Abstract
Although sometimes considered a “house-keeping” function, multiple aspects of protein synthesis are regulated differently among somatic cells, including stem cells, and can be modulated in a cell-type specific manner. These differences are required to establish and maintain differences in cell identity, cell function, tissue homeostasis, and tumor suppression.
Much of what is known about protein synthesis has been learned from studies in yeast and mammalian cell lines (Hinnebusch and Lorsch, 2012; Lorsch and Dever, 2010). Moreover, analysis of uncultured tissues or tumors provides an aggregate picture of protein synthesis across the tissue without revealing differences among individual cells. Nonetheless, it is clear that protein synthesis differs among mammalian somatic cells. For instance, cells of the exocrine pancreas display some of the highest rates of protein synthesis of any adult cell type (Case, 1978; Logsdon and Ji, 2013). Protein production rates vary greatly among liver, kidney and muscle cells (Allfrey et al., 1953; Garlick, 1972; Garlick et al., 1980; Garlick et al., 1991). Related cells within the same lineage can also exhibit different rates of mRNA translation. For example, in the immune system primed T cells experience a global attenuation of mRNA translation, which is subsequently reversed upon restimulation (Scheu et al., 2006). However, we are only beginning to appreciate the ways in which these differences in protein synthesis are necessary for tissue development and homeostasis. This is because new approaches are making it possible to more generally compare protein synthesis and its regulation among different kinds of cells, revealing an unexpected richness in the biology.
Studies of stem cell function and tissue homeostasis offer the opportunity to better understand differences in protein synthesis among somatic cells and their physiological significance. The balance between stem cell self-renewal and differentiation must be tightly regulated: self-renewal without differentiation leads to tumorigenesis while differentiation without self-renewal depletes stem cells and tissue regenerative capacity. Great effort has been invested in determining the transcriptional and epigenetic networks that govern stem cell identity and function, but gene expression programs are ultimately governed at the level of mRNA translation (Schwanhausser et al., 2011). Cell type-specific differences in translation regulate development, differentiation, and responses to stresses such as nutrient deprivation. Differences in the regulation of translation among cells may actually help to establish and maintain differences in cell identity and function.
In this review we focus on differences in the regulation of translation among cells and the physiological consequences. Some well-characterized mechanisms that regulate differences in protein synthesis among cells, such as differences in microRNA expression, are beyond the scope of this review. We focus instead on differences in protein synthesis, protein stability and ribosome assembly among cells that are critical for tissue development and homeostasis. The mechanisms that underlie these differences are only beginning to be elucidated but this is providing fundamental new insights into development, tissue regeneration, and how these processes go wrong in degenerative diseases and cancer.
Dynamic protein synthesis among embryonic cells
Cellular function depends upon proteostasis - appropriate regulation of protein synthesis, protein folding, and protein degradation. Each of these are likely to exhibit cell type-specific differences in regulation that influence stem cell function, tissue development, and homeostasis (Vilchez et al., 2014). However, differences among somatic cells in protein folding and protein degradation are not as well characterized as differences in protein synthesis. We will thus focus mainly on protein synthesis even though this is only one component of the proteostasis network.
Developmental studies have begun to reveal the extent to which related cells exhibit functionally significant changes in protein synthesis as they differentiate. In mammals, these changes can first be seen immediately after fertilization. Most proteins exhibit two-fold or greater changes in abundance as mouse embryos transition from the 1-cell to 2-cell stage (Latham et al., 1991). Studies of Drosophila have shown that the changes in protein levels during the oocyte-to-embryo transition largely occur at the level of translation, not transcription (Kronja et al., 2014). Translational control mechanisms also help to promote germ cell differentiation in Drosophila males (Insco et al., 2012). Building upon these studies, the functional importance of differences in global protein production levels between undifferentiated cells and their progeny has recently begun to come into focus.
mRNA translation changes on a global scale as mammalian embryonic stem cells (ESCs) differentiate to form embryoid bodies (Ingolia et al., 2011; Sampath et al., 2008). Continuously dividing cells tend to synthesize more protein than non-dividing cells, but ESCs are distinct in that they maintain lower levels of bulk mRNA translation and protein accumulation than their differentiated progeny despite continuous cell division. ESCs display a marked reduction in overall translational efficiency (a lower fraction of mRNAs associate with actively translating polysomes) relative to other cells. Induction of differentiation increases global transcript levels and leads to more efficient loading of mRNAs into polysomes, increasing protein synthesis (Ingolia et al., 2011; Sampath et al., 2008).
Differences in protein synthesis among ESCs and their differentiating progeny correlate with global translational changes. For example, ESCs exhibit increased translation of upstream open reading frames (uORFs) (Ingolia et al., 2011). uORFs form when translation begins at initiation sites within the 5’UTRs of mRNAs that are upstream of the initiation sites of recognized coding sequences. While uORFs can promote reinitiation at downstream ORFs in specific cases, many act to decrease the translation of the actual coding sequence by impeding scanning by the pre-initiation complex (Barbosa et al., 2013). uORFs can be found in 49% of human transcripts and influence protein expression in a variety of different contexts, including during stress responses (Barbosa et al., 2013). Additional studies will be required to determine whether the global change in uORF translation during ESC differentiation commonly occurs during the differentiation of other stem cells and whether it influences cell fate.
Global regulation of protein degradation also controls protein levels within ESCs and ESC maintenance. Human ESCs express high levels of the 19S proteasome subunit PSMD11 and exhibit increased proteasome capacity relative to a variety of differentiated progeny (Vilchez et al., 2012). Over-expression of PSMD11 increases proteasome capacity while decreased expression of PSMD11 reduces ubiquitin-dependent protein degradation (Vilchez et al., 2012). Reduced proteasome activity reduces the expression of pluripotency markers (Vilchez et al., 2012), suggesting that ESCs depend on elevated proteasome activity for their maintenance. Given that protein synthesis increases during ESC differentiation (Ingolia et al., 2011; Sampath et al., 2008), these findings suggest that low protein levels, achieved through reduced translation and increased degradation, promote ESC maintenance (Figure 1A).
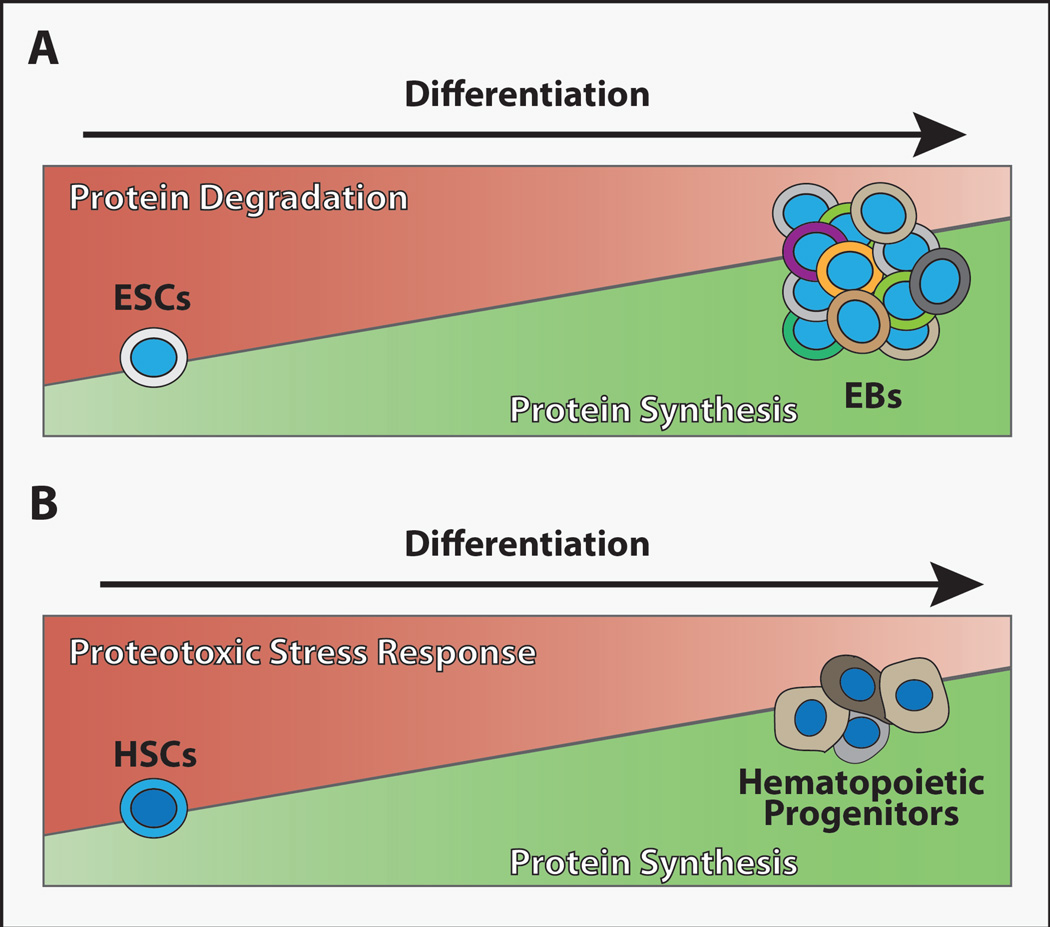
Figure 1. Proteostasis differs between stem cells and their daughters.
(A) Global protein translation levels remain low in embryonic stem cells (ESCs) but increase as these cells differentiate to form embryoid bodies (EBs) (Ingolia et al., 2011; Sampath et al., 2008). By contrast, the activity of protein degradation mechanisms appears high in ESCs relative to EBs (Vilchez et al., 2012).
(B) Adult HSCs display low levels of global protein synthesis relative to differentiating hematopoietic progenitor cells (Signer et al., 2014), and proteotoxic stress response appears enhanced in umbilical cord blood (fetal) HSCs (van Galen et al., 2014). Whether these trends in proteostasis regulatory mechanisms will be seen in other stem cell populations remains unclear, but keeping overall protein levels low in long-lived stem cell populations may promote their fitness and longevity.
Dynamic protein synthesis among stem/progenitor cells
To understand the extent to which differences in protein synthesis among cells regulate normal tissue development and homeostasis, it is necessary to study fetal and adult tissues in vivo. One limitation has been the dependence upon pulsed metabolic labeling to mark the synthesis of nascent polypeptides. Amino acid analogues, such as methionine analogues, do not compete with endogenous amino acids, limiting this approach to cells that survive and proliferate in culture media depleted for the relevant amino acid (Beatty et al., 2006). This has impeded efforts to directly measure differences in protein synthesis among cells in vivo. Moreover, the tissue culture environment differs from the in vivo environment in many ways that influence cellular properties (Joseph and Morrison, 2005), including protein synthesis, at least in hematopoietic stem cells (HSCs) (Signer et al., 2014). Other techniques for measuring protein synthesis can be applied to uncultured cells, but depend upon stable isotope labeling in vivo followed by mass spectrometry (Kruger et al., 2008; Xiao et al., 2008) or on ribosome profiling (Li et al., 2014). These techniques have so far only been used when cells are abundant and have not been scaled down to analyze purified rare cell populations, such as somatic stem cells. Single cell assays to measure protein synthesis have been developed to look at the translation of mRNAs containing specific translation regulatory motifs (Han et al., 2014), but not at global protein synthesis rates. Consequently, there is a general need to develop more assays that enable quantitation of protein synthesis in small numbers of cells in vivo.
A recent advance that facilitates quantitation of protein synthesis in individual cells in vivo is the synthesis of O-propargyl-puromycin (OP-Puro) (Liu et al., 2012). OP-Puro enters the acceptor site of ribosomes and is covalently incorporated into nascent polypeptide chains. Isolation and fixation of OP-Puro-treated cells, followed by exposure to an azide-conjugated fluorophore, leads to the fluorescination of all polypeptides that incorporate OP-Puro via a click chemistry reaction (Liu et al., 2012). The amount of protein synthesis per hour in individual cells in vivo can then be quantitated by flow cytometry (Signer et al., 2014).
Administration of OP-Puro in vivo shows that cells within somatic stem cell lineages exhibit dynamic regulation of protein synthesis. Adult HSCs exhibit less protein synthesis than restricted hematopoietic progenitors (Signer et al., 2014). We observed up to 10-fold differences in the amount of protein synthesized per cell per hour in different hematopoietic stem/progenitor cell populations. Although most adult HSCs are quiescent, their lower levels of protein synthesis are independent of cell cycle status, cell size, and RNA content (Signer et al., 2014). Genetic perturbations that modestly increase or decrease protein synthesis both impair HSC function. A 30% decrease in global protein synthesis in Rpl24Bst/+ mutant HSCs impairs their ability to reconstitute the hematopoietic system of irradiated mice. Likewise, increasing protein synthesis by deleting the PI3K pathway inhibitor Pten depletes HSCs and promotes leukemia development (Signer et al., 2014; Yilmaz et al., 2006). Introducing the Rpl24Bst/+ mutation onto the Pten-deficient background rescues both of these phenotypes, demonstrating that PTEN maintains HSCs and suppresses leukemia mainly by attenuating protein synthesis. Thus, HSCs and restricted hematopoietic progenitors synthesize different amounts of protein per hour and these differences are necessary for normal HSC function.
Experimental interventions or mutations that reduce protein synthesis extend lifespan in an evolutionarily conserved manner (reviewed in (Taylor and Dillin, 2011)). This has been proposed to occur at least partly because reduced protein synthesis increases protein quality and reduces the burden imposed on the protein-folding chaperone system by misfolded protein aggregates. Certain long-lived mitotic cells, including some stem cells, may maintain proteome quality by limiting protein synthesis (Signer et al., 2014) or increasing proteasome activity (Vilchez et al., 2012)(Vilchez et al., 2014). The increased longevity associated with decreased protein synthesis may partly reflect the increased fitness of certain mitotic cells or increased tissue regenerative capacity during aging as a consequence of reduced proteotoxic stress, though this has not yet been tested.
Proteotoxic stress occurs under physiological conditions, such as in pancreatic β cells that increase protein synthesis to produce insulin in response to blood glucose spikes (Back et al., 2009). Proteotoxic stress reduces protein synthesis by inhibiting mRNA translation, typically at the level of initiation. However, proteotoxic stress can also pause ribosomes during elongation (Liu et al., 2013). This pausing typically occurs at sites near where the nascent polypeptide chain emerges from the ribosome and depends on the activity of various chaperone proteins. Given the cell-specific differences in the expression and activity of various chaperones, the response to proteotoxic stress likely varies from cell to cell (Morimoto, 2008). Human cord blood HSCs are particularly sensitive to proteotoxic stress, undergoing apoptosis in response to a PERK-mediated unfolded protein response (van Galen et al., 2014). The data suggest that protein misfolding influences HSC function under physiological conditions and support the idea that hematopoietic cells vary in their sensitivity to proteotoxic stress.
TORC1 regulates stem cells through protein synthesis
mTORC1 signaling represents a major mechanism by which cells integrate nutrient availability, growth factor signaling and developmental cues to regulate protein synthesis (Figure 2A) (Laplante and Sabatini, 2012). mTORC1 activation promotes protein synthesis through inhibition of 4E-BPs and activation of S6K1, which acts on a number of translation initiation proteins and other factors involved in translational elongation and ribosome biogenesis (Browne and Proud, 2002; Ma and Blenis, 2009; Mayer et al., 2004; Shahbazian et al., 2006) (Figure 2B).
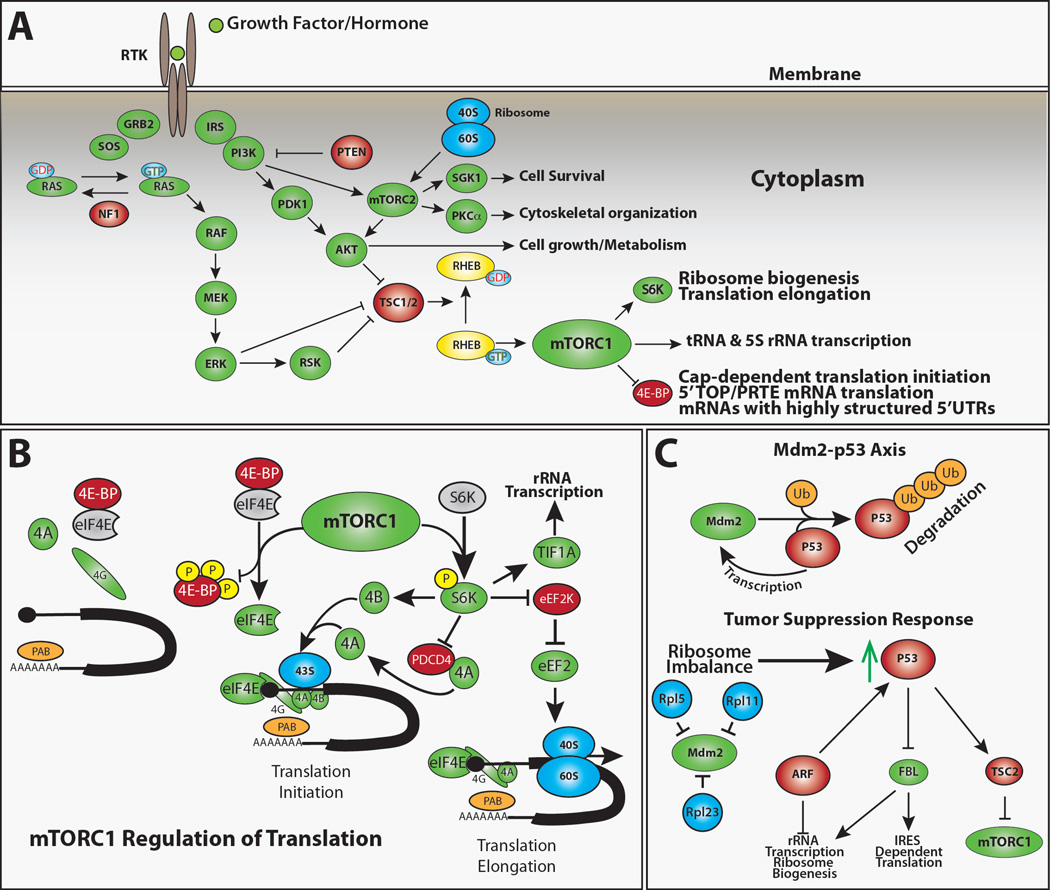
Figure 2. Signaling pathways that regulate protein synthesis.
(A) Growth factors, hormones, nutrients, and stress all influence protein synthesis through complex signaling pathways (Laplante and Sabatini, 2012; Topisirovic and Sonenberg, 2011). mTORC1 preferentially promotes the translation of a subset of mRNAs that contain complex 5’UTRs and clusters of 5’ terminal oligopyrimidines (TOP) or similar pyrimidine-rich sequences (Hsieh et al., 2012; Jefferies et al., 1994; Thoreen et al., 2012). These TOP genes primarily encode proteins that promote translation, including ribosomal proteins, initiation factors, and elongation factors (Thoreen et al., 2012). While many proteins have been proposed to regulate the translation of TOP mRNAs (Damgaard and Lykke-Andersen, 2011; Tcherkezian et al., 2014) and the translation of specific TOP containing mRNAs is regulated in a cell-specific manner (Avni et al., 1997; Ivanov et al., 2011), the roles of these elements in stem cell maintenance and tissue homeostasis remain unclear.
(B) The regulation of translation initiation represents the major mechanism by which cells regulate protein synthesis. The eIF4F complex, which includes eIF4E, eIF4G and eIF4A, promotes the translational initiation of virtually all cellular mRNAs (Sonenberg and Hinnebusch, 2009) and mRNAs with highly structured 5’UTRs are particularly sensitive to eIF4E activity (Feoktistova et al., 2013; Koromilas et al., 1992). Activated mTORC1 phosphorylates 4E-BPs, preventing their inhibition of eIF4E. In the absence of 4E-BPs, eIF4E binds to the m7G mRNA cap and recruits eIF4G, which in turn acts as a scaffold for the RNA helicase eIF4A. The formation of this eIF4F complex on the 5’-end of mRNA promotes the initiation of cap-dependent translation. Activated mTORC1 also phosphorylates and activates S6K, which phosphorylates eIF4B, eEF2K and TIF1A. Phosphorylated eIF4B enhances the helicase activity of eIF4A to unwind secondary structures in mRNA, which promotes translational initiation. Phosphorylation of eEF2K by S6K interferes with its ability to block the activity of the translational elongation factor eEF2. Activation of TIF1A promotes ribosomal RNA transcription.
(C) The E3 ubiquitin ligase Mdm2 promotes the degradation of the tumor suppressor p53. p53 acts in a feedback loop to promote the transcription of the Mdm2 gene. Thus under normal conditions, p53 levels remain low. Imbalances in ribosome biogenesis induce a tumor suppressor response in which free ribosomal proteins, particularly Rpl5 and Rpl11, bind and inhibit Mdm2 (Bhat et al., 2004; Dai and Lu, 2004; Dai et al., 2006; Zhang et al., 2003). This results in the accumulation of p53, which promotes the activity of TSC2, thereby repressing the activity of mTORC1. p53 and other tumor suppressors, such as ARF, inhibit rRNA transcription and ribosome biogenesis (Lessard et al., 2010; Sloan et al., 2013; Sugimoto et al., 2003).
The importance of mTORC1-mediated regulation of translation initiation during development has recently begun to emerge. For example, increases in protein synthesis that occur during early sea urchin embryogenesis correlate with decreased inhibition of eIF4E by 4E-BP as a consequence of increased 4E-BP degradation (Cormier et al., 2001; Salaun et al., 2003). The increase in 4E-BP degradation is blocked by rapamycin treatment, suggesting that it reflects mTORC1 signaling. Phosphorylated 4E-BP1 levels increase as ESCs differentiate into EBs, potentially explaining the increase in protein synthesis in EBs (Sampath et al., 2008). While loss of S6K1 has little or no effect on the viability and proliferation of ESCs, activation of S6K1 promotes the differentiation of these cells (Easley et al., 2010; Kawasome et al., 1998). mTORC1 signaling likely promotes increased protein synthesis during ESC differentiation.
Multiple adult tissues depend upon appropriate mTORC1 signaling to regulate stem cell maintenance, proliferation, and tumor suppression (Gan et al., 2008; Groszer et al., 2001; Yilmaz et al., 2006; Zhang et al., 2006b). Increased activation of mTORC1 in vivo quickly depletes adult neural stem cells (Bonaguidi et al., 2011), epidermal stem cells (Castilho et al., 2009) and HSCs (Yilmaz et al., 2006; Zhang et al., 2006b). The stem cell depletion that occurs after Pten deletion occurs largely due to a deleterious increase in protein synthesis, at least in the hematopoietic system (Signer et al., 2014). Decreasing mTORC1 activity or constitutively expressing 4E-BP1 in neonatal neural stem cells enhances self-renewal and prevents differentiation (Hartman et al., 2013). While mTORC1 signaling regulates the growth of many cells, the level of mTORC1 activation varies among different cells (Betschinger et al., 2013; Sampath et al., 2008; Signer et al., 2014) and in the same cells over time (Chen et al., 2009; Magee et al., 2012). This raises the possibility that changes in mTORC1 signaling, and modulation of global protein synthesis rates, may determine developmental changes, rather than simply arising as a consequence of those changes.
Ribosome assembly influences tissue homeostasis
Given that differences among cells in protein synthesis, even among lineally related cells in the same tissue, are required for normal tissue homeostasis (e.g. (Signer et al., 2014)), one key question is how these differences among cells are regulated. Beyond differences in mTORC1 signaling, additional mechanisms involving modulation of ribosomal subunit expression and ribosome assembly also contribute to differences in protein synthesis among cells. The heterogeneity in these mechanisms among cells is illustrated by the cell- and tissue-specific defects that arise as a consequence of defects in ribosome assembly.
Genes that encode individual ribosome components exhibit great differences in expression among tissues (Kondrashov et al., 2011) and even among cell types within the same tissue (Signer et al., 2014). Consistent with this, mutations in genes that encode ribosome components often have phenotypes that are surprisingly cell-type specific. In zebrafish, mutations in the ubiquitously expressed ribosome proteins, Rpl22 and Rpl22-like (Rpl22l), have distinct developmental phenotypes, affecting T lineage progenitors and HSCs respectively (Zhang et al., 2013). Both Rpl22 and Rpl22l compete for binding to smad1 mRNA and have opposing effects on its translation, which partially accounts for their distinct phenotypes. Similarly, Rps19-deficient zebrafish embryos display specific defects in erythropoiesis and cartilage development (Danilova et al., 2008), while reduced expression of Rps29 results in erythropoietic defects and ectopic cell death in the head (Taylor et al., 2012). The Tail short (Ts) mutation in mouse Rpl38 causes specific skeletal defects marked by reduced expression of 8 of 39 Hox proteins (Kondrashov et al., 2011). Rpl38Ts mutations reduce the levels of 80S monosome formation on specific Hox gene transcripts. By contrast, mutations in multiple genes that encode other ribosome components (Rps19DSK3/+, Rps20Dsk4/+, Rpl29+/−, Rpl29−/−, Rpl24Bst/+) do not cause similar skeletal phenotypes or effects on Hox gene expression (Kondrashov et al., 2011). Thus, Rpl38 plays a specialized role in regulating the translation of specific mRNAs, perhaps in a tissue-specific manner.
Human ribosome-related diseases, collectively referred to as ribosomopathies, further reflect the preferential dependence of specific cells on specific ribosome components. Human ribosomopathies include Diamond-Blackfan anemia, 5q syndrome, Treacher Collins syndrome, Blooms and Werner syndromes, Dyskeratosis Congenita, and Cartilage Hair Hypoplasia (Armistead and Triggs-Raine, 2014). These disorders are caused by mutations in genes that encode ribosome components or genes that affect Pol I transcription or ribosomal RNA processing (Armistead and Triggs-Raine, 2014). Each ribosomopathy exhibits different spectrums of defects in tissue homeostasis, though hematopoietic and craniofacial/neural crest defects are commonly observed (Narla and Ebert, 2010). It remains unclear why hematopoietic cells and neural crest cells exhibit particular sensitivity to loss-of-function mutations in ribosome components or factors required for their synthesis.
The hematopoietic defects observed as a result of mutations in certain ribosomal components likely reflect a combination of defects in stem cells and restricted hematopoietic progenitors. For example, HSC function is impaired in Dyskeratosis Congenita (Friedland et al., 1985; Marsh et al., 1992) and in 5Q syndrome (Nilsson et al., 2000) whereas Diamond-Blackfan anemia appears to more strongly reflect defects in erythroid restricted progenitors (Abkowitz et al., 1991; Lipton et al., 1986).
Phenotypes caused by ribosomal defects reflect a variable combination of tumor suppressor induction along with changes in the synthesis of key proteins (Armistead and Triggs-Raine, 2014; Barkic et al., 2009; Bellodi et al., 2010; Danilova et al., 2008; Ludwig et al., 2014; Narla and Ebert, 2010; Taylor et al., 2012; Zhang and Lu, 2009). Ribosome proteins can also exhibit important functions outside the ribosome, complicating the interpretation of at least some mutant phenotypes (Xue and Barna, 2012). For example, Rps13 regulates the splicing of its own message (Malygin et al., 2007) and ribosomes can bind mTORC2 and promote its signaling in a translation independent manner (Zinzalla et al., 2011). It remains unclear whether extra-ribosomal functions are unusual properties pertaining to a minority of ribosome components or whether many ribosome components have unrecognized extra-ribosomal functions, including cell type-specific functions.
Ribosome assembly is also regulated differently in different cells. Disruption of Utp4 causes North American Indian childhood cirrhosis (Armistead and Triggs-Raine, 2014). Utp4 is a component of the small subunit processome, a large ribonucleoprotein complex that promotes the maturation of 18S rRNA (Freed and Baserga, 2010). Unique among ribosomapathies in only affecting a single tissue, North American Indian childhood cirrhosis, first presents as childhood jaundice and progresses to biliary cirrhosis. Knockdown of Utp4 in zebrafish also leads to specific defects in the biliary system (Wilkins et al., 2013), suggesting that the function of Utp4 in this tissue has been conserved across species. By contrast, disruption of Notchless, a murine ortholog of the yeast 60S subunit maturation factor Rsa4, depletes HSCs but not more mature hematopoietic cells (Le Bouteiller et al., 2013).
Asymmetric segregation of ribosome processing and rRNA transcription factors influence fate determination in stem cells and their daughter cells. The ribosomal RNA processing factor Wicked, a member of the conserved rRNA processing U3 snoRNP complex, exhibits asymmetric segregation during Drosophila germline stem cell divisions (Fichelson et al., 2009). The asymmetric distribution of Wicked during mitosis ensures that presumptive germline stem cells inherit more of this factor than daughters fated to differentiate. This same mode of inheritance is also observed in several other stem cell populations (Fichelson et al., 2009). More recent work shows that a Drosophila Pol I regulatory factor, under-developed (udd), also becomes enriched in germline stem cells relative to their differentiating daughters (Zhang et al., 2014a). Loss of both wicked and udd compromises germline stem cell maintenance (Fichelson et al., 2009; Zhang et al., 2014a). The drop in Pol I activity in udd mutants promotes some aspects of differentiation while expanded rRNA transcription as a result of increased TIF-1A expression delays differentiation (Zhang et al., 2014a). Interestingly, down-regulation of rRNA synthesis in cultured mammalian hematopoietic progenitors can also promote differentiation (Hayashi et al., 2014). Further work will be needed to explore whether mammalian lineages also exhibit asymmetric inheritance of Pol I regulatory and rRNA processing factors.
It will be important to compare ribosome biogenesis and translational regulation among stem cells and their daughters at different stages of development, in different tissues, and in different stem cell activation states. It remains unclear whether stem cells exhibit consistent differences relative to their differentiating progeny or whether there is as much diversity in regulatory mechanisms among stem cells as among differentiated cells.
Regulated protein synthesis control suppresses neoplastic proliferation
The regulation of protein synthesis promotes tissue homeostasis partly by preventing inappropriate proliferation. Most cells increase their rate of protein synthesis during cell division. Cancer cells generally synthesize protein more rapidly and more efficiently than normal cells (Ruggero, 2013). Translational control mechanisms regulate the expression and function of a variety of oncogenes and tumor suppressors through diverse mechanisms (Topisirovic and Sonenberg, 2011) and cancer cells dysregulate translation initiation to increase protein synthesis and sustain neoplastic proliferation (Ruggero, 2013). Mechanisms that increase or deregulate protein synthesis can thus promote the development of cancer.
Mechanisms that negatively regulate protein synthesis often suppress tumorigenesis. Key oncogenic signals, such as Myc or PI3K pathway activation, increase protein synthesis and change the efficiency with which subsets of mRNAs are translated (Barna et al., 2008; Hsieh et al., 2012). This is critically important because the regulation of translation initiation, the efficiency with which mRNAs are recruited to active ribosomes, represents the major mechanism by which cells regulate protein synthesis. Based on studies of cancer cells (Hsieh et al. 2012), it seems likely that there are differences among stem cells and restricted progenitors in terms of how they regulate translation initiation, though this has not yet been studied.
Genetic changes or pharmacological agents that reduce protein synthesis can impede cancer development and progression (Barna et al., 2008; Hsieh et al., 2010; Hsieh et al., 2012; Signer et al., 2014). Moreover, a number of tumor suppressors negatively regulate protein synthesis, for example by negatively regulating mTORC1 signaling (Laplante and Sabatini, 2012). Tumor suppressors can thus regulate protein synthesis through a variety of mechanisms.
Mutations in ribosome biogenesis factors or ribosome components impair tissue homeostasis and promote the development of cancer through diverse mechanisms. For example, in X-linked dyskeratosis congenita, disruption of dyskerin (DKC1), an enzyme that pseudouridylates rRNA (Ni et al., 1997), leads to bone marrow failure, skin abnormalities, various carcinomas, and acute myeloid leukemia (Heiss et al., 1998; Ruggero et al., 2003). DKC1 mutations do not affect global protein synthesis but do reduce the translation of a subset of mRNAs with IRES elements in their 5’ UTRs (Yoon et al., 2006), including the p53 and p27 tumor suppressors (Bellodi et al., 2010). Thus, one mechanism by which mutations in ribosome components and biogenesis factors promote the development of cancer is through effects on the translation of tumor suppressors. To the extent that ribosomopathies induce proteotoxic stress or a tumor suppressor response, they may also create a selective pressure to inactivate tumor suppressors in normal cells that are engaged in tissue regeneration, increasing the probability of subsequent transformation.
Tumor suppressor responses may be induced by sustained oncogenic stimuli as a consequence of their effects on protein synthesis. Diverse oncogenic stimuli induce tumor suppressor responses in a wide range of normal cells (Collado and Serrano, 2010). A series of studies suggest that cells may sense oncogenic stimuli based on their effects on protein synthesis. Multiple distinct mutations in ribosomal proteins induce the accumulation of p53, as do defects in ribosomal biogenesis and nucleolar stress (Armistead and Triggs-Raine, 2014; Barkic et al., 2009; Bellodi et al., 2010; Danilova et al., 2008; Narla and Ebert, 2010; Taylor et al., 2012; Zhang and Lu, 2009). For example, disruption of Rps14 activates p53 and loss of p53 rescues the hematopoietic progenitor cell depletion observed in these mutants (Barlow et al., 2010; Dutt et al., 2011). p53 expression increases in response to defects in ribosome biogenesis, partly as a consequence of Mdm2 binding by ribosomal components (such as Rpl5 and Rpl11) (Bhat et al., 2004; Dai and Lu, 2004; Dai et al., 2006; Zhang et al., 2003), suggesting that disruption of the normal stoichiometry of ribosome assembly leads to the sequestration of Mdm2 by free ribosomal proteins (Figure 2C).
The efficiency of reprogramming of somatic cells to pluripotency by Sox2, Myc, KLF4 and Oct4 (all of which are oncogenes) is limited partly by p53 and p16Ink4a/p19Arf tumor suppressor expression (Kawamura et al., 2009; Li et al., 2009; Marion et al., 2009; Utikal et al., 2009). An important recent paper by Sonenberg and colleagues demonstrates that loss of 4E-BP1/2 impairs the reprogramming of fibroblasts to pluripotent stem cells, partly by inducing the expression of the p21cip1 tumor suppressor (Tahmasebi et al., 2014). However, in the absence of p53, loss of 4E-BP1/2 promotes reprogramming. Since 4E-BPs are negative regulators of translation, these data suggest protein synthesis must increase for successful reprogramming, but that this induces a tumor suppressor response that reduces reprogramming efficiency.
Tumor suppressors can also be induced in response to changes in the quality of the proteome, such as in an unfolded protein response. Increased protein synthesis can overwhelm the chaperone system and lead to the accumulation of misfolded proteins that aggregate and impede diverse cellular processes (Back et al., 2009; Schroder and Kaufman, 2005). Induction of an unfolded protein response reduces protein synthesis and increases proteasome activity and tumor suppressor expression. The unfolded protein response reduces the number of active polysomes through eIF2α phosphorylation, allowing free ribosomal proteins to associate with Mdm2, stabilize p53, and induce cell cycle arrest (Zhang et al., 2006a). Thus, cellular surveillance of protein synthesis and protein quality is achieved through diverse mechanisms that induce tumor suppressors, influencing tissue homeostasis and neoplastic proliferation.
Perspective
Stem cell function and tissue homeostasis are regulated by networks of proto-oncogenes and tumor suppressors that feature key transcriptional regulators that distinguish stem cells from their progeny (He et al., 2009). While these networks have been understood to regulate cell identity, signal transduction, transcription, and cell cycle progression, other areas of cellular physiology continue to be of uncertain relationship to cellular identity. It is likely that many of these aspects of cellular physiology exhibit much more cellular specificity in their regulation than currently appreciated. Consistent with this, protein synthesis is regulated differently in different kinds of cells and these differences are critical for fate determination and the maintenance of tissue homeostasis (Figure 3). These functions are necessarily and intimately intertwined with the role of translational regulation in tumor suppression. The ability of translational regulation to suppress the development of cancer reflects its role in preventing inappropriate proliferation by stem cells and other cells within normal tissues. As our understanding of translational regulation matures, new biology will emerge, marked by unanticipated regulatory mechanisms.
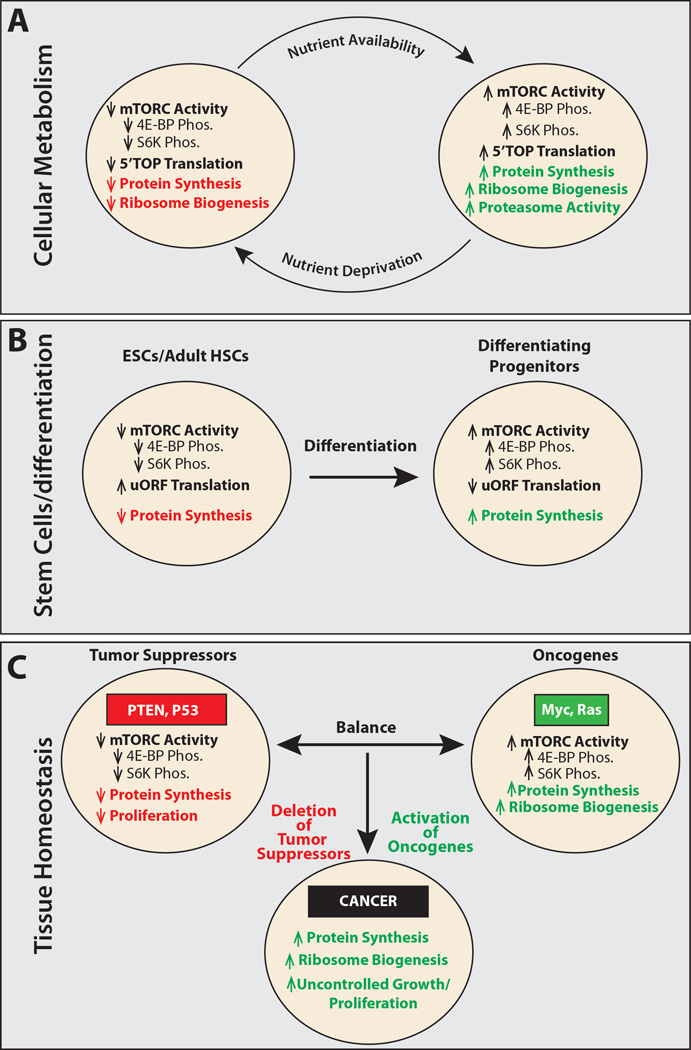
Figure 3. Differences in the regulation of protein synthesis among cells.
(A) Cells respond to changes in nutrient levels by altering protein synthesis, ribosome biogenesis (Laplante and Sabatini, 2012), and protein degradation (Zhang et al., 2014b) by modulating mTORC1 signaling.
(B) Embryonic stem cells (ESCs) and adult hematopoietic stem cells (HSCs) exhibit lower levels of protein synthesis than their differentiating progeny. Whether other stem cell lineages display similar changes in protein synthesis upon differentiation will require further analysis. One intriguing possibility is that this property may vary among stem cells, depending on their cell cycle kinetics and whether they are long-lived or short-lived in vivo.
(C) Tissue homeostasis depends on a balance between proto-oncogenes and tumor suppressors. Tumor suppressors such as PTEN and p53 reduce cellular growth and proliferation through a number of different mechanisms, including decreasing mTORC1 activity and reducing global protein synthesis (Laplante and Sabatini, 2012). Proto-oncogenes, including Myc and Ras, increase protein translation and ribosome biogenesis, fueling growth and proliferation. Cells transform to cancer when mutations reduce tumor suppressor function or increase oncogene function (van Riggelen et al., 2010).
Acknowledgements
SJM is a Howard Hughes Medical Institute Investigator, the Mary McDermott Cook Chair in Pediatric Genetics, and the director of the Hamon Laboratory for Stem Cells and Cancer. This work was supported by the Cancer Prevention and Research Institute of Texas and the National Institute on Aging (R37 AG024945). M.B. was supported by the National Institute of General Medical Sciences (R01 GM086647). R.A.J.S. was supported by fellowships from the Leukemia & Lymphoma Society (5541-11) and the Canadian Institutes of Health Research (MFE-106993).
References
- Abkowitz JL, Sabo KM, Nakamoto B, Blau CA, Martin FH, Zsebo KM, Papayannopoulou T. Diamond-blackfan anemia: in vitro response of erythroid progenitors to the ligand for c-kit. Blood. 1991;78:2198–2202. [PubMed] [Google Scholar]
- Allfrey V, Daly MM, Mirsky AE. Synthesis of protein in the pancreas. II. The role of ribonucleoprotein in protein synthesis. The Journal of General Physiology. 1953;37:157–175. doi: 10.1085/jgp.37.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armistead J, Triggs-Raine B. Diverse diseases from a ubiquitous process: The ribosomopathy paradox. FEBS Letters. 2014;588:1491–1500. doi: 10.1016/j.febslet.2014.03.024. [DOI] [PubMed] [Google Scholar]
- Avni D, Biberman Y, Meyuhas O. The 5' terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Research. 1997;25:995–1001. doi: 10.1093/nar/25.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back SH, Scheuner D, Han J, Song B, Ribick M, Wang J, Gildersleeve RD, Pennathur S, Kaufman RJ. Translation attenuation through eIF2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metabolism. 2009;10:13–26. doi: 10.1016/j.cmet.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa C, Peixeiro I, Romao L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genetics. 2013;9:e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkic M, Crnomarkovic S, Grabusic K, Bogetic I, Panic L, Tamarut S, Cokaric M, Jeric I, Vidak S, Volarevic S. The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Molecular and Cellular Biology. 2009;29:2489–2504. doi: 10.1128/MCB.01588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, Lane AL, Jolin HE, Pannell R, Middleton AJ, Wong SH, Warren AJ, Wainscoat JS, Boultwood J, McKenzie AN. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nature Medicine. 2010;16:59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty KE, Liu JC, Xie F, Dieterich DC, Schuman EM, Wang Q, Tirrell DA. Fluorescence visualization of newly synthesized proteins in mammalian cells. Angewandte Chemie. 2006;45:7364–7367. doi: 10.1002/anie.200602114. [DOI] [PubMed] [Google Scholar]
- Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. The EMBO Journal. 2010;29:1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153:335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. The EMBO Journal. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. European journal of biochemistry / FEBS. 2002;269:5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- Case RM. Synthesis, intracellular transport and discharge of exportable proteins in the pancreatic acinar cell and other cells. Biological Reviews of the Cambridge Philosophical Society. 1978;53:211–354. doi: 10.1111/j.1469-185x.1978.tb01437.x. [DOI] [PubMed] [Google Scholar]
- Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell. 2009;5:279–289. doi: 10.1016/j.stem.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu Y, Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Science Signaling. 2009;2:ra75. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nature Reviews Cancer. 2010;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier P, Pyronnet S, Morales J, Mulner-Lorillon O, Sonenberg N, Belle R. eIF4E association with 4E-BP decreases rapidly following fertilization in sea urchin. Developmental Biology. 2001;232:275–283. doi: 10.1006/dbio.2001.0206. [DOI] [PubMed] [Google Scholar]
- Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. Journal of Biological Chemistry. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- Dai MS, Shi D, Jin Y, Sun XX, Zhang Y, Grossman SR, Lu H. Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. Journal of Biological Chemistry. 2006;281:24304–24313. doi: 10.1074/jbc.M602596200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Lykke-Andersen J. Translational coregulation of 5'TOP mRNAs by TIA-1 and TIAR. Genes & Development. 2011;25:2057–2068. doi: 10.1101/gad.17355911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, Wilson FH, Currie T, Khanna-Gupta A, Berliner N, Kutok JL, Ebert BL. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117:2567–2576. doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easley CAt, Ben-Yehudah A, Redinger CJ, Oliver SL, Varum ST, Eisinger VM, Carlisle DL, Donovan PJ, Schatten GP. mTOR-mediated activation of p70 S6K induces differentiation of pluripotent human embryonic stem cells. Cellular Reprogramming. 2010;12:263–273. doi: 10.1089/cell.2010.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13339–13344. doi: 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichelson P, Moch C, Ivanovitch K, Martin C, Sidor CM, Lepesant JA, Bellaiche Y, Huynh JR. Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nature Cell Biology. 2009;11:685–693. doi: 10.1038/ncb1874. [DOI] [PubMed] [Google Scholar]
- Freed EF, Baserga SJ. The C-terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis. Nucleic Acids Research. 2010;38:4798–4806. doi: 10.1093/nar/gkq185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland M, Lutton JD, Spitzer R, Levere RD. Dyskeratosis congenita with hypoplastic anemia: a stem cell defect. American Journal of Hematology. 1985;20:85–87. doi: 10.1002/ajh.2830200112. [DOI] [PubMed] [Google Scholar]
- Gan B, Sahin E, Jiang S, Sanchez-Aguilera A, Scott KL, Chin L, Williams DA, Kwiatkowski DJ, DePinho RA. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19384–19389. doi: 10.1073/pnas.0810584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlick PJ. Measurement of liver protein-synthetic rate. The Biochemical Journal. 1972;126:23P. doi: 10.1042/bj1260023pa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlick PJ, McNurlan MA, Preedy VR. A rapid and convenient technique for measuring the rate of protein synthesis in tissues by injection of [3H]phenylalanine. The Biochemical Journal. 1980;192:719–723. doi: 10.1042/bj1920719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlick PJ, Wernerman J, McNurlan MA, Heys SD. Organ-specific measurements of protein turnover in man. The Proceedings of the Nutrition Society. 1991;50:217–225. doi: 10.1079/pns19910031. [DOI] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- Han K, Jaimovich A, Dey G, Ruggero D, Meyuhas O, Sonenberg N, Meyer T. Parallel measurement of dynamic changes in translation rates in single cells. Nature Methods. 2014;11:86–93. doi: 10.1038/nmeth.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman NW, Lin TV, Zhang L, Paquelet GE, Feliciano DM, Bordey A. mTORC1 targets the translational repressor 4E-BP2, but not S6 kinase 1/2, to regulate neural stem cell self-renewal in vivo. Cell Reports. 2013;5:433–444. doi: 10.1016/j.celrep.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Kuroda T, Kishimoto H, Wang C, Iwama A, Kimura K. Downregulation of rRNA transcription triggers cell differentiation. PloS One. 2014;9:e98586. doi: 10.1371/journal.pone.0098586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annual Review of Cell and Developmental Biology. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nature Genetics. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harbor Perspectives in Biology. 2012;4 doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, Wang S, Ren P, Martin M, Jessen K, Feldman ME, Weissman JS, Shokat KM, Rommel C, Ruggero D. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insco ML, Bailey AS, Kim J, Olivares GH, Wapinski OL, Tam CH, Fuller MT. A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell Stem Cell. 2012;11:689–700. doi: 10.1016/j.stem.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Kedersha N, Anderson P. Stress puts TIA on TOP. Genes & Development. 2011;25:2119–2124. doi: 10.1101/gad.17838411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the "polypyrimidine tract" mRNA family. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph NM, Morrison SJ. Toward an understanding of the physiological function of Mammalian stem cells. Developmental Cell. 2005;9:173–183. doi: 10.1016/j.devcel.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasome H, Papst P, Webb S, Keller GM, Johnson GL, Gelfand EW, Terada N. Targeted disruption of p70(s6k) defines its role in protein synthesis and rapamycin sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5033–5038. doi: 10.1073/pnas.95.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5' non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO Journal. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I, Yuan B, Eichhorn SW, Dzeyk K, Krijgsveld J, Bartel DP, Orr-Weaver TL. Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell Reports. 2014;7:1495–1508. doi: 10.1016/j.celrep.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham KE, Garrels JI, Chang C, Solter D. Quantitative analysis of protein synthesis in mouse embryos. I. Extensive reprogramming at the one- and two-cell stages. Development. 1991;112:921–932. doi: 10.1242/dev.112.4.921. [DOI] [PubMed] [Google Scholar]
- Le Bouteiller M, Souilhol C, Beck-Cormier S, Stedman A, Burlen-Defranoux O, Vandormael-Pournin S, Bernex F, Cumano A, Cohen-Tannoudji M. Notchless-dependent ribosome synthesis is required for the maintenance of adult hematopoietic stem cells. Journal of Experimental Medicine. 2013;210:2351–2369. doi: 10.1084/jem.20122019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard F, Morin F, Ivanchuk S, Langlois F, Stefanovsky V, Rutka J, Moss T. The ARF tumor suppressor controls ribosome biogenesis by regulating the RNA polymerase I transcription factor TTF-I. Molecular Cell. 2010;38:539–550. doi: 10.1016/j.molcel.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton JM, Kudisch M, Gross R, Nathan DG. Defective erythroid progenitor differentiation system in congenital hypoplastic (Diamond-Blackfan) anemia. Blood. 1986;67:962–968. [PubMed] [Google Scholar]
- Liu B, Han Y, Qian SB. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Molecular Cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon CD, Ji B. The role of protein synthesis and digestive enzymes in acinar cell injury. Nature Reviews Gastroenterology & Hepatology. 2013;10:362–370. doi: 10.1038/nrgastro.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Dever TE. Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. Journal of Biological Chemistry. 2010;285:21203–21207. doi: 10.1074/jbc.R110.119743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig LS, Gazda HT, Eng JC, Eichhorn SW, Thiru P, Ghazvinian R, George TI, Gotlib JR, Beggs AH, Sieff CA, Lodish HF, Lander ES, Sankaran VG. Altered translation of GATA1 in Diamond-Blackfan anemia. Nature Medicine. 2014;20:748–753. doi: 10.1038/nm.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nature Reviews Molecular Cell Biology. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Magee JA, Ikenoue T, Nakada D, Lee JY, Guan KL, Morrison SJ. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012;11:415–428. doi: 10.1016/j.stem.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malygin AA, Parakhnevitch NM, Ivanov AV, Eperon IC, Karpova GG. Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic Acids Research. 2007;35:6414–6423. doi: 10.1093/nar/gkm701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JC, Will AJ, Hows JM, Sartori P, Darbyshire PJ, Williamson PJ, Oscier DG, Dexter TM, Testa NG. "Stem cell" origin of the hematopoietic defect in dyskeratosis congenita. Blood. 1992;79:3138–3144. [PubMed] [Google Scholar]
- Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes & Development. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes & Development. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Astrand-Grundstrom I, Arvidsson I, Jacobsson B, Hellstrom-Lindberg E, Hast R, Jacobsen SE. Isolation and characterization of hematopoietic progenitor/stem cells in 5q-deleted myelodysplastic syndromes: evidence for involvement at the hematopoietic stem cell level. Blood. 2000;96:2012–2021. [PubMed] [Google Scholar]
- Ruggero D. Translational control in cancer etiology. Cold Spring Harbor Perspectives in Biology. 2013;5:a012336. doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- Salaun P, Pyronnet S, Morales J, Mulner-Lorillon O, Belle R, Sonenberg N, Cormier P. eIF4E/4E-BP dissociation and 4E-BP degradation in the first mitotic division of the sea urchin embryo. Developmental Biology. 2003;255:428–439. doi: 10.1016/s0012-1606(02)00099-4. [DOI] [PubMed] [Google Scholar]
- Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2:448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Scheu S, Stetson DB, Reinhardt RL, Leber JH, Mohrs M, Locksley RM. Activation of the integrated stress response during T helper cell differentiation. Nature Immunology. 2006;7:644–651. doi: 10.1038/ni1338. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annual Review of Biochemistry. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JW, Blenis J, Pende M, Sonenberg N. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. The EMBO Journal. 2006;25:2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Bohnsack MT, Watkins NJ. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Reports. 2013;5:237–247. doi: 10.1016/j.celrep.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Kuo ML, Roussel MF, Sherr CJ. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Molecular cell. 2003;11:415–424. doi: 10.1016/s1097-2765(03)00057-1. [DOI] [PubMed] [Google Scholar]
- Tahmasebi S, Alain T, Rajasekhar VK, Zhang JP, Prager-Khoutorsky M, Khoutorsky A, Dogan Y, Gkogkas CG, Petroulakis E, Sylvestre A, Ghorbani M, Assadian S, Yamanaka Y, Vinagolu-Baur JR, Teodoro JG, Kim K, Yang XJ, Sonenberg N. Multifaceted Regulation of Somatic Cell Reprogramming by mRNA Translational Control. Cell Stem Cell. 2014;14:606–616. doi: 10.1016/j.stem.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Humphries JM, White RM, Murphey RD, Burns CE, Zon LI. Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Experimental Hematology. 2012;40:228–237. e225. doi: 10.1016/j.exphem.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harbor perspectives in biology. 2011;3:a004440. doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5'TOP mRNA translation. Genes & Development. 2014;28:357–371. doi: 10.1101/gad.231407.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I, Sonenberg N. mRNA translation and energy metabolism in cancer: the role of the MAPK and mTORC1 pathways. Cold Spring Harbor Symposia on Quantitative Biology. 2011;76:355–367. doi: 10.1101/sqb.2011.76.010785. [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Galen P, Kreso A, Mbong N, Kent DG, Fitzmaurice T, Chambers JE, Xie S, Laurenti E, Hermans K, Eppert K, Marciniak SJ, Goodall JC, Green AR, Wouters BG, Wienholds E, Dick JE. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510:268–272. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nature Reviews Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT, Gage FH, Dillin A. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Simic MS, Dillin A. Proteostasis and aging of stem cells. Trends in Cell Biology. 2014;24:161–170. doi: 10.1016/j.tcb.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Wilkins BJ, Lorent K, Matthews RP, Pack M. p53-mediated biliary defects caused by knockdown of cirh1a, the zebrafish homolog of the gene responsible for North American Indian Childhood Cirrhosis. PloS One. 2013;8:e77670. doi: 10.1371/journal.pone.0077670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao GG, Garg M, Lim S, Wong D, Go VL, Lee WN. Determination of protein synthesis in vivo using labeling from deuterated water and analysis of MALDI-TOF spectrum. Journal of Applied Physiology. 2008;104:828–836. doi: 10.1152/japplphysiol.00976.2007. [DOI] [PubMed] [Google Scholar]
- Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nature Reviews Molecular Cell Biology. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- Zhang F, Hamanaka RB, Bobrovnikova-Marjon E, Gordan JD, Dai MS, Lu H, Simon MC, Diehl JA. Ribosomal stress couples the unfolded protein response to p53-dependent cell cycle arrest. Journal of Biological Chemistry. 2006a;281:30036–30045. doi: 10.1074/jbc.M604674200. [DOI] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, Wu H, Li L. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006b;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Shalaby NA, Buszczak M. Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science. 2014a;343:298–301. doi: 10.1126/science.1246384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Duc AC, Rao S, Sun XL, Bilbee AN, Rhodes M, Li Q, Kappes DJ, Rhodes J, Wiest DL. Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Developmental Cell. 2013;24:411–425. doi: 10.1016/j.devcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, Kwiatkowski DJ, Manning BD. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014b doi: 10.1038/nature13492. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Molecular and Cellular Biology. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]