Abstract
During postnatal development, neuronal activity controls the remodeling of initially imprecise neuronal connections through the regulation of gene expression. MeCP2 binds to methylated DNA and modulates gene expression during neuronal development and MECP2 mutation causes the autistic disorder Rett syndrome. To investigate a role for MeCP2 in neuronal circuit refinement and to identify activity-dependent MeCP2 transcription regulations, we leveraged the precise organization and accessibility of olfactory sensory axons to manipulation of neuronal activity through odorant exposure in vivo. We demonstrate that olfactory sensory axons failed to develop complete convergence when Mecp2 is deficient in olfactory sensory neurons (OSNs) in an otherwise wild-type animal. Furthermore, we demonstrate that expression of selected adhesion genes was elevated in Mecp2-deficient glomeruli, while acute odor stimulation in control mice resulted in significantly reduced MeCP2 binding to these gene loci, correlating with increased expression. Thus, MeCP2 is required for both circuitry refinement and activity-dependent transcriptional responses in OSNs.
INTRODUCTION
The murine olfactory system exhibits intricate and precise neuronal connectivity in which olfactory sensory neuron (OSN) axons expressing the same odorant receptor (OR) target a common region or glomerulus in the olfactory bulb (OB) (1). Each mouse OSN expresses only one of the ∼1000 OR genes found in the mouse genome (2). OSNs expressing the same OR are widely dispersed in the main olfactory epithelium (MOE) but their axons navigate along different paths from the MOE and eventually converge onto the same glomerulus (1,3). Similar to the development of other neuronal connections, olfactory connectivity is initially exuberant then refined postnatally to achieve OR-specific glomerular targeting (4,5). Neuronal activity plays a critical role in the maintenance and refinement of olfactory connectivity. Though many gene expression changes have been associated with neuronal activity and glomerular convergence in the OSNs (6–8), the regulatory mechanisms of activity-dependent gene expression in the MOE controlling this process are not entirely clear. MeCP2 is one of the key regulators linking neuronal activity to gene expression (9). MeCP2 binds to methylated Cytosine within DNA and regulates gene expression through chromatin remodeling and promoter-mediated transcriptional regulation (10–12). Activity-dependent modification of MeCP2 and specific activity-dependent MeCP2 target genes have been shown in neurons in vitro (9,13,14), but the precise mechanism of MeCP2 regulation of activity-dependent gene expression in vivo is still elusive (15).
Therefore, to determine the role of MeCP2 in neuronal circuitry refinement and activity-dependent transcriptional regulation, we took advantage of the unique precision and genetic traceability of olfactory sensory axon connectivity and the accessibility of OSNs to activity manipulation in vivo. It has been established that MeCP2 is expressed in the MOE and plays a role in the regulation of OSN maturation (16). Though MeCP2 does not appear to regulate olfactory axon growth and targeting into its general glomerular location in the OB, the requirement of Mecp2 in olfactory circuitry refinement within OSNs are less clear (17,18). In this study, we investigate the hypothesis that MeCP2 is required within OSNs for the refinement of olfactory axon convergence through regulation of cell adhesion molecule gene expression. To study further the mechanisms by which MeCP2 mediates neuronal activity-dependent gene expression, we exposed both wild-type and Mecp2−/y mice to odor stimulation and subsequently analyzed selected target gene expression in MOE. The effects of odor stimulation on MeCP2-binding genome wide were further determined by ChIP-seq analysis. Through comparison of gene expression changes in wild-type and Mecp2−/y MOE and integration with MeCP2 ChIP-seq data we sought to determine the role of MeCP2 in the regulation of neuronal activity-dependent gene expression in vivo.
RESULTS
Mecp2 is required in OSNs for the refinement of olfactory axon convergence
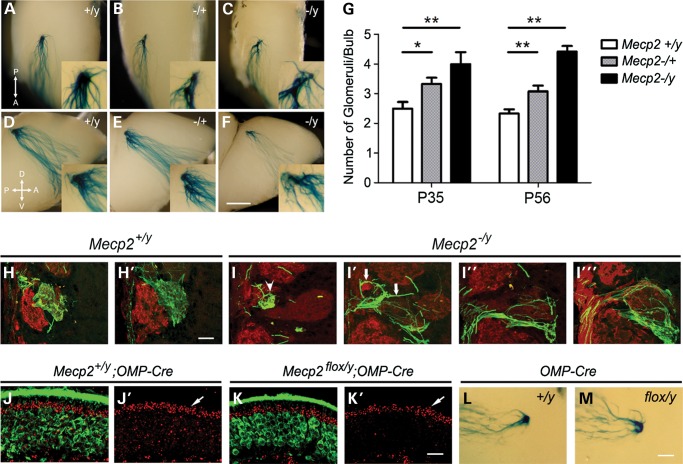
Previous studies demonstrated that Mecp2 regulates OSN differentiation and glomerular organization in the OB during early postnatal stages (17,18). It has also been shown that olfactory axons target to their respective location in the adult OB (18). In this study, we directly investigated whether Mecp2/MeCP2 is required for the refinement of olfactory axon convergence during postnatal development. Since Mecp2 is an X-linked gene, M72-IRES-taulacZ mice were bred with Mecp2−/+ mice to allow visualization of M72 OSN axons in the OB in wild-type compared with Mecp2−/y (hemizygous null) littermates. Consistent with previous reports, the locations that M72-IRES-taulacZ axons converge onto in the OB were comparable between Mecp2−/y and Mecp2+/y mice (17). In wild-type 8-week-old animals, the majority of olfactory axons expressing M72 and taulacZ coalesce into two glomeruli: one on the medial side and one on the lateral side of the OB (Fig. 1A and D). In both Mecp2−/+ heterozygous and Mecp2−/y hemizygous mice, we observed that M72 axons target onto multiple glomeruli in each OB (Fig. 1B, C, E and F). Supernumerary glomeruli appear to be close to the M72 glomerular location on both the medial and lateral side of the OB.
Figure 1.
Mecp2 is required in OSNs for the refinement of olfactory sensory axon convergence. M72-IRES-taulacZ axons converge in wild-type adult OB, one on the lateral side (A) and one on the medial side (D). In adult Mecp2−/+ heterozygous (B, E) and Mecp2−/y hemizygous OB (C, F). (G). Lack of complete M72 olfactory sensory axon convergence persisted into adulthood in (P56) Mecp2 mutants (G). Serial sections of wild-type OB show M72-IRES-taulacZ axons (green) travel in the olfactory nerve layer and terminate into glomeruli delineated by OMP IF (red) (H, H′). Abnormal fasciculation (arrows) and mistargeting of M72 axons (arrowhead) are observed in Mecp2−/y OSN axons (I–I′′′). Mecp2 expression is ablated in mature OSNs via OMP-Cre-mediated events. In control mice, MeCP2 expression (red in J and J′) is present in OMP expressing (green in J) OSNs. In OMP-Cre-mediated conditional knockout OE, Mecp2 expression (red in K and K′) is not detectable in mature OSNs but is at the same level in sustentacular cells compared with control (arrows in J′ and K′). Supernumerary M72 glomeruli were detected in P56 Mecp2flox/y;OMP-Cre OB (M) but not in Mecp2+/y;OMP-Cre OB (L). Bar = 500 µm in (A)–(F), 50 µm in (H)–(I), 25 µm in (J)–(K) and 250 µm in (L)–(M). Student's t-test *P < 0.05, **P < 0.01.
Normally M72 glomeruli refine from multiple, heterogeneous glomeruli to a single mature homogenous glomerulus in each hemi-bulb between postnatal days (PD) 20–40 (4). Therefore, the presence of supernumerary glomeruli in Mecp2−/y deficient mice could be the result of a lack of glomerular refinement, or a deficiency of maintaining the convergence. We evaluated whether M72 glomeruli refine normally during postnatal development by examining the average glomerular number at PD35, when refinement is completed, in wild-type and Mecp2−/y mice. The average M72 glomerular numbers per bulb at PD35 were 2.5 ± 0.54 (n = 8) in Mecp2+/y wild type; 3.33 ± 0.51 (n = 8) in Mecp2−/+ heterozygous and 4.0 ± 0.82 (n = 6) in Mecp2−/y mice. At PD56 (8 weeks old), the average M72 glomerular numbers per bulb were 2.33 ± 0.49 (n = 12) in Mecp2+/y wild type; 3.08 ± 0.66 (n = 12) in Mecp2−/+ heterozygous and 4.41 ± 0.66 (n = 12) in Mecp2−/y mice. These data demonstrate a lack of axon refinement phenotype in Mecp2 mutant OB that persisted into adulthood (Fig. 1G).
To better characterize OSN axon organization and OB glomerular targeting axon localization studies were performed in serial OB sections. M72 axons travelled through the olfactory nerve layer and targeted into a single glomerulus per half bulb in Mecp2+/y wild-type control mice. M72 axons also distributed uniformly and occupied the entire glomerulus (Fig. 1H). Few M72 axons were detected wandering through the glomerular layer in wild-type animals. In Mecp2−/y mice, M72 axons appeared to form multiple fasciculate bundles while traveling from the nerve layer into the glomerular layer. Multiple OB glomerular trespassing events were observed and axons traveling through the glomerulus were often found in abnormal bundles (Fig. 1I). Perhaps most indicative of disrupted activity-based pruning, M72 axons appear to terminate into multiple glomeruli consistent with the observations made with the whole-mount staining (Supplementary Material, Fig. S1). These analyses indicate that olfactory axon fasciculation and refinement defects in OB are due to loss of Mecp2.
The specific requirement for Mecp2 function in the refinement of olfactory axon convergence was further investigated by the specific genetic deletion of Mecp2 in OSNs using an olfactory marker protein (OMP) promoter driving Cre recombinase. For this purpose, heterozygous Mecp2tm1.1Bird/J females were mated with OMP-Cre expressing males carrying M72-IRES-taulacZ alleles to produce males with either retained Mecp2 expression or the absence of MeCP2 solely in OMP expressing OSNs but not in other OB cell types. The resulting loss of MeCP2 in mature OSNs was validated in male Mecp2flox/y mice carrying the allele bearing the floxed Mecp2 exons 3 and 4 by immunofluorescence (IF) analysis (Fig. 1J and K). As expected, the supernumerary glomeruli phenotype was detected in all OSN-specific Mecp2flox/y knockout animals by x-gal staining (n = 8) (Fig. 1L and M). Together, these results establish that MeCP2 is specifically required in the mature OSNs for the completion of olfactory axon convergence.
MeCP2 and differential expression of Kirrel2, Kirrel3 and Pcdh20 in OSNs
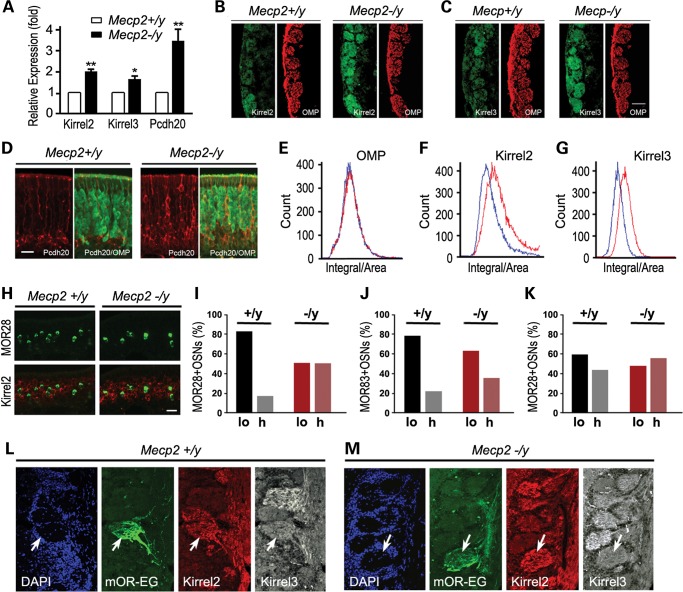
Differential expression patterns of key OSN cell adhesion molecules were found to be variable among different OSN populations. In combination, these gene expression differences could constitute the OSN identity that guides and maintains homotypic OSN axons (6,7,19). First, we investigated whether MeCP2 regulates expression of Kirrel2, Kirrel3, Pcdh20 and Cntn4, genes encoding surface cell adhesion molecule that play important roles in defining olfactory axon identities. Fortuitously, these genes are specifically expressed only in OSNs but not in supporting cells and other cell types within the olfactory epithelium (6,7,19,20). This fact allowed us to analyze the entire MOE for transcript levels of these genes expressed in OSNs. Kirrel2, Kirrel3 and Pcdh20 mRNA transcript levels were determined by quantitative real-time qRT-PCR (Fig. 2A). Relative transcript levels were normalized against β-III tubulin expression. Kirrel2 and Kirrel3 transcripts in Mecp2−/y MOE were significantly increased 2.03-fold (SD = 0.19, P < 0.001) and 1.65 ± 0.23-fold (P < 0.02) compared with their respective wild-type control expression levels. Also, Cntn4 transcript levels were significantly elevated albeit the increase was smaller (1.26-fold, SD = 0.11) between wild-type control and Mecp2−/y mice (P < 0.02) (Supplementary Material, Fig. S2A). Pcdh20 transcript level in Mecp2−/y MOE was notably 3.50-fold (SD = 0.50, P > 0.001) of that in the wild-type control (Fig. 2A). These results indicate that Mecp2 functions to repress Kirrel2, Kirrel3, Pcdh20 and Cntn4 adhesion molecule expression in OSNs.
Figure 2.
Mecp2 suppresses Kirrel2, Kirrel3 and Pcdh20 expression. (A) Relative transcript levels of Kirrel2, Kirrel3 and Pcdh20 in Mecp2−/y OE to wild type. (B) Kirrel2 IF signals (green) in Mecp2−/y (right panel) and wild-type OB (left panel). OMP (red) signal outlines the location of the glomerular layer. (C) Kirrel3 IF signals (green) in Mecp2−/y glomeruli (right panel) compared with Mecp2+/y control (right panel). (D) Pcdh20 expression (red) in the MOE localizes to OSNs below the OMP-positive (green) OSN layer in the Mecp2+/y control OB (left panel) while in Mecp2−/y mice, Pcdh20-positive OSNs (red) also appear in the OMP cell layers (right panel). (E) OMP signal levels are similar between wild-type control (blue line) and Mecp2−/y (red line) within the OB glomeruli. (F, G) Kirrel2 (F) and Kirrel3 (G) signal levels (red lines) in Mecp2−/y glomeruli increase significantly compared with wild-type control levels (blue lines)) (t-test: P < 1e−30). (H) Double in situ hybridization for Kirrel2 expression (red) and MOR28-positive (green) OSNs in wild-type control Mecp2+/y (left panel) and Mecp2−/y OB (right panel). (I) 83.5% of MOR28-positive OSNs in wild-type OE (left panel) have low levels of Kirrel2 expression (lo) and 16.5% had high levels of Kirrel2 (hi while in Mecp2−/y OE, MOR28-positive OSNs roughly equal proportions of Kirrel2 low and high populations were detected (50%) in OE. n = 200. (J) Kirrel3 expression is low in 78.8% of MOR83-positive OSNs in wild-type control versus 64.2% in Mecp2−/y OE. (K) 42.3% of MOR28-positive neurons express Pcdh20 in wild-type control OE versus 53.8% in Mecp2−/y, n = 100 for (I)–(K). (L, M) mOR-EG glomeruli are shown in mOR-EG-IRES-tauGFP OB (green). Kirrel2 and Kirrel3 immunoreactivity levels are visualized in both wild-type control (left panel) and Mecp2−/y OB (right panel). Scale bars = 100 µm in (B), (C), (L) and (M); 35 µm in (D) and (H). Student's t-test *P < 0.01, **P < 0.001.
Kirrel2 and Kirrel3 proteins are localized at the olfactory axon terminals within the glomeruli (7). To examine whether the levels of Kirrel2 and Kirrel3 are regulated by MeCP2, we performed IF analysis to assay the products of these two genes in OB. Consistent with their transcript expression results, we observed increased expression of Kirrel2, Kirrel3 and Pcdh20 in the glomerular layer of Mecp2−/y OB compared with that of the Mecp2+/y wild-type OB (Fig. 2B and C). We further quantified IF signals of Kirrel2 and Kirrel3 within the glomeruli by laser scanning cytometry (LSC). For this sensitive, quantitative assay serial sections of the OBs were immunostained and each glomerulus was first identified based on OMP staining. Then fluorescence intensities of Kirrel2 and Kirrel3 IF signals were determined within each individual glomerulus. While the distribution of OMP signal intensities did not significantly change in glomeruli of Mecp2−/y OB, both Kirrel2 and Kirrel3 levels were significantly higher in Mecp2−/y OB compared with the wild-type control OBs (P < 1e−30, Fig. 2E–G). In agreement with the lower magnitude of transcript expression change, a small but significant increase in Cntn4 staining intensity between Mecp2−/y OB and wild-type OB glomeruli was observed (P < 0.02, Supplementary Material, Fig. S2B). Thus, Kirrel2, Kirrel3, Pcdh20 and Cntn4 gene expression levels in the OB are significantly altered by loss of MeCP2.
Kirrel2 and Kirrel3 exhibit differential expression patterns among glomeruli with different OR identities. In general, high expression of either protein is correlated with low expression of the other within individual OB glomeruli (7). Therefore, whether or not the alteration of Kirrel2/3 expression patterns by loss of MeCP2 distorts the expression diversity of these genes among OB glomeruli was examined. IF signals of each glomerulus were extracted from LSC datasets and plotted according to their staining intensity ranking (Supplementary Material, Fig. S2D–E). Both Kirrel2 and Kirrel3 glomerular staining showed a wide range of intensity with the majority of the glomeruli having intermediate signal levels and fewer glomeruli exhibiting either very low or very high levels of expression. In Mecp2−/y OB, the glomerular staining intensity of Kirrel2 and Kirrel3 was overall higher than in the Mecp2+/y wild-type control (Supplementary Material, Fig. S2D–E). However, the absolute differential expression range for the glomeruli remained the same. Specifically, the glomerular signal intensity plot was shifted upward but maintained the same general pattern of distribution for both Kirrel2 and Kirrel3 (Supplementary Material, Fig. S2D–E). The glomerular signal for Cntn4 while also elevated did not show a significant difference between Mecp2−/y OB and wild-type littermates (Supplementary Material, Fig. S2C). Therefore, MeCP2 does not appear to determine the range of diversity of cell surface protein expression levels among different OSN populations.
To further evaluate whether the inverse relationship between Kirrel2 and Kirrel3 expression in the glomerulus is influenced by MeCP2, we immunostained and compared the expression of Kirrel2 and Kirrel3 in mouse eugenol odorant receptor (mOR-EG) glomeruli (Fig. 2L and M). In wild-type mOR-EG glomeruli, Kirrel2 expression is high and Kirrel3 expression is relatively low. Thus, the relative expression of Kirrel2 and Kirrel3 protein levels in mOR-EG glomeruli is preserved in Mecp2−/y OB as well.
Kirrel2 and Kirrel3 expression within OSN subpopulations was further examined by double in situ hybridization to determine whether MeCP2 defines OR-specific expression levels of Kirrel2 and Kirrel3. Consistent with protein expression results, Kirrel2 and Kirrel3 transcript levels are higher in Mecp2−/y OE than that of wild-type OE (Fig. 2H). It has previously been established that Kirrel2 and Kirrel3 are expressed in a complementary manner in different OSN populations. In MOR28-positive OSN populations, Kirrel2 is expressed at relatively low levels and Kirrel3 is at relatively high levels. In MOR83-positive OSN populations, Kirrel3 is expressed at low levels, whereas Kirrel2 is expressed at higher levels (7). We first confirmed that the majority of MOR28 OSNs (83.5%) express low or non-detectable levels of Kirrel2 in the wild-type OE. With an overall increase in Kirrel2 expression in the olfactory epithelium, MOR28 OSNs with identifiable Kirrel2 in situ transcript signals increased from 26.5% in wild type to 50% in Mecp2−/y OE (Fig. 2I). An increase in the ratio of Kirrel3 transcript-positive OSNs was also observed within MOR83 population in Mecp2−/y OE with 78.8% positive compared with 64.2% in the wild-type OE (Fig. 2J).
Pcdh20 protein is localized in the OSN cell bodies and along their axons. In wild-type adult MOE, Pcdh20 is expressed by immature OSNs located below OMP-positive OSNs (20). Increases in the number of Pcdh20-positive OSNs was observed and more Pcdh20-positive neurons were seen within OMP-positive mature OSN layer in Mecp2−/y mice (Fig. 2D). Compared with the wild-type controls, more Pcdh20-positive OSN axons were observed within the glomeruli of Mecp2−/y OBs. Instead of an overall increase of Pcdh20 IF signal, we often observe individual Pcdh20-positive olfactory axon fibers within the glomeruli. Pcdh20 IF levels, although trending higher, in the glomeruli quantified by LSC were not significantly different (Supplementary Material, Fig. S2C). However, an increase in the ratio of Pcdh20-positive OSNs was identified within MOR28 population from 42.3% in Mecp2+/y control OB to 53.8% in Mecp2−/y OB (Fig. 2K).
Odor-evoked activity alters MeCP2 binding to Kirrel2 and Pcdh20 in OSNs
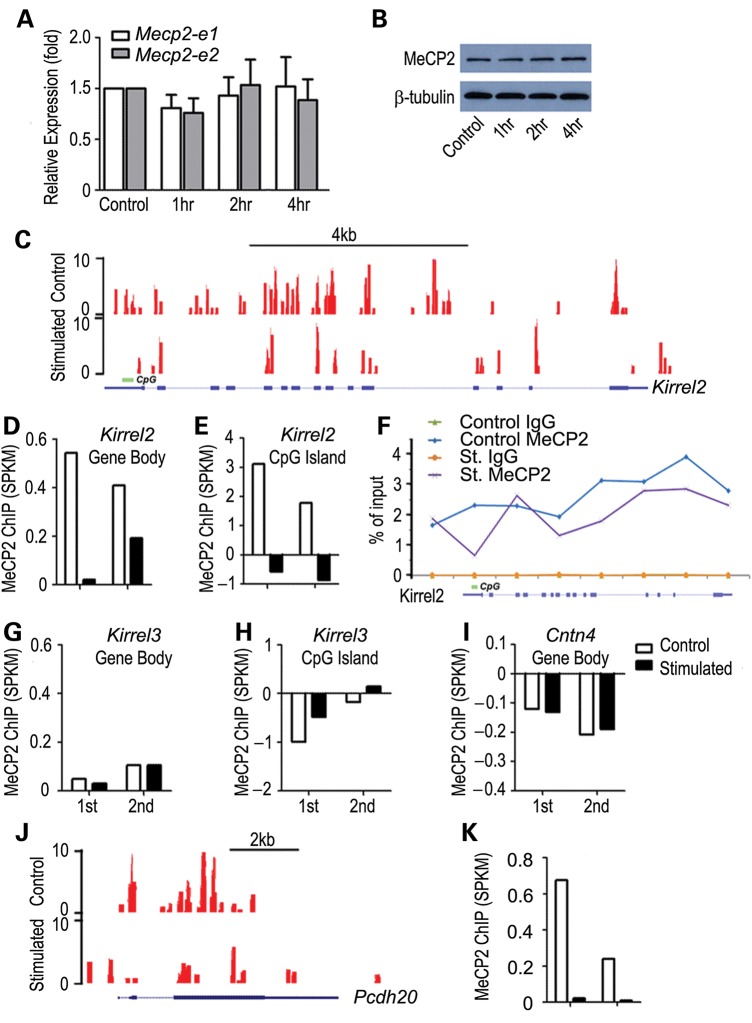
Kirrel2 in OSNs is expressed in an activity-dependent manner. Long-term blocking of odor-evoked activity results in a decrease in Kirrel2 expression (7). To determine a possible role of Mecp2 in the regulation of activity-dependent gene expression in OSNs, we first compared Mecp2 expression levels under odor stimulation with that of the control air flow condition. Under the control condition, test mice were maintained with a constant flow of filtered air. Odor stimulation was given using a mixture of complex odorants in a laminar flow exposure. After 4 h of odor stimulation, no change in Mecp2 transcript levels was observed by real-time RT-PCR (Fig. 3A). Consistent with qRT-PCR data, western blot analysis indicated no significant change in MeCP2 protein levels in the MOE with odor stimulation (Fig. 3B).
Figure 3.
Odor stimulation results in decreased MeCP2 binding within Kirrel2 and Pcdh20 loci. (A) Mecp2 transcript levels for both Mecp2_e1 and Mecp2_e2 isoforms showed no significant change in either transcript between filtered air control and odor stimulation after 4 h. t-test P > 0.1 (A). Western blotting analysis showed no change in MeCP2 protein in the MOE after acute odor stimulation (B). ChIP-seq analysis of MeCP2 binding in the control filtered air exposed and 4 h complex odor mixture stimulated animals are shown. Example UCSC genome browser tracks of MeCP2 binding to Kirrel2 locus show a widespread binding pattern throughout the gene body (C). Duplicate sets of ChIP-seq reads (1st and 2nd) are mapped and normalized against input. While control mouse datasets show significant MeCP2 binding, odor stimulation results in dramatically reduced MeCP2 binding to the Kirrel2 locus (D). MeCP2 binding to the promoter CpG island of Kirrel2 also shows a sharp drop (E). Eight regions along Kirrel2 locus, including 5′ upstream, promoter CpG, exon and intronic regions, were chosen for quantitative ChIP-PCR analysis. MeCP2 binding to six out of the eight regions decreases consistent with the ChIP-seq analysis (F). No binding was observed with IgG control (green and brown lines in F). No detectable change in MeCP2 binding to the Kirrel3 locus, both throughout the entire locus (G) and at the CpG island (H) is observed. No significant MeCP2 binding to the Cntn4 locus is detected (I). Odorant stimulation does not alter MeCP2 binding within these two gene loci (G and I). MeCP2 binding to the Pcdh20 locus under control condition is also distributed along the entire locus (J). A significant decrease in MeCP2 binding to the Pcdh20 locus was observed under odorant stimulation (J and K). Read counts are shown on the Y-axis in (C) and (J).
To study MeCP2 binding to the chromatin of our target gene regions, we performed chromatin immunoprecipitation high-throughput sequencing (ChIP-seq) analysis for the MOE. Genomic DNA fragments bound to MeCP2 were isolated by immunoprecipitation (Supplementary Material, Fig. S4). High-throughput Illumina sequencing was performed to identify these MeCP2 interacting DNA fragments. Input genomic DNA fragments were sequenced in parallel for normalization of MeCP2 ChIP-seq signals. As has been described in prior ChIP-seq analyses of cerebellar neurons (21), MeCP2 binding was broadly distributed across the genome. At the Kirrel2 locus, MeCP2 binding was observed throughout the promoter and the gene body region including both exons and introns (Fig. 3C). Averaged MeCP2 binding across the Kirrel2 gene body, represented by segments per kilobase per million mapped reads (SPKM), was 0.54 SPKM (Fig. 3D) compared with 3.12 SPKM for the CpG islands located within the Kirrel2 promoter (Fig. 3E) after normalization to input. Following odor-stimulation, MeCP2 binding was dramatically decreased across the Kirrel2 locus, with the promoter CpG island showing a de-enrichment of MeCP2 binding in odor-stimulated versus control MOE (Fig. 3C–E). Similar results were obtained with an independent experiment replicate (2nd in Fig. 3D and E). These results suggest that MeCP2 binding to Kirrel2 is reduced and redistributed with odorant stimulation.
In addition, eight regions along the Kirrel2 locus were selected for further ChIP-qPCR validation. The level of MeCP2 binding in each region was compared between control and odor-stimulated conditions. MeCP2 binding to six different regions was decreased after odor stimulation when compared with the control (Fig. 3F). The other two regions, one localized 5′ upstream and one spanning the third exon of Kirrel2, showed no change in MeCP2 binding with odor stimulation. IgG ChIP-qPCRs show no detectable product in both control and stimulated samples (Fig. 3F). The large decrease in MeCP2 binding at the Kirrel2 promoter CpG island after odor stimulation (from 2.31% in control to 0.67%) confirmed the ChIP-seq result that odor stimulation triggers a rapid decrease in MeCP2 binding in the Kirrel2 locus.
Though Kirrel3 expression was also affected by Mecp2 deficiency, MeCP2 binding at the Kirrel3 locus was at relatively low baseline levels according to ChIP-seq analysis compared with Kirrel2. Averaged MeCP2 binding over the Kirrel3 gene body (547 kb) was <0.1 SPKM under the control condition and no detectable MeCP2 binding to the Kirrel3 CpG island promoter was observed (Fig. 3G and H). In contrast to the dynamic change in MeCP2 binding at Kirrel2, odor stimulation did not alter MeCP2 binding levels at the Kirrel3 locus. Furthermore, no significant MeCP2 binding at the Cntn4 locus was detected (Fig. 3I). However, ChIP-seq analysis showed widespread MeCP2 binding throughout the Pcdh20 locus including at the promoter region (Fig. 3J). Like Kirrel2, following odor stimulation, MeCP2 binding to the Pcdh20 locus decreased dramatically and was redistributed (SPKM: 0.03) compared with the level in wild-type control OSNs (SPKM: 0.68). A reproducible decrease in MeCP2 binding was identified along the Pcdh20 locus after odor stimulation (Fig. 3K). In summary, among the four selected activity-dependent OSN expressed genes, MeCP2 demonstrated high binding levels throughout the gene body regions of Kirrel2 and Pcdh20, whereas little MeCP2 binding was observed for the entire Kirrel3 and Cntn4 loci. Significantly, upon odor stimulation, MeCP2 binding to the entire locus of Kirrel2 and Pcdh20 was consistently reduced.
MeCP2 is a key regulator for activity-dependent Kirrel2 and Pcdh20 expression
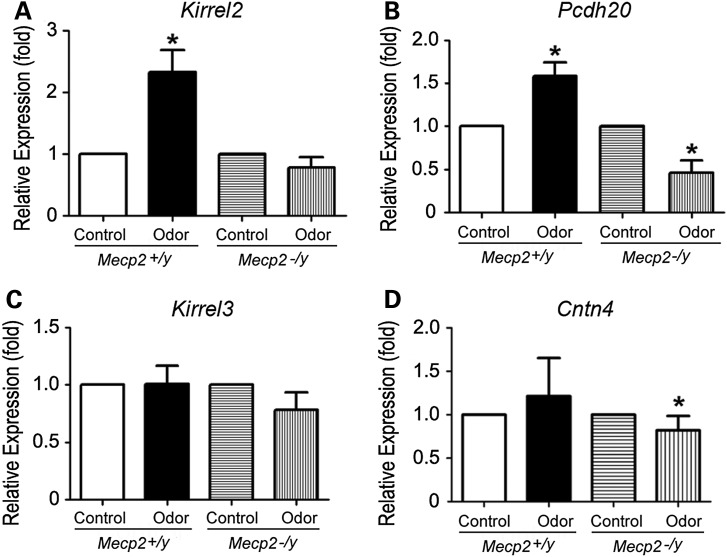
To evaluate the relationship between MeCP2 binding changes to the gene locus and changes in gene expression under neuronal activity, we next examined whether transcript levels of the selected genes was influenced by a short duration of odor stimulation. For these studies, transcript levels of Kirrel2, Kirrel3, Cntn4 and Pcdh20 were examined by real-time qRT-PCR and normalized against β-III tubulin transcript levels which remain constant in neuronal populations present in the MOE tissue. After odor stimulation, Kirrel2 levels increased 2.33-fold (SD = 0.80, n = 5, P < 0.02) and Pcdh20 levels increased 1.58-fold (1.58 ± 0.36, n = 5, P < 0.02, Fig. 4A and B). In contrast, no significant change in Kirrel3 (1.01 ± 0.35, n = 5, P > 0.5) or Cntn4 (0.78 ± 0.21; n = 5, P > 0.1) level was observed in wild-type mice (Fig. 4C and D). These qRT-PCR results provide further evidence that the direct MeCP2 target genes Kirrel2 and Pcdh20 are rapidly upregulated by odor-evoked activity in OSNs.
Figure 4.
Mecp2 is the key regulator for odor-evoked Kirrel2 and Pcdh20 expression. Quantitative RT-PCRs for MOE tissues from mice under control filtered air exposure or after 4 h of odor mixture stimulation are performed to compare relative levels of gene expression after normalization against β-III tubulin. In wild-type OE, with control defined as 1, Kirrel2 expression increases significantly after 4 h of odor stimulation (*P < 0.02) compared with the control filtered air exposure. (A) This change in Kirrel2 expression is absent in Mecp2−/y MOE after odor stimulation (P > 0.1, in A). Pcdh20 expression levels decreased (*P < 0.02) after odor stimulation in Mecp2−/y when compared with the control OE, suggesting that the increases in Pcdh20 expression triggered by odor stimulation are regulated by MeCP2 (B). No significant change in Kirrel3 transcript expression is observed after odor stimulation in wild type (P > 0.1) and in Mecp2−/y (C). Though Cntn4 transcript level does not change after odor stimulation in wild type (P > 0.1), there is a decrease after odor stimulation compared with control in Mecp2−/y (*P < 0.02 Student's t-test).
If the upregulation of Kirrel2 and Pcdh20 transcript levels triggered by odor stimulation is controlled by Mecp2, we would expect a lack of odor-induced upregulation of these transcripts in Mecp2−/y OSNs. qRT-PCR results in Mecp2−/y mice reveal that Kirrel2 transcript levels following odor stimulation were 0.78-fold of the control (SD = 0.29, n = 3, P > 0.1), whereas Pcdh20 transcript levels were significantly lower than the control (0.46 ± 0.24, n = 3, P < 0.02). Thus, these results are consistent with the prediction that the absence of Mecp2 results in the failure of Kirrel2 and Pcdh20 upregulation in the odor-stimulation activation paradigm (Fig. 4A). Therefore, these results provide independent evidence that MeCP2 is the primary regulator of activity-dependent Kirrel2 and Pcdh20 expression. Our data support a mechanism in which MeCP2 acts to maintain a low transcript level at baseline but also promotes higher transcript levels following neuronal activity for Kirrel2 and Pcdh20.
DISCUSSION
In this study, we investigated the mechanisms underlying MeCP2 regulation of activity-dependent gene expression in MOE and the requirement for Mecp2 within OSNs during the refinement of olfactory axon convergence in mice. In the MOE, OSNs are the sole neuronal population, which allows a clearer understanding of the effects of Mecp2 deficiency compared with prior observations made in brain regions where diverse cellular and neuronal populations are present. Though different OR expression subdivides OSNs into a large number of homotypic subpopulations, these neurons have similar morphology, gene expression profiles and connectivity patterns. Consistent with recent reports in other neuronal subtypes, we found that MeCP2 binding in MOE OSNs was widespread genome wide, occurring in gene body, promoter and intergenic regions (14,21). Our data show that MeCP2 functions to repress levels of key connectivity genes expressed in the MOE. Among the selected genes regulated by MeCP2, we noticed that two of these genes, Kirrel2 and Pcdh20, have distinct MeCP2-binding patterns but two others, Kirrel3 and Cntn4, did not. Although MeCP2 clearly affects expression of the four genes examined, it is possible that both direct binding to specific gene loci (Kirrel2 and Pchd20) and more globally to chromosome regions other than gene loci per se (Kirrel3 and Cntn4) are involved in this transcriptional regulation paradigm.
Alterations in MeCP2 binding have significant effects on olfactory targeting gene expression. Our data showing that MeCP2 functions to repress transcript levels of Kirrel2 in the MOE, while also being required for activity-dependent upregulation of Kirrel2 levels upon odor stimulation is consistent with data in cultured neurons showing that MeCP2 is required at multiple stages during the kinetics of activity-dependent transcription (22). While MeCP2-mediated repression maintains a low baseline level of neuronal activity-dependent gene expression before and after a response, it also activates expression of the same genes by enabling higher inducible transcription levels in response to neuronal activity. However, here we also demonstrate specificity and dynamic activity-dependent changes of MeCP2 binding within two gene loci (Kirrel2 and Pcdh20) that were not observed at two additional gene loci (Kirrel3 and Cntn4). The reduction of MeCP2 binding at the promoter CpG island of Kirrel2 and the global changes to MeCP2 binding and transcription level changes upon odor stimulation are consistent with site-specific activity-dependent changes in MeCP2 binding. While Pcdh20 does not have a CpG island in its promoter, it also showed a reproducible decrease in MeCP2 binding with odor-induced activity. Furthermore, both Kirrel2 and Pcdh20 were transcriptionally upregulated in response to odor only in wild-type control but not Mecp2−/y OSNs, consistent with an activating role for MeCP2 in the transcriptional responsiveness of these genes. Our data support a mechanism in which MeCP2 acts to maintain a low transcript level at baseline but also promotes higher transcript levels following neuronal activity for Kirrel2 and Pcdh20.
Odor-evoked activity regulates the expression of many genes in OSNs (6–8). Most of the activity-dependent genes have been identified in either naris closure or CNGA2 genetic deletion models. Both of these models cause long-term loss of odor-evoked activity in OSNs. However, it is not clear whether acute odor stimulation, a more physiological relevant condition, could recapitulate the gene expression changes predicted from the lack of activity model (8). Activity-dependent gene expression in cortical neurons is typically identified by acute activation through application of extracellular potassium. While these experimental approaches are useful to synchronize neuronal activity in vitro, changes in transcription need to be further validated in vivo under physiological levels of neuronal stimulation. OSNs in the nasal cavity can be activated synchronously and naturally in vivo by odor stimulation. Thus, the olfactory system provides an ideal, biologically relevant model system to examine activity-dependent gene expression. Our data reveals a complex picture with MeCP2 not participating (Cntn4), partially regulating (Kirrel3) or primarily regulating (Kirrel2 and Pcdh20) odor-evoked changes in gene expression. With the widespread binding profile of MeCP2 throughout the genome, further genomic analysis is necessary to identify additional target genes that are influenced by MeCP2 for activity-dependent gene expression. Despite the varying effect of MeCP2 binding on gene expression these studies have significant implications. KIRREL3 is an autism-associated gene (23,24) and Kirrel genes (or Neph) are widely expressed in neuronal tissues and are required for synaptogenesis and synaptic specificity (25,26). Alterations of Kirrel gene expression results in mistargeting of axons within the OB (7). Differential expression of Kirrel genes in vomeronasal neurons is required for proper axon targeting and circuitry formation (27). These model system studies, together with our findings, indicate key functions of Kirrel genes in circuitry formation and neuronal plasticity in general.
Mecp2 is expressed in mature OSNs. Mecp2 causes a transient differentiation delay, but does not appear to alter OSN populations in the MOE in adult stages (16). Olfactory sensory axons in the MOE appear to target their respective loci in the OB in Mecp2−/y mice (17). Although Mecp2 is expressed in both OSNs and their OB target, by conditionally deleting Mecp2 in OSNs, we determined that the requirement for olfactory axon convergence refinement is in the OSNs. In Drosophila, it has been shown that intrinsic mechanisms in OSNs play critical roles in their axon targeting and convergence (28). Although the OB is important to provide an axon guidance substrate, several lines of evidence support the model of intrinsic sorting and targeting functions of olfactory sensory axons (28,29).
In this study, we identified that Mecp2 is required in OSNs for the refinement of olfactory axon connectivity. Though MeCP2 demonstrated widespread genome wide chromatin binding, its binding dynamics at axonal refinement genes in the MOE is dependent upon odor-evoked activity and appears to be the primary regulator for activity-dependent expression of these genes. MeCP2-dependent transcription regulation may be important for balancing a large number of neuronal plasticity gene expressions. Identification of activity-dependent cell adhesion genes as MeCP2 target genes are important for linking gene-specific changes to Mecp2 deficiency and better understanding RTT syndrome. The results of this study in OSNs have high relevance to human disease as defects in CNTN4 and KIRREL3 have been correlated with autistic disorders (23,24).
MATERIALS AND METHODS
Animals
All procedures were approved by IACUC of University of California at Davis. Mecp2-deficient Mecp2(tm1.1Bird) was generated by Bird and colleagues and M72-IRES-taulacZ and OMP-Cre strains were generated by Mombaerts and colleagues. These mutants were acquired from Jackson Laboratories (The Jackson Laboratory, Bar Harbor, ME, USA). All control animals were wild-type littermates of the mutant mice. Odor stimulation was provided by laminar flow of a complex odor mixture for 4 h (details in Supplementary Material).
Chromatin immunoprecipitation and sequencing
ChIP-seq was performed based on methods established previously (11,30). Two independent chromatin immunoprecipitation assays were performed from wild-type adult MOE using MAGnify chromatin immunoprecipitation kit (Life Technologies, Grand Island, NY, USA). Detailed methods and analysis of ChIP-seq and other technical details of this study are described in Supplementary Material.
SUPPLEMENTARY MATERIAL
FUNDING
This study is supported by NIH (NIDCD) grants DC10237 and DC11346 to Q.G. and NS081913 to J.L.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr T. Cutforth for providing β-gal and Kirrel2 antibodies, Mr J. Estep, G. Wong and Ms I Yin for technical assistance and Dr R.P. Tucker for critical reading of the manuscript.
Conflict of Interest statement: None.
REFERENCES
- 1.Mombaerts P., Wang F., Dulac C., Chao S.K., Nemes A., Mendelsohn M., Edmondson J., Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- 2.Serizawa S., Miyamichi K., Nakatani H., Suzuki M., Saito M., Yoshihara Y., Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- 3.Ressler K.J., Sullivan S.L., Buck L.B. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 4.Zou D.J., Feinstein P., Rivers A.L., Mathews G.A., Kim A., Greer C.A., Mombaerts P., Firestein S. Postnatal refinement of peripheral olfactory projections. Science. 2004;304:1976–1979. doi: 10.1126/science.1093468. [DOI] [PubMed] [Google Scholar]
- 5.Goodman C.S., Shatz C.J. Developmental mechanisms that generate precise patterns of neuronal connectivity. Cell. 1993;72:77–98. doi: 10.1016/s0092-8674(05)80030-3. [DOI] [PubMed] [Google Scholar]
- 6.Williams E.O., Sickles H.M., Dooley A.L., Palumbos S., Bisogni A.J., Lin D.M. Delta Protocadherin 10 is Regulated by Activity in the Mouse Main Olfactory System. Front. Neural Circuits. 2011;5:9. doi: 10.3389/fncir.2011.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serizawa S., Miyamichi K., Takeuchi H., Yamagishi Y., Suzuki M., Sakano H. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell. 2006;127:1057–1069. doi: 10.1016/j.cell.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 8.Bennett M.K., Kulaga H.M., Reed R.R. Odor-evoked gene regulation and visualization in olfactory receptor neurons. Mol. Cell. Neurosci. 2010;43:353–362. doi: 10.1016/j.mcn.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z., Hong E.J., Cohen S., Zhao W.N., Ho H.Y., Schmidt L., Chen W.G., Lin Y., Savner E., Griffith E.C., et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis J.D., Meehan R.R., Henzel W.J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 11.Yasui D.H., Peddada S., Bieda M.C., Vallero R.O., Hogart A., Nagarajan R.P., Thatcher K.N., Farnham P.J., Lasalle J.M. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc. Natl. Acad. Sci. USA. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng H.H., Bird A. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 13.Martinowich K., Hattori D., Wu H., Fouse S., He F., Hu Y., Fan G., Sun Y.E. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 14.Cohen S., Gabel H.W., Hemberg M., Hutchinson A.N., Sadacca L.A., Ebert D.H., Harmin D.A., Greenberg R.S., Verdine V.K., Zhou Z., et al. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Zhong X., Chau K.F., Williams E.C., Chang Q. Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nat. Neurosci. 2011;14:1001–1008. doi: 10.1038/nn.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matarazzo V., Cohen D., Palmer A.M., Simpson P.J., Khokhar B., Pan S.J., Ronnett G.V. The transcriptional repressor Mecp2 regulates terminal neuronal differentiation. Mol. Cell. Neurosci. 2004;27:44–58. doi: 10.1016/j.mcn.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Degano A.L., Pasterkamp R.J., Ronnett G.V. MeCP2 deficiency disrupts axonal guidance, fasciculation, and targeting by altering Semaphorin 3F function. Mol. Cell. Neurosci. 2009;42:243–254. doi: 10.1016/j.mcn.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer A., Qayumi J., Ronnett G. MeCP2 mutation causes distinguishable phases of acute and chronic defects in synaptogenesis and maintenance, respectively. Mol. Cell. Neurosci. 2008;37:794–807. doi: 10.1016/j.mcn.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Kaneko-Goto T., Yoshihara S., Miyazaki H., Yoshihara Y. BIG-2 mediates olfactory axon convergence to target glomeruli. Neuron. 2008;57:834–846. doi: 10.1016/j.neuron.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Lee W., Cheng T.W., Gong Q. Olfactory sensory neuron-specific and sexually dimorphic expression of protocadherin 20. J. Comp. Neurol. 2008;507:1076–1086. doi: 10.1002/cne.21569. [DOI] [PubMed] [Google Scholar]
- 21.Skene P.J., Illingworth R.S., Webb S., Kerr A.R., James K.D., Turner D.J., Andrews R., Bird A.P. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzales M.L., Adams S., Dunaway K.W., LaSalle J.M. Phosphorylation of distinct sites in MeCP2 modifies cofactor associations and the dynamics of transcriptional regulation. Mol. Cell. Biol. 2012;32:2894–2903. doi: 10.1128/MCB.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talkowski M.E., Rosenfeld J.A., Blumenthal I., Pillalamarri V., Chiang C., Heilbut A., Ernst C., Hanscom C., Rossin E., Lindgren A.M., et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerin A., Stavropoulos D.J., Diab Y., Chenier S., Christensen H., Kahr W.H., Babul-Hirji R., Chitayat D. Interstitial deletion of 11q-implicating the KIRREL3 gene in the neurocognitive delay associated with Jacobsen syndrome. Am. J. Med. Genet. A. 2012;158A:2551–2556. doi: 10.1002/ajmg.a.35621. [DOI] [PubMed] [Google Scholar]
- 25.Shen K., Fetter R.D., Bargmann C.I. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell. 2004;116:869–881. doi: 10.1016/s0092-8674(04)00251-x. [DOI] [PubMed] [Google Scholar]
- 26.Gerke P., Benzing T., Hohne M., Kispert A., Frotscher M., Walz G., Kretz O. Neuronal expression and interaction with the synaptic protein CASK suggest a role for Neph1 and Neph2 in synaptogenesis. J. Comp. Neurol. 2006;498:466–475. doi: 10.1002/cne.21064. [DOI] [PubMed] [Google Scholar]
- 27.Prince J.E., Brignall A.C., Cutforth T., Shen K., Cloutier J.F. Kirrel3 is required for the coalescence of vomeronasal sensory neuron axons into glomeruli and for male-male aggression. Development. 2013;140:2398–2408. doi: 10.1242/dev.087262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komiyama T., Carlson J.R., Luo L. Olfactory receptor neuron axon targeting: intrinsic transcriptional control and hierarchical interactions. Nat. Neurosci. 2004;7:819–825. doi: 10.1038/nn1284. [DOI] [PubMed] [Google Scholar]
- 29.St John J.A., Clarris H.J., McKeown S., Royal S., Key B. Sorting and convergence of primary olfactory axons are independent of the olfactory bulb. J. Comp. Neurol. 2003;464:131–140. doi: 10.1002/cne.10777. [DOI] [PubMed] [Google Scholar]
- 30.Yasui D.H., Xu H., Dunaway K.W., Lasalle J.M., Jin L.W., Maezawa I. MeCP2 modulates gene expression pathways in astrocytes. Mol. Autism. 2013;4:3. doi: 10.1186/2040-2392-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.