ABSTRACT
Marine sponges are the most primitive metazoan and host symbiotic microorganisms. They are crucial components of the marine ecological system and play an essential role in pelagic processes. Copper pollution is currently a widespread problem and poses a threat to marine organisms. Here, we examined the effects of copper treatment on the composition of the sponge-associated bacterial community and the genetic features that facilitate the survival of enriched bacteria under copper stress. The 16S rRNA gene sequencing results showed that the sponge Haliclona cymaeformis harbored symbiotic sulfur-oxidizing Ectothiorhodospiraceae and photosynthetic Cyanobacteria as dominant species. However, these autotrophic bacteria decreased substantially after treatment with a high copper concentration, which enriched for a heterotrophic-bacterium-dominated community. Metagenomic comparison revealed a varied profile of functional genes and enriched functions, including bacterial motility and chemotaxis, extracellular polysaccharide and capsule synthesis, virulence-associated genes, and genes involved in cell signaling and regulation, suggesting short-period mechanisms of the enriched bacterial community for surviving copper stress in the microenvironment of the sponge. Microscopic observation and comparison revealed dynamic bacterial aggregation within the matrix and lysis of sponge cells. The bacteriophage community was also enriched, and the complete genome of a dominant phage was determined, implying that a lytic phage cycle was stimulated by the high copper concentration. This study demonstrated a copper-induced shift in the composition of functional genes of the sponge-associated bacterial community, revealing the selective effect of copper treatment on the functions of the bacterial community in the microenvironment of the sponge.
IMPORTANCE
This study determined the bacterial community structure of the common sponge Haliclona cymaeformis and examined the effect of copper treatment on the community structure and functional gene composition, revealing that copper treatment had a selective effect on the functions of the bacterial community in the sponge. These findings suggest that copper pollution has an ecological impact on the sponge symbiont. The analysis showed that the untreated sponges hosted symbiotic autotrophic bacteria as dominant species, and the high-concentration copper treatment enriched for a heterotrophic bacterial community with enrichment for genes important for bacterial motility, supplementary cellular components, signaling and regulation, and virulence. Microscopic observation showed obvious bacterial aggregation and a reduction of sponge cell numbers in treated sponges, which suggested the formation of aggregates to reduce the copper concentration. The enrichment for functions of directional bacterial movement and supplementary cellular components and the formation of bacterial aggregates and phage enrichment are novel findings in sponge studies.
INTRODUCTION
Copper, an essential trace metal for organisms, can be highly toxic to organisms at excessive concentrations (1). The toxicity of copper is attributed to its harmful effects on cell membranes and nucleic acid structure together with its ability to alter enzyme specificity and disrupt cellular functions (2). Currently, the frequent use of copper as antifouling paint has led to marine organisms being exposed to high concentrations of copper (3). Thus, the effect of copper pollution on marine ecology is an important area of study.
Marine sponges serve as an important component of the benthic environment, with a considerable biomass and an essential influence on pelagic processes such as the food chain and elemental cycling (4, 5). Sponges establish close relationships with microbes, and this process occurs in the body of sponges as a result of symbiosis (6). Sponge-specific microbial clusters have been defined and constitute a 16S rRNA gene database for search purposes (7). The biomass of microbes in sponges is very high, constituting up to 60% of the total biomass of the sponge (8). Until now, more than 35 microbial phyla have been detected in sponges by denaturing gradient gel electrophoresis, 16S rRNA gene sequencing, and clone library sequencing. The majority of these bacteria were found to belong to the phyla Proteobacteria, Cyanobacteria, Firmicutes, Bacteroidetes, Acidobacteria, Actinobacteria, and Chloroflexi (9). Symbiotic microorganisms interact with the host by providing carbohydrates (10), vitamins (11), and antibiotics (12); oxidizing toxic metabolites such as ammonia (13) and sulfide (14); and participating in carbon, sulfur, and nitrogen cycles, all of which result in a relatively independent ecosystem in the sponge body. In return, the microorganisms acquire from the host a living shield and some metabolic substrates.
Sessile and filter-feeding characteristics have made sponges efficient predators of food bacteria (a marine sponge of 1 kg may filter up to 24,000 liters of seawater per hour in the field) (15) but also susceptible to the bioconcentration and bioaccumulation of pollutants (16). The harmful effects of heavy metal on sponges have been studied, and the results showed that the survival, growth, shape, water motion, and reproduction of sponges are all affected by copper pollution (17). At the cellular level, cadmium, copper, and mercury may have adverse effects on sponges in terms of the shape, motility, and aggregation of isolated cells (18). Moreover, high concentrations due to long-term copper and cadmium treatments also can lead to settler mortality and larval settlement failure (17).
In contrast, knowledge regarding the effect of heavy metals on the sponge-associated microbial community is limited. A previous study using copper, cadmium, and mercury to treat bacteria isolated from the sponge Fasciospongia cavernosa defined a group of heavy-metal-resistant bacteria in sponges as indicators of aquatic pollution (19). Specifically, copper exposure shifted the microbial community of the sponge Rhopaloeides odorabile with respect to morphology and taxonomy (20).
In the present study, we investigated the effect of copper treatment on the sponge-associated bacterial community in terms of community structure and functional gene composition to determine how copper affects the symbiotic bacterial community (and even bacteriophages). We also examined the genetic features that facilitated the survival of the bacterial community that was enriched in response to copper stress.
RESULTS
Pyrosequencing of 16S rRNA gene amplicons.
In total, 189,119 raw reads were acquired from the 20 samples by using Roche 454 FLX titanium sequencing; 117,015 high-quality reads were included in the analysis after quality control. After the removal of chloroplast and mitochondrial rRNA, 35,668 reads were identified for operational taxonomic unit (OUT) clustering. A total of 2,459 OTUs were selected on the basis of 3% sequence dissimilarity, and representative sequences of OTUs were taxonomically assigned. Twenty-one bacterial phyla were found in the samples, and the dominant phyla were Proteobacteria, Cyanobacteria, Firmicutes, and Bacteroidetes. The numbers of reads and OTUs, as well as the diversity estimation (Shannon index) are listed in Table S1 in the supplemental material. Each sample consisted of an average of 1,877 reads, and the rarefaction curve for the 16S rRNA gene reads showed that the sample diversity (Shannon index) had reached a plateau (see Fig. S1 in the supplemental material).
Shift of the bacterial community in response to copper treatment as revealed by amplicon sequencing.
Unweighted UniFrac clustering (determined by the unweighted-pair group method using average linkages [UPGMA]) of bacterial communities in the samples based on pyrosequencing indicated that the 2- and 4-day high-dose treatments changed the bacterial community composition in the sponge. In the jackknife tree in Fig. 1, the negative controls and low- and medium-dose-treated samples (N0, N2, L2, M2, N4, L4, and M4) clustered together in one group with similar pairwise distances. However, the high-dose-treated samples were distributed in a different group, which indicated that the bacterial community structure in the high-dose-treated sponges was different (H2 and H4). Principal-coordinate analysis (PCoA) of the bacterial communities revealed a similar pattern (see Fig. S2 in the supplemental material). The negative-control and low- and medium-dose-treated samples formed an independent group (group 1) that was distinct from the 2- and 4-day high-dose treatment groups, which were separated by PC1 (explaining 23.7% of the variance).
FIG 1 .
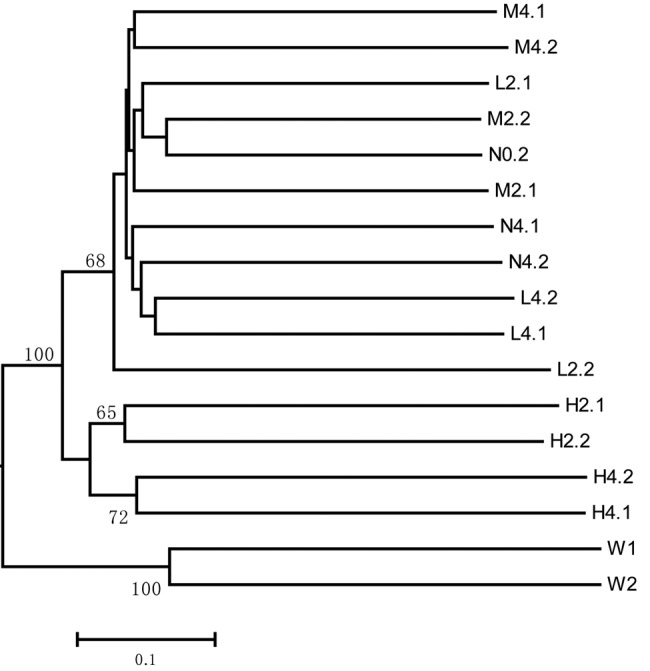
UPGMA clustering of unweighted UniFrac distances between bacterial communities of copper-treated sponges and controls based on the distribution of bacterial 16S rRNA gene sequences. The letters N, L, M, and H refer to sponge samples from the negative-control and low-, medium-, and high-dose treatments, respectively. The numbers 0, 2, and 4 indicate 0, 2, and 4 days of treatment, respectively, and the numbers after the dots are replicate numbers. W1 and W2 are the two replicates of the seawater sample.
The bacterial community of the untreated sponge was dominated by autotrophic symbiotic bacteria.
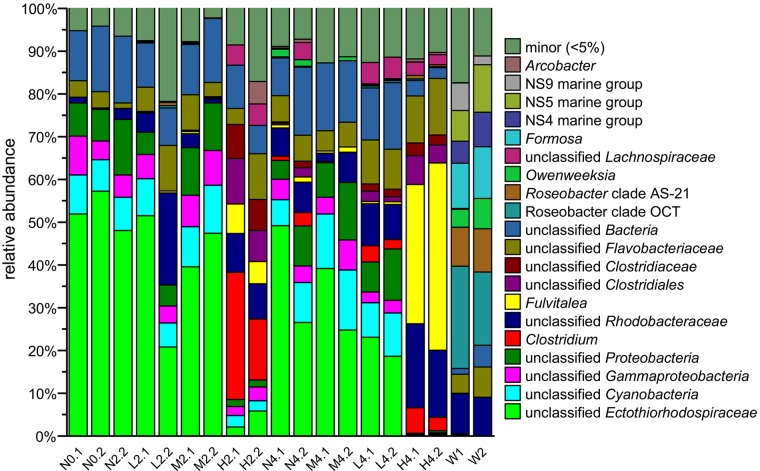
At the phylum level, the bacterial community of the negative control on day 0 (N0) was dominated by Proteobacteria (71.8%), Cyanobacteria (9.0%), and Bacteroidetes (5.9%) (see Fig. S3 in the supplemental material). The taxonomic profile at the genus level showed the dominance of autotrophic bacteria; an unclassified Ectothiorhodospiraceae bacterium (sulfur-oxidizing bacteria [SOB] accounted for 54.6% of the bacterial community in N0) and an unclassified cyanobacterium (photosynthetic bacteria accounted for 8.2% of the bacterial community in N0). The Ectothiorhodospiraceae bacteria were found only in sponge samples and not in seawater samples (Fig. 2). Moreover, the phylogenetic analysis revealed that the dominant bacterium of the unclassified Ectothiorhodospiraceae was located in sponge-specific clusters (see Fig. S4 in the supplemental material; the dominant OTU sequence accounted for 82.9% of the bacterial community), which indicated their coevolution and close relationship with the host sponge. The dominance of thioautotrophic and photosynthetic bacteria in the untreated samples suggested an important symbiotic function of sulfur oxidation and photosynthesis in the sponge (see Discussion).
FIG 2 .
The bacterial community composition at the genus level of sponge samples treated with copper, controls, and seawater samples. The y axis indicates the relative abundance of taxa, and the x axis indicates samples. The letters N, L, M, and H refer to sponge samples from the negative-control and low-, medium-, and high-dose treatments, respectively. The numbers 0, 2, and 4 indicate 0, 2, and 4 days of treatment, respectively, and the numbers after the dots are replicate numbers. W1 and W2 are two replicates of the seawater sample.
Copper treatment decreased the autotrophic symbiotic bacterial community and enriched for heterotrophic bacteria.
At the phylum level, the low- and medium-dose treatments had no obvious effects on the bacterial community (see Fig. S3 in the supplemental material). However, the 2- and 4-day high-dose treatments, respectively, notably decreased the proportions of Proteobacteria (to 22.9 and 24.9% of the bacterial community) and Cyanobacteria (to 2.6 and 0.2% of the bacterial community) and enriched for Firmicutes (to 44.9 and 16.8% of the bacterial community) and Bacteroidetes (to 21.1 and 54.8% of the bacterial community).
At the genus level, copper treatment caused a sharp decrease in autotrophic symbiotic bacteria and an increase in heterotrophic bacteria (Fig. 2). The proportions of autotrophs, including the unclassified Ectothiorhodospiraceae and the unclassified Cyanobacteria, were significantly decreased (t test, P < 0.05) by the treatment. The proportion of unclassified Ectothiorhodospiraceae was decreased to 4.0 and 0.5%, while that of the unclassified Cyanobacteria was decreased to 2.5 and 0.2% in H2 and H4, respectively. Other significantly decreased bacteria (t test, P < 0.01) included an unclassified proteobacterium and an unclassified gammaproteobacterium (Fig. 2), which were also sponge specific and were not detected in the bacterial community of the seawater samples (Fig. 2). In contrast, heterotrophic bacteria were largely increased in H2 and H4, including Clostridium (to 22.0 and 4.5%), unclassified Rhodobacteraceae (8.6 and 17.7%), Fulvitalea (6.1 and 38.2%), unclassified Clostridiales (8.9 and 5.5%), and unclassified Clostridiaceae (7.6 and 2.7%). All of the above enriched bacteria in the sponge were not found in the seawater (Fig. 2), except the unclassified Gammaproteobacteria and unclassified Rhodobacteraceae (further taxonomic analysis revealed that the OTUs of these bacteria differed in sponge and seawater samples, indicating different species).
Metagenomic sequencing and analysis.
To investigate the genetic features that facilitate the survival of the enriched bacteria under copper-induced stress, we compared the functional gene compositions of H4 and N4 by metagenomic sequencing. In total, we acquired 58 million and 46 million DNA reads (8.8 and 6.9 Gb) for the three biological replicates of N4 and H4, respectively. After quality filtration, 57 million and 44 million high-quality reads (7.2 and 5.6 Gb) for N4 and H4, respectively, were included in the following analysis. After assembly with the merged data, 103,414 contigs (>500 bp; maximum contig, 179 kb; N50, 1.7 kb; total length, 143 Mb) were used for gene identification, resulting in 59,582 bacterial, 601 archaeal, and 24,834 eukaryotic protein sequences according to the Clusters of Orthologous Groups (COG) database annotation.
Selective effect of copper treatment on the functions of the microbial community.
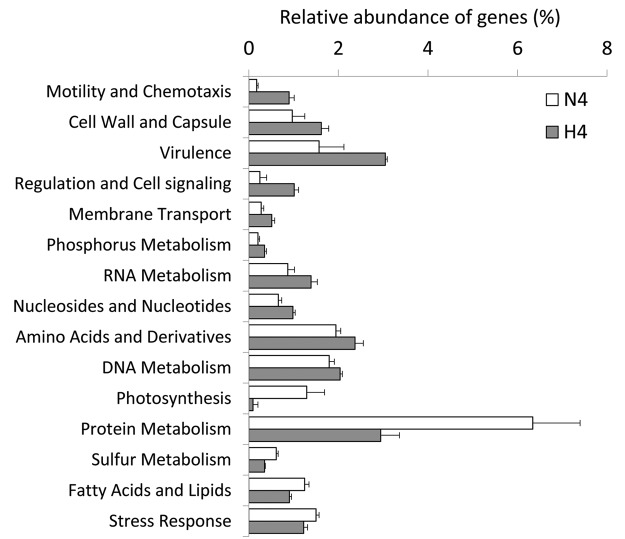
The functional gene composition profile showed that the main functions in N4 and H4 were carbohydrate metabolism, cofactor metabolism, virulence, protein metabolism, amino acid metabolism, and cell wall- and capsule-related functions, among others (see Fig. S5 in the supplemental material). Genes with significantly different (t test, P < 0.05) relative abundances in N4 and H4 were further screened out.
The abundance of functional genes involved in photosynthesis and sulfur metabolism decreased to 7 and 57% of their relative abundances in N4, respectively, consistent with the sharp decrease in symbiotic autotrophic bacteria in H4 (unclassified Ectothiorhodospiraceae and unclassified Cyanobacteria, Fig. 2). The remarkable decrease in the two autotrophic bacteria and in related functional genes involved in sulfur metabolism and photosynthesis might abrogate the supply of carbohydrates and detoxification of sulfides in the sponge.
In contrast, some other functional genes were significantly increased in response to the treatment. The enriched functional genes reflected the genetic features that might benefit survivors under copper treatment stress. Comparative analyses revealed that motility and chemotaxis, regulation and cell signaling, virulence, cell wall and capsule synthesis, membrane transport, phosphorus metabolism, and RNA metabolism, among others, were largely enriched (t test, P < 0.05) by the copper treatment (Fig. 3). Among them, the most remarkable increase in functional genes were those responsible for motility and chemotaxis, regulation and cell signaling, virulence, and cell wall and capsule synthesis, which were enriched >1.5-fold and accounted for >0.8% of the bacterial population in the H4 samples. These enriched functional genes can be summarized as cellular components (flagellum, cell wall, capsule, and extracellular polysaccharides; see Fig. S6a and b in the supplemental material), environmental signal sensing and response (chemotaxis, a two-component regulatory system, orphan regulatory proteins, and cyclic AMP [cAMP] signaling; see Fig. S6a and c in the supplemental material), and virulence-related membrane proteins (a virulence secretion system, a Ton and Tol transport system, and adhesion and invasion proteins; see Fig. S6d in the supplemental material). The selected functional genes suggested the involvement of multiple strategies of the enriched microorganisms in surviving the stress induced by copper treatment (see Discussion).
FIG 3 .
Functional genes with significant relative abundances (t test, P < 0.05) changed (decreased or enriched) in response to copper treatment. N4 denotes negative-control samples, and H4 indicates high-dose-treated samples. The x axis shows the normalized relative abundance of genes (as a percentage), and the y axis shows the functional categories at the first level of the SEED classification system.
Bacterial aggregation and reduced sponge cell numbers in response to copper treatment.
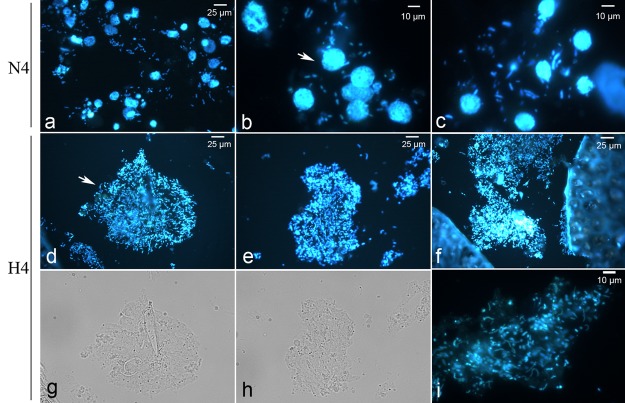
Microscopic observation and comparison revealed an obvious aggregation of bacteria (with changed bacterial morphotypes) and a reduction of sponge cell numbers in the treated sponges. Bacteria dispersed around the cells of control sponges (N4) and included mainly cocci and bacilli (see Fig. 5a to c). However, in the treated sponges (H4), bacteria aggregated and formed globular clusters throughout all of the fields (see Fig. 5d to i), and a considerable number of vibrios appeared in the aggregates (see Fig. 5i). The aggregated bacteria were distributed in the sponge matrix, which had different colors compared to the surroundings. Comparison of the microscopic images of the 2- and 4-day samples from the medium- and high-dose treatments to the corresponding control images revealed a dynamic decrease in the proportion of sponge cells and the aggregation of bacteria (see Fig. S7 in the supplemental material). Samples N2 and N4 showed intact sponge cells and dispersed bacteria, while samples M4 and H2 demonstrated intermediate sponge cell lysis and bacterial aggregation. In H4, sponge cells were almost absent and the bacteria had aggregated into clusters. Similar results for changes in bacterial morphotype and sponge cell lysis have also been reported in a previous copper treatment study of the sponge Rhopaloeides odorabile (20).
FIG 5 .
Microscopic observation of bacteria and sponge cells in control (N4, a to c) and treated (H4, d to f) samples following DAPI staining. Sponge cells (indicated by the arrow in panel b) and bacteria were stained blue. Bacteria in the treated samples aggregated (indicated by the arrow in panel d) and were distributed in the matrix (i). Novel bacterial morphotypes (vibrios) appeared in the bacterial aggregates (i). Panels g and h show bright-field images corresponding to those in panels d and e, respectively.
Enrichment of the bacteriophage community by copper treatment.
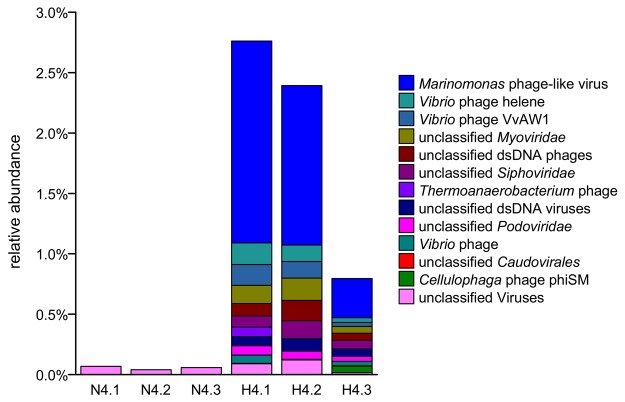
Interestingly, the relative abundance of bacteriophages among the total prokaryotes was highly enhanced from 0.06 to 1.98% in response to copper treatment (Fig. 4; t test, P < 0.05). Previous studies have also shown that heavy-metal pollutants induce lytic bacteriophages in marine bacteria (21, 22). The remarkable enrichment of phages caused by copper treatment suggested the multiplication of phages and lysis of their hosts.
FIG 4 .
Relative abundance and composition of the bacteriophage community in high-dose-treated sponge (H4) and control (N4) samples based on metagenomic data.
Further analyses of phages revealed that the most dominant species was a Marinomonas phage-like virus that accounted for up to 55.7% of the phages detected (Fig. 4). Because of the relatively high sequencing coverage of the genome of the Marinomonas phage-like virus, we were able to determine the whole genome sequence (34,074 bp, circular) with a sequencing coverage of 273× in the metagenomic data. A genomic comparison (see Fig. S8 in the supplemental material) revealed a colinear arrangement of genes with the closest relative Marinomonas bacteriophage P12026, which can lytically infect the free-living marine bacterium Marinomonas sp. strain IMCC12026 (23). However, many genes did not provide significant BLAST results (E value cutoff, 1e-5) in the NR database, suggesting that the phage was sponge specific. The Pfam database annotated only 18 out of 53 proteins from the phage (Table 1), including fundamental phage components (phage tail, portal, phage head-tail joining, capsid, and adhesion protein), replication-related enzymes (terminase, protease, Rec family, and nuclease), and other phage proteins. The low similarity (average nucleotide identity of 65.9%) to Marinomonas bacteriophage P12026 suggested that the host specificity of the phage in the sponge may be to a host other than Marinomonas. The alignment of short reads to the genome of the phage showed that the genome is circular (data not shown), and the circular form of the phage genome together with the greatly increased phage abundance suggested a lytic outbreak triggered by the high concentration of copper.
TABLE 1 .
Functions of the 18 genes present in the Marinomonas phage in the sponge annotated by the Pfam database
| Pfam ID | Gene product |
|---|---|
| PF06199.6 | Phage major tail protein 2 |
| PF04883.7 | HK97-gp10, putative tail component |
| PF04860.7 | Phage portal protein |
| PF03592.11 | Terminase small subunit |
| PF01510.20 | N-Acetylmuramoyl-l-alanine amidase |
| PF05521.6 | Phage head-tail joining protein |
| PF01145.20 | SPFH domain/band 7 family |
| PF11836.3 | Unknown function (DUF3356) |
| PF04586.12 | Caudovirus prohead protease |
| PF03837.9 | RecT family |
| PF05065.8 | Phage capsid family |
| PF04233.9 | Phage Mu protein F-like protein |
| PF05268.6 | Phage tail fiber adhesin Gp38 |
| PF12728.2 | Helix-turn-helix domain |
| PF12705.2 | PD-(D/E)XK nuclease superfamily |
| PF14549.1 | DNA-binding transcriptional regulator Cro |
| PF04466.8 | Phage terminase large subunit |
| PF11367.3 | Unknown function (DUF3168) |
DISCUSSION
In the present study, we investigated the shift of the sponge-associated bacterial community in response to copper treatment and the mechanisms underlying the functional selection of the treatment of the bacterial community. The results revealed that the dominant species of the bacterial community harbored by the sponge belonged to the symbiotic sulfur-oxidizing Ectothiorhodospiraceae and photosynthetic Cyanobacteria. However, the dominant symbiotic bacteria decreased substantially in response to the high concentration of copper, which enriched for a heterotrophic-bacterium-dominated community. A metagenomic comparison revealed the functional selective effect of the treatment on the enriched bacterial community. Additionally, the bacteriophage community was also enriched, suggesting that a lytic cycle was stimulated by copper.
Potential roles of autotrophic bacteria in the sponge.
In the present study, the natural sponge was dominated by an unclassified member of the family Ectothiorhodospiraceae and an unclassified cyanobacterium, which are sulfur-oxidizing and photosynthetic bacteria, respectively. Ectothiorhodospiraceae is a family that includes numerous anoxygenic photosynthetic SOB and nonphotosynthetic SOB. Previous studies have shown that symbiotic SOB in the sponge can oxidize sulfide (produced by anaerobic sulfur-reducing bacteria [SRB]) in the sponge body as an energy source and detoxify sulfide, which is toxic to sponges (9, 14, 24). Sponge-specific Ectothiorhodospiraceae also have been shown to be dominant in the sponge Axinella corrugata, accounting for more than 34.5% of the entire microbial community (25). In the present study, the unclassified member of the family Ectothiorhodospiraceae (accounting for 54.6% of the bacterial community) was phylogenetically sponge specific and was not detected in the seawater samples. Our analyses of the draft genome of the unclassified Ectothiorhodospiraceae revealed a complete sulfur oxidation pathway (reverse dissimilatory sulfate reduction) and Calvin cycle for CO2 fixation (26). The symbiotic role of the sponge-specific Ectothiorhodospiraceae could be fixation of CO2 and detoxification of sulfide generated by anaerobic SRB in the sponge body. The dominance of bacteria in the sponge, sulfide oxidation, and symbiotic features, including the streamlining of virulence-associated genes and enrichment for symbiosis-related genes (26), suggested an important role for Ectothiorhodospiraceae in the sponge. Symbiotic Cyanobacteria have been found in many sponges that perform photosynthesis (27–29). They are known to play beneficial roles, providing carbohydrates and oxygen for the host via photosynthesis. The roles of the Cyanobacteria herein could be fixation of CO2 and provision of carbohydrates to the host.
Unique functional selective effect of copper treatment on enriched bacteria in the sponge.
Copper may damage cells beyond their physiological limits, inducing the release of reactive oxygen species (30, 31) and destabilization of the iron-sulfur cluster and thiol group proteins. Previous studies examining individual cultured bacteria have revealed mechanisms responsible for resistance to high copper concentrations and for maintenance of cellular homeostasis. The mechanism of resistance is conserved in most Gram-negative bacteria and even in Gram-positive bacteria (30). Escherichia coli contains a P-type ATPase that efficiently pumps Cu (32). The efflux of periplasmic Cu is due to huge multicomponent protein complexes such as CusCBA (33). In Salmonella, the Cus system is replaced by another periplasmic Cu defense protein termed CueP (34). Eleven metal-translocating P-type ATPases of class IB have been described in Mycobacterium tuberculosis, and only CtpV was directly associated with Cu homeostasis (35). These mechanisms are utilized by individual bacteria for resistance to a high concentration of copper ions. However, in the present study, no copper efflux pump genes were significantly enriched in the bacterial community by copper treatment.
The enriched bacteria might employ a quick and efficient mechanism for protection against copper in the unique environment of the sponge body. The functional genes that were enriched by copper treatment, including those that play a role in bacterial motility and chemotaxis, bacterial capsule synthesis, virulence, and bacterial signaling and regulation, reflected the functions required by the enriched bacteria for surviving the stress induced by copper treatment in the sponge. Functional genes responsible for bacterial motility and chemotaxis (including the flagellum and chemotactic proteins) together with signaling and regulation (a two-component regulatory system, orphan regulatory proteins, and cAMP signaling) might facilitate the transport of bacteria to areas (such as bacterial aggregates) with a low copper concentration in the matrix of the sponge body. The bacterial capsule and extracellular polysaccharides might act as a barrier, preventing copper from entering bacterial cells. Morphological observation of the bacterial aggregates in the treated sponge (see Fig. S7 in the supplemental material) demonstrated a directional migration of the enriched bacteria to the lysed sponge cells (which may require the functions of bacterial motility, chemotaxis, signal regulation, and polysaccharide synthesis, which were enriched in the treated sponges according to the metagenomic analysis) and suggested the bacterial strategy of forming aggregates to escape the stress of the copper treatment. The enrichment for functions of directional bacterial movement and supplementary cellular components, together with the observation of bacterial aggregation, are novel in sponge studies. Additionally, functional genes responsible for bacterial virulence (type IV pilus, Ton and Tol transport system, invasion and intracellular resistance, and adhesion genes, among others) might facilitate the adherence and invasion of host cells by potentially pathogenic bacteria, as suggested by the remarkably reduced numbers of sponge cells in the treatment (postulated to be caused by the attack of pathogenic bacteria). The enriched bacteria were not detected in the reference seawater samples. Thus, copper treatment might have enriched for rare species in the sponge rather than causing invasion of the sponge by seawater bacteria.
The change in sponge health status also may have affected the bacterial community composition and functional gene composition. As shown in Fig. 5 (see also Fig. S7 in the supplemental material), the sponge cell number was reduced by the treatment (time and concentration related), suggesting damage to the health of the sponge (although no obvious necrosis was observed). The dynamics of the reduction in sponge cell numbers (see Fig. S7 in the supplemental material) indicated a loss of control of the sponge cells (potential copper intoxication or virus invasion, followed by consumption by the enriched bacteria) over the bacterial community. The normal interaction between the bacterial community and sponge host may have been disrupted, and exposure to copper may have been the major factor shaping the bacterial community.
The lack of related functions and the subsequent sharp decrease in Ectothiorhodospiraceae and Cyanobacteria confirmed the functional selection induced by copper treatment. Genomic analysis of the unclassified Ectothiorhodospiraceae by the binning method revealed a substantial lack of genes encoding the flagellum, chemotactic protein, and virulence-associated functions compared with its closest free-living relative, Thioalkalivibrio (26). Although the genome of the unclassified cyanobacterium was not determined, it is well known that most cyanobacteria lack a flagellum and are unable to move freely. The weakness in core functions selected by copper treatment may contribute to the failure of the symbiotic bacteria to survive following exposure to copper.
In conclusion, copper treatment could shift the bacterial community of the sponge, decreasing the number of symbiotic autotrophs and enriching for heterotrophs. The treatment selected for the following functions: bacterial motility, chemotaxis, capsule and polysaccharide synthesis, signal transduction and regulation, and virulence, which would facilitate the survival of bacteria under copper stress. The damage to sponge cells and shift of the symbiotic bacterial community reflected the harmful effects of copper on the symbiont system.
MATERIALS AND METHODS
Sponge collection and treatments.
The sponge H. cymaeformis (see Fig. S9a in the supplemental material) was collected from shallow water of a bay in Hong Kong (22°16′32.96″N, 114°17′39.95″E). Three colonies similar in size were transported to the laboratory and maintained in a water tank with running seawater for 1 day prior to treatment. Aquariums (2 liters) N, L, M, and H in triplicate were filled with fresh, sand-filtered seawater (1.5 liters) with corresponding concentrations of CuSO4 for the negative-control and low (10 µg/liter)-, medium (100 µg/liter)-, and high (1 mg/liter)-dose treatments, respectively (in total, 12 glass aquariums were presaturated with the corresponding concentrations of CuSO4). Because the sponge colonies consisted of relatively independent gemmules resulting from asexual reproduction, the bacterial communities in different gemmules were highly similar (as indicated by 16S rRNA gene pyrosequencing [data not shown]). Each of the three sponge colonies was separated into small colonies, placed in glass aquariums (see Fig. S9b in the supplemental material), and provided with aeration and light. At the 0-, 2-, and 4-day time points, colonies were sampled in triplicate (biological replicates from the three colonies), of which ~2 cm3 of tissue was stored in DNA extraction buffer (500 mM NaCl, 50 mM Tris-HCl [pH 8], 40 mM EDTA, 0.75 M sucrose) at −20°C and ~1 cm3 of the tissue was fixed in 4% formaldehyde overnight. Seawater samples collected from the same place as the sponges were used as references (1 liter of water from each sample was filtered through a 0.22-µm filter and then stored in DNA extraction buffer).
DNA extraction and metagenomic sequencing.
After homogenization with a sterilized mortar and pestle to release the microorganisms, the samples were centrifuged at 100 × g for 1 min to pellet the sponge debris. The supernatant was then filtered through a 12-µm polycarbonate membrane (GE Water & Process Technologies) to remove eukaryotic cells. The filtrate was observed by safranin staining (Bacto Laboratories Pty. Ltd.) at a magnification of ×100 under an Olympus BX51 microscope. Most of the microbial cells were ~1 µm in size, which indicated effective removal of the eukaryotic cells. Filtrates were then collected by centrifugation at 10,000 × g for 5 min and stored in 800 µl of DNA extraction buffer. DNA was extracted as previously described (36). Briefly, 10 µl of lysozyme (100 mg/ml) was used to lyse the microbial cell wall, and then 80 µl of 20% SDS and 8 µl of proteinase K (10 µg/µl) were used to digest the protein. The DNA was extracted with a mixture of chloroform-isoamyl alcohol (24:1) and then precipitated with an equal volume of 100% isopropanol. The precipitated DNA was then washed with cold 75% ethanol. The quantity of the extracted DNA was evaluated with a NanoDrop ND-1000 device (Thermo Fisher) and agarose gel electrophoresis.
Metagenomic DNA from sponges treated with a large dose of copper and the corresponding control were sequenced in triplicate with an Illumina Hiseq2000 platform (500-bp library) according to the manufacturer’s instructions.
16S rRNA gene amplification and sequencing.
Primers 341F (5′ CCT ACG GGA GGC AGC AG 3′) and 907R (5′ CCG TCA ATT CCT TTR AGT TT 3′), targeting the hypervariable V3-to-V5 region of the 16S rRNA gene, were used for PCR amplification. Unique 8-nucleotide bar codes were added to the 5′ ends of the primers for multiplexed pyrosequencing by BARCRAWL (37). For the PCR, each 20-µl reaction mixture consisted of 4 µl of 5 × Phusion HF buffer (M0530S; New England BioLabs Inc.), 1.6 µl of deoxynucleoside triphosphates (each at 2.5 µM), 1 µl of each primer (10 µM), 0.6 µl of dimethyl sulfoxide, 10 ng of template DNA, 0.2 µl of Phusion High-Fidelity DNA polymerase (0.4 U), and 10.6 µl of pure water. A thermal cycler (Bio-Rad, USA) was used according to the following program: initial denaturation at 98°C for 1 min; 25 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 20 s; and a final extension at 72°C for 5 min. Amplicons were mixed in equal amounts and pyrosequenced with a ROCHE 454 FLX Titanium platform.
Taxonomy and diversity of the 16S rRNA gene sequencing data.
The Quantitative Insights Into Microbial Ecology pipeline (38) was used for analysis of the pyrosequencing data. The following quality controls for the sequences were implemented: removal of sequences with a quality score of <25, homopolymers with a length of >6 bp, a length of <100 or >1,000 bp, >6 ambiguous bases (qualified sequences included 0.0094 ambiguous bases per read), >3 mismatches in the primer, and 1 mismatch in the bar codes. High-quality sequences were assigned to their corresponding samples according to the bar codes. OTUs with 3% dissimilarity were selected and assigned with the Ribosomal Database Project Classifier (version 2.2) (39) by using the Silva111 database (40) at a confidence level of 80%. Representative sequences of each OTU were aligned by using PyNAST (41). Chimeric sequences were identified by ChimeraSlayer (42) and removed. Similarities among the sample bacterial communities were determined by using UniFrac PCoA. An unweighted UniFrac jackknife clustering was constructed to display the relatedness of the bacterial communities. Phylogenetic trees of potentially symbiotic bacteria (such as the unassigned Ectothiorhodospiraceae) and other sponge-specific bacteria were constructed with Mega (version 6.05; maximum-likelihood tree constructed with the General Time Reversible model and 500 bootstrap replications) (43) by using the partial 16S rRNA genes obtained in this study and reference sequences from NCBI (BLAST hits from the NT database with an identity of >95%, including mainly unculturable bacteria, and the 16S rRNA sequence database with an identity of >85% including identified species). Statistical analysis of differences in relative abundances (expressed as a percentage) of taxa in different samples was performed by using the t test with a P value threshold of 0.05.
Metagenomic sequencing and analysis.
Quality control was performed on the raw Illumina pair-end reads (2 × 150 bp) by using the NGS QC Toolkit (version 2.3) (44). Sequences with an average quality score of <20 were removed. The first 10 bases of each read (abnormal bases indicated by nucleotide composition) and homopolymers (>6 bases), together with the subsequent three end regions, were trimmed. High-quality reads of all six samples were merged for assembly by using the CLC Genomics Workbench (version 6.5; CLC bio, Boston, MA) with parameters of automatic word size, automatic bubble size, a minimum contig length of 200 bp, an insert size of 450 to 550 bp (established during import), and map reads back to the contigs (a mismatch cost of 2, a insertion cost of 3, a deletion cost of 3, a length fraction of 0.5, and a similarity fraction of 0.98 with selection of the update contigs option).
Contigs with a length >500 bp were used for gene prediction analysis by Prodigal (45) by the meta method. Translated amino acids of each gene were used for BLASTp analysis against the COG database, version 9.1 (http://string-db.org/). The best hits with a threshold E value of 1e-5, a cutoff score of 100, and an alignment length covering >30% of the query were selected for functional annotation of the proteins. Prokaryotic protein sequences were extracted and used for BLASTp analysis against the NR database with an E value cutoff of 1e-5 and a maximum hit number of 20. In contrast, qualified short reads from the six samples were aligned to the above assembled contigs (>500 bp) by using Bowtie2 (46), and the sequencing coverage of the contigs in each sample was calculated by using SAMtools (47). A Perl script was used to integrate information regarding the contig coverage (hence, the coverage of protein sequences) in the BLASTp outputs for the six samples, which were then imported in Megan 5 (48) with the lowest common ancestor (LCA) parameter of minimum support (49), minimum score (49), maximum expect (0.01), and top percent (10) applied. Functional classification system SEED hierarchy was applied for the functional comparison of the six samples. The number of reads from the six samples was normalized to the smallest number of reads in sample C2 (control sample 2), and the relative abundance (as a percentage) of functions was calculated. A t test was conducted to compare the treated and control sponges by using a P value threshold of 0.05.
Gene annotation and comparison of the Marinomonas phage-like virus genome.
The contig of the bacteriophage (annotated as the Marinomonas phage-like virus) was extracted, and genes were predicted with Prodigal (50) by the single-genome method. Translated protein sequences were used to perform the HMM search against the Pfam database (49) by using hmmsearch 3.0 with the trusted cutoff for each protein family. The contig was used to conduct the BLASTn search against the reference genome Marinomonas bacteriophage P12026 (AFM54896) (23) with an E value of 1e-5. The output was used as the input for the Artemis comparison tool (51) for the genomic comparison.
Microscopic observation of sponge sections.
Sponge tissues were fixed in 4% formaldehyde overnight in a freezer (4°C) and stored in phosphate-buffered saline and ethanol (1:1) in the freezer (−80°C) until further processing. Fixed samples were dehydrated in 75, 80, 95, and 100% ethanol, followed by xylene (three times) in sequence. Melted paraffin was used to remove the xylene and embed the tissues. A microtome (Leica, Germany) was used to cut sections of 5 µm, which were then mounted on 0.01% poly-l-lysine-coated slides. The sections were deparaffinized twice in xylene, and then serial concentrations of ethanol (95, 80, and 75%) were used to rehydrate the sections. 4′,6-Diamidino-2-phenylindole (DAPI; 5 ng/µl) was added for 10 min of incubation to stain the cells in the sections, which were then washed with distilled water. The sections were observed under an Olympus BX51 fluorescence microscope.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequencing and metagenomic sequencing data obtained in this study were deposited in the NCBI Sequence Read Archive under accession numbers SRP043404 and SRP038115, respectively.
SUPPLEMENTAL MATERIAL
Rarefaction curve of the 16S rRNA gene reads from each sample showing the diversity of the bacterial community in sponge and seawater samples. The x axis represents the number of rarified reads, and the y axis represents the diversity indicated by the Shannon index Download
PCoA of the bacterial communities of copper-treated sponge, control, and seawater samples based on the distribution of the 16S rRNA genes. Cluster 1 refers to samples of N0, N2, N4, L2, L4, M2, and M4. Cluster 2 refers to samples of H2, and cluster 3 refers to samples of H4. Cluster 4 denotes the seawater samples. Download
Bacterial community composition of sponge samples treated with copper, controls, and seawater samples at the phylum level. The y axis shows the relative abundance of the taxa, and the x axis shows the samples. The letters N, L, M, and H refer to sponge samples from the negative-control and low-, medium-, and high-dose treatments, respectively. The numbers 0, 2, and 4 denote 0, 2, and 4 days of treatment, and the numbers after the dots are replicate numbers. W1 and W2 are two replicates of the seawater sample. Download
Phylogenetic tree including the Ectothiorhodospiraceae bacteria and their closest relatives. The bacterium (●) is located in a sponge-specific cluster and is phylogenetically distant from the free-living relatives. A bootstrap confidence of >70 is indicated. Download
Compositions of functional genes in the bacterial communities in sponge samples treated with a high concentration of copper (1 mg/liter) for 4 days (H4) and the negative controls (N4). Download
Comparison of the relative abundances of functional genes involved in bacterial motility and chemotaxis (a), bacterial cell wall and capsule synthesis (b), regulation and cell signaling (c), and virulence (d) in negative-control (N4) and high-dose-treated (H4) samples. The x axis shows the normalized relative abundances of genes (as percentages), and the y axis shows the functional categories at level 3 of the SEED classification system. Download
Comparison of microscopic images of the 2- and 4-day samples of medium- and high-dose treatments and corresponding controls showing the dynamics of damage, the decrease in sponge cell numbers, and the aggregation of bacteria. Each column includes two pictures with different scale bars. The negative controls N2 and N4 show the initial status, and M4 and H2 show the intermediate stages of sponge cell lysis (arrows indicate residual sponge cells) and bacterial aggregation. The H4 shows completely lysed sponge cells and bacterial aggregates (with novel bacterial morphotypes). Download
Comparison of the whole genome of the Marinomonas phage-like virus in the sponge and the Marinomonas phage P12026 (reference number AFM54896) reference genome. BLAST alignments (with an E value of <1e-5) are indicated by red (plus/plus) and blue (plus/minus) bars. Download
The sponge, H. cymaeformis (Demospongiae, Haplosclerida), used in this study and the treatment process. The sponge (a) consists of relatively independent gemmules (each with one osculum) resulting from asexual reproduction. The schematic (b) shows the treatments at different concentrations (low, 10 µg/liter; medium, 100 µg/liter; high, 1 mg/liter) and sampling at different time points (0, 2, and 4 days). Download
Sample information including the number of sequences, the total number of OTUs, and the diversity indicated by the Shannon index with a normalized number of sequences of 200.
ACKNOWLEDGMENTS
This research was supported by the Nature Science Foundation of China (U1301232), the National Basic Research Program of China (973 Program, 2012CB417304), the State Key Laboratory of Marine Pollution, the Seed Collaborative Research Fund (CITYU12SC01), and an award (SA-C0040/UK-C0016) from the King Abdullah University of Science and Technology to P. Y. Qian.
Footnotes
Citation Tian R, Wang Y, Bougouffa S, Gao Z, Cai L, Zhang W, Bajic V, Qian P. 2014. Effect of copper treatment on the composition and function of the bacterial community in the sponge Haliclona cymaeformis. mBio 5(6):e01980-14. doi:10.1128/mBio.01980-14.
REFERENCES
- 1. Peña MM, Lee J, Thiele DJ. 1999. A delicate balance: homeostatic control of copper uptake and distribution. J. Nutr. 129:1251–1260. [DOI] [PubMed] [Google Scholar]
- 2. Bruins MR, Kapil S, Oehme FW. 2000. Microbial resistance to metals in the environment. Ecotoxicol. Environ. Saf. 45:198–207. 10.1006/eesa.1999.1860. [DOI] [PubMed] [Google Scholar]
- 3. Claisse D, Alzieu C. 1993. Copper contamination as a result of antifouling paint regulations. Mar. Pollut. Bull. 26:395–397. 10.1016/0025-326X(93)90188-P. [DOI] [Google Scholar]
- 4. Dayton PK. 1989. Interdecadal variation in an Antarctic sponge and its predators from oceanographic climate shifts. Science 245:1484–1486. 10.1126/science.245.4925.1484. [DOI] [PubMed] [Google Scholar]
- 5. Maldonado M, Carmona C, Velasquez Z, Puig A, Cruzado A, Lopez A, Young CM. 2005. Siliceous sponges as a silicon sink: an overlooked aspect of benthopelagic coupling in the marine silicon cycle. Limnol. Oceanogr. 50:799–809. 10.4319/lo.2005.50.3.0799. [DOI] [Google Scholar]
- 6. Lee YK, Lee JH, Lee HK. 2001. Microbial symbiosis in marine sponges. J. Microbiol. 39:254–264. [Google Scholar]
- 7. Simister RL, Deines P, Botté ES, Webster NS, Taylor MW. 2012. Sponge-specific clusters revisited: a comprehensive phylogeny of sponge-associated microorganisms. Environ. Microbiol. 14:517–524. 10.1111/j.1462-2920.2011.02664.x. [DOI] [PubMed] [Google Scholar]
- 8. Wilkinson CR. 1978. Microbial associations in sponges. I. Ecology, physiology and microbial populations of coral reef sponges. Mar. Biol. 49:161–167. [Google Scholar]
- 9. Taylor MW, Radax R, Steger D, Wagner M. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295–347. 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilkinson CR. 1983. Net primary productivity in coral-reef sponges. Science 219:410–412. 10.1126/science.219.4583.410. [DOI] [PubMed] [Google Scholar]
- 11. Thomas T, Rusch D, DeMaere MZ, Yung PY, Lewis M, Halpern A, Heidelberg KB, Egan S, Steinberg PD, Kjelleberg S. 2010. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J 4:1557–1567. 10.1038/ismej.2010.74. [DOI] [PubMed] [Google Scholar]
- 12. Anand TP, Bhat AW, Shouche YS, Roy U, Siddharth J, Sarma SP. 2006. Antimicrobial activity of marine bacteria associated with sponges from the waters off the coast of South East India. Microbiol. Res. 161:252–262. 10.1016/j.micres.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 13. Preston CM, Wu KY, Molinski TF, DeLong EF. 1996. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen nov, sp, nov. Proc. Natl. Acad. Sci. U. S. A. 93:6241–6246. 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Imhoff JF, Trüper HG. 1976. Marine sponges as habitats of anaerobic phototrophic bacteria. Microb. Ecol. 3:1–9. 10.1007/BF02011449. [DOI] [PubMed] [Google Scholar]
- 15. Vogel S. 1977. Current-induced flow through living sponges in nature. Proc. Natl. Acad. Sci. U. S. A. 74:2069–2071. 10.1073/pnas.74.5.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hansen IV, Weeks JM, Depledge MH. 1995. Accumulation of copper, zinc, cadmium and chromium by the marine sponge Halichondria-Panicea Pallas and the implications for biomonitoring. Mar. Pollut. Bull. 31:133–138. 10.1016/0025-326X(94)00228-2. [DOI] [Google Scholar]
- 17. Cebrian E, Martí R, Uriz JM, Turon X. 2003. Sublethal effects of contamination on the Mediterranean sponge Crambe crambe: metal accumulation and biological responses. Mar. Pollut. Bull. 46:1273–1284. 10.1016/S0025-326X(03)00190-5. [DOI] [PubMed] [Google Scholar]
- 18. Cebrian E, Uriz MJ. 2007. Contrasting effects of heavy metals on sponge cell behavior. Arch. Environ. Contam. Toxicol. 53:552–558. 10.1007/s00244-006-0257-2. [DOI] [PubMed] [Google Scholar]
- 19. Selvin J, Priya SS, Kiran GS, Thangavelu T, Bai NS. 2009. Sponge-associated marine bacteria as indicators of heavy metal pollution. Microbiol. Res. 164:352–363. 10.1016/j.micres.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 20. Webster NS, Webb RI, Ridd MJ, Hill RT, Negri AP. 2001. The effects of copper on the microbial community of a coral reef sponge. Environ. Microbiol. 3:19–31. 10.1046/j.1462-2920.2001.00155.x. [DOI] [PubMed] [Google Scholar]
- 21. Lee LH, Lui D, Platner PJ, Hsu SF, Chu TC, Gaynor JJ, Vega QC, Lustigman BK. 2006. Induction of temperate cyanophage AS-1 by heavy-metal-copper. BMC Microbiol. 6:17. 10.1186/1471-2180-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sode K, Oonari R, Oozeki M. 1997. Induction of a temperate marine cyanophage by heavy metal. J. Mar. Biotechnol. 5:178–180. [Google Scholar]
- 23. Kang I, Jang H, Oh HM, Cho JC. 2012. Complete genome sequence of Marinomonas bacteriophage P12026. J. Virol. 86:8909–8910. 10.1128/JVI.01328-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoffmann F, Larsen O, Thiel V, Rapp HT, Pape T, Michaelis W, Reitner J. 2005. An anaerobic world in sponges. Geomicrobiol. J. 22:1–10. 10.1080/01490450590922505. [DOI] [Google Scholar]
- 25. White JR, Patel J, Ottesen A, Arce G, Blackwelder P, Lopez JV. 2012. Pyrosequencing of bacterial symbionts within Axinella corrugata sponges: diversity and seasonal variability. PLOS ONE 7:e38204. 10.1371/journal.pone.0038204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tian RM, Wang Y, Bougouffa S, Gao ZM, Cai L, Bajic V, Qian PY. 2014. Genomic analysis reveals versatile heterotrophic capacity of a potentially symbiotic sulfur-oxidizing bacterium in sponge. Environ. Microbiol. 10.1111/1462-2920.12586. [DOI] [PubMed] [Google Scholar]
- 27. Arillo A, Bavestrello G, Burlando B, Sara M. 1993. Metabolic integration between symbiotic Cyanobacteria and sponges—a possible mechanism. Mar. Biol. 117:159–162. 10.1007/BF00346438. [DOI] [Google Scholar]
- 28. Cheshire AC, Wilkinson CR. 1991. Modelling the photosynthetic production by sponges on Davies Reef, Great Barrier Reef. Mar. Biol. 109:13–18. 10.1007/BF01320226. [DOI] [Google Scholar]
- 29. Wilkinson CR. 1987. Interocean differences in size and nutrition of coral-reef sponge populations. Science 236:1654–1657. 10.1126/science.236.4809.1654. [DOI] [PubMed] [Google Scholar]
- 30. Orell A, Navarro CA, Arancibia R, Mobarec JC, Jerez CA. 2010. Life in blue: copper resistance mechanisms of bacteria and archaea used in industrial biomining of minerals. Biotechnol. Adv. 28:839–848. 10.1016/j.biotechadv.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 31. Dupont CL, Grass G, Rensing C. 2011. Copper toxicity and the origin of bacterial resistance—new insights and applications. Metallomics 3:1109–1118. 10.1039/c1mt00107h. [DOI] [PubMed] [Google Scholar]
- 32. Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. 2000. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U. S. A. 97:652–656. 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Outten FW, Huffman DL, Hale JA, O’Halloran TV. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670–30677. 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 34. Osman D, Waldron KJ, Denton H, Taylor CM, Grant AJ, Mastroeni P, Robinson NJ, Cavet JS. 2010. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J. Biol. Chem. 285:25259–25268. 10.1074/jbc.M110.145953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. 2010. CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol. Microbiol. 77:1096–1110. 10.1111/j.1365-2958.2010.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee OO, Wong YH, Qian PY. 2009. Inter- and intraspecific variations of bacterial communities associated with marine sponges from San Juan Island, Washington. Appl. Environ. Microbiol. 75:3513–3521. 10.1128/AEM.00002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frank DN. 2009. BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinformatics 10:362. 10.1186/1471-2105-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Petrosino JF, Knight R, Birren BW, Human Microbiome Consortium 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504. 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30:2725–2729. 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLOS One 7:e30619. 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. 2012. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 28:2223–2230. 10.1093/bioinformatics/bts429. [DOI] [PubMed] [Google Scholar]
- 46. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21:1552–1560. 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer ELL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301. 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rarefaction curve of the 16S rRNA gene reads from each sample showing the diversity of the bacterial community in sponge and seawater samples. The x axis represents the number of rarified reads, and the y axis represents the diversity indicated by the Shannon index Download
PCoA of the bacterial communities of copper-treated sponge, control, and seawater samples based on the distribution of the 16S rRNA genes. Cluster 1 refers to samples of N0, N2, N4, L2, L4, M2, and M4. Cluster 2 refers to samples of H2, and cluster 3 refers to samples of H4. Cluster 4 denotes the seawater samples. Download
Bacterial community composition of sponge samples treated with copper, controls, and seawater samples at the phylum level. The y axis shows the relative abundance of the taxa, and the x axis shows the samples. The letters N, L, M, and H refer to sponge samples from the negative-control and low-, medium-, and high-dose treatments, respectively. The numbers 0, 2, and 4 denote 0, 2, and 4 days of treatment, and the numbers after the dots are replicate numbers. W1 and W2 are two replicates of the seawater sample. Download
Phylogenetic tree including the Ectothiorhodospiraceae bacteria and their closest relatives. The bacterium (●) is located in a sponge-specific cluster and is phylogenetically distant from the free-living relatives. A bootstrap confidence of >70 is indicated. Download
Compositions of functional genes in the bacterial communities in sponge samples treated with a high concentration of copper (1 mg/liter) for 4 days (H4) and the negative controls (N4). Download
Comparison of the relative abundances of functional genes involved in bacterial motility and chemotaxis (a), bacterial cell wall and capsule synthesis (b), regulation and cell signaling (c), and virulence (d) in negative-control (N4) and high-dose-treated (H4) samples. The x axis shows the normalized relative abundances of genes (as percentages), and the y axis shows the functional categories at level 3 of the SEED classification system. Download
Comparison of microscopic images of the 2- and 4-day samples of medium- and high-dose treatments and corresponding controls showing the dynamics of damage, the decrease in sponge cell numbers, and the aggregation of bacteria. Each column includes two pictures with different scale bars. The negative controls N2 and N4 show the initial status, and M4 and H2 show the intermediate stages of sponge cell lysis (arrows indicate residual sponge cells) and bacterial aggregation. The H4 shows completely lysed sponge cells and bacterial aggregates (with novel bacterial morphotypes). Download
Comparison of the whole genome of the Marinomonas phage-like virus in the sponge and the Marinomonas phage P12026 (reference number AFM54896) reference genome. BLAST alignments (with an E value of <1e-5) are indicated by red (plus/plus) and blue (plus/minus) bars. Download
The sponge, H. cymaeformis (Demospongiae, Haplosclerida), used in this study and the treatment process. The sponge (a) consists of relatively independent gemmules (each with one osculum) resulting from asexual reproduction. The schematic (b) shows the treatments at different concentrations (low, 10 µg/liter; medium, 100 µg/liter; high, 1 mg/liter) and sampling at different time points (0, 2, and 4 days). Download
Sample information including the number of sequences, the total number of OTUs, and the diversity indicated by the Shannon index with a normalized number of sequences of 200.