Abstract
Background
TP53 defects, i.e. 17p13 deletion and/or nucleotide mutations, associate with short survival and chemorefractoriness in chronic lymphocytic leukemia (CLL). In this context, since direct sequencing of the TP53 gene does not evaluate TP53 functionality, a functional assessment of TP53 pathway may be of interest to identify high risk CLL. By taking advantage of a training cohort of 100 CLL and a validation cohort of 40 CLL with different patterns of TP53 mutation/deletion by FISH and sequencing, we propose an in-vitro assay in which the modulation of TP53 protein and CDKN1A mRNA were investigated upon 24-hour exposure of CLL cells to Nutlin-3.
Methods
The functional assay was set-up on cell lines recapitulating all TP53 genotypes (EHEB, TP53wt/wt; RAJI, TP53mut/wt; MEC-1 and MAVER1, TP53mut/del; HL-60, TP53del/del) and evaluated in two multi-institutional cohorts, purposely enriched in CLL bearing TP53 disruption: a training cohort of 100 cases and a validation cohort of 40 cases, both characterized by FISH and TP53 direct sequencing. Cells were exposed to 10 μM Nutlin-3 for 24 hours; TP53 accumulation was evaluated by Western blotting; TP53 transcriptional activity was determined by quantitative realtime PCR (qRT-PCR) of the TP53 target gene CDKN1A.
Results
According to TP53 protein modulation, in the training cohort we identified: i) 63 cases (51 TP53wt/wt, 12 TP53del/wt) with absence of basal TP53 and induction after treatment (normal pattern); ii) 18 cases (3 TP53mut/wt, 15 TP53mut/del) with high basal TP53 without increase after treatment (mutant pattern); iii) 19 cases (5 TP53mut/wt; 3 TP53mut/del; 11 TP53wt/wt) with basal TP53 that increases upon treatment (intermediate pattern). Evaluation of CDKN1A mRNA levels upon Nutlin-3 exposure showed that the 26 TP53 mutated (TP53mut/del or TP53mut/wt) cases had lower induction levels than the majority (57/63) of cases with normal pattern, and 10/12 cases with intermediate pattern without evidence of TP53 derangement by FISH and sequencing. These results were confirmed in the independent validation cohort of 40 cases (13 TP53wt/wt, 3 TP53del/wt, 12 TP53mut/del, 12 TP53mut/wt).
Conclusions
The proposed functional assay may integrate the conventional analyses for the identification of TP53 dysregulated CLL.
Keywords: CLL, TP53, Prognosis
Background
Chronic lymphocytic leukemia (CLL) is a heterogeneous disease with highly variable clinical courses with survivals ranging from months to decades [1]. In particular, a subset of patients is affected by a high-risk CLL form that rapidly progresses and develops a symptomatic disease requiring treatment. Over-represented in this group are patients bearing either a chromosomal deletion of 17p13.1, location of the tumor suppressor gene TP53, and/or carrying mutations of the TP53 gene [2-4]. Over 80% of CLL with a deletion at 17p13 also present a TP53 mutation in the remaining allele, whereas TP53 mutations in absence of a concomitant deletion at 17p13 occur in 5-10% of CLL cases [5-9].
The TP53 protein is a transcription factor with a short half-life, present at low levels under resting conditions and that becomes activated following DNA damage. Activation, occurring predominantly by phosphoryilation, prolongs the half-life of the protein and allows it to accumulate into the nucleus where it induces apoptosis, cell cycle arrest, and DNA repair [10], thus playing a pivotal role in limiting clonal expansion, maintaining genomic stability, eventually mediating the action of DNA damaging chemotherapy [11-15]. Conventional treatment of CLL is usually based on cytotoxic chemotherapy using alkylating agents or nucleoside analogues. The group of patients bearing a TP53 disruption (i.e. deletion of 17p13 and mutations of the TP53 gene, or TP53 gene mutations alone) has been shown to respond particularly poorly to chemotherapy [2-4,16,17]. Therefore, although deletion or mutation of TP53 gene in previously untreated CLL patients are reported to be 10-15% [7,8,18], the frequency of TP53 dysfunction increases to nearly 50% of patients when the disease progresses following initial therapies [19,20], suggesting that DNA damaging therapies exert a selective pressure that may lead to TP53 inactivation and subsequent resistance to commonly used chemotherapeutic agents.
In the last decade, functional assays in primary CLL cells have been developed [20-29], with the aim: i) to avoid large time- and money-consuming screenings of TP53 gene mutations in non-17p deleted CLL cases; ii) to detect defects in the TP53 pathway escaping fluorescence-in situ-hybridization (FISH) for 17p deletions or mutational analysis by direct sequencing. In particular, in-vitro exposure of CLL cells to the small non-genotoxic molecule Nutlin-3, a potent and selective inhibitor of TP53/MDM2 interaction, has been proposed to evaluate TP53 functionality [21,26,30].
By applying a training/validation strategy using a cohort of 140 CLL cases with known TP53 status, we propose a short term in vitro functional assay, based on the exposure of CLL cells to the non-genotoxic TP53 activator Nutlin-3, as a tool to identify CLL cases with dysregulated TP53 in a clinical setting.
Results
Set up of a western blot assay to detect TP53 dysfunctions
A series of 5 cell lines encompassing all the types of TP53 dysfunction was employed: i) EHEB cells lacking both 17p deletion and TP53 mutations (TP53wt/wt); ii) RAJI cells carrying TP53 mutations in the absence of concomitant 17p deletion (TP53mut/wt); iii) MAVER-1 and MEC-1 cells carrying both 17p deletion and TP53 mutations (TP53del/mut); iv) HL-60 cells bearing 17p deletion in both alleles (TP53del/del). The TP53 status of the employed cell clones was re-confirmed in the present study by both FISH and direct sequencing approaches in agreement with data reported by the IARC TP53 Mutation Database [31] (Additional file 1: Table S1).
Cells from cell lines bearing or not TP53 dysfunction were treated for 24 hours with 10 μM Nutlin-3, and the levels of TP53 were evaluated by western blotting. As summarized in Figure 1A, EHEB cells (TP53wt/wt) showed an absence of basal TP53 and a marked induction upon in vitro Nutlin-3 exposure (“normal” pattern). An AnnexinV/7-AAD assay verified that Nutlin-3 exposure was capable to effectively induce apoptosis in EHEB cells, as documented by the death of the majority of the population within 24 hours (Additional file 2: Figure S1). In contrast, MAVER-1 (TP53del/mut), RAJI (TP53mut/wt) and MEC-1 (TP53del/mut) cells showed a pattern characterized by comparable TP53 levels between Nutlin-3 treated and untreated cells (“mutant” pattern; Figure 1A), mapping at the conventional weight for MAVER-1 and RAJI, or at lower molecular weight for MEC-1 cells, which expressed a truncated TP53 protein, as reported [31]. Finally, a total absence of TP53 levels before and upon in vitro Nutlin-3 exposure was detected in HL-60 cells (TP53del/del; “null” pattern; Figure 1A). Consistently, comparable mortality rates between Nutlin-3 treated and untreated conditions were observed by AnnexinV/7-AAD assay in cell lines expressing mutant/null TP53 statuses by western blot (Additional file 2: Figure S1).
Figure 1.

Set up of a western blot assay to detect TP53 dysfunction. A) Figure shows results from western blot assay on the series of the 5 cell lines, in particular EHEB (TP53wt/wt) with a normal pattern, MEC-1 (TP53del/mut), MAVER-1 (TP53del/mut), RAJI (TP53mut/wt) with a mutant pattern, and HL60 (TP53del/del) with a null pattern. B) Figure shows prototypic results from western blot assay of CLL cases with a normal pattern, with an intermediate pattern and with a mutant pattern. C) Histograms show the functional classification according to western blot assay of CLL cases of the training cohort subdivided into the main genetic subgroups. For each functional category the subdivision in genetic subgroup is indicated.
To evaluate the sensitivity of the western blot assay for TP53 detection, RAJI cells, expressing TP53 in basal conditions, were mixed with EHEB cells, not expressing TP53 in basal conditions, in order to obtain samples containing 1%, 5%, 10%, 20%, 50% and 100% of RAJI cells. A conservative estimate based on western blot results suggested a sensitivity capable to detect as low as 5% of TP53-expressing cells (Additional file 2: Figure S1B).
Functional characterization of TP53 dysfunction in CLL cases
In order to test the capability of the western blot assay to detect TP53 functionality, we employed a training cohort of 100 CLL cases. This cohort was purposely enriched in cases bearing TP53 disruption (38 out of 100; 38%), including 17 p deletion alone (TP53del/wt, 12 cases; 12%), 17p deletion associated with concomitant TP53 mutations (TP53del/mut, 18 cases; 18%), and a TP53 mutated status in the absence of 17p deletion (TP53mut/wt, 8 cases; 8%), as indicated by FISH and direct sequencing approaches (Additional file 1: Table S2). In particular, FISH analyses revealed a percentage of 17p deleted nuclei ranging from 14% to 90% in the TP53 mutated cases (median percentage 53% of deleted nuclei) and from 5% to 70% in deleted only cases (median percentage 10% of deleted nuclei, p = 0.003). Considering the evaluation of TP53 mutational status by direct sequencing, all TP53 mutations clustered in exons 5–8 with the only exclusion of a case, in which a mutation in intron 3 was detected (Additional file 1: Table S2).
By performing western blot analysis of TP53 expression on CLL cells from the training cohort exposed or not to Nutlin-3, 63 out of 100 (63%) cases revealed a normal pattern. These cases included all the 12 cases bearing 17p deletion in the absence of detectable concomitant TP53 mutations, and all the 14 CLL cases of the analyzed cohort bearing a 11q deletion in absence of concomitant TP53 disruption (Figure 1B,C and Additional file 2: Figure S2). Consistently, Nutlin-3 treated CLL cells from all these 63 cases displayed a high mortality in vitro by the Annexin V/7-AAD assay (not shown).
Eighteen out of 100 CLL cases (18%) displayed a mutant TP53 pattern (i.e. high basal level of TP53 without increase upon Nutlin-3 exposure; Figure 1B,C and Additional file 2: Figure S2). All these cases showed a TP53 mutated status by direct sequencing, associated (15/18) or not with a concomitant 17p deletion (Figure 1C and Additional file 2: Figure S2). Consistently, all these cases had low mortality rates in vitro by Annexin V/7-AAD assay without differences between Nutlin-3 treated and untreated conditions (not shown).
Finally, 19 out of 100 CLL cases (19%) showed an intermediate pattern, characterized by an important basal accumulation of TP53, increased upon incubation with Nutlin-3 (Figure 1B,C and Additional file 2: Figure S2). Patients belonging to this third group presented different mortality levels by Annexin V assay (not shown). Eight out of 19 CLL cases with an intermediate pattern showed a mutated TP53 status (3 cases with concomitant 17p deletion) whereas the remaining 11 cases did not show a mutated TP53 status by using direct sequencing (Figure 1C and Additional file 2: Figure S2). Of note, among the 11 cases with an intermediate pattern not showing a TP53 mutated status by direct sequencing, none was 17p deleted.
Evaluation of TP53 target genes
No differentially expressed genes were found between Nutlin-3 treated and untreated CLL cells by utilizing a global gene expression profiling approach comparing Nutlin-3 treated versus untreated samples of 7 TP53del/mut CLL cases (not shown). Consistently, when TP53del/mut CLL cells, exposed or not to Nutlin-3, were tested for the modulation of the genes previously identified to represent the signature of Nutlin-3 exposed TP53wt/wt CLL cells [32], none of these genes were found to be differentially expressed upon Nutlin-3 exposure (Additional file 2: Figure S3), including the TP53 target genes CDKN1A, BAX and PUMA[32].
According to these results, we evaluated the expression levels of CDKN1A, BAX and PUMA upon Nutlin-3 treatment in CLL cases with an unmutated TP53 status (TP53wt/wt or TP53del/wt genotype, overall accounting for 74 cases) versus cases with a mutated TP53 status (TP53mut/del or TP53mut/wt genotype, overall accounting for 26 cases), as defined by direct sequencing. As shown in Figure 2A, CLL cases with an unmutated TP53 status had a marked induction upon Nutlin-3 exposure for all the three genes (0.0031 versus 0.0318, CDKN1A; 0.0155 versus 0.0641, BAX; 0.0047 versus 0.0274, PUMA; p < 0.001 for all the comparisons). This also held true for CLL cases carrying a 11q deletion, which were characterized by a marked induction for all the three genes, not dissimilar from that of CLL cases with a normal karyotype or carrying 13q deletion or trisomy 12 (not shown). On the contrary, in CLL cases with a mutated TP53 status, CDKN1A expression levels of Nutlin-3 treated and untreated samples barely reached a significant difference (0.042 versus 0.0083, p = 0.043) and, in a similar manner, failed to reach a significant difference for BAX and PUMA (0.0164 versus 0.0354, p = 0.103, BAX; 0.0033 versus 0.0049, p = 0.947, PUMA). In keeping with previous reports [26], upon Nutlin-3 treatment, CDKN1A expression levels showed the greatest difference of increases between cases with an unmutated and a mutated TP53 status (14.73 vs 2.42, p < 0.0001) and the greatest amplitude of induction in cases with an unmutated TP53 status when compared to induction of BAX (5.93. versus 2.68, p = 0.0015) and PUMA (8.98 versus 3.80, p = 0.0021). Altogether, we chose to employ CDKN1A as TP53 target gene in the context of an assay for the functional evaluation of TP53 status (Figure 2B).
Figure 2.
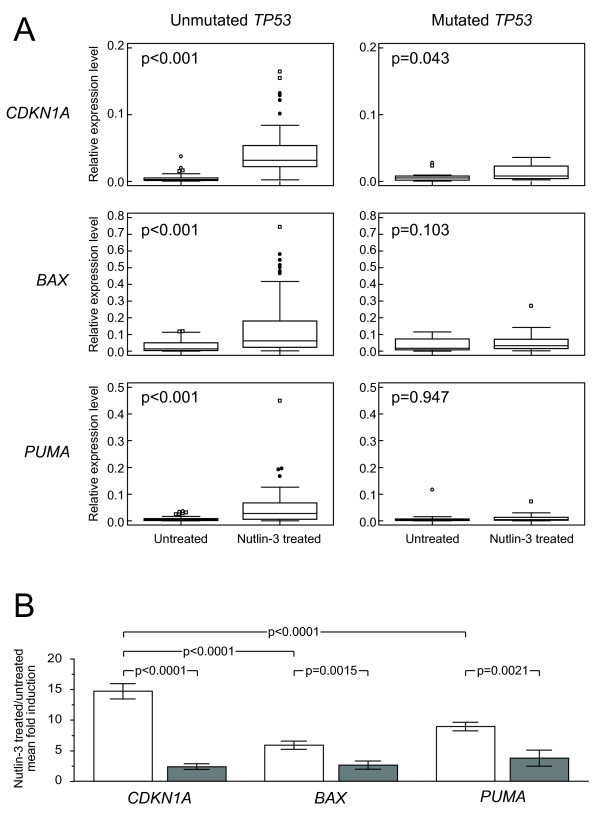
Induction of the TP53 target genes CDKN1A, BAX and PUMA in CLL cases of the training cohort with a TP53 mutated status (TP53mut/del or TP53mut/wt genotypes) or aTP53 wild type status evaluated by direct sequencing. A) Box-and-whisker plots show data obtained by qRT-PCR evaluation of CDKN1A, BAX and PUMA expression levels in untreated or Nutlin-3 treated samples of CLL cases of the training cohort with a mutated or with an unmutated TP53 status. The corresponding p value (Student’s t-test) is reported. B) Histograms represent Nutlin-3 treated/untreated mean fold induction in TP53 wild type (white bar) and TP53 mutated (TP53mut/del or TP53mut/wt genotypes, grey bar) CLL cases. Reported p values refer to Student’s t-test. Error bars represent SD.
As shown in Figure 3, when the whole cohort of 100 cases was plotted according to the evaluation of CDKN1A expression level increases, all the 26 cases with a TP53 mutated status (TP53del/mut or TP53mut/wt cases) clustered in the left part of the graph by expressing CDKN1A levels below (23/26) or slightly above 5-fold increase (3/26). Consistently, the great majority (57/63, 90.5%) of cases with a normal pattern in vitro had CDKN1A values clearly above the 5-fold increase. Finally, as many as 10/12 cases with intermediate pattern in vitro without evidence of TP53 derangement by FISH and direct sequencing had a CDKN1A increase above the 5-fold threshold.
Figure 3.
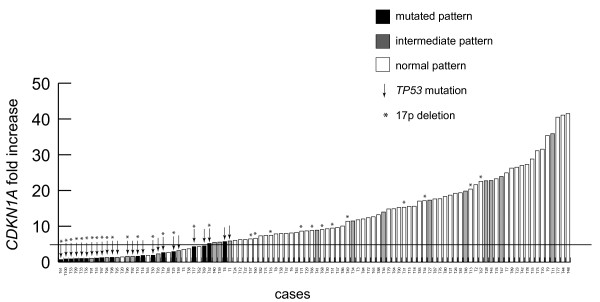
Functional classification of CLL cases of the training cohort according to both western blot assay and evaluation of CDKN1A induction by qRT-PCR. Histograms show data, obtained by qRT-PCR amplification, of CDKN1A fold increase expression for each CLL case of the training cohort classified according to western blot assay. Arrows indicate CLL cases with a TP53 gene mutated status as evaluated by direct sequencing, asterisks indicate CLL cases with a 17p deletion.
Functional characterization of TP53 dysfunction in an independent validation cohort
In order to validate the functional assay here proposed, we analyzed a second independent cohort of 40 patients, purposely enriched in CLL cases with a TP53 disrupted status evaluated by FISH and direct sequencing, composed by: i) 24 cases with TP53 mutation with concomitant 17p deletion (TP53del/mut, 12 cases) or without concomitant 17p deletion (TP53mut/wt, 12 cases); ii) 3 cases bearing a 17p deletion in absence of a concomitant TP53 mutation (TP53del/wt); iii) 4 cases bearing a 11q deletion in absence of concomitant 17p deletion and/or TP53 mutation; iv) 9 cases without TP53 mutation, 17p deletion and/or 11q deletion (Additional file 1: Table S3). As shown in Additional file 2: Figure S4, for each case of this cohort, both western blotting for TP53 and qRT-PCR for CDKN1A upon in vitro Nutlin-3 treatment of CLL cells were performed. Results of these analyses on the validation cohort were separately evaluated “in blind” by 5 independent data analyzers. The sensitivity of the proposed functional assay resulted 0.9 (0.78-1, 95% Confidence Interval, CI), whilst the specificity resulted 0.875 (0.713-1, 95% CI). In detail, for 35 out of 40 cases the 5 analyzers concordantly defined a TP53 status in keeping with the TP53 genotype, as evaluated by FISH and direct sequencing (Additional file 1: Table S4). Although apparently not expressing TP53 mutations by direct sequencing, two cases (e.g. V6, V16) were concordantly defined as with a dysfunction by the 5 data analyzers. Nevertheless, given the low CDKN1A increases found for these two cases (Additional file 2: Figure S4B), the concordant readouts of the 5 data analyzers could be considered in keeping with the documented higher sensitivity of the functional test compared with the TP53 direct sequencing. Finally, for 3 out of 40 cases (e.g. V29, V40, V42), the TP53 pathway functionality was not correctly defined, although in 1 case (e.g. V29) 2 out of 5 data analyzers attributed a functional status consistent with the TP53 genotype (Additional file 1: Table S4).
Comparison with alternative TP53 functional assays
A series of 10 CLL cases composed by 4 11q deleted/TP53wt/wt cases, 1 TP53del/mut cases, 2 TP53mut/wt and 3 TP53wt/wt cases was evaluated for the presence of ATM alterations by performing alternative treatments with etoposide, etoposide plus Nutlin-3 or Nutlin-3, as previously reported [21] (Additional file 1: Table S5). The alternative treatments were set up on the 4 cell lines EHEB, MEC-1, RAJI, and HL60 (Additional file 2: Figure S5A). As shown in Additional file 2: Figure S5B, with the only exclusion of 2 out of 4 11q deleted cases, no relevant differences were detected in terms of type of response among experiments with the alternative treatments.
The same series of 10 CLL cases was also employed to perform a comparison between the approach by western blot/qRT-PCR and an alternative approach by FACS evaluating the modulation of TP53 and CDKN1A protein expression [26,33,34]. Again, FACS analysis was set up on the series of 4 cell lines EHEB, MEC-1, RAJI, and HL60. As shown in Additional file 2: Figure S6A, the obtained patterns were consistent with those obtained for the western blot/qRT-PCR approach and in keeping with the cell line TP53 genotype (Additional file 2: Figure S6A). In the same manner, results of FACS analysis for evaluation of TP53 and CDKN1A protein expression on the series of 10 CLL cases did not significantly diverge from those obtained for the western blot/qRT-PCR approach here proposed (Additional file 1: Table S6 and Additional file 2: Figure S6B).
Discussion
Impaired TP53 function through mutation and/or deletion is the most characterized factor associated with chemoresistance in CLL [15]. Currently, FISH is a widely used technique to detect chromosomal abnormalities such 17p deletion in CLL but there is not a complete overlap between 17p deletion and TP53 mutation, although the deletion of one TP53 allele is frequently accompanied by mutation of the other allele. Direct sequencing is considered the standard technique for the detection of TP53 mutations, with a sensitivity of about 15%-20% of mutated DNA, but it is a time consuming test, that does not take in account TP53 functionality. In this context, a functional assessment of TP53 pathway becomes of interest in high risk CLL patient, especially in the subgroup that may escape FISH or mutational analysis due to low 17p deleted/TP53 mutated clone size. To address this issue in the last decade several TP53 functional assays have been proposed (Additional file 1: Table S7) [20-26,28,29,34].
By applying a training/validation strategy on a cohort of 140 CLL, largely enriched for cases with a TP53 disrupted status, we compared a TP53 functional assay, based on the combined evaluation of TP53 protein expression levels by western blotting and of CDKN1A transcript expression levels by qRT-PCR, with the canonical evaluation of TP53 mutational status by direct sequencing. Cases identified as with a normal pattern by western blot, and with high induction of CDKN1A expression level, consistently presented an unmutated TP53 status by direct sequencing, also irrespective of the presence of a 17p deletion (see for example T13, T15, T51, T71 in Additional file 2: Figure S2). Of note, this type of response was also shared by all the cases of the training and of the validation cohorts in which an 11q deletion was not associated with a concomitant TP53 disrupted status. Cases identified as with a mutant pattern by western blot and with low induction of CDKN1A expression level were consistently TP53 mutated by direct sequencing. Low CDKN1A increases characterized cases with an intermediate pattern in which TP53 mutations were detectable by direct sequencing. On the contrary, cases with an intermediate pattern and with high induction of CDKN1A expression level were consistently TP53 unmutated by direct sequencing. Since there is a tight correlation between the size of the TP53 mutant clone and the impaired response to evaluation of TP53 functionality, the latter could be cases with a small TP53 disrupted clone size, that can be unveiled by western blot analysis but not by qRT-PCR where the lack of CDKN1A induction by the TP53 mutated sub-clone is overcome by its up regulation occurring in the normal TP53 component. Finally, cases with a normal pattern by western blot but low CDKN1A increases (i.e. T20, T63, T85 in Additional file 2: Figure S2) could be characterized by defects on DNA damage pathway other than TP53 defects such as type C defects [23].
The assay here proposed, being based on the use of Nutlin-3 as TP53 activator, is not specifically focused on the detection of ATM mutation as they are other assays based on the combinatorial use of etoposide plus Nutlin-3 [21]. In our comparison series, in 2 out of 4 11q deleted cases, differences were detected in terms of type of response between experiments carried out by exposing CLL cells to Nutlin-3 and experiments performed by utilizing the combinatorial strategy (Additional file 2: Figure S5). In this context, a lack of the revelation of a dysfunctional response in the case of for 11q deleted cases could depend on the residual function of the remaining allele [29,35], although ATM sequencing was not performed in this study and, therefore, a loss of heterozygosis was not specifically investigated in these cases. This could be viewed as a limit of the proposed western blot/qRT-PCR approach although, from a clinical point of view, ATM defects, associated with a failure of TP53 activation, are no longer to be considered a high risk category [35]. These aspects could indicate different mechanisms for TP53 activation between etoposide or irradiation and alkylating agents or purine analogs [35]. Also in the light of the consideration mentioned above, we chose to employ a relatively pure TP53 activator such as Nutlin-3, given its non-genotoxic features that simplify its handling.
Differently from the TP53 functional assay here proposed, an approach by using FACS could be applied to samples with low tumor load without a previous step of purification of the neoplastic component [26,33,34]. On the other hand, a downside of the FACS approach could be the experimental variability due to the efficiency of the permeabilization procedure and intra-cytoplasmic staining of TP53 and CDKN1A proteins. In fact, in the comparison with western blotting, the FACS methodology showed lower signals, especially for the evaluation of TP53 expression levels (Additional file 2: Figure S6). Therefore, evaluation of TP53 expression levels by western blot seems in our hands a more powerful approach, particularly useful in cases with low TP53 dysfunctional sub-clones (Additional file 2: Figure S6). In addition, the FACS approach does not allow to highlight TP53 frameshift mutations that are, instead, easily defined by western blot (Figure 1, Additional file 2: Figure S2, Figure S5 and Figure S6).
According to the above mentioned results, we propose a diagnostic flowchart based on the investigation for 17p deletion by FISH and the evaluation of TP53 pathway functionality by the assay combining western blotting for TP53 and qRT-PCR for CDKN1A upon a 24-hour in-vitro incubation of CLL cells with the non-genotoxic TP53 activator Nutlin-3. As shown in Figure 4, the proposed functional assay might be particularly useful in CLL cases in which a significant amount of 17p deleted nuclei are not detected by the conventional FISH analysis. In particular, the proposed functional assay is able to identify cases with dysfunctional responses to TP53 activators when a mutant pattern (always associated with a reduced up regulation of CDKN1A) or an intermediate pattern by western blot is documented, or when a normal/intermediate western blot pattern is associated with a reduced CDKN1A up regulation. In this context, the higher sensitivity of western blot if compared with direct sequencing might make the proposed assay of potential utility as a tool for the detection of chemoresistant cases bearing dysfunctional TP53 at a sub-clonal level. Clinically, the capability to detect TP53 dysfunctional sub-clones with small cell size (i.e. <15-20% of leukemic cell population) could be very important at diagnosis or at early disease phases to anticipate disease aspects such as chemorefractoriness and relapse, as well as to detect a minimal residual clone (Additional file 1: Table S7). However, further studies are needed to collect definitive information on the clinical relevance of TP53 dysfunction, as evaluated by the western blot/qRT-PCR assay in the proposed diagnostic flowchart, in comparison with the conventional and widely adopted combination of FISH analysis and TP53 direct sequencing [2-4,7,8,18-29]. Finally, it is noteworthy that the assay here proposed, differently from the conventional FISH analysis/TP53 direct sequencing approach, by evaluating CDKN1A expression levels by qRT-PCR, could be of relevant utility to define cases with a type C defect that has been associated both with short progression free survival intervals and with early relapse [35].
Figure 4.
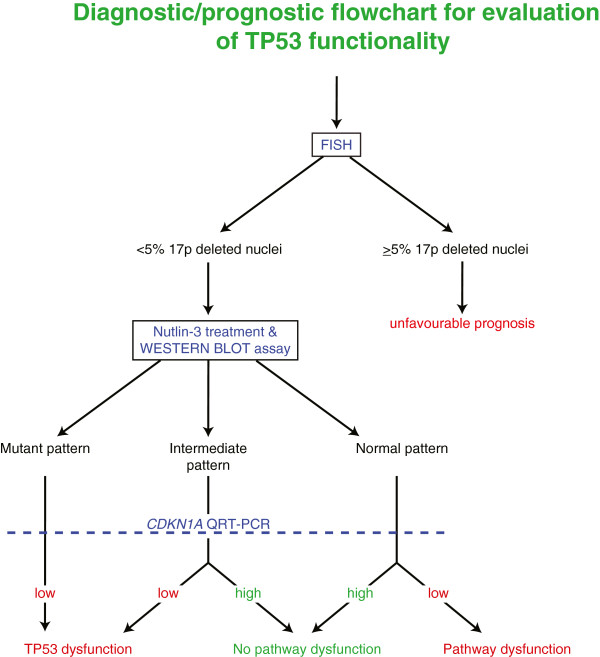
Diagnostic/prognostic flowchart for evaluation of TP53 functionality of CLL cases.
Conclusions
The functional assay here proposed has the advantage of a relatively low cost, as compared to direct sequencing, and might contribute: i) to define dysfunctional cases that may escape FISH and TP53 mutational analysis by direct sequencing due to low 17p deleted/TP53 mutated clone size; ii) to define dysfunctional cases not exhibiting 17p deletion or TP53 mutations (e.g. dysfunctional cases with a type C defect). The flowchart here proposed could represent a valuable contribution to diagnostic and prognostic criteria for CLL patients, especially for those affected by a high risk disease form.
Methods
Patients and cell lines
This study was performed on a multi-institutional cohort of 100 CLL utilized to set-up the flow chart for TP53 status evaluation in CLL (training cohort), and a second multi-institutional independent cohort of 40 CLL, employed to validate the TP53 status flowchart utilizing a “blind” approach (validation cohort), diagnosed according to the IWCLL-NCI criteria [36]. All patients were provided informed consent in accordance with local Institution Review Board requirements and the Declaration of Helsinki. The main clinical and biological features of the two cohorts are summarized in Additional file 1: Table S2 (training cohort) and in Additional file 1: Table S3 (validation cohort). In this context, in 123/140 cases the neoplastic component was more than 80% of cells; in the remaining 17 cases the in vitro experiments (see below) were performed upon B cell purification by CD19+ conjugated columns (Miltenyi Biotec). Sequences of IGHV genes were performed as previously reported, using the ImMunoGeneTics (IMGT) directory for the identification of IGHV-D-J rearrangements and a 2% cut off of for mutated/unmutated discrimination [37]. Detection of CD38, CD49d and ZAP70 expression was performed as previously reported using the cut offs of 30% positive cells (CD38, CD49d expression) or 20% of positive cells (ZAP70 expression) [37].
EHEB, MEC-1, RAJI and HL-60 cell lines were obtained from DSMZ (Germany); MAVER-1 cell line was kindly provided by Alberto Zamò (Department of Pathology, University of Verona, Italy). The most relevant molecular features of cell lines to address the study are summarized in Additional file 1: Table S1.
Analysis of cytogenetic aberrations
Cytogenetic abnormalities were detected as previously reported [37] by FISH for deletion on chromosomes 11, 13, and 17 using the 3 locus specific probes LSI-ATM (SpectrumGreen), LSI-D13S319 (SpectrumOrange), and LSI-p53 (SpectrumOrange), and for aneuploidy of chromosome 12 using an alpha satellite DNA probe CEP12 (SpectrumGreen) (Vysis). In all cases, at least 200 interphase cells with well delineated fluorescent spots were examined. A cut-off of 5% of nuclei was applied to discriminate between negative cases and cases bearing a specific chromosomal abnormality [37]. In the case of 17p deletion, all cases with deletion detectable in 5-10% of nuclei were re-evaluated with a second different specific LSI-p53 probe (Metasystems).
Analysis of TP53 mutations
In CLL cases and cell lines, mutation analysis of TP53 exons 2 to 11 was done by DNA direct sequencing on an ABI Prism 3130 automated DNA sequence analyzer (Applied Biosystems) according to the IARC guidelines (http://www.p53.iarc.fr) and analyzed by Sequencing Analysis v5.2 software (Applied Biosystems) [7]. Mutations were confirmed on both strands on independent amplimers and validated by the IARC TP53 Mutation Database R15 [31].
Cell culture conditions
Primary CLL were obtained from peripheral blood samples by Ficoll-Hypaque (Pharmacia) density gradient centrifugation and cryopreserved until use. After thawing and upon evaluation of the percentage of CLL cells by flow cytometry, CLL cells were further purified by immunomagnetic negative selection as described [38].
Cell lines and CLL cells were cultured (1 × 107 cells/ml) in RPMI-1640 (Biochrom) supplemented with 10% heat-inactivated fetal bovine serum (Biochrom), 100 U/ml penicillin, 0.1 mg/ml streptomycin and 2 mM L-glutamine (Invitrogen) in the presence or not of 10 μM Nutlin-3 (Cayman Chemical) or 50 μM etoposide, or 50 μM etoposide plus 10 μM Nutlin-3 for 24 hours (CLL cases) [21,32], or up to 48 hours for cell lines in case of Nutlin-3 treatment, as previously reported [32]. Cell viability was assayed by AnnexinV and 7-amino-actinomycin-D (7-AAD, both from Becton-Dickinson) double staining (AnnexinV/7-AAD assay) and data were acquired on a FACSCanto flow cytometer and analyzed by Diva software (Becton-Dickinson) as previously reported [38].
Western blot
Total proteins were extracted in RIPA lysis buffer (Santa Cruz) from cultured cells, collected 24 hours after Nutlin-3 treatment, quantified with Bradford assay (Bio-Rad) and ran in 10% SDS-PAGE gels prior to transfer to nitrocellulose membranes (GE Healtcare) for immunoblot analysis and detection by ECL (GE Healtcare) or Immobilon (Millipore Corporation). 1:1000 mouse-anti-TP53 (clone DO1, sc-126) was used for protein detection (Santa Cruz). 1:100.000 mouse-anti-β-actin antibody (clone AC-74, Sigma-Aldrich) was used as loading control.
TP53 and CDKN1A flow cytometry
Fixation and permeabilization of cells were performed with Fix and Perm (Caltag) with methanol modification according to producer guidelines. Cell staining was performed using direct anti-p53(clone DO7)-FITC (sc-47698, Santa Cruz) or indirect anti-p21 (clone Ab-1, OP64, Calbiochem) with secondary antimouse IgG-FITC (sc-2010, Santa Cruz) or anti-CD19-APC (clone HIB19, 555415, BD-Pharmingen) or specific isotype controls. Data were acquired on a FACSCanto flow cytometer and analyzed by Diva software (Becton-Dickinson) as previously reported [38].
Gene expression profile experiments
Gene expression profile experiments (GEP) were performed in a cohort of 20 CLL cases (Additional file 1: Table S8). Total RNA was extracted from purified CLL cells and normal peripheral blood B cells of healthy donors using Trizol reagent (Invitrogen) and validated for integrity and purity using the Agilent 2100 Bioanalyzer (Agilent Tecnologies). GEP was performed using the whole-human genome (4×44K) oligo microarray platform (Agilent Tecnologies) as previously described [39]. Microarray slides were analyzed as previously described [39]. Bioinformatic analyses were performed using SAM algorithm.
Quantitative real-time PCR (qRT-PCR)
Expression of specific genes of interest (i.e. CDKN1A, BAX, PUMA and B2M) was evaluated with the TaqMan Gene Expression assay kit (Applied Biosystem, Life Tecnologies); the relative amount of each gene was calculated utilizing the expression of B2M as internal control using the equation 2-ΔCt where ΔCt = (Ctgene-CtB2M). Fold changes between classes were calculated as reported [39]. All qRT-PCR experiments were performed on an Applied Biosystem 7700 Sequence Detection System (Applied Biosystem).
Statistical analysis
Data were compared using Student’s t-test for independent or paired samples. All statistical analyses were performed using the MedCalc software (MedCalc Software).
Abbreviations
FISH: Fluorescence in situ hybridization; qRT-PCR: Quantitative real time PCR; FACS: Fluorescence activated cell sorting; GEP: Gene expression profiling; 7-AAD: 7-amino-actinomycin.
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
FP contribute to write the manuscript, analyzed the data and performed the research, MDB wrote the manuscript and analyzed the data, NP, RB, AZ, FMR, MD, PS contributed to perform the research, AC, AG, FDR, FZ, GP, DR, GG, GDP, GZ, RF, AG provided well characterized biological samples and contributed to write the manuscript, VG designed the study and contributed to write the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Additional tables: Table S1. TP53 mutational status of the cell lines employed in the study. Table S2. Characterization of CLL cases of the training cohort. Table S3. Characterization of CLL cases of the validation cohort. Table S4. Validation cohort: evaluation “in blind” of 5 independent data analyzers. Table S5. Characterization of CLL cases used for comparison experiments. Table S6. Median fluorescence intensity values for TP53 and CDKN1A protein expression by FACS analysis. Table S7. Overview of the peculiar features of the different TP53 functional assays described by literature. Table S8. Characterization of CLL cases used for microarray experiments.
Additional figures: Figure S1. Evaluation of apoptosis levels by Annexin V-7AAD and sensitivity of western blot analysis upon Nutlin-3 treatment. Figure S2. Western blot for CLL cases of the training cohort. Figure S3. Lack of the gene expression signature associated with Nutlin-3 treatment in TP53del/mut CLL samples. Figure S4. Western blot/qRT PCR assay for CLL cases of the validation cohort. Figure S5. Comparison with alternative TP53 functional assays using combinatorial strategies with etoposide. Figure S6. Comparison with evaluation of TP53 and CDKN1A protein expression levels by FACS analysis.
Contributor Information
Federico Pozzo, Email: pozzo.federico@gmail.com.
Michele Dal Bo, Email: micheledalbo@gmail.com.
Nadia Peragine, Email: peragine@bce.uniroma1.it.
Riccardo Bomben, Email: rbomben@cro.it.
Antonella Zucchetto, Email: zucchetto.soecs@cro.it.
Francesca Maria Rossi, Email: frossi@cro.it.
Massimo Degan, Email: mdegan@cro.it.
Davide Rossi, Email: rossidav@med.unipmn.it.
Annalisa Chiarenza, Email: annalisa.chiarenza@tin.it.
Alberto Grossi, Email: alberto_grossi@libero.it.
Francesco Di Raimondo, Email: diraimon@unict.it.
Francesco Zaja, Email: zaja.francesco@aoud.sanita.fvg.it.
Gabriele Pozzato, Email: g.pozzato@fmc.units.it.
Paola Secchiero, Email: paola.secchiero@unife.it.
Gianluca Gaidano, Email: gaidano@med.unipmn.it.
Giovanni Del Poeta, Email: g.delpoeta@tin.it.
Giorgio Zauli, Email: zauli@burlo.trieste.it.
Robin Foà, Email: rfoa@bce.uniroma1.it.
Anna Guarini, Email: guarini@bce.uniroma1.it.
Valter Gattei, Email: vgattei@cro.it.
Acknowledgments
Supported in part by: Ministero della Salute (Ricerca Finalizzata I.R.C.C.S., “Alleanza Contro il Cancro”; Rete Nazionale Bio-Informatica Oncologica/RN-BIO; Progetto Giovani Ricercatori no. GR-2010-2317594, no GR-2009-1475467, and no GR-2008-1138053, Ministero della salute, Rome, Italy; Fondazione Internazionale di Ricerca in Medicina Sperimentale (FIRMS); Associazione Italiana contro le Leucemie, linfomi e mielomi (AIL), Venezia Section, Pramaggiore Group, Italy; Ricerca Scientifica Applicata, Regione Friuli Venezia Giulia (“Linfonet” Project), Trieste, Italy; the Associazione Italiana Ricerca Cancro (AIRC), Special Program Molecular Clinical Oncology, 5x1000, no 10007; Investigator Grant IG-13227, and MFAG-10327, Milan, Italy; “5x1000 Intramural Program”, Centro di Riferimento Oncologico, Aviano, Italy.
References
- Chiorazzi N, Rai KR, Ferrarini M. Chronic Lymphocytic Leukemia. N Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- Catovsky D, Richards S, Matutes E, Oscier D, Dyer MJ, Bezares RF, Pettitt AR, Hamblin T, Milligan DW, Child JA, Hamilton MS, Dearden CE, Smith AG, Bosanquet AG, Davis Z, Brito-Babapulle V, Else M, Wade R, Hillmen P. Assessment of fludarabine plus cyclophosphamide for patients with chronic lymphocytic leukaemia (the LRF CLL4 Trial): a randomised controlled trial. Lancet. 2007;370:230–239. doi: 10.1016/S0140-6736(07)61125-8. [DOI] [PubMed] [Google Scholar]
- Dohner H, Fischer K, Bentz M, Hansen K, Benner A, Cabot G, Diehl D, Schlenk R, Coy J, Stilgenbauer S. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood. 1995;85:1580–1589. [PubMed] [Google Scholar]
- Grever MR, Lucas DM, Dewald GW, Neuberg DS, Reed JC, Kitada S, Flinn IW, Tallman MS, Appelbaum FR, Larson RA, Paietta E, Jelinek DF, Gribben JG, Byrd JC. Comprehensive assessment of genetic and molecular features predicting outcome in patients with chronic lymphocytic leukemia: results from the US Intergroup Phase III Trial E2997. J Clin Oncol. 2007;25:799–804. doi: 10.1200/JCO.2006.08.3089. [DOI] [PubMed] [Google Scholar]
- Dicker F, Herholz H, Schnittger S, Nakao A, Patten N, Wu L, Kern W, Haferlach T, Haferlach C. The detection of TP53 mutations in chronic lymphocytic leukemia independently predicts rapid disease progression and is highly correlated with a complex aberrant karyotype. Leukemia. 2009;23:117–124. doi: 10.1038/leu.2008.274. [DOI] [PubMed] [Google Scholar]
- Gonzalez D, Martinez P, Wade R, Hockley S, Oscier D, Matutes E, Dearden CE, Richards SM, Catovsky D, Morgan GJ. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011;29:2223–2229. doi: 10.1200/JCO.2010.32.0838. [DOI] [PubMed] [Google Scholar]
- Rossi D, Cerri M, Deambrogi C, Sozzi E, Cresta S, Rasi S, De PL, Spina V, Gattei V, Capello D, Forconi F, Lauria F, Gaidano G. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clin Cancer Res. 2009;15:995–1004. doi: 10.1158/1078-0432.CCR-08-1630. [DOI] [PubMed] [Google Scholar]
- Zenz T, Krober A, Scherer K, Habe S, Buhler A, Benner A, Denzel T, Winkler D, Edelmann J, Schwanen C, Dohner H, Stilgenbauer S. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood. 2008;112:3322–3329. doi: 10.1182/blood-2008-04-154070. [DOI] [PubMed] [Google Scholar]
- Zenz T, Eichhorst B, Busch R, Denzel T, Habe S, Winkler D, Buhler A, Edelmann J, Bergmann M, Hopfinger G, Hensel M, Hallek M, Dohner H, Stilgenbauer S. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28:4473–4479. doi: 10.1200/JCO.2009.27.8762. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, Wyllie AH. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Lotem J, Sachs L. Hematopoietic cells from mice deficient in wild-type p53 are more resistant to induction of apoptosis by some agents. Blood. 1993;82:1092–1096. [PubMed] [Google Scholar]
- Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- Zenz T, Benner A, Dohner H, Stilgenbauer S. Chronic lymphocytic leukemia and treatment resistance in cancer: the role of the p53 pathway. Cell Cycle. 2008;7:3810–3814. doi: 10.4161/cc.7.24.7245. [DOI] [PubMed] [Google Scholar]
- Cang S, Mukhi N, Wang K, Liu D. Novel CD20 monoclonal antibodies for lymphoma therapy. J Hematol Oncol. 2012;5:64. doi: 10.1186/1756-8722-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Wang X. Therapeutic advancement of chronic lymphocytic leukemia. J Hematol Oncol. 2012;5:55. doi: 10.1186/1756-8722-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, Dohner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- Lozanski G, Heerema NA, Flinn IW, Smith L, Harbison J, Webb J, Moran M, Lucas M, Lin T, Hackbarth ML, Proffitt JH, Lucas D, Grever MR, Byrd JC. Alemtuzumab is an effective therapy for chronic lymphocytic leukemia with p53 mutations and deletions. Blood. 2004;103:3278–3281. doi: 10.1182/blood-2003-10-3729. [DOI] [PubMed] [Google Scholar]
- Pospisilova S, Gonzalez D, Malcikova J, Trbusek M, Rossi D, Kater AP, Cymbalista F, Eichhorst B, Hallek M, Dohner H, Hillmen P, van OM, Gribben J, Ghia P, Montserrat E, Stilgenbauer S, Zenz T. ERIC recommendations on TP53 mutation analysis in chronic lymphocytic leukemia. Leukemia. 2012;26:1458–1461. doi: 10.1038/leu.2012.25. [DOI] [PubMed] [Google Scholar]
- Best OG, Gardiner AC, Majid A, Walewska R, Austen B, Skowronska A, Ibbotson R, Stankovic T, Dyer MJ, Oscier DG. A novel functional assay using etoposide plus nutlin-3a detects and distinguishes between ATM and TP53 mutations in CLL. Leukemia. 2008;22:1456–1459. doi: 10.1038/sj.leu.2405092. [DOI] [PubMed] [Google Scholar]
- Chiaretti S, Tavolaro S, Marinelli M, Messina M, Del GI, Mauro FR, Santangelo S, Piciocchi A, Peragine N, Truong S, Patten N, Ghia EM, Torrente I, De Propris MS, Nanni M, Lawrence J, Guarini A, Foa R. Evaluation of TP53 mutations with the AmpliChip p53 research test in chronic lymphocytic leukemia: Correlation with clinical outcome and gene expression profiling. Genes Chromosomes.Cancer. 2011;50:263–274. doi: 10.1002/gcc.20852. [DOI] [PubMed] [Google Scholar]
- Johnson GG, Sherrington PD, Carter A, Lin K, Liloglou T, Field JK, Pettitt AR. A novel type of p53 pathway dysfunction in chronic lymphocytic leukemia resulting from two interacting single nucleotide polymorphisms within the p21 gene. Cancer Res. 2009;69:5210–5217. doi: 10.1158/0008-5472.CAN-09-0627. [DOI] [PubMed] [Google Scholar]
- Kringen P, Bergamaschi A, Due EU, Wang Y, Tagliabue E, Nesland JM, Nehman A, Tonisson N, Borresen-Dale AL. Evaluation of arrayed primer extension for TP53 mutation detection in breast and ovarian carcinomas. Biotechniques. 2005;39:755–761. doi: 10.2144/000112000. [DOI] [PubMed] [Google Scholar]
- Mackus WJ, Kater AP, Grummels A, Evers LM, Hooijbrink B, Kramer MH, Castro JE, Kipps TJ, van Lier RA, van Oers MH, Eldering E. Chronic lymphocytic leukemia cells display p53-dependent drug-induced Puma upregulation. Leukemia. 2005;19:427–434. doi: 10.1038/sj.leu.2403623. [DOI] [PubMed] [Google Scholar]
- Mohr J, Helfrich H, Fuge M, Eldering E, Buhler A, Winkler D, Volden M, Kater AP, Mertens D, Te RD, Dohner H, Stilgenbauer S, Zenz T. DNA damage-induced transcriptional program in CLL: biological and diagnostic implications for functional p53 testing. Blood. 2011;117:1622–1632. doi: 10.1182/blood-2010-08-300160. [DOI] [PubMed] [Google Scholar]
- Marinelli M, Peragine N, Di MV, Chiaretti S, De Propris MS, Raponi S, Tavolaro S, Mauro FR, Del GI, Guarini A, Foa R. Identification of molecular and functional patterns of p53 alterations in chronic lymphocytic leukemia patients in different phases of the disease. Haematologica. 2013;98:371–375. doi: 10.3324/haematol.2012.069906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt AR, Sherrington PD, Stewart G, Cawley JC, Taylor AM, Stankovic T. p53 dysfunction in B-cell chronic lymphocytic leukemia: inactivation of ATM as an alternative to TP53 mutation. Blood. 2001;98:814–822. doi: 10.1182/blood.V98.3.814. [DOI] [PubMed] [Google Scholar]
- Te Raa GD, Malcikova J, Pospisilova S, Trbusek M, Mraz M, Le Garff-Tavernier M, Merle-Beral H, Lin K, Pettitt AR, Merkel O, Stankovic T, van Oers MH, Eldering E, Stilgenbauer S, Zenz T, Kater AP. Overview of available p53 function tests in relation to TP53 and ATM gene alterations and chemoresistance in chronic lymphocytic leukemia. Leuk: Lymphoma; 2013. [DOI] [PubMed] [Google Scholar]
- Saha MN, Qiu L, Chang H. Targeting p53 by small molecules in hematological malignancies. J Hematol Oncol. 2013;6:23. doi: 10.1186/1756-8722-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- Zauli G, di Iasio MG, Secchiero P, Dal Bo M, Marconi D, Bomben R, Del Poeta G, Gattei V. Exposure of B cell chronic lymphocytic leukemia (B-CLL) cells to nutlin-3 induces a characteristic gene expression profile, which correlates with nutlin-3-mediated cytotoxicity. Curr Cancer Drug Targets. 2009;9:510–518. doi: 10.2174/156800909788486777. [DOI] [PubMed] [Google Scholar]
- Carter A, Lin K, Sherrington PD, Pettitt AR. Detection of p53 dysfunction by flow cytometry in chronic lymphocytic leukaemia. Br J Haematol. 2004;127:425–428. doi: 10.1111/j.1365-2141.2004.05223.x. [DOI] [PubMed] [Google Scholar]
- Le Garff-Tavernier M, Blons H, Nguyen-Khac F, Pannetier M, Brissard M, Gueguen S, Jacob F, Ysebaert L, Susin SA, Merle-Beral H. Functional assessment of p53 in chronic lymphocytic leukemia. Blood Cancer J. 2011;1:e5. doi: 10.1038/bcj.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Adamson J, Johnson GG, Carter A, Oates M, Wade R, Richards S, Gonzalez D, Matutes E, Dearden C, Oscier DG, Catovsky D, Pettitt AR. Functional analysis of the ATM-p53-p21 pathway in the LRF CLL4 trial: blockade at the level of p21 is associated with short response duration. Clin Cancer Res. 2012;18:4191–4200. doi: 10.1158/1078-0432.CCR-11-2936. [DOI] [PubMed] [Google Scholar]
- Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, Hillmen P, Keating MJ, Montserrat E, Rai KR, Kipps TJ. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattei V, Bulian P, Del Principe MI, Zucchetto A, Maurillo L, Buccisano F, Bomben R, Dal-Bo M, Luciano F, Rossi FM, Degan M, Amadori S, Del PG. Relevance of CD49d protein expression as overall survival and progressive disease prognosticator in chronic lymphocytic leukemia. Blood. 2008;111:865–873. doi: 10.1182/blood-2007-05-092486. [DOI] [PubMed] [Google Scholar]
- Zucchetto A, Vaisitti T, Benedetti D, Tissino E, Bertagnolo V, Rossi D, Bomben R, Dal Bo M, Del Principe MI, Gorgone A, Pozzato G, Gaidano G, Del PG, Malavasi F, Deaglio S, Gattei V. The CD49d/CD29 complex is physically and functionally associated with CD38 in B-cell chronic lymphocytic leukemia cells. Leukemia. 2012;26:1301–1312. doi: 10.1038/leu.2011.369. [DOI] [PubMed] [Google Scholar]
- Bomben R, Dal-Bo M, Benedetti D, Capello D, Forconi F, Marconi D, Bertoni F, Maffei R, Laurenti L, Rossi D, Del Principe MI, Luciano F, Sozzi E, Cattarossi I, Zucchetto A, Rossi FM, Bulian P, Zucca E, Nicoloso MS, Degan M, Marasca R, Efremov DG, Del PG, Gaidano G, Gattei V. Expression of mutated IGHV3-23 genes in chronic lymphocytic leukemia identifies a disease subset with peculiar clinical and biological features. Clin Cancer Res. 2010;16:620–628. doi: 10.1158/1078-0432.CCR-09-1638. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional tables: Table S1. TP53 mutational status of the cell lines employed in the study. Table S2. Characterization of CLL cases of the training cohort. Table S3. Characterization of CLL cases of the validation cohort. Table S4. Validation cohort: evaluation “in blind” of 5 independent data analyzers. Table S5. Characterization of CLL cases used for comparison experiments. Table S6. Median fluorescence intensity values for TP53 and CDKN1A protein expression by FACS analysis. Table S7. Overview of the peculiar features of the different TP53 functional assays described by literature. Table S8. Characterization of CLL cases used for microarray experiments.
Additional figures: Figure S1. Evaluation of apoptosis levels by Annexin V-7AAD and sensitivity of western blot analysis upon Nutlin-3 treatment. Figure S2. Western blot for CLL cases of the training cohort. Figure S3. Lack of the gene expression signature associated with Nutlin-3 treatment in TP53del/mut CLL samples. Figure S4. Western blot/qRT PCR assay for CLL cases of the validation cohort. Figure S5. Comparison with alternative TP53 functional assays using combinatorial strategies with etoposide. Figure S6. Comparison with evaluation of TP53 and CDKN1A protein expression levels by FACS analysis.


