Abstract
During development of the CNS, neurons and glia are generated in a sequential manner. The mechanism underlying the later onset of gliogenesis is poorly understood, although the cytokine-induced Jak-STAT pathway has been postulated to regulate astrogliogenesis. Here, we report that the overall activity of Jak-STAT signaling is dynamically regulated in mouse cortical germinal zone during development. As such, activated STAT1/3 and STAT-mediated transcription are negligible at early, neurogenic stages, when neurogenic factors are highly expressed. At later, gliogenic periods, decreased expression of neurogenic factors causes robust elevation of STAT activity. Our data demonstrate a positive autoregulatory loop whereby STAT1/3 directly induces the expression of various components of the Jak-STAT pathway to strengthen STAT signaling and trigger astrogliogenesis. Forced activation of Jak-STAT signaling leads to precocious astrogliogenesis, and inhibition of this pathway blocks astrocyte differentiation. These observations suggest that autoregulation of the Jak-STAT pathway controls the onset of astrogliogenesis.
During embryonic development, the generation of three major neural cell types (neurons, astrocytes, and oligodendrocytes) in the CNS occurs sequentially, whereby almost all neurons are generated before the appearance of glial cells1,2, with the exception of a few sites of postnatal and adult neurogenesis such as the subgranular zone (SGZ) of the hippocampus and the subventricular zone (SVZ) of the forebrain3. This strategy of building the CNS through sequential production of neurons and glia has become more comprehensible, as recent findings have demonstrated that glial cells are important in critical neuronal maturation processes such as axonal path finding and synapse formation4–6. It is conceivable that delayed or precocious production of glial cells may lead to inappropriate wiring, disorganization, and eventually, dysfunction of the CNS.
The ‘neurons-first, glia-second’ differentiation theme for neural progenitors can be recapitulated in culture. Cortical neural progenitor stem cells isolated from relatively early embryonic stages (for example, mouse embryonic day (E) 10–11) give rise to neurons, not glial cells, after short-term culturing (fewer than 4 d), whereas cortical progenitors isolated from perinatal stages tend to differentiate into astrocytes under the same culture conditions7. In addition, both E10–11 cortical progenitors and embryonic stem cell–derived neural stem or progenitor cells (NSCs or NPCs) switch from being neurogenic to gliogenic over time in vitro8,9, suggesting that the molecular switch for the transition from neurogenesis to gliogenesis may be internally programmed in neural progenitors.
Such internal programs presumably not only orchestrate the order of neuronal and glial differentiation but also dictate cellular differentiation responses of NPCs to extracellular factors. For instance, bone morphogenetic proteins (BMPs) induce neuronal differentiation in primary cultured early (E11–12), neurogenic cortical progenitors, but induce astroglial differentiation in relatively late (E16–17) progenitors10,11. The presence or absence of a single proneural basic helix-loop-helix (bHLH) transcription factor, neurogenin-1 (Ngn1), which specifies neuronal fate and is highly expressed only in the cortical germinal zone during the neurogenic period, determines whether BMPs promote neuronal or glial differentiation12. Other astroglial-inducing factors, such as leukemia inhibitory factor (LIF), Notch-Delta, and basic fibroblast growth factor (bFGF) can initiate the astrocyte differentiation program only in E15 or older cortical progenitors, not in early (for example, E11), neurogenic progenitors8,13–19. In E11 CNS neural progenitors, both Notch-Delta and bFGF function instead as pro-proliferation and anti-differentiation factors15,18,20–22.
LIF-induced Jak-STAT signaling is a critical part of the astrogliogenic machinery23,24. Mouse knockout studies have demonstrated that genetic deficiency in major components of this pathway, including LIF, its receptors LIFRβ and gp130, or the signaling molecules STAT1/3, leads to impaired astroglial differentiation25–27 (Supplementary Fig. 1). Moreover, factors that promote astrogliogenesis, including BMPs, bFGF, and Notch signaling, all require pre-activation of the Jak-STAT pathway15,16,19,28. It was previously postulated that the Jak-STAT pathway is fully active in early neural progenitors. The failure of activated STAT1/3 to initiate glial gene transcription in neurogenic progenitors has been attributed to STAT-independent inhibitory mechanisms8,13. Evidence against this hypothesis has come from the findings that factors inhibiting gliogenesis during the neurogenic period, such as the pro-neural bHLH factors (for example, Ngn1 and Ngn2) and DNA methylation, not only inhibit STAT1/3 from activating glial genes, but also suppress the phosphorylation and activation of the Jak-STAT pathway12 (data not shown). Moreover, forced overexpression of the epidermal growth factor receptor (EGFR) in early progenitors induces precocious astrocyte differentiation in response to LIF by increasing STAT3 protein levels14, which suggests that during the neurogenic period, the Jak-STAT pathway is only weakly activated, if at all.
Here, we report that the Jak-STAT pathway, as part of the astrogliogenic machinery, is indeed dynamically regulated in cortical progenitors during development in vivo. This pathway is inactive during the early neurogenic phase, when neurogenic bHLH factors are highly expressed, but becomes robustly activated during the perinatal (gliogenic) period as the expression of the neurogenic factors is reduced. Forced activation of this pathway can lead to precocious astrocyte differentiation, whereas inhibition of this pathway blocks astrogliogenesis. Notably, we discovered an autoregulatory loop of the Jak-STAT pathway, in which STAT1/3 directly activates the expression of many components of this pathway. This feed-forward loop provides a positive mechanism to direct the progenitor stem cells towards astrogliogenesis. We hypothesize that sufficient activation of the Jak-STAT pathway functions as a switch for the transition from neurogenesis to gliogenesis, which essentially determines the timing for astrocyte differentiation.
RESULTS
Inhibition of astrogliogenesis in neurogenic progenitors
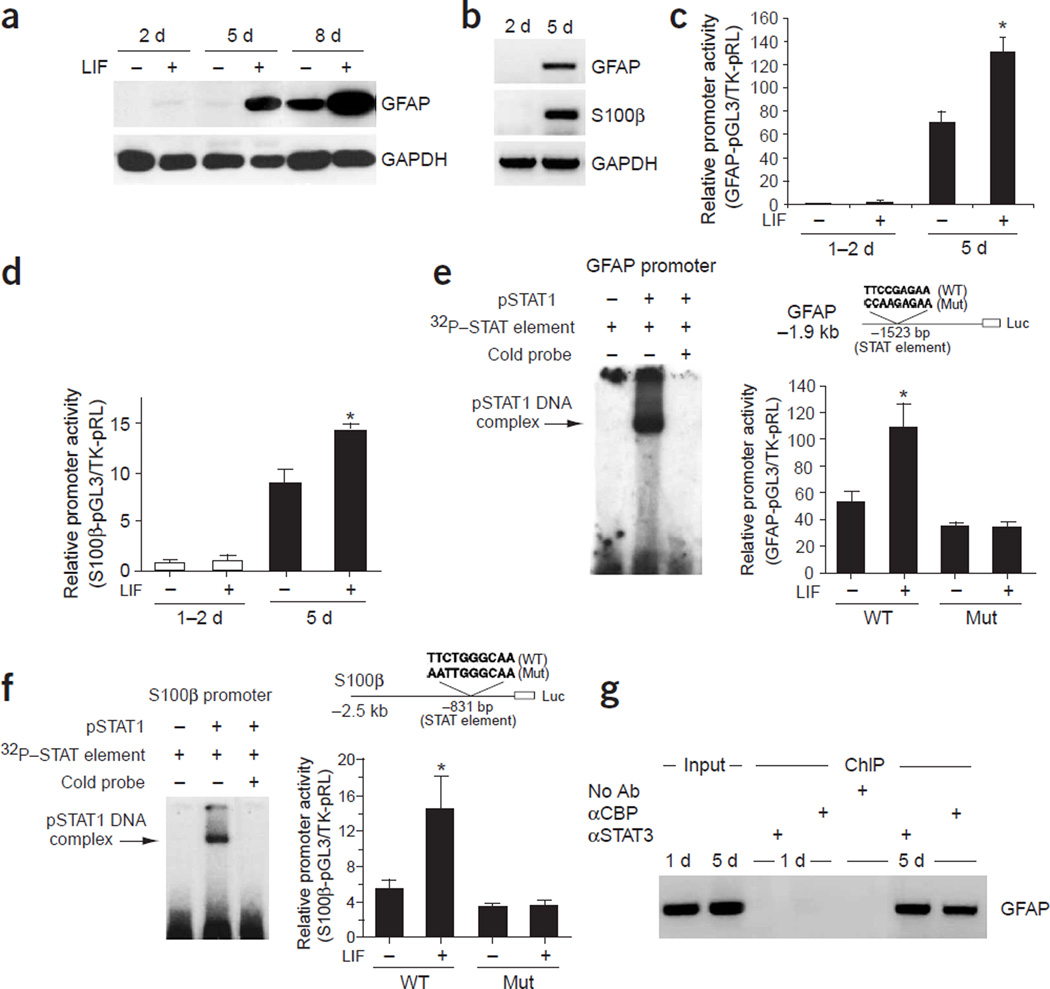
The production of neurons and astrocytes in the developing cerebral cortex, as well as elsewhere in the CNS, occurs in a sequential manner7. At early embryonic stages (for example, mouse E11), cortical progenitors generate only neurons, not glial cells. At perinatal stages, cells that are immunoreactive for the astrocyte marker glial fibrillary acidic protein (GFAP) start to appear throughout the cerebral cortex. Sequential differentiation of neurons and glia can be recapitulated in cultures derived from freshly isolated E11.5mouse cortices. Short-term (<3 d) cultured E11.5 cortical NPCs did not differentiate into astrocytes (Fig. 1a,b). However, with prolonged culturing (>5 d), robust astrogliogenesis did occur, as indicated by RT-PCR, western blot analyses and immunocytochemistry (data not shown), which measured the expression levels of astroglial markers such as GFAP and S100β (Fig. 1a,b).
Figure 1.
The late onset of astrocyte differentiation in vitro. (a) Western blot analysis of primary mouse E11 cortical cells cultured for 2, 5, or 8 DIV with or without LIF (100 ng ml−1) continuous treatment. GAPDH signals were used as loading controls. (b) RT-PCR analysis of the expression of astrocytic markers GFAP and S100β, in 2-DIV or 5-DIV cultured E11 cortical cells. GAPDH RT-PCR signals were used as controls. (c,d) 1.9-kb GFAP and 2-kb S100β promoter luciferase reporter constructs were introduced into E11 cortical cells, either untreated or treated with LIF (100 ng ml−1) for 1 d. Luciferase assays were performed between 1 to 2 DIV or at 5 DIV (with transfection and LIF treatment starting 1 d before harvesting) (*, P < 0.05 as compared to the non-LIF treated group, n ≥ 6). (e,f) Bacterially expressed phosphorylated STAT1 (pSTAT1) binds to the STAT binding site (DNA cis-element) within the GFAP and S100β promoters as determined by electrophoretic mobility shift assays (EMSA). Mutation studies of the STAT binding site indicated that STAT cis-elements are required for the activation of glial specific genes by LIF (*, P < 0.05 compared with non-LIF treated group, n ≥ 6). (g) ChIP assay showing that the association of the STAT3/CBP complex with the GFAP promoter is dynamically regulated, which correlates with the gliogenic potential of the cells. The STAT3/CBP complex associates with the GFAP promoter in 5-DIV, but not in 1-DIV cultured E11 cortical cells with 1 d of LIF treatment before cell harvesting. Ab, antibody. α, antibody against (in all figures).
To investigate the mechanism underlying glial gene regulation, we used a 2-kb proximal S100β promoter and a 1.9-kb proximal GFAP promoter to drive a luciferase reporter. Using luciferase assays, we found that both the GFAP and S100β promoters were not responsive to LIF in 1- to 2-d cultured (1–2 DIV)mouse E11 cortical progenitors, but they became responsive to LIF at 5 DIV (Fig. 1c,d). Within both the GFAP and S100β promoters there are STAT binding cis-elements (Fig. 1e,f) that can bind recombinant tyrosine-phosphorylated STAT1 (pSTAT1) in electrophoretic mobility shift assays (EMSAs). In addition, nucleotide substitution mutations of these two cis-elements abolished LIF responsiveness of the promoters in late (gliogenic) cortical progenitors (Fig. 1e,f). Together, these findings suggest that STAT-mediated transcription is important for astrogliogenesis; however, in early cortical progenitors, STAT-activated transcription of glial genes does not occur. Chromatin immunoprecipitation (ChIP) assays with antibodies against STAT3 or CREB binding protein (CBP), a transcriptional coactivator for STAT1/3 (ref. 28), further indicated that the endogenous GFAP promoter did not associate with STAT3 or CBP in early (neurogenic) cortical progenitors even after LIF treatment. At 5 DIV, in the presence of LIF, both STAT3 and its cofactor, CBP, became robustly associated with the GFAP promoter, presumably activating astroglial gene expression (Fig. 1g).
Dynamic regulation of Jak-STAT in the developing CNS
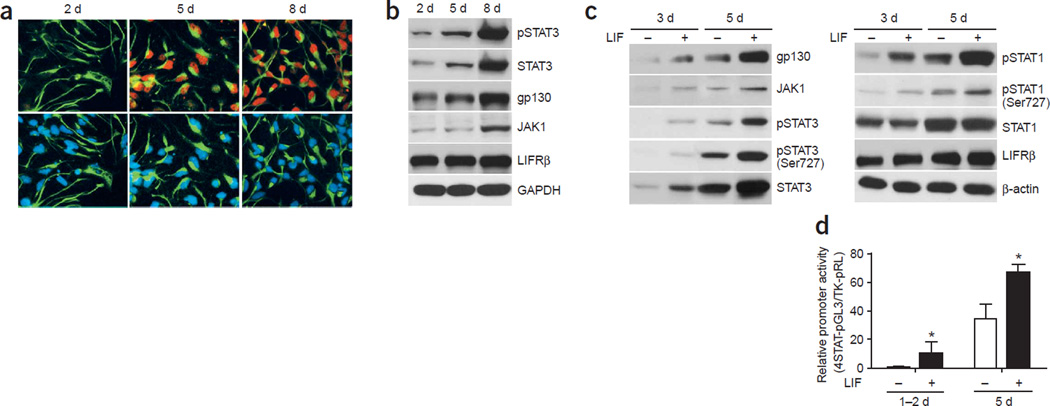
The lack of STAT3 association with the glial genes in early NPCs may result from overall low activity of Jak-STAT signaling (Supplementary Fig. 2) or, alternatively, from other inhibitory mechanisms specific for glial genes, in which case cellular STATs might be fully active8. To distinguish between these two possibilities, we measured the levels of STAT1/3 phosphorylation as well as protein levels of major components of the LIF-induced Jak-STAT pathway in E11 neural progenitors. Immunocytochemical studies with 2-, 5- and 8-d cultured cortical progenitors using a STAT3 Tyr705 phospho-specific antibody24 and an antibody against the neural progenitor marker nestin indicated that LIF did not induce strong STAT3 phosphorylation in short-term cultured E11 cortical cells (Fig. 2a). However, activation and phosphorylation of STAT3 upon LIF treatment became obvious at 5 DIV, and we observed robust activation of STAT3 at 8 DIV in nestin-positive neural progenitors. As noted, the active form of STAT3 (pSTAT3) colocalizes with nestin, suggesting that pSTAT3 signals observed in western blot analyses come primarily from nestin-positive cells in E11 cortical cultures.
Figure 2.
Dynamic regulation of the Jak-STAT pathway during development. (a) Immunocytochemical studies of tyrosine-phosphorylated STAT3 (red) and nestin (green), a neural progenitor cell marker, in 2-, 5-, and 8-DIV cultured E11 cortical cells treated with LIF (100 ng ml−1) for 20 min. Cell nuclei are indicated by DAPI (blue) staining. (b) Western blot analysis of the Jak-STAT pathway of E11 neural progenitor cells at different culturing time periods with 20-min LIF treatment before sample harvesting. (c) Long-term treatment with LIF seems to upregulate the expression of components in the Jak-STAT pathway (LIF was added on the first day of culturing). (d) Activation of a synthetic STAT reporter over time in culture. Luciferase analysis of a synthetic promoter that contains four tandem repeats of the STAT binding elements was used to evaluate the overall STAT-mediated transcription activity in E11 cortical progenitor cells cultured for 1–2 d or 5 d with or without 1-d treatment of LIF (*, P < 0.05 as compared with non–LIF treated group, n ≥ 6).
Our western blot analyses of the NPCs at different time periods during culturing further supported our results from immunocytochemistry by demonstrating that upon LIF stimulation, various components of the Jak-STAT pathway (such as STAT3, the receptor gp130 and the kinase Jak1) were poorly expressed in short-term cultured neurogenic progenitors and gradually became highly expressed after prolonged culturing, as progenitors switched to a gliogenic mode (Fig. 2b). This, in turn, led to robust cytokine-induced activation and phosphorylation of STAT3 (pSTAT3; Fig. 2b). In addition, continuous treatment with exogenous LIF seemed to upregulate the expression of multiple components of the Jak-STAT pathway, including gp130, Jak1, and STAT3, accelerating the gradual strengthening of this pathway (Fig. 2c). As LIF treatment in 3-DIV cultures could partially mimic the effects of long-term (5-DIV) culturing, and as STAT1/3 proteins became phosphorylated in 5 DIV cultures even without LIF treatment (Fig. 2c), it is likely that some endogenously secreted IL-6 cytokine family members gradually became available and were stimulating these cultures to achieve the transition from neurogenesis to gliogenesis. It is noteworthy that in addition to tyrosine phosphorylation of STAT1 (Tyr701) and STAT3 (Tyr705), LIF also induced Ser727 phosphorylation on both STAT1 and STAT3 in 3- to 5-DIV cultures (Fig. 2c), which was important for tyrosine-phosphorylated (and thus dimerized) STAT1/3 to interact with transcriptional cofactors such as CBP, subsequently turning on gene transcription29. As serine phosphorylation of STAT1/3 can be induced by many other cell signaling pathways such as MAPK, PI3K–AKT, and mTOR30,31, the Ser727 phosphorylation process allows for potential cross-talk between the Jak-STAT pathway and other intracellular signaling events.
To further characterize whether the overall low expression of components of the Jak-STAT pathway as well as reduced STAT phosphorylation levels in early NPCs led to low activity of STAT1/3-mediated transcription, we used a synthetic promoter that contains four tandem repeats of the STAT binding element to evaluate overall STAT-mediated transcription activity in early NPCs cultured for different time periods. The promoter-luciferase reporter assays indicated that, similar to what was seen with the GFAP promoter, the synthetic STAT reporter had low activity in early NPCs and became more active and LIF-responsive after prolonged culturing (Fig. 2d). Unlike the GFAP promoter, which could be activated or repressed by either STAT-dependent or STAT-independent mechanisms, the synthetic STAT reporter is a direct readout of STAT trans-activation function. Thus, this result provides direct evidence that the overall activity of the Jak-STAT pathway is low in early (neurogenic) NPCs and gradually becomes elevated as NPCs gain competence for astrogliogenesis in vitro.
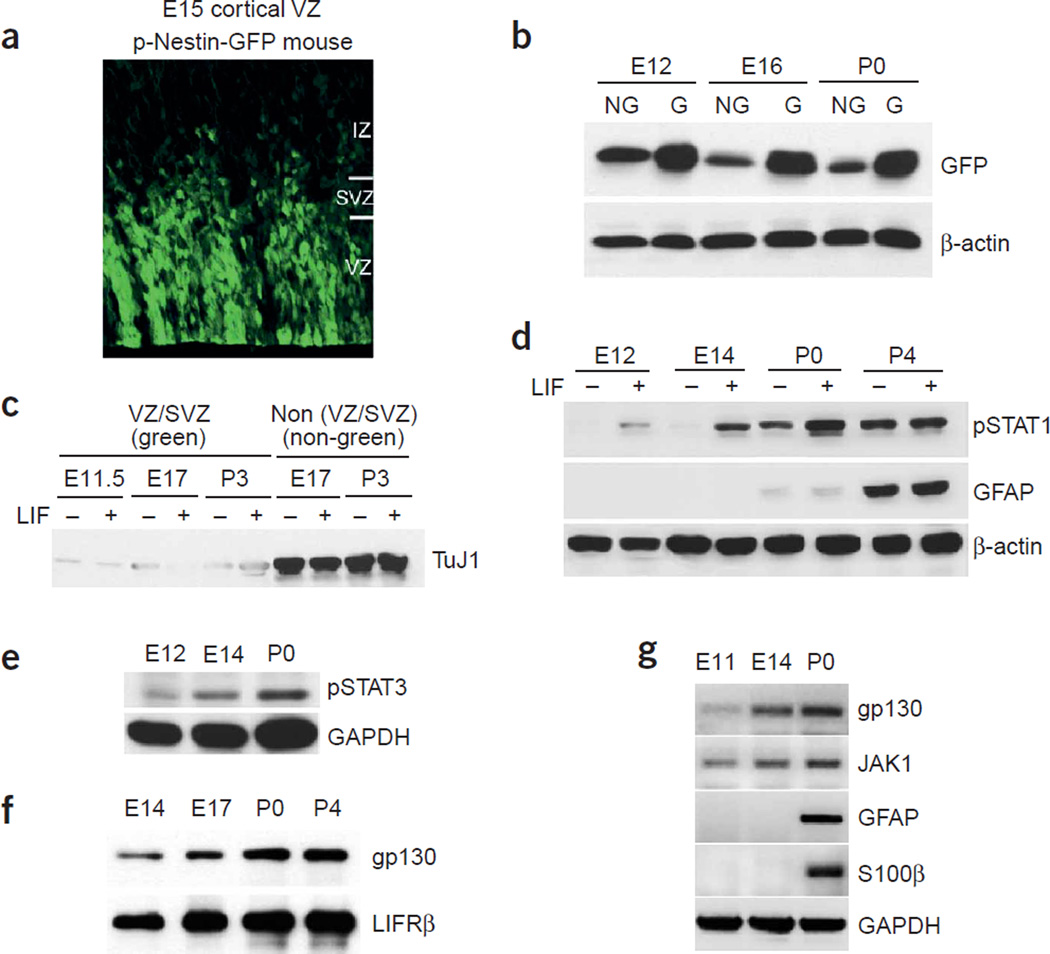
To determine whether dynamic regulation of Jak-STAT signaling also occured in cortical progenitor zones in vivo, we used transgenic mice carrying green fluorescent protein (GFP) driven by the regulatory region of the neural progenitor gene nestin32 (pNestin-GFP mice). The neural progenitor zone of the pNestin-GFP mice emitted green fluorescence under ultraviolet light (Fig. 3a). We performed fluorescence-guided microdissection to isolate tissues from the ventricular zone and/or SVZ (the ‘VZ/SVZ’ tissues) at different developmental stages. In addition, we dissected overlying ‘non-green’ regions to compare cellular compositions (see below). Immediately after dissection, tissues were either snap-frozen or treated with 100 ng ml−1 LIF for 20 min to enhance STAT phosphorylation. When the VZ/SVZ (green) tissues and the non-VZ/SVZ (non-green) tissues were lysed and blotted for GFP and a neuronal marker, the neuron-specific type III β-tubulin, we found enrichment of GFP in the VZ/SVZ tissues (Fig. 3b). Moreover, neurons were essentially depleted from the dissected VZ/SVZ tissues, as indicated by the substantially reduced TuJ1 immunoreactivity in western blot analyses (Fig. 3c), suggesting that GFP fluorescence–assisted dissection of the VZ/SVZ provided enriched populations of cortical progenitors at different developmental time points.
Figure 3.
Sequential activation of the Jak-STAT pathway in vivo correlates with the timing of astrogliogenesis. (a) A fluorescent image of E15 cortical ventricular area from pNestin-GFP mice, demonstrating enriched GFP expression in the ventricular zone (VZ). (b,c) The ventricular zone (green) and non–ventricular zone (non-green) tissues were dissected from different developmental stages (E12, E16 or postnatal day (P) 0 from the pNestin-GFP transgenic mice under a fluorescent dissection microscope. Western blot analyses used a TuJ1 antibody that labels neuronal specific βIII tubulin, and a GFP antibody. β-actin blot indicates the loading control. (d–f) Western blot analyses of STAT activation and astrocyte differentiation in vivo at different developmental stages. After incubation with or without LIF (100 ng ml−1) for 20 min, lysates of green VZ/SVZ tissues from various developmental stages (E12, E14, P0 and P4) were probed with antibodies against tyrosine-phosphorylated STAT1 or STAT3 (d,e), astrocyte marker GFAP (d), gp130, LIFRβ (f), GAPDH (e), and β-actin (d). Lysates from P0 or P4 without 20-min LIF treatment were also enriched for active forms of STAT1 and STAT3. (g) RT-PCR analysis of gp130, Jak1 and astrocytic markers GFAP and S100β at different developmental stages.
We probed the VZ/SVZ tissues for components of the Jak-STAT pathway, phosphorylation of STAT1/3, and expression of astroglial genes and found that the overall activity of the Jak-STAT machinery was low in progenitors during the neurogenic period. Jak-STAT activity became robustly elevated at perinatal stages, when astrogliogenesis was actively ongoing (Fig. 3d–f). The 20-min treatment of exogenous LIF was aimed at determining the responsiveness of the pathway to cytokines, whereas signals from the unstimulated tissues were reflective of endogenous events. mRNA levels of components of the Jak-STAT pathway were correlated with their protein levels (Fig. 3g). Taken together, these findings suggest that the transcriptional regulation of various components of the Jak-STAT pathway is one of the regulatory processes of Jak-STAT signaling, the central control of the astrogliogenic machinery.
A positive autoregulatory loop of the Jak-STAT machinery
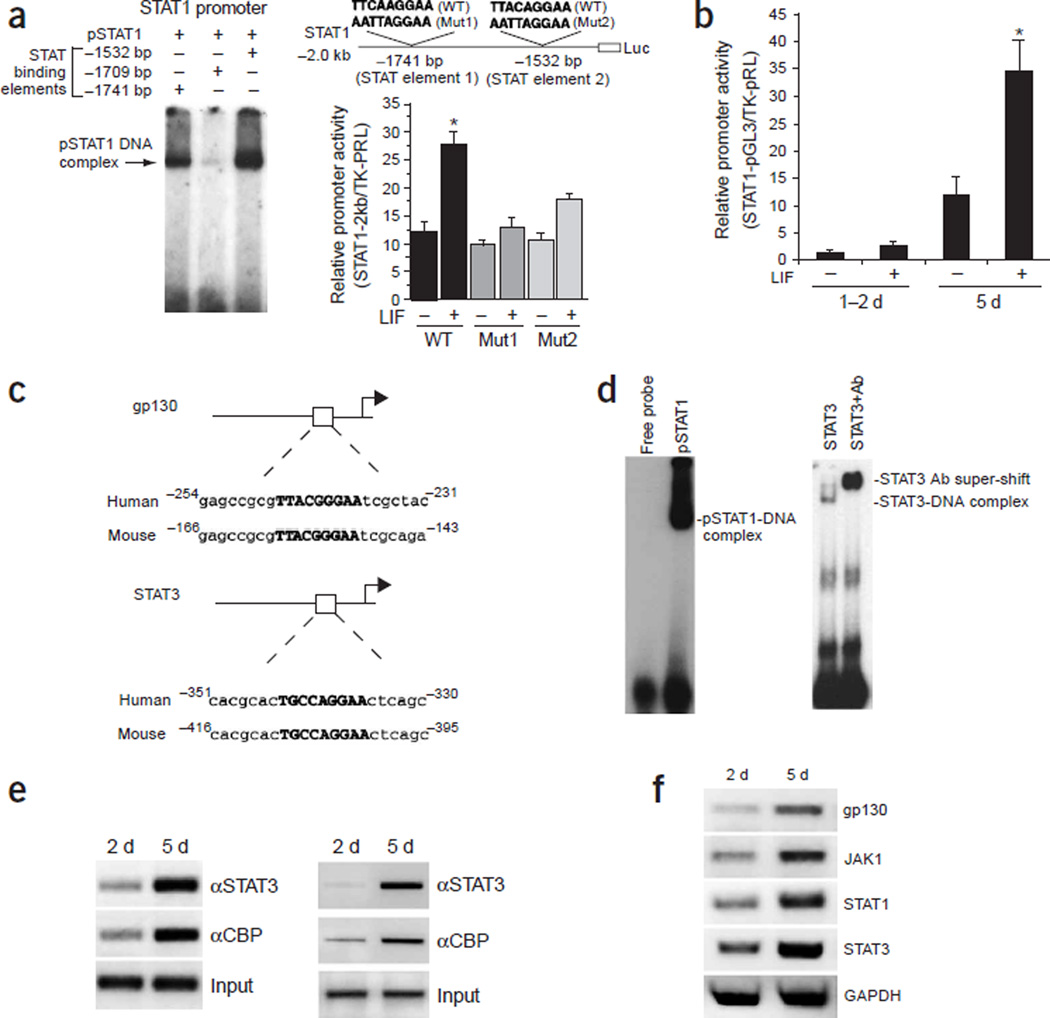
As exogenous LIF treatment can mimic the developmental process by increasing the expression of various components of the Jak-STAT pathway, and as the Jak-STAT pathway is the major signaling cascade mediating the effect of LIF on astrocyte differentiation, we decided to explore whether STAT1/3 could directly upregulate the expression of components of the Jak-STAT pathway. Using the mouse genome database, we performed sequence analyses on the promoter regions of STAT1, STAT3, gp130 and Jak1 in addition to two astrocytic marker genes, GFAP and S100β. We found that all of these promoters contained canonical STAT1/3-binding cis-elements, which are also conserved between mouse and human, suggesting that these promoters could be directly regulated by STAT1/3. Two of the three STAT binding elements within the STAT1 promoter were capable of binding to recombinant pSTAT1 in EMSA assays (Fig. 4a, left). Furthermore, both sites are functional, because nucleotide substitution mutations of either of these two sites led to substantially reduced STAT1 promoter activity in late cortical NPCs in promoter-luciferase reporter assays (Fig. 4a, right). Consistent with the finding that the Jak-STAT pathway has overall lower activity in early (neurogenic) NPCs, the 2.0-kb STAT1 promoter, which contains two functional STAT binding sites and should be responsive to LIF, had very low transcription activity in 1- to 2-DIV E11 cortical cells (Fig. 4b). After prolonged culturing (5 DIV), this promoter became more active and gained LIF responsiveness, just as the GFAP and S100β promoters did. In addition to STAT1, the STAT3 promoter has a functional STAT binding site from −409 bp to –401 bp (5′-TGCCAGGAA-3′) as well33, which is conserved between mouse and human (Fig. 4c). The mouse gp130 promoter also contains a conserved STAT binding site from −158 bp to –150 bp (5′-TTACGGGAA-3′), which bound to recombinant pSTAT1 and STAT3 in EMSA (Fig. 4c,d)34. ChIP assays further indicated that STAT3 could indeed associate with the STAT1 and the gp130 promoters in 5-DIV E11 cortical NPCs (Fig. 4e). In addition, the transcriptional coactivator for STAT1/3 and other transcription factors such as CBP, which contains histone acetyltransferase activity, was more robustly associated with the STAT1 (Fig. 4e, left) and the gp130 promoters (Fig. 4e, right) in 5-DIV NPCs, consistent with the idea that STAT1/3 directly regulates the STAT1 and gp130 genes and strongly activates them when cells become more competent for astrogliogenesis. Similar to gp130 and STAT1, the Jak1 promoter also has a STAT binding element conserved between mouse (from –636 bp to –628 bp; 5′-TTCCTTAAA-3′) and human (from −682 bp to –674 bp; 5′-TTCCTAAAA-3′) in human; (Supplementary Fig. 3). Therefore, this promoter might be regulated in amanner similar to gp130, STAT1, GFAP and S100β. As predicted by the coordinated regulation of the Jak-STAT machinery, gradually increased levels of gp130, Jak1, STAT1 and STAT3 were observed in 1-DIV and 5-DIV E11 cortical NPCs (Fig. 4f).
Figure 4.
The positive autoregulatory loop of the Jak-STAT machinery. (a) STAT1/3 regulates the expression of STAT1. Two (−1741 bp, −1532 bp) of the three STAT binding elements within the 2-kb STAT1 promoter are capable of binding to recombinant pSTAT1 in EMSA assays. Luciferase analysis of the mutagenized STAT1 promoter indicates that both cis-elements are required for LIF induction of the promoter in 5-DIV cultured E11 cortical cells. Mut1, STAT element 1 mutant; Mut2, STAT element 2 mutant. (b) Luciferase analysis of the STAT1 promoter in E11 neural progenitor cell cultures treated with LIF for 1 d at different culturing time points. The STAT1 promoter becomes much more active in the 5-DIV long-term cultures than in 1- to 2-DIV cultures (*, P < 0.05 compared with the rest of the group, n ≥ 6). (c) STAT binding sites within the promoters of gp130 and STAT3 are conserved between human and mouse. (d) STAT1/3 can bind to the putative STAT-responsive element from −158 bp to −150 bp(5′-TTACGGGAA-3′) within the gp130 promoter, as shown by EMSA assay. (e) ChIP analyses showing the developmentally regulated association between STAT3 and CBP with STAT1 (left) and gp130 (right) promoters in E11 primary cortical cell culture at 2 and 5 DIV. (f) RT-PCR of the Jak-STAT signaling components in E11 primary cortical cells cultured for 2 and 5 DIV.
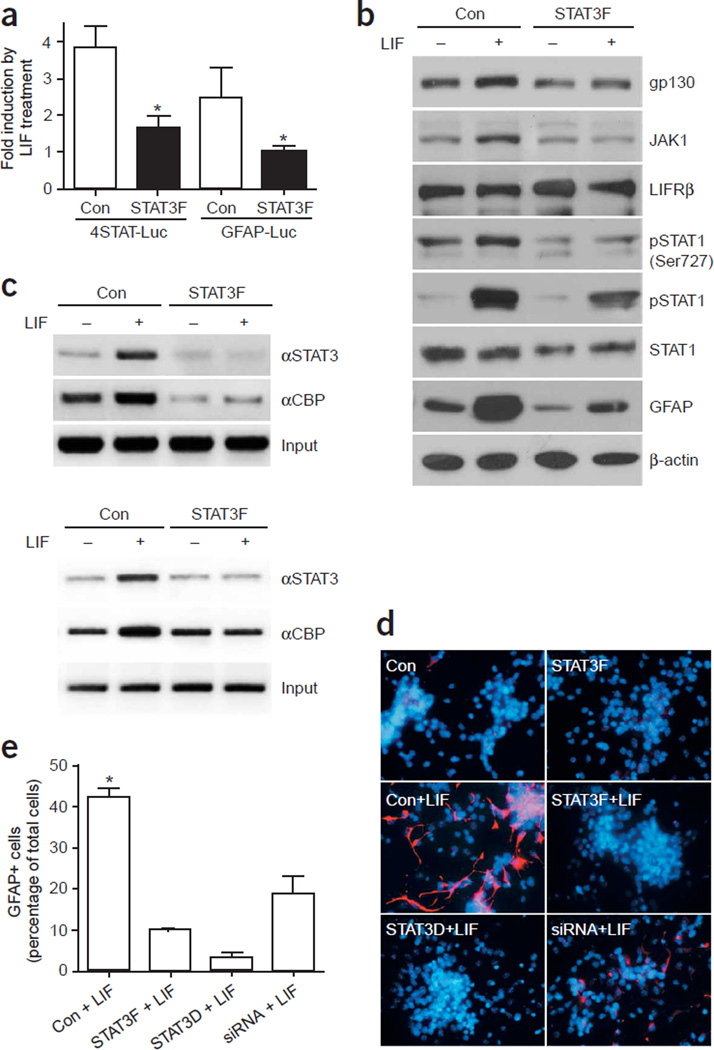
To further test the hypothesis that overall elevated Jak-STAT activity augments the astrogliogenic machinery, we performed both loss- and gain-of-function studies. For loss-of-function studies, we used a dominant interfering form of STAT3, STAT3F, which harbors a mutation of Tyr705 to phenylalanine. STAT3F can be recruited to gp130 and LIFRβ once these receptor subunits are phosphorylated upon LIF stimulation24. However, as STAT3F cannot be phosphorylated on amino acid 705 by Jak1, it cannot dissociate from the receptor and therefore blocks endogenous STAT1/3 from docking to the receptor and becoming phosphorylated by Jak. When we introduced STAT3F into 3-DIV E14.5 cortical cells, it significantly suppressed LIF-triggered activation of the synthetic STAT reporter (Fig. 5a). In addition, STAT3F expression reduced protein levels of gp130, Jak1, and STAT1, as well as both serine and tyrosine phosphorylation of STAT1 (Fig. 5b), suggesting that the overall activity of the Jak-STAT pathway is suppressed by overexpression of the dominant negative STAT3F. As a result, less STAT3 and its coactivator CBP were associated with the GFAP and gp130 promoters, as indicated by ChIP analyses (Fig. 5c). Consistent with the results of the ChIP analyses, the GFAP promoter was less active when STAT3F was expressed (Fig. 5a). Finally, as expected, STAT3F substantially suppressed astroglial differentiation in E14.5 cortical NPCs at 4 DIV (Fig. 5d). Another dominant interfering form of STAT3 is STAT3D, which harbors amino acid mutations in the DNA binding domain that render the mutant STAT3 unable to bind DNA while maintaining the ability to dimerize with wild-type STAT1/3. STAT3D inhibited astrocyte differentiation (Fig. 5d,e). Astroglial differentiation was also inhibited when STAT3 siRNA was introduced into the NPCs (Fig. 5d,e). Moreover, the importance of this LIF-triggered Jak-STAT pathway in astrocyte differentiation in vivo has been further substantiated by knockout studies25–27 (Supplementary Fig. 1), placing this pathway at the center of the astrogliogenic machinery.
Figure 5.
Inhibition of the Jak-STAT pathway suppresses astrogliogenesis. (a) STAT3F (STAT3 Y705F) suppresses LIF-triggered activation of both the synthetic STAT reporter (4STAT-Luc) and the GFAP promoter (GFAP-Luc) in 3-DIV E14.5 cortical cells that were treated with LIF for 1 d (*, P < 0.05 as compared with the rest of the groups, n ≥ 6). (b) Western blot analysis showing protein levels of the components of the Jak-STAT pathway after overexpression of STAT3F in E14.5 cortical cells either left untreated or treated with LIF for 2 d before harvesting at 3 DIV. (c) ChIP analyses demonstrate the association of the STAT/CBP complex with the GFAP (upper panel) and gp130 (lower panel) promoters with or without STAT3F expression in 4-DIV E11 primary neural progenitors treated with LIF for 2 d. (d) Inhibition of astroglial differentiation by various dominant interfering forms of STAT3. Cultures were either left untreated or treated with LIF (100 ng ml−1) for 24 h before fixation at 4 DIV. Cells were stained with an antibody recognizing GFAP (red). Nuclei are shown by Hoechst staining (blue). (e) Quantification of GFAP-positive cells as a percentage of total cells in E14.5 cortical neural progenitor cells after 4 DIV in the presence of LIF, after transfection with control, STAT3F, STAT3D and STAT3 siRNA (*, P < 0.05 as compared with the rest of the groups using one-way ANOVA and Fisher’s post hoc test, n = 6).
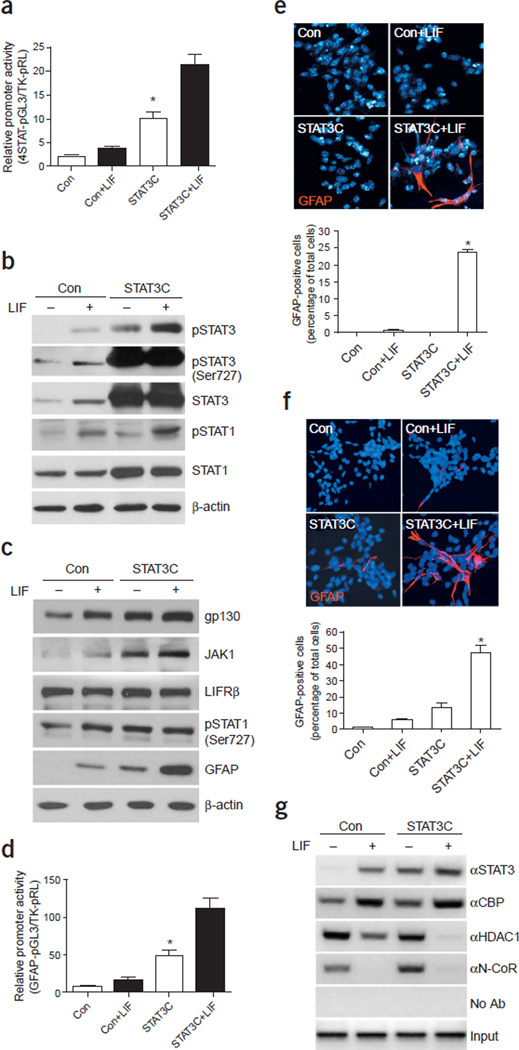
Based on the aforementioned findings, it is conceivable that if the overall activity of the Jak-STAT pathway determines the onset of astrogliogenesis, precocious activation of this pathway using gain-of-function experiments should induce an earlier onset of astrogliogenesis. As the receptor gp130 and the kinase Jak1 are expressed at low levels in early cortical NPCs, overexpression of only STAT1 or 3 may not elevate the overall activity of the pathway immediately, as these factors need to be phosphorylated in order to be functional14. To achieve rapid activation of the pathway, we used a tyrosine phosphorylation–independent, constitutively dimerizing form of STAT3, STAT3C, which was engineered to contain cystine residues within the C-terminal domain, allowing for dimerization through disulfide bonds35. The STAT3C dimer can translocate to the nucleus in a ligand-independent manner. However, its transactivation function still relies on Ser727 phosphorylation35.
The effect of STAT3C on the synthetic STAT reporter is shown in Figure 6a. This reporter had low activity and was not very responsive to LIF in 1-DIV E11 cortical cultures. STAT3C significantly increased the activity of this synthetic promoter (Fig. 6a). LIF stimulation further increased the activity of the promoter, potentially by increasing STAT1/3 serine phosphorylation or by recruiting more endogenous pSTAT1/3 or CBP to the transcriptional machinery36. Western blot analyses indicated that overexpression of STAT3C in 3-DIV E11 cortical NPCs increased the overall activity of the Jak-STAT machinery as reflected by increased tyrosine (Tyr701 or Tyr705) and serine (Ser727) phosphorylation of STAT1/3 as well as increased protein levels of STAT1 and receptor gp130 (Fig. 6b,c). LIF alone caused an increase in serine phosphorylation of both STAT1/3; however, in the presence of STAT3C overexpression, we did not observe an obvious additional increase in S727 phosphorylation. The combination of LIF and STAT3C treatment had a marked effect on GFAP expression as measured by western blotting, luciferase, and immunocytochemical assays (Fig. 6c–f, Supplementary Fig. 4). It is possible that LIF may influence glial gene expression by enhancing the association between STAT3 and CBP, as previously reported36. Consistent with this hypothesis, ChIP analyses indicated that LIF enhanced the recruitment of CBP to the GFAP promoter, likely through STAT3C (Fig. 6g). In addition, LIF also caused a marked decrease in the association between the GFAP promoter and a nuclear corepressor, N-CoR, and a histone deacetylase, HDAC1 (Fig. 6g). Decreased association of both factors may help to open up the chromatin structure, allowing STAT1/3 to successfully activate transcription of glial genes37. Taken together, our data support the notion that the autoregulatory mechanism of the Jak-STAT pathway is central to astroglial differentiation, even though additional factors can influence the activity of this central machinery.
Figure 6.
Constitutively active STAT3C leads to precocious astrocyte differentiation in the presence of LIF. (a) Luciferase assay of the synthetic STAT reporter in the presence of STAT3C with or without LIF for 1 d in E11 cortical culture at 2 DIV (*, P < 0.05 as compared to the rest of the groups, n ≥ 6). (b,c) STAT3C leads to precocious activation of the Jak-STAT pathway. E11 cortical cell culture infected with STAT3C virus for 3 d were analyzed by western blot using antibodies against various components of the Jak-STAT pathway. (d) GFAP promoter activity is increased after overexpression of STAT3C. The combination of 2-d LIF and STAT3C treatment leads to a marked increase in GFAP promoter activity (*, P < 0.05 as compared with the rest of the groups, n ≥ 6). (e) Immunocytochemistry of GFAP expression of E11 cortical cell culture at 2 DIV in the absence or presence of STAT3C expression (adenoviral infection at 1 DIV) with or without 2-d LIF treatment. Glial cells were labeled with antibodies against GFAP (red) and nuclei were stained by Hoechst (blue). Lower panel shows quantification of GFAP-positive cells as a percentage of total cells in the immunostaining experiments (*, P < 0.05 as compared with the rest of the groups, one-way ANOVA, n = 6). (f) GFAP expression of E12 cortical cell culture at 4 DIV in the absence or presence of STAT3C (added at 1 DIV) with or without LIF longterm treatment were analyzed as in e. Quantification of the immunostaining experiments is shown in lower panel (*, P < 0.05 as compared with the rest of the groups, one-way ANOVA, n = 6). (g) ChIP analysis indicates association of STAT3/CBP, HDAC1 and N-CoR with the GFAP promoter in cultured 3-DIV E11 neural progenitor cells infected with the STAT3C virus (viral infection at day 1 and LIF treatment for 2 d).
Reduced expression of proneural genes derepresses Jak-STAT
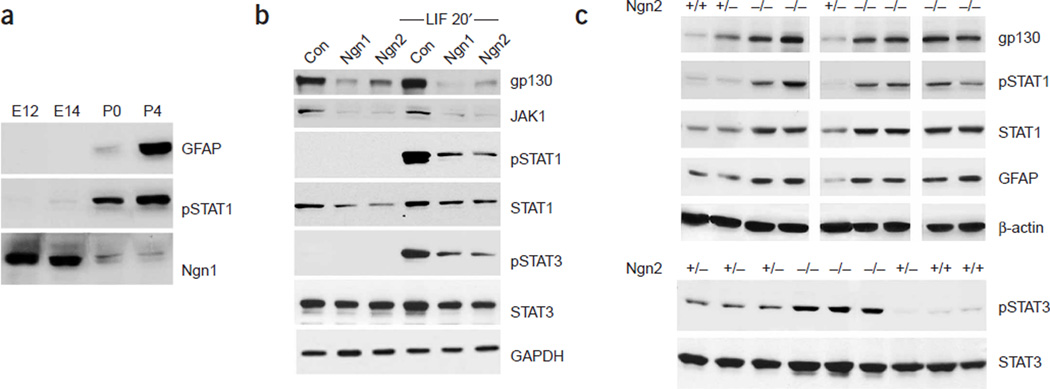
The autoregulatory assembly of the Jak-STAT astrogliogenic pathway is modulated by many intracellular and extracellular factors. The overall activity of the Jak-STAT pathway is suppressed during the neurogenic period but is elevated during the gliogenic phase. We have previously reported that proneural bHLH genes such as Ngn1 are potent inhibitors of the Jak-STAT pathway induced by the IL-6 family of cytokines12. As the expression of the proneural bHLH genes (Ngn1 and Ngn2) in the developing cortical ventricular zone is in reverse correlation with both STAT activity and the appearance of an astroglial marker, GFAP (Fig. 7a, Supplementary Fig. 5), we hypothesized that reduced expression of proneural bHLH genes at later developmental stages would be involved in the activation and derepression of the Jak-STAT pathway. Consistent with this hypothesis, both Ngn1 and Ngn2, when overexpressed in NPCs, strongly suppressed the astrogliogenic Jak-STAT machinery (Fig. 7b). In addition, in Ngn2 knockout mouse cortices, where Ngn1 expression is also reduced because of Ngn2 deficiency38, we observed precocious activation of the astrogliogenic machinery and early onset of astroglial gene expression (Fig. 7c, Supplementary Fig. 6). Taken together, these results indicate that the Jak-STAT pathway, although suppressed during the neurogenic period, was never completely shut off. Our study indicates that reduced expression of the proneural bHLH genes is involved in the activation and derepression of this pathway. In addition to the decreased proneural gene expression, DNA demethylation of some of the components of the Jak-STAT pathway was also involved in derepression of the pathway (data not shown). Thus, although we cannot exclude the possibility that there may be extracellular factors that are involved in activating the Jak-STAT machinery during the neurogenic to gliogenic switch, derepression of this pathway is at least one of the major mechanisms for reactivation of the gliogenic machinery.
Figure 7.
De-repression of the Jak-STAT pathway owing to reduced neurogenin-1 and neurogenin-2 expression during the switch from neurogenesis to gliogenesis. (a) Reciprocal expression of proneural bHLH genes and astroglial differentiation factors in cortical germinal zones during development as shown by western blot analyses of the proneural bHLH gene, Ngn1, astrocyte differentiation–related factors such as tyrosine phosphorylated STAT1 (pSTAT1), and an astrocyte marker, GFAP, in cortical nestin-positive neural epithelial cells at different developmental stages. (b) Gain-of-function experiments demonstrate the suppression of the Jak-STAT signaling components by proneural bHLH genes Ngn1 and Ngn2, as shown by western blot. E11 mouse cortical neural stem cells cultures were infected with control, Ngn1, or Ngn2 adenoviruses, and 24–36 h after infection, the cells were either left untreated or treated with LIF for 20 min before harvesting. (c) Loss-of-function experiments using Ngn2 knockout mice indicate precocious activation of the Jak-STAT machinery and astroglial differentiation in the absence of Ngn2 expression. E14 mouse wild type, Ngn2+/− and Ngn2−/− cortices were dissected, followed by 20 min LIF treatment, and lysed for western blot analyses.
DISCUSSION
The development of the mammalian CNS, including the cerebral cortex, is well organized temporally and spatially39. Previous studies have shown that during cortical development, the neurogenic machinery is largely composed of a cascade of transcriptional activation events mediated by neurogenic bHLH factors. This cascade starts with the proneural genes Neurog1 and Neurog2 (also known as Ngn1 and Ngn2), which activate transcription of downstream neurogenic bHLH factors Neurod1 and Neurod6 (also known as Math2). In turn, Neurod1 and Neurod6 activate Nsc1, another bHLH factor that continues the neurogenic process by turning on terminal neuronal markers40–42. Our studies suggest that the LIF-triggered Jak-STAT pathway is at the center of the astrogliogenic machinery (Supplementary Fig. 7).We have further demonstrated an autoregulatory loop whereby STAT1/3 directly induces the expression of various components of the cytokine-induced Jak-STAT pathway. This autoregulatory loop coordinates different parts of the pathway to synchronize the elevation or suppression of Jak-STAT signaling. Although our data demonstrate a transcriptional activation mechanism, it is likely that post-transcriptional mechanisms could also be a part of the autoregulatory loop.
Previous loss-of-function studies (for instance, gene knockout experiments knocking out ligands, receptors, or signaling molecules) have demonstrated the importance of Jak-STAT signaling in glial differentiation in vivo25–27(Supplementary Fig. 1). However, the Jak-STAT pathway has not been placed at the center of the astrogliogenic machinery, because additional factors and pathways also seem to regulate astrogliogenesis8,11,17,37. When we investigated the mechanisms by which these additional gliogenic factors regulate astrocyte differentiation, we found (either in the literature or by experimentation) that almost all of them cooperate with the Jak-STAT pathway to regulate the astrogliogenic process. For example, BMPs have been shown to synergize with LIF to induce astrogliogenesis through the formation of a potent transactivation complex composed of STAT1/3, CBP and Smad1, which is a signaling molecule downstream of the BMP receptors28. In addition, the Notch canonical pathway has to cooperate with STAT1/3 to enhance astroglial differentiation15. It has also been proposed that a gene downstream of Notch, Hes1, can promote activation and phosphorylation of STAT3 (ref. 16). Moreover, EGFR overexpresssion leads to precocious astroglial differentiation in part by inducing STAT3 expression14.
The neurogenic and the astrogliogenic machinery have an intricate relationship to ensure the sequential differentiation of neurons and glia. During the early (neurogenic) period, the neurogenic bHLH factors inhibit the overall activity of the Jak-STAT pathway, decreasing protein levels of gp130, Jak1, and STAT1 (Supplementary Fig. 7) and thus inhibiting cytokine-triggered STAT1/3 tyrosine phosphorylation12,43. Additionally, the proneural bHLH genes inhibit the transactivation function of STAT1/3 by sequestering the transcriptional coactivator CBP12. At later developmental periods, it has been postulated that neurons may secrete factors to promote astrogliogenesis44. Furthermore, robustly activated Jak-STAT signaling at later stages may, in turn, help to shut off the neurogenic program, completing the switch from neurogenesis to gliogenesis in most CNS regions except for the SVZ of the forebrain and the SGZ of the hippocampus, which harbor ongoing postnatal neurogenesis throughout life45–47. We hypothesize that in these regions, immature astroglial cells generated through Jak-STAT signaling do not go on to terminally differentiate. Instead, they become relatively quiescent and maintain the potential to transform back to neural progenitors. However, during the transition from GFAP-positive ‘stem cells’ to neuronal progenitors48, the postnatal hippocampal proneural gene Neurod1 can also inhibit the Jak-STAT pathway (Supplementary Fig. 8), thus shutting down astroglial-associated genes such as GFAP before the cells can initiate the neuronal differentiation process.
Placing the Jak-STAT pathway at the center of the astrogliogenic machinery and uncovering an autoregulatory loop of this pathway help to set up a framework for understanding the control of astrogliogenesis during development. The dynamic regulation of the Jak-STAT pathway from a weak activation state at the neurogenic phase to a strong activation state at the perinatal stages clearly demonstrates the switching process of progenitors from being neurogenic to gliogenic. However, this autoregulatory loop of Jak-STAT signaling does not operate solely on its own during development. It is already clear that many intracellular and extracellular factors either positively (for example, EGFR) or negatively (for example, Ngn1 or Ngn2 or DNA methylation) regulate this pathway, promoting or suppressing astroglial differentiation in order to achieve the highly ordered generation of neurons and glia to appropriately build the functional CNS.
METHODS
Cell culture and reagent
Timed pregnant Balb/c mice were used to prepare E11 (occasionally, E12 and E14) cortical progenitor cell cultures as described previously15. LIF (100 ng ml−1, R&D Systems) was used for astrocyte differentiation (2-, 4-, or 7-d long-term treatment). Short-term LIF (100 ng ml−1) treatment (20 min) was used to detect STAT1/3 phosphorylation.
Mice
Nestin-GFP transgenic mice on a C57BL/6 background were obtained from J.d.V. as reported previously32. Nestin-GFP positive cells were isolated from different developmental stages (E12, E14, E16, P0, and P4) from the ventricular regions using a fluorescent-dissection microscope (Nikon). After dissection, cells were treated with or without LIF (100 ng ml−1) for 20 min to induce STAT phosphorylation and activation. Neurogenin-2 knockout mice were generated by F.G.’s group38. Mice were handled in accordance with the Animal Research Committee of the University of California, Los Angeles.
Immunocytochemistry
Cell cultures were fixed for 2 min with methanol/acetone (vol/vol 1:1) at room temperature (20–25 °C) and processed for immunofluorescence as described previously15. Primary antibody incubation was performed in a dilution buffer (TBS with 0.02% Tween-20 plus 3% BSA) at 4 °C overnight. Astrocytes were labeled with a mouse monoclonal or a rabbit anti-GFAP antibody (Sigma), neural stem cells were labeled with a mouse monoclonal anti-nestin antibody (Pharmingen) and pSTAT3 was labeled with a rabbit anti-phospho-STAT3 (Cell Signaling). All secondary antibodies (Cy2 and Cy3) were from Jackson Immunoresearch. DAPI or Hoechst staining were used to label nuclei. Images were captured on an Olympus fluorescent microscope.
Immunoblotting
For western assays, cells were rinsed in PBS, lysed in 0.7% NP40 lysis buffer (50 mM Tris-HCl (pH 8.0), 0.1 mM EDTA (pH 8.0), 250 mM NaCl, 10% glycerol, 0.2 mM Na3VO4, 50 mM NaF, 1 mM PMSF, 10 mM DTT and a cocktail of protease inhibitors) and centrifuged at 13,000 rpm for 15 min at 4 °C. After the determination of protein concentration (Bio-Rad), the resulting supernatants were size-separated by 10% SDS-PAGE and transferred to nitrocellulose membranes (BioRad). Western blotting was performed using standard protocols. The antibodies used for western were as follows: mouse monoclonal anti-GFAP (Sigma), mouse monoclonal anti-Jak1 (BD Transduction), rabbit anti-gp130 (Santa Cruz, C-20), rabbit anti-LIFRβ (Santa Cruz, C-19), rabbit anti-STAT1 (gift from K. Shuai, University of California at Los Angeles), mouse monoclonal anti-STAT3 (BD Transduction), mouse monoclonal anti-phosphotyrosine STAT1 (BD Transduction), rabbit anti-phosphotyrosine STAT3 (Cell Signaling), rabbit anti-phosphoserine STAT1 (Biosource), rabbit anti-phosphoserine STAT3 (Biosource), mouse monoclonal TuJ1 (Covance), mouse monoclonal anti-GFP (Roche), mouse monoclonal anti-β-actin (Sigma), mouse monoclonal anti-GAPDH (Abcam), and rabbit anti–neurogenin-1 (gift from J.E. Johnson). Secondary goat anti-mouse or antirabbit IgG-horseradish antibodies (CalBiochem) were used, and detection was performed using the ECL plus chemiluminescence system (PerkinElmer) on X-Omat Blue films (Kodak).
Adenovirus
The STAT3C construct (obtained from J.F. Bromberg, Memorial Sloan Kettering Cancer Center, and J.E. Darnell, Jr., Rockefeller University) was cloned into the BamHI site of an adenoviral shuttle vector pMZL6 containing a GFP expression cassette. STAT3F, a dominant-negative mutant of STAT3, was inserted into the EcoRI site of pMZL6 (ref. 12). Recombinant adenoviruses were made by cotransfection of the shuttle plasmids with the plasmid pBHG10 into HEK293 cells. Viruses were amplified by infecting HEK293 cells and supernatants were harvested, titered, and frozen at −80 °C until infection.
Luciferase reporter assay
The 1.9 kb GFAP promoter–luciferase reporter construct (GFAP-pGL3), its STAT binding site mutant form and dominant-negative forms of STAT3 expressing constructs (STAT3D or STAT3F) have been previously described24. The 4STAT-pGL3 luciferase reporter plasmid contains four copies of the high-affinity binding site for STAT1 or STAT3 followed by the luciferase gene. The mouse S100β and STAT1 promoter were cloned into the pGL3 Basic Vector (Promega). All mutant constructs of S100β-pGL3 or STAT1-pGL3 were created by PCR and verified by DNA sequencing. The TK-pRL Renilla luciferase constructs (Promega) were used as transfection controls in the dual luciferase assays. Transient transfection into neural progenitor cells at different stages was performed using Fugene-6 reagent (Roche) in accordance with the manufacturer’s instructions. STAT3C adenoviruses or LIF (100 ng ml−1) were given 2 h after transfection. Luciferase assays were performed at 24–36 h after transfection using the dual-luciferase reporter system (Promega), and all the results shown indicate luciferase activities normalized against an internal control luciferase reporter of Renilla luciferase (Promega).
RNA analysis
Total RNA was extracted using Trizol reagent (Invitrogen). Reverse transcriptions were performed with 1 µg DNase I–treated RNA as templates using the Omniscript RT Kit (Qiagen). PCR was then performed using primers synthesized by IDT (Supplementary Methods).
Electrophoretic mobility shift assay (EMSA)
Purified bacterially expressed tyrosine-phosphorylated STAT1 was obtained from K. Shuai49. pSTAT1 was incubated with a 32P-labeled STAT1 binding site within the mouse GFAP, S100β, gp130 and STAT1 promoters. Competitive inhibition experiments were performed using a 100-fold molar excess of unlabeled oligonucleotides. Complexes were resolved on 5% non-denaturing polyacrylamide gels, and radiolabelled bands were visualized by autoradiography.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation was performed on neural progenitor cells based on the protocol generated by Upstate Biotechnologies. Neural progenitor cells grown on 100-mm dishes were left untreated or were treated with LIF (100 ng ml−1) for indicated durations and cross linked using 1% formaldehyde for 10 min at room temperature (20–25 °C). ChIP primers are: 5′-GAC TAA GCT GTT TCC TCG GC-3′, 5′-TGA GGT CAC TGT ACC CAG AG-3′ (for the GFAP promoter CBF site); 5′-TTG TGC CCA CTG AAT GAC TC-3′, 5′-GCA GTA CAA GCT CCC AGC TC-3′ (for the GFAP promoter STAT site); 5′-GCG ACC CAT TAC CCT AGA GA-3′, 5′-GCT CAT TGG CTC TGG TCA GT-3′ (for the gp130 promoter STAT site); 5′-GAC AGA GGG ATG TCC TGC-3′, 5′-CTT CGG ACC TCC ACT GAC-3′ (for the STAT1 promoter STAT sites). Antibodies used for chromatin immunoprecipitation assays were rabbit anti-STAT3 (Santa Cruz, C-20), rabbit anti-CBP (Santa Cruz, A-22), rabbit anti-HDAC1 (Upstate), rabbit anti-N-CoR (gift from J. Wong, Baylor College of Medicine).
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank L. Zipursky, H. Herschman and K. Shuai at UCLA for critical reading of the manuscript and providing suggestions, and L. Hutnick at UCLA for cloning the S100β promoter. We would like to acknowledge J. Wong (Baylor College of Medicine), J.E. Johnson (University of Texas South western), D. Levy (New York University), K. Shuai (University of California at Los Angeles), J.E. Darnell, Jr. (Rockefeller University), J. Bromberg (Memorial Sloan-Kettering Cancer Center) and S.C. Landis (National Institute of Neurological Disorders and Stroke) for sharing critical reagents. This work is supported by National Institutes of Health (NIH) RO1 grant (MH066196), a Beckman Young Investigator Award, a Sloan Research Fellowship, a Klingenstein award and a National Alliance for Research on Schizophrenia and Depression award to Y.E.S., NIH program project grant (HD006576) to J.d.V. and Y.E.S. and NIH NS44405 to G.F.
Footnotes
Note: Supplementary information is available on the Nature Neuroscience website.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Sauvageot CM, Stiles CD. Molecular mechanisms controlling cortical gliogenesis. Curr. Opin. Neurobiol. 2002;12:244–249. doi: 10.1016/s0959-4388(02)00322-7. [DOI] [PubMed] [Google Scholar]
- 2.Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 3.Temple S, Alvarez-Buylla A. Stem cells in the adult mammalian central nervous system. Curr. Opin. Neurobiol. 1999;9:135–141. doi: 10.1016/s0959-4388(99)80017-8. [DOI] [PubMed] [Google Scholar]
- 4.Barres BA, Smith SJ. Neurobiology. Cholesterol—making or breaking the synapse. Science. 2001;294:1296–1297. doi: 10.1126/science.1066724. [DOI] [PubMed] [Google Scholar]
- 5.Ullian EM, Sapperstein SK, Christopherson KS, Barres BA. Control of synapse number by glia. Science. 2001;291:657–661. doi: 10.1126/science.291.5504.657. [DOI] [PubMed] [Google Scholar]
- 6.Shu T, Richards LJ. Cortical axon guidance by the glial wedge during the development of the corpus callosum. J. Neurosci. 2001;21:2749–2758. doi: 10.1523/JNEUROSCI.21-08-02749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian X, et al. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- 8.Takizawa T, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 9.Sun YE, Martinowich K, Ge W. Making and repairing the mammalian brain–signaling toward neurogenesis and gliogenesis. Semin. Cell Dev. Biol. 2003;14:161–168. doi: 10.1016/s1084-9521(03)00007-7. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Cogswell CA, LoTurco JJ. Neuronal differentiation of precursors in the neocortical ventricular zone is triggered by BMP. J. Neurosci. 1998;18:8853–8862. doi: 10.1523/JNEUROSCI.18-21-08853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mabie PC, Mehler MF, Kessler JA. Multiple roles of bone morphogenetic protein signaling in the regulation of cortical cell number and phenotype. J. Neurosci. 1999;19:7077–7088. doi: 10.1523/JNEUROSCI.19-16-07077.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y, et al. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- 13.Molne M, et al. Early cortical precursors do not undergo LIF-mediated astrocytic differentiation. J. Neurosci. Res. 2000;59:301–311. doi: 10.1002/(sici)1097-4547(20000201)59:3<301::aid-jnr3>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 14.Viti J, Feathers A, Phillips J, Lillien L. Epidermal growth factor receptors control competence to interpret leukemia inhibitory factor as an astrocyte inducer in developing cortex. J. Neurosci. 2003;23:3385–3393. doi: 10.1523/JNEUROSCI.23-08-03385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge W, et al. Notch signaling promotes astrogliogenesis via direct CSL-mediated glial gene activation. J. Neurosci. Res. 2004;69:848–860. doi: 10.1002/jnr.10364. [DOI] [PubMed] [Google Scholar]
- 16.Kamakura S, et al. Hes binding to STAT3 mediates crosstalk between Notch and Jak-STAT signalling. Nat. Cell Biol. 2004;6:547–554. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- 17.Tanigaki K, et al. Notch1 and Notch3 instructively restrict bFGF-responsive multipotent neural progenitor cells to an astroglial fate. Neuron. 2001;29:45–55. doi: 10.1016/s0896-6273(01)00179-9. [DOI] [PubMed] [Google Scholar]
- 18.Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 19.Song MR, Ghosh A. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat. Neurosci. 2004;7:229–235. doi: 10.1038/nn1192. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn HG, Winkler J, Kempermann G, Thal LJ, Gage FH. Epidermal growth factor and fibroblast growth factor-2 have different effects on neural progenitors in the adult rat brain. J. Neurosci. 1997;17:5820–5829. doi: 10.1523/JNEUROSCI.17-15-05820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartlett PF, et al. Regulation of neural stem cell differentiation in the forebrain. Immunol. Cell Biol. 1998;76:414–418. doi: 10.1046/j.1440-1711.1998.00762.x. [DOI] [PubMed] [Google Scholar]
- 22.Chambers CB, et al. Spatiotemporal selectivity of response to Notch1 signals in mammalian forebrain precursors. Development. 2001;128:689–702. doi: 10.1242/dev.128.5.689. [DOI] [PubMed] [Google Scholar]
- 23.Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 1996;10:3129–3140. doi: 10.1101/gad.10.24.3129. [DOI] [PubMed] [Google Scholar]
- 24.Bonni A, et al. Regulation of gliogenesis in the central nervous system by the Jak-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- 25.Bugga L, Gadient RA, Kwan K, Stewart CL, Patterson PH. Analysis of neuronal and glial phenotypes in brains of mice deficient in leukemia inhibitory factor. J. Neurobiol. 1998;36:509–524. doi: 10.1002/(sici)1097-4695(19980915)36:4<509::aid-neu5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 26.Koblar SA, et al. Neural precursor differentiation into astrocytes requires signaling through the leukemia inhibitory factor receptor. Proc. Natl. Acad. Sci. USA. 1998;95:3178–3181. doi: 10.1073/pnas.95.6.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakashima K, et al. Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J. Neurosci. 1999;19:5429–5434. doi: 10.1523/JNEUROSCI.19-13-05429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakashima K, et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- 29.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 30.Yokogami K, Wakisaka S, Avruch J, Reeves SA. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr. Biol. 2000;10:47–50. doi: 10.1016/s0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- 31.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi M, Saito H, Suzuki M, Mori K. Visualization of neurogenesis in the central nervous system using nestin promoter-GFP transgenic mice. Neuroreport. 2000;11:1991–1996. doi: 10.1097/00001756-200006260-00037. [DOI] [PubMed] [Google Scholar]
- 33.Ichiba M, Nakajima K, Yamanaka Y, Kiuchi N, Hirano T. Autoregulation of the Stat3 gene through cooperation with a cAMP-responsive element-binding protein. J. Biol. Chem. 1998;273:6132–6138. doi: 10.1074/jbc.273.11.6132. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien CA, Manolagas SC. Isolation and characterization of the human gp130 promoter. Regulation by STATS. J. Biol. Chem. 1997;272:15003–15010. doi: 10.1074/jbc.272.23.15003. [DOI] [PubMed] [Google Scholar]
- 35.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 36.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 37.Hermanson O, Jepsen K, Rosenfeld MG. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- 38.Nieto M, Schuurmans C, Britz O, Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- 39.Bayer SA. Neocortical Development. New York: Raven; 1991. [Google Scholar]
- 40.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 41.Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- 42.Mattar P, et al. A screen for downstream effectors of Neurogenin2 in the embryonic neocortex. Dev. Biol. 2004;273:373–389. doi: 10.1016/j.ydbio.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 43.Turnley AM, Faux CH, Rietze RL, Coonan JR, Bartlett PF. Suppressor of cytokine signaling 2 regulates neuronal differentiation by inhibiting growth hormone signaling. Nat. Neurosci. 2002;5:1155–1162. doi: 10.1038/nn954. [DOI] [PubMed] [Google Scholar]
- 44.Morrow T, Song MR, Ghosh A. Sequential specification of neurons and glia by developmentally regulated extracellular factors. Development. 2001;128:3585–3594. doi: 10.1242/dev.128.18.3585. [DOI] [PubMed] [Google Scholar]
- 45.Doetsch F. The glial identity of neural stem cells. Nat. Neurosci. 2003;6:1127–1134. doi: 10.1038/nn1144. [DOI] [PubMed] [Google Scholar]
- 46.Doetsch F. A niche for adult neural stem cells. Curr. Opin. Genet. Dev. 2003;13:543–550. doi: 10.1016/j.gde.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 47.Alvarez-Buylla A, Garcia-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat. Rev. Neurosci. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- 48.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 49.ten Hoeve J, et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell. Biol. 2002;22:5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.