Abstract
ALOX5 is implicated in chronic myeloid leukemia development in mouse leukemic stem cells, but its importance in human chronic myeloid leukemia is unknown. Functional ALOX5 was assessed using an LTB4 ELISA and ALOX5, and LTB4R1 mRNA expression was determined via a TaqMan gene expression assay. LTB4R1 and 5-LOX protein levels were assessed by cell surface flow cytometry analysis. At diagnosis ALOX5 was below normal in both blood and CD34+ stem cells in all patients. On treatment initiation, ALOX5 levels increased in all patients except those who were destined to progress subsequently to blast crisis. LTB4 levels were increased despite low ALOX5 expression, suggesting that the arachidonic acid pathway is functioning normally up to the point of LTB4 production. However, the LTB4 receptor (BLT1) protein in newly diagnosed patients was significantly lower than after a period of treatment (P<0.0001). The low level of LTB4R1 at diagnosis explains the downregulation of ALOX5. In the absence of LTB4R1, the arachidonic acid pathway intermediates (5-HEPTE and LTA4) negatively regulate ALOX5. ALOX5 regulation is aberrant in chronic myeloid leukemia patients and may not be important for the development of the disease. Our data suggest caution when extrapolating mouse model data into human chronic myeloid leukemia.
Introduction
Chronic myeloid leukemia (CML) is a malignant disease of a primitive hematologic cell, characterized by inappropriate expansion of myeloid cells. Although the disease is readily controlled by imatinib, approximately one-third of patients will eventually fail treatment,1,2 and a significant proportion of these will progress towards blast crisis which is usually fatal. However, the factors that contribute towards CML growth and progression are not well understood. Recently, Chen et al.3 reported that Arachidonate 5-lipoxygenase (ALOX5) is up-regulated in mouse leukemic stem cells, and this upregulation is not inhibited by imatinib treatment. Furthermore, mice transplanted with ALOX5 deficient BCR-ABL1 positive bone marrow cells were resistant to CML induction. ALOX5 deficiency had no effect on the growth of BCR-ABL1 negative cells, suggesting that it may be essential for malignant but not normal hematopoietic stem cell growth, through an unknown mechanism.
The ALOX5 gene encodes a member of the lipoxygenase gene family and plays a role in the synthesis of leukotrienes from arachidonic acid (Figure 1). ALOX5 catalyses the conversion of arachidonic acid to 5(S)-hydroperoxy-6-trans-8,11,14-cis-eicosatetraenoic acid (5-HEPTE), and subsequently to the allylic epoxide 5(S)-trans-7,9-trans-11,14-cis-eicosatetraenoic acid (leukotriene A4/LTA4). LTA4 is unstable and is converted to LTB4 that is more stable. 5-HEPTE and LTA4 negatively regulate ALOX5 expression; positive regulation of ALOX5 occurs when LTB4 binds to its receptor LTB4R1 (also known as BLT1) to mediate its positive feedback on ALOX5.4–9
Figure 1.
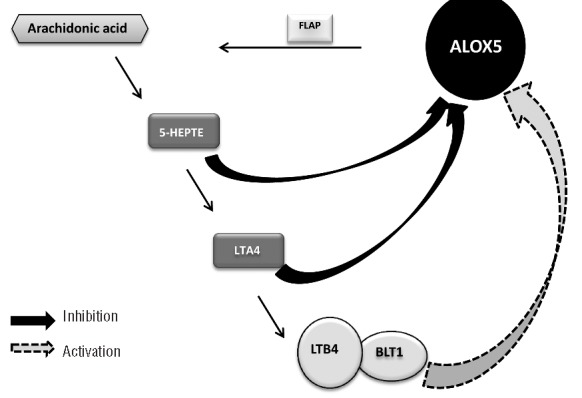
Arachidonic acid metabolism pathway. 5-LOX is encoded by the ALOX5 gene and is activated by the enzyme FLAP. Arachidonic acid is converted to 5-HEPTE by 5-LOX which is further converted to LTA4 by 5-LOX. Both 5-HEPTE and LTA4 negatively regulate ALOX5 expression. LTA4 is unstable and is further converted to LTB4. The binding of LTB4 to its receptor LTB4R1 positively regulates ALOX5 expression.
The role of ALOX5 in human CML is unknown. The hypothesis proposed in this study was that CML cells have high levels of ALOX5 and that the level of ALOX5 predicts a patient’s response to imatinib treatment. If this hypothesis were correct, then ALOX5 may be a new therapeutic target.
Methods
Sample collection
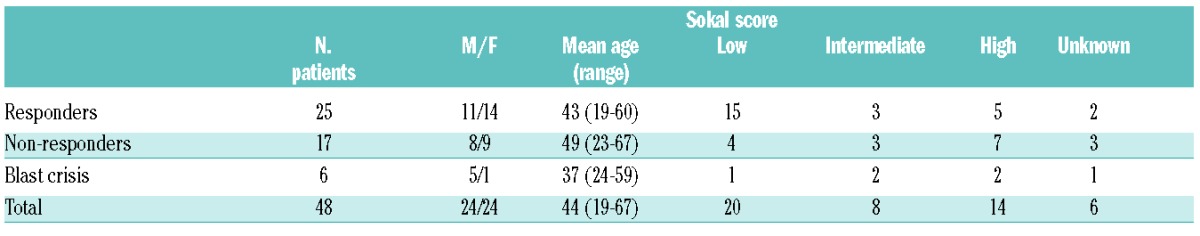
Blood samples were collected from 48 patients at initial chronic phase diagnosis and at three, six and 12 months following the commencement of imatinib treatment. Plasma and total leukocytes were prepared. Samples were enriched for CD34+ cells using CliniMACS (Miltenyi Biotec, Surrey, UK) according to the manufacturer’s instructions. The study was approved by the Liverpool Central Research Ethics Committee and all patients gave informed consent. Patients’ characteristics are shown in Table 1. Peripheral blood from healthy volunteers were used as normal controls (n=10)
Table 1.
Patients’ characteristics.
Clinical response
Following 12 months of imatinib treatment, patients were stratified into three clinical outcomes:
Responders – patients who achieve a complete cytogenetic response (CCR) defined as no Philadelphia positive metaphases amongst at least 20 marrow metaphases. In some cases, serial cytogenetic data were not available and achievement of CCR is based on a BCR-ABL1/ABL1 transcript ratio of less than 1%, which we have previously shown to be tightly correlated with cytogenetically defined CCR.10
Non-responders – patients who had achieved a complete hematologic response but not a complete cytogenetic response and who had not progressed.
Blast crisis – patients who presented in chronic phase but who subsequently progressed into blast crisis.
Plasma preparation
For plasma preparation, 5 mL of peripheral blood was collected into EDTA, and centrifuged at 770 g for 15 min. Plasma was collected from samples, aliquoted and stored at −20°C prior to use.
Leukotriene B4 ELISA
Leukatriene B4 (LTB4) assay was performed according to the manufacturer’s protocol (Cambridge Biosciences, Cambridge, UK). Briefly, 100 μL of the specific standard, sample or assay buffer was added in duplicate to appropriate wells of a 96-well plate and a further 50 μL of assay buffer to the negative control wells. We then added 50 μL of LTB4 conjugate and 50 μL of LTB4 antibody, supplied in the kit, were added to the appropriate wells. The plate was incubated at room temperature, with shaking, for 2 h. Following incubation, the plate was washed three times with wash solution (provided in the kit). Then 5 μL L of conjugate was added to the total activity wells, followed by 200 μL of pNpp substrate solution to every well. The plate was incubated at 37°C with shaking, for 2 h. Then 50 μL of 2M HCl was then added to all wells. The absorbance was immediately read at 405 nm using a BioTeK μQuant plate reader. The mean optical density of the negative controls was subtracted from the optical density reading of all wells to eliminate ‘background noise’.
ALOX5 and LTB4R1 expression
Quality real-time polymerase chain reaction (qRT-PCR) was performed using cDNA from either total leukocytes or purified CD34+ cells. Pre-designed TaqMan RT-PCR assays were used for ALOX5 Hs01095330_m1, BLT1 (LTB4R1) Hs019388704_s1 and GAPDH Hs99999905_m1 (Applied Biosciences, UK). In evaluating the mRNA expression data, the comparative Ct method was used, with the 2−ΔΔCt formula to achieve results for relative quantification (RQ).10 A pool of cDNA from 4 normal individuals was used as calibrator and all the samples were normalized to GAPDH. The RQ Manager software supported by the ABI Prism 7900HT System was used for data analysis.
Measurement of LTB4 receptor 1
5mL of peripheral blood was collected into EDTA from CML patients attending the CML clinic. Samples were processed within 4 h of the sample being taken and cells were used fresh. Briefly, erythrocytes were depleted using red cell lysis buffer (0.1 M ammonium chloride, 10 mM sodium bicarbonate and 1.3 mM EDTA; Sigma-Aldrich, Dorset, UK). Total leukocytes were then incubated with Leukotriene B4 Receptor 1 (BLT1/LTB4R1) (R&D systems, Abingdon, UK) and anti -mouse IgG1 antibody (BD) as the control antibody, in the dark for 30 min with shaking. Cells were then washed in PBS containing 0.5% BSA and analyzed by FACS.
Measurement of ALOX5 protein by FACS
5-lipoxygenase (5-Lox) protein was assessed by FACS using the method previously described using 5-LOX antibody (Santa Cruz Biotechnology, California, USA) and mouse IgG1 (Becton Dickinson, Oxford, UK) as an isotypic control.11
Results
ALOX5 mRNA expression and clinical outcome
In order to determine whether ALOX5 expression was elevated in CML patients, and if the levels were related to clinical outcome, ALOX5 expression was measured at diagnosis (chronic phase) and following three, six and 12 months of imatinib treatment. Patients’ details are shown in Table 1. At diagnosis the expression level of ALOX5 was below the level observed in normal healthy volunteers’ cDNA pool. This was the case in Responders, Non-responders and those who subsequently progress to blast crisis, as defined in the Methods section (Figure 2). No difference was seen between patients according to their Sokal score (data not shown). In the Responder cohort, once imatinib treatment had commenced, the expression level of ALOX5 increased and remained high throughout treatment. The increase in ALOX5 expression compared with that at diagnosis was statistically significant at three, six and 12 months (P=0.003, P=<0.001 and P=0.024, respectively). These data suggest that ALOX5 expression in patients who subsequently achieve a CCR is restored to a level observed in the normal calibrator pool upon treatment (Figure 2A). A similar trend in ALOX5 expression was observed in the Non-responder group (Figure 2B), with ALOX5 expression increasing once treatment had commenced. Again, the increase in ALOX5 expression compared with diagnosis was statistically significant at three, six and 12 months (P=0.008, P=0.03 and P=0.013, respectively). Conversely, in patients destined to subsequently progress to blast crisis, imatinib treatment failed to increase ALOX5 expression (Figure 2C).
Figure 2.
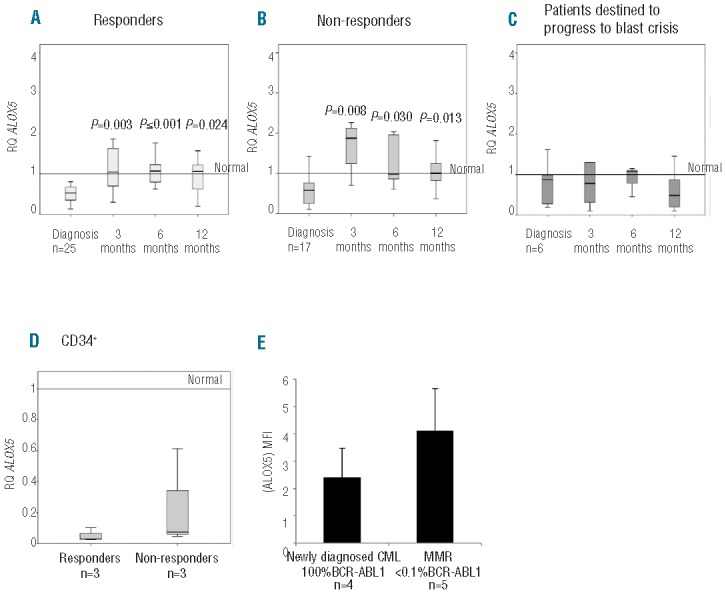
ALOX5 expression. (A) ALOX5 expression levels in the Responder group (n=25). (B) ALOX5 expression levels in the Non-responders group (n=17). (C) ALOX5 expression levels in patients destined to progress into blast crisis (n=6). (D) ALOX5 expression levels in CD34+ cells taken at diagnosis and stratified by the patients’ eventual clinical response. The normal level was determined using 4 healthy volunteers (n=6). (E) 5-LOX protein expression. 5-LOX protein was assessed by FACS using fresh leukocytes (n=9). 5-LOX expression is higher in CML patients in major molecular response as determined by qRT-PCR.
These data suggest that a failure to increase ALOX5 expression following three months of imatinib treatment may indicate a higher risk of disease progression (Figure 2C). No difference in ALOX5 expression was observed at diagnosis between those patients destined to progress to blast crisis and the Responder and Non-responder groups, suggesting that ALOX5 expression at diagnosis is not predictive of the outcome of imatinib-treated patients.
When the same experiment was repeated on diagnostic CD34+ stem cells (from the same patients), no difference in ALOX5 expression was observed between Responders and Non-responders. ALOX5 expression was lower in CD34+ cells than in total leukocytes when comparing results in Figure 2A and D, although similar overall results were obtained, confirming that ALOX5 is down-regulated in CML patients in both CD34+ and chronic phase MNC (P=0.01) (Figure 2 A and D).
To determine whether 5-LOX protein levels were a function of ALOX5 gene expression, 5-LOX protein expression was assessed by FACS. As patients respond to imatinib treatment and achieve a major molecular response (MMR), there is a trend for an increase in the amount of detectable ALOX5 protein (Figure 2E); this is consistent with the increase in mRNA expression observed in Figure 2A–C.
ALOX5 function: LTB4 levels in CML patients
Plasma levels of LTB4 can be used as a marker of ALOX5 functional activity (as previously described by Chen et al.3). LTB4 levels were measured at diagnosis, and at three and six months post imatinib treatment in 27 patients of known subsequent clinical outcome (18 Responders, 7 Non-responders and 2 patients whom subsequently progressed to blast crisis; Figure 3A). LTB4 levels increased in all three patient groups following imatinib treatment. At diagnosis, the LTB4 level in patients destined to progress to blast crisis was the lowest of the three patient groups. Conversely, once imatinib treatment commenced, the LTB4 level increased to the highest level compared to the other two clinical groups (Figure 3A). The levels of LTB4 in the CML patients were in general higher than normal; this may indicate an accumulation of LTB4 and a potential block in the arachidonic acid pathway.
Figure 3.
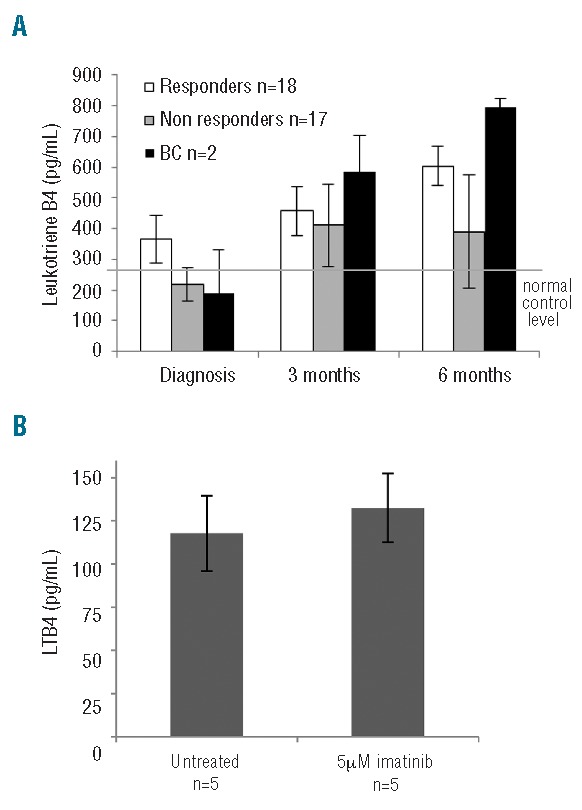
LTB4 levels following imatinib treatment. (A) Mean plasma LTB4 levels of 27 patients stratified by clinical outcome; 18 responders, 7 non-responders and 2 blast crisis. Samples were taken during chronic phase at diagnosis, 3 and 6 months after commencement of imatinib treatment. Normal level was determined using plasma from 15 healthy volunteers. (B) MNC from five healthy volunteers were treated with 5 μM imatinib for 24 hours and the changes in LTB4 levels were measured. No significant difference was observed between the untreated and imatinib treated normal samples.
Since LTB4 levels increased in all patients assessed, it was necessary to confirm whether this was a direct physiological response to imatinib treatment, or if it was due to the disease. To address this issue, MNC from 5 healthy volunteers were treated with 5 μM imatinib for 24 h and the changes in LTB4 levels were measured. No significant difference was observed between the untreated and imatinib-treated normal samples, suggesting that changes in LTB4 observed in CML patients may be attributed to the leukemia and not a physiological response to imatinib treatment (Figure 3B). ALOX5 function was assessed using samples from patients diagnosed in chronic phase and from patients actually in blast crisis. LTB4 levels were found to be elevated in samples taken at blast crisis compared to chronic phase samples, but this difference was not statistically significant. The observed increase in LTB4 observed coincided with a decrease in ALOX5 expression as shown is Figure 2C.
LTB4 receptor 1 is low at diagnosis
The original hypothesis proposed in this study was that CML patients would have high levels of ALOX5. The data presented thus far are in complete contrast to both this hypothesis and the study presented by Chen et al.3 which demonstrated that mice transplanted with BCR-ABL1 positive bone marrow cells lacking the ALOX5 gene were resistant to the induction of CML, suggesting that ALOX5 was essential for the induction and development of CML.3 However, that study was performed using a mouse model, and the experiments have not yet been carried out on human CML samples, which could explain the discrepancies between the present data and those of Chen et al.3 In order to further explain the differences between the two studies, the LTB4 pathway was interrogated further. Figure 1 demonstrates the arachidonic acid metabolism pathway. Prior to conversion to LTB4, arachidonic acid is converted into two intermediate products, 5-HEPTE and then LTA4. Both 5-HEPTE and LTA4 negatively regulate ALOX5 expression, while LTB4 is known to positively regulate ALOX5 expression.12 In order for LTB4 to mediate its positive effect on ALOX5 expression it must bind to the LTB4 receptor.
In the CML clinical samples investigated it is apparent that ALOX5 expression is suppressed below normal. Furthermore, the LTB4 levels in the CML samples were elevated compared to normal healthy controls, suggesting an accumulation of LTB4. Since LTB4 needs to bind to its receptor LTB4R1 to positively regulate ALOX5 expression, these data suggest that LTB4R1 expression or function may be abnormal in CML.
LTB4R1 protein expression was measured by cell surface FACS analysis. LTB4R1 protein was found to be low in newly diagnosed chronic phase CML samples compared to normal (P=<0.0001) (Figure 4A). LTB4R1 protein was even lower in a blast crisis sample. We next compared the levels of LTB4R1 protein in patients being treated by imatinib and stratified these data by the patients BCR-ABL1/ABL1 percentage. LTB4R1 protein levels were lower than normal in those patients partially responding to imatinib treatment (defined as a BCR-ABL1/ABL1 ratio of 1–10%) (P=0.04). LTB4R1 levels increased from the diagnosis level once treatment had commenced. Patients partially responding and those patients whom had achieved a complete cytogenetic response (defined as a BCR-ABL1/ABL1 ratio of <1%13) had significantly higher levels of LTB4R1 protein compared to the level observed at diagnosis (P=0.02 and P=<0.0001, respectively). These data suggest that there is an inverse relationship between LTB4R1 protein and BCR-ABL1 mRNA expression. LTB4R1 protein is very low at diagnosis but increases towards normal as the BCR-ABL1 mRNA expression decreases.
Figure 4.
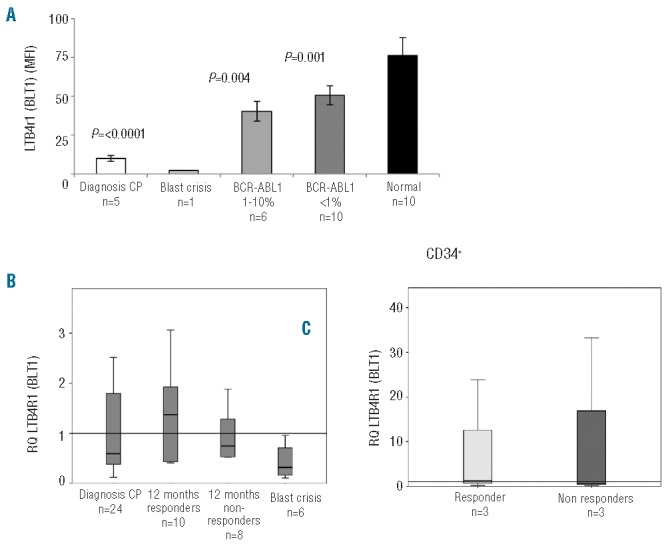
LTB4 (BLT1) receptor expression. (A) LTB4R1 surface protein and mRNA expression was measured in newly diagnosed chronic phase CML patients (n=5), a patient in blast crisis (n=1), patients currently responding to imatinib treatment (n=6), patients in CCR (n=10) and healthy volunteers (n=10). LTB4R1 cell surface expression was low in newly diagnosed chronic phase CML patients compared to patients responding to treatment or healthy volunteers. *P values in the comparison made with normal blood. (B) LTB4R1 mRNA expression was measured in newly diagnosed chronic phase CML patients (n=24), patients treated with imatinib for twelve months who achieved a CCR (n=10) and Non-responders (n=8), as well as patients who were in blast crisis (n=6). (C) LTB4R1 mRNA expression in diagnostic CD34+ cells stratified by the patient’s clinical outcome.
LTB4R1 mRNA expression
LTB4R1 mRNA expression was found to be low in chronic phase diagnostic samples compared to patients who had received 12 months of imatinib treatment (Figure 4B). In the Responder group, the LTB4R1 expression increased more than in the Non-responder group, although this was not statistically significant. When LTB4R1 mRNA expression was assessed in samples from patients in blast crisis at the time of sampling, LTB4R1 mRNA expression was much lower than the diagnostic chronic phase group and the treatment samples taken at 12 months. LTB4R1 mRNA expression was not statistically significant at diagnosis, nor did it predict a patient’s clinical outcome (data not shown).
As patients progress to blast crisis, ALOX5 gene expression is suppressed, possibly due to an accumulation of pathway intermediates (5-HEPTE and LTA4) and a decrease in LTB4R1. This idea is supported by the LTB4R1 protein data and the accumulation of LTB4 in blast crisis. LTB4R1 mRNA expression was also assessed in CD34+ stem cells taken at diagnosis (Figure 4C). No difference in LTB4R1 expression was observed between those patients who later achieved a CCR and those who did not.
Conclusions
The identification of proteins that discriminate leukemic from normal stem cells is a challenging concept. Successful identification and therapeutic targeting of leukemic stem cells remains the ultimate goal in eradicating CML.
The present data suggest that although ALOX5 has been identified as being essential for the development of CML in mice, this is not the case in human CML. Initial assessment of ALOX5 expression before and after imatinib treatment in human CML demonstrated that ALOX5 was down-regulated below normal, unlike the mouse model of CML which showed that ALOX5 was up-regulated. Following imatinib treatment, ALOX5 mRNA expression increased in the Responder and Non-responder groups. In patients who subsequently progressed to blast crisis, no change in ALOX5 expression was observed. The failure to increase ALOX5 expression following three months of imatinib treatment may identify patients at high risk of disease progression. However, whether changes in ALOX5 expression offer any additional value beyond those obtained from measuring early cytogenetic or molecular responses remains unknown and would need to be tested further as part of a clinical trial.
ALOX5 function was assessed by measuring plasma LTB4. LTB4 was found to increase in all three groups of patients following imatinib treatment. LTB4 was also found to be increased in blast crisis compared to chronic phase. These data suggest that the arachidonic acid pathway (Figure 1) is functionally active to the point of LTB4 production.
To determine the differences between the ALOX5 pathway observed in mice and the clinical samples studied, the LTB4 receptor (BLT1) was investigated. LTB4 acts by binding to LTB4R1 in order to mediate its positive role on ALOX5. At diagnosis of CML, patients had very low levels of LTB4R1 protein compared to normal samples. Following imatinib treatment, LTB4R1 levels increased towards normal, and in one case in blast crisis LTB4R1 protein was further suppressed. Using the Bayesian model averaging (BMA) method on a large microarray data set, Radich et al.14 identified six genes that discriminate between early chronic phase, late chronic phase, accelerated phase and blast crisis; one of these genes was LTB4R1. The authors found this to be down-regulated during blast crisis which is in agreement with our data. These findings help to explain the data seen in CML patients. At diagnosis of CML, ALOX5 gene expression was found to be suppressed and there was an accumulation of LTB4. The accumulation of LTB4 is likely due to lack of the LTB4 receptor. A lack of LTB4R1 allows the 5-HEPTE and LTA4 intermediates of the pathway to suppress ALOX5 gene expression, and therefore LTB4 accumulates as it has no receptor to bind to. Following imatinib treatment, LTB4R1 protein increases; thus LTB4 can bind to its receptor and positively regulate ALOX5, as demonstrated by an increase in 5-LOX protein and gene expression.
In patients with atherosclerosis, the binding of LTB4 to LTB4R1 induces the rapid phosphorylation of mitogen activated protein kinases (MAPK, ERK1/2 and JNK1/2) and PI3K/AKT, and also increases NF-κB activation,15 which are all targets of BCR-ABL1. It is, therefore, interesting to speculate that CML cells attempt to switch off excessive signaling via these pathways by down-regulating LTB4R1 in order to try and maintain cellular homeostasis. Our conclusions are based on work carried out using both MNC and CD34+ cells. We would like to extend these observations by assessing ALOX5 and LTB4R1 expression in primitive CD34+CD38−or CD34+CD38−CD90+ stem progenitor cells; but like most laboratories we do not have access to such sorted primitive subpopulations. We hope these can be the subject of future studies.
In conclusion, it is apparent that, regarding ALOX5, the CML mouse model and CML patients represent two different pathways. This accounts for the difference between the clinical samples studied herein and the results of the Chen et al.3 paper. These data suggest caution when extrapolating mouse model data into human CML.
Acknowledgments
We would like to thank the Alison Holcroft, Rachael Fowler and Andrea Davies for their help and support during this project. AG was supported by a generous donation in memory of a CML patient.
Footnotes
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Lucas CM, Wang L, Austin GM, Knight K, Watmough SJ, Shwe KH, et al. A population study of imatinib in chronic myeloid leukaemia demonstrates lower efficacy than in clinical trials. Leukemia. 2008; 22(10):1963–6. [DOI] [PubMed] [Google Scholar]
- 2.de Lavallade H, Apperley JF, Khorashad JS, Milojkovic D, Reid AG, Bua M, et al. Imatinib for Newly Diagnosed Patients With Chronic Myeloid Leukemia: Incidence of Sustained Responses in an Intention-to-Treat Analysis. J Clin Oncol. 2008;26(20):3358–63. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Hu Y, Zhang H, Peng C, Li S. Loss of the Alox5 gene impairs leukemia stem cells and prevents chronic myeloid leukemia. Nat Genet. 2009;41(7):783–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manev H, Manev R. 5-Lipoxygenase (ALOX5) and FLAP (ALOX5AP) gene polymorphisms as factors in vascular pathology and Alzheimer’s disease. Medical Hypotheses. 2006;66(3):501–3. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Li D, Li S. The Alox5 gene is a novel therapeutic target in cancer stem cells of chronic myeloid leukemia. Cell Cycle. 2009;8(21):3488–92. [DOI] [PubMed] [Google Scholar]
- 6.Yokomizo T, Uozumi N, Takahashi T, Kume K, Izumi T, Shimizu T. Leukotriene A4 hydrolase and leukotriene B4 metabolism. J Lipid Mediat Cell Signal. 1995;12:321–32. [DOI] [PubMed] [Google Scholar]
- 7.Drazen JM, Yandava CN, Dube L, Szczerback N, Hippensteel R, Pillari A, et al. Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. Nat Genet. 1999;22(2):168–70. [DOI] [PubMed] [Google Scholar]
- 8.Dwyer JH, Allayee H, Dwyer KM, Fan J, Wu H, Mar R, et al. Arachidonate 5-Lipoxygenase Promoter Genotype, Dietary Arachidonic Acid, and Atherosclerosis. N Engl J Med. 2004;350(1):29–37. [DOI] [PubMed] [Google Scholar]
- 9.Hoque A, Lippman SM, Wu T, Xu Y, Liang ZD, Swisher S, et al. Increased 5-lipoxygenase expression and induction of apoptosis by its inhibitors in esophageal cancer: a potential target for prevention. Carcinogenesis. 2005;26(4):785–91. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Pearson K, Pillitteri L, Ferguson JE, Clark RE. Serial monitoring of BCR-ABL by peripheral blood real-time polymerase chain reaction predicts the marrow cytogenetic response to imatinib mesylate in chronic myeloid leukaemia. Br J Haematol. 2002;118(3):771–7. [DOI] [PubMed] [Google Scholar]
- 11.Lucas CM, Harris RJ, Giannoudis A, Davies A, Knight K, Watmough SJ, et al. Chronic myeloid leukaemia patients with the e13a2 BCR-ABL fusion transcript have inferior responses to imatinib than e14a2 patients. Haematologica. 2009;94(10):1362–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyons MA, Wittenburg H. Susceptibility to cholesterol gallstone formation: Evidence that LITH genes also encode immune-related factors. Biochim Biophys Acta. 2006; 1761(10):1133–47. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Pearson K, Pillitteri L, Ferguson JE, Clark RE. Serial monitoring of BCR-ABL by peripheral blood real-time polymerase chain reaction predicts the marrow cytogenetic response to imatinib mesylate in chronic myeloid leukaemia. Br J Haematol. 2002;118(3):771–7. [DOI] [PubMed] [Google Scholar]
- 14.Oehler VG, Yeung KY, Choi YE, Bumgarner RE, Raftery AE, Radich JP. The derivation of diagnostic markers of chronic myeloid leukemia progression from microarray data. Blood. 2009;114(15):3292–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sánchez-Galán E, Gómez-Hernández A, Vidal C, Martín-Ventura JL, Blanco-Colio LM, Muñoz-García B, et al. Leukotriene B4 enhances the activity of nuclear factor-κB pathway through BLT1 and BLT2 receptors in atherosclerosis. Cardiovasc Res. 2009; 81(1):216–25. [DOI] [PubMed] [Google Scholar]


