Abstract
Synaptotagmins are a large gene family in animals that have been extensively characterized due to their role as calcium sensors to regulate synaptic vesicle exocytosis and endocytosis in neurons, and dense core vesicle exocytosis for hormone secretion from neuroendocrine cells. Thought to be exclusive to animals, synaptotagmins have recently been characterized in Arabidopsis thaliana, in which they comprise a five gene family. Using infectivity and leaf-based functional assays, we have shown that Arabidopsis SYTA regulates endocytosis and marks an endosomal vesicle recycling pathway to regulate movement protein-mediated trafficking of the Begomovirus Cabbage leaf curl virus (CaLCuV) and the Tobamovirus Tobacco mosaic virus (TMV) through plasmodesmata (Lewis and Lazarowitz, 2010). To determine whether SYTA has a central role in regulating the cell-to-cell trafficking of a wider range of diverse plant viruses, we extended our studies here to examine the role of SYTA in the cell-to-cell movement of additional plant viruses that employ different modes of movement, namely the Potyvirus Turnip mosaic virus (TuMV), the Caulimovirus Cauliflower mosaic virus (CaMV) and the Tobamovirus Turnip vein clearing virus (TVCV), which in contrast to TMV does efficiently infect Arabidopsis. We found that both TuMV and TVCV systemic infection, and the cell-to-cell trafficking of the their movement proteins, were delayed in the Arabidopsis Col-0 syta-1 knockdown mutant. In contrast, CaMV systemic infection was not inhibited in syta-1. Our studies show that SYTA is a key regulator of plant virus intercellular movement, being necessary for the ability of diverse cell-to-cell movement proteins encoded by Begomoviruses (CaLCuV MP), Tobamoviruses (TVCV and TMV 30K protein) and Potyviruses (TuMV P3N-PIPO) to alter PD and thereby mediate virus cell-to-cell spread.
Keywords: movement protein, cell-to-cell movement, synaptotagmin, Turnip vein clearing virus, Cabbage leaf curl virus, Cauliflower mosaic virus, Turnip mosaic virus
Introduction
To successfully infect their hosts, plant viruses must overcome the hurdle of the plant cell wall, which is a barrier to viral exit from and entry into cells. Viruses accomplish this by encoding movement proteins to transport their genomes cell-to-cell within a leaf and on into the vascular system, through which the virus will systemically invade its host. Cell-to-cell movement occurs via a common pathway: viruses exploit plasmodesmata (PD), complex trans-wall channels that regulate the transport of solutes and key regulatory molecules between cells (Zambryski and Crawford, 2000; Oparka and Roberts, 2001; Benitez-Alfonso et al., 2010; Verchot-Lubicz et al., 2010). Movement proteins alter PD permeability to enable transport of viral genomes into adjacent cells (Waigmann et al., 2004; Ueki and Citovsky, 2011). Beyond this key role in cell-to-cell transport, movement proteins coordinate replication of viral genomes with their directed transport to PD to ensure that replication precedes virus cell-to-cell movement (Lazarowitz and Beachy, 1999). This intracellular trafficking requires interactions with the viral genome per se, and with ER membrane, vesicle trafficking proteins, the cytoskeleton and/or nuclear proteins, depending on the virus and its replication strategy. Some plant viruses encode additional “movement proteins” to bind the viral genome and coordinate this intracellular trafficking with the cell-to-cell movement protein. For example, Geminiviruses have DNA genomes and replicate in the nucleus. Those with two genome components (Begomoviruses), such as Cabbage leaf curl virus (CaLCuV) and Squash leaf curl virus (SqLCV) encode a movement protein that acts as a nuclear shuttle protein (NSP) to bind and transport viral genomes between the nucleus and cytoplasm. The cell-to-cell movement protein (MP) traps NSP-genome complexes in the cytoplasm and redirects them to PD and across the wall (Sanderfoot and Lazarowitz, 1996; Sanderfoot et al., 1996). In contrast, many viruses encode a single cell-to-cell movement protein (MP)1 to execute all these intracellular and intercellular functions, as typified by the 30-kDa MP encoded by the Tobamovirus Tobacco mosaic virus (TMV). TMV, like most RNA viruses, multiplies solely in the cytoplasm of infected cells, and it has been the model for understanding how a single MP functions (Lazarowitz and Beachy, 1999; Nelson and Citovsky, 2005; Verchot-Lubicz et al., 2010). Despite much effort in the past 25 years to delineate the strategies employed by different plant viruses to transport their genomes within and between cells (Harries et al., 2010; Verchot-Lubicz et al., 2010; Harries and Ding, 2011), the key challenge in the field remains to define how movement proteins transport virus genomes to PD and, once there, how they mechanistically alter PD gating. Recent studies in our lab identified the plant synaptotagmin SYTA as a potential key regulator of MP action in modifying PD permeability (Lewis and Lazarowitz, 2010).
Synaptotagmins (SYTs) are a large family of evolutionarily conserved single-pass transmembrane proteins that have been well characterized in animals due to their essential roles in regulating neurotransmitter release and hormone secretion in nerve or neuroendocrine cells (Chapman, 2008; Moghadam and Jackson, 2013). Encoded by a family of at least 17 genes in mammals, and with three synaptotagmins (SYTs 1, 4, and 7) also present in C. elegans and Drosophila, all SYTs have a characteristic conserved domain structure: an uncleaved signal peptide that overlaps with a short N-terminal transmembrane domain (TM), followed by a variable domain (VD) and a cytoplasmic C-terminal region that contains tandem C2 Ca2+/lipid-binding domains called C2A and C2B (Chapman, 2008; Moghadam and Jackson, 2013). Mammals also encode three related extended synaptotagmins (E-SYTs2), which have additional C2 domains and are ubiquitously expressed. While yeast do not encode classical SYTs, they do encode tricalbins, which are E-SYT orthologs that have three C2 domains. Recent studies in mammalian cells and yeast show that E-SYTs act as tethers, along with other proteins, to mediate the formation of contact sites between the endoplasmic reticulum (ER) and plasma membrane (Manford et al., 2012; Toulmay and Prinz, 2012; Giordano et al., 2013).
Mammalian SYTs bind Ca2+ and interact with acidic phospholipids and the core SNARE proteins of the membrane fusion machinery to regulate vesicle fusion at the plasma membrane for exocytosis of neurotransmitters or hormones. A variety of studies, mainly on mammalian SYT1, the first identified family member, suggest that synaptotagmins act as Ca2+ sensors to regulate fusion pore stability and thereby mediate rapid and synchronous exocytosis (Chapman, 2008; Moghadam and Jackson, 2013). As additional family members were characterized, it became clear that SYTs vary in their affinity for Ca2+, binding Ca2+ with low (SYTs 1, 2, and 3), intermediate (SYTs 5, 6, 9, and 10) or high affinity (SYT7), or not binding Ca2+ at all (>50% of the metazoan SYTs, including SYT4). Although a complete mechanistic understanding is still lacking, the emerging picture is that this functional variation among SYTs, coupled with their likely acting as dimers, may be important in SYTs regulating the choice between transient pore opening to favor the rapid release of small molecules, and full fusion of the vesicle and plasma membranes to release larger molecules (Chapman, 2008; Moghadam and Jackson, 2013). Such functional diversity could be important in fine tuning cellular response to Ca2+ signals of varying strength and duration. It is also now apparent that SYTs are found in a variety of cell types, where they can regulate exo/endocytic events beyond synaptic vesicle exocytosis and retrieval, or hormone release, such as Ca2+-dependent release of lysosomal proteins or Ca2+-triggered exocytosis-mediated membrane repair in non-specialized cell types (Chakrabarti et al., 2005; Becker et al., 2009).
Metazoan SYT1 is the most extensively characterized synaptotagmin. SYT1 is widely expressed in neurons and neuroendocrine cells, where it localizes to synaptic vesicles or large dense core vesicles to tightly couple Ca2+ entry with vesicle docking and fusion to trigger rapid exocytosis of neurotransmitters or hormones (so-called synchronous release) (Chapman, 2008; Moghadam and Jackson, 2013). Key to its functions are the tandem C2 domains, which bind Ca2+ and interact with effector SNAREs and phospholipids to trigger membrane fusion (Hui et al., 2006; Lee et al., 2010). Studies suggest that the SYT1 C2A and C2B domains, in response to binding Ca2+, shallowly insert into the inner leaflet of the plasma membrane to induce local membrane curvature, which favors lipid mixing and leads to membrane fusion (Martens et al., 2007; Hui et al., 2009; McMahon et al., 2010). In addition to exocytosis, SYT1 and SYT4 also regulate endocytosis, in a Ca2+-dependent manner, to retrieve membrane from the cell surface and maintain homeostasis (Yao et al., 2012b). Detailed studies of SYT1 activity using missense mutations in C2A and C2B that uncouple exocytosis and endocytosis show that Ca2+ binding has different roles in the two processes (Yao et al., 2012b). In exocytosis, Ca2+ binding by both C2A and C2B is required for the electrostatic switch to allow the surface loops of both domains to interact with SNAREs and cooperatively penetrate the plasma membrane to trigger efficient synchronous neurotransmitter release (Hui et al., 2006; Striegel et al., 2012). However, either C2A or C2B can function as a Ca2+ sensor for endocytosis without apparently penetrating the membrane (Yao et al., 2012a).
We and others recently identified synaptotagmins in plants. The model plant Arabidopsis thaliana encodes five synaptotagmins (SYT A, B, C, D, and E), all of which are predicted to have the characteristic domain structure conserved among their animal orthologs (Craxton, 2001, 2004; Schapire et al., 2008; Yamazaki et al., 2008; Lewis and Lazarowitz, 2010). Of these, SYTA is the best studied, and has been shown to have important roles in biotic and abiotic stress (Schapire et al., 2008; Yamazaki et al., 2008; Lewis and Lazarowitz, 2010). Our group identified Arabidopsis SYTA in a yeast interactive screen as a protein that interacts with the Begomovirus CaLCuV cell-to-cell MPCaLCuV, and we established its role in regulating endocytosis and MP-mediated cell-to-cell transport using both in vitro and in vivo approaches (Lewis and Lazarowitz, 2010).
When tested in an in vitro pulldown assay, SYTA directly interacts with MPCaLCuV, and with MPSqLCV encoded by the Begomovirus SqLCV and the 30-kDa MPTMV encoded by the Tobamovirus TMV (Lewis and Lazarowitz, 2010). To establish the role of SYTA in virus movement, we used an Arabidopsis Col-0 SYTA knockdown line syta-1, and a SYTA dominant-negative mutant SYTAΔC2B, in which the C2B domain is deleted (Lewis and Lazarowitz, 2010). Consistent with a role in virus movement, CaLCuV systemic infection is significantly delayed and virus disease symptoms are attenuated in syta-1 vs. wild type Col-0 plants (Lewis and Lazarowitz, 2010). To show that SYTA regulates MP-mediated virus cell-to-cell transport, we used transient expression assays: MPCaLCuV and MPTMV cell-to-cell trafficking via PD is significantly inhibited when each movement protein, tagged with GFP, is biolistically bombarded into leaves from syta-1 vs. Col-0 plants, or is co-expressed with SYTAΔC2B in Nicotiana benthamiana leaf epidermal cells using agroinfiltration (Lewis and Lazarowitz, 2010). Using GFP fusions of SYTA and SYTAΔC2B, we also showed that SYTA localizes to plasma membrane-derived endosomes in N. benthamiana protoplasts, and we and others found SYTA on endosomes and at discrete areas along the plasma membrane in leaf epidermal cells (Schapire et al., 2008; Lewis and Lazarowitz, 2010). In contrast, SYTAΔC2B localizes to the plasma membrane in protoplasts, and is not found on endosomes; and it blocks the formation of plasma membrane-derived endosomes and endosomal recycling (Lewis and Lazarowitz, 2010). Our functional studies further demonstrate that SYTAΔC2B must be at the plasma membrane to interfere with MPTMV or MPCaLCuV cell-to-cell trafficking. Thus, SYTA regulates endocytosis and MP-mediated trafficking of plant virus genomes through PD, and our findings suggest that distinct virus movement proteins transport their cargos to PD for cell-to-cell spread via an endocytic recycling pathway (Lewis and Lazarowitz, 2010).
While all plant viruses exploit PD for virus genome cell-to-cell transport, research on a range of diverse viruses has led to a variety of models for how different plant viruses transport their genomes within and between cells, and how this intracellular and intercellular genome trafficking may be coordinated (Harries et al., 2010; Verchot-Lubicz et al., 2010; Harries and Ding, 2011). Our finding that SYTA regulates MP-mediated transport via PD for viruses as distinct as CaLCuV and TMV suggests that SYTA may have a central role in regulating cell-to-cell movement for a range of diverse plant viruses. To test this hypothesis, we have used infectivity assays and MP-based cell-to-cell trafficking assays in our Arabidopsis Col-0 syta-1 knockdown line vs. wild type (wt) plants to examine the role of SYTA in the cell-to-cell movement of additional plant viruses that employ different modes of movement, namely the Potyvirus Turnip mosaic virus (TuMV), the Caulimovirus Cauliflower mosaic virus (CaMV) and the Tobamovirus Turnip vein clearing virus (TVCV), which, unlike TMV, efficiently infects Arabidopsis. As reported here, we found that both TuMV and TVCV systemic infection were delayed, and the cell-to-cell trafficking of the their respective movement proteins P3N-PIPO and MPTVCV, each tagged with GFP, were inhibited in syta-1 vs. wild type Col-0 plants. In contrast, CaMV systemic infection was not inhibited in syta-1. Our studies show that SYTA is a key regulator of plant virus intercellular movement, being necessary for the ability of diverse cell-to-cell movement proteins encoded by Begomoviruses (MPCaLCuV), Tobamoviruses (30K MPTVCV and MPTMV) and Potyviruses (P3N-PIPOTuMV), but not Caulimoviruses (MPCaMV), to alter PD to mediate virus cell-to-cell spread.
Materials and methods
Plant material and viruses
A. thaliana ecotype Col-0 plants were grown in growth rooms or chambers at 22C under 16 h/8 h light/dark cycle for CaLCuV, TVCV, and TuMV infectivity assays, and 8 h/16 h light/dark cycle for CaMV infectivity and for movement protein trafficking assays (Lewis and Lazarowitz, 2010). For transient expression studies using agroinfiltration, N. benthamiana plants were grown and maintained under greenhouse conditions of 25C with 16 h supplemental lighting (Levy et al., 2013). Genomic clones of CaLCuV A and B components, TuMV (pGreen-TuMV), TVCV (p50TVCV) and CaMV (pCa122) have all been described, as has the Col-0 T-DNA insertion line syta-1 (Kobayashi et al., 2002; Carvalho et al., 2004; Dunoyer et al., 2004; Lewis and Lazarowitz, 2010; Levy et al., 2013).
Infectivity assays
Agroinoculation of Arabidopsis Col-0 plants with CaLCuV A and B genomic components, each cloned in the binary vector pCB301, has been described (Carvalho et al., 2004). We used the same protocol for TuMV infectivity assays. In brief, cultures of Agrobacterium tumefaciens GV3101 containing the appropriate infectious clones were grown in LB media at 28C for 16 h, diluted 1:20 into fresh LB and incubated an additional ~20 h. The first two true leaves of A. thaliana 12-da-old seedlings were cut transversally with a razor blade and dipped for 5 min into the A. tumefaciens GV3101 cultures harboring CaLCuV (A and B clones mixed) or pGreen-TuMV, each of which had been concentrated to OD600 of 0.5. Plants were maintained at 22C under 16 h/8 h light/dark cycle and scored for viral disease symptoms. For TVCV infectivity assays, Col-0 plants at the 8-leaf stage were rub inoculated with 10 μl of a standardized amount of leaf extract from N. benthamiana plants that had been infected with RNA transcripts of p50TVCV, as previously described (Levy et al., 2013). Infectivity was determined based on the levels of TVCV CP in extracts of systemic leaves (0.5 g tissue/ml buffer), as analyzed by SDS-PAGE and Coomassie blue staining (Levy et al., 2013). CaMV infectivity assays were done by bombarding leaves of 3-wk-old Arabidopsis plants with pCa122 as previously described (Kobayashi et al., 2002). To quantify CaMV DNA levels in systemic leaves, primers 5′-ATGGCCGAATCAATTTTAGACAGGACC-3′ and 5′-CGATGTTGAGCATTCCCATAGACGTTT-3′ were used for semi-quantitative PCR (sqPCR) analysis to amplify the first 455 nucleotides of the CaMV coat protein (CP) gene. For an internal loading control, we used primers 5′-CATGGAGTTGTCAGTGAAATGGGC-3′ and 5′-GTCTTTGGCCATAGGTACATG-3′ to sqPCR amplify nucleotides 992–1554 of the SYTB gene sequence. DNA was extracted from mock-inoculated control plants or from CaMV systemically infected leaves at 21 da post inoculation, as previously described (Lewis and Lazarowitz, 2010).
Movement protein trafficking assays
Cell-to-cell movement of MPCaLCuV, MPTVCV and TuMV P3N-PIPO, and tubule formation by MPCaMV, was assayed by biolistic bombardment of Arabidopsis leaves with plasmids that expressed GFP-fusions of each movement protein under the control of the CaMV 35S promoter. The plasmids pTEX::MPCaLCuV-GFP, and p35S::P3N-PIPO-GFP cloned in the plant expression vector pJ4GFP-XB, have been described (Lewis and Lazarowitz, 2010; Vijayapalani et al., 2012). We used Gateway technology to clone MPCaMV and MPTVCV C-terminal fusions to GFP (MPCaMV-GFP and MPTVCV-GFP). The coding sequence for MPCaMV was PCR amplified using primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTTATGGATTTGTATCCAGAAG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTCTCCACAGATTTCTTTTAA-3. Primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTTATGTCGATAGTCTCGTACGAACC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGCATTGGTATGGGCTCTGC-3′ were used to PCR amplify the MPTVCV coding sequence. PCR products were cloned into the entry vector pDONR207 and then recombined into the destination vector pSITE2NB (Chakrabarty et al., 2007) to create pSITE2::MPCaMV-GFP and pSITE2::MPTVCV-GFP. To assess movement protein cell-to-cell trafficking or tubule formation, detached leaves from 4-wk-old Arabidopsis wild type Col-0 or syta-1 plants were biolistically bombarded with pTEX::MPCaLCuV-GFP, TuMV p35S::P3N-PIPO-GFP, pSITE2::MPCaMV-GFP or pSITE2::MPTVCV-GFP using the Helios Gene Gun (Bio-Rad, Hercules, CA), as described (Ueki et al., 2009). We used confocal imaging to count or examine cells in individual fluorescent foci at different time points between 8 and 48 h post bombardment.
Statistical analyses
These were done using JMP Pro 0.0.2 and SAS software (SAS, Cary, NC). Local cell-to-cell spread was analyzed using X2 analysis, and the Wilcoxon test was applied to infectivity assays.
Microscopy
Confocal laser scanning microscopy (CLSM) was performed using a Leica TCS-SP5 microscope (Leica Microsystems, Exton, PA) with a 63x/NA1.2 water immersion objective. GFP fluorescence was excited with a 488-nm argon laser, and emission was detected at 500–530 nm.
Results
TuMV and TVCV infection is delayed in a SYTA knockdown line
We previously reported that Begomovirus CaLCuV systemic infection is significantly delayed in the Arabidopsis Col-0 T-DNA insertion mutant line syta-1 (Sail 775A08) (Lewis and Lazarowitz, 2010). The T-DNA in this line is inserted just downstream of the SYTA C2B coding region. This insertion produces a functional chimeric SYTA protein (the signal peptide, and TM, VD, and C2A and C2B domains all remain intact) that is slightly smaller (59.1 kDa) than wild type SYTA (61.7 kDa), and this truncated form of SYTA protein in syta-1 accumulates to only ~10% the level of full-length SYTA produced in wild type Col-0 (Lewis and Lazarowitz, 2010). Despite this, our homozygous syta-1 lines do not exhibit overt phenotypes, which makes this SYTA knockdown mutant ideally suited for virus infectivity and movement protein trafficking studies (Lewis and Lazarowitz, 2010). CaLCuV systemic spread is delayed from 1 to 3 days, and viral disease symptoms are attenuated, in syta-1 vs. wild type Col-0 plants, based on the onset of appearance of viral disease symptoms and PCR analysis to assess viral genome accumulation in systemic leaf extracts. CaLCuV infectivity levels are also inhibited ~10 to ~30% in syta-1 (Lewis and Lazarowitz, 2010).
To assess whether SYTA is more generally required for infection and spread of diverse plant viruses, we inoculated wild type Col-0 and two separate homozygous syta-1 segregant lines [lines 1 and 16, see Lewis and Lazarowitz (2010)] with the Potyvirus TuMV or the Tobamovirus TVCV, and analyzed virus infectivity levels and the spread of systemic infection. CaLCuV is a single-stranded DNA (ssDNA) virus that replicates in the nucleus. Both TuMV and TVCV are positive-sense RNA (+RNA) viruses that replicate in the cytoplasm (Lazarowitz, 2001), and our recent BiFC studies show that MPTVCV and SYTA directly interact in planta (A. Levy, J. Zheng and SGL, submitted). TuMV infectivity and systemic spread were monitored by the development of viral disease symptoms. Because TVCV produces mild disease symptoms (variable degrees of stunted and bent stems) on Arabidopsis Col-0, we assessed TVCV infectivity and spread by SDS-PAGE analysis to determine the accumulation of viral coat protein (CP) in systemic leaves at different days following inoculation (Levy et al., 2013).
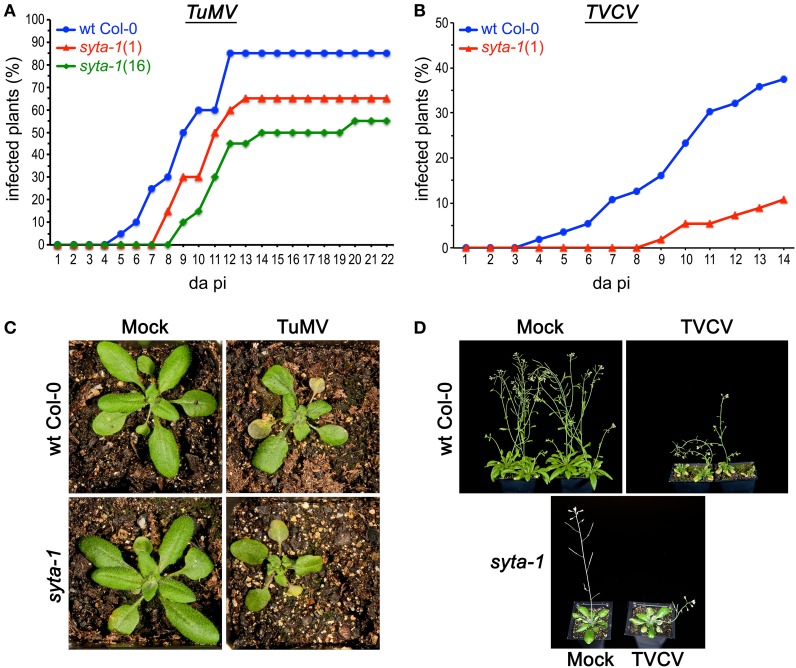
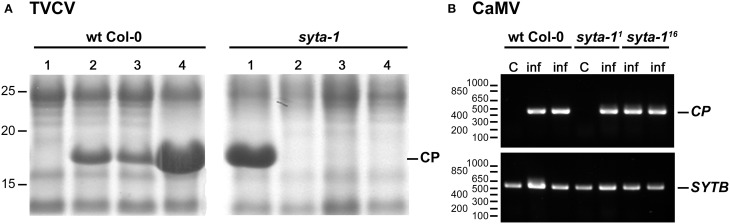
As we found for CaLCuV, infection by both TuMV and TVCV progressed more slowly in syta-1 when compared to wild type Col-0 plants: the onset of systemic infection was delayed, and infectivity levels were lower in syta-1 lines (Figure 1A; Tables 1, 2). TuMV systemic disease symptoms in wild type Col-0 were first evident at 5–6 da post inoculation, and 75–100% of inoculated wild type plants became infected. In contrast, the onset of TuMV disease symptoms was delayed from 1 to 3 da in our syta-1 knockdown lines, and infectivity was decreased to ~60–85% of the levels in wild type Col-0 (50–85% of the inoculated plants became infected), although there was no apparent attenuation of viral disease symptoms (Figures 1A,C; Table 1). TVCV systemic infection, based on the accumulation of viral CP, was also first evident at 5–6 da post inoculation in wild type Col-0, with ~30–45% of inoculated wild type plants being systemically infected by 16 da post-inoculation. In contrast, the onset of TVCV systemic infection was delayed from 1 to 4 da in syta-1 plants and infectivity was decreased to only ~30–50% of the levels in wild type Col-0, based again on the accumulated levels of TVCV CP in systemic leaf extracts (11–20% of inoculated plants became systemically infected) (Figures 1B,D, 2A; Table 2). Furthermore, the levels of TVCV CP in systemic leaf extracts were, on average, lower in syta-1 vs. wt Col-0 plants on the same days post-inoculation (Figure 2A, and data not shown). As with TuMV, TVCV disease symptoms appeared to be the same in wild type Col-0 and syta-1 plants at all stages of infection (Figure 1D and data not shown). Thus, as we had found for CaLCuV, SYTA was necessary for TuMV and TVCV systemic spread and infection in Arabidopsis, consistent with SYTA being important for the cell-to-cell movement of these two +RNA viruses.
Figure 1.
TuMV and TVCV infections are delayed in syta-1. (A) Time course of appearance of disease symptoms on wild type (wt) Arabidopsis Col-0 plants (blue), and syta-1 mutant lines 1 (red) and 16 (green) inoculated with equal amounts of TuMV (Table 1, Trial 2). (B) Development of systemic disease as assessed by viral CP accumulation in Col-0 plants (blue) and syta-1 mutant line 1 (red) inoculated with equal amounts of TVCV (Table 2, Trial 3). (C) TuMV or mock inoculated wild type Col-0 or syta-1 plants at 16 da post inoculation (pi). (D) TVCV or mock inoculated wild type Col-0 plants at 30 da post inoculation or syta-1 plants at 14 da post inoculation (pi).
Table 1.
TuMV Infectivity on syta-1 and wild type Arabidopsis Col-0a.
| da post infection (pi) | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 23 | 24 | 25 | ||
| 1 | syta-1 | 0 | 0 | 5 | 15 | 30 | 40 | 55 | 60 | 70 | 75 | 80 | 80 | 85 | 85 | 85 | 85 | 85 | ||||
| Col-0 | 0 | 0 | 21 | 53 | 74 | 95 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |||||
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 2 | syta-1 (1) | 0 | 0 | 0 | 0 | 15 | 30 | 30 | 50 | 60 | 65 | 65 | 65 | 65 | 65 | 65 | 65 | 65 | 65 | |||
| syta-1 (16) | 0 | 0 | 0 | 0 | 0 | 10 | 15 | 30 | 45 | 45 | 50 | 50 | 50 | 50 | 50 | 50 | 55 | 55 | ||||
| Col-0 | 0 | 5 | 10 | 25 | 30 | 50 | 60 | 60 | 85 | 85 | 85 | 85 | 85 | 85 | 85 | 85 | 85 | 85 | ||||
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| 3 | syta-1 | 0 | 0 | 0 | 2 | 11 | 30 | 45 | 47 | 49 | 57 | 57 | 60 | 66 | 70 | 72 | 72 | 74 | 74 | 74 | 74 | 74 |
| Col-0 | 0 | 0 | 8 | 19 | 46 | 56 | 67 | 71 | 77 | 81 | 81 | 81 | 81 | 81 | 83 | 83 | 83 | 88 | 88 | 88 | 88 | |
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 4 | syta-1 | 0 | 0 | 0 | 0 | 0 | 5 | 30 | 30 | 41 | 43 | 43 | 48 | 48 | 52 | 52 | 52 | 52 | 57 | 57 | ||
| Col-0 | 0 | 0 | 2 | 2 | 2 | 2 | 25 | 32 | 50 | 57 | 61 | 68 | 70 | 70 | 73 | 73 | 73 | 73 | 73 | |||
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| 5 | syta-1 | 0 | 0 | 0 | 13 | 16 | 22 | 28 | 38 | 38 | 38 | 44 | 47 | 47 | 50 | 50 | 50 | 53 | ||||
| Col-0 | 0 | 0 | 18 | 52 | 61 | 67 | 67 | 67 | 70 | 73 | 73 | 73 | 76 | 79 | 82 | 85 | 85 | |||||
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
Symptomatic plants as percentage of total inoculated plants (40–60 plants of each genotype per assay). Nonparametric survival analysis with right censoring (Wilcoxon test statistic) is p < 0.001 for the five trials shown. Unless noted otherwise, syta-1 (line 1) was used for infectivity assays.
Table 2.
TVCV Infectivity on syta-1 and wild type Arabidopsis Col-0a.
| da post infection (pi) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | ||
| 1 | syta-1 | 0 | 0 | 0 | 0 | 2 | 2 | 2 | 4 | 4 | 7 | 7 | 9 | 11 | 11 |
| Col-0 | 0 | 0 | 0 | 4 | 4 | 7 | 7 | 11 | 14 | 18 | 21 | 25 | 27 | 29 | |
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 2 | syta-1 | 0 | 0 | 0 | 0 | 0 | 2 | 4 | 4 | 5 | 5 | 11 | 11 | 14 | 20 |
| Col-0 | 0 | 0 | 2 | 4 | 4 | 8 | 12 | 18 | 24 | 26 | 32 | 36 | 38 | 42 | |
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 3 | syta-1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 5 | 5 | 7 | 9 | 11 |
| Col-0 | 0 | 0 | 0 | 2 | 4 | 5 | 11 | 13 | 16 | 23 | 30 | 32 | 36 | 38 | |
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
Symptomatic plants as percentage of total inoculated plants (50–56 plants of each genotype per assay). Nonparametric survival analysis with right censoring (Wilcoxon test statistic) is p < 0.05 for the three trials shown.
Figure 2.
TVCV, but not CaMV, systemic spread is delayed in syta-1. (A) Coomassie blue-stained SDS-PAGE gels of systemic leaf extracts from four TVCV-inoculated Arabidopsis wt Col-0 or syta-1 (line 1) plants at 10 da post-inoculation. Positions of TVCV CP (17.5 kDa) and molecular weight markers (kDa) are indicated. (B) Semi-quantitative PCR analyses of CaMV genome DNA in systemic leaf extracts from mock inoculated control (C) or CaMV infected (inf) plants at 21 da post-inoculation. Shown are agarose gel analyses of extracts from Arabidopsis wt Col-0 plants and the two independent mutant lines syta-1 (line 1) (syta-11) and syta-1 (line 16) (syta-116). Positions of PCR products of CaMV CP gene and SYTB gene (internal loading control), and of molecular weight markers (nt) are indicated. Equal amounts of systemic leaf extracts were analyzed in each gel in panels (A) and (B).
SYTA regulates the cell-to-cell trafficking of the TuMV and TVCV movement proteins
The Begomovirus MPCaLCuV and MPSqLCV, and the Tobamovirus MPTMV, directly bind to SYTA in vitro, which suggests a potential role for SYTA in virus movement, and we have established that SYTA regulates the cell-to-cell trafficking of MPCaLCuV and MPTMV, when each is transiently expressed as functional fluorescently-tagged proteins in N. benthamiana or Arabidopsis leaf epidermal cells (Lewis and Lazarowitz, 2010). Given our previous findings, the delay we observed in the onset of TVCV disease symptoms and the inhibition of infectivity in syta-1 plants were not unexpected. To directly determine whether SYTA regulated TVCV and TuMV cell-to-cell movement, we used our biolistic bombardment assay to transiently express functional GFP fusions of MPTVCV and TuMV P3N-PIPO (hereafter P3N-PIPOTuMV) in Arabidopsis syta-1 compared to wild type Col-0 plants (Lewis and Lazarowitz, 2010). We used the Golgi marker t-TM-GFP, which contains the transmembrane domain from rat sialotransferase and does not traffic between cells, to determine an optimal amount of plasmid DNA that would produce foci in which ≥95% contained a single fluorescent cell and ≤5% contained only 2 fluorescent cells at 16–20 h post bombardment (Table 3) (Lewis and Lazarowitz, 2010). We also determined the earliest time (~8–20 h post bombardment, depending on the MP) at which each MP would be expressed in isolated single cells (≥95% of foci, with ≤5% having two adjacent GFP-labeled cells) and then assessed MP cell-to-cell trafficking over time using CLSM. We included GFP-MPCaLCuV as a positive control (Lewis and Lazarowitz, 2010). As we previously reported (Lewis and Lazarowitz, 2010; Levy et al., 2013), we did not observe any qualitative differences at the imaging resolution of these MP trafficking assays in wt Col-0 vs. syta-1 plants for each MP tested. Our higher resolution studies of MPTVCV also show quantitative, but no qualitative differences, for MPTVCV-GFP expressed in wt Col-0 vs. syta-1 plants (A. Levy, J. Zheng and SGL, submitted).
Table 3.
MP cell-to-cell trafficking in syta-1 and wild type Col-0.
| MP | Trial | Line | Total foci (cells) | 1 cells (%)a | ≥2cells (%)a | P(χ2)b |
|---|---|---|---|---|---|---|
| GFP-MPCaLCuVc | 1 | wt | 45 (165) | 28.9 | 71.1 | <0.0001 |
| syta-1 | 46 (136) | 50.0 | 50.0 | |||
| 2 | wt | 33 (158) | 21.2 | 78.8 | ||
| syta-1 | 32 (94) | 46.9 | 53.1 | |||
| 3 | wt | 33 (150) | 12.1 | 87.8 | ||
| syta-1 | 30 (74) | 56.7 | 43.3 | |||
| MPTVCV-GFPc | 1 | wt | 25 (124) | 8.0 | 92.0 | <0.0001 |
| syta-1 | 30 (97) | 30.0 | 70.0 | |||
| 2 | wt | 15 (92) | 6.7 | 93.3 | ||
| syta-1 | 25 (68) | 58.8 | 41.2 | |||
| 3 | wt | 17 (77) | 13.8 | 86.2 | ||
| syta-1 | 20 (64) | 30.0 | 70.0 | |||
| 4 | wt | 29 (117) | 13.8 | 86.2 | ||
| syta-1 | 26 (69) | 34.6 | 65.4 | |||
| P3N-PIPOTuMV-GFPc | 1 | wt | 21 (44) | 38.1 | 61.9 | <0.0001 |
| syta-1 | 17 (23) | 64.7 | 35.3 | |||
| 2 | wt | 26 (48) | 42.3 | 57.7 | ||
| syta-1 | 28 (35) | 75.0 | 25.0 | |||
| 3 | wt | 27 (58) | 40.7 | 59.3 | ||
| syta-1 | 29 (33) | 89.7 | 10.3 | |||
| 4 | wt | 27 (47) | 48.1 | 51.9 | ||
| syta-1 | 26 (30) | 88.5 | 11.5 | |||
| t-TM-GFPd | 1 | wt | 57 (60) | 94.7 | 5.3d | ---- |
| 2 | wt | 58 (60) | 96.6 | 3.4d | ||
| 3 | wt | 40 (41) | 97.5 | 2.5d | ||
| 4 | wt | 40 (40) | 100.0 | 0.0d |
Percent fluorescent cells where each MP was in a single cell (1 cell), or moved to 2 or more cells (≥2).
χ2 test for cell-to-cell movement of each MP bombarded into syta-1 vs. wild type Col-0 leaves for all three or four trails shown.
Each MP was assayed for cell-to-cell movement at the following times post bombardment: MPCaLCuV 36–46 h, MPTVCV 24 h, P3N-PIPOTuMV 48 h, MPCaMV 24 h.
Golgi marker: transmembrane domain (TM) of rat sialotranferase fused to GFP. No difference was found in the percentage of foci with 1 vs. 2 labeled cells for syta-1 vs. wild type Col-0. Foci with >2 adjacent t-TM-GFP-labeled cells were never observed. Foci were imaged at 12–24 h post bombardment.
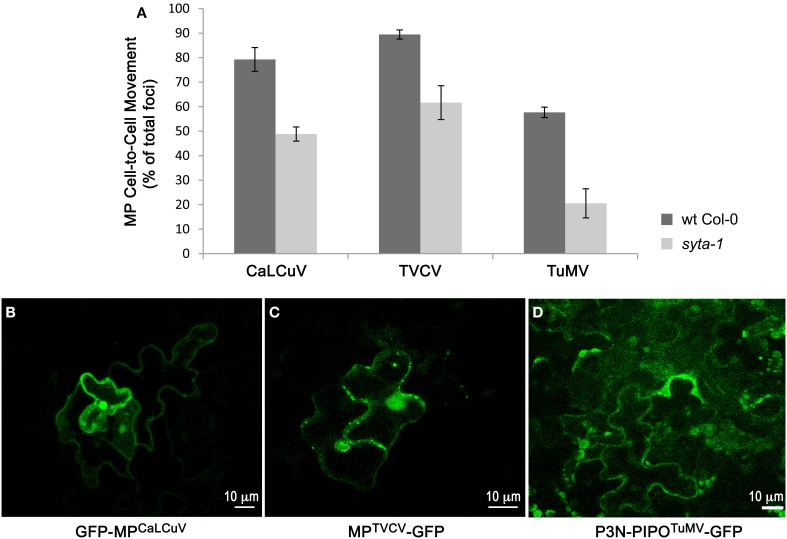
The cell-to-cell trafficking of MPTVCV-GFP and P3N-PIPOTuMV-GFP were both significantly inhibited in syta-1 knockdown lines as compared to wild type Arabidopsis Col-0 (Figure 3, Table 3). Similar to MPTMV-GFP, MPTVCV-GFP moved into surrounding cells in 86–93% of the foci in wild type Col-0 plants. In contrast, MPTVCV-GFP spread to surrounding cells only ~40–70% of the time in syta-1 (Figures 3A,C, Table 3). P3N-PIPOTuMV-GFP moved into surrounding cells in ~50–60% of the foci in wild type plants, similar to what was previously reported in N. benthamiana (Vijayapalani et al., 2012). In contrast, P3N-PIPOTuMV-GFP spread to surrounding cells only 10–35% of the time in syta-1 (Figures 3A,D, Table 3). Similar to what we previously reported, GFP-MPCaLCuV moved into surrounding cells in ~70–88% of foci in wild type Col-0 compared to only ~43–50% of foci in syta-1 plants (Figures 3A,B, Table 3). Consistent with our infectivity studies, these results show that SYTA regulates the cell-to-cell trafficking of MPTVCV and P3N-PIPOTuMV via PD, as well as of MPCaLCuV and MPTMV (Lewis and Lazarowitz, 2010).
Figure 3.
Cell-to-cell trafficking of MPCaLCuV, MPTVCV and P3N-PIPOTuMV is inhibited in syta-1. (A) Percent of total foci in which MPCaLCuV, MPTVCV or P3N-PIPOTuMV moved to 2 or more cells in wild type (wt) Col-0 or syta-1 (line 1) plants. Shown are averages from 3 (MPCaLCuV) or 4 (MPTVCV and P3N-PIPOTuMV) independent trials (Table 3). (B–D) Images of typical individual foci on wt Col-0 bombarded with GFP-MPCaLCuV, MPTVCV-GFP and P3N-PIPOTuMV-GFP show cell-to-cell trafficking at 40 h (MPCaLCuV), 24 h (MPTVCV) or 48 h (P3N-PIPOTuMV) post bombardment.
SYTA is not required for CaMV infectivity
We further examined whether SYTA might have a role in the infectivity and movement of the Caulimovirus CaMV, which is a pararetrovirus. The CaMV dsDNA genome is transcribed into a 35S polycistronic mRNA in the nucleus by host cell RNA polymerase II. Encapsidation in the cytoplasm of this 35S +RNA and the viral-encoded reverse transcriptase activates the reverse transcriptase to synthesize the CaMV dsDNA genome within assembled virus particles. Thus, unlike the Begomoviruses, Tobamoviruses, and Potyviruses, CaMV moves cell-to-cell as virus particles, aided by a Virion-Associated Protein (VAP) that associates with viral CP to translocate virions across PD through tubules formed by MPCaMV (Lazarowitz, 2001; Stavolone et al., 2005).
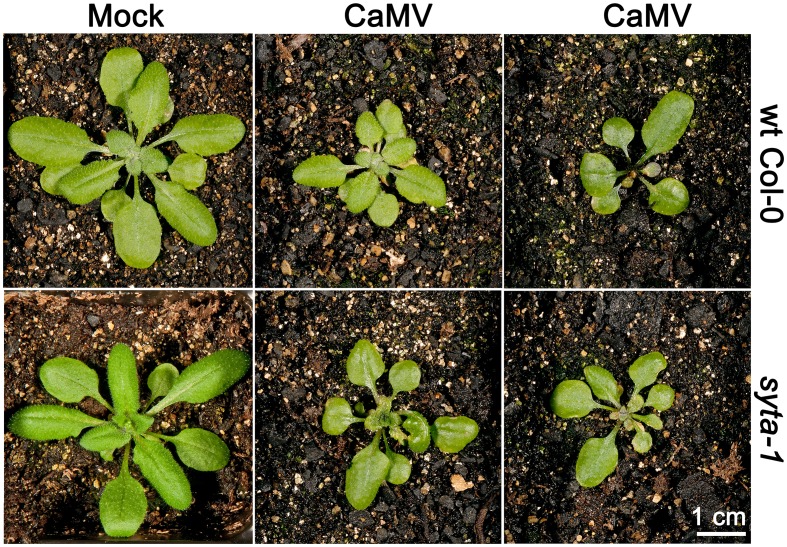
We tested CaMV infectivity on our syta-1 knockdown lines compared to wild type Col-0 plants. Based on the appearance of typical viral systemic disease symptoms and quantifying CaMV DNA levels in systemically infected leaves, CaMV infectivity levels and systemic spread on syta-1 compared to wild type Col-0 plants were not statistically different (Figures 2B, 4, Table 4). In contrast to TuMV and TVCV (Figure 1, Tables 1, 2), and to CaLCuV (Lewis and Lazarowitz, 2010), CaMV systemic disease symptoms first appeared at the same time on both syta-1 and wild type plants, and CaMV infection progressed at the same rate, attaining levels up to 100% infectivity in both syta-1 and wt Col-0 plants. Fitting with this, CaMV DNA accumulated to the same levels in systemically infected leaves from both syta-1 and wt Col-0 plants.
Figure 4.
CaMV produces typical disease symptoms in syta-1 plants. CaMV or mock inoculated wild type Col-0 or syta-1 plants at 16 da post inoculation. Two typical examples of CaMV-infected plants are shown for wt Col-0 and syta-1 plants.
Table 4.
CaMV Infectivity on syta-1 and wild type Arabidopsis Col-0a.
| da post infection (pi) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | ||
| 1 | syta-1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 47 | 49 | 53 | 67 | 77 | 77 | 79 | 84 | 86 | 86 | 91 | 91 |
| Col-0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 9 | 43 | 57 | 77 | 91 | 95 | 95 | 95 | 98 | 98 | 98 | 100 | 100 | |
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 2 | syta-1 | 0 | 5 | 24 | 38 | 74 | 88 | 88 | 98 | 100 | |||||||||||
| Col-0 | 0 | 20 | 45 | 55 | 90 | 95 | 95 | 100 | 100 | ||||||||||||
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||||||
| 3 | syta-1 | 0 | 0 | 0 | 5 | 5 | 13 | 21 | 32 | 37 | 50 | 53 | 55 | 61 | 61 | 61 | 66 | 66 | |||
| Col-0 | 0 | 0 | 0 | 3 | 3 | 16 | 21 | 29 | 39 | 47 | 66 | 68 | 74 | 79 | 79 | 79 | 82 | ||||
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
Symptomatic plants as percentage of total inoculated plants (40–44 plants of each genotype per assay). Nonparametric survival analysis with right censoring (Wilcoxon test statistic) is p = 0.87 for the three trials shown.
MPCaMV forms tubules and does not traffic cell to cell itself. While MPCaMV with an N- or C-terminal GFP tag can form tubules in plant protoplasts, the GFP tag interferes with the formation of the MPCaMV-containing tubules required for CaMV cell-to-cell movement when GFP-MPCaMV or MPCaMV-GFP is expressed in insect cells or in planta, probably due to steric hindrance (Thomas and Maule, 2000). This in vivo tubule formation can be restored by mixing GFP-tagged and untagged native MPCaMV; but, it is difficult to quantify tubule formation at PD due to the background of GFP-tagged MPCaMV aggregates in the cytoplasm and the low efficiency of tubule formation (Thomas and Maule, 2000; Amari et al., 2010). We attempted to assess whether SYTA affected MPCaMV tubule formation by co-bombarding Arabidopsis leaves with GFP-MPCaMV or MPCaMV-GFP mixed with native MPCaMV at ratios of 1:2, 1:1, and 2:1. We observed what appeared to be a small number of MPCaMV-labeled tubules formed at the cell periphery in each of the co-bombarded samples, which were similar in appearance to GFP-MPGFLV tubules in leaf epidermal cells (Thomas and Maule, 2000; Amari et al., 2010). We also observed cytoplasmic MPCaMV that likely was the previously reported GFP-tagged MPCaMV aggregates (data not shown) (Thomas and Maule, 2000; Amari et al., 2010). Consistent with our CaMV infectivity assays, we found no obvious differences when we co-bombarded a 1:1 mix of MPCaMV-GFP/MPCaMV into leaves from syta-1 and wild type Col-0 plants. Take together, our findings suggest that, unlike Begomoviruses, Tobamoviruses and Potyviruses, SYTA does not regulate CaMV cell-to-cell movement.
Discussion
Viral movement proteins are essential to coordinate replication of viral genomes with their directed transport to PD, and to facilitate their transport through PD for cell-to-cell spread. A single MP alone may execute these intracellular and intercellular genome transport functions, as is the case for the Tobamoviruses TMV and TVCV; or the cell-to-cell MP may cooperate with additional viral-encoded proteins to coordinate these events, such as occurs for the Begomovirus CaLCuV, the Potyvirus TuMV, and the Caulimovirus CaMV. Whichever the case, the ultimate path through the cell wall is a common one: all plant viruses exploit PD. Yet, how viral genomes reach PD and, once there, how MPs mechanistically alter PD gating, remain unknown. Our previous studies identified Arabidopsis SYTA as a plant synaptotagmin that interacts with the MPs encoded by the Begomovirus CaLCuV and the Tobamovirus TMV, and showed that SYTA regulates endocytosis and marks an endosomal recycling pathway that we proposed traffics these MPs to PD (Lewis and Lazarowitz, 2010). We have now expanded our analyses to include viruses from other distinct families, namely the Potyvirus TuMV and the Caulimovirus CaMV, as well as a Tobamovirus that infects Arabidopsis (TVCV), in order to test the hypothesis that SYTA has a central role in regulating cell-to-cell movement for a range of diverse plant viruses. Our findings show that SYTA regulates the cell-to-cell movement of TVCV and TuMV, in addition to CaLCuV, despite their distinct modes of movement; but, it does not regulate CaMV movement.
What is the precise role of SYTA in regulating virus movement? Although the mechanistic details remain to be elucidated, the known functions of synaptotagmins, and current knowledge about the different strategies for cell-to-cell movement employed by the plant viruses used in our studies, do provide some clues. While all plant viruses move cell to cell via PD, a variety of schemes have been described for how plant viruses from different families accomplish this feat. These models reflect different viral replication strategies, and whether the virus moves in an encapsidated form (i.e., is CP required for movement) or its MP interacts with additional viral proteins to traffic progeny viral genomes from replication sites to, and through, PD. Based on biophysical considerations, such MP-genome complexes, regardless of whether they contain additional viral-encoded proteins, are too large to freely diffuse from sites of viral replication, be these nuclear or cytoplasmic, through the cytosol to reach PD. Hence, trafficking from replication sites to PD must be an active process that involves the endomembrane network and vesicle transport, and/or the cytoskeletal system (Oparka, 2004; Schoelz et al., 2011).
Tobamoviruses
The Tobamoviruses TMV and TVCV, with their +RNA genomes that are replicated in the cytoplasm, represent potentially the simplest scenario. These viruses encode a single 30-kDa MP and do not require CP for local cell-to-cell spread. In principle, moving simply the +RNA genome into new cells would suffice to initiate infection. Consistent with this, MPTMV and MPTVCV can bind RNA and localize to PD, and MPTMV was the first movement protein shown to alter PD gating (Wolf et al., 1989; Citovsky et al., 1990; Heinlein et al., 1998; Levy et al., 2013). These findings, together with detailed studies of the subcellular distribution of MPTMV and MPTVCV during the time course of infection, are consistent with the model that the Tobamovirus 30-kDa MP binds progeny +RNA genomes at their site(s) of replication and traffics them to PD, where MP then acts to increase PD permeability and thereby facilitate the transport of viral genomes into neighboring cells to initiate new infections.
A range of studies underscore the importance of the ER and endomembrane system in this process. TMV and TVCV replicate in the cytoplasm at specific replication sites, which are inclusions of viral RNA, MP and replicase that form at cortical ER membrane sites (Heinlein et al., 1998; Schoelz et al., 2011; Levy et al., 2013). Forming viral replication sites in association with host membranes is a common feature of +RNA viruses that infect animals and plants, and is probably a strategy to protect viral genomes from recognition and destruction by host innate defenses (den Boon and Ahlquist, 2010). Both MPTMV and MPTVCV have been shown to associate with ER membranes (Heinlein et al., 1998; Fujiki et al., 2006; Levy et al., 2013), and a variety of studies indicate that this association is necessary for these Tobamovirus MPs to target viral genomes to PD for cell-to-cell spread. Expression of a dominant-negative form of the GTPase Sar1 or low concentrations of Brefeldin A (BFA), both of which disrupt vesicle transport from the ER to the Golgi, do not inhibit MPTMV trafficking to PD or virus infection; but, high concentrations of BFA that disrupt the ER network do prevent MPTMV from reaching PD (Heinlein et al., 1998; Tagami and Watanabe, 2007; Wright et al., 2007). These findings, and mutational analyses of MPTMV, lead to the conclusion that although MPTMV does not traffic through the secretory pathway to reach PD, the ER and endomembrane system are necessary for MPTMV-genome complexes to target to PD and for TMV cell-to-cell spread (Schoelz et al., 2011). Our demonstration that SYTA and endosomal recycling to the plasma membrane regulate the ability of MPTMV and MPTVCV to reach and alter PD, and TVCV cell-to-cell movement (Lewis and Lazarowitz, 2010) (and our findings here), together with the subcellular distribution of MPTVCV during the course of virus infection and interaction of MPTVCV and SYTA in planta (Levy et al., 2013; A. Levy, J. Zheng, and SGL, submitted), lead to the same conclusion for MPTVCV. This unifies the findings for TMV and TVCV by showing that the ER and vesicle trafficking are required for both MPTMV and MPTVCV to traffic to PD and for TVCV, as well as TMV, intercellular movement. In addition, functional studies using our dominant negative mutant SYTAΔC2B show that the SYTA-regulated vesicle trafficking pathway is distinct from the secretory pathway (Lewis and Lazarowitz, 2010). Thus, neither MPTVCV nor MPTMV traffic to PD via the secretory pathway.
In contrast to the common requirement for the ER and vesicle trafficking, TMV and TVCV differ in their requirements for the cytoskeleton in virus movement. The use of pharmacological agents has shown that TMV requires intact microfilaments for virus cell-to-cell movement, although there are disagreements about the role of microtubules in TMV intercellular spread (Harries et al., 2009b; Niehl and Heinlein, 2010; Schoelz et al., 2011). The TMV 126-kDa replicase forms cytoplasmic inclusions that associate with actin microfilaments and, thus, it has been proposed that TMV replication complexes that contain both MPTMV and replicase traffic via the ER-actin network to PD for virus cell-to-cell spread (Harries et al., 2009b). In contrast to TMV, disrupting either the actin cytoskeleton or microtubules has no effect on TVCV cell-to-cell spread, nor does TVCV replicase associate with microfilaments. Thus, the cytoskeleton per se is not necessary for TVCV infection and spread (Harries et al., 2009b). In agreement with this, we have shown that the actin cytoskeleton is disrupted and becomes disorganized during TVCV infection, and MPTVCV does not associate with microtubules (Levy et al., 2013). This further underscores the importance of the ER and endomembrane system in TVCV intracellular and intercellular trafficking. Indeed, a striking distinction between TMV and TVCV is that MPTVCV, in addition to localizing to ER membrane sites and PD, is also found in novel F-actin-containing nuclear filaments that associate with chromatin, and this nuclear function of MPTVCV is necessary to promote efficient TVCV cell-to-cell movement (Levy et al., 2013).
Begomoviruses
Like Tobamoviruses, the Begomoviruses (Geminivirus family) CaLCuV and SqLCV do not require CP to move cell to cell (Gardiner et al., 1988; Ingham et al., 1995; Pooma et al., 1996; Qin et al., 1998). However, in contrast to TMV and TVCV, CaLCuV and SqLCV encode a nuclear shuttle protein NSP in addition to the cell-to-cell movement protein (MPCaLCuV or MPSqLCV), both of which are essential for virus intercellular transport and systemic infection (Lazarowitz and Beachy, 1999). This requirement for two proteins to traffic newly replicated progeny genomes to and through PD is the consequence of Begomoviruses having single strand DNA (ssDNA) genomes that are replicated in the nucleus and, thus, need to be efficiently transported across the nuclear envelope, as well as to and through PD (Pascal et al., 1994; Sanderfoot and Lazarowitz, 1995; Sanderfoot et al., 1996; Ward et al., 1997). Mutational analyses of CaLCuV or SqLCV MP and NSP, combined with transient expression studies in cultured insect cells or plant protoplasts, subcellular fractionation studies and electron microscopy lead to a model in which Begomovirus NSP and MP cooperatively act to move progeny viral genomes from their nuclear sites of replication to PD for intercellular transport (Pascal et al., 1994; Sanderfoot and Lazarowitz, 1995; Sanderfoot et al., 1996; Ward et al., 1997; Ward and Lazarowitz, 1999; Carvalho and Lazarowitz, 2004). According to this model, NSP binds progeny ssDNA genomes in the nucleus and transports them between the nucleus and cytoplasm. MP traps these NSP-genome complexes in the cytoplasm and redirects them to and through PD into neighboring cells, following which NSP is proposed to efficiently target the viral ssDNA genome into the nucleus to initiate new rounds of infection (Sanderfoot and Lazarowitz, 1996).
Despite these differences from Tobamoviruses in terms of replicating in a distinct compartment (the nucleus) and requiring an additional protein (NSP) to bind progeny genomes, MPCaLCuV and MPSqLCV, like MPTMV and MPTVCV, act to direct newly replicated viral genomes from their sites of replication to PD; and, also like TMV and TVCV, a variety of studies demonstrate the importance of the ER and endomembrane system in this process. MPSqLCV co-fractionates with ER membranes in extracts from infected pumpkin plants and, based on immunolabeling studies, it localizes to ER-derived tubules that extend toward and cross the walls of SqLCV-infected cells at what may be immature PD (Ward et al., 1997). Fitting with this, MPSqLCV and MPCaLCuV each target to cortical regions of the ER when transiently expressed in insect cells and/or plant protoplasts (Sanderfoot and Lazarowitz, 1995, 1996; Ward et al., 1997; Carvalho and Lazarowitz, 2004). Mutational analyses show that this targeting to cortical ER is essential for virus infection and systemic spread; yet, both MPSqLCV and MPCaLCuV lack signal sequences and behave like peripheral membrane proteins (Sanderfoot and Lazarowitz, 1995; Sanderfoot et al., 1996; Carvalho and Lazarowitz, 2004; Lewis and Lazarowitz, 2010). Furthermore, as we demonstrated for MPTMV and MPTVCV, SYTA regulates the ability of MPCaLCuV to traffic to and modify PD, and CaLCuV cell-to-cell movement, via an endosomal recycling pathway that is distinct from the secretory pathway (Lewis and Lazarowitz, 2010). Hence, just like the Tobamoviruses TMV and TVCV, the ER and vesicle trafficking are necessary for MPCaLCuV to traffic viral genomes to PD for cell-to-cell movement, and MPCaLCuV does not appear to reach PD via the secretory pathway.
Potyviruses
The Potyviruses have a single +RNA genome that is replicated in the cytoplasm. Thus, like the Tobamoviruses, moving simply the +RNA genome into new cells should, in principle, suffice to initiate infection. Nevertheless, studies mainly on Tobacco etch virus (TEV) and TuMV have shown that the mechanism employed by Potyviruses to replicate and target viral genomes to PD is more complex as it involves several viral-encoded proteins: CI; the coat protein CP; and the recently identified cell-to-cell movement protein PIPO (pretty interesting Potyvirus ORF), which is translated by ribosomal frameshifting within the Potyvirus polyprotein as P3N-PIPO (Dolja et al., 1995; Carrington et al., 1998; Wei et al., 2010a; Vijayapalani et al., 2012). CI is an RNA-binding protein with helicase and ATPase activities, and is essential for Potyvirus genome replication (Fernandez et al., 1995, 1998; Merits et al., 1998). CI also forms cylindrical inclusions (hence its name) in the cytoplasm adjacent to PD in infected cells, and conical structures that extend into PD (Rodriguez-Cerezo et al., 1997; Roberts et al., 1998; Sorel et al., 2014). Mutational analyses of TEV CP and CI (Dolja et al., 1994, 1995; Carrington et al., 1998); immunolabeling of CP and CI in cells infected with Tobacco vein mottle virus or Pea Seed-borne mosaic virus (Rodriguez-Cerezo et al., 1997; Roberts et al., 1998); and transient expression studies of fluorescently-tagged TuMV CI, CP and P3N-PIPO in N. benthamiana leaf cells (Wei et al., 2010a; Vijayapalani et al., 2012) lead to a model in which newly replicated viral CP-genome complexes move cell-to-cell and CI cooperatively interacts with P3N-PIPO, similar to NSP and MP in Begomovirus movement, to traffic these CP-genome complexes from viral replication sites to, and through, PD. According to this model, CI at perinuclear viral replication sites interacts with viral CP-genomes complexes, which may or may not be virions, and transports these to P3N-PIPO-modified PD. P3N-PIPO alters PD permeability and interacts with CI within inclusions to coordinate the formation of conical structures that are anchored within PD via P3N-PIPO, and through which viral CP-genome complexes are transported into adjacent cells (Sorel et al., 2014). In support of this model, Potyvirus CP is associated with the cylindrical inclusions, and viral CP and RNA have been localized to the CI conical structures that extend from and within PD (Rodriguez-Cerezo et al., 1997; Roberts et al., 1998; Sorel et al., 2014). CI also interacts with P3N-PIPO, and P3N-PIPO localizes to PD and affects PD gating (Wei et al., 2010a; Vijayapalani et al., 2012).
As for Tobamoviruses and Begomoviruses, a range of studies point to the importance of the ER and endomembrane system in the intracellular and intercellular trafficking of the Potyvirus genome. Potyvirus replication involves the sequential recruitment of ER-derived vesicles and the chloroplast outer membrane to create what has been described as viral replication factories supported by a globular structure in the perinuclear region of the cell (Wei and Wang, 2008; Wei et al., 2010b; Grangeon et al., 2012). In considering the coordination of virus replication and movement, and the multiple roles of CI, it is important to note that Potyviruses are in the Picornavirus superfamily and encode a single ORF, which is translated into a polyprotein that is proteolytically processed by three viral-encoded proteinases. The efficiencies of cleavage at the different processing sites vary, and precursors and their fully processed individual proteins can carry out different functions (Carrington et al., 1993; Lazarowitz, 2001). In particular, evidence supports the existence of CI-6K-VPg-NIa(Pro) and 6K-VPg-NIa(Pro) precursors, in which NIa(Pro) is the proteinase that processes these precursors, and the cleavage of CI from 6K-VPg-NIa(Pro) occurs slowly. The 6K-VPg-NIa(Pro) precursor targets the viral genome to initiate the replication process at ER sites, and CI would also be targeted to these sites through its linkage to 6K prior to cleavage (Carrington et al., 1993; Merits et al., 2002). The TEV and TuMV 6K protein (a.k.a. 6K2), which is an integral membrane protein, induces the proliferation of ER vesicles at ER exit sites that associate with cis-Golgi stacks (Restrepo-Hartwig and Carrington, 1994; Schaad et al., 1997; Wei and Wang, 2008). These 6K-associated ER-derived vesicles are proposed to then traffic to the outer chloroplast membrane, where they induce membrane invagination to form sites for potyvirus replication (Wei et al., 2010b). Transient expression studies of fluorescently-tagged TuMV 6K in N. benthamiana leaf cells and the localization of 6K in infected cells, the expression of compartment-specific markers and of dominant-negative mutants that disrupt COPI- or COPII-dependent vesicle trafficking, and the use of BFA, have lead to the suggestion that Potyviruses initiate translation of the viral genome on the ER and form 6K-vesicles at ER exit sites, and these vesicles then traffic via the Golgi to chloroplasts for virus replication. Thus, the early secretory pathway appears to be important for the formation of Potyvirus replication complexes (Wei and Wang, 2008; Wei et al., 2010b). In addition, 6K-YFP-labeled vesicles were observed to traffic along actin filaments, and this was inhibited by latrunculin B or a dominant-negative “tail” mutant of myosin XI-K. Thus, the actin cytoskeleton and myosin motors also appear to be required for 6K vesicles to traffic from the ER to chloroplast sites of virus replication (Wei et al., 2010b).
The ER and vesicle trafficking have also been implicated in CI-mediated transport of viral CP-genome complexes from these perinuclear replication sites to PD-associated inclusions, and in the targeting of P3N-PIPO to PD as well. Expression of a dominant-negative Sar1 mutant, or a high concentration of BFA, both inhibited the trafficking of fluorescently-tagged P3N-PIPOTuMV to PD, and of tagged PDLP1, a plasmodesmal type I membrane receptor-like protein that traffics to PD via the secretory pathway (Amari et al., 2010; Wei et al., 2010a). However, in contrast to PDLP1, which was retained in the reticulate ER network in the presence of the Sar1 mutant, P3N-PIPOTuMV was cytoplasmic when co-expressed with this Sar1 mutant (Wei et al., 2010a). This fits with the fact that P3N-PIPO is not predicted to contain a membrane-binding or membrane-spanning domain (Wei et al., 2010a; Vijayapalani et al., 2012), and suggests that although the ER and secretory pathway are necessary for P3N-PIPOTuMV to target to PD, P3N-PIPOTuMV itself does not traffic via the secretory pathway to reach PD. Thus, it appears that P3N-PIPOTuMV may depend on a protein(s) that traffics through the secretory pathway in order to reach PD. Consistent with CI also not having predicted membrane-binding or membrane-spanning domains, and CI requiring P3N-PIPOTuMV to localize to PD, neither BFA nor the SAR1 mutant inhibit CI self-association (aggregates) in the cytoplasm; but, both do inhibit CI localizing to PD when co-expressed with P3N-PIPOTuMV (Wei et al., 2010a). Further underscoring the importance of the ER and vesicle trafficking in P3N-PIPOTuMV targeting to PD, as well as the differences from PDLP1 trafficking, the actomyosin system does not appear to be required, at least for P3N-PIPOTuMV to reach PD: latrunculin B or the expression of dominant-negative myosin XI-K or VIII-1 tail mutants did not inhibit P3N-PIPOTuMV targeting to PD, although they did inhibit PDLP1 targeting to PD (Wei et al., 2010a).
As to the suggestion that P3N-PIPOTuMV would interact with another protein(s) to reach PD, a recent study showed that P3N-PIPOTuMV interacts in planta with Arabidopsis PCaP1, a hydrophilic cation-binding protein that stably associates with the plasma membrane via N-myristoylation (Vijayapalani et al., 2012). Although its function is unknown, PCaP1 interacts with Ca2+-bound calmodulin and phosphatidylinositol phosphates, and has been proposed to have a role in cell signaling (Nagasaki et al., 2008). BiFC studies show that P3N-PIPOTuMV interacts with PCaP1 at the plasma membrane and in PD. Furthermore, TuMV cell-to-cell spread and systemic infection are severely inhibited in an Arabidopsis pcap1 knockout line; thus, PCaP1 is necessary for TuMV infection (Vijayapalani et al., 2012). Based on these findings, it has been proposed that PCaP1 interacts with P3N-PIPO to potentially anchor the movement complex to the plasma membrane, from which PCaP1-bound P3N-PIPO can direct CI-bound viral RNPs (CP-genome complexes) to PD to move the viral RNPs into neighboring cells (Vijayapalani et al., 2012). The pathway by which CI-bound viral RNPs traffic to the cell periphery is still unknown, as is how PCaP1-bound P3N-PIPO would relocate from the plasma membrane to PD. In this regard, a recent study using pharmacological agents and dominant-negative mutants has implicated both the secretory pathway and the actomyosin system in TuMV cell-to-cell spread (Agbeci et al., 2013).
Caulimoviruses
Based on their proposed modes of cell-to-cell movement, plant viruses have historically been characterized as “non-tubule forming,” which do not morphologically alter PD in a radical manner, or “tubule forming,” which drastically alter PD morphology by forming MP-containing large tubules that traverse PD and eliminate the desmotubule, which is the appressed ER at the PD core (Maule, 2008; Schoelz et al., 2011). The so-called tubule-forming viruses, as typified by CaMV, the Tospovirus Tomato spotted wilt virus (TSWV), and the Secovirus Grape fanleaf virus (GFLV), among others, move the viral genome as assembled icosahedral virions (CaMV, GFLV) or subviral particles (nucleocapsids and associated viral RNA-dependent RNA polymerase, e.g., TSWV). However, based on current molecular and cellular studies, we suggest that Tobamoviruses, Begomoviruses and Potyviruses can be viewed as representing a continuum of increasing complexity in virus movement. For TMV and TVCV, MP directly binds the viral genome to target it to PD and facilitate its intercellular movement. PD may appear to be swollen, but are not morphologically altered. In the case of SqLCV and CaLCuV, MP binds and directs NSP-ssDNA complexes, which are not virus particles, to and through PD, potentially guiding these complexes along tubules that may be modified desmotubule, although this latter point requires further investigation for viruses other than the phloem-limited SqLCV. For TEV and TuMV, CP-genome complexes, which may or may not be virions, are bound to CI, and MP (P3N-PIPO) targets CI to PD and enables CI to form cone-like extensions within PD, through which CP-genome complexes are proposed to move. PD are still recognizable as such, and MP is proposed to anchor the CI-containing cones to the PD. The state of the desmotubule is not clear; but, the fact that P3N-PIPO, like other viral MPs, can alter PD gating suggests that PD integrity is maintained. The tubule-forming viruses such as CaMV would represent the next and most extreme step in this progression: MP dramatically alters PD structure, including eliminating the desmotubule, to create large tubules that extend between cells and through which assembled virions or subviral particles tunnel into adjacent cells (Niehl and Heinlein, 2010; Schoelz et al., 2011). These differences, to a large extent, appear to reflect the different replication strategies of TVCV, CaLCuV, TuMV, and CaMV.
How the so-called tubule-forming MPs target to PD and form tubules is not understood, although the plasmodesmal integral membrane protein PDLP1 has been shown to have a role in the assembly of both MPCaMV and MPGFLV tubules at PD (Amari et al., 2010). Given this, it is not surprising that studies have implicated the endomembrane system in MPCaMV and MPGFLV tubule formation, although not in MPCaMV associating with the plasma membrane when expressed in plant cell protoplasts (Huang et al., 2000; Carluccio et al., 2014). Transient expression of fluorescently-tagged MPCaMV or other tubule-forming MPs in plant cell protoplasts is an established model for investigating the requirements for tubule formation. MPCaMV, although it does not have a transmembrane domain, accumulates at localized areas of the plasma membrane in protoplasts, from which tubules eventually extend outwards at the cell surface (Huang et al., 2000; Carluccio et al., 2014). In this model system, cytoskeletal assembly inhibitors do not affect tubule formation by MPCaMV or MPGFLV, or the accumulation of MPCaMV at localized areas of the plasma membrane; but, high concentrations of BFA that disrupt the endomembrane system do inhibit tubule formation (Huang et al., 2000; Laporte et al., 2003).
Interactions of MPCaMV with pectin methylesterase (Chen et al., 2000) and PRA1, a Rab GTPase receptor that localizes to prevacuolar compartments (PVCs) (Huang et al., 2000), have been cited as further implicating the endomembrane system in tubule formation, and a recent study provides evidence for MPCaMV trafficking in the endocytic pathway (Carluccio et al., 2014). In this study, fluorescently-tagged MPCaMV, when transiently expressed in protoplasts, was found on endosomal vesicles at ~10 h post transfection, shortly after MPCaMV was observed to accumulate at localized areas of the plasma membrane and tubule elongation from the cell surface began (~6–8 h post transfection). Labeling with the lipophilic dye FM4-64 suggested that a subset of the MPCaMV-labeled vesicles are derived from the plasma membrane (Carluccio et al., 2014). The authors identified three conserved YXXΦ Tyr-sorting motifs in MPCaMV. Such motifs are recognized by the μ-subunit of adaptor protein (AP) complexes for cargo selection on clathrin-coated vesicles, and have previously been identified in MPGFLV and in one of the so-called triple gene block movement proteins encoded by the Pomovirus Potato mop-top virus (Laporte et al., 2003; Haupt et al., 2005). MPCaMV YXXΦ motifs appeared to be essential for infectivity and, consistent with endocytic trafficking being important for infection, CaMV did not infect ben1(min7) and ben2(vps45) plants, two Arabidopsis mutant lines with defects in the trafficking of early endosomes through the TGN/EE, a dynamic tubular-vesicular trans-Golgi network in plant cells that is the main sorting hub for secretory and vacuolar traffic, as well as the destination for endocytic cargo (Zeenko et al., 2002). Based on mutational studies, at least one YXXΦ motif is needed for MPCaMV to form tubules at the protoplast surface, to target to PD in leaf epidermal cells, and to interact in vitro with the μ1 adaptin subunit of Arabidopsis AP-1, which is mainly at the trans-Golgi; but, MPCaMV lacking all three YXXΦ motifs still interacts with PDLP1 in vitro. Interestingly, the MPCaMV triple YXXΦ mutant still appeared to associate with the plasma membrane at 8 h post transfection, but did not subsequently localize to endosomes; rather, it appeared in cytoplasmic aggregates, at this and later times post-transfection, that did not label with FM4-64 (Carluccio et al., 2014). Further suggesting a role for endocytic trafficking in MPCaMV function, blocking AP-2-mediated endocytosis with tyrphostin A23 caused MPCaMV to remain at the plasma membrane of protoplasts in a diffuse pattern and not appear on endosomes. In addition, a high concentration of BFA caused MPCaMV to accumulate in large FM4-64 labeled internal patches, and a subset of MPCaMV-marked endosomes co-localized with RabF2b GTPase (ARA7), a marker for MVBs/PVCs (Carluccio et al., 2014). Based on these findings, MPCaMV was proposed to reach the plasma membrane by an unknown mechanism, following which it traffics via an endocytic recapture pathway to reach PD and interact with PDLP1 to form tubules for virus intercellular movement. It was also proposed that excess MPCaMV is targeted for degradation via MVBs (Carluccio et al., 2014). Verification of this model will require more direct studies and further characterization of endosomal trafficking pathways in plant cells, which are complex and, at times, controversial (Chen et al., 2000; Ryabova et al., 2002, 2004; Ueki and Citovsky, 2005; Craxton, 2007; Thiebeauld et al., 2009). Nevertheless, our findings show that whatever endocytic route MPCaMV may use to reach PD, this is distinct from the SYTA-regulated recapture pathway that our studies have identified (Lewis and Lazarowitz, 2010) (Levy, Zheng and SGL, submitted).
A working model for SYTA action in virus movement
As underscored by these four and other proposed schemes for virus movement, progress has been made in identifying viral proteins required for movement and their interacting partners, but we still do not understand the mechanisms by which MPs reach PD and alter their permeability or the biophysics of moving viral genome-protein complexes. Our goal here was to examine whether SYTA function might be a general requirement for plant virus cell-to-cell movement. We found that SYTA acts as a common mechanism to regulate plant virus the cell-to-cell movement, but not a universal one. Synaptotagmins are important for membrane trafficking in eukaryotic cells, acting to regulate vesicle fusion at the plasma membrane for exocytosis and endocytosis (Chapman, 2008; Moghadam and Jackson, 2013). Consistent with the established roles of synaptotagmins in animal cells, SYTA localizes to the plasma membrane in plant cells (Schapire et al., 2008; Lewis and Lazarowitz, 2010), and our studies have shown that SYTA regulates endocytosis and an endocytic recycling pathway to the plasma membrane (Lewis and Lazarowitz, 2010). Importantly, SYTA directly interacts with MPTMV and MPTVCV, and with MPCaLCuV and MPSqLCV, and our functional studies show that the dominant-negative mutant SYTAΔC2B, which is defective in mediating endocytosis, must be at the plasma membrane in order to interfere with MPTMV and MPCaLCuV cell-to-cell trafficking (Lewis and Lazarowitz, 2010; A. Levy, J. Zheng, and SGL, submitted). Hence, the SYTA-regulated recycling pathway at the plasma membrane is necessary for these distinct Tobamovirus and Begomovirus MPs to alter PD gating and transport virus genomes through PD. This leads us to propose that SYTA acts to target viral MPs and their genome cargos to PD via this endocytic recapture pathway. This model does not preclude an additional role for SYTA in the mechanism by which MPs alter PD permeability. Without an available syta null mutant, we cannot conclude whether SYTA is both necessary and sufficient for MPCaLCuV, MPTVCV, and P3N-PIPOTuMV to reach and alter PD or other Arabidopsis SYTs may contribute in an overlapping manner to this process. Our characterization of the remaining four Arabidopsis SYTs shows that SYTA, SYTB and SYTC each have distinct patterns of expression, and that neither SYTB nor SYTC appears to mediate plant virus cell-to-cell movement via PD (H. Shimada-Beltran and SGL, unpublished; A. Uchiyama, J. Zheng, P. Javia, and SGL, in preparation). Our studies further show that SYTE, like SYTA, is highly expressed in all cell types and at all stages of development in Arabidopsis. Thus, it remains possible that SYTE could have an overlapping function with SYTA in regulating virus intercellular movement. Additional studies are required to clarify this point.
How can our finding that SYTA is needed for TVCV, CaLCuV, and TuMV MP function and cell-to-cell movement, but not for CaMV infectivity and spread, inform our thinking? SYTA regulates the cell-to-cell trafficking of MPTVCV and P3N-PIPOTuMV via PD (Figure 3, Table 3), as well as that of MPCaLCuV and MPTMV (Lewis and Lazarowitz, 2010). P3N-PIPOTuMV interacts with PCaP1, and this interaction is proposed to anchor P3N-PIPOTuMV to the plasma membrane, from which PCaP1-bound P3N-PIPOTuMV with its cargo (CI-bound viral RNPs) is directed to PD. SYTA directly interacts with MPCaLCuV, MPTMV and MPTVCV (Lewis and Lazarowitz, 2010; Levy, Zheng, and SGL, submitted), but we do not know whether it interacts with P3N-PIPOTuMV. However, PCaP1 is not required for Tobamovirus infection: Oilseed rape mosaic virus, which like TVCV is a subgroup 3 Tobamovirus, is equally infectious on an Arabidopsis pcap1 knockout line and wild type Col-0 plants (Vijayapalani et al., 2012). Hence, the common link here is SYTA. P3N-PIPOTuMV requires the secretory pathway to reach PD, although it does not itself traffic via the secretory pathway to PD (Wei et al., 2010a). PCaP1 is a hydrophilic protein that stably associates with the plasma membrane via N-myristoylation (Vijayapalani et al., 2012) and, thus, it would not reach the plasma membrane via the secretory pathway. However, SYTA does traffic to the plasma membrane via the secretory pathway (Lewis and Lazarowitz, 2010). Therefore, fitting with our model for SYTA targeting MPTMV and MPCaLCuV and their genome cargos to PD, we propose that P3N-PIPOTuMV binds to PCaP1 at the plasma membrane, following which the SYTA-regulated recycling pathway directs this complex, with its cargo, to PD.
MPCaMV is proposed to reach the plasma membrane by an unknown mechanism and then traffic via an endocytic recapture pathway to reach PD, where it interacts with PDLP1 to form tubules for virus intercellular movement (Amari et al., 2010; Carluccio et al., 2014). Our results (Figure 2B, Table 4) suggest that if MPCaMV uses an endosomal recycling pathway to traffic from the plasma membrane to PD, this pathway is distinct from the SYTA-regulated recycling pathway. The key distinctive features of MPCaMV and CaMV movement—compared to TVCV, CaLCuV, and TuMV—are that MPCaMV assembles into large tubules that dramatically alter PD by eliminating the desmotubule; MPCaMV interacts with the PD transmembrane protein PDLP1, which is important for tubule assembly within PD; and MPCaMV tubules dramatically widen what was the PD channel so that CaMV virions can traffic cell to cell, but it does not direct these virus particles, and hence the viral genome, to PD (Harries et al., 2009a; Schoelz et al., 2011). Given the large size of CaMV icosahedral virus particles, it seems that a specialized machinery and cytoskeletal-guided movement, rather than a vesicle trafficking pathway, would be better suited to transporting the encapsidated genome to PD. Indeed, the CaMV P6 protein interacts with both viral CP and MPCaMV (Himmelbach et al., 1996; Hapiak et al., 2008), and the available evidence suggests that aggregates or inclusions of P6 may transport CaMV virions along microfilaments to the assembled MPCaMV tubules, where viral-encoded VAP, interacting with CP and MPCaMV, mediates guidance of virions through the tubules and into neighboring cells (Stavolone et al., 2005; Harries et al., 2009a). Neither the endomembrane system or an intact cytoskeleton appear to be required for MPCaMV to reach the plasma membrane, at least in protoplasts, and MPCaMV appears to be recruited onto endosomes through an interaction of its Tyr-sorting motifs with AP-2 at the plasma membrane (Huang et al., 2000; Carluccio et al., 2014). Thus, it seems that SYTA is not involved in MPCaMV reaching the plasma membrane, nor would an interaction with SYTA be needed to direct MPCaMV onto endosomes, although, in principle, SYTA could mediate the fusion at the plasma membrane to create these endosomes. However, all of our data indicate that the important step is the SYTA-regulated endosomal recycling pathway directing MP-containing complexes from the plasma membrane to the PD channel for genome transport into neighboring cells. Thus, since MPCaMV interacts with a PD transmembrane protein (PDLP1) and becomes a physical component of the channel, it may be that MPCaMV trafficking to PD involves pathways that are independent from the SYTA-regulated endosomal recapture pathway. It is possible that MPCaMV tubules, by so drastically altering the channel, including removing the desmotubule, render the channel unrecognizable as a “PD,” and hence the SYTA-regulated endosomal recycling pathway does not traffic macromolecular complexes to these sites. A corollary of this is that recognition of the appropriate vesicle docking site(s) at PD may require an intact desmotubule and/or its continuity with the cortical ER. Finally, an intriguing, and not mutually exclusive, possibility is that beyond simply delivering MPs and their cargos to PD, the interaction with SYTA may also be involved in MP activity to alter PD permeability. Our current studies are exploring these possibilities.
Author contributions
Asako Uchiyama, Harumi Shimada-Beltran, Amit Levy, Judy Y. Zheng and Sondra G. Lazarowitz designed research and analyzed data. Asako Uchiyama performed the TVCV and CaMV infectivity assays, and all of the movement assays, and statistical analyses of the data. For the P3N-PIPOTuMV movement assays, Asako Uchiyama worked with and supervised Parth A. Javia. Harumi Shimada-Beltran and Judy Y. Zheng carried out TuMV infectivity assays and statistical analyses of the data. Judy Y. Zheng was supervised by Asako Uchiyama and Sondra G. Lazarowitz. Amit Levy performed statistical analyses of P3N-PIPOTuMV movement assays and sqPCR analyses of CaMV DNA in systemic leaf extracts. Sondra G. Lazarowitz supervised all aspects of this work. Amit Levy and Sondra G. Lazarowitz made the figures and tables for this manuscript. Sondra G. Lazarowitz wrote the manuscript with contributions from Amit Levy and Harumi Shimada-Beltran.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Andrew Maule, Dr. Thomas Hohn and Dr. Vitaly Citovsky for graciously providing the infectious clones of TuMV (pGreenTuMV), CaMV (pCa122), and TVCV (pTVCV50), respectively. We also thank Dr. W. Alan Miller for generously providing p35S::P3N-PIPO-GFP cloned in the plant expression vector pJ4GFP-XB for the P3N-PIPOTuMV movement assays. This research was supported by Public Health Service grant AI-066054 from the National Institute of Allergy and Infectious Disease to Sondra G. Lazarowitz, and Ruth L. Kirschstein Postdoctoral National Research Service Award AI-084436 from the National Institute of Allergy and Infectious Disease to Asako Uchiyama.
Footnotes
1Hereafter, MP refers to the virus cell-to-cell movement protein that gates PD, with the virus name in superscript. Virus-specific names denote additional movement proteins involved in intracellular transport.
2By convention, the nomenclature for animal and yeast proteins capitalizes only the first letter, whereas in the plant nomenclature all 3 letters are capitalized. We will use the plant convention here, with the species specifically designated.
References
- Agbeci M., Grangeon R., Nelson R. S., Zheng H., Laliberte J. F. (2013). Contribution of host intracellular transport machineries to intercellular movement of turnip mosaic virus. PLoS Pathog. 9:e1003683. 10.1371/journal.ppat.1003683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amari K., Boutant E., Hofmann C., Schmitt-Keichinger C., Fernandez-Calvino L., Didier P., et al. (2010). A family of plasmodesmal proteins with receptor-like properties for plant viral movement proteins. PLoS Pathogens. 6:e1001119. 10.1371/journal.ppat.1001119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S. M., Delamarre L., Mellman I., Andrews N. W. (2009). Differential role of the Ca(2+) sensor synaptotagmin VII in macrophages and dendritic cells. Immunobiology 214, 495–505. 10.1016/j.imbio.2008.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Faulkner C., Ritzenthaler C., Maule A. J. (2010). Plasmodesmata: gateways to local and systemic virus infection. Mol. Plant Microbe Interact. 23, 1403–1412. 10.1094/MPMI-05-10-0116 [DOI] [PubMed] [Google Scholar]
- Carluccio A. V., Zicca S., Stavolone L. (2014). Hitching a ride on vesicles: cauliflower mosaic virus movement protein trafficking in the endomembrane system. Plant Physiol 164, 1261–1270. 10.1104/pp.113.234534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J. C., Haldeman R., Dolja V. V., Restrepo-Hartwig M. A. (1993). Internal cleavage and trans-proteolytic activities of the VPg-proteinase (NIa) of tobacco etch potyvirus in vivo. J. Virol. 67, 6995–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J. C., Jensen P. E., Schaad M. C. (1998). Genetic evidence for an essential role for potyvirus CI protein in cell-to-cell movement. Plant J. 14, 393–400. 10.1046/j.1365-313X.1998.00120.x [DOI] [PubMed] [Google Scholar]
- Carvalho C. M., Pouwels J., van Lent J. W., Bisseling T., Goldbach R. W., Wellink J. (2004). The movement protein of cowpea mosaic virus binds GTP and single-stranded nucleic acid in vitro. J. Virol. 78, 1591–1594. 10.1128/JVI.78.3.1591-1594.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho M. F., Lazarowitz S. G. (2004). Interaction of the movement protein NSP and the Arabidopsis acetyltransferase AtNSI is necessary for Cabbage leaf curl geminivirus infection and pathogenicity. J. Virol. 78, 11161–11171. 10.1128/JVI.78.20.11161-11171.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Andrade L. O., Andrews N. W. (2005). Trypanosoma cruzi invades synaptotagmin VII-deficient cells by a PI-3 kinase independent pathway. Mol. Biochem. Parasitol. 141, 125–128. 10.1016/j.molbiopara.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Chakrabarty R., Banerjee R., Chung S. M., Farman M., Citovsky V., Hogenhout S. A., et al. (2007). PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol. Plant Microbe. Interact. 20, 740–750. 10.1094/MPMI-20-7-0740 [DOI] [PubMed] [Google Scholar]
- Chapman E. R. (2008). How does synaptotagmin trigger neurotransmitter release? Annu. Rev. Biochem. 77, 615–641. 10.1146/annurev.biochem.77.062005.101135 [DOI] [PubMed] [Google Scholar]
- Chen M. H., Sheng J. S., Hind G., Handa A. K., Citovsky V. (2000). Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 19, 913–920. 10.1093/emboj/19.5.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Knorr D., Schuster G., Zambryski P. (1990). The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60, 637–647. 10.1016/0092-8674(90)90667-4 [DOI] [PubMed] [Google Scholar]
- Craxton M. (2001). Genomic analysis of synaptotagmin genes. Genomics 77, 43–49. 10.1006/geno.2001.6619 [DOI] [PubMed] [Google Scholar]
- Craxton M. (2004). Synaptotagmin gene content of the sequenced genomes. BMC Genomics 5:43. 10.1186/1471-2164-5-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craxton M. (2007). Evolutionary genomics of plant genes encoding N-terminal-TM-C2 domain proteins and the similar FAM62 genes and synaptotagmin genes of metazoans. BMC Genomics 8:259. 10.1186/1471-2164-8-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J. A., Ahlquist P. (2010). Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64, 241–256. 10.1146/annurev.micro.112408.134012 [DOI] [PubMed] [Google Scholar]
- Dolja V. V., Haldeman-Cahill R., Montgomery A. E., Vandenbosch K. A., Carrington J. C. (1995). Capsid protein determinants involved in cell-to-cell and long distance movement of tobacco etch potyvirus. Virology 206, 1007–1016. 10.1006/viro.1995.1023 [DOI] [PubMed] [Google Scholar]
- Dolja V. V., Haldeman R., Robertson N. L., Dougherty W. G., Carrington J. C. (1994). Distinct functions of capsid protein in assembly and movement of tobacco etch potyvirus in plants. EMBO J. 13, 1482–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Thomas C., Harrison S., Revers F., Maule A. (2004). A cysteine-rich plant protein potentiates Potyvirus movement through an interaction with the virus genome-linked protein VPg. J. Virol. 78, 2301–2309. 10.1128/JVI.78.5.2301-2309.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A., Lain S., Garcia J. A. (1995). RNA helicase activity of the plum pox potyvirus CI protein expressed in Escherichia coli. Mapping of an RNA binding domain. Nucleic Acids Res. 23, 1327–1332. 10.1093/nar/23.8.1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez I., Ubach J., Dulubova I., Zhang X., Sudhof T. C., Rizo J. (1998). Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell 94, 841–849. 10.1016/S0092-8674(00)81742-0 [DOI] [PubMed] [Google Scholar]
- Fujiki M., Kawakami S., Kim R. W., Beachy R. N. (2006). Domains of tobacco mosaic virus movement protein essential for its membrane association. J. Gen. Virol. 87, 2699–2707. 10.1099/vir.0.81936-0 [DOI] [PubMed] [Google Scholar]
- Gardiner W. E., Sunter G., Brand L., Elmer J. S., Rogers S. G., Bisaro D. M. (1988). Genetic analysis of tomato golden mosaic virus: the coat protein is not required for systemic spread or symptom development. EMBO J. 7, 899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F., Saheki Y., Idevall-Hagren O., Colombo S. F., Pirruccello M., Milosevic I., et al. (2013). PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153, 1494–1509. 10.1016/j.cell.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeon R., Jiang J., Laliberte J. F. (2012). Host endomembrane recruitment for plant RNA virus replication. Curr. Opin. Virol. 2, 683–690. 10.1016/j.coviro.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapiak M., Li Y., Agama K., Swade S., Okenka G., Falk J., et al. (2008). Cauliflower mosaic virus gene VI product N-terminus contains regions involved in resistance-breakage, self-association and interactions with movement protein. Virus Res. 138, 119–129. 10.1016/j.virusres.2008.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries P. A., Palanichelvam K., Yu W., Schoelz J. E., Nelson R. S. (2009a). The cauliflower mosaic virus protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules. Plant Physiol. 149, 1005–1016. 10.1104/pp.108.131755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries P. A., Park J. W., Sasaki N., Ballard K. D., Maule A. J., Nelson R. S. (2009b). Differing requirements for actin and myosin by plant viruses for sustained intercellular movement. Proc. Natl. Acad. Sci. U.S.A. 106, 17594–17599. 10.1073/pnas.0909239106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries P. A., Schoelz J. E., Nelson R. S. (2010). Intracellular transport of viruses and their components: utilizing the cytoskeleton and membrane highways. Mol. Plant Microbe. Interact. 23, 1381–1393. 10.1094/MPMI-05-10-0121 [DOI] [PubMed] [Google Scholar]
- Harries P., Ding B. A. (2011). Cellular factors in plant virus movement: at the leading edge of macromolecular trafficking in plants. Virology 411, 237–243. 10.1016/j.virol.2010.12.021 [DOI] [PubMed] [Google Scholar]
- Haupt S., Cowan G. H., Ziegler A., Roberts A. G., Oparka K. J., Torrance L. (2005). Two plant-viral movement proteins traffic in the endocytic recycling pathway. Plant Cell 17, 164–181. 10.1105/tpc.104.027821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M., Padgett H. S., Gens J. S., Pickard B. G., Casper S. J., Epel B. L., et al. (1998). Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10, 1107–1120. 10.1105/tpc.10.7.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A., Chapdelaine Y., Hohn T. (1996). Interaction between cauliflower mosaic virus inclusion body protein and capsid protein: implications for viral assembly. Virology 217, 147–157. 10.1006/viro.1996.0102 [DOI] [PubMed] [Google Scholar]
- Huang Z., Han Y., Howell S. H. (2000). Formation of surface tubules and fluorescent foci in Arabidopsis thaliana protoplasts expressing a fusion between the green fluorescent protein and the cauliflower mosaic virus movement protein. Virology 271, 58–64. 10.1006/viro.2000.0292 [DOI] [PubMed] [Google Scholar]
- Hui E., Bai J., Chapman E. R. (2006). Ca2+-triggered simultaneous membrane penetration of the tandem C2-domains of synaptotagmin I. Biophys. J. 91, 1767–1777. 10.1529/biophysj.105.080325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui E., Johnson C. P., Yao J., Dunning F. M., Chapman E. R. (2009). Synaptotagmin-mediated bending of the target membrane is a critical step in Ca(2+)-regulated fusion. Cell 138, 709–721. 10.1016/j.cell.2009.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham D. J., Pascal E., Lazarowitz S. G. (1995). Both bipartite geminivirus movement proteins define viral host range, but only BL1 determines viral pathogenicity. Virology 207, 191–204. 10.1006/viro.1995.1066 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Tsuge S., Stavolone L., Hohn T. (2002). The cauliflower mosaic virus virion-associated protein is dispensable for viral replication in single cells. J. Virol. 76, 9457–9464. 10.1128/JVI.76.18.9457-9464.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte C., Vetter G., Loudes A. M., Robinson D. G., Hillmer S., Stussi-Garaud C., et al. (2003). Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY-2 cells. Plant Cell 15, 2058–2075. 10.1105/tpc.013896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz S. G. (2001). The plant viruses, in Fields' Virology, eds Knipe D. M., Howley P. M. (Philadelphia, PA: Lippincott, Williams and Wilkins; ), 533–598. [Google Scholar]
- Lazarowitz S. G., Beachy R. N. (1999). Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11, 535–548. 10.1105/tpc.11.4.535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. K., Yang Y., Su Z., Hyeon C., Lee T. S., Lee H. W., et al. (2010). Dynamic Ca2+-dependent stimulation of vesicle fusion by membrane-anchored synaptotagmin 1. Science 328, 760–763. 10.1126/science.1187722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A., Zheng J. Y., Lazarowitz S. G. (2013). The tobamovirus Turnip vein clearing Virus 30-kilodalton movement protein localizes to novel nuclear filaments to enhance virus infection. J. Virol. 87, 6428–6440. 10.1128/JVI.03390-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Lazarowitz S. G. (2010). Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. U.S.A. 107, 2491–2496. 10.1073/pnas.0909080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford A. G., Stefan C. J., Yuan H. L., Macgurn J. A., Emr S. D. (2012). ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell 23, 1129–1140. 10.1016/j.devcel.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Martens S., Kozlov M. M., McMahon H. T. (2007). How synaptotagmin promotes membrane fusion. Science 316, 1205–1208. 10.1126/science.1142614 [DOI] [PubMed] [Google Scholar]
- Maule A. J. (2008). Plasmodesmata: structure, function and biogenesis. Curr. Opin. Plant Biol. 11, 680–686. 10.1016/j.pbi.2008.08.002 [DOI] [PubMed] [Google Scholar]
- McMahon H. T., Kozlov M. M., Martens S. (2010). Membrane curvature in synaptic vesicle fusion and beyond. Cell 140, 601–605. 10.1016/j.cell.2010.02.017 [DOI] [PubMed] [Google Scholar]
- Merits A., Guo D., Saarma M. (1998). VPg, coat protein and five non-structural proteins of potato a potyvirus bind RNA in a sequence-unspecific manner. J. Gen. Virol. 79, 3123–3127. [DOI] [PubMed] [Google Scholar]
- Merits A., Rajamaki M. L., Lindholm P., Runeberg-Roos P., Kekarainen T., Puustinen P., et al. (2002). Proteolytic processing of potyviral proteins and polyprotein processing intermediates in insect and plant cells. J. Gen. Virol. 83, 1211–1221. [DOI] [PubMed] [Google Scholar]
- Moghadam P. K., Jackson M. B. (2013). The Functional Significance of Synaptotagmin Diversity in Neuroendocrine Secretion. Front. Endocrinol. 4:124. 10.3389/fendo.2013.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki N., Tomioka R., Maeshima M. (2008). A hydrophilic cation-binding protein of Arabidopsis thaliana, AtPCaP1, is localized to plasma membrane via N-myristoylation and interacts with calmodulin and the phosphatidylinositol phosphates PtdIns(3,4,5)P(3) and PtdIns(3,5)P(2). FEBS J. 275, 2267–2282. 10.1111/j.1742-4658.2008.06379.x [DOI] [PubMed] [Google Scholar]
- Nelson R. S., Citovsky V. (2005). Plant viruses. Invaders of cells and pirates of cellular pathways. Plant Physiol. 138, 1809–1814. 10.1104/pp.104.900167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehl A., Heinlein M. (2010). Cellular pathways for viral transport through plasmodesmata. Protoplasma 248, 75–99. 10.1007/s00709-010-0246-1 [DOI] [PubMed] [Google Scholar]
- Oparka K. J. (2004). Getting the message across: how do plant cells exchange macromolecular complexes? Tr Plant Sci. 9, 33–41. 10.1016/j.tplants.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Oparka K. J., Roberts A. G. (2001). Plasmodesmata. A not so open-and-shut case. Plant Physiol. 125, 123–126. 10.1104/pp.125.1.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal E., Sanderfoot A. A., Ward B. M., Medville R., Turgeon R., Lazarowitz S. G. (1994). The geminivirus BR1 movement protein binds single-stranded DNA and localizes to the cell nucleus. Plant Cell 6, 995–1006. 10.1105/tpc.6.7.995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooma W., Gillette W. K., Jeffrey J. L., Petty I. T. (1996). Host and viral factors determine the dispensability of coat protein for bipartite geminivirus systemic movement. Virology 218, 264–268. 10.1006/viro.1996.0189 [DOI] [PubMed] [Google Scholar]
- Qin S., Ward B. M., Lazarowitz S. G. (1998). The bipartite geminivirus coat protein aids BR1 function in viral movement by affecting the accumulation of viral single-stranded DNA. J. Virol. 72, 9247–9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo-Hartwig M. A., Carrington J. C. (1994). The tobacco etch potyvirus 6-kilodalton protein is membrane associated and involved in viral replication. J. Virol. 68, 2388–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts I. M., Wang D., Findlay K., Maule A. J. (1998). Ultrastructural and temporal observations of the potyvirus cylindrical inclusions (CIs) show that the CI protein acts transiently in aiding virus movement. Virology 245, 173–181. 10.1006/viro.1998.9132 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Cerezo E., Findlay K., Shaw J. G., Lomonossoff G. P., Qiu S. G., Linstead P., et al. (1997). The coat and cylindrical inclusion proteins of a potyvirus are associated with connections between plant cells. Virology 236, 296–306. 10.1006/viro.1997.8736 [DOI] [PubMed] [Google Scholar]
- Ryabova L. A., Pooggin M. M., Hohn T. (2002). Viral strategies of translation initiation: ribosomal shunt and reinitiation. Prog. Nucleic Acid Res. Mol. Biol. 72, 1–39. 10.1016/S0079-6603(02)72066-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabova L., Park H. S., Hohn T. (2004). Control of translation reinitiation on the cauliflower mosaic virus (CaMV) polycistronic RNA. Biochem. Soc. Trans. 32, 592–596. 10.1042/BST0320592 [DOI] [PubMed] [Google Scholar]
- Sanderfoot A. A., Ingham D. J., Lazarowitz S. G. (1996). A viral movement protein as a nuclear shuttle. The geminivirus BR1 movement protein contains domains essential for interaction with BL1 and nuclear localization. Plant Physiol. 110, 23–33. 10.1104/pp.110.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A. A., Lazarowitz S. G. (1995). Cooperation in viral movement: the geminivirus BL1 movement protein interacts with br1 and redirects it from the nucleus to the cell periphery. Plant Cell 7, 1185–1194. 10.1105/tpc.7.8.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A. A., Lazarowitz S. G. (1996). Getting it together in plant virus movement: cooperative interactions between bipartite geminivirus movement proteins. Trends Cell Biol. 6, 353–358. 10.1016/0962-8924(96)10031-3 [DOI] [PubMed] [Google Scholar]
- Schaad M. C., Jensen P. E., Carrington J. C. (1997). Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16, 4049–4059. 10.1093/emboj/16.13.4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapire A. L., Voigt B., Jasik J., Rosado A., Lopez-Cobollo R., Menzel D., et al. (2008). Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 20, 3374–3388. 10.1105/tpc.108.063859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoelz J. E., Harries P. A., Nelson R. S. (2011). Intracellular transport of plant viruses: finding the door out of the cell. Mol. Plant 4, 813–831. 10.1093/mp/ssr070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorel M., Garcia J. A., German-Retana S. (2014). The Potyviridae cylindrical inclusion helicase: a key multipartner and multifunctional protein. Mol. Plant Microbe. Interact. 27, 215–226. 10.1094/MPMI-11-13-0333-CR [DOI] [PubMed] [Google Scholar]
- Stavolone L., Villani M. E., Leclerc D., Hohn T. (2005). A coiled-coil interaction mediates cauliflower mosaic virus cell-to-cell movement. Proc. Natl. Acad. Sci. U.S.A. 102, 6219–6224. 10.1073/pnas.0407731102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striegel A. R., Biela L. M., Evans C. S., Wang Z., Delehoy J. B., Sutton R. B., et al. (2012). Calcium binding by synaptotagmin's C2A domain is an essential element of the electrostatic switch that triggers synchronous synaptic transmission. J. Neurosci. 32, 1253–1260. 10.1523/JNEUROSCI.4652-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami Y., Watanabe Y. (2007). Effects of brefeldin A on the localization of Tobamovirus movement protein and cell-to-cell movement of the virus. Virology 361, 133–140. 10.1016/j.virol.2006.11.008 [DOI] [PubMed] [Google Scholar]
- Thiebeauld O., Schepetilnikov M., Park H. S., Geldreich A., Kobayashi K., Keller M., et al. (2009). A new plant protein interacts with eIF3 and 60S to enhance virus-activated translation re-initiation. EMBO J. 28, 3171–3184. 10.1038/emboj.2009.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. L., Maule A. J. (2000). Limitations on the use of fused green fluorescent protein to investigate structure-function relationships for the cauliflower mosaic virus movement protein. J. Gen. Virol. 81, 1851–1855. [DOI] [PubMed] [Google Scholar]
- Toulmay A., Prinz W. A. (2012). A conserved membrane-binding domain targets proteins to organelle contact sites. J. Cell Sci. 125, 49–58. 10.1242/jcs.085118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki S., Citovsky V. (2005). Identification of an interactor of cadmium ion-induced glycine-rich protein involved in regulation of callose levels in plant vasculature. Proc. Natl. Acad. Sci. U.S.A. 102, 12089–12094. 10.1073/pnas.0505927102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki S., Citovsky V. (2011). To gate, or not to gate: regulatory mechanisms for intercellular protein transport and virus movement in plants. Mol. Plant 4, 782–793. 10.1093/mp/ssr060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki S., Lacroix B., Krichevsky A., Lazarowitz S. G., Citovsky V. (2009). Functional transient genetic transformation of Arabidopsis leaves by biolistic bombardment. Nat. Protoc. 4, 71–77. 10.1038/nprot.2008.217 [DOI] [PubMed] [Google Scholar]
- Verchot-Lubicz J., Torrance L., Solovyev A. G., Morozov S. Y., Jackson A. O., Gilmer D. (2010). Varied movement strategies employed by triple gene block-encoding viruses. Mol. Plant Microbe. Interact. 23, 1231–1247. 10.1094/MPMI-04-10-0086 [DOI] [PubMed] [Google Scholar]
- Vijayapalani P., Maeshima M., Nagasaki-Takekuchi N., Miller W. A. (2012). Interaction of the trans-frame potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog. 8:e1002639. 10.1371/journal.ppat.1002639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E., Ueki S., Trutnyeva K., Citovsky V. (2004). The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. Crit. Rev. Plant Sci. 23, 195–250 10.1080/07352680490452807 [DOI] [Google Scholar]
- Ward B. M., Lazarowitz S. G. (1999). Nuclear export in plants. Use of geminivirus movement proteins for a cell-based export assay. Plant Cell 11, 1267–1276. 10.1105/tpc.11.7.1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B. M., Medville R., Lazarowitz S. G., Turgeon R. (1997). The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J. Virol. 71, 3726–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T. Y., Huang T. S., McNeil J., Laliberte J. F., Hong J., Nelson R. S., et al. (2010b). Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 84, 799–809. 10.1128/JVI.01824-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T. Y., Wang A. M. (2008). Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI- and COPII-dependent manner. J. Virol. 82, 12252–12264 10.1128/JVI.01329-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Zhang C., Hong J., Xiong R., Kasschau K. D., Zhou X., et al. (2010a). Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 6:e1000962. 10.1371/journal.ppat.1000962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Deom C. M., Beachy R. N., Lucas W. J. (1989). Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246, 377–379. 10.1126/science.246.4928.377 [DOI] [PubMed] [Google Scholar]
- Wright K. M., Wood N. T., Roberts A. G., Chapman S., Boevink P., Mackenzie K. M., et al. (2007). Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: evidence from FRAP. Traffic 8, 21–31. 10.1111/j.1600-0854.2006.00510.x [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Kawamura Y., Minami A., Uemura M. (2008). Calcium-dependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1. Plant Cell 20, 3389–3404. 10.1105/tpc.108.062679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Kwon S. E., Gaffaney J. D., Dunning F. M., Chapman E. R. (2012a). Uncoupling the roles of synaptotagmin I during endo- and exocytosis of synaptic vesicles. Nat. Neurosci. 15, 243–249. 10.1038/nn.3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L. H., Rao Y., Varga K., Wang C. Y., Xiao P., Lindau M., et al. (2012b). Synaptotagmin 1 is necessary for the Ca2+ dependence of clathrin-mediated endocytosis. J. Neurosci. 32, 3778–3785. 10.1523/JNEUROSCI.3540-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski P., Crawford K. (2000). Plasmodesmata: gatekeepers for cell-to-cell transport of developmental signals in plants. Ann. Rev. Cell Dev. Biol. 16, 393–421. 10.1146/annurev.cellbio.16.1.393 [DOI] [PubMed] [Google Scholar]
- Zeenko V. V., Ryabova L. A., Spirin A. S., Rothnie H. M., Hess D., Browning K. S., et al. (2002). Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J. Virol. 76, 5678–5691. 10.1128/JVI.76.11.5678-5691.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]