Abstract
Our understanding of the pathogenesis of most primary glomerular diseases, including IgA nephropathy, membranous nephropathy and focal segmental glomerulosclerosis, is limited. Advances in molecular technology now permit genome-wide, high-throughput characterization of genes and gene products from biological samples. Comprehensive examinations of the genome, transcriptome, proteome and metabolome (collectively known as omics analyses), have been applied to the study of IgA nephropathy, membranous nephropathy and focal segmental glomerulosclerosis in both animal models and human patients. However, most omics studies of primary glomerular diseases, with the exception of large genomic studies, have been limited by inadequate sample sizes and the lack of kidney-specific data sets derived from kidney biopsy samples. Collaborative efforts to develop a standardized approach for prospective recruitment of patients, scheduled monitoring of clinical outcomes, and protocols for sampling of kidney tissues will be instrumental in uncovering the mechanisms that drive these diseases. Integration of molecular data sets with the results of clinical and histopathological studies will ultimately enable these diseases to be characterized in a comprehensive and systematic manner, and is expected to improve the diagnosis and treatment of these diseases.
Introduction
Primary glomerulonephritides—glomerular diseases that are not caused by a systemic disease—are the third leading cause of end-stage renal disease (ESRD) in the USA and remain major causes of chronic kidney disease in China and worldwide.1,2 The underlying molecular basis of most primary glomerulonephritides is, however, still largely unknown. As a result, the current classification of primary glomerulonephritides still relies on renal histologic features, and disease-specific therapies are lacking. Current therapeutic options for primary glomerulonephritides are limited to corticosteroids and immunosuppressive agents, which mainly target the immune system but also exert antiproteinuric effects by acting directly on the glomerular podocytes.3 Long-term use of these medications is, however, fraught with adverse effects, treatment resistance and loss of response.
Conventional studies, using hypothesis-driven and candidate-gene approaches, have identified various genes, growth factors and signalling pathways that are involved in primary glomerulonephritides, although these approaches do not fully capture the entire picture of disease and have not been successful in identifying the underlying molecular basis of these diseases. With advances in molecular medicine, however, we can now profile the molecular signature of a large number of biological samples on a genome-wide scale, and do so faster than ever before. This ability to capture the molecular signature of an affected organ with novel high-throughput methods and to integrate these molecular findings with laboratory and clinical parameters will facilitate examination of the primary glomerulonephritides as an interacting network of genes, proteins and biochemical reactions that affect the function of cells and organs (Box 1).
Box 1. An overview of molecular genetics.
The essential hereditary information of a living organism is encoded along DNA-based nucleotide sequences. DNA is transcribed into RNA-based sequences, which are then decoded and translated into the proteins that ultimately carry out most of the cellular functions. Products of biological reactions catalysed by proteins are known as metabolites.
The entire repertoire of DNA-based genetic information, RNA transcripts, proteins and metabolic products in a collection of cells or organisms are known as the genome, transcriptome, proteome and metabolome, respectively. Experimental methods that are capable of measuring these changes on the genome-wide scale are referred to as ‘omics’ technologies.
With the invention and rapid development of molecular technologies for measuring these biomolecules over the past two decades, it is now possible to examine the molecular signature of disease in a comprehensive and high-throughput manner.
During the past decade, several investigators have applied this systems biology approach to the study of the primary glomerulonephritides. The general applications of systems biology for studying kidney diseases (molecular techniques, analytical tools and the interpretation of these large data sets),4 interpreting the results of genetic studies by linking genotypes to phenotypes,5 and studying diabetic kidney disease have been discussed previously.6–8 In this Review, therefore, we focus on studies that have examined the genome, transcriptome, proteome or metabolome of patients and animal models of IgA nephropathy (IgAN), membranous nephropathy and focal segmental glomerulosclerosis (FSGS). Results from these studies have enriched our understanding of this group of complex diseases in a way that would not have been possible using traditional hypothesis-driven and candidate-gene approaches.
Genome-wide analyses
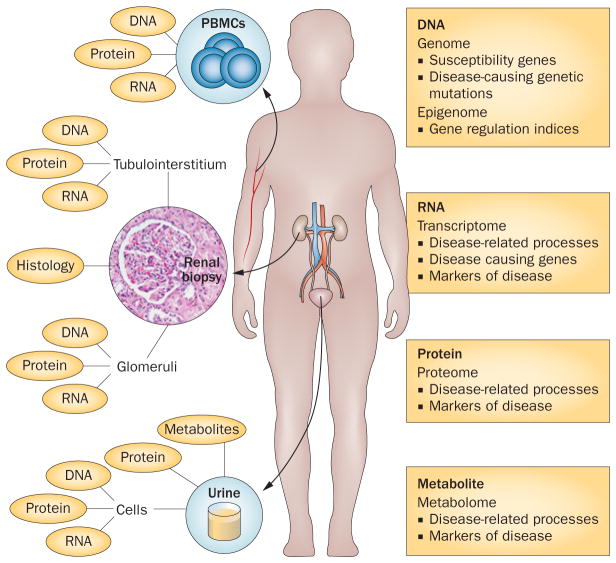
Several genomic data sets have been generated for IgAN, membranous nephropathy and FSGS over the past decade, and the interpretation of this information is fundamental to understanding the mechanisms of these diseases and the development of targeted therapies. The genome of all cell types in a given organism is essentially the same, although the transcriptome, proteome and metabolome can differ greatly. The use of various sources of biological samples could, therefore, provide detailed information about the molecular characteristics of primary glomerulonephritides (Figure 1). Of note, quantitative analyses of the genome, transcriptome, epigenome, proteome and metabolome of a single cell have been described in other organisms and systems, but have not yet been applied to the study of kidney disease.
Figure 1.
Sources of biological material for use in ‘omics’ studies of primary glomerulonephritides. The three major sources of material for characterization of glomerular diseases are PBMCs, renal biopsy samples and urine. DNA extracted from the patient’s cells can be used to analyse genomic information, such as susceptibility genes and disease-causing mutations and variants, and epigenetic factors that regulate gene expression through DNA methylation, changes to chromatin structure and genomic imprinting. RNA transcripts, including messenger RNA, microRNA and other noncoding RNAs, can modulate disease-related processes or the expression of disease-causing genes in ways that are specific to individual tissues or cells. Proteins extracted from cells and soluble proteins in the urine can inform us about disease-related process or serve as markers of disease. Renal histologic features of disease, when integrated with molecular signatures of disease, can serve as diagnostic and predictive biomarkers. The transcriptome, proteome and epigenome are likely to be specific to the tissue (kidney versus nonkidney) and cellular compartment (glomerular versus tubulointerstitial) analysed. Tissue-specific, renal-compartment-specific and even cell-specific collection of these omics-level data are, therefore, important for understanding disease processes. Abbreviation: PBMC, peripheral blood mononuclear cell.
Genomic studies
High-density oligonucleotide microarray technology enables the simultaneous interrogation of hundreds of thousands of single nucleotide polymorphisms (SNPs). The pattern of SNPs across the genome is different for every individual. Genome-wide association studies (GWAS) and family-based linkage disequilibrium studies both rely on SNP genotyping to map genetic loci related to a disease phenotype. However, the two approaches differ in their experimental design. GWAS correlate SNPs with a disease phenotype in a population, whereas linkage disequilibrium studies examine the segregation of SNPs within a family that includes several affected individuals, to identify disease-related loci. For single-gene diseases, mapping of the disease-related loci in a family of affected individuals has traditionally relied on linkage disequilibrium mapping. By contrast, risk alleles for polygenic diseases are now usually identified by GWAS. The advantage of GWAS is that common disease genes can be identified. The disadvantage of this approach, however, is that the aggregated risk of disease conferred by all identified disease-related loci usually accounts for only a small proportion of the risk of developing the disease. An important caveat to consider in the interpretation of results from GWAS is that identification of an association between a gene variant and a disease phenotype does not necessarily imply a causal relationship between the two. Loci identified in GWAS are, in fact, rarely the causal variants. Instead, the identified disease-associated polymorphisms might segregate along with the causal gene simply because they occur in the same piece of chromosome (linkage disequilibrium). Alternatively, the identified loci could have a permissive role; that is, they facilitate the manifestation of the phenotype, but by themselves are not sufficient to cause the disease.
Sequencing an individual’s entire genome has become much faster and more affordable with the introduction of next-generation DNA sequencing technology. However, it is still not practical to analyse the whole genome of every participant in GWAS. Instead, researchers continue to rely on mapping of SNPs spread out across the genome. Other approaches are, however, becoming feasible. For example, sequencing the entire coding regions of the human genome, known as whole-exome sequencing, has changed the way we study rare Mendelian disorders. As 85% of all genetic mutations with a Mendelian pattern of inheritance occur within the exome, direct sequencing of all 180,000 protein-encoding human exons will greatly facilitate the discovery of causative genes for single-gene disorders, most of which are rare and difficult to study by SNP-based techniques. Indeed, among the estimated 5,400 known diseases that follow a Mendelian pattern of inheritance, the disease-causing gene has been identified in only about 2,600.9 Whole-exome sequencing has already identified several genes related to FSGS, for example COQ6,10 MYO1E11 and NEIL2.12 Sequence-level data from whole-exome sequencing experiments is expected to provide a deep layer of information that is not available from SNP genotyping.
Transcriptomic studies
The transcriptome encompasses all RNA-based molecules, including mRNA, ribosomal RNA, transfer RNA and other noncoding RNAs, such as the microRNAs (miRNAs), a family of RNAs that are approximately 22 nucleotides long. The miRNAs are endogenously produced transcripts that are involved in the fine regulation of gene expression.
The transcriptome is particularly pertinent for contextualizing the molecular basis of disease processes. The transcriptome of cells and organs directly implicated in, or affected by a disease, is generally considered most informative for understanding its pathogenesis. In the primary glomerulonephritides, the cells involved in disease processes could be kidney cells or circulating immune cells, and the affected cells are podocytes, parietal epithelial cells, mesangial cells, endothelial cells and renal tubular cells. However, glomeruli occupy less than 5% of the total kidney mass, and the molecular signatures of the kidney cortex are likely to be very different from those of the glomerular compartment or of specific cell types within the glomerulus. Several approaches, including glomerular sieving and laser-capture microdissection,13 have been developed to isolate glomeruli from biological specimens, such as nephrectomy samples or renal biopsy cores. The cellular yield from these approaches is sufficient for transcriptomic analysis.
One of the earliest attempts to fully characterize the regulation of genes in human renal biopsies was undertaken by investigators who established the European Renal cDNA Bank (ERCB)–Kroener-Fresenius Biopsy Bank.14 The Nephrotic Syndrome Study Network (NEPTUNE) is another major multicentre initiative that is funded by the NIH to prospectively enrol patients with minimal change disease (MCD), membranous nephropathy and FSGS, and to collect clinical data and biological samples from these patients.15 One of the goals of NEPTUNE is to correlate clinical parameters with molecular signatures in biological samples, including the kidney transcriptome. Many published data sets of the kidney transcriptome are curated and accessible through Nephromine.16
Proteomic, metabolomic and histologic analyses
Proteins and metabolites excreted in the urine provide a unique opportunity to examine the proteome and metabolome of the kidney in a noninvasive manner. The clinical utility of the urinary proteome is widely recognised for diagnosis and predicting the prognosis of patients with kidney disease.17–19 However, metabolomic studies are currently scarce in kidney research.20–22 Systems pathology represents the integration of molecular and imaging data with patients’ clinical histories.23 The goal of such analysis is to identify morphometric features and molecular biomarkers that correlate with a clinical event—an aspect of data integration for studying glomerular diseases that has been proposed by the NEPTUNE group.
Histologic features and clinical presentation still form the basis of the diagnostic criteria for most human diseases, and subsets of these characteristics can have known prognostic value. For instance, patients with the collapsing variant of FSGS tend to fare poorly compared with those who have tip lesions. However, whether proteomic and metabolomic signatures exist that correspond to these FSGS histologic variants is not yet known.
IgA nephropathy
IgAN is a major cause of kidney failure worldwide. The prevalence of this disease is highest in Asian populations, intermediate in European populations, and lowest in African populations, although whether these differences reflect the influence of genetic or environmental factors is unknown. The clinical presentation of IgAN is variable, ranging from haematuria, with or without nephrotic syndrome, to a rapidly progressive loss of renal function. Histological features of IgAN run the gamut of mesangial proliferation to glomerular extracapillary proliferation with crescent formation. Overproduction of IgA1 molecules that are hypogalactosylated in the hinge region is associated with IgAN, although this feature alone does not cause renal injury.24 The formation of glycan-specific IgA and IgA that recognize these hypogalactosylated IgA1 molecules, with the resulting deposition of IgA1 in the glomerular mesangium, is required for disease development. The exact mechanism that precipitates each of these stages of disease development is not well defined, although information from a number of omics studies are now further enhancing our understanding of IgAN pathogenesis. Mechanistic studies are still needed to further dissect how innate immunity and inflammation contribute to the pathogenesis of IgAN. Results from genomic studies have, however, generated a shortlist of plausible candidate genes that are likely to be linked with, or directly responsible for, the development of IgAN.
Genomic studies
Several lines of evidence suggest that the pathogenesis of IgAN has a considerable genetic component—for example, the prevalence of IgAN differs greatly across ethnic groups, and the prevalence of disease is higher in relatives of affected individuals. A region of chromosome 6p that encodes the major histocompatibility complex (MHC) has been identified through GWAS as containing the strongest common susceptibility alleles predisposing patients of European population to IgAN.25 Interestingly, three independent susceptibility loci in this MHC region were also subsequently identified as associated with IgAN in the Han Chinese population, as well as a common deletion in the genes encoding complement factor H−related proteins 1 and 3 on chromosome 1q32, and a locus on chromosome 22q12 that encompasses several genes.26 In another GWAS of IgAN in the Han Chinese population,27 two novel susceptibility genes, TNFSF13 (encoding tumour necrosis factor ligand superfamily member 13, also known as APRIL) and DEFA1 (encoding neutrophil defensin 1) were also linked to IgAN in Chinese patients, and an association between the TNFSF13 gene and high serum IgA levels in smokers has been reported in a southern Chinese population.28 Whether TNFSF13 variants have any clinical utility in the diagnosis of IgAN will need to be studied in a larger cohort of patients. Susceptibility loci for IgAN identified by GWAS are often near to genes involved in innate immunity and inflammation (Table 1). Taken together, these genetic studies strongly suggest a heritable component for IgAN.
Table 1.
Summary of GWAS results from IgAN and membranous nephropathy cohorts
| Study | Cohort | dbSNP locus | Locus
|
P value | Odds ratio (95% CI) per allele | ||
|---|---|---|---|---|---|---|---|
| Chromosome | Minor allele | Nearest gene | |||||
| Yu et al. (2012)27 | 4,137 Han Chinese patients with IgAN and 7,734 population controls | rs2738048 | 8 | A | DEFAs | 3.18×10−14 | 0.79 (0.74–0.84) |
| rs3803800 | 17 | G | TNFSF13 | 9.40×10−11 | 1.21 (1.14–1.28) | ||
| rs4227 | 17 | T | MPDU1 | 4.31×10−10 | 1.23 (1.16–1.32) | ||
| rs12537 | 22 | C | MTMR3 | 1.17×10−11 | 0.78 (0.72–0.84) | ||
| rs2523946 | 6 | T | HLA-A | 1.74×10−11 | 1.21 (1.15–1.28) | ||
| rs660895 | 6 | A | HLA-DRB1 | 4.13×10−20 | 1.34 (1.26–1.42) | ||
| rs1794275 | 6 | G | HLA-DQA/B | 3.43×10−13 | 1.30 (1.21–1.39) | ||
|
| |||||||
| Feehally et al. (2010)25 | 244 European patients with IgAN and 4,980 population controls | rs3115573 | 6 | A | NLNHP4 | 2.00×10−6 | 1.55 (1.29–1.85) |
| rs3130315 | 6 | G | NLNHP4 | 2.00×10−6 | 1.55 (1.29–1.85) | ||
|
| |||||||
| Gharavi et al. (2011)26 | 3,144 Han Chinese patients with IgAN and 2,822 population controls* | rs3766404 | 1 | C | CFHR1,3 | 4.24×10−5 | 0.77 |
| rs6677604 | 1 | A | CFHR1,3 | 2.96×10−10 | 0.68 | ||
| rs2856717 | 6 | T | HLA-DQB1 | 8.44×10−16 | 0.73 | ||
| rs9275596 | 6 | C | HLA-DQB1 | 1.59×10−26 | 0.63 | ||
| rs9357155 | 6 | A | PSMB8 | 2.11×10−12 | 0.71 | ||
| rs2071543 | 6 | A | PSMB8 | 5.77×10−12 | 0.73 | ||
| rs1883414 | 6 | T | HLA-DPB2 | 4.84×10−9 | 0.78 | ||
| rs3129269 | 6 | T | HLA-DPB2 | 8.54×10−9 | 0.79 | ||
| rs2412971 | 22 | A | HORMAD2 | 1.86×10−9 | 0.80 | ||
| rs2412973 | 22 | A | HORMAD2 | 4.46×10−9 | 0.80 | ||
|
| |||||||
| Stanescu et al. (2011)51 | 556 white European patients with membranous nephropathy and 2,338 population controls | rs4664308 | 2 | A | PLA2R1 | 8.6×10−29 | 2.28 (1.96–2.64) |
| rs2187668 | 6 | G | HLA-DQA1 | 8.0×10−93 | 4.32 (3.73–5.01) | ||
All values are given for the Beijing discovery cohort, plus both the Shanghai and European replication cohorts, combined. Abbreviations: dbSNP, database of single nucleotide polymorphisms; GWAS, genome-wide association study; IgAN, IgA nephropathy.
Transcriptomic studies
IgAN is generally believed to be related to presence of a systemic immune response, and a number of transcriptomic studies have examined gene expression levels in patients with IgAN (Table 2). Histologic alterations in the glomeruli are thought to be indirect sequelae of the primary systemic immune response and, therefore, most studies of the IgAN transcriptome have focused on gene expression patterns in peripheral blood mononuclear cells (PBMCs). A gene expression microarray study of PBMCs isolated from 12 patients with IgAN and eight healthy control individuals identified 210 genes that were differentially expressed.29 Further bioinformatics analysis of these 210 genes revealed that canonical Wnt–β-catenin and PI3K–Akt signalling are the two pathways that are most dysregulated in patients with IgAN. Dysregulation of these two pathways was subsequently confirmed by the finding of reduced levels of inversin and PTEN proteins (which are key regulators of Wnt–β-catenin and PI3K–Akt signalling pathways, respectively) in PBMCs from patients with IgAN. A comparison of gene expression levels in PBMCs from white European patients with IgAN, FSGS, antineutrophil cytoplasmic autoantibody (ANCA)-associated glomerulonephritis and MCD versus PMBCs from healthy volunteers30 identified 15 genes that were significantly correlated with disease activity and kidney function in the 22 patients with IgAN. In another study, gene expression profiles of PBMCs from seven patients with IgAN were compared to those of seven healthy individuals and six patients with MCD.31 The profiles correlated with various clinical indices in the patients with IgAN. For example, expression of urotensin-2 (encoded by UTS2) positively correlated with blood pressure in patients with IgAN, and retinoic acid receptor α (encoded by RARA) was significantly downregulated in samples from patients with IgAN, although expression of this protein was not significantly correlated with any clinical indices.31
Table 2.
Transcriptomics studies in IgAN
| Author | Tissue | Cohort | Differentially regulated genes or pathways | P value* | Other changes |
|---|---|---|---|---|---|
| Cox et al. (2010)29 | Human PBMCs | 12 patients with IgAN 8 healthy controls |
Wnt–β-catenin and PI3K–Akt signalling | 0.0039 0.0040 |
NR |
| Preston et al. (2004)30 | Human PBMCs | 8 patients with IgAN 32 healthy controls 24 patients with ANCA-associated glomerulonephritis 18 patients with SLE 5 patients with FSGS 6 patients with MCD |
BTG2, PMAIP1, B3GNT5, UBE2J1, SRPK1, SOCS3, LYZ, TRIB1, CSRNP1, PTGS2, EGR1, G0S2, LGALS3, IL8 | NR | Expression of these genes correlated with patients’ serum creatinine levels and disease activity |
| Woo et al. (2010)31 | Human PBMCs | 7 patients with IgAN 6 patients with MCD 7 healthy controls |
URT2 (3.9-fold upregulated in IgAN) RARA (0.41-fold downregulated in IgAN) |
<0.05 <0.005 |
Urotensin−2 (encoded by URT2) levels positively correlated with systolic and diastolic blood pressure |
| Tokunaga et al. (2010)33 | Mouse kidney | HIGA mice ddY mice |
IGFBP1 | NR | Serum IGFBP−1 levels were significantly higher in patients than controls, and correlated with renal function, mesangial cell proliferation and interstitial fibrosis in patients with IgAN |
| Serino et al. (2012)42 | Human PBMCs | 7 patients with IgAN 7 healthy controls |
miR-148b‡, miR-188-5p§, miR-886-3p‡, miR-let-7b‡, miR-let-7d§ (upregulated in IgAN) miR-361-3p‡ (downregulated in IgAN) |
<0.01‡ <0.03§ |
Results validated in an independent cohort of 10 patients with IgAN and 10 healthy controls |
Versus controls.
P <0.01 relates to these microRNAs.
P <0.03 relates to these microRNAs.
Abbreviations: ANCA, antineutrophil cytoplasmic autoantibody; ddY, a mouse strain that can serve as a spontaneous animal model for IgAN; FSGS, focal segmental glomerulosclerosis; HIGA, high IgA; IgAN, IgA nephropathy; MCD, minimal change disease; NR, not reported; PBMC, peripheral blood mononuclear cell; SLE, systemic lupus erythematosus.
In IgAN, as well as in other glomerular diseases, an increased glomerular filtration rate (GFR) exposes renal tubular cells to excessive amounts of filtered albumin and other proteins. The pattern of gene expression in the tubulointerstitium of kidney biopsy samples taken from patients with IgAN is different to that of healthy individuals. In normal cultured renal tubular cells, 231 genes were identified as differentially regulated under conditions of albumin exposure.32 Expression of these genes in the tubulointerstitium of kidney biopsy samples from patients with IgAN was compared with that in renal tissue from healthy kidney donors, and the two groups could be distinguished solely on the expression pattern of these 231 albumin-regulated genes. Furthermore, profiling of a subset of 11 of the original 231 genes provided a distinctive expression signature that could distinguish patients with FSGS, MGN, MCD and IgAN from healthy controls. However, the expression signature of this 11-gene subset is not specific to IgAN per se, but instead is shared across these different forms of primary glomerulonephritis. Thus, whether a characteristic gene expression profile exists in the glomerular or the tubulointerstitial compartment that is specific to IgAN and could be important for the manifestation of this renal disease remains to be determined.
Insulin-like growth factor binding protein 1 (IGFBP-1) is highly expressed in the kidney of HIGA mice (a mouse model of IgAN with high serum IgA levels).33 In addition, serum IGFBP-1 levels are significantly higher in patients with IgAN than in healthy individuals and correlate with estimated GFR as well as with the degree of mesangial cell proliferation and interstitial fibrosis in these patients.33 These observations suggest that IGFBP-1 levels reflect the severity of IgAN and could, therefore, be useful as a marker of IgAN progression. Of note, IGFBPs are produced by a variety of tissues, and might also contribute to the development of glomerulosclerosis.34,35
miRNAs are attractive molecules for biomarker research because they are present in multiple body fluids and are stable in storage and handling.36,37 Accumulating evidence suggests that miRNAs are involved in the pathogenesis of several kidney diseases.38,39 A comparison of the urinary and intrarenal expression of several miRNAs in patients with IgAN40,41 and healthy volunteers indicated that the levels of urinary miR-200a, miR-200b and miR-429 (but not those of miR-200c, miR-141, miR-205 or miR-192) were downregulated in patients with IgAN. The degree of reduction correlated with both disease severity and progression.40 Intrarenal and urinary levels of miR-146a and miR-155 (which have important roles in the regulation of innate and adaptive immune responses) were assessed in samples from 43 patients with IgAN and compared with their levels in samples from 33 healthy controls.41 Intrarenal and urinary levels of miR-146a and miR-155 were significantly elevated in patients with IgAN, and the level of elevation correlated with the clinical and histological severity of the disease. High-throughput miRNA profiling of PBMCs from patients with IgAN and healthy controls identified 37 differentially expressed miRNAs.42 miR-148b, one of the significantly upregulated miRNAs, targets the product of C1GALT1 (glycoprotein-N-acetylgalactosamine 3-β-galactosyltransferase 1) and contributes to the abnormal glycosylation of IgA1. Reduced expression of this glycoprotein was confirmed in PBMCs of patients with IgAN and correlated with miR-148b expression. Overexpression of miR-148b in PBMCs from healthy individuals reduced endogenous mRNA levels of this glycoprotein by threefold. Conversely, inhibition of miR-148b in PBMCs of patients with IgAN increased both mRNA and protein levels of glycoprotein-N-acetylgalactosamine 3-β-galactosyltransferase 1 to match those observed in healthy persons. Furthermore, upregulation of miR-148b directly correlated with levels of galactose-deficient IgA1. Taken together, these data suggest a role for miRNAs, specifically miR-148b, in the pathogenesis of IgAN. More studies are needed, however, to confirm whether miRNAs are robust biomarkers for diagnosis and prediction of the prognosis of IgAN.
Proteomic studies
Several studies examining the urinary proteome have been published.43–45 A panel of 10 urinary proteins that have different expression levels in patients with IgAN and healthy individuals has been identified (of which, eight are upregulated and two are downregulated in patients with IgAN), for example. Moreover, this panel distinguished patients with severe IgAN from those with mild IgAN with 90.48% sensitivity and 96.77% specificity, respectively.46 Distinct urinary protein profiles that can distinguish between healthy individuals and patients with IgAN have also been identified by several other groups, although none of the findings has been applied in a clinical setting to definitively demonstrate the clinical utility of urinary protein profiling (Table 3).
Table 3.
Proteomics studies in IgAN and passive Heymann nephritis
| Author | Sample source | Participants | Method | Results |
|---|---|---|---|---|
| Park et al. (2006)43 | Urine | 13 patients with IgAN 12 healthy controls |
2D-GE | 59 proteins were differentially expressed |
| Yokota et al. (2007)44 | Urine | 17 patients with IgAN 10 healthy controls |
2D-DIGE | 10 proteins (albumin, transferrin, α1-antitrypsin, β-globin, α1-globin, carbonic anhydrase I, cystatin C, retinol-binding protein 4 and 1-microglobulin) were differentially expressed* |
| He et al. (2012)46 | Urine | 56 patients with IgAN‡ 14 healthy controls |
MALDI-TOF-MS | 21 peaks distinguished mild and severe groups§ 50 peaks distinguished mild and normal groups|| 50 peaks distinguished severe and normal groups¶ |
| Rocchetti et al. (2008)47 | Urine | 18 patients with IgAN 20 healthy controls |
2D PAGE and nano-HPLC-ESI-MS/MS | Among patients with IgAN, kininogen, ITI-HC1 and transthyretin levels were different in responders and nonresponders to ACE inhibitors Low levels of urine kininogen predicted inadequate or absent clinical response to ACE inhibitors in 20 patients with biopsy-proven IgAN |
| Ngai et al. (2006)53 | Urine | Control rats Rats with PHN assessed at postinduction days 0, 10, 20, 30, 40 and 50# |
2D-PAGE | 37 differentially expressed proteins across all time points |
| Wang et al. (2012)54 | Kidney tissue | Control rats Rats with PHN assessed at postinduction days 14 and 21** |
LC–MS/MS | 160 proteins were significantly different across all three groups The endoplasmic reticulum stress proteins GRP78 and GRP94 and the autophagy marker LC3 were upregulated in PHN rats versus controls |
All except 1-microglobulin were higher in patients with IgAN than controls.
Of whom 23 had a severe and 33 had a mild presentation.
For a subgroup of 10 peaks selected as biomarkers, sensitivity was 90.48% and specificity 96.77%.
For a subgroup of 10 peaks selected as biomarkers, sensitivity was 93.55% and specificity 85.71%.
For a subgroup of 20 peaks selected as biomarkers, sensitivity was 100% and specificity 92.86%.
6 rats per group.
8 rats per group.
Abbreviations: ACE, angiotensin-converting enzyme; DIGE, difference gel electrophoresis; GE, gel electrophoresis; HPLC-ESI, high performance liquid chromatography with electrospray ionization; IgAN, IgA nephropathy; LC, liquid chromatography; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; MS, mass spectrometry; MS/MS, tandem mass spectrometry; PAGE, polyacrylamide gel electrophoresis; PHN, passive Heymann nephritis.
Proteomic studies can also yield potential predictive markers. For example, the urinary proteomic profile of patients with IgAN predicts their response to angiotensin-converting enzyme (ACE) inhibitors; urine levels of kininogen-1, inter-α-trypsin inhibitor heavy chain H4 and transthyretin differed significantly between responders and nonresponders to ACE inhibitor therapy.47 Very low urinary levels of kininogen-1 correlated with a poor response to this treatment.47
Studies with large sample sizes are needed to evaluate the clinical applicability of these urinary protein markers. Furthermore, whether examination of the urine proteome will provide an insight into the pathogenesis of primary glomerular diseases is unclear, because differential excretion of urinary proteins is more likely to result from the urinary loss of serum proteins (via glomerular dysfunction) rather than from a disease-specific process.
Membranous nephropathy
Membranous nephropathy is the second or third most common type of primary glomerulonephritis resulting in ESRD in the USA, Europe and Asia.48 This antibody-mediated autoimmune glomerular disease is characterized by the presence of immune deposits on the epithelial aspect of the glomerular capillary wall. Our understanding of the pathogenesis of membranous nephropathy is mostly derived from studies in rats with passive Heymann nephritis (PHN), in which a glomerular disease that closely resembles human membranous nephropathy is induced by a single injection of heterologous antiserum or IgG against renal tubular cell antigens.49 One of the major success stories of molecular medicine in glomerular diseases was the identification of secretory phospholipase A2 receptor (PLA2R) as the target antigen of the autoantibodies produced in the majority (~70%) of patients with primary or idiopathic membranous nephropathy.50 Western blotting was used to compare the proteins extracted from the glomeruli and sera of healthy individuals and patients with idiopathic membranous nephropathy, and other glomerular diseases. A 185 kDa band (which was detectable only when nonreducing blotting conditions were used) was present in the sera from patients with membranous nephropathy. Candidate antigens within this 185 kDa band were analysed by mass spectrometry, and the identity of the target antigen was confirmed using available antibodies and recombinant proteins.
Despite this advance in our understanding of membranous nephropathy, many questions remain unanswered. Firstly, it is not clear why PLA2R (which is expressed on podocytes) becomes a target for circulating autoantibodies only in these patients, nor why anti-PLA2R antibodies are detected in only some patients with membranous nephropathy. Whether the limitations of current antibody assays contribute to the lack of detectable anti-PLA2R antibodies in some patients remains to be investigated. Most importantly, the role of PLA2R in the pathogenesis of membranous nephropathy remains to be defined. However, with the availability of genomic, transcriptomic, proteomic and metabolomic data from patients with membranous nephropathy, we can reasonably expect that further clinically relevant biomarkers and treatment targets for this disease will be developed in the near future.
Genomic studies
GWAS have identified associations between idiopathic membranous nephropathy and the HLA-DQA1 locus on chromosome 6p21 and the PLA2R1 locus on chromosome 2q24, in white patients of European ancestry (Table 1).51 Interestingly, the risk alleles of these two genes have an additive effect; patients carrying all four risk alleles have an odds ratio of 78.46 for developing membranous nephropathy compared to individuals who have the protective variants of all four alleles. More studies are, however, needed to confirm whether these risk loci also exist in other populations and to determine how genes in these two loci interact with each other to increase susceptibility to membranous nephropathy.
Results from GWAS confirmed the association between PLA2R1 and membranous nephropathy, but the pathogenetic role of PLA2R remains unresolved. Some researchers postulate that genes that influence the development of primary membranous nephropathy might do so by altering the immune response or by enhancing podocyte PLA2R expression, configuration or both. However, how these risk alleles precipitate the development of idiopathic membranous nephropathy remains unclear.
Transcriptomic studies
The examination of kidney gene expression in rats with PHN identified 234 transcripts that were differentially expressed compared with control animals.52 Bioinformatics analysis of the differentially expressed genes, based on prior knowledge of known protein–protein interactions, revealed that biological processes involved in DNA damage and repair, changes in the extracellular matrix, dysregulation of cytokines and growth factors, and rearrangements of the cytoskeleton are represented among the differentially expressed genes. How these processes are specifically related to the development of membranous nephropathy is currently not clear and, to our knowledge, the transcriptome of human membranous nephropathy has not yet been published. However, one of the aims of NEPTUNE is to identify the renal gene expression profiles of participants with membranous nephropathy and we anticipate that the human renal transcriptome of membranous nephropathy will be available in the near future. In the future, this data set could be linked to genomic data, to facilitate genotype–phenotype analysis of membranous nephropathy.5
Proteomic studies
Serial analysis of the urinary proteome profile of rats, in urine samples collected at days 0, 10, 20, 30, 40 and 50 after the induction of PHN,53 revealed that 37 proteins were differentially expressed across all these time points. The differentially expressed proteins were classified into several categories: proteins that decrease in amount during PHN; proteins that increase in amount during PHN; proteins that increase during the early phase of PHN but return to basal levels in the late phase; proteins with undetectable levels during PHN; and proteins that were detectable in the urine only during PHN. Most of the differentially expressed proteins have functions related to signalling pathways, protein trafficking and the regulation of glomerular permeability.
A study of the kidney proteome in rats with PHN, using label-free quantitative proteomic analysis, found a total of 160 proteins that were differentially expressed between control rats and rats with PHN assessed at days 14 and 21 after induction of nephritis.54 These two time points were selected because proteinuria in rats with PHN is marked by day 14 and peaks on day 28. However, only 30% of patients with membranous nephropathy develop severe renal interstitial damage. Interestingly, GRP78 and GRP94 proteins (which are associated with endoplasmic reticulum stress) as well as the autophagy marker LC3 were upregulated on both days 14 and 21, which suggests that endoplasmic reticulum stress and autophagy have an important role in the pathogenesis of membranous nephropathy. Such examinations of the dynamic changes that occur in the evolution of PHN should guide future studies in patients with membranous nephropathy. To our knowledge, the kidney and podocyte proteomes associated with human membranous nephropathy have not yet been published, and investigations of protein-based urinary biomarkers for diagnosis and prediction of the prognosis of human membranous nephropathy are lacking. Although measurements of serum levels of anti-PLA2R antibody are already used to guide the diagnosis and monitor the response to treatment of patients with membranous nephropathy,50,55,56 the identification of additional biomarkers with clinical utility would be useful for management of these patients.57
Metabolomic studies
A parallel metabolomic study conducted on serum and urine samples from patients with membranous nephropathy58 showed that citric acid and four amino acids (L-asparagine, L-serine, L-threonine and pyroglutamic acid) were markedly increased in serum samples from patients with severe proteinuria (>3.5 g/24 h). Levels of dicarboxylic acids, phenolic acids and cholesterol were also substantially elevated in the urine of patients with severe proteinuria.58 The clinical relevance of these findings remains to be determined.
Focal segmental glomerulosclerosis
FSGS is a major cause of proteinuria and renal failure.59 The disease comprises a number of clinical and pathological syndromes that share a common glomerular lesion, including primary (or idiopathic) FSGS and adaptive FSGS (mediated by glomerular hypertension and hyperfiltration) as well as genetic, virus-associated and drug-induced forms.60 Histologically, FSGS has been classified into several subtypes, including tip variant, perihilar variant, cellular variant, collapsing variant, and FSGS not otherwise specified.61,62 HIV-associated nephropathy (HIVAN) is histologically classified as the collapsing variant of FSGS.
Genomic studies
Genetic forms of FSGS can be either monogenetic or polygenetic. Several mutations or polymorphisms, all of which involve genes expressed in podocytes,60 have been linked to monogenic forms of FSGS (Table 4). These genes encode podocyte structural proteins, or enzymes required for processes associated with podocyte homeostasis (such as cytoskeletal organization, cell energetics and cell–cell and cell–matrix interactions).63 Genetic variants have been identified in up to two-thirds of patients with monogenic forms of FSGS who present in the first year of life. For example, a locus at chromosome 14q is associated with increased susceptibility to autosomal-dominant FSGS,64 and sequencing of this chromosomal region identified nine independent missense mutations in INF2 (which encodes inverted formin-2, a member of the formin family of actin-regulating proteins). The observation that alterations in this podocyte-expressed formin can cause FSGS emphasizes the importance of fine regulation of actin polymerization in podocyte function. Interestingly, mutations in INF2 also seem to underlie the increased incidence of FSGS in patients with Charcot–Marie–Tooth neuropathy, which suggests that inverted formin-2 is involved in the functioning of both the glomerulus and the peripheral nervous system.65
Table 4.
Genes associated with FSGS in humans
| Gene | Location | Gene product | Disease | Inheritance mode | OMIM number |
|---|---|---|---|---|---|
| ACTN4 | 19q13 | α-Actinin-4 | FSGS type1 | AD | 603278 |
| NPHS1 | 19q13 | Nephrin | Finnish-type congenital nephropathy, SRNS | AR, sporadic | 256300 |
| NPHS2 | 1q25.32 | Podocin | SRNS type 2 | AR, sporadic | 600995 |
| TRPC6 | 11q21.22 | Short transient receptor potential channel 6 | FSGS type 2 | AD | 603965 |
| CD2AP | 6q12 | CD2-associated protein | SRNS type 4 | AR, AD, sporadic | 600995 |
| PLCE1 | 10q23.24 | Phospholipase Cε-1 | SRNS type 3 | AR | 610725 |
| WT1 | 11p13 | Wilms tumour protein | Denys–Drash syndrome, Frasier syndrome, nephrotic syndrome type 4 | AD | 194080, 136680, 256370 |
| LMX1B | 9q34.1 | LIM homeobox transcription factor 1β | Nail-patella syndrome | AD | 161200 |
| tRNA-Leu | Mitochondria | NA | NA | Maternally inherited | 590050 |
| COQ2 | 4q21.22 | 4-Hydroxybenzoate polyprenyltransferase, mitochondrial | Primary coenzyme Q10 deficiency type 1 | AR | 607426 |
| ITGB4 | 17q25.1 | Integrin β4 | NA | AR | NA |
| LAMB2 | 3p21 | Laminin subunit β2 | Pierson syndrome | AR | 609049 |
| INF2 | 14q32 | Inverted formin-2 | Charcot–Marie–Tooth disease (dominant intermediate type E), FSGS type 5 | AD | 614455, 613237 |
| MYH9 | 22q13.1 | Myosin-9 | Fechtner syndrome, Epstein syndrome | AR | 153640, 153650 |
| MYO1E | 15q21 | Unconventional myosin Ie | FSGS type 6 | AR | 614131 |
| APOL1 | 22q13.1 | Apolipoprotein L1 | Nondiabetic causes of end-stage renal disease, FSGS | Sporadic | 612551 |
| COQ6 | 14q24 | Ubiquinone biosynthesis mono-oxygenase COQ6 | Primary coenzyme Q10 deficiency type 6 | Sporadic | 614647 |
| PTPRO | 12p12.3 | Receptor-type tyrosine-protein phosphatase O | Nephrotic syndrome type 6 | AR | 614196 |
Abbreviations: AD, autosomal dominant; AR, autosomal recessive; FSGS, focal segmental glomerulosclerosis; OMIM, online Mendelian inheritance in man; NA, not available; SRNS, steroid-resistant nephrotic syndrome.
Mutations in the MYO1E gene (which encodes unconventional myosin Ie) can cause childhood-onset, familial, steroid-resistant FSGS.11,12 Whole-genome linkage disequilibrium analysis followed by high-throughput sequencing of the positive-linkage area in a family with autosomal recessive FSGS led to identification of a missense mutation in MYO1E that resulted in the amino acid substitution Ala159Pro.11 Sequencing the MYO1E gene in a second family with FSGS led to identification of another mutation in MYO1E that causes protein truncation (Tyr695X).12 Patients with FSGS who were homozygous for either of these mutations did not respond to glucocorticoid therapy.12 Unconventional myosin Ie is a member of the class 1 myosins—small proteins that interact with cell membranes via their C-terminus, and with actin filaments via their N-terminus motor domain. Unconventional myosin Ie increases actin function, which is likely to be important for podocytes to maintain stretch tension against pulsatile glomerular perfusion pressure.
Six different mutations in COQ6 (which encodes ubiquinone biosynthesis mono-oxygenase COQ6) have been identified in 13 individuals with early onset of steroid-resistant nephrotic syndrome (SRNS) and sensorineural deafness, from seven families.10 COQ6 is required for synthesis of coenzyme Q10, which acts as a redox carrier in the mitochondrial respiratory chain. Knockdown of COQ6 in podocyte cell lines and zebrafish embryos results in apoptosis, an effect that can be partially reversed with coenzyme Q10 treatment. Molecular diagnosis of coenzyme-Q10-related forms of SRNS and hearing loss is feasible and clinically relevant because these patients could potentially benefit from coenzyme Q10 supplementation. The effects of coenzyme Q10 therapy have not been systematically studied in this setting, owing to the rarity of coenzyme-Q10-related SRNS, but anecdotal reports suggest that coenzyme Q10 supplementation has beneficial effects on renal disease in these patients.10
Mice with the FVB/N genetic background are more susceptible than other inbred mouse strains to developing the HIVAN phenotype after infection with HIV-1.66 A locus on chromosome 3 is strongly associated with the HIVAN phenotype in mice. The expression level of podocin, a key functional podocyte protein, is regulated by this locus, as well as by a second susceptibility locus on chromosome 13. These findings in the HIVAN mouse model indicate that HIV-1 infection alters a network of genes (including genes that encode key functional proteins in the podocyte) to cause renal injury in a host with a susceptible genetic background. Similarly, in humans, individuals of African ancestry have a strong predilection to develop HIVAN after infection with HIV-1.
Patients of African ancestry are also predisposed to develop FSGS and hypertensive kidney disease. Two large genetic studies initially identified a strong association between a region on chromosome 22 and increased susceptibility to HIVAN, idiopathic FSGS and hypertensive kidney disease in individuals of African ancestry.67,68 This region of chromosome 22 contains APOL1 (which encodes apolipoprotein L1) as well as MYH9 (which encodes myosin-9). MYH9 was an initially attractive candidate gene for kidney disease, because MYH9 mutations cause several rare diseases associated with autosomal dominant FSGS, including Fechtner syndrome and Epstein syndrome.69,70 In further support of this association, GWAS conducted in African American and European American patients with FSGS showed that genetic variants in the region of chromosome 22 containing both MYH9 and APOL1 were strongly associated with increased risk of FSGS in African American individuals but not in European American individuals.71 However, when the DNA sequence of this region was examined in a larger, worldwide cohort, two APOL1 variants (named G1 and G2), rather than any variants in the MYH9 gene, accounted for the association with FSGS and hypertension-related kidney disease.71 These two APOL1 variants are common in African patients, but absent from European patients with FSGS.
APOL1 encodes a serum factor that lyses Trypanosoma brucei, a protozoan that causes sleeping sickness in humans. Two subspecies of this organism (T. brucei rhodesiense and T. brucei gambiense) have evolved the ability to infect humans because they produce an APOL1 inhibitor. However, the parasite-derived APOL1 inhibitor is not able to bind to the G1 and G2 APOL1 variants. Thus, individuals with these APOL1 variants are resistant to infection with both T. brucei rhodesiense and T. brucei gambiense. Resistance against this parasitic infection could conceivably have conferred positive selection pressure for these two APOL1 variants in African populations. By contrast, how the G1 and G2 APOL1 variants contribute to an increased susceptibility to FSGS, hypertensive kidney disease and HIVAN in patients of African descent remains to be determined.
An association between the presence of the G1 APOL1 allele and a young age at the initiation of haemodialysis in African American individuals has been reported.72 This association is important because early intervention to prevent the progression of kidney disease might benefit this high-risk population. APOL1 genotyping of 271 African American patients with HIVAN, 168 European American patients with HIVAN and 939 healthy control individuals indicated that homozygosity for the APOL1 variants conferred an odds ratio of 17 for developing FSGS and an odds ratio of 29 for developing HIVAN (in a recessive model).73 FSGS in individuals with two APOL1 risk alleles was characterized by an early age of onset and accelerated progression to ESRD, but a similar sensitivity to steroid therapy, when compared with FSGS in patients with no or one APOL1 risk allele. Individuals with two APOL1 risk alleles have an estimated 4% lifetime risk of developing FSGS, and untreated HIV-infected individuals with two APOL1 risk alleles have a 50% risk of developing HIVAN. The effect of carrying two APOL1 risk alleles explains 18% of cases of FSGS and 35% of cases of HIVAN in African populations.73
The exact role of APOL1 in podocyte injury and in the development of FSGS and ESRD, however, remains unknown. Clearly, however, the genetic locus containing MYH9 and APOL1 is associated with development of FSGS and hypertensive kidney disease, and could at least partially account for the increased incidence of these two kidney diseases in patients of African ethnic groups. Even though the G1 and G2 alleles of APOL1 are strongly associated with kidney disease, the reasons why 95% of people with two copies of the G1 or G2 alleles do not develop FSGS remain to be addressed.74 The findings from these genetics studies will, however, be useful in clinical practice to diagnose familial forms of FSGS, to predict response to steroid treatment, and to select patients for coenzyme Q10 treatment. However, we are still far from being able to understand the mechanism of disease in the majority of patients who develop FSGS. To further dissect how disease-associated genes (such as APOL1) are able to induce kidney injury, mechanistic studies in model organisms are required.
Transcriptomic studies
Microarray transcriptomic analysis of mRNA extracted from glomeruli, using archived, formalin-fixed and paraffin-embedded renal biopsy samples from patients with primary classic FSGS, collapsing FSGS, MCD and healthy controls, identified two distinct gene clusters delineating classic FSGS and collapsing FSGS from patients with MCD and controls.75 Comparison of the combined group of individuals with classic FSGS and collapsing FSGS versus the combined group of controls and patients with MCD revealed 316 genes that were significantly differentially regulated (134 were upregulated and 182 were downregulated). Many of these differentially expressed genes encode proteins that are part of the slit diaphragm junctional complex or related to the podocyte. The processes of development, differentiation and morphogenesis, cell motility and migration, cytoskeleton organization and signal transduction are also over-represented among the functions of the differentially regulated genes, according to the results of an ontological analysis. Transcription factors associated with developmental processes are also substantially over-represented in patients with classic and collapsing FSGS, highlighting the importance of reactivation of developmental pathways in the pathogenesis of the disease.
The functional role of the genes, processes and pathways identified by microarray analysis and predicted by bioinformatics to be involved in the development of FSGS need to be experimentally verified using transgenic or knockout animal models.
Novel uses for systems biology
Drug discovery
Steroids and immunosuppressive agents are currently the mainstay of treatment for primary glomerulonephritis. However, owing to the adverse effects associated with long-term use of these agents, the identification of novel drugs or therapeutic targets is urgently needed. The first major hurdle in relation to drug discovery for these kidney diseases is the lack of understanding of the underlying molecular mechanisms. The second major challenge is that most pharmacological agents target only one pathway, whereas most kidney diseases are caused by the activation of multiple pathways or disease processes.
Systems pharmacology (which involves the application of systems biology approaches combining large-scale experimental studies with computational analyses to the study of drugs, their targets, and beneficial as well as adverse effects), is an alternative approach to discovering new drugs to treat complex diseases.76,77 Our research group has used a systems-based approach to identify novel therapeutic targets for HIVAN using Tg26 (HIV-1 transgenic) mice as a model.78 Microarray studies on the kidney cortex of Tg26 mice and their age-matched and sex-matched wild-type littermates identified differentially expressed genes. Bioinformatics analyses led to identification of the upstream signalling pathways, transcription factors and protein kinases that are responsible for regulation of these differentially expressed genes, and highlighted homeodomain-interacting protein kinase 2 (HIPK2) as a master kinase that regulates the altered gene expression in the diseased kidney. We verified that HIPK2 expression was indeed higher in kidneys from mice and humans with glomerular diseases, and showed that knockout mice that do not express HIPK2 are protected from various forms of kidney injury. Our results suggest that a systems biology approach could be useful in the identification of new targets for drug treatment of kidney diseases.
The renal-disease-specific transcriptome can be used to define disease-specific subnetworks within the entire repertoire of molecular interactions in human cells, which can serve as the basis for identifying new drug targets. The Nephromine resource16 already includes an integrated function to screen for potential genes and molecules that could correct the altered gene expression pattern observed in the diseased kidney. In addition, the Connectivity Map database79 has been used to screen for drugs that could mitigate kidney disease. This database contains gene expression microarray signatures from approximately 6,000 experiments testing the effect of about 1,300 (mostly FDA-approved) individual drugs at different concentrations on several human cancer cell lines. In these data, our research group identified pairs of drugs that optimally rectified the altered pattern of gene expression observed in Tg26 mice. We computationally predicted that the combination of an ACE inhibitor and a histone deacetylase inhibitor could maximally normalize the pattern of gene expression in Tg26 mice with HIVAN towards that of healthy mice. Testing of this drug combination in Tg26 mice revealed that the combination of both agents provided additional renal protection compared to treatment with either drug alone, as assessed in terms of the reduction of proteinuria, improvement of renal function and attenuation of kidney injury.80 Our findings indicate that drug-induced changes in expression signatures can be used for predicting the effects of combination drug therapy.
Identifying new targets
A systems-based approach to understanding disease processes also has the potential to discover novel drug targets, which would not be possible with traditional hypothesis-driven approaches solely based on prior knowledge. However, although some systems biology approaches are mostly hypothesis-free (that is, requiring no prior information on which genes are associated with increased disease risk and expression of which proteins or RNA transcripts are altered in disease states), most approaches blend systems biology and hypothesis-based approaches. In most instances, hypothesis-directed interrogation of biological processes using omic-level analysis is followed by interpretation of the resulting data set in the light of prior knowledge. For example, a combination of a hypothesis-based approach (the postulation that serum from patients with membranous nephropathy must contain antibodies directed against the target antigen) and a systems-based approach (mass spectrometry, which generated a list of candidate target antigens corresponding to the 185 kDa band) resulted in identification of the PLA2R antigen involved in membranous nephropathy.50 Importantly, it is very challenging to interpret the results of (and to frame the large number of data points generated from) omics-level analysis without some prior knowledge.
Conclusions
An improved understanding of the molecular basis of kidney disease is needed for the development of disease-specific treatments, the identification of noninvasive molecular markers for the diagnosis and monitoring of affected patients and the formulation of targeted and personalized therapy. Many large omics data sets have been generated for IgAN, membranous nephropathy and FSGS over the past decade. However, the interpretation of these data and their clinical application will require further functional studies to characterize candidate genes and therapeutic targets. Transcriptomic and proteomic data sets for the primary glomerular diseases are currently scarce, primarily because of the limited number of kidney samples that are available for study. Furthermore, the sample size of most published studies (excluding genomic studies) has been small, which limits the interpretation of data and predisposes the analyses to multiple testing biases.
To overcome these deficits, it is critical that we develop national and international consortia to promote strict disease classification criteria, clear criteria for recruitment of patients into clinical trials, standardized protocols for collection of tissue and fluid (kidney biopsy, urine and blood) samples, and for prospective collection of detailed clinical data. Collaborative efforts, such as the ERCB, NEPTUNE and the European Renal Genomic Project, are already underway. In collaboration with NEPTUNE, for example, a biobank containing a large number of kidney, blood, and urine samples from patients with various glomerular diseases (including IgAN, FSGS and membranous nephropathy) is being built in Jinling Hospital, Nanjing, China, and a detailed electronic medical record system has been developed to link clinical information to all samples in the biobank. Samples and clinical data collected from these efforts will be an invaluable resource for future investigations as newer technologies become available.
The ultimate aim of integrating clinical, pathological and molecular information is to develop approaches that further characterize the pathogenetic mechanisms underlying glomerular diseases and to facilitate the development of improved biomarkers and drug targets for the diagnosis and treatment of glomerular disease. To achieve these objectives, a joint collaborative effort of physicians, epidemiologists, molecular biologists, statisticians and systems biologists with computer science and mathematical backgrounds is needed.
Key points.
Novel systems biology techniques that permit comprehensive molecular characterization of biological samples are improving our understanding of the molecular basis underlying primary glomerular diseases
Large-scale genomic studies of glomerular diseases have identified genetic loci that are associated with the development of these diseases and have enriched our understanding of their pathogenesis
Several novel monogenic forms of focal segmental glomerulosclerosis have been identified, which has improved the classification of hereditary and spontaneous variants of this disease and could potentially influence therapeutic decisions
Polymorphisms in APOL1 that increase the risk of developing focal segmental glomerulosclerosis and HIV-associated nephropathy have been identified in African America populations
Clinical application of the findings generated from systems-biology-based analysis is currently limited
Collaborative efforts for recruiting patients, monitoring clinical outcomes and developing standardized protocols for kidney tissue sampling, are needed to study rare primary glomerular diseases
Review criteria.
A search of the English-language literature on applications of systems biology in the primary glomerulonephritides between January 2005 and January 2013 was performed using PubMed. Key terms searched were “nephropathy”, “IgA nephropathy”, “membranous nephropathy”, “FSGS”, “transcriptomics”, “gene expression”, “metabolomics”, “proteomics”, and “systems biology”.
Acknowledgments
S. Jiang, Z.−H. Liu and J. C. He are supported by Chinese 973 fund 2012CB517601NIH. J. C. He is supported by NIH grant numbers 1R01DK088541, 1R01DK078897, P01DK56492 and 1RC4DK090860, and a Veterans Administration Merit Award; P. Y. Chuang is supported by NIH grant number 5K08DK082760.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
S. Jiang, P. Y. Chuang, Z.−H. Liu and J. C. He contributed equally to writing the article. Z.−H. Liu and J. C. He also reviewed and edited the manuscript before submission. All authors contributed to researching data for the article and discussions of its content.
Contributor Information
Song Jiang, Research Institute of Nephrology, Jinling Hospital, Nanjing University School of Medicine, 305 East Zhongshan Road, Nanjing 210002, China.
Peter Y. Chuang, Division of Nephrology, Icahn School of Medicine at Mount Sinai, One Gustave L. Levy Place, New York, NY 10029-6574, USA
Zhi-Hong Liu, Research Institute of Nephrology, Jinling Hospital, Nanjing University School of Medicine, 305 East Zhongshan Road, Nanjing 210002, China.
John C. He, Division of Nephrology, Icahn School of Medicine at Mount Sinai, One Gustave L. Levy Place, New York, NY 10029-6574, USA
References
- 1.United States Renal Data System. 2012 Atlas of CKD and ESRD [online] 2012 http://www.usrds.org/atlas.aspx.
- 2.Zuo L, Wang M. Current burden and probable increasing incidence of ESRD in China. Clin Nephrol. 2010;74 (Suppl 1):S20–S22. [PubMed] [Google Scholar]
- 3.Greenbaum LA, Benndorf R, Smoyer WE. Childhood nephrotic syndrome—current and future therapies. Nat Rev Nephrol. 2012;8:445–458. doi: 10.1038/nrneph.2012.115. [DOI] [PubMed] [Google Scholar]
- 4.He JC, Chuang PY, Ma’ayan A, Iyengar R. Systems biology of kidney diseases. Kidney Int. 2012;81:22–39. doi: 10.1038/ki.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keller BJ, Martini S, Sedor JR, Kretzler M. A systems view of genetics in chronic kidney disease. Kidney Int. 2012;81:14–21. doi: 10.1038/ki.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komorowsky CV, Brosius FC. 3rd, Pennathur, S. & Kretzler, M. Perspectives on systems biology applications in diabetic kidney disease. J Cardiovasc Transl Res. 2012;5:491–508. doi: 10.1007/s12265-012-9382-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jim B, Santos J, Spath F, Cijiang He J. Biomarkers of diabetic nephropathy, the present and the future. Curr Diabetes Rev. 2012;8:317–328. doi: 10.2174/157339912802083478. [DOI] [PubMed] [Google Scholar]
- 8.Starkey JM, Tilton RG. Proteomics and systems biology for understanding diabetic nephropathy. J Cardiovasc Transl Res. 2012;5:479–490. doi: 10.1007/s12265-012-9372-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hildebrandt F. Genetic kidney diseases. Lancet. 2010;375:1287–1295. doi: 10.1016/S0140-6736(10)60236-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heeringa SF, et al. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest. 2011;121:2013–2024. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mele C, et al. MYO1E mutations and childhood familial focal segmental glomerulosclerosis. N Engl J Med. 2011;365:295–306. doi: 10.1056/NEJMoa1101273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanna-Cherchi S, et al. Exome sequencing identified MYO1E and NEIL1 as candidate genes for human autosomal recessive steroid-resistant nephrotic syndrome. Kidney Int. 2011;80:389–396. doi: 10.1038/ki.2011.148. [DOI] [PubMed] [Google Scholar]
- 13.De Spiegelaere W, et al. Quantitative mRNA expression analysis in kidney glomeruli using microdissection techniques. Histol Histopathol. 2011;26:267–275. doi: 10.14670/HH-26.267. [DOI] [PubMed] [Google Scholar]
- 14.Cohen CD, Frach K, Schlondorff D, Kretzler M. Quantitative gene expression analysis in renal biopsies: a novel protocol for a high-throughput multicenter application. Kidney Int. 2002;61:133–140. doi: 10.1046/j.1523-1755.2002.00113.x. [DOI] [PubMed] [Google Scholar]
- 15.Gadegbeku CA, et al. Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int. 2013;83:749–756. doi: 10.1038/ki.2012.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Compendia Bioscience & Personalized Molecular Nephrology Research Laboratory. Nephromine [online] 2012 http://www.nephromine.org.
- 17.Zurbig P, et al. Urinary proteomics for early diagnosis in diabetic nephropathy. Diabetes. 2012;61:3304–3313. doi: 10.2337/db12-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullen W, Delles C, Mischak H. Urinary proteomics in the assessment of chronic kidney disease. Curr Opin Nephrol Hypertens. 2011;20:654–661. doi: 10.1097/MNH.0b013e32834b7ffa. [DOI] [PubMed] [Google Scholar]
- 19.Thongboonkerd V. Biomarker discovery in glomerular diseases using urinary proteomics. Proteomics Clin Appl. 2008;2:1413–1421. doi: 10.1002/prca.200800036. [DOI] [PubMed] [Google Scholar]
- 20.Portilla D, Schnackenberg L, Beger RD. Metabolomics as an extension of proteomic analysis: study of acute kidney injury. Semin Nephrol. 2007;27:609–620. doi: 10.1016/j.semnephrol.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart DS. Metabolomics: a complementary tool in renal transplantation. Contrib Nephrol. 2008;160:76–87. doi: 10.1159/000125935. [DOI] [PubMed] [Google Scholar]
- 22.Hirayama A, et al. Metabolic profiling reveals new serum biomarkers for differentiating diabetic nephropathy. Anal Bioanal Chem. 2012;404:3101–3109. doi: 10.1007/s00216-012-6412-x. [DOI] [PubMed] [Google Scholar]
- 23.Donovan MJ, Costa J, Cordon-Cardo C. Systems pathology: a paradigm shift in the practice of diagnostic and predictive pathology. Cancer. 2009;115:3078–3084. doi: 10.1002/cncr.24353. [DOI] [PubMed] [Google Scholar]
- 24.Lai KN. Pathogenesis of IgA nephropathy. Nat Rev Nephrol. 2012;8:275–283. doi: 10.1038/nrneph.2012.58. [DOI] [PubMed] [Google Scholar]
- 25.Feehally J, et al. HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol. 2010;21:1791–1797. doi: 10.1681/ASN.2010010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gharavi AG, et al. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet. 2011;43:321–327. doi: 10.1038/ng.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu XQ, et al. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet. 2012;44:178–182. doi: 10.1038/ng.1047. [DOI] [PubMed] [Google Scholar]
- 28.Yang C, et al. Genome-wide association study identifies TNFSF13 as a susceptibility gene for IgA in a South Chinese population in smokers. Immunogenetics. 2012;64:747–753. doi: 10.1007/s00251-012-0636-y. [DOI] [PubMed] [Google Scholar]
- 29.Cox SN, et al. Altered modulation of Wnt-β-catenin and PI3K/Akt pathways in IgA nephropathy. Kidney Int. 2010;78:396–407. doi: 10.1038/ki.2010.138. [DOI] [PubMed] [Google Scholar]
- 30.Preston GA, et al. Gene expression profiles of circulating leukocytes correlate with renal disease activity in IgA nephropathy. Kidney Int. 2004;65:420–430. doi: 10.1111/j.1523-1755.2004.00398.x. [DOI] [PubMed] [Google Scholar]
- 31.Woo KT, et al. Urotensin 2 and retinoic acid receptor α (RARA) gene expression in IgA nephropathy. Ann Acad Med Singapore. 2010;39:705–709. [PubMed] [Google Scholar]
- 32.Reich HN, et al. A molecular signature of proteinuria in glomerulonephritis. PLoS ONE. 2010;5:e13451. doi: 10.1371/journal.pone.0013451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokunaga K, et al. Insulin-like growth factor binding protein-1 levels are increased in patients with IgA nephropathy. Biochem Biophys Res Commun. 2010;399:144–149. doi: 10.1016/j.bbrc.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 34.Flyvbjerg A. Role of growth hormone, insulin-like growth factors (IGFs) and IGF-binding proteins in the renal complications of diabetes. Kidney Int Suppl. 1997;60:S12–S19. [PubMed] [Google Scholar]
- 35.Doublier S, et al. Glomerulosclerosis in mice transgenic for human insulin-like growth factor-binding protein-1. Kidney Int. 2000;57:2299–2307. doi: 10.1046/j.1523-1755.2000.00090.x. [DOI] [PubMed] [Google Scholar]
- 36.Weber JA, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patnaik SK, Mallick R, Yendamuri S. Detection of microRNAs in dried serum blots. Anal Biochem. 2010;407:147–149. doi: 10.1016/j.ab.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li JY, Yong TY, Michael MZ, Gleadle JM. Review: the role of microRNAs in kidney disease. Nephrology (Carlton) 2010;15:599–608. doi: 10.1111/j.1440-1797.2010.01363.x. [DOI] [PubMed] [Google Scholar]
- 39.Kato M, Arce L, Natarajan R. MicroRNAs and their role in progressive kidney diseases. Clin J Am Soc Nephrol. 2009;4:1255–1266. doi: 10.2215/CJN.00520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G, et al. Expression of microRNAs in the urinary sediment of patients with IgA nephropathy. Dis Markers. 2010;28:79–86. doi: 10.3233/DMA-2010-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang G, et al. Elevated levels of miR-146a and miR-155 in kidney biopsy and urine from patients with IgA nephropathy. Dis Markers. 2011;30:171–179. doi: 10.3233/DMA-2011-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serino G, Sallustio F, Cox SN, Pesce F, Schena FP. Abnormal miR-148b expression promotes aberrant glycosylation of IgA1 in IgA nephropathy. J Am Soc Nephrol. 2012;23:814–824. doi: 10.1681/ASN.2011060567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park MR, et al. Establishment of a 2D human urinary proteomic map in IgA nephropathy. Proteomics. 2006;6:1066–1076. doi: 10.1002/pmic.200500023. [DOI] [PubMed] [Google Scholar]
- 44.Yokota H, et al. Absence of increased α1-microglobulin in IgA nephropathy proteinuria. Mol Cell Proteomics. 2007;6:738–744. doi: 10.1074/mcp.M600336-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Haubitz M, et al. Urine protein patterns can serve as diagnostic tools in patients with IgA nephropathy. Kidney Int. 2005;67:2313–2320. doi: 10.1111/j.1523-1755.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- 46.He Q, et al. Urinary proteome analysis by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry with magnetic beads for identifying the pathologic presentation of clinical early IgA nephropathy. J Biomed Nanotechnol. 2012;8:133–139. doi: 10.1166/jbn.2012.1364. [DOI] [PubMed] [Google Scholar]
- 47.Rocchetti MT, et al. Urine protein profile of IgA nephropathy patients may predict the response to ACE-inhibitor therapy. Proteomics. 2008;8:206–216. doi: 10.1002/pmic.200700492. [DOI] [PubMed] [Google Scholar]
- 48.Maisonneuve P, et al. Distribution of primary renal diseases leading to end-stage renal failure in the United States, Europe, and Australia/New Zealand: results from an international comparative study. Am J Kidney Dis. 2000;35:157–165. doi: 10.1016/S0272-6386(00)70316-7. [DOI] [PubMed] [Google Scholar]
- 49.Kerjaschki D, Farquhar MG. The pathogenic antigen of Heymann nephritis is a membrane glycoprotein of the renal proximal tubule brush border. Proc Natl Acad Sci USA. 1982;79:5557–5561. doi: 10.1073/pnas.79.18.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beck LH, Jr , et al. M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N Engl J Med. 2009;361:11–21. doi: 10.1056/NEJMoa0810457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanescu HC, et al. Risk HLA-DQA1 and PLA2R1 alleles in idiopathic membranous nephropathy. N Engl J Med. 2011;364:616–626. doi: 10.1056/NEJMoa1009742. [DOI] [PubMed] [Google Scholar]
- 52.Hauser PV, et al. Microarray and bioinformatics analysis of gene expression in experimental membranous nephropathy. Nephron Exp Nephrol. 2009;112:e43–e58. doi: 10.1159/000213505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ngai HH, et al. Serial changes in urinary proteome profile of membranous nephropathy: implications for pathophysiology and biomarker discovery. J Proteome Res. 2006;5:3038–3047. doi: 10.1021/pr060122b. [DOI] [PubMed] [Google Scholar]
- 54.Wang L, et al. Autophagy can repair endoplasmic reticulum stress damage of the passive Heymann nephritis model as revealed by proteomics analysis. J Proteomics. 2012;75:3866–3876. doi: 10.1016/j.jprot.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 55.Beck LH, Jr , et al. Rituximab-induced depletion of anti-PLA2R autoantibodies predicts response in membranous nephropathy. J Am Soc Nephrol. 2011;22:1543–1550. doi: 10.1681/ASN.2010111125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin W, et al. Anti-phospholipase A2 receptor antibody in membranous nephropathy. J Am Soc Nephrol. 2011;22:1137–1143. doi: 10.1681/ASN.2010090967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cravedi P, Ruggenenti P, Remuzzi G. Circulating anti-PLA2R autoantibodies to monitor immunological activity in membranous nephropathy. J Am Soc Nephrol. 2011;22:1400–1402. doi: 10.1681/ASN.2011060610. [DOI] [PubMed] [Google Scholar]
- 58.Gao X, et al. Systematic variations associated with renal disease uncovered by parallel metabolomics of urine and serum. BMC Syst Biol. 2012;6 (Suppl 1):S14. doi: 10.1186/1752-0509-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kitiyakara C, Kopp JB, Eggers P. Trends in the epidemiology of focal segmental glomerulosclerosis. Semin Nephrol. 2003;23:172–182. doi: 10.1053/snep.2003.50025. [DOI] [PubMed] [Google Scholar]
- 60.D’Agati VD, Kaskel FJ, Falk RJ. Focal segmental glomerulosclerosis. N Engl J Med. 2011;365:2398–2411. doi: 10.1056/NEJMra1106556. [DOI] [PubMed] [Google Scholar]
- 61.Thomas DB, et al. Clinical and pathologic characteristics of focal segmental glomerulosclerosis pathologic variants. Kidney Int. 2006;69:920–926. doi: 10.1038/sj.ki.5000160. [DOI] [PubMed] [Google Scholar]
- 62.D’Agati V. Pathologic classification of focal segmental glomerulosclerosis. Semin Nephrol. 2003;23:117–134. doi: 10.1053/snep.2003.50012. [DOI] [PubMed] [Google Scholar]
- 63.Lowik MM, Groenen PJ, Levtchenko EN, Monnens LA, van den Heuvel LP. Molecular genetic analysis of podocyte genes in focal segmental glomerulosclerosis—a review. Eur J Pediatr. 2009;168:1291–1304. doi: 10.1007/s00431-009-1017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown EJ, et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010;42:72–76. doi: 10.1038/ng.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boyer O, et al. INF2 mutations in Charcot–Marie–Tooth disease with glomerulopathy. N Engl J Med. 2011;365:2377–2388. doi: 10.1056/NEJMoa1109122. [DOI] [PubMed] [Google Scholar]
- 66.Gharavi AG, et al. Mapping a locus for susceptibility to HIV-1-associated nephropathy to mouse chromosome 3. Proc Natl Acad Sci USA. 2004;101:2488–2493. doi: 10.1073/pnas.0308649100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kopp JB, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kao WH, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghiggeri GM, et al. Genetics, clinical and pathological features of glomerulonephritis associated with mutations of nonmuscle myosin IIA (Fechtner syndrome) Am J Kidney Dis. 2003;41:95–104. doi: 10.1053/ajkd.2003.50028. [DOI] [PubMed] [Google Scholar]
- 70.Sekine T, et al. Patients with Epstein–Fechtner syndromes owing to MYH9 R702 mutations develop progressive proteinuric renal disease. Kidney Int. 2010;78:207–214. doi: 10.1038/ki.2010.21. [DOI] [PubMed] [Google Scholar]
- 71.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kanji Z, et al. Genetic variation in APOL1 associates with younger age at hemodialysis initiation. J Am Soc Nephrol. 2011;22:2091–2097. doi: 10.1681/ASN.2010121234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kopp JB, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22:2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnstone DB, et al. APOL1 null alleles from a rural village in India do not correlate with glomerulosclerosis. PLoS ONE. 2012;7:e51546. doi: 10.1371/journal.pone.0051546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hodgin JB, et al. A molecular profile of focal segmental glomerulosclerosis from formalin-fixed, paraffin-embedded tissue. Am J Pathol. 2010;177:1674–1686. doi: 10.2353/ajpath.2010.090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berger SI, Iyengar R. Role of systems pharmacology in understanding drug adverse events. Wiley Interdiscip Rev Syst Biol Med. 2011;3:129–135. doi: 10.1002/wsbm.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao S, Iyengar R. Systems pharmacology: network analysis to identify multiscale mechanisms of drug action. Annu Rev Pharmacol Toxicol. 2012;52:505–521. doi: 10.1146/annurev-pharmtox-010611-134520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jin Y, et al. A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nat Med. 2012;18:580–588. doi: 10.1038/nm.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 80.Zhong Y, et al. Renoprotective effect of combined inhibition of angiotensin-converting enzyme and histone deacetylase. J Am Soc Nephrol. 2013;24:801–811. doi: 10.1681/ASN.2012060590. [DOI] [PMC free article] [PubMed] [Google Scholar]