Abstract
Databases are, at their core, abstractions of data and their intentionally derived relationships. They serve as a central organizing metaphor and repository, supporting or augmenting nearly all bioinformatics. Behavioral domains provide a unique stage for contemporary databases, as research in this area spans diverse data types, locations, and data relationships. This chapter provides foundational information on the diversity and prevalence of databases, how data structures support the various needs of behavioral neuroscience analysis and interpretation. The focus is on the classes of databases, data curation, and advanced applications in bioinformatics using examples largely drawn from research efforts in behavioral neuroscience.
1. INTRODUCTION
It is difficult to imagine modern neuroscience research without the supporting infrastructure provided by bioinformatics databases. Consistent with the broader view of informatics, a bioinformatics renders a formalized representation of information, placing empirical observations within the context of the larger subdiscipline and augmenting the impact of local observations and experimentation. The ultimate goal is to allow other researchers from a variety of tangential disciplines to share a common lexicon and classification framework to bridge the data-mining gap, automating the process of knowledge discovery. With mature bioinformatics, for example, the broad implications of behavioral neuroscience can be measured against the convergent functional genomics of several model organisms, opening up avenues of validation previously hidden behind isolated or contextually limited data. Additionally, in contrast to reductionists views of physical models, there is no true interpretation of biological data (Birney & Clamp, 2004) and well-conceived database implementations can move semi-quantitative phenotypes or behavioral observations toward a more tightly structured quantitative result without limiting the scope of analysis to domains where the researcher has deep knowledge.
Behavioral neuroscience databases are required to harness the rapid and accelerating volume of new data and to integrate an incredibly diverse set of traditional and high-throughput technologies. The latter use of databases is of particular interest as behavioral neuroscience spans countless experimental designs and geographic locations, but suffers from the universal lack of an organic data format. For example, the Society For Neuroscience has 42,000 members (www.sfn.org), working with a variety of model organisms and focused on an innumerable array of differing physiological depth and developmental timescales. Gaining a mastery of a common literature within this diverse group is daunting, but managing the integration of 42,000 individual lab notebooks in countless formats is not feasible. Without a common data format or meaningful translational key, the intractable density of information within individual data silos can paralyze analytics, causing researchers to shift focus away from the painful difficulty of knowledge discovery within disassociated data and focus on previously explored areas where data types and structures have been well-documented.
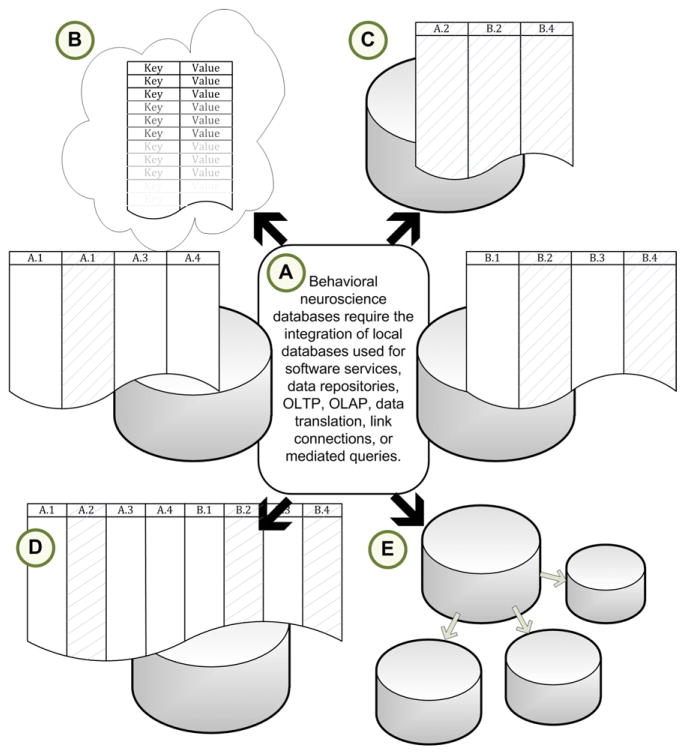
Modern open-source database management systems (DBMS) are used by bioinformatics specialists to mediate potential information bottlenecks. Biological databases serve to shift the burden of data management from the researcher onto a generalizable platform, effectively placing information in a layer that performs local information management duties while making itself transparently accessible to analysis tools and other databases (Fig. 2.1). An interesting consequence of database effectiveness and interaction transparency is that researchers have become desensitized to their deep complexities. There is often a failure to recognize the intimate relationship between types of databases, their intended use, and the landscape and provenance of the underlying data. In behavioral neuroscience research, the depth of these relationships is uniquely important because of the underlying breadth of subdomains, the interaction of vastly arrayed qualitative and quantitative data types, and layers of non-overlapping and often ambiguous semantics ranging from molecular to behavioral observations.
Figure 2.1.
Databases interact with nearly all aspects of biological science. The ubiquitous and transparent nature of relational databases places them near the center of numerous bioinformatics functions in neuroscience. (A) They serve as local and community data repositories, the backend for numerous software services, and data sources for translating information between domains. Convergence of relational databases may be through (B) non-strict NoSQL databases, (C) federated databases, or (C) data warehouses. (D) Each approach can use either local or distributed database architectures.
This survey of the types and scope of databases useful to behavioral neuroscience illustrates the connections between the varying types of underlying data and the purpose of the database. While there certainly is no singular biological database model that defines the entire granularity implicit within the domain, there does exist an emerging understanding of the opportunities and limitations of neuroscience related biological databases.
2. NEUROSCIENCE DATABASES
Researchers interested in understanding, collating, and analyzing the information of neuroscience have numerous hurdles. From a practical perspective, within the biological database community there is a vacillation between infrastructure building and scholarship, creating competing incentives for finding publishable hypotheses within the tangle of existing databases and the creation of new databases (Altman, 2004). As a result, many life science databases in general and behavioral neuroscience databases in particular have grown out of a single research lab to mediate a particular tactical need. For example, neuroscience databases and data management tools include those seeking to manage transcriptional data (Shepherd et al., 1998), complex images such as fMRI scans (Marcus et al., 2007), laboratory information management systems (LIMS) and data management (Baker, Galloway, Jackson, Schmoyer, & Snoddy, 2004), formal collaborations and federated repositories (Gardner et al., 2008), publication data (Ruttenberg, Rees, Samwald, & Marshall, 2009), protein interaction (Colland et al., 2004; Shoemaker et al., 2012) and mass spec data (Horai et al., 2010), behavioral data (Maddatu, Grubb, Bult, & Bogue, 2012), electrophysiological measurements (Günay et al., 2009), and a series of disorder related repositories (Goodman et al., 2003; Matuszek & Talebizadeh, 2009). While not necessarily in conflict with the strategic goals of the greater behavioral neuroscience community, the ad hoc collection of boutique databases, analysis tools and information repositories that exist on the local level are often incompatible with comprehensive data mining. This incompatibility arises from an inability to accurately communicate and translate between individual repositories and the lack of a globally definable workflow that can be used to shape a universal strategy.
Even within behavioral neuroscience, multiple data mining strategies exist to identify the causative molecular profile of a given disease model, leading the community to recognize the need to maximize data mining flexibility across all information sources in order to support the iterative hypothesis generation, testing, and observation cycle implicit in the scientific method of life science. The goal of rapidly identifying putative and testable hypotheses about genes or proteins as they relate to behavioral neuroscience disorders has shaped the way next-generation bioinformatics databases integrate data across domains. Some, such as the NeuroCommons Project, attempt to create open-source knowledge frameworks that can integrate diverse data sets at the level of semantics and natural language processing (Ruttenberg et al., 2009). Others, such as GeneWeaver (Baker, Jay, Bubier, Langston, & Chesler, 2012) and GeneNetwork (Wang, Williams, & Manly, 2003), rely almost wholly on the semi-automated integration of primary and secondary data across broad genomics or genetics data sets. Still others, like the Neuroscience Information Framework (NIF; see Chapter 3), attempt to federate data and information across an entire range of databases and independent data sets (Gardner et al., 2008).
Regardless of which strategic approach to database integration the behavioral neuroscience community converges upon, individual researchers or collaborations at the local level should be focused on keeping data in a self-consistent structured and annotated format. Databases, with their ubiquitous presentation, provide the best option for the broadest range of data structures. While numerous strategies exist to integrate databases at several levels, a minimal understanding of how databases function can help guide the discussion of these infrastructure options. More importantly, the landscape of databases available to novice and expert users continues to grow, providing numerous new options for managed data access and integration of intra- and interdisciplinary data.
3. DATABASES: UNDER THE HOOD
3.1. A generalized solution
A database can be generalized to include any intentional system used to structure data for purposes of storage or retrieval. While all sorts of trivial items fit this definition, including phone books, excel spreadsheets, and this very publication, the idealized notion of a database is often thought of as the ubiquitous electronic repository providing data support for specific domains. The primary discerning difference between the former and latter examples are that the latter has a programmatically managed software layer that interacts with the underlying data and data structure, optimizing both the physical and virtual placement of data to expedite data retrieval, increase fault tolerance, and minimize data redundancy. The overarching management layer, or DBMS, uses, in one way or another, a well-indexed snapshot of its managed data to direct the search, retrieval, import, and annotation of stored information. In practical terms, a successful database would enhance data portability, compatibility (translation), extensibility (ease of annotation and curation), and, importantly, data interoperability and querying.
The concept of a database is deeply and correctly coupled with the concept of data querying. This concept should be familiar to cognitive scientists who consider the processes of memory storage and retrieval. For example, databases are analogous to memory recall (retrieval without cues) and recollection (memory reconstruction) but require sophisticated DBMS systems and structured schemas to optimize the query models. More complex types of memory retrieval, such as recognition or relearning, might be loosely synonymous to concepts of data browsing and data mining, respectively, where complex patterns can be dynamically detected and internalized for future reference. Unlike organisms, however, mechanistic approaches to these advanced data recovery processes require highly efficient data organizing structures and are tightly coupled to procedural algorithms. The analogy of behavioral neuroscience, in general, to information technology is often locally correct but globally insufficient. Fundamentally, for example, living organisms perform better and more efficiently on increasingly complex tasks while information technology becomes increasingly slow and hopelessly deficient as task complexity increases. Young children can manage the intricate semantics of language but have a difficult time multiplying four digit numbers together; computers are optimized to solve the inverse set of problems (Von Foerster, 1967). The limitation of databases, in many ways, is our expectation of precise calculations given the fuzzy inconsistencies of data.
3.2. The database explosion
The explosion of biological database adoption among researchers, many in laboratories without dedicated informatics infrastructures, is driven in large part by need as the types and scope of data produced by modern technologies far outpaces our ability to properly collate the data. To illustrate this point, the Human Genome alone would occupy over 180,000 pages when printed out at a 4.5-point font, and finding meaningful information within it would require equally inefficient volumes of indexed data. Compounding the obviously unmanageable scale of data, there is the need to articulate an endless variety of data types, spanning character-based data, images and proprietary data types. The generic notion of a database is designed explicitly to mediate the centrality of these issues.
The drastic increase in database requirements coincided with the emergence of sophisticated open-source relational DBMS, such as MySQL and PostgreSQL. These systems brought free, robust, and flexible relational databases into the realm of the average biologist, effectively removing the need of costly unsupportable informatics overhead associated with proprietary systems such as Oracle or DB2. Biologists, in turn, began to effectively spread boutique bioinformatics databases with minimal entry requirements. The emergence of need and the ubiquitously standardized relational database has pushed researchers to adopt practices that only a decade ago seemed insurmountable. They have embraced a digitized life; gained an appreciation, albeit a subconscious one, of atomic data types; have rationalized the benefits of extensible data models; and have structured future experimentation planning around compatibility.
3.3. Relational databases
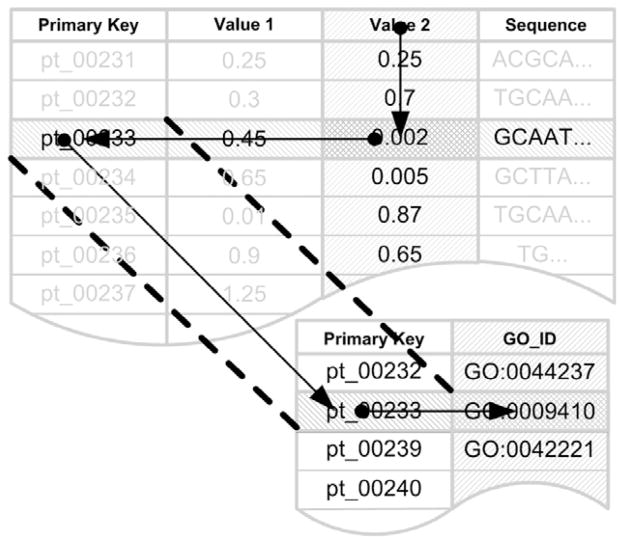
The most common incarnation of a DBMS is based on a relational structure. This can be referred to as a Relational DBMS, or RDBMS, where data are structured according to rows and columns. The most common metaphor for visualizing this type of data structure is the spreadsheet, where rapid look-ups are performed by identifying data at the intersection of rows and columns of interest (Fig. 2.2). In both RDBMS and spreadsheets, there is a requirement that data types must be atomic, meaning that they must have a finite scope of values interpretable by computation systems. Any given spreadsheet cell must be either referenced as a number or character, not as both. In many nonbiological databases adherence to atomic data types is easily achieved. This is not necessarily the case with biological data, which can often be described as fuzzy, making it difficult to find items that have continuous similarity with other items. For example, the spectrum of observable phenotypes, characterized by complex disorders like autism, alcoholism, or drug addiction, do not by themselves reference the full spectrum of underlying functional processes motivating their presentation. As a result, the vast majority of continuous biological data needs to be extracted from bioinformatics databases and manipulated by independent algorithms.
Figure 2.2.
The semantic of a relational database. Relational databases rely on strict schemas and data types layered two-dimensional metaphors, where data can be found at the intersection of rows and columns of interest. Strict schematic rules and the use of primary keys ensure a minimization of data redundancy and provides for a mathematically based approach to data querying (SQL).
Finding synergy between diverse data types is often overcome through the creation of elaborate data schemas that attempt to either gather a wide range of very granular data to produce strict data types, or manage only very high-level metadata connections, effectively eliminating the internal database optimizations that are at the core of modern database robustness. In behavioral neuroscience, this is analogous to the pros and cons of losing information within a subset of molecular functions versus losing information about the relationship between the biological processes occupied by those molecular functions.
One major distinction between flat-file data representations, like row by column spreadsheets and NoSQL (Not-only SQL) databases, and RDBMS is that data in relational database schemas are built around a unique identifier for each record, called a primary key. A primary key ensures that one and only one instance of an entity or relationship exists and allows database schemas to be optimized to reduce redundancy and query time through a process called normalization. Interestingly, this powerful aspect of a relational database can often serve to complicate their application in biological domains. For example, the word hypothalamus can be used as an implicit organizing metaphor for objects relating to stress response, diurnal cycles, metabolism, and thermoregulation, among others, but it does not uniquely reference any given atomic (non-divisible) object. Unfortunately, the application of semantic terms, such as “hypothalamus”, is wholly ineffective in life science because of the plasticity of language and redundancy of function in biology. While ontologies are useful to relate shared relationships based on collaborative annotations and can substitute, at times, as contextual primary keys, they do not wholly replace the normative database definition of a primary key. In fact, from a strict database perspective, there is a noticeable lack of primary keys in biology, as there exists no emergent or organic descriptor that can reference every known and unknown biological object in perpetuity. As a result, many existing behavioral neuroscience databases use as their reference points objects that may change over time or between contexts. The alcohol-related gene CREB, for example, has references to 77 unique accession numbers in NCBI-related databases, making it nearly impossible to pinpoint a canonical definition.
Another consequence of the structure imposed by RDBMS is the creation of a standardized declarative query language. Based on mathematical concepts of relational algebra and tuple relational calculus, SQL (Structured Query Language) provides set logical and procedural ways to interact with data in a context that is independent of the relational database vendor (see Berenson et al., 1995 for a review). While modern RDBMs shoulder much of the burden for query optimization and load balancing, the concepts driving relational databases are formative to understanding the numerous database variants employed to overcome shortcomings in this approach. Ultimately, the choice of an underlying biological database is a trade off between costs, speed, redundancy, and complexity, all driven by the types of data to be stored.
3.4. Analytical databases
Analytical databases are typically read-only databases that are specifically designed to support data mining on an underlying, mostly static, set of information. They are not designed solely to distribute or house data. Community data repositories that fall into this category are the result of efforts to bring both data and tools that operate on that data under the same information structure. Researchers in behavioral neuroscience interested in sharing a stable set of data while providing interactive tools for integrating primary or secondary data to create new knowledge may gravitate toward these types of resources. Examples in behavioral neuroscience include the Comparative Toxicogenomics Database (Davis, Murphy, Rosenstein, Wiegers, & Mattingly, 2008), MuTrack (Baker et al., 2004), GeneWeaver (Baker et al., 2012), or NCBI’s GEO and CDART (Sayers et al., 2012). As information processing becomes more seamlessly integrated with database infrastructures there is a trend to include analytics at the user interface level, but this trend is limited by the complexity of the analytics and the scope of the information to be mined. Dynamic analytics at the user interface level, for example, do not perform well in complex (or genome-scale) tasks that require prolonged periods of time to accomplish. Advances in high-performance computing algorithms are mitigating this challenge (Chesler & Langston, 2006).
3.5. Data warehouse
Data warehouses are effective for behavioral scientists desiring to integrate and distribute data without embedding an analytics framework (Keator, 2009). As the name indicates, data warehouses are explicitly designed to store data under a common framework. Individual operation systems, located locally or disparately, contribute information through a shared integration layer to a central repository. Through this process of integration, data is cleansed, or transformed to meet homogeneous criteria. Unfortunately, the process of data cleansing often leads to lossy data constructs, where the original data may not be recapitulated. On the other hand, centralized data repositories can easily be subdivided into functional domains of interest, referred to as “data marts,” like BioMart (Haider et al., 2009). In neuroscience, data warehouses are manifested in several efforts to collect and unify data under consistent schemas. There are domain-specific data centers, such as BrainMap (www.brainmap.org), which stores functional neuroimaging literature, and PubBrain (www.pubbrain.org), which communicates directly with the PubMed data warehouse, and broader community efforts. The NIF is an example of a community data warehouse that contains a registry of over 4800 individual data or metadata resources (Gardner et al., 2008).
3.6. Federated databases
Federated databases were originally described as a set of autonomous databases that promote unified access through a set of structured meta-data fields (see Heimbigner & McLeod, 1985). This approach has been more loosely applied to include composite databases, which are transparent integrations of autonomous database systems under a globally mandated schema. In both cases, integration is done at the level of common meta-data architecture. Federated databases can be either locally centralized or geographically distributed, and occupy a level autonomy that ranges from loosely coupled to tightly coupled federated schemas. Good examples in behavioral neuroscience include NIF (Gardner et al., 2008) and the Biomedical Informatics Research Network (Ashish, Ambite, Muslea, & Turner, 2010). While the vast majority of behavioral neuroscience laboratories lack the technical skills to navigate the implementation of their own federated databases, they can mediate the exchange of their data with these robust repositories by intentional efforts of data standardization. Minimal Information Standards can be used to provide a common framework to integrate data. Minimum Information for Biological and Biomedical Investigations (Taylor et al., 2008) or Minimal Information About Neural Electromagnetic Ontologies (Frishkoff et al., 2011; see also Chapter 15) are two examples.
3.7. Laboratory information management systems
The most prevalent type of data resource within behavioral neuroscience is the LIMS. These predominantly local systems are developed over time to meet the specific needs of a given laboratory or research group and are often not designed de novo to integrate data with external resources. In many cases, several LIMS coexist to capture varying parts of the information landscape. Wikis, for example, provide an excellent means for capturing the free-form concepts of an electronic laboratory notebook, where students and investigators can collaborate and develop institutional memory about protocols and experimental results (Waldrop, 2008). Larger collaborations may choose highly structured to LIMS to track samples and provide a layer of analytics (Baker et al., 2004). These types of LIMS systems often require dedicated informatics objectives and resources but can be built upon readily available technologies. While no single resource exists to satisfy the LIMS needs of every situation, domain-specific LIMS can address the management of particular technologies. The BioArray Software Environment is designed to manage microarray data (Saal et al., 2002), while the BioGRID is a general purpose repository for interaction datasets (Stark et al., 2006). Commercial solutions exist, as well, but they can limit researchers into a proprietary framework than does not necessarily promote flexibility.
3.8. Knowledge bases
Many consortium projects, programs, model organism communities, and collaborative efforts bring together widely diverse research approaches and resources around a particular area of investigation. These specialized databases are designed to logically represent information repositories to aid in decision-making processes and can include white papers, FAQs, user manuals, tutorials, encyclopedias, dictionaries, and other forms of flat files. Wikiomics (Waldrop, 2008), in neuroscience, for example, provides a good example for this type of free-form data organized around intuitive or pre-identified relationships. Machine-readable databases attempt to make logical connections between data and data types by relying on the semi-structured annotation of the underlying data. Ontologies in neuroscience can leveraged for annotation of unstructured data. The NCBO annotator, for example, can be used to automate the context of free-form data by attaching semantic meaning to ontological frameworks (Jonquet, Shah, & Musen, 2009). Similarly, the NIF have leveraged Texpresso for similar purposes to locate and extract data from the literature (Bandrowski et al., 2012; Müller et al., 2008). Machine and human-driven knowledge bases can therefore be successfully combined to navigate data using both approaches.
4. BEYOND RELATIONAL DATABASES
As the scope and depth of data within behavioral neuroscience databases rapidly expands, the commensurate increase in relational database complexity and size consequently limits retrieval times, restricts exhaustive integration, and requires increasingly more overhead and expertise to manage. Since early 2009, there has been an intentional effort to circumvent these complexity drawbacks by implementing a type of database referred to as NoSQL databases. These databases, while not technically relational databases since they lack traditional mechanisms that would allow for normalization, have the benefit of being natively optimized for popular cloud-based and multicore computer architectures. They are designed to discover data in extremely large data sets at speeds that rival and surpass the performance of large parallel databases without many of the drawbacks (Stonebraker et al., 2010). Since NoSQL databases lack traditional schemas, there are few limiting requirements for time-consuming database administration and can be managed through low-level application programming interfaces instead of optimized SQL queries.
4.1. Wide column and key-value stores
The removal of tightly controlled data schemas, which effectively denormalizes data structures and therefore greatly increases the risk of redundancy, is compensated for by creating operations that are (1) easily deployed and (2) natively distributed. Hadoop (Shvachko, Kuang, Radia, & Chansler, 2010), an open-source implementation of MapReduce (Dean & Ghemawat, 2008), is an example of a key-value long table. Similar to Google’s BigTable implementation (Chang et al., 2008), Hadoop relies exclusively on the qualities of well-indexed data to very rapidly discover values associated with particular keys, called key-value pairs. When implemented properly and for purposes of finding one-to-one or one-to-many associations with a key of interest, Hadoop delivers the power of large and expensive parallel RDBMS without any of the overhead. Other popular implementations of MapReduce include Cassandra and Amazon’s SimpleDB. While they may perform extremely well in data location and retrieval, they sub-perform under a range of scenarios, including determining data consistency and transaction control, which are pushed back to the user or the interface controller. Regardless, the future of these types of data structures is very bright in areas of biological databases where querying specific entities within voluminous data stores is a common task.
4.2. Document stores
The contemporary version of the flat-file database is referred to as a document-oriented NoSQL database, sometimes known as the document store. Here, databases such as MongoDB, CouchDB, and OrientDB, among others, are optimized specifically for indexed JSON-styled documents (Banker, 2012; Wei, Sicong, Qian, & Amiri, 2009). They form the backbone of many web services required to rapidly distribute large numbers of records, including increasingly popular web streaming content. While not used in any current large-scale behavioral neuroscience effort, the document store’s reliance on NoSQL’s key-value relationship schema places it in the unique position of being able to satisfy growing data needs without costly infrastructure support. Indeed, schemas in document stores are dynamically generated and can scale to meet nearly all data types.
4.3. Graph databases
Systems biology, largely centered on the analysis of biological networks, is becoming increasingly widely applied in neuroscience. There exists no shortage of topological life science domains that currently incorporate networks (and therefore the underlying graph theory) for the elucidation of specific processes. Behavioral neuroscience, for example, is interested in the descriptive and predictive potentials of how the underlying gene, protein or metabolic network relationships effect complex traits (Spanagel, 2009). Of paramount importance is the discovery of unifying principles mediating network topology and their biological relevance. There is a need to understand how large-scale interacting dynamical systems, such as those found in systems biology, behave collectively (Strogatz, 2001); empirical studies have shed light on the topology of cellular and metabolic networks (Bhalla & Iyengar, 1999; Hartwell, Hopfield, Leibler, & Murray, 1999; Veeramani & Bader, 2010) and neural networks (Kim, 2004). The extension of graph theory into the collective analysis of behavioral neuroscience networks provides a tremendous reservoir of qualitative insight into the function of biological systems under equilibrium and dynamic stresses.
This has led to an urgent need to refine computational models for graph pattern mining and a robust means for storing, collating, and translating across immense genome-scale graphs in a way that supports the global application of appropriate analysis tools. Because there exists no relational database model applicable across large heterogeneous data representations (and, consequently, repositories) of graph/network-based approaches to biological data, several NoSQL models have made rapid progress to close the gap. These approaches use key-value relationships to generalize pairwise and tripartite relationships between unbounded numbers of biological data types, creating general graph-based schemas that are optimized for generically applied networks and semantic web information. These include Neo4j (and its biology relative, Bio4j), AllegroGraph, sones, infogrid, and trinity, among others. Other graph-based efforts are focusing on compatible labeled graph formats represented by the web-based RDF schemas (Belleau, Nolin, Tourigny, Rigault, & Morissette, 2008; Mironov et al., 2012). The NIF and semantic enterprise wiki from the Allen Institute rely, in part, on graph databases.
5. LIVING WITH HETEROGENEITY
5.1. Integrating primary data
The hierarchical complexities and layered dependencies underlying the continuum of observable processes in behavioral neuroscience result in an inability of a single researcher to encapsulate an effective scope of knowledge. Perhaps the paramount success of bioinformatics is the recognition that deep understanding is found at the intersection of multiple data domains and data types across physiological, developmental, and evolutionary time scales. This can be done by articulating primary data across numerous domains and has led to several emergent realities: (1) structured vocabularies and experimental protocols provide a foundational framework designed to enhance integration, (2) federated databases operate more efficiently on highly structured data, and (3) data needs to be valued as open-source resources (Chesler & Baker, 2010).
Structured vocabularies and ontologies are well-defined controlled vocabularies designed to formalize interactions within the broad scope of experimental observations. However, for each approach to structured integration, there is a tradeoff between prescription and flexibility. As data attributes become more highly structured, the underlying database becomes more accurate and efficient, but at the same time more narrowly defined. In life science, this is the tension between a narrow scope that returns false negatives and articulations that are too broadly defined to be informative. Compounding this tension is a competing tradeoff between the often labor-intensive process required to hand-curate narrowly defined domains and the computationally efficiency associated with automated or semi-automated data management. These manifest themselves in the type of connections established between data sets, from low-level link connections (SRS; Etzold, Ulyanov, & Argos, 1996) and mediated queries (TAMBIS (Stevens et al., 2000) and Kleisli (Davidson et al., 2001)) to full integration. For example, domain-specific and generalized ontologies, such as NIF’s NeuroLEX or GO (Ashburner et al., 2000), respectively, are intended to provide translational flexibility at the interface of databases and analysis tools and are excellent pivot objects in mediated data sources. However, ontologies are not error free and may be considered too sparse or biased to cover an appropriate range of represented system states in a completely automated fashion. The significant challenges in the construction of an ontology that spans all behavioral neuroscience is representative of this problem.
One interesting core aspect of RDBMS is their definitional use of primary keys for the purposes of normalization and uniquely defining relationships of interest, ideally allowing for the harmonization of data between data sources. A primary key uniquely defines an object and remains temporally and contextually constant. Life science is unique in that there exists no global organic primary key. While genes are often used as a core organizing metaphor, they do not have the benefit of remaining contextually constant. The concept that even trusted biological objects shift in both meaning and value over time is a well-known and primary distinction between biological databases and other enterprise level databases (Birney & Clamp, 2004). Thus, primary data is often organized around relative relationships between objects or data types of interest. Automating the discovery of relative relationships between databases is a difficult task that requires the constant curation of information, even in federated environments where strict rules are applied, and often relies heavily on ontological relationships. NoSQL data stores have the benefit of not having to contend with primary keys or strict schemas, lowering the difficulty of dealing with shifting definitions of reference sources.
5.2. Managing secondary data
One approach to reduce confounding background clatter of data with low information content is to focus database integration efforts on published or peer-reviewed data sets. Since these data sets are often representative and significant subsets of larger primary data pools, they are referred to as secondary data. In many ways, the neuroscience bioinformatics is a leader in this area, with efforts like the Neuroinformatics Framework (NIF) (Gardner et al., 2008) and GeneWeaver (Baker et al., 2012), where data stores are integrated at the most granular level of discrete object relationships.
As efforts to collect and collate neuroscience data have discovered, there is a clear imperative to scraping secondary data from published material. Printed academic journals have been slow to standardize the format of primary and supplemental content. For example, while most journals accept Microsoft-based publication standards, reading in data from a table requires both the digitized access to the document and a curator to determine the context of the information. One strong argument for the tacit use of ontologies and structured vocabularies is to further enforce a machine-readable context for published secondary data to the extent that biological databases will eventually merge with journals to seamlessly integrate data. The use of Uniform Resource Identifiers for uniquely referencing particular entities will further such capabilities. Capturing digitized primary and secondary data in a NoSQL-Journal hybrid approach, for example, also allows for the capture of data provenance. While there is a high-level practical need to track data, there is a cultural need to indicate data generation and sourcing in order to encourage researchers to share, and ultimately enhance, knowledge production and aggregation.
Another interesting phenomenon of secondary data analysis is that data aggregation over these sets indicates a strong asymmetry in data density. This means that observable associations between certain biological objects are consistent over a wide range of data sets. This observation, known as a scale-free network in graph theory (Wolf, Karev, & Koonin, 2002), is a well-recognized phenomenon of primary data interactions in biological data, but was unfamiliar in broad secondary or federated data sets. The observation of data sparsity over data scarcity has implications in how neuroscience databases should think about internal schemas. For example, if a database is tasked with storing data about molecular networks in behavioral neuroscience and discovering information about the shortest path between objects of interest, then storing data in an edge list is much better for handling algorithms associated with shortest path problems in sparse networks. It also indicates that in the practical and esoteric world of database, the volume of data does not always relate to information or importance of that data.
6. CONCLUSION
Bioinformatics is fundamentally about the information of biology. Information, in turn, is buried within a cacophony of data produced by a wide swath of molecular techniques. In neuroscience, the breadth of data is exceptionally large as it spans genomics, proteomics, metabolomics, image analysis, and behavioral science, among other protocols, and requires researchers to store data with due diligence based on the data types, data scope and depth, and underlying querying requirements. Traditional relational databases can effectively manage data but require in-depth domain knowledge and strong database expertise to produce schemas robust enough to handle scope and integration. The emergence of NoSQL databases in the recent years has caused researchers to reexamine how data is structured and explore flexible alternatives for viewing relationships among differing data types typically encountered in behavioral neuroscience.
References
- Altman RB. Building successful biological databases. Briefings in Bioinformatics. 2004;5:4–5. doi: 10.1093/bib/5.1.4. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashish N, Ambite JL, Muslea M, Turner JA. Neuroscience data integration through mediation: an (F)BIRN case Study. Frontiers in Neuroinformatics. 2010;4:118. doi: 10.3389/fninf.2010.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Galloway L, Jackson B, Schmoyer D, Snoddy J. MuTrack: A genome analysis system for large-scale mutagenesis in the mouse. BMC Bioinformatics. 2004;5:11. doi: 10.1186/1471-2105-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Jay JJ, Bubier JA, Langston MA, Chesler EJ. GeneWeaver: A web-based system for integrative functional genomics. Nucleic Acids Research. 2012;40:D1067–D1076. doi: 10.1093/nar/gkr968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandrowski AE, Cachat J, Li Y, Muller HM, Sternberg PW, Ciccarese P, et al. A hybrid human and machine resource curation pipeline for the Neuroscience Information Framework. Database. 2012;2012:bas005. doi: 10.1093/database/bas005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker K. MongoDB in Action. Shelter Island, NY: Manning; 2012. [Google Scholar]
- Belleau F, Nolin MA, Tourigny N, Rigault P, Morissette J. Bio2RDF: Towards a mashup to build bioinformatics knowledge systems. Journal of Biomedical Informatics. 2008;41:706–716. doi: 10.1016/j.jbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Berenson H, Bernstein P, Gray J, Melton J, O’Neil E, O’Neil P. A Critique of ANSI SQL Isolation Levels. ACM Press; 1995. pp. 1–10. [Google Scholar]
- Bhalla US, Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381–387. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M. Biological database design and implementation. Briefings in Bioinformatics. 2004;5:31–38. doi: 10.1093/bib/5.1.31. [DOI] [PubMed] [Google Scholar]
- Chang F, Dean J, Ghemawat S, Hsieh WC, Wallach DA, Burrows M, et al. Bigtable. ACM Transactions on Computer Systems. 2008;26:1–26. [Google Scholar]
- Chesler EJ, Baker EJ. The importance of open-source integrative genomics to drug discovery. Current Opinion in Drug Discovery & Development. 2010;13:310–316. [PubMed] [Google Scholar]
- Chesler E, Langston M. Combinatorial genetic regulatory network analysis tools for high throughput transcriptomic data. In: Eskin E, Ideker T, Raphael B, Workman C, editors. Systems Biology and Regulatory Genomics. Berlin/Heidelberg: Springer; 2006. pp. 150–165. [Google Scholar]
- Colland F, Jacq X, Trouplin V, Mougin C, Groizeleau C, Hamburger A, et al. Functional proteomics mapping of a human signaling pathway. Genome Research. 2004;14:1324–1332. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SB, Crabtree J, Brunk BP, Schug J, Tannen V, Overton GC, et al. K2/Kleisli and GUS: Experiments in integrated access to genomic data sources. IBM Systems Journal. 2001;40:512–531. [Google Scholar]
- Davis AP, Murphy CG, Rosenstein MC, Wiegers TC, Mattingly CJ. The Comparative Toxicogenomics Database facilitates identification and understanding of chemical-gene-disease associations: Arsenic as a case study. BMC Medical Genomics. 2008;1:48. doi: 10.1186/1755-8794-1-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean J, Ghemawat S. MapReduce. Communications of the ACM. 2008;51:107. [Google Scholar]
- Etzold T, Ulyanov A, Argos P. SRS: Information retrieval system for molecular biology data banks. Methods in Enzymology (Elsevier) 1996;266:114–128. doi: 10.1016/s0076-6879(96)66010-8. [DOI] [PubMed] [Google Scholar]
- Frishkoff G, Sydes J, Mueller K, Frank R, Curran T, Connolly J, et al. Minimal Information for Neural Electromagnetic Ontologies (MINEMO): A standards-compliant method for analysis and integration of event-related potentials (ERP) data. Standards in Genomic Sciences. 2011;5:211–223. doi: 10.4056/sigs.2025347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner D, Akil H, Ascoli GA, Bowden DM, Bug W, Donohue DE, et al. The neuroscience information framework: a data and knowledge environment for neuroscience. Neuroinformatics. 2008;6:149–160. doi: 10.1007/s12021-008-9024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman N, McCormick K, Goldowitz D, Hockly E, Johnson C, Kristal B, et al. Plans for HDBase—A research community website for Huntington’s Disease. Clinical Neuroscience Research. 2003;3:197–217. [Google Scholar]
- Günay C, Edgerton JR, Li S, Sangrey T, Prinz AA, Jaeger D. Database analysis of simulated and recorded electrophysiological datasets with PANDORA’s tool-box. Neuroinformatics. 2009;7:93–111. doi: 10.1007/s12021-009-9048-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A. BioMart Central Portal—Unified access to biological data. Nucleic Acids Research. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- Heimbigner D, McLeod D. A federated architecture for information management. ACM Transactions on Information Systems. 1985;3:253–278. [Google Scholar]
- Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, et al. MassBank: A public repository for sharing mass spectral data for life sciences. Journal of Mass Spectrometry. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- Jonquet C, Shah NH, Musen MA. The open biomedical annotator. Summit on Translatation Bioinformatics. 2009;2009:56–60. [PMC free article] [PubMed] [Google Scholar]
- Keator DB. Management of information in distributed biomedical collaboratories. Methods in Molecular Biology. 2009;569:1–23. doi: 10.1007/978-1-59745-524-4_1. [DOI] [PubMed] [Google Scholar]
- Kim BJ. Performance of networks of artificial neurons: The role of clustering. Physical Review. E, Statistical, Nonlinear, and Soft Matter Physics. 2004;69:045101. doi: 10.1103/PhysRevE.69.045101. [DOI] [PubMed] [Google Scholar]
- Maddatu TP, Grubb SC, Bult CJ, Bogue MA. Mouse Phenome Database (MPD) Nucleic Acids Research. 2012;40:D887–D894. doi: 10.1093/nar/gkr1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. Journal of Cognitive Neuroscience. 2007;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- Matuszek G, Talebizadeh Z. Autism Genetic Database (AGD): A comprehensive database including autism susceptibility gene-CNVs integrated with known noncoding RNAs and fragile sites. BMC Medical Genetics. 2009;10:102. doi: 10.1186/1471-2350-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov V, Seethappan N, Blondé W, Antezana E, Splendiani A, Kuiper M. Gauging triple stores with actual biological data. BMC Bioinformatics. 2012;13(Suppl 1):S3. doi: 10.1186/1471-2105-13-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller HM, Rangarajan A, Teal TK, Sternberg PW. Textpresso for neuroscience: Searching the full text of thousands of neuroscience research papers. Neuroinformatics. 2008;6:195–204. doi: 10.1007/s12021-008-9031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttenberg A, Rees JA, Samwald M, Marshall MS. Life sciences on the Semantic Web: The Neurocommons and beyond. Briefings in Bioinformatics. 2009;10:193–204. doi: 10.1093/bib/bbp004. [DOI] [PubMed] [Google Scholar]
- Saal LH, Troein C, Vallon-Christersson J, Gruvberger S, Borg A, Peterson C. BioArray Software Environment (BASE): A platform for comprehensive management and analysis of microarray data. Genome Biology. 2002;3:SOFTWARE0003. doi: 10.1186/gb-2002-3-8-software0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd GM, Mirsky JS, Healy MD, Singer MS, Skoufos E, Hines MS, et al. The Human Brain Project: Neuroinformatics tools for integrating, searching and modeling multidisciplinary neuroscience data. Trends in Neurosciences. 1998;21:460–468. doi: 10.1016/s0166-2236(98)01300-9. [DOI] [PubMed] [Google Scholar]
- Shoemaker BA, Zhang D, Tyagi M, Thangudu RR, Fong JH, Marchler-Bauer A, et al. IBIS (Inferred Biomolecular Interaction Server) reports, predicts and integrates multiple types of conserved interactions for proteins. Nucleic Acids Research. 2012;40:D834–D840. doi: 10.1093/nar/gkr997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvachko K, Kuang H, Radia S, Chansler R. The Hadoop Distributed File System. IEEE 26th Symposium On Mass Storage Systems and Technologies (MSST); 2010. pp. 1–10. [Google Scholar]
- Spanagel R. Alcoholism: A systems approach from molecular physiology to addictive behavior. Physiological Reviews. 2009;89:649–705. doi: 10.1152/physrev.00013.2008. [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: A general repository for interaction datasets. Nucleic Acids Research. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R, Baker P, Bechhofer S, Ng G, Jacoby A, Paton NW, et al. TAMBIS: Transparent access to multiple bioinformatics information sources. Bioinformatics. 2000;16:184–186. doi: 10.1093/bioinformatics/16.2.184. [DOI] [PubMed] [Google Scholar]
- Stonebraker M, Abadi D, DeWitt DJ, Madden S, Paulson E, Pavlo A, et al. MapReduce and parallel DBMSs: Friends or foes? Communications of the ACM. 2010;53:64–71. [Google Scholar]
- Strogatz SH. Exploring complex networks. Nature. 2001;410:268–276. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- Taylor CF, Field D, Sansone SA, Aerts J, Apweiler R, Ashburner M, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: The MIBBI project. Nature Biotechnology. 2008;26:889–896. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramani B, Bader JS. Predicting functional associations from metabolism using bi-partite network algorithms. BMC Systems Biology. 2010;4:95. doi: 10.1186/1752-0509-4-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Foerster H. Biological principles of information storage and retrieval. In: Kent A, Taubee OE, Beltzer J, Goldstein GD, editors. Electronic Handling of Information: Testing and Evaluation. London: Academic Press; 1967. pp. 123–147. [Google Scholar]
- Waldrop M. Big data: Wikiomics. Nature. 2008;455:22–25. doi: 10.1038/455022a. [DOI] [PubMed] [Google Scholar]
- Wang J, Williams RW, Manly KF. WebQTL: Web-based complex trait analysis. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- Wei K, Sicong T, Qian X, Amiri H. Most. 2009. An Investigation of No-SQL Data Stores. [Google Scholar]
- Wolf YI, Karev G, Koonin EV. Scale-free networks in biology: New insights into the fundamentals of evolution? BioEssays. 2002;24:105–109. doi: 10.1002/bies.10059. [DOI] [PubMed] [Google Scholar]