knight R. 2014. The intestinal metabolome: an intersection between microbiota and host. Gastroenterology 146:1470–1476. 10.1053/j.gastro.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pastille E, Didovic S, Brauckmann D, Rani M, Agrawal H, Schade FU, Zhang Y, Flohé SB. 2011. Modulation of dendritic cell differentiation in the bone marrow mediates sustained immunosuppression after polymicrobial sepsis. J. Immunol. 186:977–986. 10.4049/jimmunol.1001147. [DOI] [PubMed] [Google Scholar]
- 29. Stäger S, Maroof A, Zubairi S, Sanos SL, Kopf M, Kaye PM. 2006. Distinct roles for IL-6 and IL-12p40 in mediating protection against Leishmania donovani and the expansion of IL-10+ CD4+ T cells. Eur. J. Immunol. 36:1764–1771. 10.1002/eji.200635937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao J, Zhao J, Van Rooijen N, Perlman S. 2009. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 5:e1000636. 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, Hansen AK. 2010. Variation in the gut microbiota of laboratory mice is related to both genetic and environmental factors. Comp. Med. 60:336–347. [PMC free article] [PubMed] [Google Scholar]
- 32. Dimitriu PA, Boyce G, Samarakoon A, Hartmann M, Johnson P, Mohn WW. 2013. Temporal stability of the mouse gut microbiota in relation to innate and adaptive immunity. Environ. Microbiol. Rep. 5:200–210. 10.1111/j.1758-2229.2012.00393.x. [DOI] [PubMed] [Google Scholar]
- 33. Yan Q, Sun L, Zhu Z, Wang L, Li S, Ye RD. 2014. Jmjd3-mediated epigenetic regulation of inflammatory cytokine gene expression in serum amyloid A-stimulated macrophages. Cell. Signal. 26:1783–1791. 10.1016/j.cellsig.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ather JL, Ckless K, Martin R, Foley KL, Suratt BT, Boyson JE, Fitzgerald KA, Flavell RA, Eisenbarth SC, Poynter ME. 2011. Serum amyloid A activates the NLRP3 inflammasome and promotes Th17 allergic asthma in mice. J. Immunol. 187:64–73. 10.4049/jimmunol.1100500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sheasley-O’Neill SL, Brinkman CC, Ferguson AR, Dispenza MC, Engelhard VH. 2007. Dendritic cell immunization route determines integrin expression and lymphoid and nonlymphoid tissue distribution of CD8 T cells. J. Immunol. 178:1512–1522. 10.4049/jimmunol.178.3.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. 2011. Human nutrition, the gut microbiome and the immune system. Nature 474:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, Laufer TM, Ignatowicz L, Ivanov II. 2014. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40:594–607. 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Khosravi A, Yáñez A, Price JG, Chow A, Merad M, Goodridge HS, Mazmanian SK. 2014. Gut microbiota promote hematopoiesis to control bacterial Infection. Cell Host Microbe 15:374–381. 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. 2014. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20:159–166. 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 40. Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, Littman DR, Ivanov II. 2011. The genome of Th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe 10:260–272. 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuwahara T, Ogura Y, Oshima K, Kurokawa K, Ooka T, Hirakawa H, Itoh T, Nakayama-Imaohji H, Ichimura M, Itoh K, Ishifune C, Maekawa Y, Yasutomo K, Hattori M, Hayashi T. 2011. The lifestyle of the segmented filamentous bacterium: a non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res. 18:291–303. 10.1093/dnares/dsr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Houpt ER, Glembocki DJ, Obrig TG, Moskaluk CA, Lockhart LA, Wright RL, Seaner RM, Keepers TR, Wilkins TD, Petri WA. 2002. The mouse model of amebic colitis reveals mouse strain susceptibility to infection and exacerbation of disease by CD4+ T cells. J. Immunol. 169:4496–4503. 10.4049/jimmunol.169.8.4496. [DOI] [PubMed] [Google Scholar]
- 43. Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. 1995. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol. 39:555–562. [DOI] [PubMed] [Google Scholar]
- 44. Roy S, Kabir M, Mondal D, Ali IK, Petri WA, Haque R. 2005. Real-time-PCR assay for diagnosis of Entamoeba histolytica infection. J. Clin. Microbiol. 43:2168–2172. 10.1128/JCM.43.5.2168-2172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative flow cytometry and baseline IL-17A and IL-23 expression in the cecum following SFB gavage. Representative gating strategies utilized for neutrophils (A) and CD11c+ cells (B) are shown. IL-17A (C) and IL-23 (D) induction in ceca (lamina propria) was measured via qPCR 7 days after gavage with PBS (SFB−) or SFB (SFB+). SFB colonization was confirmed via qPCR and microscopy. Download