Abstract
Parent-of-origin-specific expression at imprinted genes is regulated by allele-specific DNA methylation at imprinting control regions (ICRs). This mechanism of gene regulation, where one element controls allelic expression of multiple genes, is not fully understood. Furthermore, the mechanism of gene dysregulation through ICR epimutations, such as loss or gain of DNA methylation, remains a mystery. We have used genetic mouse models to dissect ICR-mediated genetic and epigenetic regulation of imprinted gene expression. The H19/insulin-like growth factor 2 (Igf2) ICR has a multifunctional role including insulation, activation and repression. Microdeletions at the human H19/IGF2 ICR (IC1) are proposed to be responsible for IC1 epimutations associated with imprinting disorders such as Beckwith–Wiedemann syndrome (BWS). Here, we have generated and characterized a mouse model that mimics BWS microdeletions to define the role of the deleted sequence in establishing and maintaining epigenetic marks and imprinted expression at the H19/IGF2 locus. These mice carry a 1.3 kb deletion at the H19/Igf2 ICR [Δ2,3] removing two of four CCCTC-binding factor (CTCF) sites and the intervening sequence, ∼75% of the ICR. Surprisingly, the Δ2,3 deletion does not perturb DNA methylation at the ICR; however, it does disrupt imprinted expression. While repressive functions of the ICR are compromised by the deletion regardless of tissue type, insulator function is only disrupted in tissues of mesodermal origin where a significant amount of CTCF is poly(ADP-ribosyl)ated. These findings suggest that insulator activity of the H19/Igf2 ICR varies by cell type and may depend on cell-specific enhancers as well as posttranslational modifications of the insulator protein CTCF.
INTRODUCTION
A relatively small number of genes are regulated by genomic imprinting in mammals and these genes are expressed exclusively or predominantly from a single parental allele. Most of these genes are localized in clusters under the control of an imprinting control region (ICR), which is marked with its parental origin during gametogenesis, largely through the use of DNA methylation (1). The central role of ICRs is demonstrated when these sequences are naturally or experimentally deleted or mutated, leading to the misexpression of multiple genes in the region. In humans, aberrant expression of imprinted genes is associated with a number of imprinting disorders, including the neurobehavioral disorders Prader-Willi and Angelman syndromes as well as the growth disorders Beckwith–Wiedemann (BWS) and Silver–Russell (SRS) syndromes (2).
One of the most widely studied imprinting clusters is the H19/insulin-like growth factor 2 (Igf2) locus, which maps to the distal part of mouse chromosome 7 and is in conserved synteny with human chromosome 11p15 (3). In both mouse and humans, the non-coding RNA gene H19 is maternally expressed and associated with growth suppression. H19 is located adjacent to the paternally expressed insulin-like growth factor 2 (Igf2) gene. In the mouse, the ∼2 kb ICR resides 2 kb upstream of the start of H19 gene transcription and exhibits epigenetic properties that are specific to each of the two parental chromosomes. The mouse ICR (also designated differentially methylated domain or DMD) is methylated on the paternal allele, while the maternal chromosome is unmethylated and bound by the insulator protein CCCTC-binding factor (CTCF). On the maternal ICR allele, CTCF binds to four consensus sites and functions as an insulator and enhancer-blocker that prevents Igf2 from accessing the shared enhancers located 3′ of the two genes, thereby allowing H19 exclusive enhancer engagement on the maternal allele. ICR methylation on the paternal allele leads to hypermethylation of the H19 promoter and repression of H19. Because CTCF cannot bind methylated DNA, Igf2 has sole access to shared enhancers. Whereas many of the aspects of regulation at this locus have been ascertained through mouse modeling [as reviewed by Ideraabdullah et al. (3)], mutations recently identified in BWS patients reveal the presence of other critical imprinting features at the human H19/IGF2 locus (4–8).
BWS is a congenital disorder associated with a number of pre- and postnatal developmental phenotypes including whole body overgrowth, enlarged organs, body asymmetry (hemihyperplasia) and an increased incidence of Wilms' tumor and hepatoblastoma. Macrosomia (excessive birth weight) and macroglossia (enlarged tongue) are two of the most common phenotypes associated with BWS and are usually present at birth, although they have been reported to develop postatally in rare circumstances (9). These overgrowth phenotypes become less apparent in most BWS patients by 7–8 years of age as does their risk for tumor development (10). Eighty-five percent of BWS cases are sporadic and not all phenotypes are present in every patient [reviewed by Choufani (11)]. Phenotypic heterogeneity can be attributed in part to somatic mosaicism and also to the suggestion that various phenotypes are correlated with different genetic/epigenetic abnormalities (11).
The majority of individuals with BWS have mutations in the KCNQ1 imprinting cluster, located adjacent to H19/IGF2 on human chromosome 11 (12). However, a subset of BWS cases are associated with DNA methylation defects at the H19/IGF2 ICR that lead to overexpression of IGF2 and/or loss of H19 expression (13). Importantly, individuals with BWS linked to H19/IGF2 epimutations are also at increased risk for developing Wilms tumors (14). In some BWS patients, microdeletions have been reported to coincide with the epimutations at the human ICR (designated IC1) (15–17). BWS microdeletions typically range from 0.8 to 2.2 kb, removing 1–3 CTCF-binding sites and the intervening sequences (5). Maternal transmission of the deletions is associated with in a mosaic gain of DNA methylation at the IC1 and biallelic IGF2 expression. Interestingly, maternal transmission of the larger 2.2 kb deletion seemingly perturbs insulator function at the locus to a lesser extent compared with the shorter 0.8–1.8 kb deletions (5,18). The source of these microdeletions and their role in the IC1 DNA methylation defects remains under investigation. However, it is proposed that these microdeletions perturb an imprinting mechanism that relies on spacing of CTCF sites (17).
The conservation in mechanism of imprinting at the H19/IGF2 locus between human and mouse is likely highly dependent on conservation of CTCF sites because the ICR size and sequence between CTCF sites is highly divergent (19,20). Not only are the sites conserved but the bipartite pattern of CTCF site spacing, 2 : 2 in the mouse and 3 : 4 in humans, is also conserved (21–23). This spacing is disrupted in BWS microdeletions at the H19/IGF2 ICR but it has yet to be experimentally determined whether this is the cause of the ICR DNA methylation defects associated with BWS.
To address this question, we generated a targeted mouse model of the BWS microdeletion. We have previously successfully modeled BWS and SRS loss of imprinting mutations in the mouse using targeted single base pair substitutions or large deletions at the ICR (24–26). To specifically model the BWS microdeletions in the mouse by disrupting CTCF spacing and assess the role of this mutation in ICR methylation perturbation, we generated a 1.3 kb deletion at the endogenous 2 kb mouse locus, designated H19ICRΔ2,3, by homologous recombination. Whereas paternal transmission of the deletion causes biallelic H19 expression regardless of the tissue, maternal transmission results in a tissue-specific loss of Igf2 imprinting such that imprinting is maintained in endodermal tissues but disrupted in tissues of mesodermal origin. CTCF protein levels are not significantly different in mesodermal and endodermal derivatives despite the observation that in mesodermal tissues, CTCF has additional posttranslational modifications and significantly reduced binding to the mutant ICR relative to wild type. Finally, maternal transmission of the Δ2,3 deletion does not cause DNA methylation defects, suggesting that the deletion alone may not be sufficient to cause the methylation defects in humans and supporting previous results demonstrating that changes in DNA methylation at ICRs are not required to perturb imprinted expression.
RESULTS
Generation and targeting of the H19ICRΔ2,3 allele
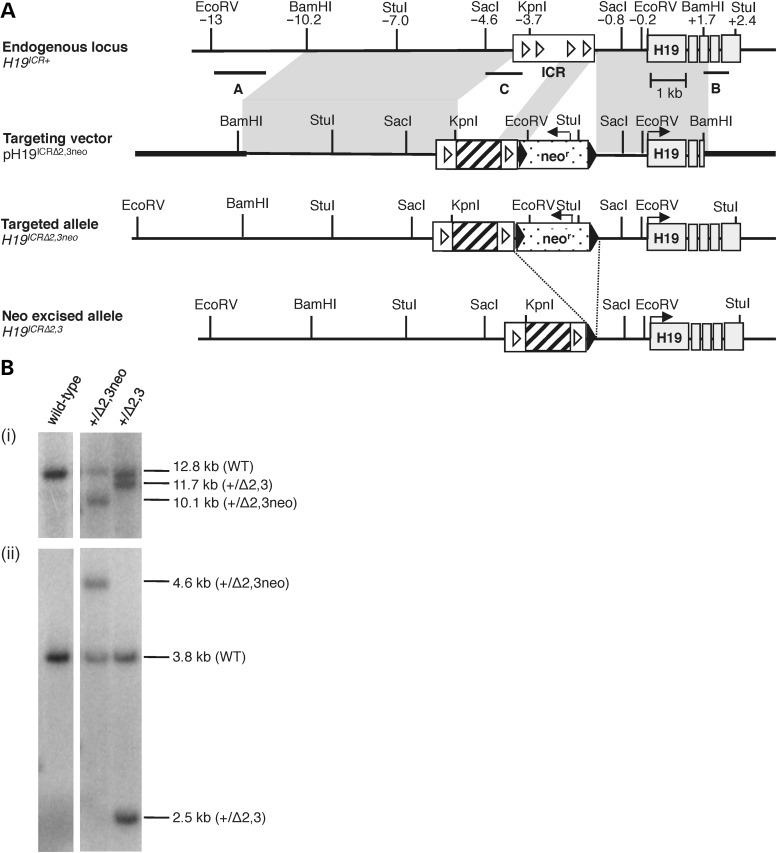
To determine the role of cis-acting microdeletions analogous to those associated with disrupted DNA methylation at the H19/IGF2 IC1 in BWS patients, we generated mice carrying a targeted deletion at the endogenous H19 ICR using homologous recombination in ES cells (Fig. 1A). This deletion reduces the size of the ICR by ∼75%, removing 1.3 kb of sequence, 70% of the CpGs and 50% of the CTCF sites (sites 2 and 3) (Table 1). Germline transmission of the H19ICRΔ2,3 mutant allele and excision of the neomycin resistance (neor) cassette in the mouse were confirmed by Southern blot analysis (Fig. 1B). Mutant progeny lacking the neor cassette were assayed for defects in H19/Igf2 imprinting when the mutant allele was either maternally or paternally inherited. Parental origin of alleles was distinguished by crossing heterozygous mutant H19ICRΔ2,3/+ mice to wild-type B6(CAST7) (C7) mice, which have a CAST (Mus musculus castaneus) chromosome 7 on a C57BL/6J (B6) (Mus musculus domesticus) background (27). For all comparisons, mutant neonatal progeny were compared with their wild-type littermates. For all genotypes, the maternal allele is listed first and the paternal allele second.
Figure 1.
Generation of targeted H19ICRΔ2,3 allele. (A) Targeting scheme at the H19/Igf2 locus. Illustrated from top to bottom are the wild-type endogenous locus (H19ICR+), the targeting vector (pH19ICRΔ2,3neo), the correctly targeted allele with the neomycin resistance cassette (H19ICRΔ2,3neo) and targeted allele after neor excision (H19ICRΔ2,3). Restriction sites and their relative positions (in kb) to the H19 TSS are indicated above the endogenous locus. Shaded region between endogenous locus and targeting vector indicates regions of homology. Southern probes (A, B and C) are indicated by horizontal lines below the endogenous locus. Also depicted is the H19/Igf2 ICR (white rectangle), CTCF sites (white triangles), H19 exons (gray rectangles), pBluescriptIIKS sequence (bold line) versus 129/Sv mouse DNA (thin line), neor cassette (polka dot box) and loxP sites (black arrowheads). Sequence deleted by the ICRΔ2,3 mutation is indicated by a cross-hatched region. Line arrows indicate direction of gene expression. (B) Southern blots using mouse tissues confirmed correctly targeted alleles using external probe A and EcoRV digest (i), external probe B and StuI digest (data not shown), and internal probe C and SacI digest (ii) as previously described (36).
Table 1.
DNA sequence differences between wild-type and H19ICRΔ2,3 alleles
| Allele | ICR size (bp)a | CTCF site no.b | CpG no.c | CpG (%)d | %Δe |
|---|---|---|---|---|---|
| Wild-type | 1745 | 4 | 51 | 5.8 | − |
| H19ICRΔ2,3 | 445 | 2 | 16 | 7.2 | +24.1 |
aNumber of base pairs from the start of the 21 bp consensus CTCF site 1 to the end of the 21 bp consensus CTCF site 4 at the ICR upstream of H19 (Chr 7:142 580 302–142 582 047, Genome Reference Consortium, GRC, Build 38).
bNumber of CTCF sites within ICR.
cNumber of CpG dinucleotides within ICR.
dPercent of CpGs calculated by dividing the number of CG dinucleotides by the total number of dinucleotides.
ePercent change in CpG density between the mutant and wild-type ICRs calculated by subtracting wild-type percent CpGs from mutant percent of CpGs and dividing the final value by the wild-type percent of CpGs.
H19ICRΔ2,3 transmission significantly alters neonatal pup weight
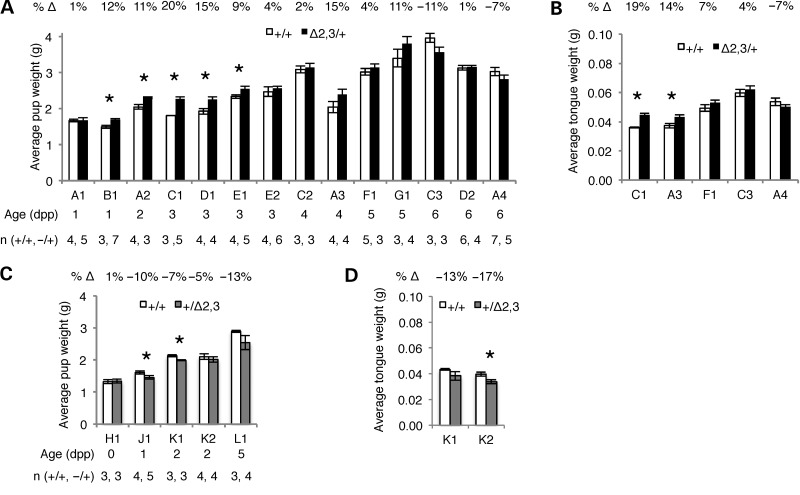
Because growth-related phenotypes are a major characteristic of H19 and IGF2 loss of imprinting in humans, we first determined whether mice carrying the H19ICRΔ2,3 mutant allele exhibited the overgrowth or growth restriction phenotypes typically observed in BWS or SRS patients, respectively. Neonatal pups ranging from Day 1 to Day 6 after birth were weighed and whole body and tongue weights were compared within litters between wild-type and heterozygous mutant offspring. We observed a consistent trend in pup and tongue weight when the H19ICRΔ2,3 allele was maternally transmitted such that average weight for mutant pups and tongues tended to be greater than their wild-type littermates (Fig. 2A). While this trend was significant for most litters <4 days old, differences were not significant for older pups (Fig. 2B). Although fewer litters were analyzed, similar trends were observed when the H19ICRΔ2,3 allele was paternally transmitted such that average weights for mutant pups and tongues tended to be lower than wild-type littermates but not always significantly so (Fig. 2C and D). We did not find a correlation between litters with weight differences and shared dam or dam's age (data not shown). The absence of significant weight increase in older maternally inheriting H19ICRΔ2,3/+ mutants compared with wild-type may reflect reduced growth rate with age as is observed with BWS individuals (10). In contrast, the lack of weight difference in older pups with the paternally transmitted deletion is not consistent with SRS patients whose weight as they age continues to be significantly lower than non-SRS individuals (28). This result is consistent with the observation that paternal transmission of BWS microdeletions does not result in SRS (5).
Figure 2.
Neonatal pup and tongue weights in H19ICRΔ2,3/+and H19ICR+/Δ2,3pups compared with wild-type littermates. Consistent trends in pup and tongue weight associated with maternal or paternal inheritance of Δ2,3. Charts show average weight in grams (y-axis) of pups with Δ2,3 deletion (filled bars, maternal transmission—black, paternal transmission—gray) compared with wild-type littermates (open bars). Standard error bars are depicted. Numbers above chart indicate average percent change (%Δ) in weight, positive numbers (increase) and negative numbers (decrease). Letter–number combinations below chart indicate dam (letter) and litter (number), e.g. A1 indicates dam “A” and litter “1”; litters are grouped by pup age but otherwise unordered. Pup age in days post parturition and number of pups analyzed per group (n) is listed in columns below dam. Asterisks indicate significant difference (P < 0.05) as determined by two-tailed t-test. (A) Average pup weight with maternal Δ2,3 inheritance. (B) Average tongue weight with maternal Δ2,3 inheritance. (C) Average pup weight with paternal Δ2,3 inheritance. (D) Average tongue weight with paternal Δ2,3 inheritance.
Maternal transmission of H19ICRΔ2,3 results in tissue-specific loss of insulator activity
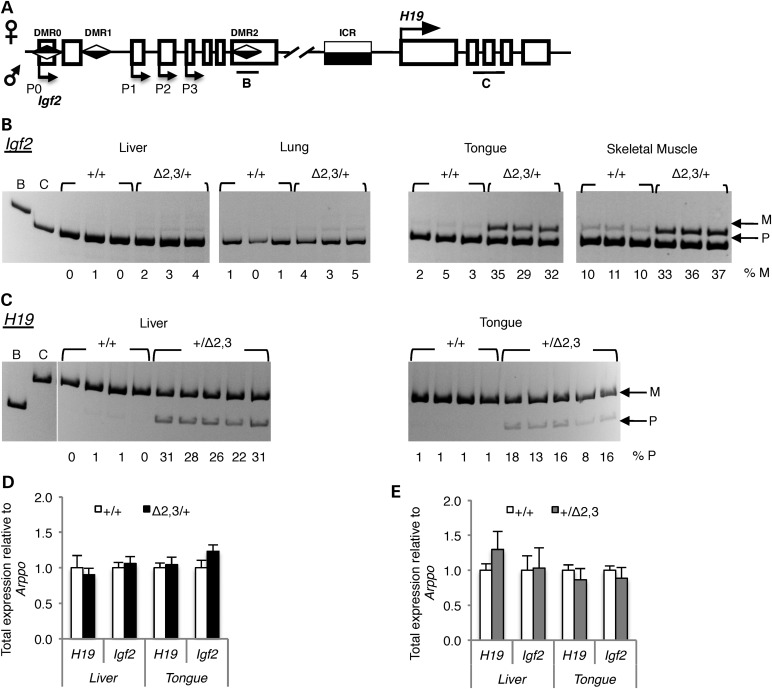
To determine whether weight differences observed between H19ICRΔ2,3 mutant and wild-type pups are due to loss of imprinted expression associated with the Δ2,3 deletion, we assayed allele-specific expression at the H19 and Igf2 locus depicted in Figure 3A. Surprisingly, we found that maternal inheritance of the H19ICRΔ2,3 allele results in activation of the normally silent maternal Igf2 allele in neonatal tissues of mesodermal origin. High levels of aberrant maternal Igf2 expression (29–37% of total) were observed in mutant H19ICRΔ2,3/+ mesodermal tissues tongue and skeletal muscle compared with tissues from wild-type H19ICRΔ+/+ littermates (Fig. 3B). In neonatal kidney and heart, aberrant maternal H19ICRΔ2,3 Igf2 expression was also observed but levels were variable (2–19% of total, data not shown). In contrast, in mutant endodermal tissues, liver and lung, levels of maternal Igf2 were very low (2–5% of total, Fig. 3B). This mesoderm-specific loss of imprinted Igf2 expression was observed as early as embryonic Day 13.5 in mutant tissues containing mesodermal cells (embryo body, yolk sac and placenta, 10–30% maternal Igf2 expression) but not endodermal-derived liver (0% maternal Igf2 expression, data not shown). This is the first example of an H19/Igf2 ICR mutation that perturbs Igf2 imprinting in mesodermal but not endodermal tissues, indicating that a crucial mesoderm-specific regulatory mechanism is acting at the ICR and is disrupted by maternal transmission of the Δ2,3 deletion.
Figure 3.
H19ICRΔ2,3 inheritance disrupts H19/Igf2 imprinted expression in neonatal tissues. (A) Illustration of allelic expression at the H19/Igf2 locus. Location of allele-specific assays in Igf2 and H19 is shown as horizontal bars B and C below the locus. (B and C) Allele-specific Igf2 or H19 expression in neonatal tissues as determined by reverse-transcriptase PCR and allele-specific digest and migration in acrylamide gel; maternal (m) and paternal (p) alleles migrate at different sizes due to polymorphic restriction digest sites within the amplicon. Tissue being assayed is listed above gel, indicating lanes containing individual mutant (Δ2,3/+ or +/ Δ2,3) and wild-type (+/+) littermate samples; mesodermal lineages (liver and lung), endodermal lineages (tongue and skeletal muscle). (B) Allele-specific Igf2 expression with maternal inheritance of H19ICRΔ2,3 as compared with wild-type littermates. Percent maternal Igf2 expression is listed below each lane for each respective sample. (C) Allele-specific H19 expression with paternal inheritance of H19ICRΔ2,3 when compared with wild-type littermates. Percent paternal H19 expression is listed below each lane for each respective sample. (D) Total H19 or Igf2 expression (relative to Arppo expression) with maternal H19ICRΔ2,3 inheritance in wild type (+/+, open bars, n = 4) or mutant (Δ2,3/+, filled bars, n = 5), with standard error bars presented. (E) Total H19 or Igf2 expression (relative to Arppo expression) with paternal H19ICRΔ2,3 inheritance in wild type (+/+, open bars, n = 4) or mutant (+/Δ2,3, filled bars, n = 5), with standard error bars presented.
In contrast to the tissue-specific loss of imprinting observed with maternal inheritance of the Δ2,3 deletion, paternal inheritance of the H19ICRΔ2,3 allele resulted in biallelic H19 expression in both endodermal and mesodermal tissues. Interestingly, here, the mesodermal tissue (tongue) is seemingly affected to a lesser extent (Fig. 3C). These data suggest that the tissue-specific loss of imprinting observed with maternal transmission of the H19ICRΔ2,3 allele is exclusive to the maternal ICR and therefore is insulator specific. Surprisingly, despite the observed loss of imprinted expression, total expression levels of H19 and Igf2 were not significantly different in affected tissues between mutant and wild-type littermates (Fig. 3D and E, data not shown). This finding demonstrates that loss of imprinted H19/Igf2 expression alone, without statistically significant differences in total transcript levels is correlated with significant phenotypic changes. Thus, measurement of total IGF2 levels without a determination of allelic expression is not a fully reliable diagnostic tool to ascertain etiology of BWS.
Sequence removed by the Δ2,3 deletion is dispensable for establishing and maintaining DNA methylation at the H19/Igf2 ICR
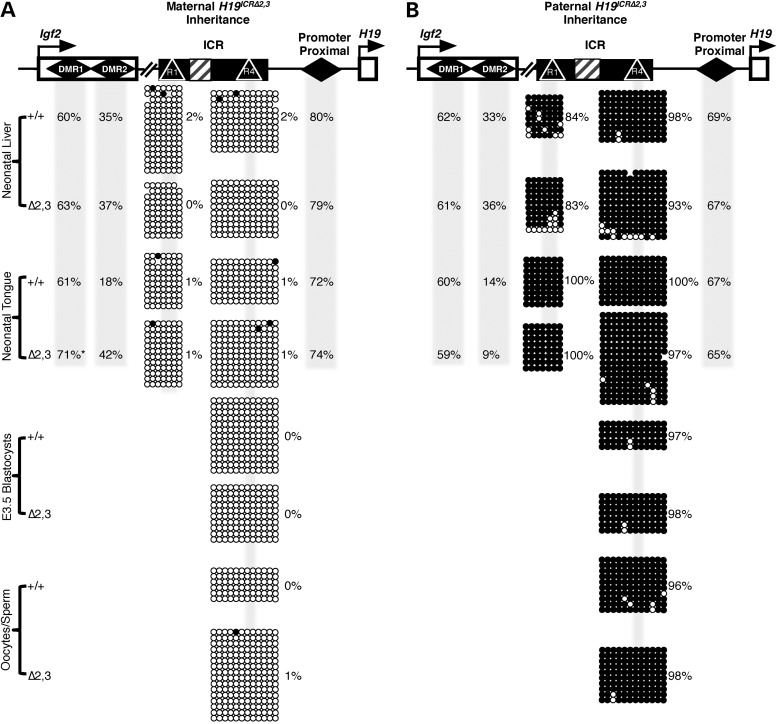
Previous studies showed that loss of imprinted expression of H19 and Igf2 is directly correlated with ICR methylation defects (24–26,29–31). Of relevance, human BWS and SRS patients are most often clinically tested for epimutations at the H19/IGF2 ICR and not changes in gene expression (12,28). As such, the absence of DNA methylation perturbation is often singly used to rule out a link between the disease/disorder and the locus. In a previous study, we found unchanged DNA methylation patterns in the presence of subtle levels (up to 18% of total) of aberrant paternal H19 expression in neonatal liver (32). Here, we tested whether higher levels of aberrant paternal H19 in neonatal liver and tongue (up to 32% of total) and maternal Igf2 in neonatal skeletal muscle and tongue (up to 37%) are associated with altered DNA methylation at the ICR in these respective tissues. As depicted in Figure 4A and B, DNA methylation was unaffected at the ICR and the H19 promoter proximal region (∼500 bp from H19 transcriptional start site) (33) in mutant liver and tongue. Furthermore, normal methylation patterns were also observed at earlier stages of development in mutant blastocysts, sperm and oocytes (Fig. 4A and B). This is true whether the deletion was maternally or paternally transmitted, despite significant changes in imprinted expression in neonatal tissues (Figs. 3 and 4). These findings demonstrate that the Δ2,3 deletion does not perturb either establishment or maintenance of parental methylation marks at the ICR and H19 promoter proximal region. Therefore, taken together, our findings signify that changes in H19/Igf2 imprinted gene expression significant enough to impact phenotypic outcome do not always coincide with DNA methylation changes at the ICR. This finding has considerable implications for clinical testing methods such that the absence of an epimutation at the H19/IGF2 ICR in BWS or SRS patients may not be sufficient to rule out involvement of the H19/IGF2 locus in the diagnosis.
Figure 4.
Effect of 70% deletion of the H19/Igf2 ICR on DNA methylation states. Percent methylation is depicted across the H19/Igf2 locus with maternal (A) or paternal (B) inheritance of the H19ICRΔ2,3 allele. (A and B) Top: the H19/Igf2 locus is depicted with the following features: Igf2 and H19 genes (open boxes with arrows), differentially methylated regions (DMRs, diamonds), ICR (filled box), CTCF sites (R1 and R2, triangles) and Δ2,3 deletion (hashed box). Total percent methylation (as determined by bisulfite conversion, PCR and pyrosequencing) is shown for Igf2 DMR1, DMR2 and the promoter proximal region in shaded columns for the cell types/tissues indicated in the left column for wild-type (+/+) and mutant samples (Δ2,3). Allele-specific methylation (A, maternal allele and B, paternal allele) at the ICR was determined by bisulfite conversion, PCR, cloning and sequencing. Methylated CpGs (filled circles) and unmethylated CpGs (unfilled circles) are depicted in rows for each clone/copy of DNA and columns indicate individual CpG sites (shaded regions indicate CpGs at CTCF sites). Percent methylation was calculated as number of methylated CpGs over total number of CpGs sequenced and is listed beside clones. Asterisks indicate significant difference (P < 0.05) in DNA methylation levels at the Igf2 DMR1 between wild-type and mutant samples as determined by two-tailed t-test, n = 3–5.
Tissue-specific loss of Igf2 imprinting is correlated with DNA methylation changes at Igf2 DMR1 and DMR2
In contrast to a lack of change in methylation at the mutant ICR, maternal H19ICRΔ2,3 transmission was associated with increases in total DNA methylation at the Igf2 DMR1 and DMR2 in neonatal H19ICRΔ2,3/+ tongue (Fig. 4A). We infer that this increase in total methylation at the Igf2 DMRs in the H19ICRΔ2,3/+ mutant tongue is the result of gain of methylation on the maternal allele because studies in wild-type mice show that the paternal allele is hypermethylated, whereas the maternal allele is hypomethylated (Fig. 3A) (34,35). Thus, gain of methylation on the maternal Igf2 DMR1 and 2 could explain the loss of silencing of maternal Igf2. Furthermore, methylation changes at the Igf2 DMR1 and DMR2 followed the tissue-specific pattern of aberrant maternal Igf2 expression, being only present in mutant tongue and not liver (Fig. 4A). Paternal H19ICRΔ2,3 transmission, which did not perturb imprinted Igf2 expression in liver or tongue, also did not alter DNA methylation at the Igf2 DMR1 and DMR2 (Fig. 4B). These findings suggest that the DNA methylation defects observed at the Igf2 DMRs are tightly linked to aberrant Igf2 transcription, although it remains unclear whether the methylation changes are causal or dependent on transcription at the locus.
To determine whether the increased Igf2 DMR1 methylation state observed in H19ICRΔ2,3/+ mutant tongue is specific to the Δ2,3 deletion or merely correlated with maternal Igf2 expression regardless of the mutation, we assayed Igf2 DMR1 methylation in neonatal tissues carrying the ΔDMD deletion (26,36). This previously generated BWS mouse model carries a deletion at the ICR of the three CTCF sites most proximal to the H19 promoter, which was shown to cause biallelic expression of Igf2 in both liver and tongue when maternally inherited (26,36). We found that maternal transmission of the H19ICRΔDMDallele is linked to hypermethylation at the Igf2 DMR1 in both liver (+/+ = 54.7 ± 1.1% versus ΔDMD/+=64.0 ± 2.2%, P = 0.02) and tongue (+/+ = 61.8 ± 1.6% versus ΔDMD/+=67.7 ± 0.9%, P = 0.03). Hence, Igf2 DMR1 methylation defects are not specific to the Δ2,3 deletion but are linked to maternal Igf2 expression. These findings are consistent with genomic analyses showing increased methylation at transcribed elements (37).
CTCF binding is disrupted in H19ICRΔ2,3/+ mesodermal but not endodermal tissues
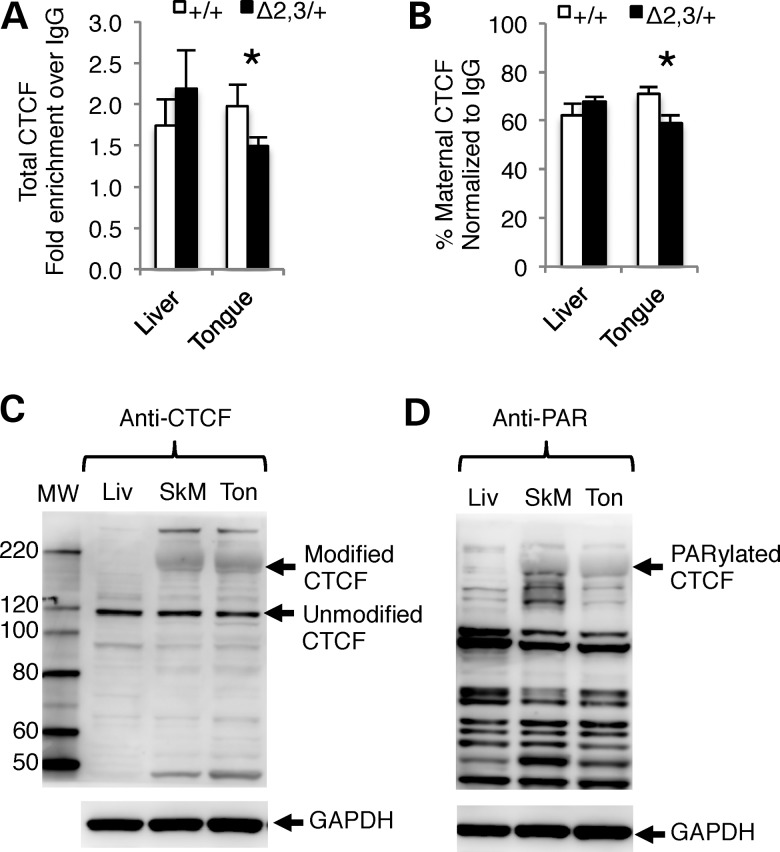
The mechanism responsible for tissue-specific loss of imprinting associated with the H19ICRΔ2,3 allele is unclear. Notably, these tissues use different enhancers (38,39), each of which may interact in a unique way with the insulator and insulator-binding protein, CTCF at the H19/Igf2 ICR. Therefore, we first used chromatin immunoprecipitation (ChIP) to investigate whether CTCF binding at the ICR differs between mesodermal and endodermal tissues when H19ICRΔ2,3 is maternally inherited. As depicted in Figure 5A, total CTCF binding at the H19/Igf2 ICR is significantly decreased in H19ICRΔ2,3/+ mutant compared with wild-type tongue. These differences are not present in matched liver samples from the same animals. Although we assayed CTCF using an amplicon at the CTCF site in the ICR most distal to the H19 promoter, the ChIP signal is likely not representative of CTCF binding at just this single site because CTCF sites are clustered together at the ICR. Of note, CTCF binding assayed at the CTCF site most proximal to the H19 promoter was affected to a lesser degree by the Δ2,3 deletion (data not shown). Next, because CTCF preferentially binds to the hypomethylated maternal allele, we tested allelic CTCF binding and also found that CTCF binding to the maternal ICR allele is significantly decreased in mutant versus wild-type tongue but not liver (Fig. 5B). Thus, CTCF binding to the H19/Igf2 ICR is disrupted by the Δ2,3 deletion in a tissue-specific manner that correlates with the observed tissue-specific expression. While this may explain the tissue-specific patterns of loss of imprinted Igf2 expression, it does not explain the origin of tissue specificity.
Figure 5.
Tissue-specific differences in CTCF binding at the mutant H19/Igf2 ICR and poly(ADP-ribosyl)ated CTCF protein. (A and B) CTCF binding to the H19/Igf2 ICR as determined by ChIP and PCR amplification of a 145 bp region overlapping the ICR CTCF site most distal to the H19 promoter. (A) Total CTCF binding and (B) percent maternal CTCF binding. Values are normalized to input and depicted as fold enrichment over IgG. n = 3 l (3–4 dpp). Within each litter, wild-type (+/+, open bars) and mutant (−/+ and +/−, filled bars) tissues were pooled separately. Asterisks indicate significant difference (P < 0.05) as determined by paired t-test. (C and D) Depicts CTCF protein levels in representative samples detected by western blot using GAPDH as loading control and protein marker to determine molecular weight (MW) of bands for identity. (C) Protein levels of CTCF isoforms in neonatal liver (Liv), skeletal muscle (SkM) and tongue (Ton) as determined by western blot probed with anti-CTCF antibody. Arrows indicate modified form of CTCF in SkM and Ton and unmodified CTCF in Liv, SkM and Ton. (D) Poly(ADP-ribosyl)ated proteins in neonatal liver (Liv), skeletal muscle (SkM) and tongue (Ton) as determined by western blot probed with anti- PAR antibody. Arrow indicates poly(ADP-ribosyl)ated band with MW identical to modified form of CTCF in (C).
Mesoderm-specific modified CTCF, corresponding to poly(ADP-ribosyl)ated CTCF, may explain tissue specificity of H19ICRΔ2,3/+ loss of imprinting
A possible source of tissue specificity and diminished binding of CTCF to the mutant maternal allele is lower levels and/or availability of CTCF in the affected tissues. Thus, we assayed by western blot total CTCF protein levels in neonatal liver, skeletal muscle and tongue. CTCF has at least seven different sized isoforms that migrate from 55 to 130 kDa on polyacrylamide gels (40). Although the open reading frame of the major isoform of CTCF would only encode an 82 kDa protein, CTCF migrates at a much higher molecular weight (∼130 kDa) (41). In our studies, we observed the most abundant form of unmodified CTCF migrating at ∼120 kDa on a NuPAGE 4–12% Bis-Tris gel (Fig. 5C). We measured similar levels of this isoform of unmodified CTCF (relative to GAPDH) between liver, skeletal muscle and tongue tissues (0.32 ± 0.12, 0.32 ± 0.03 and 0.29 ± 0.01, respectively, for all pairwise comparisons P > 0.05) (Fig. 5C). Surprisingly, we also detected a larger form of modified CTCF in tongue and skeletal muscle but not in liver, migrating at a size of ∼180 kDa (Fig. 5C).
CTCF is reported to be posttranslationally modified in some tissues, becoming phosphorylated on C terminus and migrating at ∼130–160 kDa (42), poly(ADP-ribosyl)ated at N terminus (43) to migrate at ∼180 kDa, and SUMOylated on both the C- and N-terminus migrating at ∼170 kDa (44). Furthermore, CTCF self-dimerizes and multimerizes and has numerous protein partners with which it forms heterodimers (45). Poly(ADP-ribosyl)ated CTCF has been shown to bind the H19/Igf2 ICR in vitro and impact imprinted expression in a mouse–human cell hybrid (43). Because the modified form of CTCF we detected (Fig. 5C) is most similar in size to poly(ADP-ribosyl)ated CTCF and because of the previous implications for its role in imprinting (43), we used western blotting to investigate whether the larger modified form of CTCF observed in mesodermal tissues is indeed poly(ADP-ribosyl)ated CTCF. As shown in Figure 5D, migration of the modified CTCF observed exclusively in mesodermal tissues coincides with migration of poly(ADP-ribosyl)ated protein, indicating that the mesoderm-specific form of CTCF is the poly(ADP-ribosyl)ated form.
DISCUSSION
Here, we have generated and characterized the Δ2,3 mouse model to mimic microdeletions at the H19/IGF2 ICR associated with BWS (18). BWS is one of the most common childhood overgrowth syndromes in humans (12). Microdeletions at the human H19/IGF2 ICR (IC1) represent one of several genetic mechanisms associated the disorder. These microdeletions, which remove 1–3 of seven CTCF sites and <50% of the IC1, are proposed to act by disrupting the spacing of CTCF sites. This in turn disrupts CTCF binding to the maternal IC1 allowing for gain of methylation on the maternal ICR and biallelic IGF2 expression (4). In contrast, the Δ2,3 ICR deletion in the mouse, described here, which disrupts CTCF site spacing, deletes two of four CTCF sites, and leaves only 25% of the ICR intact, does not promote even minimal maternal ICR hypermethylation. The maternal Δ2,3 ICR deletion does, however, result in loss of imprinted expression and changes in pup weight similar to that observed in BWS, indicating that the molecular mechanisms of ICR-mediated imprinting are compromised. This dissimilarity in methylation defects between the mouse model and BWS patients could be explained by one of two possibilities: (i) disruption of CTCF spacing alone is not sufficient for IC1 methylation defects and an additional yet unidentified genetic or environmental factor is involved or (ii) some of the sequences conferring ICR function are not conserved between mice and humans. Further research is required to resolve these two possibilities; however, existing data support the latter possibility.
Whereas the overall mechanism of ICR-mediated imprinting at the H19/Igf2 locus is seemingly conserved between mouse and human, it is likely that regulatory sequences required for imprinting may differ between the mouse and human ICRs. First, there are almost twice as many CTCF sites at the human IC1 (seven sites) as at the mouse ICR (four sites). Second, the sequence between CTCF sites is highly divergent between mouse and human. Third, the human IC1 consists of repetitive elements, whereas the mouse ICR does not. As discussed above, CTCF number and spacing have been implicated in BWS methylation changes. Deletion of all four CTCF sites in the mouse did result in gain of methylation on the maternal allele in somatic tissues (24); however, disruption of CTCF site spacing failed to result in ICR methylation defects in our previously generated ΔIVS mouse model (32) or in the Δ2,3 model presented here. Thus, if CTCF site spacing plays a role in methylation state at the human IC1, this role is not conserved in the mouse. Below we describe additional important differences in regulatory sequences between the mouse and human H19/Igf2 ICR.
OCT-binding sites are present at the both the human and mouse ICRs and have also been implicated in BWS to play a role in maintaining the hypomethylated state of the maternal IC1. Mutations in IC1 OCT-binding sites were detected in BWS patients with ICR methylation defects (6,7,46). While mutation of these OCT-binding sites in in vitro (47) and in vivo (ectopic loci in transgenic mice) (48) results in hypermethylation of the ICR, our deletion of both OCT-binding sites in the ΔIVS (32) and Δ2,3 mouse models did not alter methylation at the endogenous ICR. These results suggest that either mutations in the OCT-binding sites observed in BWS patients are the result of a gain of function or the mouse carries regulatory sequence with complementary function to the OCT-binding sites that is absent in humans. Alternatively, the imprinting function of the OCT-binding sites at the locus is not conserved between mice and humans.
In contrast to the postulated hypomethylating properties of the OCT-binding sites, ZFP57-binding sites at the human and mouse ICR are proposed to be required for maintaining hypermethylation (49) on the paternal allele. Consistently, H19/Igf2 ICR methylation was not affected in a Zfp57 deletion model in the mouse (49,50). Furthermore, the Δ2,3 mouse model described here, which deletes one of four ZFP57-binding motifs at the ICR (49), does not perturb ICR methylation. Nucleosome positioning sites are also proposed to play a role in methylation states at the H19/Igf2 ICR. Deletions that remove nucleosome positioning sites may indirectly alter ICR methylation by altering chromatin conformation at the locus, which has been reported to be critical to maintaining imprinted expression (51,52). Our Δ2,3 targeted mouse model deletes two of three proposed nucleosome positioning sites (53). Six nuclear hormone receptor sites are also deleted, although previous studies do not implicate their role in maintaining insulator function at the locus (31,54). Thus aside from the number of CTCF-binding sites, none of the sequences proposed to maintain allele-specific methylation at the IC1 translate to our findings in the mouse, supporting the concept that the ICRs in mouse and human differ in regulatory sequences.
It is important to note that recent studies show that the extent of methylation defects observed in association with BWS microdeletions (5) and OCT-binding site mutations (6) increased over the course of three generations. The H19ICRΔ2,3/+ neonatal tissues and H19ICRΔ2,3 Δ2,3 sperm and oocytes analyzed here for expression and methylation changes were also generated after at least three generations of maternal germline transmission. While the number of generations is comparable, we cannot account for differences in the length of time between each generation (∼6 weeks–1 year in mice versus 12–50 years in humans) or the relative age of the mother at conception. In addition to regulatory sequence and environmental exposure differences, this difference in timing could also very well explain the differences we observe between methylation perturbation at the H19/Igf2 ICR in mice versus humans.
Despite a lack of methylation changes, imprinted expression of H19 and Igf2 was disrupted in mice inheriting the Δ2,3 mutation either paternally or maternally, respectively (Fig. 3). These results demonstrate that the deleted sequences, while not required for maintaining the methylated state at the ICR, are required for maintaining imprinted expression at the locus. In concordance with this loss of imprinted expression, maternal transmission of the Δ2,3 deletion resulted in significant increase in pup and tongue weights (Fig. 2). In contrast, the ΔIVS deletion at the H19/Igf2 ICR, which removed the intervening sequence without disrupting CTCF sites, did not perturb imprinted Igf2 expression or result in neonatal pup weight increases (32). Comparison of these two models, Δ2,3 and ΔIVS, suggests that disruption of CTCF binding at the maternal H19/Igf2 ICR is required for a significant enough loss of H19/Igf2 expression to cause phenotypic changes such as those observed in BWS. Indeed, we observed that both total and allelic CTCF binding was disrupted in mice inheriting a maternal Δ2,3 ICR deletion (Fig. 5). However, this disruption in CTCF binding to the ICR did not significantly perturb ICR methylation states. This finding is very surprising considering CTCF binding is thought to be responsible for blocking methylation on the maternal ICR allele, thus if perturbed it should allow gain of methylation defects to occur. Overall, if we assume the mechanism of ICR-mediated imprinted expression is conserved between mouse and human, these findings bring to question the reliance on DNA methylation defects as a sole means of diagnosing BWS. The possibility that loss of H19/IGF2 imprinting significant enough to perturb fetal–neonatal growth may occur in the absence of DNA methylation defects should be considered when determining genetic mechanisms of BWS. The potential for tissue specificity of loss of imprinted expression should also be considered.
We found that the loss of imprinted Igf2 expression associated with maternal transmission of the Δ2,3 ICR deletion only occurred in mesodermal tissues (Fig. 3). This is the first deletion at the H19/Igf2 ICR to result in tissue-specific loss of imprinting where only mesodermal tissues are affected. Interestingly, while previously analyzed ICR-specific deletions resulted in variable degrees of loss of imprinting in different tissues, none resulted in mesoderm-specific effects. For example, neither the SiLK mutation (55), which deletes the two CTCF sites most distal to Igf2, nor the ΔDMD mutation (26,36) which overlaps the Δ2,3 deletion and deletes the three CTCF sites most distal to Igf2 and the intervening region, resulted in mesoderm-specific loss of Igf2 imprinting. These findings suggest that insulator function at the ICR relies on a complex cell-type-specific mechanism that requires more than the presence or absence of a specific number of CTCF-binding sites or the intervening sequence and regulatory sites therein. On the other hand, maternal transmission of the Δ2,3 ICR deletion did not significantly alter maternal H19 RNA levels as is observed with the ΔDMD deletion (36). This finding suggests that the sequence removed by the ΔDMD deletion but maintained in the Δ2,3 deletion (0.3 kb of sequence including the CTCF site most proximal to the H19 transcription start site, TSS) plays a role in maternal H19 activation. While paternal transmission of the Δ2,3 ICR deletion resulted in loss of H19 imprinted expression in both liver and tongue, lower levels of aberrant paternal H19 expression were detected in tongue compared with liver (on average, 14.2 ± 1.7 versus 28 ± 1.8, respectively).
Although the tissue-specific methylation changes at the Igf2 DMR1 and DMR2 are correlated with tissue-specific expression of Igf2, these methylation differences do not fully explain disruption of insulator function at the ICR. Our findings that a significant amount of poly(ADP-ribosyl)ated CTCF is present in the affected mesodermal tissues (tongue and skeletal muscle) but absent in endodermal tissues (liver), in conjunction with mesoderm-specific loss of imprinting associated with the Δ2,3 ICR deletion, strongly imply that CTCF-mediated insulator function in these two tissues is regulated differently. H19 and Igf2 expression in mesoderm versus endoderm relies on different enhancers that reside ∼20 kb apart (38,39,56,57). The tissue specificity may be due to sequence differences or that mesoderm enhancer is 20 kb further from its targets, spacing that may confer differences in the ICR-mediated interactions between these enhancers and gene promoters. Alternatively, the mesoderm-specific poly(ADP-ribosyl)ated CTCF may be solely responsible for the mesoderm-specific phenotype. In support of the latter hypothesis, it has recently been demonstrated in Drosophila that poly(ADP-ribosyl)ation regulates insulator function (58). In addition, Farrar et al. demonstrated that mutating sites of poly(ADP-ribosyl)ation on the CTCF N-terminal domain resulted in loss of optimal CTCF-mediated insulator function at the H19/Igf2 ICR (59). Caiafa and Zlatanova propose a model whereby CTCF and PARP interactions inhibit DNMT1-mediated DNA methylation and poly(ADP-ribosyl)ation of CTCF is required to form the repressive chromatin loop necessary to silence maternal Igf2 (60). Of note, Yu and colleagues use an insulator trap assay to indicate a role for poly(ADP-ribosyl)ated CTCF in insulator function at the H19/Igf2 ICR in liver cells (43). However, these experiments failed to detect poly(ADP-ribosyl)ated CTCF in liver by western blot bringing into question the relevance of their findings (43).
Based on the data presented here and previous work by others, we propose a tissue-specific model of ICR-mediated imprinting at H19/Igf2 that is dependent on the presence or absence of poly(ADP)-ribose (PAR)-ylated CTCF. The previous studies discussed above suggest that PAR-ylation of CTCF mediates CTCF interactions with other proteins and subsequent intrachromosomal interactions (looping) such as those implicated in ICR-mediated insulator function. We propose that such looping mechanisms may require a sufficient/threshold level of PAR-ylated CTCF binding to the ICR in mesodermal tissues for complete maternal Igf2 silencing in mesodermal tissues and that the Δ2,3 deletion disrupts PAR-ylated CTCF binding to the ICR to levels below this threshold. This tissue-specific ICR-mediated function regulated by PAR-ylated CTCF may allow for a more fine-tuned degree of Igf2 regulation due to its importance as a growth factor in mesodermal tissues that is not required in endodermal tissues. This higher degree of regulation seemingly increases sensitivity of the mechanism to genetic perturbation in cis. Alternatively, it may have evolved alongside unrelated mechanisms. Further studies are necessary to confirm these hypotheses, including determining whether both modified and unmodified CTCF bind to the ICR in mesodermal tissues, how either form directly affects CTCF insulator function, and how these forms mediate differences in insulator function between mesodermal and endodermal tissues. Moreover, experiments assessing the role of mesodermal enhancer sequences in tissue-specific insulator activity are required.
Overall, our findings have several important implications for understanding the molecular basis of BWS. Most importantly, clinical testing for epimutations at the H19/IGF2 IC1 alone may not be sufficient for BWS diagnosis because our study shows significant changes in Igf2 imprinting and pup weight in the absence of DNA methylation changes. Furthermore, loss of IGF2 imprinting may occur in a tissue-specific manner, which may explain the organ-specific overgrowth occurring in BWS patients. Finally, microdeletions at the IC1 may disrupt imprinting in a manner independent of changes in DNA methylation and therefore tests for microdeletions should be performed in diagnosis. Further studies elucidating the cause and role of these microdeletions in ICR-mediated regulation will likely provide essential information regarding the complex mechanism of CTCF-mediated control of gene expression at both imprinted and non-imprinted loci. More specifically, experiments including the exchange of the mouse and human ICR in the mouse and the analysis of human iPS cells should address these questions.
MATERIALS AND METHODS
Targeting vector
The pH19ICRΔ2,3neo targeting vector (Fig. 1A) was generated to contain a 1.3 kb deletion between CTCF sites 1 and 4 at the H19/Igf2 ICR. Approximately 0.4 kb of sequence (from −2.06 to −2.46 5′ upstream of the H19 TSS, including CTCF site 4) was amplified by PCR from the previously generated pH19ICRΔIVSneo (32) using forward primer 5′-CTCACTAAAGGGAACAAAAGC-3′ and reverse primer 5′-GGTACCAGTGGCTGGTAAGAC-3′. Sequencing was used to confirm accuracy and the amplicon was inserted at the endogenous KpnI site, −3.7 kb upstream of the H19 TSS in the previously described targeting vector [TVΔDMDneo (26), modified to remove KpnI site in pBSIIKSM]. The TVΔDMDneo targeting vector contained a neomycin resistance (neor) gene cassette (for selection) flanked by loxP sites and pBSIIKSM. Informative restriction enzyme digest reactions and sequencing (using primers 1F and 1R as previously described) (32) were used to confirm insertion and orientation of the mutated fragment within the targeting vector.
Embryonic stem cells and mouse generation
The pH19ICRΔ2,3neo targeting vector was linearized and electroporated into E14.1 ES cells (61) as described previously (26). Clones were assayed for G418 resistance and positive clones were isolated. Targeting to the H19/Igf2 locus was confirmed by restriction digestion followed by Southern hybridization as described previously (26). Correctly targeted ES cell clones were injected into B6 blastocysts and mice were generated by the Transgenic and Chimeric Mouse Facility at the University of Pennsylvania. Chimeras were obtained and mated to B6 mice and germline transmission of the targeted mutant alleles was confirmed in the agouti progeny by DNA isolated from ear punches subjected to Southern blot as described below. All studies adhered to procedures approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania.
Mouse breeding and genotyping
Germline transmission of the H19ICRΔ2,3neo allele (Fig. 1A) was confirmed by PCR-based genotyping on DNA isolated from ear punches using primers that flank the deletion (primers 1F and 1R, as described above). The neor cassette (flanked by loxP sites) was excised by crossing heterozygous mutant mice to mice expressing Cre recombinase under the control of the human cytomegalovirus promoter on a B6 genetic background. DNA was isolated from offspring tissues by phenol chloroform extraction and neor cassette excision was confirmed in the progeny by Southern analysis (Fig. 1A and B) and PCR analysis using primers that flank the neor cassette, H19-2.3F and H19-2.0F, as described previously (36). Mutant lines lacking the neor cassette were maintained by crossing to B6 mice and selecting for progeny carrying the mutation as determined by PCR analysis. To distinguish parental alleles for all allelic assays described in this study, heterozygous mutant mice, H19ICRΔ2,3neo/+, were crossed to C7 mice. C7 mice are homozygous on chromosome 7 for wild-type alleles from the Mus musculus castaneus (CAST) strain on a mostly B6 background (27). Unless otherwise indicated, for all experiments, mutant mice are compared with their wild-type littermates.
Gene expression analysis
Total RNA was extracted from mouse tissues using Trizol Reagent (Invitrogen) following the manufacturer's protocol. RNA was DNAse treated using RQ1 DNAse (Promega) and reverse transcription (RT) was performed using Superscript III (Invitrogen) according to the manufacturer's protocol. For each RNA sample, RT reactions included both negative (no transcriptase added to confirm cDNA-specific amplification) and positive (transcriptase added) reactions. Approximately 2.5 ng of cDNA was used for all assays. Allele-specific expression was assayed by PCR followed by restriction digestion specified by polymorphisms between the B6 and CAST alleles. Igf2 was amplified using primers Igf2-18 and Igf2-20 and digested with restriction enzyme Tsp4091, as previously described (32,62). H19 was amplified using primers HE2 and HE4 and digested with restriction enzyme Cac8I, as previously described (62). Digested RT-PCR fragments were resolved on either 15% (Igf2) or 12% (H19) polyacrylamide gels and band intensities were measured using ImageJ software (rsb.info.nih.gov/ij). For quantitative (q) RT-PCR, total Igf2 levels were measured relative to Arpp0 (acidic phosphoprotein P0 subunit) as previously described (63). P-values were calculated using a two-tailed Student's t-test with two-sample unequal variance.
DNA methylation analysis
DNA methylation was determined by bisulfite mutagenesis (BSM), PCR and either pyrosequencing or cloning and Sanger sequencing. First, DNA was isolated from neonatal tissues and sperm by phenol chloroform isolation as described previously (64) and converted by BSM using the Epitect kit (Qiagen) according to the manufacturer's protocol. DNA from pooled oocytes (30–100) or blastocysts (2–5) from individual mice was isolated and bisulfite converted using the Epitect plus kit (Qiagen) according to the manufacturer's protocol. 50 ng of BSM DNA (sperm and neonatal tissues), 10 oocytes equivalent or half blastocyst equivalent of BSM DNA was used per PCR. The Igf2 DMR1 region was amplified using forward primer 5′-TGAGGTTAGATTAGGTTGTAAGTT-3′ and 5′ biotinylated reverse primer 5′-CTTCCCTACCCCTTAAACC-3′ using Pyromark reagents (Qiagen) according to the manufacturer's protocol. The Igf2 DMR1 amplicon was pyrosequenced using primers 5′-TTTTAGAGGTTTTTGGAGAA-3′ and 5′-GGATTTTGTTAGGTAGGA-3′, which sequence two CpGs each for a total of 4 CpGs analyzed. The Igf2 DMR2 region was amplified using nested PCR with forward primer 3′-GGGAGTTTAGGTTAATATGATATTTTG-3′ and reverse primer 5′-ACTATCCCTACTCAAAAAAAAATCAC-3′ followed by second PCR with the initial forward and reverse primers 5′-GGGTAAGTTTTTTTAATATGATATTTGG-3′. The H19 promoter proximal region was amplified using forward primer 5′-TTTGGTGTTTTGATTTGTGGATGTTGA-3′ and 5′ biotinylated reverse primer 5′-ATCTCCCTATCCTACTAAAATACTACTATC-3′ and pyrosequenced using primer 5′-TTTTAATAGAAGTTAAGAGTAATG-3′ to assay three CpGs. The H19/Igf2 ICR at R1 (region including CTCF site 1) and R4 (region including CTCF site 4) was assayed by nested PCR: heterozygous mutant neonatal samples with forward primer 5′-GAGTATTTAGGAGGTATAAGAATT-3′ and reverse primer 5′-TAAATACACAAATACCTAATCCCT-3′ followed by forward primer 5′-GTAAGGAGATTATGTTTTATTTTTGG-3′ and reverse primer 5′-TAAATACACAAATACCTAATCCCT-3′; and homozygous wild-type and homozygous mutant samples at R1 with forward primer 5′-GAGTATTTAGGAGGTATAAGAATT-3′ and reverse primer 5′-ATCAAAAACTAACATAAACCCCT-3′ followed by forward primer 5′-GTAAGGAGATTATGTTTTATTTTTGG-3′ and reverse primer 5′-CTAACCTCATAAAACCCATAACTAT-3′ or at R4 with forward primer 5′-GGTAAATTTATGGGTTATTTAAGG-3′ and reverse primer 5′-CCTAAATTACCTAAAACATTACAA-3′ followed by forward primer 5′-AATGTTTATAAGGGTTATGGGGTGG-3′ and reverse primer 5′-CCCAACCTCTACTTTTATAAC-3′. Igf2 DMR2 and H19/Igf2 ICR amplicons were cloned using the TOPO TA cloning kit (Life Technologies) and individual positive clones were selected and sequenced. Methylation levels were determined by comparing cloned sequences using the Bisulfite Sequencing and Methylation Analysis software (http://services.ibc.uni-stuttgart.de/BDPC/BISMA/). Maternal versus paternal alleles were distinguished on the basis of single-nucleotide polymorphisms present when comparing B6 with CAST sequences.
ChIP assays
ChIP was performed using pooled and matched neonatal livers or tongues from equal numbers of H19ICRΔ2,3neo/+ or H19ICR+/+ littermates. Chromatin from finely chopped fresh neonatal livers was cross-linked in Dulbeccos' modified Eagles media, containing 10% fetal bovine serum, protease inhibitors (100 μm phenylmethanesulfonylfluoride, PMSF, 0.5 mm dithiothreitol, DTT and 10 mm sodium butyrate, NaBu) and 1% formaldehyde at room temperature for 10 min. The cross-linking reaction was quenched with 0.125 m glycine for 5 min at room temperature. Cross-linked tissues were washed three times in cold phosphate-buffered saline (PBS) containing protease inhibitors (100 μm PMSF, 0.5 mm DTT and 10 mm NaBu) followed by manual homogenization using a 1 ml dounce homogenizer (Wheaton). Homogenized, cross-linked samples were lysed in SDS lysis buffer containing protease inhibitors (100 μm PMSF, 0.5 mm DTT, 10 mm NaBu and 1/1000 dilution of Protease Inhibitor Cocktail, Roche Diagnostics GmbH) for 1 h on ice followed by sonication in ice slurry using the Biorupter 4CD-200 (Diagenode) pulsing 30 s on and 30 s off for 20–25 min per 500 mg of cells. Successful sonication was determined by reversing cross-links in an aliquot of chromatin incubated overnight at 65°C with RNAse A followed by a 4 h incubation at 50°C with proteinase K. Reversed cross-linked samples were run on 2% agarose gel to confirm equal degree of sonication between samples and sizes equivalent to 1–1000 kb. DNA was measured on reversed cross-linked samples using a nanodrop spectrophotometer (Thermo Scientific). Protein A sepharose beads (GE) were washed twice in PBS and twice in dilution buffer (0.01% sodium dodecyl sulfate, SDS, 1.1% Triton X-100, 1.2 mm ethylenediaminetetraacetic acid, EDTA, 16.7 mm Tris, 167 mm sodium chloride, NaCl) before being resuspended as a 50% slurry in dilution buffer. Beads were then preblocked using 55 μg of sonicated salmon sperm DNA per 100 μl of bead slurry at 4°C for 30 min. Chromatin was resuspended in dilution buffer and precleared in 45 μl of preblocked protein A sepharose bead slurry at 4°C for 30 min. Four percent of each precleared sample was aliquoted and used as input control. Thirty micrograms of precleared chromatin per IP was incubated at 4°C overnight with 2.5 μg of antibody (CTCF, Millipore, IgG Santa Cruz) followed by incubation for 3 h at 4°C with 22 μl preblocked protein A sepharose bead slurry. Chromatin bound beads were then washed once each in the following buffers with protease inhibitors: (A) 0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris, 150 mm NaCl; (B) 0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris, 500 mm NaCl; and (C) 250 mm lithium chloride, LiCl, 1% NP-40, 1% Na-deoxycholate, 1 mm EDTA, 10 mm Tris. Chromatin bound beads were then washed twice in Tris-EDTA (TE) with protease inhibitors. Chromatin was eluted off of beads by incubating at 65°C for 15 min in TE (5 mm EDTA) and 1% SDS. Cross-links were reversed in eluted chromatin and input control samples by incubation at 65°C overnight followed by incubation at 50°C for 4 h with proteinase K. DNA was isolated by phenol chloroform extraction followed by precipitation with glycogen and sodium acetate, NaOAc, in ethyl alcohol, EtOH. Precipitated DNA was washed in 70% EtOH and resuspended in 50 μl of 0.1× TE.
Quantitative PCRs were performed on a LightCycler real-time PCR machine (Roche) using the LightCycler FastStart DNA MasterPlus SYBR Green I kit (Roche). Quantitative PCRs were carried out using 5 μl of ChIPed DNA (diluted 1 : 5) and primers: Rp1-forward primer 5′-GACCATGCCCTATTCTTGGA-3′ and Rp1-reverse primer 5′-ACAGCATTGCCATTTGTGAA-3′. Allele-specific assays were performed using the same primers and DNA concentration used for qPCR but using GoTaq reagents (Promega) and 0.1 μCi of [α32P] dCTP with 32 amplification cycles as was determined to be in the linear range of amplification. Alleles were distinguished by restriction enzyme digestion with NCiI (New England Biolabs), which digests the paternal CAST allele and leaves the maternal B6 allele uncut. Band sizes were resolved on 12% polyacrylamide gel, which were exposed to phosophimager screens and scanned using the Typhoon scanner (GE). Relative band intensities were quantitated using ImageJ software (rsb.info.nih.gov/ij). For both total- and allele-specific assays, levels of DNA immunoprecipitated by CTCF and IgG were determined relative to input controls. Student's paired one-tailed t-test was used to determine whether differences between H19ICRΔ2,3neo/+ or H19ICR+/+ samples were statistically significant (P < 0.05).
Western blotting
Neonatal tissues were collected from heterozygous H19ICRΔ2,3neo/+ pups and flash frozen in liquid nitrogen. Tissues were homogenized in lysis buffer (10% glycerol, 2% SDS, 63 mm Tris–HCl, pH 6.8) with protease inhibitor cocktail (Roche Diagnostics) in the cold room using a Polytron homogenizer at medium speed for 1 min and lysates were cleared by centrifugation. Protein concentrations were quantified by BCA assay (Pierce) and 20 μg of protein lysate was used per sample. 4× sample buffer (NuPAGE SDS–PAGE gel system, Life Technologies) was added to protein lysate, denatured at 70°C for 10 min and run in duplicate on a 4–12% Bis-Tris gel (Life Technologies) alongside protein marker MagicMark XP Western Protein Standard (Life Technologies). The gel was transferred for 2 h at 200 mA constant current onto a nitrocellulose membrane (Millipore). The membrane was blocked in 5% milk, cut to separate samples run in duplicate and each section was probed with primary antibodies for GAPDH (1: 20 000, Cell Signalling) and either CTCF (1:2500, Millipore) or PAR (1:1000, Tulip Biolabs) at 4°C overnight. Membranes were subsequently probed with secondary antibody for 1 h at room temperature (anti-rabbit for CTCF and GAPDH blots and with anti-mouse IgG3-subtype-specific antibody for the PAR blot, Jackson ImmunoResearch). Membranes were then exposed to ECL for 5 min. Bands were visualized using ImageQuant LAS-4000 (Fujifilm) on the standard exposure setting and quantified using ImageJ software (rsb.info.nih.gov/ij). Levels of unmodified CTCF were determined relative to GAPDH levels and compared between tissues from three samples (n = 3). Statistical significance was determined using the pairwise Wilcoxon rank-sum test run under the R software for computing (http://www.r-project.org).
FUNDING
This work was supported by National Institutes of Health (NIH) postdoctoral training grant (T32HD007516), Ruth L. Kirschstein National Research Service Award (F32GM085999) and the NIH funded University of Pennsylvania Center of Excellence in Environmental Toxicology Mentored Scientist Transition Award (1P30 ES013508-05) to F.Y.I. and GM051279 to M.S.B.
ACKNOWLEDGEMENTS
We thank Edward E. Morrisey for providing the Cre mice, Jean Richa and the University of Pennsylvania Transgenic Core Facility for the production of chimeric mice, Sebastien Vigneau for assistance with the ChIP assays, Mirella Meyer-Ficca with the westerns, Gabriel Otte for statistical assistance and Christopher Krapp for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Barlow D.P. Genomic imprinting: a mammalian epigenetic discovery model. Annu. Rev. Genet. 2011;45:379–403. doi: 10.1146/annurev-genet-110410-132459. [DOI] [PubMed] [Google Scholar]
- 2.Lee J.T., Bartolomei M.S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Ideraabdullah F.Y., Vigneau S., Bartolomei M.S. Genomic imprinting mechanisms in mammals. Mutat. Res. 2008;647:77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Crescenzo A., Coppola F., Falco P., Bernardo I., Ausanio G., Cerrato F., Falco L., Riccio A. A novel microdeletion in the IGF2/H19 imprinting centre region defines a recurrent mutation mechanism in familial Beckwith-Wiedemann syndrome. Eur. J. Med. Genet. 2011;54:e451–e454. doi: 10.1016/j.ejmg.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beygo J., Citro V., Sparago A., De Crescenzo A., Cerrato F., Heitmann M., Rademacher K., Guala A., Enklaar T., Anichini C., et al. The molecular function and clinical phenotype of partial deletions of the IGF2/H19 imprinting control region depends on the spatial arrangement of the remaining CTCF-binding sites. Hum. Mol. Genet. 2013;22:544–557. doi: 10.1093/hmg/dds465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berland S., Appelback M., Bruland O., Beygo J., Buiting K., Mackay D.J., Karen Temple I., Houge G. Evidence for anticipation in Beckwith-Wiedemann syndrome. Eur. J. Hum. Genet. 2013;21:1344–1348. doi: 10.1038/ejhg.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poole R.L., Leith D.J., Docherty L.E., Shmela M.E., Gicquel C., Splitt M., Temple I.K., Mackay D.J. Beckwith-Wiedemann syndrome caused by maternally inherited mutation of an OCT-binding motif in the IGF2/H19-imprinting control region, ICR1. Eur. J. Hum. Genet. 2012;20:240–243. doi: 10.1038/ejhg.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demars J., Shmela M.E., Rossignol S., Okabe J., Netchine I., Azzi S., Cabrol S., Le Caignec C., David A., Le Bouc Y., et al. Analysis of the IGF2/H19 imprinting control region uncovers new genetic defects, including mutations of OCT-binding sequences, in patients with 11p15 fetal growth disorders. Hum. Mol. Genet. 2010;19:803–814. doi: 10.1093/hmg/ddp549. [DOI] [PubMed] [Google Scholar]
- 9.Chitayat D., Rothchild A., Ling E., Friedman J.M., Couch R.M., Yong S.L., Baldwin V.J., Hall J.G. Apparent postnatal onset of some manifestations of the Wiedemann-Beckwith syndrome. Am. J. Med. Genet. 1990;36:434–439. doi: 10.1002/ajmg.1320360412. [DOI] [PubMed] [Google Scholar]
- 10.Shuman C., Beckwith J.B., Smith A.C., Weksberg R. Beckwith-Wiedemann syndrome. In: Pagon R.A., Adam M.P., Bird T.D., editors. GeneReviews. WA: University of Washington, Seattle; 2010. http://www.ncbi.nlm.nih.gov/books/NBK1394/ [PubMed] [Google Scholar]
- 11.Choufani S., Shuman C., Weksberg R. Beckwith-Wiedemann syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:343–354. doi: 10.1002/ajmg.c.30267. [DOI] [PubMed] [Google Scholar]
- 12.Choufani S., Shuman C., Weksberg R. Molecular findings in Beckwith-Wiedemann syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2013;163C:131–140. doi: 10.1002/ajmg.c.31363. [DOI] [PubMed] [Google Scholar]
- 13.Weksberg R., Smith A.C., Squire J., Sadowski P. Beckwith-Wiedemann syndrome demonstrates a role for epigenetic control of normal development. Hum. Mol. Genet. 2003;12(Spec No 1):R61–R68. doi: 10.1093/hmg/ddg067. [DOI] [PubMed] [Google Scholar]
- 14.Bliek J., Maas S.M., Ruijter J.M., Hennekam R.C., Alders M., Westerveld A., Mannens M.M. Increased tumour risk for BWS patients correlates with aberrant H19 and not KCNQ1OT1 methylation: occurrence of KCNQ1OT1 hypomethylation in familial cases of BWS. Hum. Mol. Genet. 2001;10:467–476. doi: 10.1093/hmg/10.5.467. [DOI] [PubMed] [Google Scholar]
- 15.Cerrato F., Sparago A., Verde G., De Crescenzo A., Citro V., Cubellis M.V., Rinaldi M.M., Boccuto L., Neri G., Magnani C., et al. Different mechanisms cause imprinting defects at the IGF2/H19 locus in Beckwith-Wiedemann syndrome and Wilms’ tumour. Hum. Mol. Genet. 2008;17:1427–1435. doi: 10.1093/hmg/ddn031. [DOI] [PubMed] [Google Scholar]
- 16.Sparago A., Cerrato F., Vernucci M., Ferrero G.B., Silengo M.C., Riccio A. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat. Genet. 2004;36:958–960. doi: 10.1038/ng1410. [DOI] [PubMed] [Google Scholar]
- 17.Sparago A., Russo S., Cerrato F., Ferraiuolo S., Castorina P., Selicorni A., Schwienbacher C., Negrini M., Ferrero G.B., Silengo M.C., et al. Mechanisms causing imprinting defects in familial Beckwith-Wiedemann syndrome with Wilms’ tumour. Hum. Mol. Genet. 2007;16:254–264. doi: 10.1093/hmg/ddl448. [DOI] [PubMed] [Google Scholar]
- 18.Prawitt D., Enklaar T., Gartner-Rupprecht B., Spangenberg C., Lausch E., Reutzel D., Fees S., Korzon M., Brozek I., Limon J., et al. Microdeletion and IGF2 loss of imprinting in a cascade causing Beckwith-Wiedemann syndrome with Wilms’ tumor. Nat. Genet. 2005;37:785–786. doi: 10.1038/ng0805-785. [DOI] [PubMed] [Google Scholar]
- 19.Frevel M.A.E., Hornberg J.J., Reeve A.E. A potential imprint control element: identification of a conserved 42 bp sequence upstream of H19. Trends Genet. 1999;15:216–218. doi: 10.1016/s0168-9525(99)01752-7. [DOI] [PubMed] [Google Scholar]
- 20.Stadnick M.P., Pieracci F.M., Cranston M.J., Taksel E., Thorvaldsen J.L., Bartolomei M.S. Role of a 461 bp G-rich repetitive element in H19 transgene imprinting. Dev. Genes Evol. 1999;209:239–248. doi: 10.1007/s004270050248. [DOI] [PubMed] [Google Scholar]
- 21.Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 22.Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 23.Szabo P.E., Tang S.-H., Rentsendorj A., Pfeifer G.P., Mann J.R. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- 24.Engel N., Thorvaldsen J.L., Bartolomei M.S. CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum. Mol. Genet. 2006;15:2945–2954. doi: 10.1093/hmg/ddl237. [DOI] [PubMed] [Google Scholar]
- 25.Engel N.I., West A.E., Felsenfeld G., Bartolomei M.S. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is revealed by CpG mutations. Nat. Genet. 2004;36:883–888. doi: 10.1038/ng1399. [DOI] [PubMed] [Google Scholar]
- 26.Thorvaldsen J.L., Duran K.L., Bartolomei M.S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mann M.R.W., Chung Y.G., Nolen L.D., Verona R.I., Latham K.E., Bartolomei M.S. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod. 2003;69:902–914. doi: 10.1095/biolreprod.103.017293. [DOI] [PubMed] [Google Scholar]
- 28.Eggermann T. Russell-Silver syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:355–364. doi: 10.1002/ajmg.c.30274. [DOI] [PubMed] [Google Scholar]
- 29.Pant V., Mariano P., Kanduri C., Mattsson A., Lobanenkov V., Heuchel R., Ohlsson R. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 2003;17:586–590. doi: 10.1101/gad.254903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoenherr C.J., Levorse J.M., Tilghman S.M. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 31.Szabo P.E., Pfeifer G.P., Mann J.R. Parent-of-origin-specific binding of nuclear hormone receptor complexes in the H19-Igf2 imprinting control region. Mol. Cell. Biol. 2004;24:4858–4868. doi: 10.1128/MCB.24.11.4858-4868.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ideraabdullah F.Y., Abramowitz L.K., Thorvaldsen J.L., Krapp C., Wen S.C., Engel N., Bartolomei M.S. Novel cis-regulatory function in ICR-mediated imprinted repression of H19. Dev. Biol. 2011;355:349–357. doi: 10.1016/j.ydbio.2011.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tremblay K.D., Duran K.L., Bartolomei M.S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Constancia M., Dean W., Lopes S., Moore T., Kelsey G., Reik W. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat. Genet. 2000;26:203–206. doi: 10.1038/79930. [DOI] [PubMed] [Google Scholar]
- 35.Eden S., Constancia M., Hashimshony T., Dean W., Goldstein B., Johnson A.C., Keshet I., Reik W., Cedar H. An upstream repressor element plays a role in Igf2 imprinting. EMBO J. 2001;20:3518–3525. doi: 10.1093/emboj/20.13.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorvaldsen J.L., Mann M.R., Nwoko O., Duran K.L., Bartolomei M.S. Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting. Mol. Cell. Biol. 2002;22:2450–2462. doi: 10.1128/MCB.22.8.2450-2462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hellman A., Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 38.Kaffer C.R., Grinberg A., Pfeifer K. Regulatory mechanisms at the mouse Igf2/H19 locus. Mol. Cell. Biol. 2001;21:8189–8196. doi: 10.1128/MCB.21.23.8189-8196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leighton P.A., Saam J.R., Ingram R.S., Stewart C.L., Tilghman S.M. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 40.Klenova E.M., Nicolas R.H., Paterson H.F., Carne A.F., Heath C.M., Goodwin G.H., Neiman P.E., Lobanenkov V.V. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klenova E.M., Nicolas R.H., U S., Carne A.F., Lee R.E., Lobanenkov V.V., Goodwin G.H. Molecular weight abnormalities of the CTCF transcription factor: CTCF migrates aberrantly in SDS-PAGE and the size of the expressed protein is affected by the UTRs and sequences within the coding region of the CTCF gene. Nucleic Acids Res. 1997;25:466–474. doi: 10.1093/nar/25.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klenova E.M., Chernukhin I.V., El-Kady A., Lee R.E., Pugacheva E.M., Loukinov D.I., Goodwin G.H., Delgado D., Filippova G.N., Leon J., et al. Functional phosphorylation sites in the C-terminal region of the multivalent multifunctional transcriptional factor CTCF. Mol. Cell. Biol. 2001;21:2221–2234. doi: 10.1128/MCB.21.6.2221-2234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu W., Ginjala V., Pant V., Chernukhin I., Whitehead J., Docquier F., Farrar D., Tavoosidana G., Mukhopadhyay R., Kanduri C., et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 44.MacPherson M.J., Beatty L.G., Zhou W., Du M., Sadowski P.D. The CTCF insulator protein is posttranslationally modified by SUMO. Mol. Cell. Biol. 2009;29:714–725. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zlatanova J., Caiafa P. CTCF and its protein partners: divide and rule? J. Cell Sci. 2009;122:1275–1284. doi: 10.1242/jcs.039990. [DOI] [PubMed] [Google Scholar]
- 46.Higashimoto K., Jozaki K., Kosho T., Matsubara K., Fuke T., Yamada D., Yatsuki H., Maeda T., Ohtsuka Y., Nishioka K., et al. A novel de novo point mutation of the OCT-binding site in the IGF2/H19-imprinting control region in a Beckwith-Wiedemann syndrome patient. Clin Genet. 2013 doi: 10.1111/cge.12318. doi:10.1111/cge.12318. [DOI] [PubMed] [Google Scholar]
- 47.Hori N., Nakano H., Takeuchi T., Kato H., Hamaguchi S., Oshimura M., Sato K. A dyad oct-binding sequence functions as a maintenance sequence for the unmethylated state within the H19/Igf2-imprinted control region. J. Biol. Chem. 2002;277:27960–27967. doi: 10.1074/jbc.M202280200. [DOI] [PubMed] [Google Scholar]
- 48.Sakaguchi R., Okamura E., Matsuzaki H., Fukamizu A., Tanimoto K. Sox-Oct motifs contribute to maintenance of the unmethylated H19 ICR in YAC transgenic mice. Hum. Mol. Genet. 2013;22:4627–4637. doi: 10.1093/hmg/ddt311. [DOI] [PubMed] [Google Scholar]
- 49.Quenneville S., Verde G., Corsinotti A., Kapopoulou A., Jakobsson J., Offner S., Baglivo I., Pedone P.V., Grimaldi G., Riccio A., et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X., Ito M., Zhou F., Youngson N., Zuo X., Leder P., Ferguson-Smith A.C. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee D.H., Singh P., Tsai S.Y., Oates N., Spalla A., Spalla C., Brown L., Rivas G., Larson G., Rauch T.A., et al. CTCF-dependent chromatin bias constitutes transient epigenetic memory of the mother at the H19-Igf2 imprinting control region in prospermatogonia. PLoS Genet. 2010;6:e1001224. doi: 10.1371/journal.pgen.1001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nativio R., Sparago A., Ito Y., Weksberg R., Riccio A., Murrell A. Disruption of genomic neighbourhood at the imprinted IGF2-H19 locus in Beckwith-Wiedemann syndrome and Silver-Russell syndrome. Hum. Mol. Genet. 2011;20:1363–1374. doi: 10.1093/hmg/ddr018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanduri M., Kanduri C., Mariano P., Vostrov A.A., Quitschke W., Lobanenkov V., Ohlsson R. Multiple nucleosome positioning sites regulate the CTCF-mediated insulator function of the H19 imprinting control region. Mol. Cell. Biol. 2002;22:3339–3344. doi: 10.1128/MCB.22.10.3339-3344.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szabo P.E., Han L., Hyo-Jung J., Mann J.R. Mutagenesis in mice of nuclear hormone receptor binding sites in the Igf2/H19 imprinting control region. Cytogenet. Genome Res. 2006;113:238–246. doi: 10.1159/000090838. [DOI] [PubMed] [Google Scholar]
- 55.Drewell R.A., Brenton J.D., Ainscough J.F., Barton S.C., Hilton K.J., Arney K.L., Dandolo L., Surani M.A. Deletion of a silencer element disrupts H19 imprinting independently of a DNA methylation epigenetic switch. Development. 2000;127:3419–3428. doi: 10.1242/dev.127.16.3419. [DOI] [PubMed] [Google Scholar]
- 56.Ainscough J.F., Dandolo L., Surani M.A. Appropriate expression of the mouse H19 gene utilises three or more distinct enhancer regions spread over more than 130 kb. Mech. Dev. 2000;91:365–368. doi: 10.1016/s0925-4773(99)00289-0. [DOI] [PubMed] [Google Scholar]
- 57.Yoo-Warren H., Pachnis V., Ingram R.S., Tilghman S.M. Two regulatory domains flank the mouse H19 gene. Mol. Cell. Biol. 1988;8:4707–4715. doi: 10.1128/mcb.8.11.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ong C.T., Van Bortle K., Ramos E., Corces V.G. Poly(ADP-ribosyl)ation regulates insulator function and intrachromosomal interactions in Drosophila. Cell. 2013;155:148–159. doi: 10.1016/j.cell.2013.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farrar D., Rai S., Chernukhin I., Jagodic M., Ito Y., Yammine S., Ohlsson R., Murrell A., Klenova E. Mutational analysis of the poly(ADP-ribosyl)ation sites of the transcription factor CTCF provides an insight into the mechanism of its regulation by poly(ADP-ribosyl)ation. Mol. Cell. Biol. 2010;30:1199–1216. doi: 10.1128/MCB.00827-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caiafa P., Zlatanova J. CCCTC-binding factor meets poly(ADP-ribose) polymerase-1. J. Cell. Physiol. 2009;219:265–270. doi: 10.1002/jcp.21691. [DOI] [PubMed] [Google Scholar]
- 61.Kuhn R., Rajewsky K., Muller W. Generation and analysis of interleukin-4 deficient mice. Science. 1991;254:707–710. doi: 10.1126/science.1948049. [DOI] [PubMed] [Google Scholar]
- 62.Thorvaldsen J.L., Fedoriw A.M., Nguyen S., Bartolomei M.S. Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus. Mol. Cell. Biol. 2006;26:1245–1258. doi: 10.1128/MCB.26.4.1245-1258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weaver J.R., Sarkisian G., Krapp C., Mager J., Mann M.R., Bartolomei M.S. Domain-specific response of imprinted genes to reduced DNMT1. Mol. Cell. Biol. 2010;30:3916–3928. doi: 10.1128/MCB.01278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartolomei M.S., Webber A.L., Brunkow M.E., Tilghman S.M. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]