Abstract
Cooperation involving Plant Growth-Promoting Rhizobacteria results in improvements of plant growth and health. While pathogenic and symbiotic interactions are known to induce transcriptional changes for genes related to plant defense and development, little is known about the impact of phytostimulating rhizobacteria on plant gene expression. This study aims at identifying genes significantly regulated in rice roots upon Azospirillum inoculation, considering possible favored interaction between a strain and its original host cultivar. Genome-wide analyzes of Oryza sativa japonica cultivars Cigalon and Nipponbare were performed, by using microarrays, seven days post-inoculation with Azospirillum lipoferum 4B (isolated from Cigalon) or Azospirillum sp. B510 (isolated from Nipponbare) and compared to the respective non-inoculated condition. A total of 7384 genes were significantly regulated, which represent about 16% of total rice genes. A set of 34 genes is regulated by both Azospirillum strains in both cultivars, including a gene orthologous to PR10 of Brachypodium, and these could represent plant markers of Azospirillum-rice interactions. The results highlight a strain-dependent response of rice, with 83% of the differentially expressed genes being classified as combination-specific. Whatever the combination, most of the differentially expressed genes are involved in primary metabolism, transport, regulation of transcription and protein fate. When considering genes involved in response to stress and plant defense, it appears that strain B510, a strain displaying endophytic properties, leads to the repression of a wider set of genes than strain 4B. Individual genotypic variations could be the most important driving force of rice roots gene expression upon Azospirillum inoculation. Strain-dependent transcriptional changes observed for genes related to auxin and ethylene signaling highlight the complexity of hormone signaling networks in the Azospirillum-rice cooperation.
Keywords: Azospirillum, hormone signaling, plant defense, plant growth-promoting rhizobacteria, rice, transcriptome
Introduction
Rhizodeposition supports growth of a wide range of microorganisms able to establish intimate interactions with plant roots. In the case of parasitism, nutritional requirements of the microbe partner are supported at the expense of plant development and reproduction (O'Brien et al., 2011; Schumacher and Tudzynski, 2012). In the case of mutualism, the interaction leads to a nutritional exchange so that costs and benefits are reciprocally shared by both microbial and plant partners (Smith and Read, 2008). Whether engaged in a parasitic or mutualistic interaction, the microbial partner is perceived as an intruder and the success of the adaptation strategy partly depends on the microbe's ability to bypass defense mechanisms and invade plant tissues (Soto et al., 2009). Then, plant immune response involves gene expression changes that mediate trade-off between defense and development to ensure plant survival through an efficient allocation of resources (Buscaill and Rivas, 2014). Cooperation involving Plant Growth-Promoting Rhizobacteria (PGPR) results in improvements of plant growth and health; however, the invasion of root tissues is not a critical step in successful interaction as several efficient strains are described as root-surface colonizers (Lugtenberg and Kamilova, 2009; Chamam et al., 2013). If mechanisms directly implicated in plant growth-promotion have been extensively studied, most of these works have assessed the impact of PGPR on plant morphological traits and little is known about changes induced at the molecular level (Bashan and de-Bashan, 2010; Galland et al., 2012; van de Mortel et al., 2012; Wisniewski-Dyé et al., 2013).
For more than 50 years, PGPR of a wide range of genera including Acetobacter, Azospirillum, Bacillus, Burkholderia, Herbaspirillum, Phyllobacterium or Pseudomonas have been known for stimulating the growth of numerous host plants (Desbrosses et al., 2009; Lugtenberg and Kamilova, 2009; Richardson et al., 2009; Kumar et al., 2011; Saharan and Nehra, 2011). In particular, the genus Azospirillum constitutes an important phytostimulator and an increasing number of field trials are undertaken, principally in India and Latin America where several Azospirillum inoculants are commercialized (Steenhoudt and Vanderleyden, 2000; Bashan et al., 2004; Fuentes-Ramirez and Caballero-Mellado, 2012). In most cases, successful inoculation results in root and shoot morphological changes, plant nutrition improvements, and yield enhancements (Richardson et al., 2009; Vacheron et al., 2013; Wisniewski-Dyé et al., 2013). If the phytostimulating effect of Azospirillum was originally attributed to its ability to fix atmospheric nitrogen, it is now admitted that the modulation of the phytohormonal balance is the most important mechanism resulting in the modification of root architecture and higher nutrient uptake by the plant (Steenhoudt and Vanderleyden, 2000; Somers et al., 2004; Prigent-Combaret et al., 2008). Besides morphological changes, Azospirillum also increases root exudation and modifies the chemical structure of root cell wall (Heulin et al., 1987; El Zemrany et al., 2007). In addition, Azospirillum was found capable of increasing the resistance of the host plant against pathogen through mechanisms independent of salicylic acid signaling (Yasuda et al., 2009). Investigation on maize secondary metabolism revealed that major qualitative and quantitative modifications occur following Azospirillum inoculation (Walker et al., 2011). Moreover, these modifications depend on bacterial strain/maize cultivar combinations suggesting that a genotype specific perception of Azospirillum occurs during the cooperation with maize. These observations were recently strengthened by a study made on two rice cultivars, Cigalon and Nipponbare, after the inoculation of two Azospirillum strains isolated from each cultivar (Chamam et al., 2013): Azospirillum lipoferum 4B isolated from Cigalon roots (Thomas-Bauzon et al., 1982) and Azospirillum sp. B510 isolated from Nipponbare (Elbeltagy et al., 2001). Profiling of secondary metabolites and morphological measurements evidenced that the impact of Azospirillum differs according to strain/cultivar combinations and that a specific interaction leading to a stronger phytostimulation occurs between a strain and its original host cultivar. In addition, the endophyte strain B510 was shown to trigger a systemic response, as metabolic changes were observed in both roots and shoots. However, whether or not perception of Azospirillum involved plant immune response remains an unanswered question and regulatory mechanisms underlying host-specific metabolic changes have to be unraveled.
In this context, our study aims at characterizing genetic determinants regulated in rice roots at an early stage of the interaction with Azospirillum, considering possible favored interaction between a strain and its original host cultivar. Thus, genome-wide analyzes of root gene expression of Oryza sativa japonica cultivars Cigalon and Nipponbare were performed 7 days post-inoculation with A. lipoferum 4B or Azospirillum sp. B510 and compared to the respective non-inoculated condition. A focus was made on genes potentially involved in plant defense and hormone signaling.
Materials and methods
Biological material
In this study, two rice (Oryza sativa L.) cultivars belonging to the japonica group, cv. Cigalon (Center Français du Riz, France) and cv. Nipponbare (J. B. Morel, BGPI, Montpellier, France) were inoculated with two diazotrophic strains of the genus Azospirillum: A. lipoferum 4B initially isolated from rice roots of the cv. Cigalon in France (Thomas-Bauzon et al., 1982) and Azospirillum sp. B510 initially isolated from surface sterilized rice stems of the cv. Nipponbare (Elbeltagy et al., 2001).
RNA samples and cDNA synthesis
Six independent experiments were performed per condition (three for microarray hybridization and three for qRT-PCR validation). Seed sterilization, plant inoculation and plant growth were performed as previously described (Chamam et al., 2013; Drogue et al., 2014). Rice seeds were surface sterilized by washing for 40 min in a sodium hypochlorite solution, rinsed 5 times in demineralized sterile water, and then chlorine traces were removed by washing 3 times in sterile-filtered 2% (w/v) sodium thiosulfate before rinsing 5 times in demineralized sterile water. Surface sterilized seeds were germinated on sterile plant agar (8 g·L−1) (Sigma Chemical Co, Saint Louis, USA) for 2 days in the dark at 28°C. Bacterial cells in late-exponential phase were mixed with 50 mL of plant agar (8 g·L−1) (to a final concentration of 2.107 cells·mL−1) and introduced into 120 × 120 × 17 mm square plates. For both rice cultivars, five disinfected germinated seeds were laid onto the plates and plates were incubated vertically, for 7 days in a growth chamber (MLR350, SANYO, UK) with a photoperiod of 16 h at 28°C (light 150 μE m−2 s−1), and 8 h at 22°C in the dark. For each experiment, 30 plant root systems were pooled and frozen using liquid nitrogen. Root cell lysis was performed by grinding root systems with a mortar and pestle under liquid nitrogen. Total RNA was isolated using the TRIzol method (Invitrogen, Carlsbad, CA, USA). RNA samples were purified using RNeasy plant mini kit (Qiagen, Courtaboeuf, France) according to the manufacturer's protocol. RNA integrity was assessed using Agilent RNA 6000 Pico Kit (Agilent Technologies, Waldbronn, Germany) and the Agilent 2100 Bioanalyzer (Agilent Technologies) device.
In order to increase mRNA representation in RNA samples, total RNA were digested with mRNA ONLYTM Procaryotic mRNA isolation kit (Epicenter Biotechnologies, Madison, WI, USA) according to the provided protocol.
The microarray cDNA (three independent samples per condition) was synthesized with the Superscript® Double-Stranded cDNA Synthesis Kit (Invitrogen), using a mix (1:1) of random primers (Promega Corporation, Madison, WI, USA) and Oligo-dT (15) primers (Promega).
Microarray hybridization and data analysis
We designed an oligo microarray, which was produced by NimbleGenTM (Madison, WI, USA) derived of one which was described previously (Picault et al., 2009). This microarray is composed of about 385,000 60 mer probes selected for their GC content, Tm, and number of cycles needed to synthesize the oligo. This chip contains 90,000 probes representing 45,000 genes (two probes per gene) of rice Oryza sativa ssp. japonica, based on the TIGR rice genome annotation version 3.1 genes (Yuan et al., 2005) and 201,691 oligomers corresponding to previously described copies of LTR retrotranposons available on the retrOryza database (www.retroryza.fr; Chaparro et al., 2007). Probes represent 1000 bp of the LTR-retrotransposon flanking regions at the 3′ and 5′ side. The oligonucleotides have been designed at the 3′ end of the genes to detect the readings of reverse transcriptase. On the other hand, the LTR-retrotransposons are represented throughout their length at the rate of a probe every 500 bp. The analysis was performed using the classification proposed by El Baidouri and Panaud (2013) that consists in 369 families and 3623 loci harboring complete elements. Among the differentially expressed oligomers, only those displaying 100% identity and which are unique in the genome were analyzed.
When it was possible, probes have been designed to be unique in the genome (i.e., locus specific) to overcome the problems of oligonucleotides redundancy on the chip. When there were three mismatches during hybridization between a cDNA and an oligonucleotide, hybridization was considered stable enough to withstand the conditions of washing after the chip hybridization. The oligonucleotides are therefore regarded as locus specific when they are not matching elsewhere, but having at most three mismatches, which represents 5% of all oligonucleotides.
For each condition, three independent cDNA samples were labeled and hybridized by Roche Nimblegen according to their standard protocol. Data analysis was performed using Bioconductor microarray packages for R software (http://www.bioconductor.org/). The robust multi-array average (RMA) method associated with quantile normalization was applied (Bolstad et al., 2003; Irizarry et al., 2003). Analysis of variance with a false discovery rate adjustment method was realized (Benjamini and Hochberg, 1995). The results of different treatment comparison were obtained in Log2-fold change. The oligonucleotides selected were those which present a two fold increase or decrease in expression, i.e., a log-fold change smaller or equal to −1 for down-regulation, and greater or equal to 1 for up-regulation. Oligonucleotides displaying P ≤ 0.05 for the statistical test were selected. For each cultivar, the respective uninoculated condition was used as control. All oligonucleotides differentially expressed were remapped to Os-Nipponbare-Reference-IRGSP version 1.0 (Rice Annotation Project et al., 2008) using BLAST.
RT-qPCR
For each condition, three independent RNA samples were used to validate gene expression level by performing reverse transcription quantitative real-time PCR (RT-qPCR). Validation was made on a group of 14 representative genes (Table 1) using LightCycler® 480 SYBR Green I Master kit (Roche Diagnostics GmbH, Mannheim, Germany) on a LightCycler® 480 Real-Time PCR System (Roche). The actin gene (Os03g0718100) showing an invariant expression was used as reference to normalize RT-qPCR values. After DNase I treatment, total RNA (800 ng) was used for cDNA synthesis using GoScriptTM Reverse transcription system (Promega) with oligo-dT (15) primer in accordance with the manufacturer's protocol.
Table 1.
Validation of microarray data.
| Gene | FC Cig_4Ba | FC Cig_B510 | FC Nip_4B | FC Nip_B510b | |||||
|---|---|---|---|---|---|---|---|---|---|
| Log2 (qPCR) | Log2 (array) | Log2 (qPCR) | Log2 (array) | Log2 (qPCR) | Log2 (array) | Log2 (qPCR) | Log2 (array) | ||
| Differentially expressed in all conditions | |||||||||
| Os09g0358000 | Tyrosine kinase domain | 2.27 | 1.77 | 2.53 | 2.46 | 1.87 | 3.01 | 2.06 | 1.32 |
| Differentially expressed in Cig_4B and Nip_4B only | |||||||||
| Os09g0417800 | WRKY transcription factor 62 | 1.49 | 1.75 | 0.66 | _ | 1.00 | 1.88 | −0.01 | _ |
| Os11g0686900 | Similar to NB-ARC domain | 0.83 | 1.16 | −0.23 | _ | 1.74 | 2.40 | nd | _ |
| Differentially expressed in Cig_B510 and Nip_B510 only | |||||||||
| Os06g0115600 | Common symbiosis signaling (SYM) pathway | −0.09 | _ | −2.00 | −1.84 | 0.01 | _ | −1.18 | −1.47 |
| Os05g0583000 | Similar to WRKY8 | 0.32 | _ | 2.92 | 1.71 | 0.16 | _ | 2.24 | 1.39 |
| Os12g0139400 | A-type response regulator, cytokinin signaling | −0.15 | _ | −2.84 | −1.78 | −0.09 | _ | −2.94 | −1.10 |
| Os11g0143300 | A-type response regulator, cytokinin signaling | 0.15 | _ | −1.09 | −1.95 | 0.04 | _ | −2.25 | −1.62 |
| Os02g0805100 | Similar to auxin-responsive protein IAA12 | −0.01 | _ | −1.15 | −1.34 | −0.23 | _ | −2.47 | −1.16 |
| Differentially expressed in Cig_4B and Cig_B510 only | |||||||||
| Os05g0196600 | Similar to ACC synthase | 0.38 | 1.33 | 0.95 | 1.63 | −0.15 | _ | 0.16 | _ |
| Differentially expressed in Cig_4B only | |||||||||
| Os08g0136100 | Homeobox-leucine zipper domain | 0.87 | 1.11 | 0.25 | _ | 0.28 | _ | −0.14 | _ |
| Significantly regulated in Cig_B510 only | |||||||||
| Os06g0179200 | Similar to Nodulin-like protein | 0.16 | _ | −1.60 | −1.78 | 0.18 | _ | 0.12 | _ |
| Os08g0499300 | WRKY transcription factor 30 | 0.10 | _ | 0.52 | 1.34 | 0.12 | _ | −0.06 | _ |
| Significantly regulated in Nip_B510 only | |||||||||
| Os01g0904700 | B-type response regulator, cytokinin signaling | −0.12 | _ | −0.07 | _ | −0.04 | _ | −1.09 | −1.21 |
| Os05g0515400 | Similar to auxin response factor 14 | −0.09 | _ | 0.10 | _ | −0.09 | _ | −1.15 | −1.42 |
| Reference gene | |||||||||
| Os03g0718100 | Actin | ||||||||
Dashes replace non-significant fold change values obtained from microarray data (|Log2(FC)| ≤ 1 and Padj > 0.05).
nd, not determined.
DNA contamination was checked with reactions that lacked reverse transcriptase as negative controls. Specific primers were designed using Light Cycler Probe design Software 2.0 (Roche) with the following criteria: product size ranges 100–400 pb, primer size comprised between 17 and 22 bases, optimal primer Tm 60°C (Additional file 1: Table S1). Real time PCR conditions were: a denaturation stage of 10 min at 95°C; an amplification stage of 45 cycles of 15 s at 94°C, 10 s at 60°C and 20 s at 72°C; and a melting curve stage of 5 s at 95°C and 1 min at 65°C increased to 97°C with a ramp rate of 0.11°C s−1. All reactions were performed in three technical replicates and carried out in LightCycler 480 Multiwell plate 96 (Roche) with adhesive sealings foils (Roche) in a final volume of 10 μl containing 1 μl of each primer (5 μM), 5 μl of master mix and 3 μl of cDNA diluted 50 times. For each cultivar, the respective uninoculated condition was used as the calibrator condition and relative gene expression was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Data were statistically validated by a correlation test using the Pearson's method.
Availability of microarray data
Microarray data are available on GEO database through the following accession number GSE59137.
Results
Transcriptomic profiles were obtained using microarrays and the four following combinations were analyzed (three independent replicates per combination): Cigalon/A. lipoferum 4B (Cig_4B) and Cigalon/Azospirillum sp. B510 (Cig_B510) compared to non-inoculated Cigalon; Nipponbare/A. lipoferum 4B (Nip_4B) and Nipponbare/Azospirillum sp. B510 (Nip_B510) compared to non-inoculated Nipponbare. Genes significantly regulated were selected using a Padjusted-value (Padj) threshold of 0.05 and a fold change cutoff of 2 (|Log2(FC)| ≥ 1). According to the cultivar of which each strain was originally isolated, Cig_4B and Nip_B510 combinations constitute interactions between a strain and its original host cultivar, which will be hereafter designed as host combinations, while Cig_B510 and Nip_4B combinations constitute interactions with non-host cultivars, which will be hereafter designed as non-host combinations.
General features of rice-root transcriptome profiling in response to Azospirillum inoculation
Microarray design was based on the genome sequence of cultivar Nipponbare. To ensure that it could be used to hybridize cDNA obtained from cultivar Cigalon, the genetic proximity between both cultivars was analyzed by sequencing eight genes (including the gene encoding actin) after PCR amplification from Cigalon DNA (at least 500 pb per gene) (Additional file 2: Table S2). For all the sequenced genes, an identity of 100% was observed with the corresponding genes of Nipponbare. In addition, when comparing the non-inoculated Cigalon transcriptome profile to the non-inoculated Nipponbare profile, only 193 genes are differentially transcribed between the two cultivars (87 up-regulated and 106 down-regulated). This represents only 0.43% of the targeted genes (i.e., 45,000 genes) and may be explained by physiological differences. Thus, the microarray was considered suitable for analysis and comparison of both Nipponbare and Cigalon transcriptomes.
When considering the four combinations, a total of 7384 genes are differentially expressed in rice roots, which represent about 16% of the entire set of rice genes. Each strain/cultivar combination displays specific expression profiles highlighting a strain-specific response of the host plant. The most important changes are observed when strain B510, isolated from Nipponbare, is inoculated on Cigalon roots (Cig_B510), with 3865 regulated genes, equally induced and repressed (1993 up-regulated; 1872 down-regulated) (Figure 1). Conversely, the inoculation of strain 4B on its original cultivar Cigalon (Cig_4B) is accompanied by the differential expression of only 1243 genes, mostly induced (1196 up-regulated; 47 down-regulated). When considering Nipponbare roots, the number of regulated genes is similar for Nip_4B and Nip_B510 combinations with respectively, 2141 and 2539 regulated genes. However, these genes are mostly induced in Nip_4B (1965 up-regulated; 176 down-regulated) while they are repressed in Nip_B510 combination (203 up-regulated; 2336 down-regulated).
Figure 1.
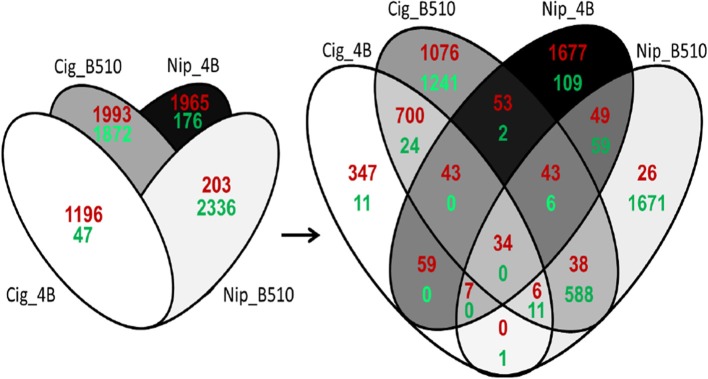
Venn diagram of genes differentially expressed in rice roots (|Log2(FC)| ≥ 1 and Padj ≤ 0.05). Oryza sativa L. japonica cultivar Cigalon and cultivar Nipponbare were inoculated with Azospirillum strains 4B and B510. For each cultivar, the respective non-inoculated condition was used as reference to evidence genes up-regulated (red) and down-regulated (green) after Azospirillum inoculation (|Log2(FC)| ≥ 1 and Padj < 0.05). Cig_4B and Nip_B510 combinations constitute the interaction between a strain and its original host cultivar (host combinations). Cig_B510 and Nip_4B combinations constitute interactions with non-host cultivars (non-host combinations).
A Venn diagram analysis conducted on expression profiles obtained for the four combinations, unveils 15 sets of genes (Figure 1). Four sets, named combination-specific genes (genes induced or repressed only in one combination), represent 83% of all differentially expressed genes, with 358 genes for Cig_4B (347 up-regulated, 11 down-regulated), 2317 genes for Cig_B510 (1076 up-regulated, 1241 down-regulated), 1786 genes for Nip_4B (1677 up-regulated, 109 down-regulated) and 1697 genes for Nip_B510 (26 up-regulated, 1671 down-regulated) (Additional file 3: Table S3). Two other sets regroup genes that are common only to Cig_4B and Cig_B510 (700 up-regulated, 24 down-regulated) or only to Nip_4B and Nip_B510 (49 up-regulated, 59 down-regulated), representing Azospirillum-regulated cultivar-specific genes (Additional File 4: Table S4). Inversely, two sets comprise strain-specific genes that display the same regulation only in both Cig_4B and Nip_4B (59 up-regulated, zero down-regulated) or only in both Cig_B510 and Nip_B510 (38 up-regulated, 588 down-regulated) (Additional File 5: Table S5). Two additional sets include genes displaying the same regulation in both host combinations, or in both non-host combinations (Additional File 6: Table S6). One set contains 34 up-regulated genes common to the four combinations and no down-regulated genes. The four remaining sets comprise genes that are common to three of the four combinations and will no longer be discussed in the current analysis. Microarray data were confirmed by analyzing expression levels of 14 representative genes using reverse transcription quantitative polymerase chain reaction (RT-qPCR) (Table 1). This includes seven up-regulated genes and seven down-regulated genes belonging to seven of the 15 categories described in Figure 1. These results show that the array data are in accordance with the RT-qPCR data (R2 = 0.83; Pvalue = 2.7.10−10).
The transcriptome survey was completed with an analysis of the LTR-retrotransposons. Indeed, LTR-retrotransposons, a particular type of transposable elements, represent 25% of the total genomic sequence of rice (Rice Annotation Project et al., 2008). The transcriptional activation of LTR-retrotransposons can lead to the activation of a flanking gene, either through the action of the enhancer regions of the element or by co-transcription (Michaud et al., 1994); conversely, LTR-retrotransposons can also act as suppressors of gene expression when they are inserted in antisense in the 3′ region of a gene. Interestingly, Azospirillum inoculation leads to a differential expression of a large number of LTR-retrotransposon families for both rice cultivars. To avoid bias related to cultivar polymorphism, the complete analysis was performed only for the cultivar Nipponbare. For this latter, differential expression is observed for 115 and 148 LTR-retrotransposon families for strains B510 and 4B, respectively. In addition, strain 4B modifies the transcription of 21.4% of all LTR-retrotransposons, among which 51% are up-regulated and 49% down-regulated. For strain B510, 17.8% of all LTR-retrotransposons are differentially expressed (37% up-regulated and 62% down-regulated). Among the differentially expressed elements, 22% are common to Nip_4B and Nip_B510 combinations. In addition, we explored if transposable elements can significantly alter the expression of adjacent genes. In this analysis, two and six LTR-retrotransposons are located in the 3 kb upstream regions of regulated genes while four and five ones are located in the 3 kb downstream regions, for Nip_B510 and Nip_4B, respectively (Additional File 7: Table S7). However, no co-transcription events between gene and LTR-retrotransposon was evidenced by using RT-PCR method (data not shown), suggesting that none of these elements have a direct impact on the expression of adjacent genes.
Impact of strain lifestyle on rice roots gene expression
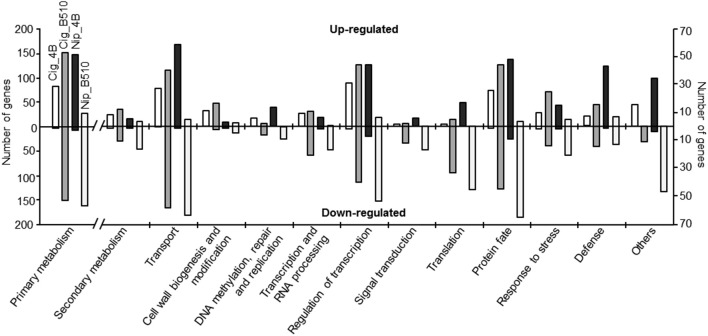
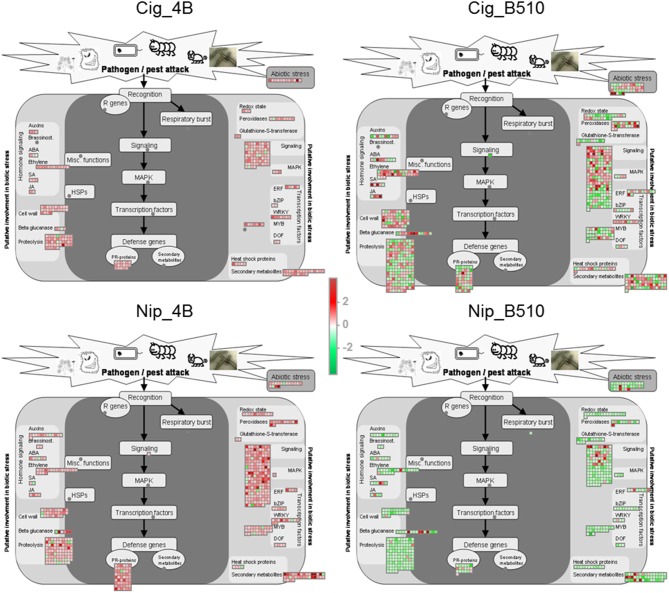
As mentioned above, most of the differentially expressed genes are induced in combinations involving strain 4B while they are mostly repressed for combinations involving strain B510 (Figure 1). In addition, when considering functional classification available for only 20% of all differentially expressed genes, several differences are evidenced at the strain level (Figure 2). Whatever the combination, the most important numbers of differentially expressed genes are observed for (i) primary metabolism, (ii) transport, (iii) regulation of transcription, and (iv) protein fate, four categories in which genes seem quasi exclusively repressed in Nip_B510 while they are quasi exclusively induced in Nip_4B. Similarly, when considering genes involved in response to stress and plant defense, two substantial categories in plant-microbe interactions, it appears that strain B510 leads to the repression of a more important number of genes than strain 4B. These results are of particular interest as they may be related to lifestyle differences between Azospirillum strains; indeed, strain 4B was shown to colonize only rice-root surface while strain B510 is able to colonize the outer layers of rice-root tissues (Elbeltagy et al., 2001; Chamam et al., 2013). To consider the impact of strain lifestyle on plant defense response, the analysis was focused on genes potentially involved in biotic stress. Identification of these genes was improved using Mapman annotation software as recently described for the Azospirillum-Arabidopsis interaction (Spaepen et al., 2014). This additional analysis, illustrated in Figure 3, confirms observations made above. Then, strain 4B leads quasi exclusively to the induction of genes related to biotic stress in Nipponbare and to a lower extent in Cigalon. On the contrary, genes involved in biotic stress response are quasi exclusively repressed in the Nip_B510 host combination, while the number of up-regulated genes and down-regulated genes is similar in the Cig_B510 non-host combination. In addition, Mapman visualization analysis highlights relevant categories of genes being differentially expressed such as genes encoding peroxidases, transcription factors of the MYB, WRKY, and ERF families as well as Pathogenesis-Related (PR) genes.
Figure 2.
Functional classification of the differentially expressed genes in Oryza sativa L. japonica in response to Azospirillum inoculation. Numbers of genes differentially expressed are shown per functional categories for each Azospirillum-rice combination: Cig_4B (white), Cig_B510 (dark gray), Nip_4B (black), and Nip_B510 (light gray). Genes were classified according to Biological Process assignation taken from the Rap-DB database. Genes with no Biological Process assignation represent about 80% of differentially expressed genes for each condition. Category “Others” includes principally genes implicated in photosynthesis, exocytosis, sexual reproduction, cytoskeleton organization, cell cycle, and cell death.
Figure 3.
Mapman software visualization of genes related to biological stress. Genes differentially expressed (|Log2(FC)| ≥ 1 and Padj ≤ 0.05) after Azospirillum inoculation on rice roots are represented according to their respective Log2 rescaled fold change. Red color represents up-regulated genes and green color represents down-regulated genes with the most intense color on the scale representing a Log2(FC) = 4.8. The dark gray rectangle includes genes directly involved in biotic stress responses and the light gray rectangle includes genes potentially involved in biotic stress response. ABA, abscisic acid; bZIP, basic region leucine zipper; DOF, DNA-binding with one finger; ERF, ethylene responsive factor; HSP, heat shock protein; JA, jasmonic acid; MAPK, mitogen activated protein kinase; MYB, myeloblast; PR, pathogenesis-related; R, resistance; SA, salicylic acid.
Another way to analyze the impact of strain lifestyle on rice roots gene expression was to consider the 4B_specific genes (59) and B510-specific genes (626) highlighted in the Venn diagram analysis (Figure 1; Additional File 5: Table S5). Among the 59 genes specifically induced by the surface-colonizing strain 4B, we identified 14 genes that could be involved in biotic stress response among which two genes encoding peroxidases, two WRKY transcription factors and one PR gene. While all the 4B-specific genes are up-regulated, 38 of the B510-specific genes are up-regulated and 588 are down-regulated including at least 14 and 102 genes associated to biotic stress response, respectively. The endophyte strain B510 triggers the induction of four genes potentially involved in hormone signaling, as well as genes encoding a peroxidase, a WRKY transcription factor and a PR protein. Moreover, a larger number of genes are down-regulated, including five PR genes, three genes encoding peroxidases but no WRKY transcription factor. Finally, the repression of a putative indole synthase (Os03g0797000), three putative auxin responsive proteins (Os02g0805100, Os08g0335600, Os11g0523800), two response regulators involved in cytokinin signaling (Os11g0143300, Os12g0139400), an isochorismate synthase (Os09g0361500) required for salicylic acid synthesis, as well as four genes potentially involved in ethylene signaling (Os03g0439500, Os04g0667400, Os09g0115500, 0s09g0309700) suggest that a B510-specific hormone signaling occurs in rice roots.
Plant markers of Azospirillum-rice cooperation and similarities with pathogen infection
As revealed in the Venn diagram analysis (Figure 1), a total of 34 genes (12 genes of unknown function) are induced in the four combinations and could be defined as plant markers of Azospirillum-rice interactions (Table 2). Interestingly, a PR gene orthologous to PR10 of Brachypodium is induced in the four conditions (Os12g0555200). In addition, two genes encoding Cys-Rich domain containing protein (Os02g0579800, Os02g0580000) and an orthologous gene to AT1G59950 of Arabidopsis thaliana (Os03g0237100), which are potentially implicated in stress response, as well as a terpene synthase involved in gibberellin synthesis (Os04g0178300), and a gene encoding a putative precursor of phytoalexin (Os11g0474800, encoding a stemar-13-ene synthase) are induced in the four combinations. Finally, the set of potential plant markers of Azospirillum-rice cooperation also includes a transcription factor (Os01g0952800) and four genes potentially involved in signal transduction (Os07g0537900, Os08g0501500, Os08g0203400, Os09g0358000, this latter being validated by RT-qPCR, Table 1).
Table 2.
List of 34 genes induced in the four combinations.
| Gene ID | Log2(FC) | Gene description | MSU Gene ID | M. oryzaea | |||
|---|---|---|---|---|---|---|---|
| Cig_4B | Nip_4B | Cig_B510 | Nip_B510 | ||||
| Os01g0191200 | 1.34 | 3.28 | 2.18 | 3.09 | Putative HAD phosphatase | LOC_Os01g09540 | |
| Os01g0495701 | 1.14 | 2.60 | 2.02 | 2.12 | Conserved hypothetical protein | None | |
| Os01g0608101 | 1.33 | 2.73 | 2.42 | 2.39 | Conserved hypothetical protein | None | |
| Os01g0647200 | 1.07 | 2.97 | 1.91 | 1.71 | Conserved hypothetical protein | LOC_Os01g45914 | |
| Os01g0952800 | 1.70 | 3.32 | 2.75 | 2.65 | Transcription factor | LOC_Os01g72370 | |
| Os02g0569400 | 1.54 | 2.42 | 2.23 | 1.78 | Cytochrome P450 family protein | LOC_Os02g36070 | |
| Os02g0579800 | 1.49 | 3.08 | 1.75 | 1.91 | Cys-rich domain containing protein | LOC_Os02g36940 | + |
| Os02g0580000 | 1.51 | 2.80 | 1.78 | 1.32 | Cys-rich domain containing protein | LOC_Os02g36950 | |
| Os02g0582900 | 2.20 | 4.95 | 3.77 | 4.98 | Conserved hypothetical protein | LOC_Os02g37190 | + |
| Os02g0594232 | 1.18 | 2.28 | 2.09 | 1.8 | Conserved hypothetical protein | None | |
| Os02g0791300 | 1.41 | 3.00 | 2.91 | 2.58 | Conserved hypothetical protein | LOC_Os02g54870 | |
| Os03g0129800 | 1.01 | 1.79 | 1.01 | 1.58 | Uncharacterized protein | LOC_Os03g03730 | + |
| Os03g0237100 | 1.42 | 3.43 | 2.40 | 2.94 | Putative NADPH-dependent codeinone reductase | LOC_Os03g13390 | |
| Os03g0307300 | 1.34 | 3.33 | 2.29 | 2.54 | Nicotianamine synthase 1 | LOC_Os03g19427 | |
| Os04g0178300 | 1.25 | 3.22 | 2.39 | 2.91 | Putative syn-copalyl diphosphate synthase | LOC_Os04g09900 | |
| Os04g0178400 | 1.92 | 3.56 | 2.60 | 2.82 | Putative cytochrome P450 | LOC_Os04g09920 | + |
| Os06g0293500 | 2.40 | 3.26 | 2.62 | 1.73 | Conserved hypothetical protein | LOC_Os06g18960 | + |
| Os06g0486800 | 1.10 | 2.31 | 1.84 | 1.42 | Putative mitochondrial formate dehydrogenase | LOC_Os06g29180 | |
| Os06g0718400 | 1.39 | 1.68 | 1.66 | 1.53 | Plastocyanin-like domain containing protein | LOC_Os06g50420 | |
| Os07g0190000 | 1.69 | 3.02 | 2.77 | 2.85 | Putative 1-deoxy-D-xylulose 5-phosphate synthase | LOC_Os07g09190 | + |
| Os07g0258400 | 1.61 | 2.25 | 2.60 | 1.70 | Putative metal transporter Nramp6 | LOC_Os07g15460 | |
| Os07g0416900 | 2.76 | 3.78 | 2.35 | 2.43 | Omega-6 fatty acid desaturase | LOC_Os07g23410 | + |
| Os07g0537900 | 1.12 | 1.24 | 1.83 | 1.62 | Ser/Thr kinase receptor domain | LOC_Os07g35340 | |
| Os07g0664000 | 1.52 | 2.24 | 2.59 | 1.85 | Putative short chain dehydrogenase/reductase | LOC_Os07g46870 | + |
| Os08g0203400 | 1.66 | 4.69 | 2.54 | 3.59 | Protein kinase/core domain containing protein | LOC_Os08g10310 | |
| Os08g0501500 | 1.03 | 1.27 | 2.26 | 1.70 | OsWAK receptor-like protein kinase | LOC_Os08g39210 | + |
| Os09g0358000 | 1.77 | 3.07 | 2.71 | 2.51 | Tyrosin kinase domain containing protein | LOC_Os09g19350 | |
| Os09g0358100 | 1.27 | 2.05 | 1.79 | 1.05 | Senescence-induced serine/threonine kinase | LOC_Os09g19360 | |
| Os10g0195250 | 1.24 | 2.08 | 2.01 | 1.68 | Conserved hypothetical protein | LOC_Os10g11889 | |
| Os11g0262600 | 1.31 | 2.45 | 2.29 | 2.29 | Conserved hypothetical protein | LOC_Os11g15624 | |
| Os11g0474800 | 2.83 | 2.81 | 3.20 | 2.57 | Putative stemar-13-ene synthase | LOC_Os11g28530 | |
| Os12g0171801 | 1.12 | 2.77 | 2.75 | 1.64 | Hypothetical protein | None | |
| Os12g0236100 | 1.17 | 2.03 | 1.70 | 1.60 | Conserved hypothetical protein | LOC_Os12g13340 | |
| Os12g0555200 | 2.22 | 3.97 | 2.77 | 3.31 | Putative pathogenesis-related Bet v family protein | LOC_Os12g36850 | + |
Genes previously shown to be induced in rice root 6 days post-inoculation with M. oryzae (Marcel et al., 2010) are indicated with the sign +.
Making use of the Rice Oligonucleotide Array Database (Cao et al., 2012), we identified experiments analyzing gene expression in rice during plant-microbe interactions. Prior to this analysis, RT-qPCR expression profiles observed in rice roots for the 14 genes used to validate our microarray data (Table 1) were compared to RT-qPCR expression profiles obtained in rice leaves and none displays differential expression when considering shoot compartment (data not shown). Then, the array database analysis was focused on rice roots and only one experiment analyzing gene expression in the root compartment was evidenced. This work considered the impact of the fungus phytopathogen Magnaporthe oryzae on rice transcriptome 6 days after infection (Marcel et al., 2010). The list of 34 potential markers of Azospirillum-rice cooperation, up-regulated in the four combinations, was compared to the list of genes differentially expressed after M. oryzae inoculation, evidencing that 10 of the potential markers (3 genes of unknown function) are also up-regulated in response to Magnaporthe infection while none of these markers are down-regulated after Magnaporthe infection (Table 2). Thus, the PR gene orthologous to PR10 of Brachypodium (Os12g0555200) discussed above, as well as a gene related to signal transduction (Os08g0501500) and a gene encoding a Cys-Rich domain containing protein (Os02g057980) are up-regulated in response to both Azospirillum and Magnaporthe.
To understand whether rice biotic stress responses could be specific to either host or non-host interactions, the analysis was focused on the comparison of host combinations and non-host combinations. According to Figure 1, the number of up-regulated genes appears to be lower when a strain is inoculated on its original host cultivar (Cig_4B and Nip_B510). In addition, fewer genes implicated in response to stress and plant defense are differentially expressed in host combinations (Figure 2), the most striking example being PR genes as highlighted in Figure 3. However, among all differentially expressed genes, only one down-regulated gene is common to both host combinations (Os07g0638600 encoding a peroxidase) and 55 genes (53 up-regulated, 2 down-regulated) are common to non-host combinations (Figure 1). Among the latter, two genes implicated in successive steps of ethylene synthesis (Os06g0524900, Os09g0451400), a NB-ARC domain containing gene (Os06g0524900) and a gene encoding an expansin (Os05g0276500) were identified. Several genes implicated in oxido-reductive processes (Os01g0327000, Os07g0164900, Os12g0260500) as well as a gene encoding a glycoside hydrolase (Os09g0395600) and a gene encoding an UDP-glucuronosyl transferase (Os01g0597800) are also induced. When comparing to the list of genes differentially expressed after Magnaporthe infection, 6 of the 53 up-regulated genes common to both Azospirillum-rice non-host combinations (Cig_B510 and Nip_4B) are also induced after Magnaporthe inoculation and none of these genes are repressed (Additional File 6: Table S6). However, these genes do not seem to be directly involved in biotic stress response of rice roots.
Combination-specific expression profiles
Many differences are observed between the four cultivar/strain combinations when comparing expression profiles, particularly for genes involved in biotic stress responses (Figure 3). This includes genes potentially involved in hormone signaling and both ethylene and auxin signaling occur to be finely regulated during Azospirillum-rice cooperation.
Indeed, if only one of the genes related to ethylene signaling was classified as Cig_4B specific, 15 of these genes were classified as Cig_B510-specific, 11 were classified as Nip_4B-specific and 13 were classified as Nip_B510-specific (Table 3). All the Cig_4B- and Nip_4B-specific genes related to ethylene signaling are up-regulated, while two of the Cig_B510-specific genes and all the Nip_B510-specific genes related to ethylene signaling are down-regulated. When considering genes related to auxin signaling, one was classified as Cig_4B specific, seven were classified as Cig_B510-specific, 12 were classified as Nip_4B-specific and eight were classified as Nip_B510-specific. In addition, nine genes involved in abscisic acid signaling and seven genes involved in salicylic acid signaling were shown to be differentially expressed in only one of the four combinations, suggesting that a wide modification of hormone signals occurs in rice roots after Azospirillum inoculation. Auxin, ethylene, abscissic acid, and salicylic acid are known to be involved in plant immunity, and many genes related to biotic stress response display a combination-specific profile in the current study. For example, 76 genes similar to PR-genes were identified to be combination-specific. As previously mentioned, fewer genes are differentially expressed in host combinations than in non-host combinations. Indeed, three and 13 of these genes were identified for Cig_4B and Nip_B510, respectively, while 25 and 35 of these genes were identified for Nip_4B and Cig_B510, respectively. Taken all together, these results suggest that Azospirillum inoculation leads to important changes in rice root hormone signaling and plant defense, depending on the strain/cultivar combination.
Table 3.
List of combination-specific genes related to ethylene signaling.
| Gene ID | Log2(FC) | Genome annotation | |||
|---|---|---|---|---|---|
| Cig_4B | Cig_B510 | Nip_4B | Nip_B510 | ||
| Os02g0594300 | 1.29 | Similar to enhancer of shoot regeneration ESR1 | |||
| Os01g0797600 | −1.77 | AP2 domain-containing ethylene responsive protein | |||
| Os01g0536400 | −1.66 | Similar to 1-aminocyclopropane-1-carboxylate oxidase | |||
| Os04g0257500 | 1.10 | Similar to ethylene-responsive transcription factor TSFR1 | |||
| Os09g0451000 | 1.17 | Similar to 1-aminocyclopropane-1-carboxylate oxidase | |||
| Os10g0523900 | 1.19 | AP2 domain containing ethylene responsive protein | |||
| Os05g0437100 | 1.26 | Ethylene-responsive transcription factor | |||
| Os01g0757200 | 1.30 | Similar to gibberellin 2-beta-dioxygenase | |||
| Os03g0860600 | 1.35 | Similar to 1-aminocyclopropane-1-carboxylate oxidase | |||
| Os02g0767300 | 1.36 | Similar to flavonol synthase | |||
| Os09g0570800 | 1.41 | Similar to 1-aminocyclopropane-1-carboxylate oxidase | |||
| Os01g0832600 | 1.66 | Similar to leucoanthocyanidin dioxygenase | |||
| Os09g0248900 | 1.70 | Similar to ethylene-responsive protein | |||
| Os02g0797100 | 1.89 | AP2 domain containing ethylene responsive protein | |||
| Os05g0127500 | 1.95 | Similar to leucoanthocyanidin dioxygenase | |||
| Os04g0182200 | 2.60 | 2OG-Fe(II) oxygenase domain containing protein | |||
| Os01g0230200 | 1.06 | Ethylene-responsive protein | |||
| Os07g0169600 | 1.12 | 2OG-Fe(II) oxygenase domain containing protein | |||
| Os03g0100900 | 1.15 | Ethylene-responsive element-binding protein | |||
| Os10g0536400 | 1.18 | 2OG-Fe(II) oxygenase domain containing protein | |||
| Os04g0407800 | 1.24 | 2OG-Fe(II) oxygenase domain containing protein | |||
| Os06g0162500 | 1.29 | Similar to Naringenin 3-dioxygenase like protein | |||
| Os02g0202000 | 1.32 | Similar to ethylene responsive protein | |||
| Os04g0548000 | 1.32 | Ethylene-responsive element-binding protein | |||
| Os02g0654700 | 1.50 | Ethylene-responsive transcription factor | |||
| Os03g0122300 | 1.53 | Similar to Flavanone 3-hydroxylase-like protein | |||
| Os08g0366100 | 1.75 | Endothelial differentiation-related factor | |||
| Os06g0177600 | −2.07 | 2OG-Fe(II) oxygenase domain containing protein | |||
| Os11g0186900 | −1.82 | Similar to 1-aminocyclopropane-1-carboxylate oxidase | |||
| Os04g0522500 | −1.58 | 2OG-Fe(II) oxygenase domain containing protein | |||
| Os05g0155200 | −1.41 | Similar to ethylene receptor | |||
| Os01g0935400 | −1.41 | 2OG-Fe(II) oxygenase domain containing protein | |||
| Os02g0520000 | −1.41 | Ethylene responsive protein | |||
| Os04g0643500 | −1.30 | 2OG-Fe(II) oxygenase domain containing protein | |||
| Os02g0276900 | −1.28 | Ethylene-responsive protein | |||
| Os04g0565900 | −1.22 | Ethylene-responsive protein | |||
| Os04g0493100 | −1.21 | Ethylene-responsive protein | |||
| Os04g0578000 | −1.18 | Similar to 1-aminocyclopropane-1-carboxylate synthase | |||
| Os03g0690500 | −1.16 | Similar to 1-aminocyclopropane-1-carboxylate oxidase | |||
| Os06g0573900 | −1.10 | Similar to 1-aminocyclopropane-1-carboxylate oxidase | |||
Besides genes related to hormone signaling, the combination-specific response also includes nine genes related to hydroxycinnamic acids metabolism (Additional File 3: Table S3). This includes seven genes encoding cinnamyl alcohol dehydrogenase (Os01g0528800, Os03g0223200, Os09g0400200, Os11g0622800 which are down-regulated; Os04g0612700, Os09g0399800, Os09g0400400 which are up-regulated), one gene encoding a cinnamoyl-CoA reductase (Os01g0828100, downregulated) and one gene encoding a coumarate-CoA synthase (Os08g0245200, downregulated). In addition, four genes related to chalcones metabolism and five genes related to anthocyanin metabolism display a combination-specific profile. Such a result is of particular interest as some secondary metabolites stemming from these pathways were previously shown to be discriminant in the analysis of root methanolic extract composition, following Azospirillum and Frankia inoculations (Popovici et al., 2011; Chamam et al., 2013).
Discussion
Unraveling the molecular basis of host-specific adaptations in PGPR-plant cooperation helps understanding frontiers between the perception of symbiotic, cooperative, and pathogenic microbes hosted by plant roots. Based on previous studies detecting metabolic and morphological changes of PGPR-inoculated plants at early stages (Cassán et al., 2009; Walker et al., 2011, 2012; Chamam et al., 2013), we analyzed the transcriptomic response of rice roots 7 days after inoculation. To our knowledge, this study constitutes the first investigation of wide transcriptomic response of rice roots to PGPR-inoculation, aiming at deciphering interaction specificity in plant-microbe cooperation.
Comparison of expression profiles obtained for Cigalon/A. lipoferum 4B (Cig_4B), Cigalon/Azospirillum sp. B510 (Cig_B510), Nipponbare/A. lipoferum 4B (Nip_4B) and Nipponbare/Azospirillum sp. B510 (Nip_B510) combinations evidences a fine-tuned transcriptomic response depending on both Azospirillum and rice genotypes. As revealed by the high percentage (83%) of genes up-regulated or down-regulated only in one of the four conditions, individual genotypic variations are the most important driving force of rice roots gene expression, in the tested conditions. Indeed, only 34 markers of the Azospirillum-rice cooperation, induced in the four combinations, were identified and further investigations should be undertaken to identify the impact of a larger range of Azospirillum strains on the regulation of these markers. Besides combination-specific traits, expression profiles showed strain-specific and cultivar-specific characteristics, highlighting potential differences in the strategies of interaction. Indeed, strain 4B causes few repressions while at least half of the genes regulated in response to strain B510 are repressed, regardless of the cultivar. In addition, only 59 genes display similar regulation in both combinations involving strain 4B whereas it represents 626 genes for those involving strain B510. Accordingly, strain-specific responses were observed when considering the respective impact of 4B and B510 on development and secondary metabolism of rice cultivars Cigalon and Nipponbare (Chamam et al., 2013). While 4B promotes shoot and root growth of both cultivars, B510 promotes development of cultivar Nipponbare exclusively. However, B510 was shown to be the only strain inducing a systemic response, as revealed by variation of secondary metabolite profiles of both shoots and roots. These strain-specific responses could be due to differences in strain lifestyle as 4B colonizes only the surface of rice roots while B510 has the ability to colonize the cortex layers in rice (Thomas-Bauzon et al., 1982; Elbeltagy et al., 2001; Chamam et al., 2013). In addition to their impact on plant growth, endophytic PGPR induce stress and defense responses and the inoculation of B510 was shown to enhance resistance against rice blast disease and rice blight disease (Miché et al., 2006; Rosenblueth and Martínez-Romero, 2006; Yasuda et al., 2009). However, whether these changes were the result of major gene induction or repression was not addressed. Recently, a study on differential gene expression of rice roots inoculated with the endophyte Herbaspirillum seropedicae evidenced a decrease in expression of defense related protein PBZ1 and thionins, suggesting that bacteria modulate plant defense to allow the establishment of an efficient cooperation (Brusamarello-Santos et al., 2011). Indeed, colonization of root tissues by bacteria depends on the balance between the plant's ability to induce efficient defenses in response to the intrusive microbe and the microorganisms' ability to bypass plant immunity (Pieterse et al., 2009).
Several genes associated to plant defense mechanisms are still regulated 7 days post-inoculation. Particularly, a gene orthologous to the PR10 gene of Brachypodium is induced in all combinations. Interestingly, this gene was previously shown to be up-regulated in rice roots 6 days after Magnaporthe infection (Marcel et al., 2010). PR genes are known to be induced during PAMPs-triggered immunity (PTI), the first step of plant defense that involves Pattern Recognition Receptors (PRRs) (Chisholm et al., 2006; Pieterse et al., 2009). PRRs recognize universal microbial determinants such as flagellin, chitin, glycoproteins, and lipoproteins designed as PAMPs/MAMPs for Pathogens/Microbes–Associated Molecular Patterns (Schwessinger and Zipfel, 2008). Even if symbiotic and pathogenic interactions exhibit a number of unique characteristics, it was suggested that similar response mechanisms were adapted to cope with different biotic and abiotic stresses (Baron and Zambryski, 1995). Surprisingly, down-regulated mechanisms seem to be more conserved than up-regulated mechanisms between pathogenic and symbiotic interactions, genes involved in plant defense and stress response being a major part of these repressions (Damiani et al., 2012). In this context, endophytic colonization being more intrusive than surface colonization, plant response to endophyte PGPR could be more similar to pathogenic response than plant response to surface colonizers. This hypothesis is supported by the fact that 30 genes common to Cig_B510 and Nip_B510 are also down-regulated during Magnaporthe infection while none of the genes common to Cig_4B and Nip_4B are down-regulated (data not shown).
Most of the genes containing a NB-ARC domain are differentially expressed on a strain/cultivar dependent manner, the most striking effect being observed for the non-host combinations. Most of NB-ARC genes regulated during interaction with strain B510 are down-regulated and repression occurs for a higher number of NB-ARC genes in the Nip_B510 host interaction. NB-ARC domain is generally associated to R genes that are involved in Effector-Triggered Immunity (ETI) (Chisholm et al., 2006; Pieterse et al., 2009). Differences observed between host and non-host combinations suggests that the way a strain is perceived by rice roots could have been subjected to long-lasting co-adaptation events between a strain and its original host cultivar, a hypothesis that has already been proposed based on secondary metabolites profiling of rice inoculated with strain 4B and B510, and on transcriptomic response of strain 4B colonizing rice (Chamam et al., 2013; Drogue et al., 2014). The involvement of plant defense systems in PGPR-plant cooperations was mainly considered in the context of biocontrol agents, their perception leading to the induction of long-lasting and broad-spectrum systemic resistance (Van Loon et al., 1998; Van Wees et al., 2008; Pieterse et al., 2009). Induced systemic resistance (ISR) is associated with priming effect for enhanced defense inducing a few reprogramming of plant transcriptome (Verhagen et al., 2004; Wang et al., 2005; Van Wees et al., 2008). In the case of phytostimulating PGPR, it was reported that members of genus Azospirillum and Burkholderia induce defense response at a lower extent than pathogens (Bashan, 1998; Bordiec et al., 2011). Moreover, induction of plant defense mechanisms was shown to control the establishment of compatible and incompatible interactions between plants and endophytic PGPR (Miché et al., 2006; Rosenblueth and Martínez-Romero, 2006; Reinhold-Hurek and Hurek, 2011). Thus, discrepancies between endophyte and surface colonizer effects on plant defense system should be further investigated by studying the impact of other Azospirillum strains that display endophytic properties, such as A. brasilense Sp245. A recent study analyzed the impact of A. brasilense Sp245 inoculation on A. thaliana gene expression and evidenced that root transcriptome undergoes significant changes on genes related to hormone signaling and plant defense (Spaepen et al., 2014) Taking into account that plant immunity involves dynamic mechanisms that lead to temporal changes in the expression of defense related genes remains an important issue to measure the sustainability of PGPR-plant cooperations.
Modulating plant hormone balance is an important trait of phytostimulating PGPR (Richardson et al., 2009; Bashan and de-Bashan, 2010; Vacheron et al., 2013). Especially, several members of the genus Azospirillum are able to produce auxin, cytokinin, and gibberellin (Richardson et al., 2009; Bashan and de-Bashan, 2010). In the case of strains 4B and B510, genes related to indole-3-acetic acid (IAA) biosynthesis pathway, ipdC/ppdC, are absent from their genomes and 1-aminocyclopropane-1 carboxylate (ACC) deamination could be a relevant mechanism for hormone modulation and plant-growth promotion (Blaha et al., 2006; Prigent-Combaret et al., 2008; Kaneko et al., 2010; Wisniewski-Dyé et al., 2011). This property is encoded by acdS found in both pathogenic and non-pathogenic bacteria (Blaha et al., 2006). ACC is a precursor of ethylene, a gaseous hormone that represses root-growth and induces systemic resistance against pathogens (Bleecker and Kende, 2000; Pieterse et al., 2009; Galland et al., 2012). It was proposed that bacterial deamination of ACC could lead to a decrease of ethylene levels in plant roots and consequently an increase in root development (Glick, 2005; Galland et al., 2012). Interestingly, the impacts of strain 4B and strain B510 on rice root morphological traits differ. Indeed, strain 4B improves total root length, principal root length and the number of roots per plant while strain B510 seems to improve mostly principal root length (Chamam et al., 2013). Then, differential impact of strain/cultivar combinations on rice root architecture could be linked to the combination-specific regulation of many genes related to ethylene biosynthesis.
Many plant hormones are involved in both plant growth and plant immunity, two physiological traits regulated by a network of interconnected signaling pathways (Pieterse et al., 2009). As such, auxin contributes to both plant development and disease resistance in a pathway interconnected with salicylic acid signaling (Wang et al., 2007). Cross-communication between plant immunity and plant development may contribute to quick adaptation in a cost-efficient manner according to numerous trade-offs reported between growth rate and disease resistance (Walters and Heil, 2007; Pieterse et al., 2009). Thus, the strain-specific effect of Azospirillum on the regulation of hormone-related genes must be taken into account to appraise the cost-benefit balance of each strain/cultivar cooperation. While these two strains display similar characteristics for hormonal production (Wisniewski-Dyé et al., 2012), regulation of rice genes related to hormone signaling, notably auxin signaling, is strikingly different when considering each strain (see above). These results highlight the complexity of hormone signaling networks involved in Azospirillum-rice cooperation.
This study aimed at identifying genetic determinants regulated in rice roots upon Azospirillum inoculation, considering possible favored interaction between a strain and its original host cultivar. Thus, two rice cultivars were inoculated with two Azospirillum strains, resulting in four strain/cultivar combinations. The wide set of genes differentially expressed only in one of the four combinations suggest that individual genotypic variations could be the most important driving force of rice root gene expression upon Azospirillum inoculation. Strain-dependent transcriptional changes observed for genes related to auxin and ethylene signaling highlight the complexity of hormone signaling networks in the Azospirillum-rice cooperation. In this context, unraveling cross-connected hormone networks involved in both growth promotion and plant defense response appears to be an important issue to understand mechanisms involved in beneficial interactions between PGPR and plants.
Author contributions
Florence Wisniewski-Dyé and Claire Prigent-Combaret conceived and designed the experiments with the help of Nathalie Picault. Amel Chamam and Hervé Sanguin performed the experiments and Benoît Drogue contributed to the preparation of rice RNA samples. Benoît Drogue and Hervé Sanguin performed the analysis of microarray data, with the help of Michael Mozar for the analysis of raw data. Christel Llauro, Nathalie Picault, and Olivier Panaud performed the qRT-PCR and the part on LTR-retrotransposons. Benoît Drogue, Hervé Sanguin, Claire Prigent-Combaret, and Florence Wisniewski-Dyé wrote the manuscript; all authors contributed to the discussion and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to C. Louvel (Center français du riz, Arles, France) and J. B. Morel (BGPI, Montpellier, France) for gift of Cigalon and Nipponbare seeds, respectively. The platforms “Serre” and DTAMB of FR41 (Université Lyon 1) were used to carry out this work. Benoît Drogue and Amel Chamam receive fellowships from, respectively ≪Région Rhône-Alpes≫ and ≪Centre National de la Recherche Scientifique≫. This work was funded by the ANR project AZORIZ (ANR-08-BLAN-0098).
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00607/abstract
References
- Baron C., Zambryski P. C. (1995). The plant response in pathogenesis, symbiosis, and wounding: variations on a common theme? Annu. Rev. Genet. 29, 107–129. 10.1146/annurev.ge.29.120195.000543 [DOI] [PubMed] [Google Scholar]
- Bashan Y. (1998). Inoculants of plant growth-promoting bacteria for use in agriculture. Biotechnol. Adv. 16, 729–770 10.1016/S0734-9750(98)00003-2 [DOI] [Google Scholar]
- Bashan Y., de-Bashan L. E. (2010). How the plant growth-promoting bacterium Azospirillum promotes plant growth—a critical assessment. Adv. Agron. 108, 77–136 10.1016/S0065-2113(10)08002-8 [DOI] [Google Scholar]
- Bashan Y., Holguin G., de-Bashan L. E. (2004). Azospirillum-plant relationships: physiological, molecular, agricultural, and environmental advances (1997–2003). Can. J. Microbiol. 50, 521–577. 10.1139/w04-035 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300. [Google Scholar]
- Blaha D., Prigent-Combaret C., Mirza M. S., Moënne-Loccoz Y. (2006). Phylogeny of the 1-aminocyclopropane-1-carboxylic acid deaminase-encoding gene acdS in phytobeneficial and pathogenic Proteobacteria and relation with strain biogeography. FEMS Microbiol. Ecol. 56, 455–470. 10.1111/j.1574-6941.2006.00082.x [DOI] [PubMed] [Google Scholar]
- Bleecker A. B., Kende H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. 10.1146/annurev.cellbio.16.1.1 [DOI] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193. 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
- Bordiec S., Paquis S., Lacroix H., Dhondt S., Ait Barka E., Kauffmann S., et al. (2011). Comparative analysis of defence responses induced by the endophytic plant growth-promoting rhizobacterium Burkholderia phytofirmans strain PsJN and the non-host bacterium Pseudomonas syringae pv. pisi in grapevine cell suspensions. J. Exp. Bot. 62, 595–603. 10.1093/jxb/erq291 [DOI] [PubMed] [Google Scholar]
- Brusamarello-Santos L. C. C., Pacheco F., Aljanabi S. M. M., Monteiro R. A., Cruz L. M., Baura V. A., et al. (2011). Differential gene expression of rice roots inoculated with the diazotroph Herbaspirillum seropedicae. Plant Soil 356, 113–125 10.1007/s11104-011-1044-z [DOI] [Google Scholar]
- Buscaill P., Rivas S. (2014). Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 20, 35–46. 10.1016/j.pbi.2014.04.004 [DOI] [PubMed] [Google Scholar]
- Cao P., Jung K.-H., Choi D., Hwang D., Zhu J., Ronald P. C. (2012). The rice oligonucleotide array database: an atlas of rice gene expression. Rice 5:17. 10.1186/1939-8433-5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassán F., Perrig D., Sgroy V., Masciarelli O., Penna C., Luna V. (2009). Azospirillum brasilense Az39 and Bradyrhizobium japonicum E109, inoculated singly or in combination, promote seed germination and early seedling growth in corn (Zea mays L.) and soybean (Glycine max L.). Eur. J. Soil Biol. 45, 28–35 10.1016/j.ejsobi.2008.08.005 [DOI] [Google Scholar]
- Chamam A., Sanguin H., Bellvert F., Meiffren G., Comte G., Wisniewski-Dyé F., et al. (2013). Plant secondary metabolite profiling evidences strain-dependent effect in the Azospirillum–Oryza sativa association. Phytochemistry 87, 65–77. 10.1016/j.phytochem.2012.11.009 [DOI] [PubMed] [Google Scholar]
- Chaparro C., Guyot R., Zuccolo A., Piegu B., Panaud O. (2007). RetrOryza: a database of the rice LTR-retrotransposons. Nucleic Acids Res. 35, D66–D70. 10.1093/nar/gkl780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S. T., Coaker G., Day B., Staskawicz B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. 10.1016/j.cell.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Damiani I., Baldacci-Cresp F., Hopkins J., Andrio E., Balzergue S., Lecomte P., et al. (2012). Plant genes involved in harbouring symbiotic rhizobia or pathogenic nematodes. New Phytol. 194, 511–522. 10.1111/j.1469-8137.2011.04046.x [DOI] [PubMed] [Google Scholar]
- Desbrosses G., Contesto C., Varoquaux F., Galland M., Touraine B. (2009). PGPR-Arabidopsis interactions is a useful system to study signaling pathways involved in plant developmental control. Plant Signal. Behav. 4, 321–323. 10.4161/psb.4.4.8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogue B., Sanguin H., Borland S., Prigent-Combaret C., Wisniewski-Dyé F. (2014). Genome wide profiling of Azospirillum lipoferum 4B gene expression during interaction with rice roots. FEMS Microbiol. Ecol. 87, 543–555. 10.1111/1574-6941.12244 [DOI] [PubMed] [Google Scholar]
- El Baidouri M., Panaud O. (2013). Comparative genomic paleontology across plant kingdom reveals the dynamics of TE-driven genome evolution. Genome Biol. Evol. 5, 954–965. 10.1093/gbe/evt025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbeltagy A., Nishioka K., Sato T., Suzuki H., Ye B., Hamada T., et al. (2001). Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 67, 5285–5293. 10.1128/AEM.67.11.5285-5293.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Zemrany H., Czarnes S., Hallett P. D., Alamercery S., Bally R., Jocteur Monrozier L. (2007). Early changes in root characteristics of maize (Zea mays) following seed inoculation with the PGPR Azospirillum lipoferum CRT1. Plant Soil 291, 109–118 10.1007/s11104-006-9178-0 [DOI] [Google Scholar]
- Fuentes-Ramirez L. E., Caballero-Mellado J. (2012). Bacterial biofertilizers, in PGPR: Biocontrol and Biofertilization, ed Siddiqui Z. A. (Berlin; Heidelberg: Springer-Verlag; ), 143–172. [Google Scholar]
- Galland M., Gamet L., Varoquaux F., Touraine B., Desbrosses G. (2012). The ethylene pathway contributes to root hair elongation induced by the beneficial bacteria Phyllobacterium brassicacearum STM196. Plant Sci. 190, 74–81. 10.1016/j.plantsci.2012.03.008 [DOI] [PubMed] [Google Scholar]
- Glick B. R. (2005). Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol. Lett. 251, 1–7. 10.1016/j.femsle.2005.07.030 [DOI] [PubMed] [Google Scholar]
- Heulin T., Guckert A., Balandreau J. (1987). Stimulation of root exudation of rice seedlings by Azospirillum strains: carbon budget under gnotobiotic conditions. Biol. Fertil. Soils 4, 9–14 10.1007/BF00280344 [DOI] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., et al. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- Kaneko T., Minamisawa K., Isawa T., Nakatsukasa H., Mitsui H., Kawaharada Y., et al. (2010). Complete genomic structure of the cultivated rice endophyte Azospirillum sp. B510. DNA Res. 17, 37–50. 10.1093/dnares/dsp026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Prakash A., Johri B. N. (2011). Bacillus as PGPR in crop ecosystem, in Bacteria in Agrobiology: Crop Ecosystems, ed Maheshwari D. K. (Berlin; Heidelberg: Springer; ), 37–59. [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lugtenberg B., Kamilova F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. 10.1146/annurev.micro.62.081307.162918 [DOI] [PubMed] [Google Scholar]
- Marcel S., Sawers R., Oakeley E., Angliker H., Paszkowski U. (2010). Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell 22, 3177–3187. 10.1105/tpc.110.078048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miché L., Battistoni F., Gemmer S., Belghazi M., Reinhold-Hurek B. (2006). Upregulation of jasmonate-inducible defense proteins and differential colonization of roots of Oryza sativa cultivars with the endophyte Azoarcus sp. Mol. Plant. Microbe Interact. 19, 502–511. 10.1094/MPMI-19-0502 [DOI] [PubMed] [Google Scholar]
- Michaud E. J., Vugt M. J., van Bultman S. J., Sweet H. O., Davisson M. T., Woychik R. P. (1994). Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 8, 1463–1472. 10.1101/gad.8.12.1463 [DOI] [PubMed] [Google Scholar]
- van de Mortel J. E., de Vos R. C. H., Dekkers E., Pineda A., Guillod L., Bouwmeester K., et al. (2012). Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 160, 2173–2188. 10.1104/pp.112.207324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien H. E., Thakur S., Guttman D. S. (2011). Evolution of plant pathogenesis in Pseudomonas syringae: a genomics perspective. Annu. Rev. Phytopathol. 49, 269–289. 10.1146/annurev-phyto-072910-095242 [DOI] [PubMed] [Google Scholar]
- Picault N., Chaparro C., Piegu B., Stenger W., Formey D., Llauro C., et al. (2009). Identification of an active LTR retrotransposon in rice. Plant J. 58, 754–765. 10.1111/j.1365-313X.2009.03813.x [DOI] [PubMed] [Google Scholar]
- Pieterse C. M. J., Leon-Reyes A., Van der Ent S., Van Wees S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- Popovici J., Walker V., Bertrand C., Bellvert F., Fernandez M. P., Comte G. (2011). Strain specificity in the Myricaceae–Frankia symbiosis is correlated to plant root phenolics. Funct. Plant Biol. 38, 682–689 10.1071/FP11144 [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C., Blaha D., Pothier J. F., Vial L., Poirier M.-A., Wisniewski-Dyé F., et al. (2008). Physical organization and phylogenetic analysis of acdR as leucine-responsive regulator of the 1-aminocyclopropane-1-carboxylate deaminase gene acdS in phytobeneficial Azospirillum lipoferum 4B and other Proteobacteria. FEMS Microbiol. Ecol. 65, 202–219. 10.1111/j.1574-6941.2008.00474.x [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B., Hurek T. (2011). Living inside plants: bacterial endophytes. Curr. Opin. Plant Biol. 14, 435–443. 10.1016/j.pbi.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Rice Annotation Project, Tanaka T., Antonio B. A., Kikuchi S., Matsumoto T., Nagamura Y., et al. (2008). The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acid Res. 36, D1028–D1033. 10.1093/nar/gkm978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A. E., Barea J.-M., McNeill A. M., Prigent-Combaret C. (2009). Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321, 305–339 10.1007/s11104-009-9895-2 [DOI] [Google Scholar]
- Rosenblueth M., Martínez-Romero E. (2006). Bacterial endophytes and their interactions with hosts. Mol. Plant. Microbe Interact. 19, 827–837. 10.1094/MPMI-19-0827 [DOI] [PubMed] [Google Scholar]
- Saharan B., Nehra V. (2011). Plant growth promoting rhizobacteria: a critical review. Life Sci. Med. Res. 2011, 1–30. [Google Scholar]
- Schumacher J., Tudzynski P. (2012). Morphogenesis and infection in Botrytis cinerea, in Morphogenesis and Pathogenicity in Fungi, eds Pérez-Martín J., Di Pietro A. (Berlin; Heidelberg: Springer; ), 225–241. [Google Scholar]
- Schwessinger B., Zipfel C. (2008). News from the frontline: recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 11, 389–395. 10.1016/j.pbi.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Smith S. E., Read D. J. (2008). Mycorrhizal Symbiosis. London: Academic Press. [Google Scholar]
- Somers E., Vanderleyden J., Srinivasan M. (2004). Rhizosphere bacterial signalling: a love parade beneath our feet. Crit. Rev. Microbiol. 30, 205–240. 10.1080/10408410490468786 [DOI] [PubMed] [Google Scholar]
- Soto M. J., Domínguez-Ferreras A., Pérez-Mendoza D., Sanjuán J., Olivares J. (2009). Mutualism versus pathogenesis: the give-and-take in plant-bacteria interactions. Cell. Microbiol. 11, 381–388. 10.1111/j.1462-5822.2009.01282.x [DOI] [PubMed] [Google Scholar]
- Spaepen S., Bossuyt S., Engelen K., Marchal K., Vanderleyden J. (2014). Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 201, 850–861. 10.1111/nph.12590 [DOI] [PubMed] [Google Scholar]
- Steenhoudt O., Vanderleyden J. (2000). Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol. Rev. 24, 487–506. 10.1111/j.1574-6976.2000.tb00552.x [DOI] [PubMed] [Google Scholar]
- Thomas-Bauzon D., Weinhard P., Villecourt P., Balandreau J. (1982). The spermosphere model. I. Its use in growing, counting, and isolating N2-fixing bacteria from the rhizosphere of rice. Can. J. Microbiol. 28, 922–928 10.1139/m82-139 [DOI] [Google Scholar]
- Vacheron J., Desbrosses G., Bouffaud M.-L., Touraine B., Moënne-Loccoz Y., Muller D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4:356. 10.3389/fpls.2013.00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon L. C., Bakker P. A., Pieterse C. M. (1998). Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483. 10.1146/annurev.phyto.36.1.453 [DOI] [PubMed] [Google Scholar]
- Van Wees S. C., Van der Ent S., Pieterse C. M. (2008). Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11, 443–448. 10.1016/j.pbi.2008.05.005 [DOI] [PubMed] [Google Scholar]
- Verhagen B. W. M., Glazebrook J., Zhu T., Chang H.-S., van Loon L. C., Pieterse C. M. J. (2004). The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol. Plant. Microbe Interact. 17, 895–908. 10.1094/MPMI.2004.17.8.895 [DOI] [PubMed] [Google Scholar]
- Walker V., Bertrand C., Bellvert F., Moënne-Loccoz Y., Bally R., Comte G. (2011). Host plant secondary metabolite profiling shows a complex, strain-dependent response of maize to plant growth-promoting rhizobacteria of the genus Azospirillum. New Phytol. 189, 494–506. 10.1111/j.1469-8137.2010.03484.x [DOI] [PubMed] [Google Scholar]
- Walker V., Couillerot O., Felten A. V., Bellvert F., Jansa J., Maurhofer M., et al. (2012). Variation of secondary metabolite levels in maize seedling roots induced by inoculation with Azospirillum, Pseudomonas and Glomus consortium under field conditions. Plant Soil 356, 151–163 10.1007/s11104-011-0960-2 [DOI] [Google Scholar]
- Walters D., Heil M. (2007). Costs and trade-offs associated with induced resistance. Physiol. Mol. Plant Pathol. 71, 3–17 10.1016/j.pmpp.2007.09.008 [DOI] [Google Scholar]
- Wang D., Pajerowska-Mukhtar K., Culler A. H., Dong X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17, 1784–1790. 10.1016/j.cub.2007.09.025 [DOI] [PubMed] [Google Scholar]
- Wang Y., Ohara Y., Nakayashiki H., Tosa Y., Mayama S. (2005). Microarray analysis of the gene expression profile induced by the endophytic plant growth-promoting rhizobacteria, Pseudomonas fluorescens FPT9601-T5 in Arabidopsis. Mol. Plant Microbe Interact. 18, 385–396. 10.1094/MPMI-18-0385 [DOI] [PubMed] [Google Scholar]
- Wisniewski-Dyé F., Borziak K., Khalsa-Moyers G., Alexandre G., Sukharnikov L. O., Wuichet K., et al. (2011). Azospirillum genomes reveal transition of bacteria from aquatic to terrestrial environments. PLoS Genet. 7:e1002430. 10.1371/journal.pgen.1002430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski-Dyé F., Drogue B., Borland S., Prigent-Combaret C. (2013). Azospirillum-plant interaction: from root colonization to plant growth promotion, in Beneficial Plant-Microbial Interactions: Ecology and Applications, eds Belén Rodelas González M., Gonzalez-López J. (Boca Raton, FL: CRC Press; ), 237–269. [Google Scholar]
- Wisniewski-Dyé F., Lozano L., Acosta-Cruz E., Borland S., Drogue B., Prigent-Combaret C., et al. (2012). Genome sequence of Azospirillum brasilense CBG497 and comparative analyses of Azospirillum core and accessory genomes provide insight into niche adaptation. Genes 3, 576–602. 10.3390/genes3040576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M., Isawa T., Shinozaki S., Minamisawa K., Nakashita H. (2009). Effects of colonization of a bacterial endophyte, Azospirillum sp. B510, on disease resistance in rice. Biosci. Biotechnol. Biochem. 73, 2595–2599. 10.1271/bbb.90402 [DOI] [PubMed] [Google Scholar]
- Yuan Q., Ouyang S., Wang A., Zhu W., Maiti R., Lin H., et al. (2005). The institute for genomic research Osa1 rice genome annotation database. Plant Physiol. 138, 18–26. 10.1104/pp.104.059063 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.