ABSTRACT
l-Cysteine is essential for virtually all living organisms, from bacteria to higher eukaryotes. Besides having a role in the synthesis of virtually all proteins and of taurine, cysteamine, glutathione, and other redox-regulating proteins, l-cysteine has important functions under anaerobic/microaerophilic conditions. In anaerobic or microaerophilic protozoan parasites, such as Entamoeba histolytica, l-cysteine has been implicated in growth, attachment, survival, and protection from oxidative stress. However, a specific role of this amino acid or related metabolic intermediates is not well understood. In this study, using stable-isotope-labeled l-cysteine and capillary electrophoresis-time of flight mass spectrometry, we investigated the metabolism of l-cysteine in E. histolytica. [U-13C3, 15N]l-cysteine was rapidly metabolized into three unknown metabolites, besides l-cystine and l-alanine. These metabolites were identified as thiazolidine-4-carboxylic acid (T4C), 2-methyl thiazolidine-4-carboxylic acid (MT4C), and 2-ethyl-thiazolidine-4-carboxylic acid (ET4C), the condensation products of l-cysteine with aldehydes. We demonstrated that these 2-(R)-thiazolidine-4-carboxylic acids serve for storage of l-cysteine. Liberation of l-cysteine occurred when T4C was incubated with amebic lysates, suggesting enzymatic degradation of these l-cysteine derivatives. Furthermore, T4C and MT4C significantly enhanced trophozoite growth and reduced intracellular reactive oxygen species (ROS) levels when it was added to cultures, suggesting that 2-(R)-thiazolidine-4-carboxylic acids are involved in the defense against oxidative stress.
IMPORTANCE
Amebiasis is a human parasitic disease caused by the protozoan parasite Entamoeba histolytica. In this parasite, l-cysteine is the principal low-molecular-weight thiol and is assumed to play a significant role in supplying the amino acid during trophozoite invasion, particularly when the parasites move from the anaerobic intestinal lumen to highly oxygenated tissues in the intestine and the liver. It is well known that E. histolytica needs a comparatively high concentration of l-cysteine for its axenic cultivation. However, the reason for and the metabolic fate of l-cysteine in this parasite are not well understood. Here, using a metabolomic and stable-isotope-labeled approach, we investigated the metabolic fate of this amino acid in these parasites. We found that l-cysteine inside the cell rapidly reacts with aldehydes to form 2-(R)-thiazolidine-4-carboxylic acid. We showed that these 2-(R)-thiazolidine-4-carboxylic derivatives serve as an l-cysteine source, promote growth, and protect cells against oxidative stress by scavenging aldehydes and reducing the ROS level. Our findings represent the first demonstration of 2-(R)-thiazolidine-4-carboxylic acids and their roles in protozoan parasites.
INTRODUCTION
In all living organisms from bacteria to higher eukaryotes, l-cysteine is implicated in a number of essential biochemical processes, including stability, structure, regulation of catalytic activity, and posttranslational modifications of various proteins (1). l-Cysteine is required for the synthesis of a variety of biomolecules, including methionine, glutathione, trypanothione, coenzyme A, hypotaurine, taurine, and cysteamine, as well as iron-sulfur (Fe-S) clusters, which are involved in electron transfer, redox regulation, nitrogen fixation, and sensing for regulatory processes (2, 3). The fact that reduced sulfur in l-cysteine (thiol, SH) is strongly nucleophilic makes it react easily with electrophilic compounds. However, the highly reactive thiol group also makes l-cysteine rather toxic to the cell (4, 5). Therefore, l-cysteine itself is maintained at relatively low levels, sufficient for protein synthesis and the production of essential metabolites but below the threshold of toxicity (5).
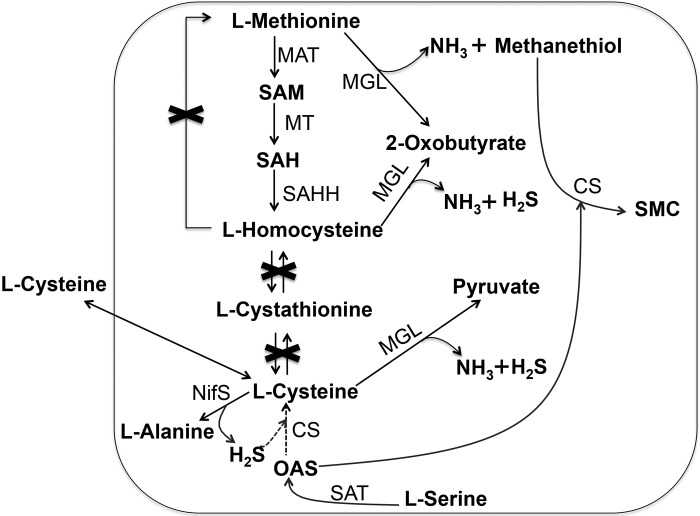
Entamoeba histolytica is an enteric protozoan parasite that causes colitis, dysentery, and extraintestinal abscesses in millions of inhabitants of areas of endemicity (6). This parasite is generally considered microaerophilic, because it consumes oxygen and tolerates low levels of oxygen pressure. However, the parasite lacks most of the components of antioxidant defense mechanisms, such as catalase, peroxidase, glutathione, and the glutathione-recycling enzymes glutathione peroxidase and glutathione reductase (7, 8). l-Cysteine is the principal low-molecular-weight thiol in E. histolytica trophozoites and is required for the survival, growth, attachment, elongation, motility, gene regulation, and antioxidative stress defense of this organism (9–12). There are a number of peculiarities in the metabolism of sulfur-containing amino acids in E. histolytica (Fig. 1). First, the organism lacks both forward and reverse transsulfuration pathways and thus is unable to interconvert l-methionine and l-cysteine (13). Second, it possesses methionine γ-lyase (MGL; EC 4.4.1.11), an enzyme that directly degrades l-methionine, l-homocysteine, and l-cysteine (14, 15). Third, E. histolytica possesses a pathway for de novo S-methylcysteine (SMC)/l-cysteine biosynthesis (16–18). Although de novo l-cysteine biosynthesis occurs in a wide range of bacteria and plants, l-cysteine production per se has not been demonstrated in E. histolytica trophozoites cultivated in vitro. Instead, the pathway is assumed to be involved primarily in the synthesis of SMC (18). Consistently with the notion that this pathway does not yield l-cysteine, amebic trophozoites require high concentrations of l-cysteine in culture for growth, which can be replaced by d-cysteine, l-cystine, or l-ascorbic acid, indicating that the extracellular cysteine/cystine, thiols, or reductants can play an interchangeable role (19). In most eukaryotes, where glutathione is the major thiol, l-cysteine is maintained at levels manyfold lower than those of glutathione (20). In contrast, E. histolytica, due to loss of glutathione metabolism, relies on l-cysteine as a major redox buffer (9, 13, 21). Therefore, the significance of l-cysteine and its metabolism in this organism remains a conundrum.
FIG 1 .
Scheme of transsulfuration, l-cysteine uptake, and sulfur-assimilatory de novo cysteine biosynthesis in E. histolytica. Abbreviations: CS, cysteine synthase (O-acetyl-l-serine sulfhydrylase, EC 2.5.1.47); MAT, methionine adenosyltransferase (S-adenosyl-l-methionine synthetase, EC 2.5.1.6); MGL, methionine γ-lyase (l-methioninase, EC 4.4.1.11); MT, various methyltransferases (EC 2.1.1.X); NifS, cysteine desulfurase (EC 2.8.1.7); OAS, O-acetylserine; SAH, S-adenosylhomocysteine; SAHH, adenosylhomocysteinase (S-adenosyl-l-homocysteine hydrolase, EC 3.3.1.1); SAM, S-adenosylmethionine; SAT, serine O-acetyltransferase (EC 2.3.1.30); SMC, S-methylcysteine.
The premise that extracellular or incorporated l-cysteine is important for cellular activities and homeostasis in E. histolytica prompted us to study the metabolic fate of extracellular l-cysteine in this parasite. Stable-isotope tracing is a powerful technique to investigate the metabolism of different carbon and nitrogen sources in microbial pathogens, such as Salmonella enterica serovar Typhimurium (22), Leishmania mexicana (23), Toxoplasma gondii (24), and Plasmodium falciparum (25). Stable-isotope labeling has provided vast improvements in both metabolite identification and pathway characterization (26). Isotopic enrichment in a wide range of intracellular and secreted metabolites can readily be measured using either mass spectrometry (MS) or nuclear magnetic resonance (NMR), providing quantitative information on metabolic networks (27–29). In this study, we have exploited this approach by using 13C3- and 15N1-labeled cysteine sources and capillary electrophoresis-time of flight MS (CE-TOFMS) to unveil the fate of l-cysteine metabolism in E. histolytica. Furthermore, we have demonstrated the physiological role of the identified l-cysteine derivatives.
RESULTS AND DISCUSSION
In vivo derivatization of stable-isotope-labeled l-cysteine by E. histolytica trophozoites.
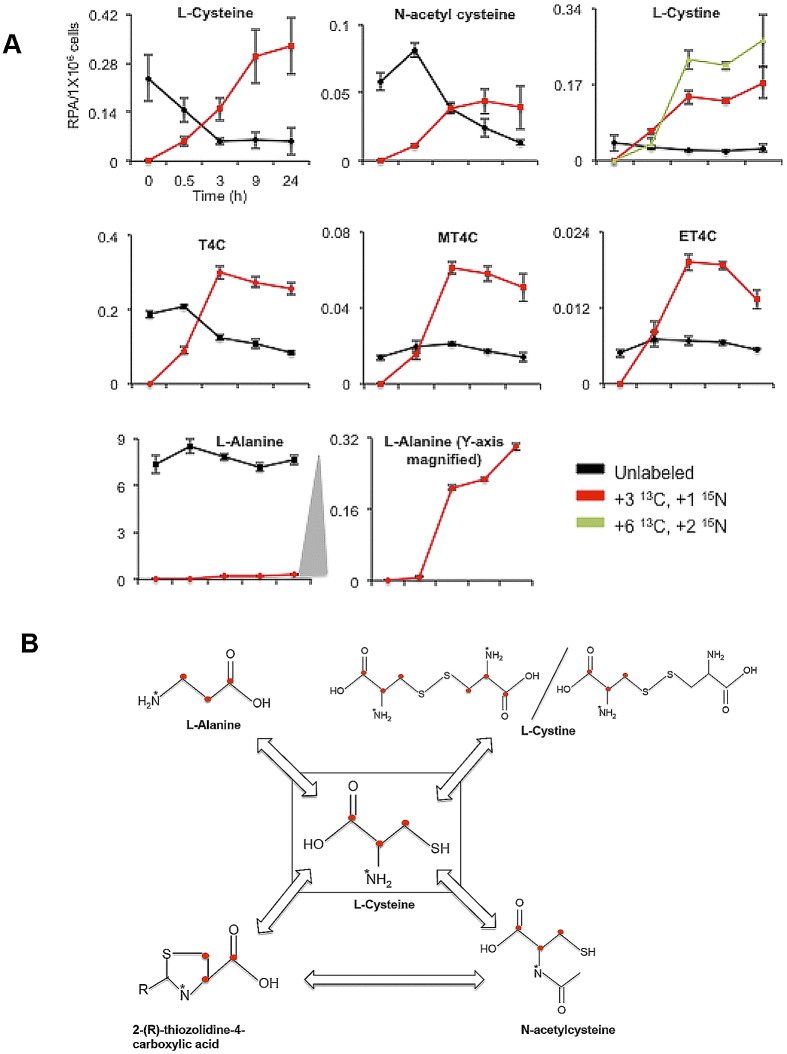
To investigate l-cysteine metabolism in E. histolytica, trophozoites were cultured in the presence of 8 mM stable-isotope (U-13C3, 15N1)-labeled l-cysteine in l-cysteine-deprived BI-S-33 medium and the turnover of intracellular metabolites was monitored by CE-TOFMS at 0.5, 3, 9, and 24 h (see Table S1 in the supplemental material). Upon the addition of [U-13C3, 15N]l-cysteine to the culture, the levels of [13C3, 15N]l-cysteine increased, and [13C3, 15N]l-cysteine replaced unlabeled l-cysteine after 3 to 9 h (Fig. 2A). l-Cysteine was metabolized into several metabolites. First, l-cysteine was derivatized into three structurally unknown metabolites (see below). Second, l-cysteine was oxidized to l-cystine. The concentration of both [13C3, 15N1]l-cystine and [13C6 15N2]l-cystine increased up to 24 h, whereas the unlabeled cystine remained constant. The slow and incomplete replacement of unlabeled l-cystine (and also l-cysteine) suggests the presence of an inaccessible pool of l-cysteine and l-cystine in the cell (Fig. 2A). Third, [13C3, 15N1]l-cysteine was metabolized into l-alanine in a reaction catalyzed by cysteine desulfurase activity, likely by NifS (30). A metabolic flow chart in Fig. 2B depicts incorporation of labels from [U-13C3, 15N1]l-cysteine into the detected metabolites in E. histolytica trophozoites.
FIG 2 .
l-Cysteine metabolism in E. histolytica. (A) Relative intracellular concentrations of various unlabeled and isotope-labeled l-cysteine-derived metabolites in E. histolytica trophozoites. Trophozoites were cultured in the presence of 8 mM stable-isotope-labeled l-cysteine (U-13C3, 15N) in l-cysteine-deprived medium for 0, 0.5, 3, 9, and 24 h. The bottom center plot is a magnified (at the y axis) plot of labeled l-alanine, shown at the bottom left. The x axis represents time in hours, whereas the y axis represents the relative peak areas (RPA) of signal detected with mass spectrometric analysis per 1 × 106 cells. Metabolite data are represented as means ± standard deviations (SD) of results from 3 biological replicates. (B) Metabolic flow chart illustrating l-cysteine metabolism in E. histolytica trophozoites. Red dots denote 13C atoms, whereas asterisks denote 15N atoms arising from [13C3, 15N1]l-cysteine.
Discovery of l-cysteine-derived T4Cs in E. histolytica trophozoites.
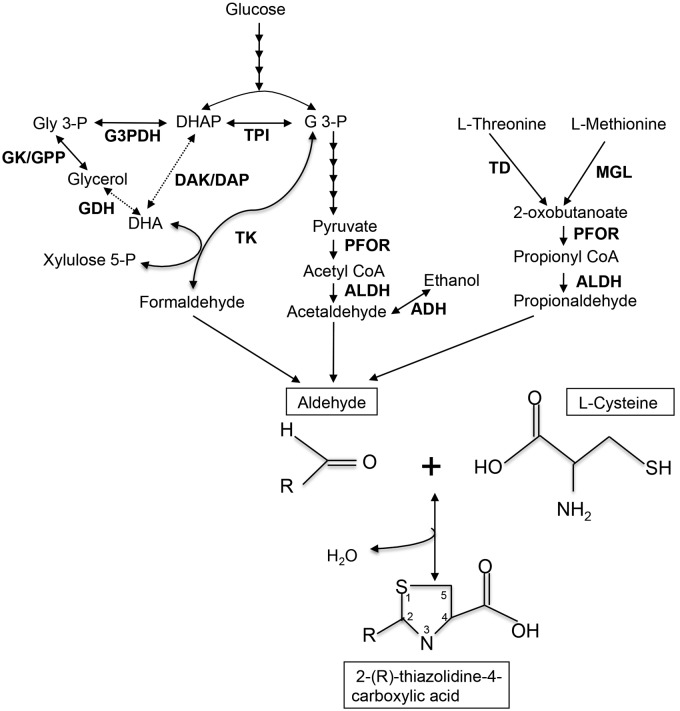
We detected three unknown labeled metabolites derived from l-cysteine. These metabolites had never been demonstrated in any protozoan parasites, including E. histolytica. Based on the accurate mass measurements, the elemental composition of the metabolites was calculated using the elemental composition calculator (analyst QS software). The elements C, N, O, H, P, and S were automatically considered. After chemical formulas were proposed (see Table S2 in the supplemental material), we searched through a number of databases for the possible compounds and structures, including PubChem (http://pubchem.ncbi.nlm.nih.gov/) and ChemSpider (http://www.chemspider.com/). Finally, their identities were confirmed by comparison to commercially available reference standards (Table S2). The three unknown metabolites were unequivocally identified as thiazolidine-4-carboxylic acid (T4C), 2-methyl thiazolidine-4-carboxylic acid (MT4C), and 2-ethylthiazolidine-4-carboxylic acid (ET4C). The changes in the profiles of these three labeled metabolites were similar; levels of these metabolites increased for up to 3 h and then slightly decreased, suggestive of further conversion or decomposition (Fig. 2A). These metabolites are most likely the condensation products of l-cysteine with aldehydes (Fig. 3). T4C is made of l-cysteine and formaldehyde (31). In Entamoeba, formaldehyde is likely produced by the action of transketolase (Fig. 3). In the E. histolytica genome database, we identified five possible transketolase genes (EHI_011410, EHI_002160, EHI_177870, EHI_157770, and EHI_082380). MT4C is the condensation product of l-cysteine with acetaldehyde. Acetaldehyde is a strongly electrophilic compound that is endogenously produced in ethanol metabolism by alcohol dehydrogenase (ADH) (32). Its high reactivity toward biogenic nucleophiles has toxicity as a consequence (33), and thus acetaldehyde needs to be immediately removed from the cell. In E. histolytica, acetaldehyde is produced from the fermentation of glucose to ethanol, with pyruvate, acetyl coenzyme A, and acetaldehyde as intermediates (34). E. histolytica possesses at least three enzymes with ADH activity. E. histolytica ADH1 (EhADH1), which is NADP dependent, shows a marked preference for branched-chain alcohols, whereas EhADH2 prefers ethanol as a substrate (35). It has been reported that EhADH2 may be solely responsible for the conversion of acetyl coenzyme A to acetaldehyde (36). ET4C is formed by condensation of propionaldehyde with l-cysteine. Propionaldehyde in Entamoeba is generated through the catabolism of the amino acids l-methionine and l-threonine (Fig. 3).
FIG 3 .
Proposed scheme of 2-(R)-thiazolidine-4-carboxylic acid biosynthesis in E. histolytica trophozoites. Solid lines represent the steps catalyzed by the enzymes whose genes are present in the genomes, whereas dashed lines indicate those likely absent in the genome or not identified so far. Abbreviations: ADH, alcohol dehydrogenase; ALDH, aldehyde dehydrogenases; CoA, coenzyme A; DAK, dihydroxyacetone kinase; DAP, dihydroxyacetone phosphatase; DHA, dihydroxyacetone; DHAP, dihydroxyacetone phosphate; GDH, glycerol dehydrogenase; GK, glycerol kinase; G 3-P, glyceraldehyde 3-phosphate; G3PDH, glycerol 3-phosphate dehydrogenase; GPP, glycerol 3-phosphate phosphatase; MGL, methionine γ-lyase; PFOR, pyruvate: ferredoxin oxidoreductase; TD, threonine dehydratase; TK, transketolase; TPI, triose phosphate isomerase.
The formation of these 2-(R)-thiazolidine-4-carboxylic acids in vivo may therefore provide a possible mechanism for the detoxification of metabolically produced aldehydes in the cell. It was previously reported that at physiological pH, the spontaneous reaction between formaldehyde and l-cysteine to form T4C is rapid and chemically favored (31) and that l-cysteine is immediately directed toward the formation of thiazolidines when these two compounds are added to isolated rat liver homogenate (37), consequently scavenging the toxicity of formaldehyde (38). In rats, T4C was shown to protect the liver against the hepatotoxic effects of ethanol, carbon tetrachloride (39), bromobenzene (40), acetoaminophene (41), tetracycline (42), and thiourea (43). The antiaging effects of T4C were demonstrated in Drosophila melanogaster (44) and mice (45), and its antitumor effect was demonstrated clinically (46). It was suggested that T4C is an effective nitrite-trapping agent in the human body and may block endogenous formation of carcinogenic N-nitroso compounds (47). Despite evidence from such studies, the metabolic fate of T4C is not well established, except in one study where the metabolic carbon atom of T4C was used as a source for the synthesis of the RNA bases guanine and uracil in Escherichia coli (48).
Oxidation and decomposition of T4C.
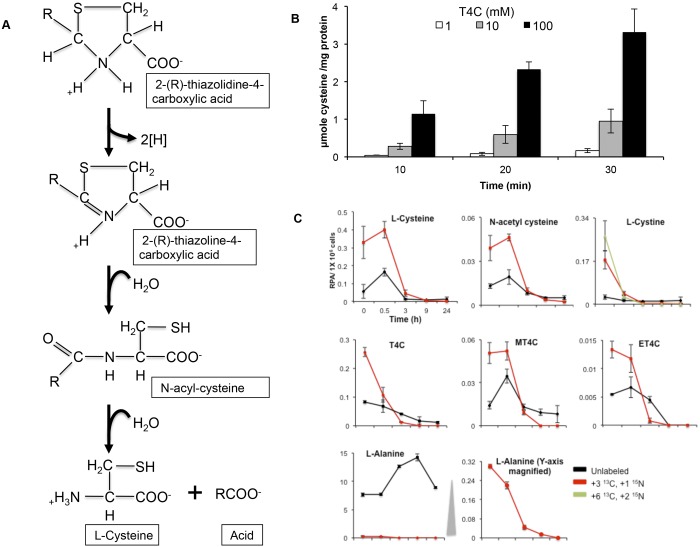
It has previously been shown that T4C is oxidized by E. coli (48), rat liver mitochondria (43), and barley (49). Oxidation of T4C by purified rat liver mitochondria yielded N-formyl-cysteine as a major end product (43). T4C is first converted to 2,3-thiazoline-4-carboxylate (Fig. 4A), 2,3-thiazolidine-4-carboxylate, and then N-acetyl (or formyl or propinyl)-l-cysteine by ring opening and finally gives rise to acetate (or formate or propionate) and l-cysteine by l-proline dehydrogenase (EC 1.5.99.8) (Fig. 4A) (50). Whether an additional enzyme is required to convert N-formyl-l-cysteine to formate and l-cysteine is still not clear (50). However, it was suggested that the hydrolysis of N-formyl-l-cysteine occurs nonenzymatically (50).
FIG 4 .
Metabolic decomposition of 2-(R)-thiazolidine-4-carboxylic acid in E. histolytica trophozoites. (A) Schematic representation of enzymatic degradation of 2-(R)-thiazolidine-4-carboxylic acids as previously proposed for Escherichia coli by Deutch (50). (B) Time course of T4C’s metabolism. The assay was performed as described in Materials and Methods. The means and SD from three independent experiments performed in triplicate are shown. (C) Relative intracellular concentrations of various unlabeled and isotope-labeled metabolites in E. histolytica trophozoites. Trophozoites were cultured in the presence of 8 mM stable-isotope-labeled l-cysteine (U-13C3, 15N) for 24 h. Then, stable-isotope-labeled l-cysteine-containing medium was replaced with l-cysteine-deprived BI-S-33 medium, and the trophozoites were harvested at 0, 0.5, 3, 9, and 24 h of cultivation. The bottom center plot is a magnified (at the y axis) plot of labeled l-alanine, shown at the bottom left. The x axis represents time in hours, whereas the y axis represents the relative peak areas per 1 × 106 cells. Metabolite data are presented as means ± SD from 3 biological replicates.
To examine whether these thiazolidine derivatives can liberate l-cysteine in amebic trophozoites, we chose T4C as an example to investigate the fate of these thiazolidine carboxylic acids. We monitored T4C degradation in mixtures of different concentrations (1 to 100 mM) of T4C and amebic lysates. When T4C was incubated with ameba lysates, their time- and dose-dependent increase in the concentration of l-cysteine was observed (Fig. 4B), suggesting that the ameba lysates contain substances such as enzyme(s) that decompose T4C. Since the structure of T4C resembles that of l-proline, with a replacement of a CH2 group in l-proline by a sulfur atom in T4C (also called thioproline), it was suggested that l-proline dehydrogenase is involved in T4C degradation (50). However, a homologous protein appears to be absent in the E. histolytica genome, although more than 55% of the genes in the E. histolytica genome remain unannotated (51).
Metabolic fate of T4C, MT4C, and ET4C.
In order to further elucidate the metabolic fate of 2-(R)-thiazolidine-4-carboxylic acids in vivo, we cultured the cell with the medium containing stable-isotope-labeled l-cysteine for 24 h, replaced the medium with the normal BI-S-33 medium lacking l-cysteine, and continued culturing for up to 24 h. A rapid decrease in the concentrations of both labeled and unlabeled MT4C, ET4C, and l-cysteine was observed after a short (0.5-h) lag period (Fig. 4C). We also found a drastic immediate decrease, without a lag period, in the concentrations of labeled T4C and l-cystine (Fig. 4C). Together with the fact that T4C is the most abundant 2-(R)-thiazolidine-4-carboxylic acid, this finding suggests that T4C is most immediately accessible and decomposed under l-cysteine deprivation. The fact that the decrease in the l-cystine concentration occurred without a lag period, unlike with l-cysteine, suggests that l-cystine was first reduced to l-cysteine. One of two atypical NADPH-dependent oxidoreductases (EhNO1/2) previously characterized, EhNO2, was shown to catalyze the NADPH-dependent reduction of l-cystine to l-cysteine (11). The changes in the concentrations of labeled and unlabeled N-acetyl-l-cysteine were similar to those of l-cysteine, MT4C, and ET4C, reinforcing the premise that MT4C is degraded via N-acetyl-l-cysteine and that these thiazolidine derivatives serve as a source of l-cysteine under l-cysteine-deficient conditions, as suggested in rat by Wlodek et al. (52). Neither labeled nor unlabeled N-formyl-l-cysteine was detected. This was most likely because their intracellular levels were too low to be detected by CE-TOFMS.
We also found that l-cysteine-derived, labeled l-alanine rapidly decreased under l-cysteine-deprived conditions but that the unlabeled l-alanine concentrations remained approximately 25- to 35-fold higher than those of labeled l-alanine (Fig. 4C). These data indicate that l-cysteine-to-l-alanine conversion by NifS, i.e., iron sulfur cluster formation, is immediately repressed under l-cysteine-deprived conditions. Alternatively, l-alanine produced from l-cysteine is rapidly secreted into the medium, as previously reported (53). It was found that E. histolytica also produces l-alanine as a major end product of energy metabolism (53). Although l-alanine may potentially be metabolized into pyruvate by alanine aminotransferase (EC 2.6.1.2), labeled pyruvate was undetectable. These data suggest that this putative alanine aminotransferase may not be functional under the culture conditions tested (data not shown). Since l-alanine is produced through the catabolism of l-cysteine and also as a major end product of energy metabolism in E. histolytica, it is conceivable that Entamoeba trophozoites excrete l-alanine to expel excess nitrogen out of the cell, as they lack a functional urea cycle (54).
Effect of T4C and MT4C on the growth of E. histolytica trophozoites.
Previous studies using rats suggested that T4C in a diet may replace l-cystine and l-cysteine to promote growth and protect the animals against oxidative stress (38, 52). In order to test this premise in Entamoeba, we monitored the growth kinetics of trophozoites in the presence and absence of l-cysteine, T4C, or MT4C. As shown in Fig. 5, 2 mM MT4C supported trophozoite growth to an extent almost comparable to that with l-cysteine, and T4C also partially supported growth. In the absence of l-cysteine, T4C, and MT4C, trophozoites showed only negligible growth. The growth-supportive effect of MT4C appears to be higher than that of T4C (Fig. 5), although the intracellular MT4C concentrations were approximately 5-fold lower than those of T4C.
FIG 5 .
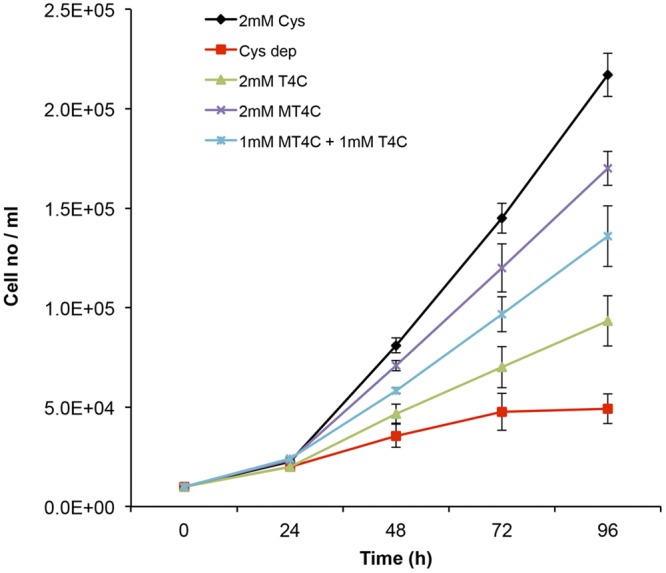
Effect of T4C, MT4C, and l-cysteine on the growth of trophozoites cultured under l-cysteine-depleted conditions. Trophozoites (104 cells/ml) were cultivated in l-cysteine-deprived BI-S-33 media with and without 2 mM T4C, MT4C, or l-cysteine or 1 mM each T4C and MT4C. The parasites were counted every 24 h on a hemocytometer. Error bars represent the standard errors of results from five independent experiments. Cys dep, absence of l-cysteine, T4C, and MT4C.
Roles of T4C and MT4C in the antioxidative-stress defense.
2-(R)-Thiazolidine-4-carboxylic acids, including T4C are cyclic-sulfur-containing amino acids that are analogous in molecular structure to l-proline. It has been shown that T4C can act as an intracellular sulfhydryl antioxidant and a scavenger of free radicals and thereby protect cellular membranes and other oxidation-prone structures in the cell from damage due to oxygen and oxygen-derived free radicals (55). It was shown that T4C stimulates oxygen uptake in rat liver mitochondria (43). As T4C plays an important role in oxidative-defense mechanisms (55), it was of interest to examine the effect of T4C and MT4C on the amount of intracellular ROS. Our previous study showed that when E. histolytica trophozoites were cultured under l-cysteine-limited conditions for 72 h, the intracellular levels of reactive oxygen species increased 4-fold (18). We cultivated trophozoites in l-cysteine-deprived BI-S-33 medium for 72 h, and the medium was replaced with l-cysteine-deprived BI-S-33 medium containing 2 mM T4C, MT4C, or l-cysteine. After 3 h, the relative level of ROS was measured using the fluorescent indicator CM-H 2DCFDA [5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester]. We found that the intracellular levels of reactive oxygen species in trophozoites cultured with 2 mM T4C, MT4C, or l-cysteine were, respectively, approximately 50, 21, or 32% lower than those in control cells (Fig. 6, bar Cys dep). These results suggest that T4C, M4C, and l-cysteine (T4C in particular) are important scavengers of reactive oxygen species in E. histolytica. The level of suppression of ROS by supplemented thiazolidine-4-carboxylic acids in the l-cysteine-deprived culture medium was only partial (<50%). This may be because, besides the thiazolidine-4-carboxylic acids described here, l-cysteine-derived metabolites that are involved in the antioxidant defense mechanism may exist in E. histolytica. Mackenzie and Harris were the first to recognize the therapeutic potential of T4C in animals (43). They noted that T4C is about five times more potent than l-cysteine in preventing massive pleural effusions and death in thiourea-treated rats. It was presumed that T4C, possessing a protected sulfur atom in its ring, opens and frees a sulfhydryl group after entering a liver cell. l-Cysteine, on the other hand, has an unprotected free sulfhydryl group, which is likely to react with oxidants before entering a cell.
FIG 6 .
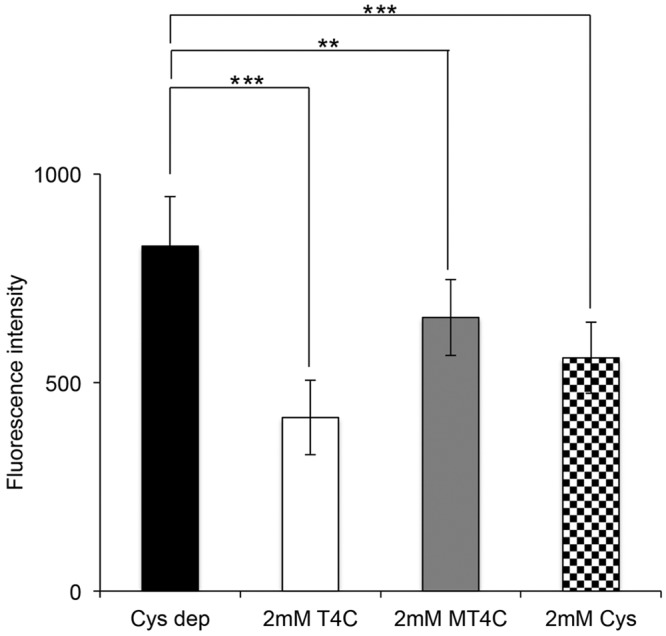
Influence of T4C, MT4C, and l-cysteine on the intracellular ROS levels. Trophozoites were cultivated in l-cysteine-deprived BI-S-33 medium for 72 h, and after that, the medium was replaced with l-cysteine-deprived BI-S-33 media containing 2 mM T4C, MT4C, or l-cysteine. After 3 h, approximately 4.0 × 105 cells were then incubated with the dye 2′,7′-DCF-DA for 20 min. The intracellular ROS levels were quantified by determination of DCF fluorescence. Results were normalized with cell numbers and are presented relative to levels in untreated control cells. The means ± SD from three independent experiments performed in triplicate are shown. Statistical comparisons were made by Student’s t test (**, P < 0.01; ***, P < 0.001).
In summary, we found that in E. histolytica, l-cysteine is utilized for the synthesis of 2-(R)-thiazolidine-4-carboxylic acid derivatives via conjugation with aldehydes. This mechanism allows regulation of the intracellular level of l-cysteine and also functions as a mechanism for detoxifying aldehydes. Our results also suggest that these thiazolidine derivatives serve as storage for l-cysteine, from which l-cysteine can be liberated when required. Furthermore, we have demonstrated that these thiazolidine derivatives, T4C in particular, can reduce the intracellular ROS levels and thus help the parasite to cope with oxidative stress. Future research is needed to determine if these thiazolidine derivatives are also present in other anaerobic/microaerophilic protozoan parasites, such as Giardia intestinalis and Trichomonas vaginalis, which also require high concentrations of extracellular l-cysteine for growth and survival, in order to verify whether common metabolic and biochemical mechanisms are shared by these parasitic protists in general.
MATERIALS AND METHODS
Chemicals and reagents.
All chemicals of analytical grade were purchased from either Wako or Sigma-Aldrich unless otherwise mentioned. 2′,7′-Dichlorodihydrofluorescein diacetate (2′,7′-DCF-DA) was purchased from Invitrogen (U-13C3, 15N). l-Cysteine was purchased from Cambridge Isotope Laboratories. Stock solutions of metabolite standards (1 to 100 mmol/liter) for CE-MS analysis were prepared in either Milli-Q water, 0.1 mol/liter HCl, or 0.1 mol/liter NaOH. A mixed solution of the standards was prepared by diluting stock solutions with Milli-Q water immediately before CE-TOFMS analysis.
Microorganisms and cultivation.
Trophozoites of the E. histolytica clonal strain HM-1:IMSS cl 6 were maintained axenically in Diamond’s BI-S-33 medium at 35.5°C, as described previously (56). Trophozoites were harvested in the late-logarithmic-growth phase 2 to 3 days after the inoculation of medium with 1/30 to 1/12 of the total culture volume.
Metabolic labeling and metabolite extraction.
E. histolytica trophozoites were cultivated in either standard BI-S-33 medium containing 8 mM l-cysteine or l-cysteine-deprived medium for 48 h. For the metabolic labeling, trophozoites were cultured in the presence of 8 mM stable-isotope-labeled l-cysteine (U-13C3, 15N) in l-cysteine-deprived medium. The reason for using 8 mM stable-isotope-labeled cysteine was because in normal BI-S-33 medium, the concentration of cysteine used to culture E. histolytica trophozoites is 8 mM cysteine. To extract metabolites, approximately 1.5 × 106 cells were harvested after 0, 0.5, 3, 9, and 24 h of cultivation in stable-isotope-labeled l-cysteine (U-13C3, 15N) medium. The cells were immediately suspended in 1.6 ml of −75°C methanol to quench metabolic activity. To ensure that experimental artifacts, such as ion suppression, did not lead to misinterpretation of metabolite levels, internal standards, namely, 2-(N-morpholino)ethanesulfonic acid, methionine sulfone, and d-camphor-10-sulfonic acid, were added to each sample (18, 57). The samples were then sonicated for 30 s and mixed with 1.6 ml of chloroform and 0.64 ml of deionized water. After being vortexed, the mixture was centrifuged at 4,600 × g at 4°C for 5 min. The aqueous layer (1.6 ml) was filtered using an Amicon Ultrafree-MC ultrafilter (Millipore Co., MA) and centrifuged at 9,100 × g at 4°C for approximately 2 h. The filtrate was dried and preserved at −80°C until mass spectrometric analysis (58). Prior to the analysis, the sample was dissolved in 20 µl of de-ionized water containing reference compounds (200 µmol/liter each of 3-aminopyrrolidine and trimesic acid).
Instrumentation and CE-TOFMS conditions.
Capillary electrophoresis-time of flight mass spectrometry (CE-TOFMS) was performed using an Agilent CE capillary electrophoresis system equipped with an Agilent 6210 time of flight mass spectrometer, Agilent 1100 isocratic high-performance liquid chromatography (HPLC) pump, Agilent G1603A CE-MS adapter kit, and Agilent G1607A CE-electrospray ionization (ESI)-MS sprayer kit (Agilent Technologies, Waldbronn, Germany). The system was controlled by Agilent G2201AA ChemStation software for CE. Data acquisition was performed by Analyst QS software for Agilent TOF (Applied Biosystems, CA; MDS Sciex, Ontario, Canada).
CE-TOFMS conditions for cationic metabolite analysis.
Cationic metabolites were separated in a fused-silica capillary column (50-µm internal diameter, 100-cm total length) filled with 1 mol/liter formic acid as the reference electrolyte (59). Sample solution (~3 nl) was injected at 5,000 Pa for 3 s, and a positive voltage of 30 kV was applied. The capillary and sample trays were maintained at 20°C and below 5°C, respectively. Sheath liquid composed of methanol-water (50%, vol/vol) that contained 0.1 µmol/liter hexakis (2,2-difluorothoxy) phosphazene was delivered at 10 µl/min. ESI-TOFMS was operated in the positive-ion mode. The capillary voltage was set at 4 kV, and a flow rate of nitrogen gas (heater temperature, 300°C) was set at 10 lb/in2 gauge. For TOFMS, the fragmentor voltage, skimmer voltage, and octopole radio frequency voltage (Oct RFV) were set at 75, 50, and 125 V, respectively. An automatic recalibration function was performed using two reference masses of reference standards, a protonated [13C]methanol dimer (m/z 66.063061) and a protonated hexakis (2,2-difluorothoxy) phosphazene (m/z 622.028963), which provided the lock mass for exact mass measurements. Exact mass data were acquired at the rate of 1.5 Hz over a 50 to 1,000 m/z range.
CE-TOFMS conditions for anionic metabolite analysis.
Anionic metabolites were separated in a cationic-polymer-coated COSMO(+) capillary column (50-µm internal diameter, 110-cm length) (Nacalai Tesque) filled with 50 mmol/liter ammonium acetate solution (pH 8.5) as the reference electrolyte (60, 61). Sample solution (~30 nl) was injected at 5,000 Pa for 30 s, and a negative voltage of −30 kV was applied. Ammonium acetate (5 mmol/liter) in methanol-water (50%, vol/vol) that contained 0.1 µmol/liter hexakis (2,2-difluorothoxy) phosphazene was delivered as sheath liquid at 10 µl/min. ESI-TOFMS was operated in the negative-ion mode. The capillary voltage was set at 3.5 kV. For TOFMS, the fragmentor voltage, skimmer voltage, and Oct RFV were set at 100, 50, and 200 V, respectively (61). An automatic recalibration function was performed using two reference masses of reference standards: a deprotonated [13C]acetate dimer (m/z 120.038339) and an acetate adduct of hexakis (2,2-difluorothoxy) phosphazene (m/z 680.035541). The other conditions were identical to those used for the cationic metabolome analysis.
CE-TOFMS data processing.
Raw data were processed using the in-house software Masterhands (62). The overall data processing flow consisted of the following steps: noise filtering, baseline removal, migration time correction, peak detection, and integration of the peak area from a 0.02-m/z-wide slice of the electropherograms. This process resembled the strategies employed in widely used data processing software for LC-MS and gas chromatography (GC)-MS data analysis, such as MassHunter (Agilent Technologies) and XCMS (63). Subsequently, accurate m/z values for each peak were calculated by Gaussian curve fitting in the m/z domain, and migration times were normalized using alignment algorithms based on dynamic programming (64, 65). All target metabolites were identified by matching their m/z values and normalized migration times with those of standard compounds in the in-house library.
Growth assay of E. histolytica trophozoites.
Approximately 6 × 104 exponentially growing trophozoites of E. histolytica clonal strain HM-1, IMSS cl 6, were inoculated in 6 ml of l-cysteine-deprived BI-S-33 medium containing 2 mM thiazolidine-4-carboxylic acid, 2 mM methyl-thiazolidine acid, and 2 mM cysteine, and the parasites were counted every 24 h on a hemocytometer.
Thiazolidine-4-carboxylate oxidation assays.
T4C oxidation activity was assayed by measuring the production of l-cysteine by ninhydrin reaction and at an absorbance at 560 nm (66). l-Cysteine contents were determined from an l-cysteine standard curve. The assay mixture contained 50 mM Tris-HCl, pH 7.5, 1 to 100 mM T4C, and appropriate amounts of the fractionated parasite lysate in 50 µl of the reaction mixture. The reaction mixture was incubated for 10 to 30 min at 37°C. The reaction was stopped with 10% trichloroacetic acid. After that, 50 µl of glacial acetic acid and 50 µl of freshly prepared ninhydrin reagent were added to each tube and the tubes were incubated at 95°C for 10 min. Finally, all tubes were cooled down on ice and the reaction mixture was diluted with 200 µl of ethanol and measured immediately at 560 nm with a UV/visible-light spectrophotometer (UV-2550; Shimadzu, Tokyo, Japan). Briefly, different concentrations (1 to 100 mM) of T4C were incubated with amebic lysates at 37°C for 10 to 30 min, and the reactions were stopped with 10% trichloroacetic acid. Aliquots of the acid-soluble material were mixed with the acidic ninhydrin reagent and heated. This sequentially resulted in the conversion of N-formylcysteine to l-cysteine and the conjugation of l-cysteine with ninhydrin to form a pink product with an absorbance maximum at 560 nm. T4C reacted with the acidic ninhydrin to form an orange product with a maximum absorbance at 430 nm and a small absorbance at 560 nm. T4C also showed some hydrolysis to l-cysteine. Control mixtures lacking amebic lysates served as controls for T4C oxidation.
Quantitation of reactive oxygen species.
Fluorescence spectrophotometry was used to measure the production of intracellular reactive oxygen species using 2′,7′-DCF-DA as a probe as previously described (67). Briefly, E. histolytica trophozoites were harvested and washed in phosphate-buffered saline (PBS), and approximately 4.0 × 105 cells were then incubated in 1 ml of PBS containing 20 µM 2′,7′-DCF-DA for 20 min at 35.5°C in the dark. The intensity of fluorescence was immediately read at excitation and emission wavelengths of 492 and 517 nm, respectively.
SUPPLEMENTAL MATERIAL
Metabolome data analyzed by capillary electrophoresis-time of flight mass spectrometry (CE-TOFMS) in this study. Abbreviations; CD, l-cysteine-deprived medium; MT, migration time; RPA, relative peak area.
Unknown cysteine-derived metabolites identified in this study by accurate CE-TOFMS mass measurements, calculation of elemental compositions, and chemical and metabolic database searches. Abbreviations: MT, migration time; m/z, mass-to-charge ratio; UK, unknown.
ACKNOWLEDGMENTS
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) (23117001, 23117005, 23390099), a grant from the Global COE Program from the MEXT, a grant for research on emerging and reemerging infectious diseases from the Ministry of Health, Labour, and Welfare of Japan, and a grant for research to promote the development of anti-AIDS pharmaceuticals from the Japan Health Sciences Foundation (KHA1101) to T.N.
Footnotes
Citation Jeelani G, Sato D, Soga T, Watanabe H, Nozaki T. 2014. Mass spectrometric analysis of l-cysteine metabolism: physiological role and fate of l-cysteine in the enteric protozoan parasite Entamoeba histolytica. mBio 5(6):e01995-14. doi:10.1128/mBio.01995-14.
REFERENCES
- 1. Nozaki T, Ali V, Tokoro M. 2005. Sulfur-containing amino acid metabolism in parasitic protozoa. Adv. Parasitol. 60:1–99. 10.1016/S0065-308X(05)60001-2. [DOI] [PubMed] [Google Scholar]
- 2. Kessler D. 2006. Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol. Rev. 30:825–840. 10.1111/j.1574-6976.2006.00036.x. [DOI] [PubMed] [Google Scholar]
- 3. Beinert H, Holm RH, Münck E. 1997. Iron-sulfur clusters: nature’s modular, multipurpose structures. Science 277:653–659. 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 4. Park S, Imlay JA. 2003. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J. Bacteriol. 185:942–950. 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stipanuk MH, Dominy JE, Jr, Lee JI, Coloso RM. 2006. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J. Nutr. 136:1652S–1659S. [DOI] [PubMed] [Google Scholar]
- 6. Stanley SL., Jr. 2003. Amoebiasis. Lancet 361:1025–1034. 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- 7. Weinbach EC, Diamond LS. 1974. Entamoeba histolytica. I. Aerobic metabolism. Exp. Parasitol. 35:232–243. 10.1016/0014-4894(74)90027-7. [DOI] [PubMed] [Google Scholar]
- 8. Mehlotra RK. 1996. Antioxidant defense mechanisms in parasitic protozoa. Crit. Rev. Microbiol. 22:295–314. 10.3109/10408419609105484. [DOI] [PubMed] [Google Scholar]
- 9. Fahey RC, Newton GL, Arrick B, Overdank-Bogart T, Aley SB. 1984. Entamoeba histolytica: a eukaryote without glutathione metabolism. Science 224:70–72. 10.1126/science.6322306. [DOI] [PubMed] [Google Scholar]
- 10. Gillin FD, Diamond LS. 1981. Entamoeba histolytica and Giardia lamblia: effects of cysteine and oxygen tension on trophozoite attachment to glass and survival in culture media. Exp. Parasitol. 52:9–17. 10.1016/0014-4894(81)90055-2. [DOI] [PubMed] [Google Scholar]
- 11. Jeelani G, Husain A, Sato D, Ali V, Suematsu M, Soga T, Nozaki T. 2010. Two atypical l-cysteine-regulated NADPH-dependent oxidoreductases involved in redox maintenance, l-cystine and iron reduction, and metronidazole activation in the enteric protozoan Entamoeba histolytica. J. Biol. Chem. 285:26889–26899. 10.1074/jbc.M110.106310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Husain A, Jeelani G, Sato D, Nozaki T. 2011. Global analysis of gene expression in response to l-cysteine deprivation in the anaerobic protozoan parasite Entamoeba histolytica. BMC Genomics 12:275. 10.1186/1471-2164-12-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, Nozaki T, Suh B, Pop M, Duchene M, Ackers J, Tannich E, Leippe M, Hofer M, Bruchhaus I, Willhoeft U, Bhattacharya A, Chillingworth T, Churcher C, Hance Z, Harris B, Harris D, Jagels K, Moule S, Mungall K, Ormond D, Squares R, Whitehead S, Quail MA, Rabbinowitsch E, Norbertczak H, Price C, Wang Z, Guillén N, Gilchrist C, Stroup SE, Bhattacharya S, Lohia A, Foster PG, Sicheritz-Ponten T, Weber C, Singh U, Mukherjee C, El-Sayed NM, Petri WA, Jr, Clark CG, Embley TM, Barrell B, Fraser CM, Hall N. 2005. The genome of the protist. Parasite Entamoeba histolytica. Nature 433:865–868. 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 14. Tokoro M, Asai T, Kobayashi S, Takeuchi T, Nozaki T. 2003. Identification and characterization of two isoenzymes of methionine γ-lyase from Entamoeba histolytica: a key enzyme of sulfur-amino acid degradation in an anaerobic parasitic protest that lacks forward and reverse transsulfuration pathways. J. Biol. Chem. 278:42717–42727. 10.1074/jbc.M212414200. [DOI] [PubMed] [Google Scholar]
- 15. Sato D, Yamagata W, Harada S, Nozaki T. 2008. Kinetic characterization of methionine gamma-lyases from the enteric protozoan parasite Entamoeba histolytica against physiological substrates and trifluoromethionine, a promising lead compound against amoebiasis. FEBS J. 275:548–560. 10.1111/j.1742-4658.2007.06221.x. [DOI] [PubMed] [Google Scholar]
- 16. Nozaki T, Asai T, Kobayashi S, Ikegami F, Noji M, Saito K, Takeuchi T. 1998. Molecular cloning and characterization of the genes encoding two isoforms of cysteine synthase in the enteric protozoan parasite Entamoeba histolytica. Mol. Biochem. Parasitol. 97:33–44. 10.1016/S0166-6851(98)00129-7. [DOI] [PubMed] [Google Scholar]
- 17. Nozaki T, Asai T, Sanchez LB, Kobayashi S, Nakazawa M, Takeuchi T. 1999. Characterization of the gene encoding serine acetyltransferase, a regulated enzyme of cysteine biosynthesis from the protist parasites Entamoeba histolytica and Entamoeba dispar. Regulation and possible function of the cysteine biosynthetic pathway in Entamoeba. J. Biol. Chem. 274:32445–32452. [DOI] [PubMed] [Google Scholar]
- 18. Husain A, Sato D, Jeelani G, Mi-ichi F, Ali V, Suematsu M, Soga T, Nozaki T. 2010. Metabolome analysis revealed increase in S-methylcysteine and phosphatidylisopropanolamine synthesis upon l-cysteine deprivation in the anaerobic protozoan parasite Entamoeba histolytica. J. Biol. Chem. 285:39160–39170. 10.1074/jbc.M110.167304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gillin FD, Diamond LS. 1981. Entamoeba histolytica and Giardia lamblia: growth responses to reducing agents. Exp. Parasitol. 51:382–391. 10.1016/0014-4894(81)90125-9. [DOI] [PubMed] [Google Scholar]
- 20. Stipanuk MH. 2004. Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Annu. Rev. Nutr. 24:539–577. 10.1146/annurev.nutr.24.012003.132418. [DOI] [PubMed] [Google Scholar]
- 21. Ali V, Nozaki T. 2007. Current therapeutics, their problems, and sulfur-containing-amino-acid metabolism as a novel target against infections by “amitochondriate” protozoan parasites. Clin. Microbiol. Rev. 20:164–187. 10.1128/CMR.00019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Enos-Berlage JL, Downs DM. 1999. Biosynthesis of the pyrimidine moiety of thiamine independent of the PurF enzyme (phosphoribosylpyrophosphate amidotransferase) in Salmonella typhimurium: incorporation of stable isotope-labeled glycine and formate. J. Bacteriol. 181:841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saunders EC, Ng WW, Chambers JM, Ng M, Naderer T, Krömer JO, Likic VA, McConville MJ. 2011. Isotopomer profiling of Leishmania mexicana promastigotes reveals important roles for succinate fermentation and aspartate uptake in tricarboxylic acid cycle (TCA) anaplerosis, glutamate synthesis, and growth. J. Biol. Chem. 286:27706–27717. 10.1074/jbc.M110.213553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Macrae JI, Sheiner L, Nahid A, Tonkin C, Striepen B, McConville MJ. 2012. Mitochondrial metabolism of glucose and glutamine is required for intracellular growth of Toxoplasma gondii. Cell Host Microbe 12:682–692. 10.1016/j.chom.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cobbold SA, Vaughan AM, Lewis IA, Painter HJ, Camargo N, Perlman DH, Fishbaugher M, Healer J, Cowman AF, Kappe SH, Llinás M. 2013. Kinetic flux profiling elucidates two independent acetyl-CoA biosynthetic pathways in Plasmodium falciparum. J. Biol. Chem. 288:36338–36350. 10.1074/jbc.M113.503557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Creek DJ, Chokkathukalam A, Jankevics A, Burgess KE, Breitling R, Barrett MP. 2012. Stable isotope-assisted metabolomics for network wide metabolic pathway elucidation. Anal. Chem. 84:8442–8447. 10.1021/ac3018795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eylert E, Schär J, Mertins S, Stoll R, Bacher A, Goebel W, Eisenreich W. 2008. Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol. Microbiol. 69:1008–1017. 10.1111/j.1365-2958.2008.06337.x. [DOI] [PubMed] [Google Scholar]
- 28. Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM. 2009. Altered regulation of metabolic pathways in human lung cancer discerned by 13 C stable isotope-resolved metabolomics (SIRM). Mol. Cancer 8:41. 10.1158/1535-7163.TARG-09-A41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eylert E, Herrmann V, Jules M, Gillmaier N, Lautner M, Buchrieser C, Eisenreich W, Heuner K. 2010. Isotopologue profiling of Legionella pneumophila. J. Biol. Chem. 285:22232–22243. 10.1074/jbc.M110.128678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ali V, Shigeta Y, Tokumoto U, Takahashi Y, Nozaki T. 2004. An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron-sulfur cluster assembly under anaerobic conditions. J. Biol. Chem. 279:16863–16874. 10.1074/jbc.M313314200. [DOI] [PubMed] [Google Scholar]
- 31. Ratner S, Clarke HT. 1937. The action of formaldehyde upon cysteine. J. Am. Chem. Soc. 59:200–209. 10.1021/ja01280a050. [DOI] [Google Scholar]
- 32. Bullock C. 1990. The biochemistry of alcohol metabolism-a brief review. Biochem. Educ. 18:62–66. 10.1016/0307-4412(90)90174-M. [DOI] [Google Scholar]
- 33. Von Wartburg JP. 1987. Acute aldehyde syndrome and chronic aldehydism. Mutat. Res. 186:249–259. 10.1016/0165-1110(87)90007-8. [DOI] [PubMed] [Google Scholar]
- 34. Lo HS, Reeves RE. 1978. Pyruvate-to-ethanol pathway in Entamoeba histolytica. Biochem. J. 171:225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang W, Li E, Kairong T, Stanley SL., Jr. 1994. Entamoeba histolytica has an alcohol dehydrogenase homologous to the multifunctional adhE gene product of Escherichia coli. Mol. Biochem. Parasitol. 64:253–260. 10.1016/0166-6851(93)00020-A. [DOI] [PubMed] [Google Scholar]
- 36. Zhang WW, Shen PS, Descoteaux S, Samuelson J. 1994. Cloning and expression of the gene for an NADP+-dependent aldehyde dehydrogenase of Entamoeba histolytica. Mol. Biochem. Parasitol. 63:157–161. 10.1016/0166-6851(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 37. Cavallini D, DeMarco C, Mondovi B, Trasarti F. 1956. Studies of the metabolism of thiazolidine carboxylic acid by rat liver homogenate. Biochim. Biophys. Acta 22:558–564. [DOI] [PubMed] [Google Scholar]
- 38. Debey HJ, Mackenzie JB, Mackenzie CG. 1958. The replacement by thiazolidine carboxylic acid of exogenous cystine and cysteine. J. Nutr. 66:607–619. [DOI] [PubMed] [Google Scholar]
- 39. Roquebert J, Dufour P, Ploux D. 1975. Action of sulfur compounds derived from cysteine on the hepatotoxic effects of carbon tetrachloride. Bull. Soc. Pharm. Bordeaux 114:7–11. [Google Scholar]
- 40. Siegers CP, Strubelt O, Völpel M. 1978. The antihepatotoxic activity of dithiocarb as compared with six other thio compounds in mice. Arch. Toxicol. 41:79–88. 10.1007/BF00351772. [DOI] [PubMed] [Google Scholar]
- 41. Strubelt O, Siegers CP, Schütt A. 1974. The curative effects of cysteamine, cysteine, and dithiocarb in experimental paracetamol poisoning. Arch. Toxicol. 33:55–64. [DOI] [PubMed] [Google Scholar]
- 42. Peres G, Dumas M. 1972. Liver protecting effects of thiazolidine carboxylic acid with respect to tetracycline. Gazz. Med. Ital. 131:276–282. [Google Scholar]
- 43. Mackenzie CG, Harris J. 1957. N-formylcysteine synthesis in mitochondria from formaldehyde and l-cysteine via thiazolidine carboxylic acid. J. Biol. Chem. 227:393–406. [PubMed] [Google Scholar]
- 44. Miquel J, Fleming J, Economos AC. 1982. Antioxidants, metabolic rate and aging in Drosophila. Arch. Gerontol. Geriatr. 1:159–165. 10.1016/0167-4943(82)90016-4. [DOI] [PubMed] [Google Scholar]
- 45. Miquel J, Economos AC. 1979. Favorable effects of the antioxidants sodium and magnesium thiazolidine carboxylate on the vitality and life span of Drosophila and mice. Exp. Gerontal 14:279–285. [DOI] [PubMed] [Google Scholar]
- 46. Brugarolas A, Gosalvez M, Gerety RJ. 1980. Treatment of cancer by an inducer of reverse transformation. Lancet ii:68–70. [DOI] [PubMed] [Google Scholar]
- 47. Kurashima Y, Tsuda M, Sugimura T. 1990. Marked formation of thiazolidine-4-carboxylic acid, an effective nitrite trapping agent in vivo, on boiling of dried shiitake mushroom (Lentinus edodes). J. Agric. Food Chem. 38:1945–1949. 10.1021/jf00100a015. [DOI] [Google Scholar]
- 48. Unger L, DeMoss RD. 1966. Metabolism of a proline analogue, l-thiazolidine-4-carboxylic acid, by Escherichia coli. J. Bacteriol. 91:1564–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Elthon TE, Stewart CR. 1984. Effects of the proline analogue l-thiazolidine-4-carboxylic acid on proline metabolism. Plant Physiol. 74:213–218. 10.1104/74.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deutch CE. 1992. Oxidation of l-thiazolidine-4-carboxylateby l-proline dehydrogenase in Escherichia coli. J. Gen. Microbiol. 138:1593–1598. 10.1099/00221287-138-8-1593. [DOI] [PubMed] [Google Scholar]
- 51. Lorenzi HA, Puiu D, Miller JR, Brinkac LM, Amedeo P, Hall N, Caler EV. 2010. New assembly, reannotation and analysis of the Entamoeba histolytica genome reveal new genomic features and protein content information. PLoS Negl. Trop Dis. 4:e716. 10.1371/journal.pntd.0000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wlodek L, Rommelspacher H, Susilo R, Radomski J, Höfle G. 1993. Thiazolidine derivatives as source of free l-cysteine in rat tissue. Biochem. Pharmacol. 46:1917–1928. 10.1016/0006-2952(93)90632-7. [DOI] [PubMed] [Google Scholar]
- 53. Zuo X, Coombs GH. 1995. Amino acid consumption by the parasitic, amoeboid protists Entamoeba histolytica and E. invadens. FEMS Microbiol. Lett. 130:253–258. 10.1111/j.1574-6968.1995.tb07728.x. [DOI] [PubMed] [Google Scholar]
- 54. Clark CG, Alsmark UC, Tazreiter M, Saito-Nakano Y, Ali V, Marion S, Weber C, Mukherjee C, Bruchhaus I, Tannich E, Leippe M, Sicheritz-Ponten T, Foster PG, Samuelson J, Noël CJ, Hirt RP, Embley TM, Gilchrist CA, Mann BJ, Singh U, Ackers JP, Bhattacharya S, Bhattacharya A, Lohia A, Guillén N, Duchêne M, Nozaki T, Hall N. 2007. Structure and content of the Entamoeba histolytica genome. Adv. Parasitol. 65:51–190. 10.1016/S0065-308X(07)65002-7. [DOI] [PubMed] [Google Scholar]
- 55. Weber HU, Fleming JF, Miquel J. 1982. Thiazolidine-4-carboxylic acid, a physiologic sulfhydryl antioxidant with potential value in geriatric medicine. Arch. Gerontol. Geriatr. 1:299–310. 10.1016/0167-4943(82)90030-9. [DOI] [PubMed] [Google Scholar]
- 56. Diamond LS, Harlow DR, Cunnick CC. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431–432. 10.1016/0035-9203(78)90144-X. [DOI] [PubMed] [Google Scholar]
- 57. Jeelani G, Sato D, Husain A, Escueta-de Cadiz A, Sugimoto M, Soga T, Suematsu M, Nozaki T. 2012. Metabolic profiling of the protozoan parasite Entamoeba invadens revealed activation of unpredicted pathway during encystation. PLoS One 7:e37740. 10.1371/journal.pone.0037740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ohashi Y, Hirayama A, Ishikawa T, Nakamura S, Shimizu K, Ueno Y, Tomita M, Soga T. 2008. Depiction of metabolome changes in histidine-starved Escherichia coli by ce-TOFMS. Mol. Biosyst. 4:135–147. 10.1039/b714176a. [DOI] [PubMed] [Google Scholar]
- 59. Soga T, Heiger DN. 2000. Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal. Chem. 72:1236–1241. 10.1021/ac990976y. [DOI] [PubMed] [Google Scholar]
- 60. Soga T, Ueno Y, Naraoka H, Ohashi Y, Tomita M, Nishioka T. 2002. Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal. Chem. 74:2233–2239. 10.1021/ac020064n. [DOI] [PubMed] [Google Scholar]
- 61. Soga T, Igarashi K, Ito C, Mizobuchi K, Zimmermann HP, Tomita M. 2009. Metabolomic profiling of anionic metabolites by CE-MS. Anal. Chem. 81:6165–6174. 10.1021/ac900675k. [DOI] [PubMed] [Google Scholar]
- 62. Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. 2010. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 6:78–95. 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78:779–787. 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 64. Baran R, Kochi H, Saito N, Suematsu M, Soga T, Nishioka T, Robert M, Tomita M. 2006. MathDAMP: a package for differential analysis of metabolite profiles. BMC Bioinformatics 7:530. 10.1186/1471-2105-7-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Soga T, Baran R, Suematsu M, Ueno Y, Ikeda S, Sakurakawa T, Kakazu Y, Ishikawa T, Robert M, Nishioka T, Tomita M. 2006. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J. Biol. Chem. 281:16768–16776. 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- 66. Gaitonde MK. 1967. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem. J. 104:627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jeelani G, Husain A, Sato D, Soga T, Suematsu M, Nozaki T. 2013. Biochemical and functional characterization of novel NADH kinase in the enteric protozoan parasite Entamoeba histolytica. Biochimie 95:309–319. 10.1016/j.biochi.2012.09.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metabolome data analyzed by capillary electrophoresis-time of flight mass spectrometry (CE-TOFMS) in this study. Abbreviations; CD, l-cysteine-deprived medium; MT, migration time; RPA, relative peak area.
Unknown cysteine-derived metabolites identified in this study by accurate CE-TOFMS mass measurements, calculation of elemental compositions, and chemical and metabolic database searches. Abbreviations: MT, migration time; m/z, mass-to-charge ratio; UK, unknown.