Summary
Subcellular biomolecular localization is critical for the metabolic and structural properties of the cell. The functional implications of the spatiotemporal distribution of protein complexes during the bacterial cell cycle have long been acknowledged; however, the molecular mechanisms for generating and maintaining their dynamic localization in bacteria are not completely understood. Here we demonstrate that the trans-envelope Tol–Pal complex, a widely conserved component of the cell envelope of Gram-negative bacteria, is required to maintain the polar positioning of chemoreceptor clusters in Escherichia coli. Localization of the chemoreceptors was independent of phospholipid composition of the membrane and the curvature of the cell wall. Instead, our data indicate that chemoreceptors interact with components of the Tol–Pal complex and that this interaction is required to polarly localize chemoreceptor clusters. We found that disruption of the Tol–Pal complex perturbs the polar localization of chemoreceptors, alters cell motility, and affects chemotaxis. We propose that the E. coli Tol–Pal complex restricts mobility of the chemoreceptor clusters at the cell poles and may be involved in regulatory mechanisms that co-ordinate cell division and segregation of the chemosensory machinery.
Introduction
Asymmetric localization is a hallmark of many different bacterial proteins, including components of cell division, virulence, adhesion, development, chemotaxis, and motility (Alley et al., 1992; Marston and Errington, 1999; Steinhauer et al., 1999; Lam et al., 2006). Genomic and proteomic approaches for identifying subcellular protein localization have expanded our understanding of asymmetric patterns of localization in bacteria (Lai et al., 2004; Kitagawa et al., 2005; Janakiraman et al., 2009; Werner et al., 2009) and revealed several classes of proteins that localize to the cell poles in rod-shaped bacteria. The canonical example of polarly localized proteins is the methyl-accepting chemotaxis proteins (MCPs, herein referred to as chemoreceptors) (Alley et al., 1992; Maddock and Shapiro, 1993). This family of transmembrane chemoreceptors is required for chemotaxis and is primarily positioned at the cell poles where it forms intercommunicating clusters that are ~ 200–400 nm in diameter (Maddock and Shapiro, 1993; Bray et al., 1998; Gestwicki and Kiessling, 2002; Zhang et al., 2007; Briegel et al., 2008; Greenfield et al., 2009; Liu et al., 2012). The asymmetric distribution of chemoreceptors in cell membranes is evolutionarily conserved in Bacteria and Archaea (Gestwicki et al., 2000), and recent cryotomographic studies demonstrate that the basic honeycomb lattice architecture of the arrays is conserved among chemotactic bacteria (Briegel et al., 2008; 2009; 2012; Liu et al., 2012). Despite a deep understanding of the central role of this machinery in controlling cell behaviour and facilitating adaptation and survival (reviewed in Wadhams and Armitage, 2004; Hazelbauer et al., 2008; Miller et al., 2009; Sourjik and Armitage, 2010; Porter et al., 2011), the mechanisms underlying its localization and the relationship between its position and function are not completely understood.
The bacterial cell poles have distinct physical features that may be involved in controlling protein positioning (reviewed in Lybarger and Maddock, 2001; Shapiro et al., 2002; Rudner and Losick, 2010). Polar localization of the chemoreceptor clusters in Escherichia coli depends partially on the histidine kinase CheA and the adaptor protein CheW (Alley et al., 1992; Maddock and Shapiro, 1993; Skidmore et al., 2000). CheA and CheW form a ternary complex with chemoreceptors and participate in chemotactic signal transduction. Imaging studies have revealed that chemoreceptors are inserted randomly into the lateral cytoplasmic membrane along the helically distributed Sec translocation machinery complexes and migrate to pre-existing clusters (Shiomi et al., 2006). In this model, it is unclear whether chemoreceptors are transported actively within the cytoplasmic membrane to the cell poles or diffuse passively and are trapped at the poles. Another model that has been hypothesized involves a stochastic nucleation mechanism in which cluster assembly and positioning at the cell poles depends on chemoreceptor concentration and on the distance of receptors from large chemoreceptor clusters (Thiem and Sourjik, 2008; Wang et al., 2008; Greenfield et al., 2009).
The cell envelope of E. coli and other Gram-negative bacteria consists of the inner membrane (IM), a ~ 2- to 4-nm-thick layer of peptidoglycan (Gan et al., 2008) positioned beyond the IM, and the outer membrane (OM). Genetic and biochemical analyses have revealed protein complexes that bridge the three layers of the cell envelope. The functions of these complexes include: performing active transport of biomolecules through the cell envelope (Skareso et al., 1993; Braun, 1995; Okuda et al., 2012), maintaining cell envelope integrity (Bouveret et al., 1995; Bernadac et al., 1998; Lazzaroni et al., 1999; Lloubès et al., 2001), and mediating the final steps of cell division (Gerding et al., 2007; Yeh et al., 2010). The Tol–Pal complex (Fig. 1A and B) is a widely conserved component of the cell envelope of Gram-negative bacteria (Sturgis, 2001) and its physiological roles include: maintaining cell wall integrity by mediating the physical connection between the OM and the peptidoglycan (Bernadac et al., 1998); expressing lipopolysaccharide surface antigens and virulence factors (Gaspar et al., 2000); promoting proper functioning of certain transport systems in the cytoplasmic membrane (Llamas et al., 2003); facilitating phage infection and translocation of group A colicins (Davies and Reeves, 1975; Click and Webster, 1997; Lazzaroni et al., 2002); and reducing cell sensitivity to detergents (Lahiri et al., 2011). In many bacteria, tol and pal mutants produce chains of cells that bleb in low-osmolarity or high-ionic-strength liquids, suggesting that the complex is part of the cell division machinery and plays a role in completing cell division under conditions of membrane stress (Bernadac et al., 1998; Gerding et al., 2007).
Fig. 1. The organization of the E. coli Tol–Pal complex.
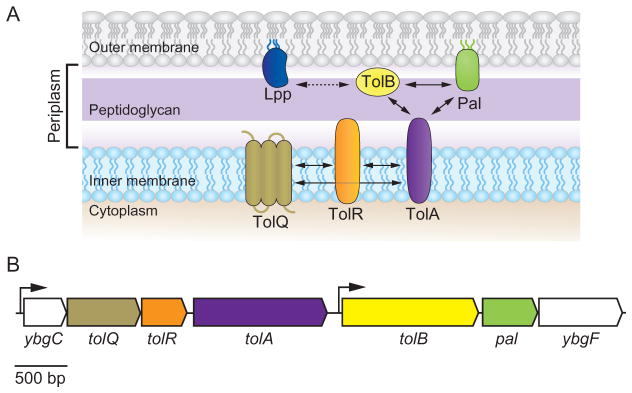
A. A cartoon depicting the predicted organization of the Tol–Pal complex in the E. coli cell envelope. TolQ, TolR, and TolA are integral inner membrane (IM) proteins that interact through their transmembrane segments (Derouiche et al., 1995; Lazzaroni et al., 1995; Germon et al., 1998). TolB is a soluble periplasmic protein and Pal is anchored to the outer membrane (OM) (Lazzaroni and Portalier, 1992; Bouveret et al., 1995; Clavel et al., 1998) (see text for details). The major OM lipoprotein Lpp is not part of the Tol–Pal complex; however, it is able to interact with TolB (indicated by a dashed arrow) (Clavel et al., 1998). Arrows indicate the interaction of the proteins in the cell envelope.
B. Schematic diagram of the gene organization of the E. coli tol–pal gene cluster. The cluster is organized in two operons (ybgC–ybgF and tolB–ybgF). The ybgC gene encodes a cytoplasmic protein whereas the ybgF gene encodes a periplasmic protein. Inactivation of YbgC and YbgF induces no obvious alteration in phenotype. Arrows indicate the direction of transcription and position of promoters (Vianney et al., 1996; Muller and Webster, 1997).
In E. coli, this trans-envelope complex consists of two subcomplexes (Fig. 1A and B). One complex is located in the IM and consists of TolA, TolQ, and TolR, which interact through their transmembrane segments with a respective stoichiometry of 4–6:2:1 (Vianney et al., 1994; Derouiche et al., 1995; Cascales et al., 2001). TolQ spans the IM three times whereas both TolR and TolA are anchored to the IM through a single membrane-spanning region located near their N-terminus (Levengood et al., 1991; Kampfenkel and Braun, 1993; Muller et al., 1993; Vianney et al., 1994; Derouiche et al., 1995). A second complex is associated with the OM and consists of TolB – a predominantly soluble periplasmic protein – and the peptidoglycan-associated lipoprotein Pal, which is anchored to the OM (Lazzaroni and Portalier, 1992; Isnard et al., 1994; Bouveret et al., 1995; 1999). The assembly of these two subcomplexes bridges the three layers of the cell envelope through multiple interactions, including the association of the C-terminal periplasmic domain of TolA with Pal and TolB (Bouveret et al., 1995; Cascales et al., 2001; Dubuisson et al., 2002; Walburger et al., 2002). Cross-linking studies and the isolation and characterization of suppressive mutations suggest interactions between individual components of the Tol–Pal complex and other components of the cell envelope. Pal is an abundant lipoprotein thought to interact with the peptidoglycan through a conserved α-helical motif (Koebnik, 1995; Bouveret et al., 1999). TolB interacts with the Lpp murein (Braun’s) lipoprotein and the porin OmpA (Bouveret et al., 1995; Clavel et al., 1998; Ray et al., 2000). Additionally, both TolA and TolB form high-molecular-weight complexes with the trimeric porins OmpF, OmpC, PhoE, and LamB (Derouiche et al., 1996; Rigal et al., 1997). The numerous interactions of the Tol and Pal components with other proteins suggest that the complex may be involved in a range of other physiological and regulatory cellular processes.
In this manuscript we demonstrate that the intact Tol–Pal complex is required for the polar localization of chemoreceptor clusters in E. coli, cell motility, and chemotaxis. We tested whether the components of the Tol–Pal complex interact with chemoreceptors to maintain the subcellular localization of polar clusters. We found that disruption of the trans-envelope Tol–Pal complex perturbs polar localization of chemoreceptor clusters without affecting the localization of other membrane-associated polar proteins. Consistent with this interaction, the absence of the Tol–Pal complex affected both swimming and swarming cell motility. Importantly, mislocalization of chemoreceptor clusters upon deletion of the Tol–Pal complex affects chemotaxis by increasing the tumbling frequency of the cells. These results add a surprising new dimension to the subcellular organization of bacteria.
Results
Chemoreceptors localize to the cell poles in E. coli mutants lacking the anionic phospholipid cardiolipin
An emerging hypothesis is that the accumulation of anionic phospholipids at the cell poles stabilizes mechanical strain that arises due to membrane curvature, influences the local physicochemical properties of phospholipid bilayers, and positions and regulates classes of membrane-associated proteins (Romantsov et al., 2007; 2008; Renner and Weibel, 2011; 2012; Renner et al., 2013). For example, diphosphatidylglycerol (also referred to as cardiolipin; ‘CL’) has been reported to accumulate at the bacterial cell poles (Mileykovskaya and Dowhan, 2000; Koppelman et al., 2001; Renner and Weibel, 2011) and interact with and regulate several membrane-associated proteins in mitochondria and bacteria (Romantsov et al., 2008; Gold et al., 2010). Due to the spatial proximity of CL and the chemoreceptors at the poles of E. coli cells, we tested whether this phospholipid was responsible for the position of the chemoreceptors.
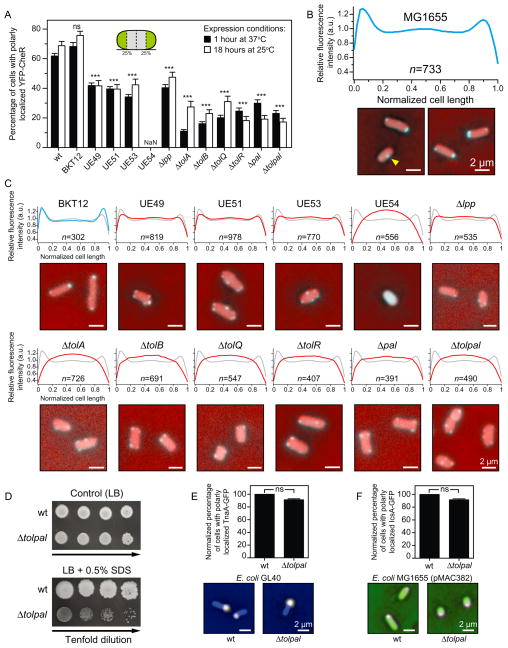
To visualize chemoreceptors in vivo we used plasmid pPA803, a derivative of pBR322 that codes for the expression of yellow fluorescent protein (YFP) fused to the N-terminal region of CheR (YFP–CheR) controlled by a lactose-inducible promoter (Zhou et al., 2011). CheR is a methyltransferase that targets a specific C-terminal pentapeptide on the major chemoreceptors Tar and Tsr (Wu et al., 1996; Shiomi et al., 2002). Importantly, YFP–CheR is functional, colocalizes with chemoreceptors, does not localize in the absence of chemoreceptors, and has been used to determine the position of chemoreceptors in vivo (Wu et al., 1996; Shiomi et al., 2002; Kentner et al., 2006; Zhou et al., 2011) (Fig. S1A–C). We used the functional YFP–CheR as a proxy for chemoreceptor localization to prevent the influence of a fluorescent tag fused to the chemoreceptors on their clustering and localization. YFP–CheR formed predominantly polar foci in cells of wild-type E. coli strain MG1655 (Fig. 2A and B). We detected 62 ± 0.01% [mean ± standard error of the mean (s.e.m.)] of the YFP–CheR clusters positioned approximately within the first and/or fourth quarters of the cell (which we defined as cell poles) after 1 h of overexpression at 37°C. Approximately 38% of the chemoreceptor clusters were distributed along the lateral length of the cell between the polar regions and particularly concentrated at the mid-cell (Fig. 2B). Only 1% ± 0.03% (mean ± s.e.m.) of the cells displayed diffused YFP–CheR signal, indicating that cluster formation was not being affected in the overexpression system (Fig. S2). Importantly, the number of polar clusters did not change significantly after 18 h of overexpression at 25°C [68 ± 0.03% (mean ± s.e.m.)] (Fig. 2A; Fig. S3A and B).
Fig. 2. Subcellular localization of chemoreceptors and other membrane-associated proteins in wild-type (wt) E. coli MG1655 and in various isogenic mutant strains.
A. Per cent of E. coli MG1655 cells and various isogenic mutants with polarly (unipolar or bipolar) localized YFP–CheR imaged after 1 h at 37°C (black bars) and after 18 h at 25°C (white bars). Inset image: A cartoon depicting the poles as the outmost quartiles of cells (coloured in green). Here and elsewhere, cells were determined to display a ‘wild-type phenotype’ if all the clusters were located within the first and/or fourth quartiles of the cell. Otherwise, cells were labelled as displaying ‘non-wild-type phenotype’. NaN, no polar clusters were detected.
B. Fluorescence intensity profile of the subcellular distribution of YFP–CheR in wild-type E. coli MG1655 cells (n = 733) imaged after 1 h at 37°C (a.u., arbitrary units). Panel below the graph: Representative fluorescence microscopy images depicting the localization pattern of YFP–CheR in wt E. coli MG1655 cells. Microscopy images are representative of three independent experiments performed on different days. The yellow arrowhead indicates the YFP–CheR foci at mid-cell. Scale bars = 2 μm.
C. Fluorescence intensity profile of the subcellular distribution of YFP–CheR in isogenic mutants of E. coli MG1655 imaged after 1 h at 37°C (a.u., arbitrary units). We superimposed the YFP–CheR fluorescence profile of the wild-type cells (in grey) on top of the profile of the isogenic mutants (in blue or red). Panel below each graph: Representative fluorescence microscopy images depicting the localization pattern of YFP–CheR in isogenic mutants. Images are representative of at least three independent experiments performed on different days. Scale bars = 2 μm.
D. Growth assay of serial dilutions (left to right) of wt E. coli MG1655 and E. coli MG1655 Δtolpal. Aliquots of 10-fold serial dilutions (5 μl, 101–104 cfu ml−1) from overnight cultures were spotted on control agar (LB) and agar containing 0.5% SDS. The plates were incubated at 37°C for 24 h and then at 25°C for additional 24 h.
E. Per cent of E. coli MG1655 (n = 464) and E. coli MG1655 Δtolpal (n = 386) cells with polarly localized TnaA–GFP. Panel below the graph: Representative images obtained in at least three independent experiments performed on different days.
F. Per cent of E. coli MG1655 (n = 601) and in E. coli MG1655 Δtolpal (n = 406) cells with polarly localized IcsA–GFP. Panel below the graph: Representative images obtained in at least three independent experiments performed on different days.
To assess differences between the localization patterns in the various E. coli strains (panels A, E, and F), we performed Chi-squared tests. All tests were two-sided and statistical significance was considered when a P-value < 0.05 was observed. ns, non-significant; *P < 0.05; **P < 0.01; ***P < 0.001 compared to the wild-type phenotype. Error bars indicate the standard error of the mean. Scale bars = 2 μm.
To test whether CL at the cell poles was responsible for the polar localization of YFP–CheR, we used a CL null strain (E. coli MG1655–BKT12) in which the three enzymes (ClsA, ClsB, and ClsC) responsible for CL biosynthesis were deleted (Tan et al., 2012). YFP–CheR displayed the same pattern of localization in E. coli strain MG1655–BKT12 as in the wild-type parent strain MG1655, suggesting that the decrease in CL content had no effect on chemoreceptor localization (Fig. 2A and C). As observed for the wild-type E. coli MG1655, the number of polar clusters in the CL null strain did not change significantly after 18 h of overexpression at 25°C (Fig. S3B and C). We observed, however, that patterns of YFP–CheR localization were altered in other E. coli strains in which phospholipid compositions were perturbed. For example, we found that E. coli UE54 expressing YFP–CheR displayed patterns of diffuse fluorescence and lacked defined YFP puncta (Fig. 2A and C and Fig. S2). E. coli UE54 is a pgsA null strain that has decreased levels of the anionic phospholipids CL and phosphatidylglycerol (PG) (Kikuchi et al., 2000; Shiba et al., 2004). We also noticed that YFP–CheR was mislocalized in all of the individual mutant strains that served as the genetic background for construction of E. coli UE54 (i.e. E. coli strains UE49, UE51, and UE53) (Fig. 2A and C). A quantitative analysis of YFP–CheR fluorescence demonstrated that the distribution of fluorescence along the length of cells in all these mutants was different from cells of wild-type E. coli strain MG1655. The only common genetic alteration in all of these E. coli UE strains in which YFP–CheR are mislocalized or unclustered is the absence of the major outer membrane lipoprotein, Lpp.
The major outer membrane lipoprotein Lpp and the Tol–Pal complex are determinants for polar localization of chemoreceptor clusters in E. coli cells
We investigated the role of the major outer membrane lipoprotein Lpp in positioning the chemoreceptors at the poles of E. coli cells. Lpp interacts both covalently and non-covalently with the peptidoglycan (Inouye et al., 1972; Braun, 1975) and contributes to the structural and functional integrity of the cell envelope (Choi et al., 1986; Shu et al., 2000). To test whether Lpp was involved in the polar localization of YFP–CheR, we expressed YFP–CheR in the Lpp null strain (E. coli MG1665 Δlpp) and observed the same pattern of localization as in the E. coli strains UE49, UE51, and UE53: that is, an increased number of YFP–CheR foci positioned along the cylindrical region of cells compared to at the poles (Fig. 2A and C). Similar results were observed in the Lpp null mutant E. coli JW1667 from the Keio collection (Baba et al., 2006) (Fig. S4A and B). This result suggests that Lpp is connected to the polar positioning of chemoreceptor clusters.
In E. coli, Lpp forms homotrimers that interact with components of the Tol–Pal complex and with the outer membrane protein A (OmpA) (Clavel et al., 1998). The E. coli Tol–Pal complex is encoded by a cluster of seven genes organized into two transcription units (Vianney et al., 1996; Muller and Webster, 1997) and consists of five proteins that form two subcomplexes (Fig. 1A and B). To assess whether the Tol–Pal proteins play a role in chemoreceptor localization, we determined the position of YFP–CheR in single-gene knockouts of tolA, tolB, tolQ, tolR, and pal derived from the wild-type MG1655 and in an E. coli MG1655 mutant lacking all the Tol and Pal proteins (we refer to this strain as Δtolpal) (Fig. 2D and Fig. S5A–D). All knockout mutants displayed characteristically mislocalized puncta, which was similar to our observation for the E. coli UE strains: fluorescent puncta positioned primarily along the cylindrical body of the cells (Fig. 2A and C). As observed for E. coli MG1655 cells, the number of polar clusters and distribution of fluorescence along the length of cells observed in most mutants did not change significantly after 18 h of overexpression at 25°C (Fig. 2A; Fig. S3B and C). Immunoblots of cell lysate showed that the YFP–CheR fusion protein was stable in UE54 and in all the tol and pal single-gene knockouts (Fig. S6). Thus, diffused fluorescence signal inside the cells is not the result of proteolysis or degradation of the YFP–CheR fusion protein liberating the fluorescent protein. With the exception of E. coli UE54, all mutants displayed insignificant or absent diffused YFP–CheR fluorescence (Fig. S2), indicating that the mutations did not affect clustering of chemoreceptors. In addition, we verified that the Δtolpal mutant still produces chemoreceptors at levels comparable to wild-type cells (Fig. S7). Thus, mislocalization of YFP–CheR was not due to absence of chemoreceptors in the Δtolpal mutant. To confirm that the localization of the chemosensory machinery was perturbed in the Δtolpal mutant, we used the additional functional reporter YFP–CheW as a proxy for chemoreceptor localization. CheW is the adaptor protein that physically couples CheA to the chemoreceptor to regulate phosphotransfer to CheY and CheB (Hazelbauer et al., 2008; Briegel et al., 2012; Liu et al., 2012). As observed for YFP–CheR localization, YFP–CheW formed predominantly polar foci in cells of wild-type E. coli strain MG1655 and characteristically mislocalized puncta in the Δtolpal mutant (Fig. S8A and B). This observation provides additional evidence that polar localization of the E. coli chemosensory machinery is significantly affected in mutants lacking proteins of the Tol–Pal complex.
There are two additional genes in the tol–pal operon (Fig. 1B): ybgF codes for a periplasmic protein and ybgC codes for a putative cytosolic protein (Vianney et al., 1996; Muller and Webster, 1997). The function of these proteins has not yet been established. In contrast to our results with the other proteins encoded in the tol–pal operon, YFP–CheR remained polarly localized in ybgC and ybgF null mutants derived from E. coli strain BW25113 (Baba et al., 2006) (Fig. S9). These results suggest that localization of YFP–CheR is exclusively dependent on the tol and pal genes within the tol–pal operon. As an additional control, we evaluated the localization of YFP–CheR in tol and pal single-gene knockout mutants from the Keio collection and in single-gene knockout mutants lacking other IM and OM lipoproteins involved in transport, stress survival, or cell wall synthesis and remodelling, and found that YFP–CheR was only mislocalized in tol and pal mutants (Fig. S9).
Interestingly, the tol–pal deletion (Δtolpal) had a large effect on the localization of YFP–CheR but did not affect the cellular localization of other proteins that are know to be enriched at the E. coli cell poles, including the E. coli IM-associated tryptophanase TnaA (Li and Young, 2012) and the heterologous OM protein IcsA from Shigella (Goldberg et al., 1993; Charles et al., 2001) (Fig. 1E and F). This observation suggests that the interaction between Tol–Pal proteins and the chemoreceptors may be unique and essential for the polar localization of the chemosensory machinery, but not for other biomolecular complexes.
Deletion of Tol–Pal components causes motility and chemotaxis defects in E. coli cells
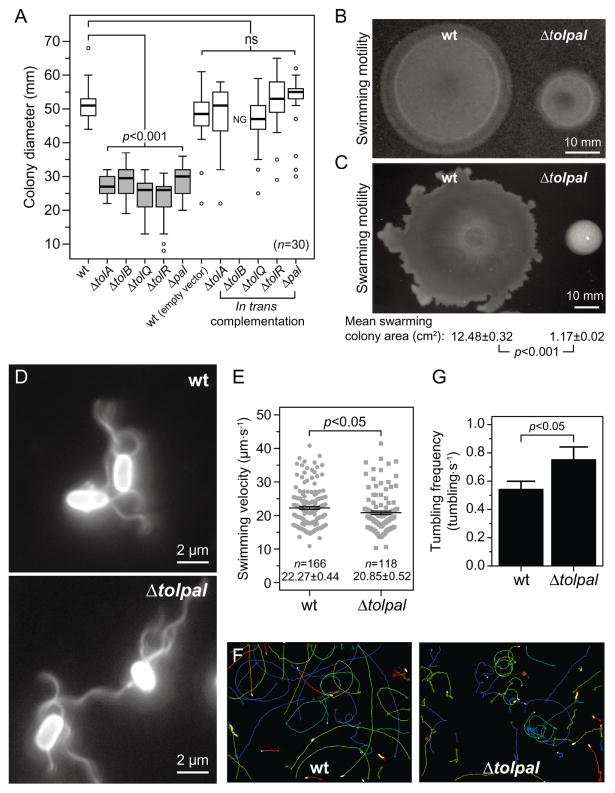
We tested whether the changes in chemoreceptor localization that accompany the absence of Tol and Pal proteins affect cell motility and chemotaxis. We assessed the motility of wild-type E. coli MG1655 and the single-gene tol and pal null mutants in motility agar. All of the single-gene mutants displayed a significant (P-value < 0.05) decrease in swimming motility compared to wild-type E. coli cells (Fig. 3A and Fig. S10A). Plasmid-based complementation of each of the tol or pal deletion mutants reverted the swimming motility defects, with the exception of tolB (Fig. 3A and Fig. S10B). As expected, cells of E. coli MG1655 Δtolpal also showed motility defects in swim plate assays compared to wild-type E. coli MG1655 cells (Fig. 3B); overexpression of individual tol and pal genes was not sufficient to revert the motility phenotype in the Δtolpal mutant (Fig. S10C and D). In all cases that we analysed, the overexpression of tolB completely impaired cell growth (Fig. S10B and C), suggesting that its accumulation may be cytotoxic. We bypassed its toxicity by decreasing the concentration of inducer and successfully reverted the motility defect of the tolB knockout strain (Fig. S11A and B).
Fig. 3. Motility phenotype and velocity of wild-type (wt) E. coli MG1655 cells and various isogenic mutants.
A. Box-and-whisker plots depicting the size of the migrating colonies (n = 30) measured for each E. coli strain. The extent of the box encompasses the interquartile range of the diameter, whiskers extend to maximum and minimum diameters, and the line within each box represents the median. Outliers are represented as open black circles. Plots represent at least three independent experiments performed on different days. For complementation experiments, 50 μg ml−1 of carbenicillin and 1 mM of IPTG were added to the motility agar. ns, non-significant; NG, no growth. All tests were two-sided and statistical significance was considered when a P-value < 0.05 was observed. Box-and-whisker plots coloured in grey represent migrating colonies that were significantly different (P < 0.001) compared to the wild-type phenotype.
B. Representative image of swimming colonies of wt E. coli MG1655 and E. coli MG1655 Δtolpal after 14.5 h of incubation at 30°C on swimming motility agar. Scale bar = 10 mm.
C. Representative image of swarming colonies of wt E. coli MG1655 and E. coli MG1655 Δtolpal after 16 h on swarming motility agar at 30°C. The area of individual swarming colonies (n = 20) was measured using ImageJ and the mean swarming colony areas were compared using an unpaired Student’s t-test of mean difference = 0. The t-test was two-sided and statistical significance was considered when a P-value < 0.05 was observed. The mean swarming colony area and the standard error of the mean for E. coli MG1655 and E. coli MG1655 Δtolpal, and the correspondent P-value are indicated below the representative image. Scale bar = 10 mm.
D. Representative immunofluorescence images of wt E. coli MG1655 and E. coli MG1655 Δtolpal cells labelled with a polyclonal anti-FliC primary antibody and an Alexa Fluor 488-conjugated secondary antibody. Scale bars = 2 μm.
E. Plot depicting velocity of individual cells (n = 166 for wt E. coli MG1655 and n = 118 for E. coli MG1655 Δtolpal). Error bars indicate the standard error of the mean. The mean velocity and standard error for each population are indicated below each plot. Statistical significance bar is shown and the P-value is the result of an unpaired Student’s t-tests of mean difference = 0. The t-test was two-sided and statistical significance was considered when a P-value < 0.05 was observed. Analyses represent the combination of at least three experiments performed on different days.
F. Representative image of cell trajectories. The images consist of an overlay of three images from two independent experiments performed on different days – each coloured line represents the trajectory of an individual cell.
G. Average swimming reversal rate of wild-type E. coli MG1655 and E. coli MG1655 Δtolpal. We performed an independent sample randomization test with 10 000 simulations of differences of means using the R software. Statistical significance was considered when an approximated P-value < 0.05 was observed.
Similarly, all single-gene tol and pal null mutants and the Δtolpal mutant displayed impaired swarming motility compared to wild-type E. coli MG1655 cells (Fig. 3C and Fig. S12A). Plasmid-based complementation of each of the tol or pal deletion mutants only reverted the swarming motility defects of mutants lacking tolQ and tolR (Fig. S12B and C). As observed in the swimming motility assay, overexpression of tolB impaired cell growth completely (Fig. S12B and C). Importantly, the growth parameters of wild-type E. coli MG1655 and all the mutants were identical in liquid media (Fig. S13), suggesting that the differences observed both in the swimming and in the swarming motility assays were not caused by growth defects associated with the deletion of the tol and pal genes. To determine whether the reduction in swimming motility was related to a decrease in flagella density, we immunolabelled flagella of wild-type E. coli MG1655 and Δtolpal mutant cells. We did not detect an apparent difference in flagella density between both cell strains that could explain the dramatic reduction in the size of the motility colonies observed for the mutant E. coli MG1655 Δtolpal (Fig. 3D). It is possible that the deletion of the tol and pal genes change the size, position, spacing, stability, structure or function of flagella (Fig. S14); however, we were unable to measure changes in these parameters using optical microscopy.
To further explore the role of the tol and pal genes in cell motility and verify whether the observed impairment of motility in the Δtolpal mutant was due to a decrease in cell velocity, we measured single cell velocities of wild-type E. coli MG1655 and E. coli MG1655 Δtolpal cells using particle-tracking algorithms. We found evidence (P-value = 0.039) of a difference in swimming velocity between these E. coli strains (Fig. 3E); however, it is unlikely that the difference accounts for the large defects in motility that we observed in the swimming and swarming motility assays (Fig. 3B and C). Further analysis of cell trajectories from the tracking experiments indicated that deleting the Tol–Pal complex may alter the trajectory of cells during runs (Fig. 3F). To determine whether deletion of the tol and pal genes alters the motion of motile E. coli cells, we tracked cells suspended in motility buffer to determine the frequency at which cells tumble. We found that the mean (± s.e.m.) tumbling frequency of E. coli MG1655 Δtolpal cells (0.75 ± 0.09 tumbles·s−1, n = 95) is significantly higher (P < 0.05) than the mean tumbling frequency observed for the wild-type strain (0.54 ± 0.05 tumbles·s−1, n = 102) (Fig. 3G and Fig. S15). This result indicates that the Δtolpal mutant displays biased clockwise flagella rotation.
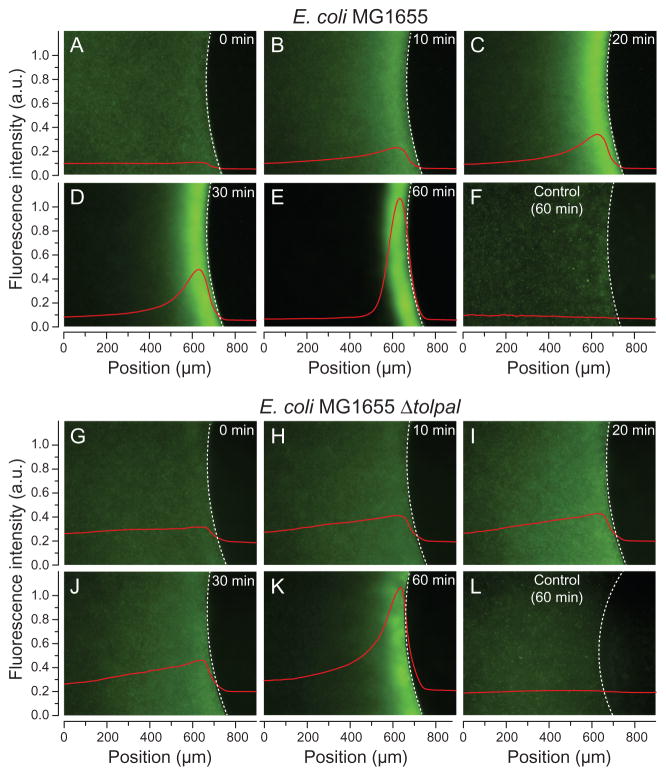
To distinguish between differences in behaviour due to motility and chemotaxis, we investigated the effect of the tol and pal genes on E. coli chemotaxis using a modified plug-in-pond assay (the μPlug assay) (Englert et al., 2009) and qualitatively measured the chemotactic response of E. coli MG1655 and E. coli MG1655 Δtolpal expressing enhanced green fluorescent protein (eGFP) (Fig. 4A–L). Both strains displayed a random distribution of cellular responses in the presence of plugs that lacked chemoattractant (Fig. 4F and L). However, when cells were exposed to a plug containing L-aspartate, both wild-type E. coli MG1655 and E. coli MG1655 Δtolpal exhibited a strong response towards the attractant, which led to the accumulation of bacteria at the agarose plug-liquid interface (Fig. 4A–E and G–K). Time-dependent quantification of the fluorescence intensity demonstrated that the community-wide chemotactic behaviour of both strains was comparable, with the Δtolpal mutant showing an apparent delay in the response. The deletion of tol and pal genes alters the location of chemoreceptors – in the case of Δtolpal, causing diffuse localization of clusters and occasionally an absence of defined foci – yet the mislocalization of these sensors did not impair chemotaxis completely. Thus, our observations of changes in the diameter of colonies of E. coli tol and pal mutants growing in swim plates – compared to the wild-type parent strain – is likely due to the bias in cell motion that accompanies these mutations.
Fig. 4.
Chemotactic response of E. coli cells to L-aspartate using the μPlug assay. Spatial distribution of eGFP-labelled E. coli MG1655 (A–E; F, control) and E. coli MG1655 Δtolpal cells (G–K; L, control) in the presence of 1 mM of L-aspartate. Panels A–E and G–K show a series of representative images obtained from the same time-lapse experiment for each strain. Fluorescence intensity profiles demonstrating the chemotactic response of cells to the μPlug (represented by red lines; a.u., arbitrary units) are overlaid on each image. A dashed white line delineates the agarose plug-liquid interface. We used IGOR Pro (Version 6.22A) to derive the total mean fluorescence intensity profiles along the entire x-axis for each image to visualize cells accumulated at the plug–liquid interface. A moving average was used to smooth the data trend in the series of images. Controls show the distribution of eGFP-labelled cells in the absence of L-aspartate. Images were false-coloured using ImageJ.
Tol–Pal complex components interact with chemoreceptors in E. coli cells
The IM protein TolA and the OM lipoprotein Pal are required for polar localization of the polar marker TipN in Caulobacter crescentus (Yeh et al., 2010). We reasoned that the intact Tol–Pal complex or its individual components may either polarly localize chemoreceptor arrays or nucleate their assembly at the cell poles. To test the hypothesis that chemoreceptors interact directly or indirectly with components of the Tol–Pal complex, we performed co-immunoprecipitation (co-IP) experiments to verify potential associations between chemoreceptors and TolA or Pal.
Initially, we used TolA and Pal null E. coli strains transformed, respectively, with a plasmid containing an N-terminal HA-tagged version of the tolA gene and a C-terminal FLAG-tagged version of the pal gene. Overexpression of the modified proteins restored cell viability in agar containing SDS (Fig. S16A and B), indicating that the tags did not extensively affect the formation and function of the native complex. However, we did not detect a band indicating an association between the chemoreceptors and the tagged versions of TolA and Pal in the immunoblot of the immunoprecipitated samples (Fig. S16C). We reasoned that the epitope tags or the levels of the overexpressed proteins that we tested could prevent interactions with the receptors. Consequently, we used wild-type cells lacking overexpression systems to verify potential interactions.
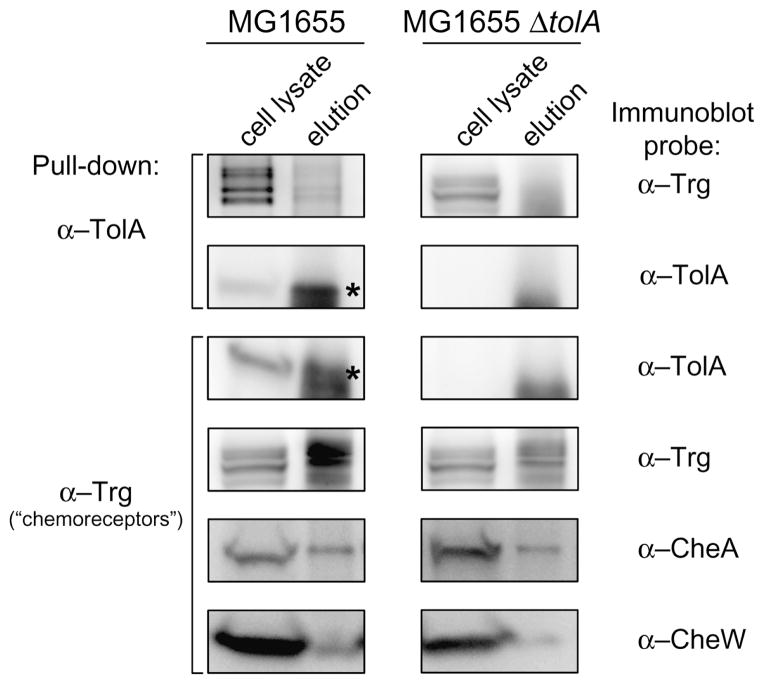
We performed co-IP experiments with wild-type E. coli MG1655 cells; as a negative control we used the same approach in the isogenic mutant lacking the IM protein TolA (E. coli MG1655 ΔtolA). Cells treated with the membrane-permeable cross-linking agent formaldehyde were lysed and suspensions were immunoprecipitated with anti-TolA-coupled beads. Analysis of the purified products by Western blot showed a band corresponding to the expected size for chemoreceptors (Fig. 5), suggesting that TolA and chemoreceptors may interact directly or indirectly in vivo. To confirm our results, we performed a reverse co-IP with whole-cell lysate and immunoprecipitated the samples with anti-Trg-coupled beads. Similarly, analysis of the purified products showed a band corresponding to the expected size for TolA (Fig. 5). As a positive control, in the cell lysate immunoprecipitated with anti-Trg-coupled beads we observed a band corresponding to CheA and CheW, which are known to interact directly with chemoreceptors and form a ternary complex that mediates transduction of the chemotactic signal in E. coli (Alley et al., 1992; Maddock and Shapiro, 1993; Skidmore et al., 2000; Hazelbauer et al., 2008; Briegel et al., 2012; Liu et al., 2012). In conjunction with our localization data, this observation supports the role of the Tol–Pal complex in polar localization of chemoreceptor clusters in E. coli.
Fig. 5.
The inner membrane protein TolA interacts with chemoreceptors. In vivo co-immunoprecipitation (co-IP) of TolA in wild-type E. coli MG1655 and in the isogenic mutant E. coli MG1655 ΔtolA. Western blots of whole-cell lysate and elution fraction of samples immunoprecipitated with anti-TolA (α-TolA) (indicated on the left) were probed with anti-Trg (α-Trg) and with α-TolA (indicated on the right). The reverse co-IP was performed with whole-cell lysate samples immunoprecipitated with α-Trg (indicated on the left) and probed with α-TolA. As a control, Western blots of whole-cell lysate and elution fraction of samples immunoprecipitated with α-Trg were separately probed with anti-CheA (α-CheA), anti-CheW (α-CheW), and α-Trg. The α-Trg is an antiserum raised to highly purified E. coli Trg; however, it also recognizes the other E. coli chemoreceptors (Tar, Tsr, and Tap) (Morgan et al., 1993). Because cross-reactivity of α-TolA with unknown components of the co-IP eluate generated another signal close to the band of interest (TolA), asterisks were added to facilitate visualization of TolA.
Discussion
Role of lipid composition in positioning of chemoreceptor clusters in E. coli cells
A growing number of proteins and complexes have been found to localize to the division plane and the poles in rod-shaped bacteria, making these regions of the cell attractive targets for studying spatial and temporal biomolecule organization. The cell poles exhibit several unique physical features and bacteria may exploit several of them to position proteins at these subcellular regions (recently reviewed in Laloux and Jacobs-Wagner, 2014). Several mechanisms have been formulated to account for the positioning of proteins at the bacterial cell poles, including localization of anionic phospholipid ‘domains’ due to curvature-induced membrane strain, and their role as landmarks for recruitment of proteins to these regions of the cell (Mileykovskaya et al., 2003; Huang and Ramamurthi, 2010; Renner and Weibel, 2011; 2012). Consistent with this model, polar localization of the E. coli osmosensory transporter (ProP) and the mechanosensitive channel (MscS) correlated with the proportion and polar localization of CL (Romantsov et al., 2007; 2008; 2010). We sought to test whether polar localization of chemoreceptor clusters was directly correlated with the presence and localization of CL. We observed that a CL null E. coli strain (E. coli MG1755–BKT12), in which the three known CL synthases are absent and no CL is detected (Tan et al., 2012), displayed the same pattern of localization observed in the wild-type parent strain MG1655. Our results indicate that CL content in the membrane does not alter chemoreceptor localization (Fig. 2A and C).
Deletion of pgsA inhibits the last committed step in the synthesis of the two major anionic phospholipids – PG and CL – in E. coli. Anionic phospholipids are involved in essential cellular processes, including DnaA reactivation (Kitchen et al., 1999), regulation of division plane formation (Renner and Weibel, 2012), and SecA-dependent protein translocation across the IM (de Vrije et al., 1988; Lill et al., 1990; Hendrick and Wickner, 1991); however, they are not essential for cell viability (Kikuchi et al., 2000; Matsumoto, 2001; Shiba et al., 2004). We observed diffuse localization of YFP–CheR in the pgsA null mutant E. coli UE54 and the absence of defined fluorescent puncta. The diffuse fluorescence pattern is most likely due to the reduced concentration or complete absence of chemoreceptor clusters (Fig. S7). Flagella immunostaining demonstrated an absence of flagella in E. coli UE54 (Fig. S17A) and an impairment in swimming motility was confirmed in a swimming motility assay (Fig. S17B and C). In contrast to the UE54 mutant, the Lpp mutant strains UE49, UE51, UE53, and the CL knockout MG1655–BKT12 were motile (Fig. S17B and C). The mutant E. coli UE54 is originally derived from a pgsA mutant isolated during the course of the construction of a strain in which anionic phospholipids content is controlled by an inducer added exogenously (Kikuchi et al., 2000; Shiba et al., 2004). Growth arrest in the E. coli pgsA null mutant is alleviated by the lack of the major outer membrane lipoprotein Lpp (Kikuchi et al., 2000). Two additional mutations, namely disruption of rcsF (to suppress thermo-sensitivity of the lpp/pgsA double-mutant) and deletion of araC (gene encoding the transcriptional regulator of the ara operon) were introduced into E. coli strain UE54. As the absence of rcsF did not perturb the polar localization of YFP–CheR in E. coli cells (Fig. S9), it is likely that both the absence of chemoreceptors and flagellar filaments in E. coli UE54 is a consequence of alterations in the levels of anionic phospholipids caused by the deletion of the pgsA gene. Previous studies have shown the connection between flagella biosynthesis and PG levels in E. coli (Tomura et al., 1993; Mizushima et al., 1995). As E. coli MG1655–BKT12 cells lack CL and do not display these chemoreceptor or flagella phenotypes, we hypothesize that these defects arise from the absence of PG. Our data indicate that CL clearly does not influence polar localization of the chemoreceptors.
Role of cell wall curvature in positioning of chemoreceptor clusters in E. coli cells
Another model for the concentration of chemoreceptors at the polar region of cells is that chemoreceptor clusters may have a shape that is sufficient to sense negatively curved regions of the membrane, such as the cell poles. Biophysical models predict that protein complexes destined for the cell poles using this mechanism need to assemble into structures that have a length scale that is sufficient to sense membrane curvature (Huang et al., 2006; Lenarcic et al., 2009; Huang and Ramamurthi, 2010). Direct sensing of cell curvature by protein complexes has been reported for SpoVM and the self-assembling protein DivIVA in Bacillus subtilis (Lenarcic et al., 2009; Ramamurthi and Losick, 2009; Ramamurthi et al., 2009). To test whether chemoreceptor clusters localize to the E. coli cell poles by sensing membrane curvature, we used microengineering approaches to artificially manipulate the bacterial cell curvature. We confined filamented cells into microchannels designed to physically impose curvature in the cylindrical region of the cell body mimicking the geometry of the bacterial poles (Renner et al., 2013) and studied the localization of YFP–CheR in cells with engineered curvature. The curvature imposed by the microchannels had no apparent influence on the localization or distribution of YFP–CheR (Fig. S18); however, we are unable to achieve a cell wall curvature that matches the cell poles. This result suggests that the geometry of the cell wall – and the subsequent strain-induced organization of phospholipids within the cell membrane – is likely not the primary cue controlling the subcellular positioning of chemoreceptor clusters.
Role of auxiliary protein complexes in positioning of chemoreceptor clusters
Another hypothesized mechanism for localizing chemoreceptors at the cell poles and the septum is by diffusion through the membrane followed by direct or indirect association with proteins or protein complexes that localize to the poles. The ‘diffusion-and-capture’ mechanism has been hypothesized for recruitment of several polarly localized proteins (Bowman et al., 2008; Ebersbach et al., 2008; Ringgaard et al., 2011; Yamaichi et al., 2012). In association with the polar transmembrane protein HubP, the ATPase ParC polarly localizes Vibrio cholerae chemoreceptors and downstream signalling proteins by actively recruiting the proteins to the cell poles before cytokinesis, ensuring that newborn cells have a cluster from the old-pole after cell division (Ringgaard et al., 2011; Yamaichi et al., 2012). Recently, the function of the Tol–Pal complex in positioning a polar protein was reported in C. crescentus (Yeh et al., 2010). The interaction of individual components of the Tol–Pal complex with the transmembrane protein TipN suggests that the complex may function as a molecular anchor to recruit proteins to the cell poles. Our observation that the disruption of the E. coli Tol–Pal complex affected polar chemoreceptor localization – and not clustering (Fig. 2A and C and Fig. S2) – prompted us to investigate the connection between the complex and the polar positioning of chemoreceptor clusters. We found evidence of an interaction between the IM transmembrane protein TolA and the chemoreceptors (Fig. 5). It is possible that the chemoreceptors interact with other IM components of the Tol–Pal components. Alternatively, cell envelope defects created by inactivation of the Tol–Pal complex may disrupt the interaction between chemoreceptor clusters and unknown positioning factors. Forward genetic screens may be useful for identifying these factors. We observed polar localization of the polar proteins TnaA and IcsA in cells containing Tol–Pal defects (Fig. 2E and F), which suggests that membrane integrity mediated by the Tol–Pal complex may be particularly required for maintaining polar localization of chemoreceptor clusters or a subset of polar proteins.
Cell motility and chemotaxis defects in Tol–Pal mutants
The impairment of cell motility in cells lacking a Tol–Pal complex has been reported previously in Pseudomonas putida (Llamas et al., 2000). Similarly, we observed that the disruption of the Tol–Pal complex significantly affects both E. coli swimming and swarming motility (Fig. 3A–C; Figs S10A–D, S11A and B and S12A–C). Single-cell tracking revealed that the Δtolpal mutant has enhanced clockwise-biased flagellar rotation (Fig. 3F and G), which may reflect a defect in the phosphorelay cascade or in the sensory adaptation system. Interestingly, although the mutants that display mislocalization of chemoreceptors caused by disruption of the Tol–Pal complex show motility and chemotaxis defects, they are still able to sense chemical gradient (Fig. 4G–L). Clustering of the ternary complexes – chemoreceptors, CheA, and CheW – is thought to play an essential role in chemotaxis signalling, and chemoeffector amplification and adaptation (Bray et al., 1998; Shimizu et al., 2000; Gestwicki and Kiessling, 2002; Sourjik and Berg, 2002; reviewed in Sourjik and Armitage, 2010). However, the role of chemoreceptor polarity on cell physiology and behaviour is not completely understood. We tested a range of concentrations of chemoattractant (50 μM to 3 mM of L-aspartate) to verify whether the sensitivity of the system was affected in Δtolpal mutants in which the chemoreceptor clusters were mislocalized. We did not observe dramatic changes in cell response over time (data not shown); however, it is possible that the lowest concentration of L-aspartate that we tested in the μPlug assay masks potential differences in sensitivity between the wild-type and Δtolpal mutant cells. The chemotactic response may be optimized for polarly localized machinery because accumulation at the poles favours the assembly of larger chemoreceptor lattices (Maddock and Shapiro, 1993; Thiem et al., 2007; Greenfield et al., 2009) that may have maximum sensitivity.
Positioning of chemoreceptors clusters mediated by the Tol–Pal complex
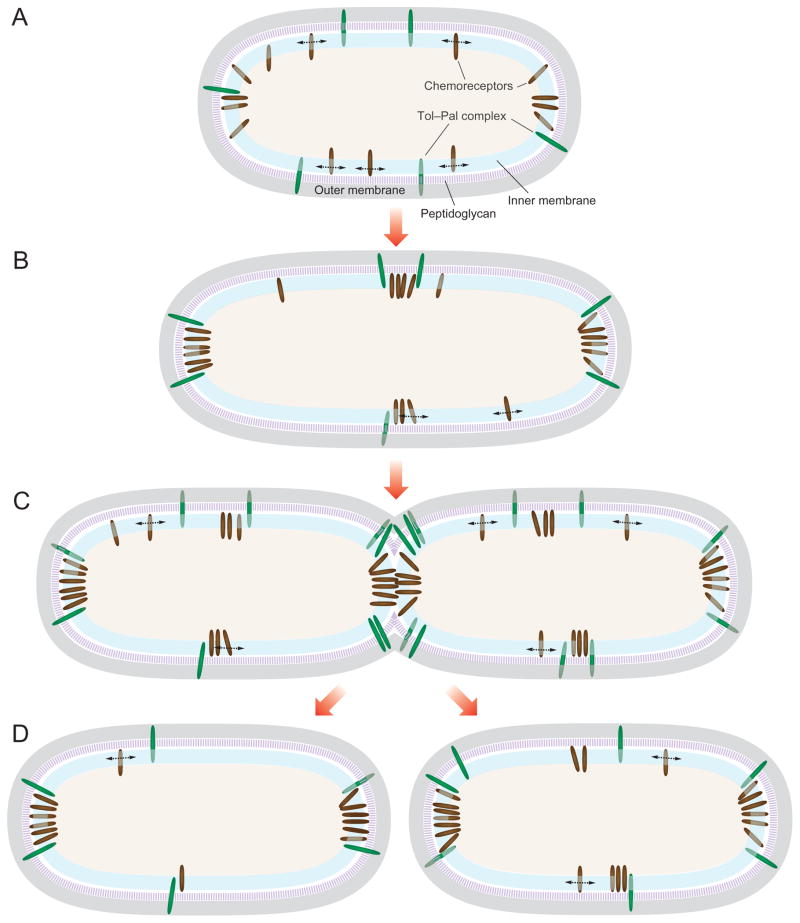
We envision a mechanism for the Tol–Pal-mediated polar localization and accumulation of E. coli chemoreceptor clusters as depicted in Fig. 6. As newly synthesized chemoreceptors are inserted into the cytoplasmic membrane, the individual receptors freely migrate within the IM phospholipid bilayer to join existing clusters or nucleate new ones (Fig. 6A). The stochastic nucleation model of clustering and positioning suggests that the distribution of receptors is distance-dependent and large clusters form in regions of the cell that are furthest from large existing clusters (Thiem et al., 2007; Thiem and Sourjik, 2008; Wang et al., 2008; Greenfield et al., 2009). Therefore, in a cell with large clusters at both poles, new clusters will preferentially form at the mid-cell, which is the location furthest from both poles (Fig. 6B). The E. coli Tol–Pal complex, in co-ordination with other structural proteins such as Lpp, may restrict the diffusion of polar and mid-cell chemoreceptor clusters. The distance-dependent chemoreceptor nucleation mechanism results in a periodic distribution of clusters along the cell body that is coincident with future division sites (Thiem et al., 2007; Thiem and Sourjik, 2008; Wang et al., 2008). We propose a mechanism in which the Tol–Pal complex imposes a physical barrier that favours the accumulation of the clusters at the poles and division site (Fig. 6C and D). Previous studies have suggested that unknown periodic structures positioned at the future cell division sites may assist in the segregation of the receptors (Thiem et al., 2007). A mechanism that co-ordinates the position of the chemosensory proteins and cell division assures that each daughter cell inherits an intact chemosensory system.
Fig. 6. Proposed model of Tol–Pal in the subcellular localization of E. coli chemoreceptor clusters.
A. As newly synthesized chemoreceptors are inserted into the cytoplasmic membrane, the individual receptors migrate freely within the IM and join existing clusters or nucleate new ones.
B. Receptor distribution is distance-dependent and clusters accumulate in regions that are furthest from existing clusters (Thiem et al., 2007; Thiem and Sourjik, 2008; Wang et al., 2008; Greenfield et al., 2009).
C and D. The Tol–Pal complex localizes to the division plane in early predivisional cells to facilitate constriction of the outer membrane during cell division (Gerding et al., 2007). This trans-envelope complex may restrict the mobility of polar and mid-cell chemoreceptor clusters by creating a physical barrier that favours their accumulation at these cellular locations. Dashed arrows denote receptor diffusion within the membrane. Green ellipses represent the Tol–Pal complex and brown ellipses represent chemoreceptors. See the text for an additional explanation and discussion.
The positioning of the lateral chemoreceptor clusters in E. coli is independent of the MinCDE division plane positioning system (Thiem et al., 2007). In support of the role of the Tol–Pal complex in the polar positioning of chemoreceptors, Tol and Pal proteins have been reported to localize dynamically during the E. coli cell cycle and form a subcomplex of the division apparatus in Gram-negative bacteria (Gerding et al., 2007). The Tol–Pal complex is held together by transient interactions near the septal ring during early stages of cell wall constriction and co-ordinates with the divisome to mediate the invagination of the OM during cytokinesis (Gerding et al., 2007). Similarly, components of the C. crescentus Tol–Pal complex localize to the division plane in early predivisional cells and remain predominantly at the new pole of swarmer and stalked progeny upon completion of division (Yeh et al., 2010). The role of the Tol–Pal complex in chemoreceptor positioning may not be a general mechanism in Gram-negative bacteria. However, a conceptually similar mechanism co-ordinating cell division, chromosome partitioning, and segregation of the chemotaxis machinery has been recently reported. In some polar-flagellated bacteria such as Vibrio, chemoreceptor cluster positioning relies on the proteins HubP and ParC, which have a cell cycle-dependent localization pattern and actively recruit chemosensory proteins to the cell poles (Ringgaard et al., 2011; Yamaichi et al., 2012). In contrast, the positioning of membrane chemoreceptor clusters in Rhodobacter sphaeroides is not correlated with future division sites, and neither FtsZ or MreB affect localization of the receptors (Chiu et al., 2013). Thus, it is apparent that different organisms may use diverse mechanisms to orchestrate the positioning and segregation of the chemosensory machinery and cell division.
In summary, our results demonstrate that the intact E. coli Tol–Pal complex is required for polar localization of chemoreceptors clusters. Polar accumulation of receptor arrays occurs independently of the lipid composition of the cytoplasmic membrane and may not be dictated by the geometry of the cell poles. Mutations that prevent the trans-envelope connections of the Tol–Pal complex and cause mislocalization of chemoreceptors affect cell motility and chemotaxis. Cumulatively, our data suggest that the E. coli Tol–Pal complex interacts with chemoreceptors and restricts the diffusion of receptor arrays. This mechanism may establish and maintain the polar localization of chemoreceptors at specific regions of the cell and may be component of a regulatory mechanism that co-ordinates cell division with segregation of the chemosensory machinery.
Experimental procedures
Bacterial strains and growth conditions
Escherichia coli cells were grown aerobically in lysogeny broth (LB) [1% (w/v) tryptone, 0.5% (w/v) yeast extract, and 1% (w/v) NaCl] or in LB medium containing 1.5% (w/v) agar (LB agar) at 37°C, unless otherwise indicated. We added ampicillin (50 μg ml−1), carbenicillin (50 μg ml−1), chloramphenicol (25 μg ml−1), and kanamycin (30 μg ml−1) to the medium or agar as needed. Chemicals and culture media reagents were obtained from BD Biosciences (San Jose, CA), Fisher Scientific (Itasca, IL) and Sigma-Aldrich (St. Louis, MO). E. coli strains and plasmids used in this work are listed in Table S1.
Genetic methods
Standard recombinant DNA procedures and manufacturer protocols were used for genomic and plasmid DNA purification, restriction endonuclease digestion, DNA amplification, gene cloning, transformation, and mutagenesis. Genomic DNA from E. coli strains was isolated with the MasterPure™ DNA Purification Kit (Epicentre, Madison, WI) according to the manufacturer’s instructions. Oligonucleotide primers were obtained from IDT (Coralville, IA) and restriction enzymes were from Promega (Madison, WI) or New England Biolabs (Ipswich, MA). We used platinum PfuUltra II Fusion HS DNA polymerase (Agilent Technologies/Stratagene, Santa Clara, CA) to produce DNA for cloning and mutagenesis, and GoTaq DNA polymerase (Promega) for general analytical PCR. Control reactions were always tested side-by-side. Plasmid constructs were purified using a QIAprep Miniprep Kit (Qiagen, Valencia, CA) and verified by PCR and DNA sequencing. We used electroporation (Sambrook and Russell, 2001) or chemical methods (Chung et al., 1989) to transform E. coli strains with plasmids. Chromosomal deletions, genetic integration, and removal of antibiotic resistance cassette FRT–kan–FRT were performed using lambda-red method recombination (Datsenko and Wanner, 2000). P1 phage transduction (Miller, 1992) was used, whenever possible, to transfer the FRT–kan–FRT cassette between E. coli strains. We verified chromosomal deletions and integrations by testing for the presence of antibiotic markers and by PCR analysis and DNA sequencing. In addition, we verified the successful deletion of tol and pal mutants by determining the susceptibility of these mutants to 0.5% SDS. All primers used in this study are listed in Table S2.
Fluorescence microscopy, image and data analysis
Escherichia coli cells harbouring the plasmid pPA803 (encoding a functional YFP–CheR fusion) (Zhou et al., 2011) were grown overnight in LB broth, diluted (1:100) in 5 ml of fresh LB broth and incubated at 37°C with agitation (200 r.p.m.) until an absorbance of 0.6 (λ = 600 nm) was reached. We induced expression of the fluorescent fusion protein by adding 100 μM of isopropyl β-D-1-thiogalactopyranoside (IPTG) and additional incubation for 1 h at 37°C with agitation (200 r.p.m.). E. coli cells harbouring a functional chromosomal TnaA–GFP fusion (Li and Young, 2012) were grown overnight in tryptone broth (TB) [1% (w/v) tryptone, 0.5% (w/v) NaCl], diluted (1:100) in 5 ml of fresh TB and incubated at 37°C with agitation (200 r.p.m.) until mid-log phase. Overnight cultures of E. coli cells harbouring the plasmid pMAC382 (encoding an IcsA–GFP fusion) (Charles et al., 2001) were grown in M9 minimal medium [1× M9 salts (17 mM NaCl, 9 mM NH4Cl, 22 mM KH2PO4, 48 mM Na2HPO4·7H2O), 0.1 mM CaCl2, 2 mM MgSO4, 0.4% (w/v) glucose] containing 0.2% (v/v) glycerol, diluted (1:100) in 5 ml of fresh M9 minimal medium and incubated at 37°C with agitation (200 r.p.m.) until an absorbance of 0.6 (λ = 600 nm) was reached. We induced expression of the fluorescent fusion protein by adding 0.2% (w/v) of L-arabinose and additional incubation in the dark for 1 h at 25°C with agitation (200 r.p.m.).
We added an aliquot (4 μl) of the induced culture on 2% (w/v) agarose pads prepared in 1× PBS buffer [137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4) (pH 7.4)], covered the samples with a glass coverslip and imaged the cells at 25°C on an inverted Nikon Eclipse TE2000 epifluorescence microscope equipped with a shuttered black and white Andor iXonEM+ DU-897 electron-multiplying charge-coupled-device (EMCCD) camera (Andor Technology, South Windsor, CT). Images were acquired using a ×100 objective (Nikon Plan Apo 100/1.40 oil DM) and the Metamorph software program (Version 7.5.6.0) (MDS Analytical Technologies, Downington, PA). Data were collected on the EMCCD using an exposure time of 33 ms; for fluorescent images we used an electron-multiplying (EM) gain of 100.
To analyse the localization of proteins in E. coli cells, we created fluorescence intensity profiles of cells using the MATLAB-based program (MathWorks, Natick, MA), Microbe-Tracker (Sliusarenko et al., 2011). Briefly, the average background fluorescence was subtracted from the specific fluorescence measured in cells. Cells were segmented, a spline was fit to the mid-point of each segment (to determine cell length), and the total fluorescence in each segment was determined. The sum of the integrated fluorescence of the segments was normalized to the total integrated fluorescence from an entire cell, which verified that segmentation did not introduce any artefacts. The relative position of the target protein was determined by comparing the fluorescence intensity of segments to their spatial position. We only analysed predivisional cells and verified the position of proteins by manual inspection. To display the images, we overlaid phase contrast and the corresponding fluorescence images and false-coloured them using Adobe Photoshop CS5; no further adjustments or other image processing was performed. To compare the differences between the localization patterns in the various E. coli strains, we performed Chi-squared tests. We considered a P-value < 0.05 to indicate statistically significant data.
Motility assay
Analysis and quantification of the swimming and swarming motility phenotypes were performed as described previously (Copeland et al., 2010), with minor modifications. We measured the diameter of swimming migrating colonies in 0.25% (w/v) agar gel infused with nutrient broth [1.0% (w/v) peptone, 0.5% (w/v) NaCl, 0.3% (w/v) beef extract], herein referred to as swimming motility agar. We measured the area of swarming migrating colonies in 0.45% (w/v) Eiken agar infused with nutrient broth [1.0% (w/v) peptone, 0.5% (w/v) NaCl, 0.3% (w/v) beef extract] and 0.5% (w/v) glucose, herein referred to as swarming motility agar. Briefly, we poured 50 ml of warm motility agar into 150 × 15 mm Petri dishes, solidified the agar at 25°C for 50 min, and removed excess liquid from the surface by storing the plates in a laminar flow hood for 20 min with the covers of the dishes ajar. We inoculated the surface of motility agar with 4 μl of a fresh E. coli cell suspension, containing approximately 105 cfu ml−1, diluted in 1× PBS buffer (pH 7.4) and incubated the plates at 30°C at ~ 90% relative humidity in a static incubator. We measured the diameter of the swimming colonies after 14.5 h and the area of the swarming colonies after 16 h. For complementation experiments, we used the same procedure described above with a minor modification: swimming or swarming motility agars were supplemented with the appropriate antibiotic and 1 mM or 0.1 mM IPTG. To test for differences between motility phenotype in the various E. coli strains, we performed one-way or two-way ANOVA followed by Tukey’s multiple-comparison post-test. We considered a P-value < 0.05 to indicate statistically significant data.
Immunofluorescence microscopy of E. coli flagella
We performed flagella immunostaining of E. coli MG1655 and of the isogenic mutant MG1655 Δtolpal cells as described previously (Tuson et al., 2013), with minor modifications. We harvested 500 μl of overnight cultures at 2500 g for 5 min at 25°C and gently resuspended the cells in the same volume of 1× PBS buffer (pH 7.4). We diluted (1:50) the cell suspension and added 2 aliquots of 15 μl to each chamber. The remaining procedure was performed according to that described by Tuson et al. (2013). Both the primary antibody (anti-FliC) and the secondary antibody (Alexa Fluor 488-conjugated goat anti-rabbit IgG) (Invitrogen, Grand Island, NY) were diluted 1:100 in blocking buffer. Cells were imaged at 25°C using an inverted Nikon Eclipse Ti microscope equipped with a Photometrics CoolSNAP HQ2 charge-coupled-device (CCD) camera (Photometrics, Tucson, AZ). Images were acquired using a ×100 oil objective (Nikon Plan Apo 100/1.40 oil Ph3 DM) and the Nikon Instruments Software (NIS)-Elements Advanced Research (AR) microscope imaging software program (Version 4.000.07) (Nikon, Melville, NY).
Cell velocity measurements
We measured the velocity of individual E. coli MG 1655 and MG1655 Δtolpal cells (in μm s−1) using a particle-tracking algorithm, as described previously (Copeland et al., 2010; Tuson et al., 2013), with minor modifications. Briefly, cell cultures were grown overnight at 37°C in TB. Saturated overnight cells cultures were diluted 1:100 into 10 ml of fresh TB and grown in 250 ml Erlenmeyer flasks at 37°C with agitation (200 r.p.m.). We harvested cells in the mid-log phase (absorbance of 0.8, λ = 600 nm), centrifuged an aliquot (1 ml) of the cell suspension for 5 min at 2000 g at 25°C, and gently resuspended the cells in the same volume of motility buffer [0.01 M KPO4, 0.067 M NaCl, 10−4 M EDTA (pH 7.0)] (Turner et al., 2000), containing 0.1 M glucose, and 0.001% (w/v) Brij-35. A 20 μl aliquot of the suspension diluted 1:1000 in motility buffer containing 0.1 M glucose, and 0.001% (w/v) Brij-35 was placed inside a ring of Apiezon M grease on pre-cleaned glass slides and sealed with a #1.5 mm glass coverslip. We imaged cell motility using a Nikon Eclipse 80i phase contrast upright microscope and a black and white Andor LucaS EMCCD camera (Andor Technology). Images were acquired using a ×40 ELWD dry objective (Nikon Plan Fluor 40/0.60 dry Ph2 DM). Motility videos consisting of 300 frames were collected with a 33 ms exposure time (30 frames per second). Microscopy data were analysed using MATLAB (MathWorks) by identifying the centre of mass of each bacterium in successive frames and grouping those points together to create a cell trajectory, from which we determined the mean cell velocity. Only tracks that had more than 30 frames (i.e. more than 1 s) – a minimal total length of 10 μm, and a minimal displacement of 5 μm – were used. Cells that moved in a constant tumbling or wobbling manner or stuck to the coverslip were manually discarded from the analysis. To compare the mean velocity of the two populations, we performed an unpaired Student’s t-test and considered a P-value < 0.05 to indicate statistically significant data.
We visually determined the reversals in swimming direction of E. coli MG1655 and MG1655 Δtolpal cells. Three individual experiments were performed and in each experiment at least 30 cells were analysed. Cells were tracked for as long as possible, the number of reversals was counted, and the reversal frequency was calculated as the number of reversals per second per cell. For each experiment, we calculated the average reversal frequency. We compared the reversal frequency using a randomization (permutation) test (10 000 simulations of differences of mean) and considered a P-value < 0.05 to indicate statistically significant data.
Chemotaxis assay
We qualitatively measured the chemotactic response of E. coli MG1655 and of MG1655 Δtolpal using a modified plug-in-pond assay (the microplug assay: μPlug) as described previously (Englert et al., 2009), with minor modifications. Briefly, E. coli containing the enhanced green fluorescent protein (eGFP)-encoding plasmid ptetEGFP were grown overnight at 37°C in LB containing 50 μg ml−1 of ampicillin. Saturated overnight cultures of cells were diluted 1:100 into 10 ml of fresh LB containing 50 μg ml−1 of ampicillin and grown in 125 ml Erlenmeyer flasks at 37°C with agitation (200 r.p.m.). When the cell cultures reached an absorbance of 0.8 (λ = 600 nm) we induced expression of the fluorescent protein by adding 200 ng ml−1 of anhydrotetracycline (aTc) for 1 h. We harvested an aliquot of the cell suspension (1 ml), centrifuged for 5 min at 2000 g at 25°C, and gently washed and resuspended the cells in the same volume of motility buffer containing 0.001% (w/v) Brij-35. We examined cells by phase contrast microscopy to ensure robust motility and normal run-tumble swimming behaviour. Cells were loaded into the microfluidic chamber containing the μPlug [0.5% (w/v) agarose dissolved in motility buffer containing 0.001% (w/v) Brij-35] infused with 1 mM of L-aspartate or with motility buffer (negative control). Cells were imaged at 25°C for 1 h with a time-lapse of approximately 1 min using an inverted Nikon Eclipse Ti microscope equipped with a Photometrics CoolSNAP HQ2 CCD camera (Photometrics, Tucson, AZ). Images were acquired using a ×4 (Nikon Plan Apo 4/0.2) dry objective and the NIS-Elements AR microscope imaging software program (Version 4.000.07) (Nikon, Melville, NY). We used IGOR Pro (Version 6.6A2) (WaveMetrics, Lake Oswego, OR) to analyse the images and calculate the total fluorescence intensity profiles.
Co-immunoprecipitation (co-IP)
Escherichia coli MG1655 and E. coli MG1655 ΔtolA cells were grown overnight in LB broth, diluted (1:100) in 500 ml of fresh LB broth, and incubated at 37°C with agitation (200 r.p.m.) for 5 h. For experiments in which we used E. coli MG1655 cells harbouring the plasmid(s) encoding the tagged versions of the proteins of interest, E. coli MG1655 ΔtolA and E. coli MG1655 Δpal cells were grown overnight in LB broth containing the adequate antibiotic, diluted (1:100) in 500 ml of fresh LB broth, and incubated at 37°C with agitation (200 r.p.m.) until reaching an absorbance of 0.6 (λ = 600 nm). We induced expression of the HA-tagged TolA and the FLAG-tagged Pal by adding 0.04% (w/v) of L-arabinose and 1 mM IPTG, respectively, for 3 h. We harvested cells by centrifuging the cultures for 20 min at 5000 g at 4°C, washed the pellets in 30 ml of co-IP buffer [20 mM HEPES (pH 7.5), 100 mM NaCl, and 20% (v/v) glycerol] by centrifuging for 10 min at 12 000 g at 4°C, and resuspended the cell pellets in 30 ml of co-IP buffer containing 0.05% (v/v) of Triton X-100. We treated the cell suspension with formaldehyde to a final concentration of 1% for 30 min at room temperature on a rotatory platform, and quenched the mixture with 0.125 M of glycine. We added 20 mg of lysozyme and 1 mM of phenylmethylsulphonyl fluoride (PMSF) to the mixture, incubated for 30 min at 4°C on a rotatory platform, and lysed the cells by one passage through a French pressure cell press (American Instrument Company, Silver Springs, MD) at 1000 lb in−2. We incubated the suspension for 30 min at 4°C on a rotatory platform and removed the insoluble material by centrifuging the cell lysate for 30 min at 12 000 g at 4°C. We collected 10 ml of the supernatant and incubated with anti-TolA (1:2000), anti-Trg (1:5000), anti-FLAG (1:2000) (Thermo Scientific, Rockford, IL), or anti-HA (1:2000) (Thermo Scientific, Rock-ford, IL) at 4°C for 18 h. Subsequently, we added agarose beads conjugated with Protein A/G (sufficient for binding 2.5 mg of IgG; Thermo Scientific) and incubated the mixture for 18 h at 4°C on a rotating platform. We pelleted the beads by centrifuging for 3 min at 2000 g at 25°C, washed them three times with 3 ml of co-IP buffer, and washed three times with 3 ml of wash buffer [50 mM Tris-HCl (pH 7.4) and 150 mM NaCl]. After the last wash, we removed the supernatant, resuspended the beads in 100 μl of 1× Laemmli sample buffer [62.5 mM Tris·Cl (pH 6.8), 25% (v/v) glycerol, 2% SDS (w/v), 0.01% (w/v) bromophenol blue] containing 5% (v/v) of β-mercaptethanol, and eluted the bound proteins by incubating the mixture for 15 min at 100°C. We centrifuged the samples at 12 000 g for 3 min to pellet the agarose beads and collected the supernatant to perform the immunoblot experiments.
SDS-PAGE and immunoblot analysis
All sample fractions were analysed on a 12.5% SDS-PAGE gel at 150 V for 60 min in protein electrophoresis buffer (25 mM Tris, 192 mM glycine, 0.1% SDS, pH 8.3). Following electrophoresis, we transferred proteins to an Amersham Hybond ECL nitrocellulose membrane (GE Healthcare, Pittsburgh, PA) for 2 h at 400 mV in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol). The nitrocellulose membrane was rinsed three times with deionized water and blocked overnight at 4°C overnight in 5% non-fat dry milk in TBST buffer [20 mM Tris, 137 mM NaCl, 1% (v/v) Tween-20] (pH 7.6). Blots were washed three times for 10 min in TBST buffer. Primary antibody was diluted in TBST and blots were incubated in this solution for 1 h. Subsequently, we washed blots three times for 10 min in TBST. Rabbit anti-mouse or goat anti-rabbit IgG secondary antibody conjugated to horseradish peroxidase (HRP) (Thermo Scientific) was diluted in TBST, and blots were incubated in this solution for 1 h. Blots were washed three times for 10 min in TBST and developed for chemiluminescence analysis using the Pierce ECL Western Blotting Substrate (Thermo Scientific). Antibody dilutions were as follows: anti-Trg at 1:2000; anti-Pal at 1:2000; anti-TolA at 1:2000; anti-FLAG monoclonal antibody (Thermo Scientific) at 1:2000, anti-HA monoclonal antibody (Thermo Scientific) at 1:2000, anti-6×His monoclonal antibody (Thermo Scientific) at 1:2000, HRP-conjugated goat anti-rabbit IgG (Thermo Scientific) at 1:5000, HRP-conjugated rabbit anti-mouse IgG (Thermo Scientific) at 1:5000; anti-GFP monoclonal antibody (Thermo Scientific) at 1:2000.
Statistical analysis
All statistic testes used in our analysis were two-sided, and P-values < 0.05 were considered significant. Statistical analyses were performed using the computing environment R (Version 3.0.1) (R Development Core Team, 2005; http://www.r-project.org/).
Supplementary Material
Acknowledgments
We acknowledge materials from Howard Berg (anti-FliC antibody), Marcia Goldberg (plasmid pMAC382); Gerald Hazelbauer (anti-Trg antibody), Roland Lloubès (anti-TolA antibody), Sandy Parkinson (plasmid pPA803, anti-CheA, and anti-CheW antibodies), Chris Raetz (E. coli strain MG1655–BKT12), Victor Sourjik (plasmid pVS102), and Kevin Young (E. coli strain GL40). We also thank the following scientists for their input: Ned Wingreen, Matthew Copeland, Ye-Jin Eun, Piercen Oliver, Lars Renner, and Hannah Tuson. This research was supported by the National Science Foundation (through award MCB-1120832 and DMR-1121288) and the National Institutes of Health (through award 1DP2OD008735-01).
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Alley MR, Maddock JR, Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubès R. Escherichia coli tol–pal mutants form outer membrane vesicles. J Bacteriol. 1998;180:4872–4878. doi: 10.1128/jb.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E, Derouiche R, Rigal A, Lloubès R, Lazdunski C, Bénédetti H. Peptidoglycan-associated lipoprotein–TolB interaction. A possible key to explaining the formation of contact sites between the inner and outer membranes of Escherichia coli. J Biol Chem. 1995;270:11071–11077. doi: 10.1074/jbc.270.19.11071. [DOI] [PubMed] [Google Scholar]
- Bouveret E, Bénédetti H, Rigal A, Loret E, Lazdunski C. In vitro characterization of peptidoglycan-associated lipoprotein (PAL) – peptidoglycan and PAL–TolB interactions. J Bacteriol. 1999;181:6306–6311. doi: 10.1128/jb.181.20.6306-6311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim Biophys Acta. 1975;415:335–377. doi: 10.1016/0304-4157(75)90013-1. [DOI] [PubMed] [Google Scholar]
- Braun V. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- Bray D, Levin MD, Morton-Firth CJ. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Dias DP, et al. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, et al. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci USA. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci USA. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Lloubès R, Sturgis JN. The TolQ–TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA–MotB. Mol Microbiol. 2001;42:795–807. doi: 10.1046/j.1365-2958.2001.02673.x. [DOI] [PubMed] [Google Scholar]
- Charles M, Pérez M, Kobil JH, Goldberg MB. Polar targeting of Shigella virulence factor IcsA in Enterobacteriacae and Vibrio. Proc Natl Acad Sci USA. 2001;98:9871–9876. doi: 10.1073/pnas.171310498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SW, Roberts MAJ, Leake MC, Armitage JP. Positioning of chemosensory proteins and FtsZ through the Rhodobacter sphaeroides cell cycle. Mol Microbiol. 2013;90:322–337. doi: 10.1111/mmi.12366. [DOI] [PubMed] [Google Scholar]
- Choi DS, Yamada H, Mizuno T, Mizushima S. Trimeric structure and localization of the major lipoprotein in the cell surface of Escherichia coli. J Biol Chem. 1986;261:8953–8957. [PubMed] [Google Scholar]
- Chung CT, Niemela SL, Miller RH. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel T, Germon P, Vianney A, Portalier R, Lazzaroni JC. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol Microbiol. 1998;29:359–367. doi: 10.1046/j.1365-2958.1998.00945.x. [DOI] [PubMed] [Google Scholar]
- Click E, Webster R. Filamentous phage infection: required interactions with the TolA protein. J Bacteriol. 1997;179:6464–6471. doi: 10.1128/jb.179.20.6464-6471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland MF, Flickinger ST, Tuson HH, Weibel DB. Studying the dynamics of flagella in multicellular communities of Escherichia coli by using biarsenical dyes. Appl Environ Microbiol. 2010;76:1241–1250. doi: 10.1128/AEM.02153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JK, Reeves P. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group A. J Bacteriol. 1975;123:102–117. doi: 10.1128/jb.123.1.102-117.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derouiche R, Bénédetti H, Lazzaroni JC, Lazdunski C, Lloubès R. Protein complex within Escherichia coli inner membrane. TolA N-terminal domain interacts with TolQ and TolR proteins. J Biol Chem. 1995;270:11078–11084. doi: 10.1074/jbc.270.19.11078. [DOI] [PubMed] [Google Scholar]
- Derouiche R, Gavioli M, Bénédetti H, Prilipov A, Lazdunski C, Lloubès R. TolA central domain interacts with Escherichia coli porins. EMBO J. 1996;15:6408–6415. [PMC free article] [PubMed] [Google Scholar]
- Dubuisson JF, Vianney A, Lazzaroni JC. Mutational analysis of the TolA C-terminal domain of Escherichia coli and genetic evidence for an interaction between TolA and TolB. J Bacteriol. 2002;184:4620–4625. doi: 10.1128/JB.184.16.4620-4625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert DL, Jayaraman A, Manson MD. Microfluidic techniques for the analysis of bacterial chemotaxis. Methods Mol Biol. 2009;571:1–23. doi: 10.1007/978-1-60761-198-1_1. [DOI] [PubMed] [Google Scholar]
- Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci USA. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar JA, Thomas JA, Marolda CL, Valvano MA. Surface expression of O-specific lipopolysaccharide in Escherichia coli requires the function of the TolA protein. Mol Microbiol. 2000;38:262–275. doi: 10.1046/j.1365-2958.2000.02094.x. [DOI] [PubMed] [Google Scholar]
- Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol–Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germon P, Clavel T, Vianney A, Portalier R, Lazzaroni JC. Mutational analysis of the Escherichia coli K-12 TolA N-terminal region and characterization of its TolQ-interacting domain by genetic suppression. J Bacteriol. 1998;180:6433–6439. doi: 10.1128/jb.180.24.6433-6439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestwicki JE, Kiessling LL. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature. 2002;415:81–84. doi: 10.1038/415081a. [DOI] [PubMed] [Google Scholar]
- Gestwicki JE, Lamanna AC, Harshey RM, McCarter LL, Kiessling LL, Adler J. Evolutionary conservation of methyl-accepting chemotaxis protein location in Bacteria and Archaea. J Bacteriol. 2000;182:6499–6502. doi: 10.1128/jb.182.22.6499-6502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold VAM, Robson A, Bao H, Romantsov T, Duong F, Collinson I. The action of cardiolipin on the bacterial translocon. Proc Natl Acad Sci USA. 2010;107:10044–10049. doi: 10.1073/pnas.0914680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MB, Bârzu O, Parsot C, Sansonetti PJ. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J Bacteriol. 1993;175:2189–2196. doi: 10.1128/jb.175.8.2189-2196.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, et al. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick JP, Wickner W. SecA protein needs both acidic phospholipids and SecY/E protein for functional high-affinity binding to the Escherichia coli plasma membrane. J Biol Chem. 1991;266:24596–24600. [PubMed] [Google Scholar]
- Huang KC, Ramamurthi KS. Macromolecules that prefer their membranes curvy. Mol Microbiol. 2010;76:822–832. doi: 10.1111/j.1365-2958.2010.07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KC, Mukhopadhyay R, Wingreen NS. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLoS Comput Biol. 2006;2:e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M, Shaw J, Shen C. The assembly of a structural lipoprotein in the envelope of Escherichia coli. J Biol Chem. 1972;247:8154–8159. [PubMed] [Google Scholar]
- Isnard M, Rigal A, Lazzaroni JC, Lazdunski C, Lloubès R. Maturation and localization of the TolB protein required for colicin import. J Bacteriol. 1994;176:6392–6396. doi: 10.1128/jb.176.20.6392-6396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janakiraman A, Fixen KR, Gray AN, Niki H, Goldberg MB. A genome-scale proteomic screen identifies a role for DnaK in chaperoning of polar autotransporters in Shigella. J Bacteriol. 2009;191:6300–6311. doi: 10.1128/JB.00833-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampfenkel K, Braun V. Membrane topologies of the TolQ and TolR proteins of Escherichia coli: inactivation of TolQ by a missense mutation in the proposed first transmembrane segment. J Bacteriol. 1993;175:4485–4491. doi: 10.1128/jb.175.14.4485-4491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentner D, Thiem S, Hildenbeutel M, Sourjik V. Determinants of chemoreceptor cluster formation in Escherichia coli. Mol Microbiol. 2006;61:407–417. doi: 10.1111/j.1365-2958.2006.05250.x. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Shibuya I, Matsumoto K. Viability of an Escherichia coli pgsA null mutant lacking detectable phosphatidylglycerol and cardiolipin. J Bacteriol. 2000;182:371–376. doi: 10.1128/jb.182.2.371-376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Kitchen JL, Li Z, Crooke E. Electrostatic interactions during acidic phospholipid reactivation of DnaA protein, the Escherichia coli initiator of chromosomal replication. Biochemistry. 1999;38:6213–6221. doi: 10.1021/bi982733q. [DOI] [PubMed] [Google Scholar]
- Koebnik R. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol Microbiol. 1995;16:1269–1270. doi: 10.1111/j.1365-2958.1995.tb02348.x. [DOI] [PubMed] [Google Scholar]
- Koppelman CM, Den Blaauwen T, Duursma MC, Heeren RM, Nanninga N. Escherichia coli minicell membranes are enriched in cardiolipin. J Bacteriol. 2001;183:6144–6147. doi: 10.1128/JB.183.20.6144-6147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri A, Ananthalakshmi TK, Nagarajan AG, Ray S, Chakravortty D. TolA mediates the differential detergent resistance pattern between the Salmonella enterica subsp. enterica serovars Typhi and Typhimurium. Microbiology. 2011;157:1402–1415. doi: 10.1099/mic.0.046565-0. [DOI] [PubMed] [Google Scholar]
- Lai EM, Nair U, Phadke ND, Maddock JR. Proteomic screening and identification of differentially distributed membrane proteins in Escherichia coli. Mol Microbiol. 2004;52:1029–1044. doi: 10.1111/j.1365-2958.2004.04040.x. [DOI] [PubMed] [Google Scholar]
- Laloux G, Jacobs-Wagner C. How do bacteria localize proteins to the cell pole? J Cell Sci. 2014;127:11–19. doi: 10.1242/jcs.138628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Lazzaroni JC, Portalier R. The excC gene of Escherichia coli K-12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein (PAL) Mol Microbiol. 1992;6:735–742. doi: 10.1111/j.1365-2958.1992.tb01523.x. [DOI] [PubMed] [Google Scholar]
- Lazzaroni JC, Vianney A, Popot JL, Bénédetti H, Samatey F, Lazdunski C, et al. Transmembrane alpha-helix interactions are required for the functional assembly of the Escherichia coli Tol complex. J Mol Biol. 1995;246:1–7. doi: 10.1006/jmbi.1994.0058. [DOI] [PubMed] [Google Scholar]
- Lazzaroni JC, Germon P, Ray MC, Vianney A. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol Lett. 1999;177:191–197. doi: 10.1111/j.1574-6968.1999.tb13731.x. [DOI] [PubMed] [Google Scholar]
- Lazzaroni JC, Dubuisson JF, Vianney A. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie. 2002;84:391–397. doi: 10.1016/s0300-9084(02)01419-0. [DOI] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, et al. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levengood SK, Beyer WF, Webster RE. TolA: a membrane protein involved in colicin uptake contains an extended helical region. Proc Natl Acad Sci USA. 1991;88:5939–5943. doi: 10.1073/pnas.88.14.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Young KD. Isolation and identification of new inner membrane-associated proteins that localize to cell poles in Escherichia coli. Mol Microbiol. 2012;84:276–295. doi: 10.1111/j.1365-2958.2012.08021.x. [DOI] [PubMed] [Google Scholar]
- Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci USA. 2012;109:E1481–E1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas MA, Ramos JL, Rodríguez-Herva JJ. Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for maintenance of outer membrane stability. J Bacteriol. 2000;182:4764–4772. doi: 10.1128/jb.182.17.4764-4772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas MA, Rodríguez-Herva JJ, Hancock REW, Bitter W, Tommassen J, Ramos JL. Role of Pseudomonas putida tol-oprL gene products in uptake of solutes through the cytoplasmic membrane. J Bacteriol. 2003;185:4707–4716. doi: 10.1128/JB.185.16.4707-4716.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloubès R, Cascales E, Walburger A, Bouveret E, Lazdunski C, Bernadac A, et al. The Tol–Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res Microbiol. 2001;152:523–529. doi: 10.1016/s0923-2508(01)01226-8. [DOI] [PubMed] [Google Scholar]
- Lybarger SR, Maddock JR. Polarity in action: asymmetric protein localization in bacteria. J Bacteriol. 2001;183:3261–3267. doi: 10.1128/JB.183.11.3261-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Marston AL, Errington J. Dynamic movement of the ParA-like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation. Mol Cell. 1999;4:673–682. doi: 10.1016/s1097-2765(00)80378-0. [DOI] [PubMed] [Google Scholar]
- Matsumoto K. Dispensable nature of phosphatidylglycerol in Escherichia coli: dual roles of anionic phospholipids. Mol Microbiol. 2001;39:1427–1433. doi: 10.1046/j.1365-2958.2001.02320.x. [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Fishov I, Fu X, Corbin BD, Margolin W, Dowhan W. Effects of phospholipid composition on MinD–membrane interactions in vitro and in vivo. J Biol Chem. 2003;278:22193–22198. doi: 10.1074/jbc.M302603200. [DOI] [PubMed] [Google Scholar]
- Miller JH. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Miller LD, Russell MH, Alexandre G. Diversity in bacterial chemotactic responses and niche adaptation. Adv Appl Microbiol. 2009;66:53–75. doi: 10.1016/S0065-2164(08)00803-4. [DOI] [PubMed] [Google Scholar]
- Mizushima T, Koyanagi R, Suzuki E, Tomura A, Kutsukake K, Miki T, Sekimizu K. Control by phosphatidylglycerol of expression of the flhD gene in Escherichia coli. Biochim Biophys Acta. 1995;1245:397–401. doi: 10.1016/0304-4165(95)00114-x. [DOI] [PubMed] [Google Scholar]
- Morgan DG, Baumgartner JW, Hazelbauer GL. Proteins antigenically related to methyl-accepting chemotaxis proteins of Escherichia coli detected in a wide range of bacterial species. J Bacteriol. 1993;175:133–140. doi: 10.1128/jb.175.1.133-140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MM, Webster RE. Characterization of the tol–pal and cyd region of Escherichia coli K-12: transcript analysis and identification of two new proteins encoded by the cyd operon. J Bacteriol. 1997;179:2077–2080. doi: 10.1128/jb.179.6.2077-2080.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MM, Vianney A, Lazzaroni JC, Webster RE, Portalier R. Membrane topology of the Escherichia coli TolR protein required for cell envelope integrity. J Bacteriol. 1993;175:6059–6061. doi: 10.1128/jb.175.18.6059-6061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Freinkman E, Kahne D. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science. 2012;338:1214–1217. doi: 10.1126/science.1228984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SL, Wadhams GH, Armitage JP. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol. 2011;9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci USA. 2009;106:13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray MC, Germon P, Vianney A, Portalier R, Lazzaroni JC. Identification by genetic suppression of Escherichia coli TolB residues important for TolB–Pal interaction. J Bacteriol. 2000;182:821–824. doi: 10.1128/jb.182.3.821-824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner LD, Weibel DB. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc Natl Acad Sci USA. 2011;2011:6264–6269. doi: 10.1073/pnas.1015757108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner LD, Weibel DB. MinD and MinE interact with anionic phospholipids and regulate division plane formation in Escherichia coli. J Biol Chem. 2012;287:38835–38844. doi: 10.1074/jbc.M112.407817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner LD, Eswaramoorthy P, Ramamurthi KS, Weibel DB. Studying biomolecular localization by engineering bacterial cell wall curvature. PLoS ONE. 2013;8:e84143. doi: 10.1371/journal.pone.0084143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigal A, Bouveret E, Lloubès R, Lazdunski C, Bénédetti H. The TolB protein interacts with the porins of Escherichia coli. J Bacteriol. 1997;179:7274–7279. doi: 10.1128/jb.179.23.7274-7279.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringgaard S, Schirner K, Davis BM, Waldor MK. A family of ParA-like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes Dev. 2011;25:1544–1555. doi: 10.1101/gad.2061811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romantsov T, Helbig S, Culham DE, Gill C, Stalker L, Wood JM. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol Microbiol. 2007;64:1455–1465. doi: 10.1111/j.1365-2958.2007.05727.x. [DOI] [PubMed] [Google Scholar]
- Romantsov T, Stalker L, Culham DE, Wood JM. Cardiolipin controls the osmotic stress response and the subcellular location of transporter ProP in Escherichia coli. J Biol Chem. 2008;283:12314–12323. doi: 10.1074/jbc.M709871200. [DOI] [PubMed] [Google Scholar]
- Romantsov T, Battle AR, Hendel JL, Martinac B, Wood JM. Protein localization in Escherichia coli cells: comparison of the cytoplasmic membrane proteins ProP, LacY, ProW, AqpZ, MscS, and MscL. J Bacteriol. 2010;192:912–924. doi: 10.1128/JB.00967-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Losick R. Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol. 2010;2:a000307. doi: 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shapiro L, McAdams HH, Losick R. Generating and exploiting polarity in bacteria. Science. 2002;298:1942–1946. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- Shiba Y, Yokoyama Y, Aono Y, Kiuchi T, Kusaka J, Matsumoto K, et al. Activation of the Rcs signal transduction system is responsible for the thermosensitive growth defect of an Escherichia coli mutant lacking phosphatidylglycerol and cardiolipin. J Bacteriol. 2004;186:6526–6535. doi: 10.1128/JB.186.19.6526-6535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu TS, Le Novére N, Levin MD, Beavil AJ, Sutton BJ, Bray D. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- Shiomi D, Zhulin IB, Homma M, Kawagishi I. Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase. J Biol Chem. 2002;277:42325–42333. doi: 10.1074/jbc.M202001200. [DOI] [PubMed] [Google Scholar]
- Shiomi D, Yoshimoto M, Homma M, Kawagishi I. Helical distribution of the bacterial chemoreceptor via colocalization with the Sec protein translocation machinery. Mol Microbiol. 2006;60:894–906. doi: 10.1111/j.1365-2958.2006.05145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W, Liu J, Ji H, Lu M. Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 Å resolution. J Mol Biol. 2000;299:1101–1112. doi: 10.1006/jmbi.2000.3776. [DOI] [PubMed] [Google Scholar]
- Skareso JT, Ahmern BMM, Seachordll CL, Darveau RP, Postle K. Energy transduction between membranes. J Biol Chem. 1993;268:16302–16308. [PubMed] [Google Scholar]
- Skidmore JM, Ellefson DD, McNamara BP, Couto MM, Wolfe AJ, Maddock JR. Polar clustering of the chemoreceptor complex in Escherichia coli occurs in the absence of complete CheA function. J Bacteriol. 2000;182:967–973. doi: 10.1128/jb.182.4.967-973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatiotemporal dynamics. Mol Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Armitage JP. Spatial organization in bacterial chemotaxis. EMBO J. 2010;29:2724–2733. doi: 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg H. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer J, Agha R, Pham T, Varga AW, Goldberg MB. The unipolar Shigella surface protein IcsA is targeted directly to the bacterial old pole: IcsP cleavage of IcsA occurs over the entire bacterial surface. Mol Microbiol. 1999;32:367–377. doi: 10.1046/j.1365-2958.1999.01356.x. [DOI] [PubMed] [Google Scholar]
- Sturgis JN. Organisation and evolution of the tol–pal gene cluster. J Mol Microbiol Biotechnol. 2001;3:113–122. [PubMed] [Google Scholar]
- Tan BK, Bogdanov M, Zhao J, Dowhan W, Raetz CRH, Guan Z. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc Natl Acad Sci USA. 2012;109:16504–16509. doi: 10.1073/pnas.1212797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiem S, Sourjik V. Stochastic assembly of chemoreceptor clusters in Escherichia coli. Mol Microbiol. 2008;68:1228–1236. doi: 10.1111/j.1365-2958.2008.06227.x. [DOI] [PubMed] [Google Scholar]
- Thiem S, Kentner D, Sourjik V. Positioning of chemosensory clusters in E. coli and its relation to cell division. EMBO J. 2007;26:1615–1623. doi: 10.1038/sj.emboj.7601610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomura A, Ishikawa T, Sagara Y, Miki T, Sekimizu K. Requirement of phosphatidylglycerol for flagellation of Escherichia coli. FEBS Lett. 1993;329:287–290. doi: 10.1016/0014-5793(93)80239-q. [DOI] [PubMed] [Google Scholar]
- Turner L, Ryu WS, Berg HC. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuson HH, Copeland MF, Carey S, Sacotte R, Weibel DB. Flagellum density regulates Proteus mirabilis swarmer cell motility in viscous environments. J Bacteriol. 2013;195:368–377. doi: 10.1128/JB.01537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vianney A, Lewin TM, Beyer WF, Lazzaroni JC, Portalier R, Webster RE. Membrane topology and mutational analysis of the TolQ protein of Escherichia coli required for the uptake of macromolecules and cell envelope integrity. J Bacteriol. 1994;176:822–829. doi: 10.1128/jb.176.3.822-829.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vianney A, Muller MM, Clavel T, Lazzaroni JC, Portalier R, Webster RE. Characterization of the tol–pal region of Escherichia coli K-12: translational control of tolR expression by TolQ and identification of a new open reading frame downstream of pal encoding a periplasmic protein. J Bacteriol. 1996;178:4031–4038. doi: 10.1128/jb.178.14.4031-4038.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrije T, de Swart RL, Dowhan W, Tommassen J, de Kruijff B. Phosphatidylglycerol is involved in protein translocation across Escherichia coli inner membranes. Nature. 1988;334:173–175. doi: 10.1038/334173a0. [DOI] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Walburger A, Lazdunski C, Corda Y. The Tol/Pal system function requires an interaction between the C-terminal domain of TolA and the N-terminal domain of TolB. Mol Microbiol. 2002;44:695–708. doi: 10.1046/j.1365-2958.2002.02895.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Wingreen N, Mukhopadhyay R. Self-Organized periodicity of protein clusters in growing bacteria. Phys Rev Lett. 2008;101:218101. doi: 10.1103/PhysRevLett.101.218101. [DOI] [PubMed] [Google Scholar]
- Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci USA. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li J, Li G, Long DG, Weis RM. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- Yamaichi Y, Bruckner R, Ringgaard S, Moll A, Cameron DE, Briegel A, et al. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev. 2012;26:2348–2360. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YC, Comolli LR, Downing KH, Shapiro L, McAdams HH. The Caulobacter Tol–Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J Bacteriol. 2010;192:4847–4858. doi: 10.1128/JB.00607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci USA. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol Microbiol. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.