Abstract
Background
The objective was to investigate the association between BMI and single nucleotide polymorphisms previously identified of obesity-related genes in two Spanish populations. Forty SNPs in 23 obesity-related genes were evaluated in a rural population characterized by a high prevalence of obesity (869 subjects, mean age 46 yr, 62% women, 36% obese) and in an urban population (1425 subjects, mean age 54 yr, 50% women, 19% obese). Genotyping was assessed by using SNPlex and PLINK for the association analysis.
Results
Polymorphisms of the FTO were significantly associated with BMI, in the rural population (beta 0.87, p-value <0.001). None of the other SNPs showed significant association after Bonferroni correction in the two populations or in the pooled analysis. A weighted genetic risk score (wGRS) was constructed using the risk alleles of the Tag-SNPs with a positive Beta parameter in both populations. From the first to the fifth quintile of the score, the BMI increased 0.45 kg/m2 in Hortega and 2.0 kg/m2 in Pizarra. Overall, the obesity predictive value was low (less than 1%).
Conclusion
The risk associated with polymorphisms is low and the overall effect on BMI or obesity prediction is minimal. A weighted genetic risk score based on genes mainly acting through central nervous system mechanisms was associated with BMI but it yields minimal clinical prediction for the obesity risk in the general population.
Keywords: Obesity, Genetics, FTO gene, Genetic score
Background
Obesity is a global pandemic and a major health concern because of the consequent morbidity and premature mortality. Changes in lifestyles resulting in energy intake and expenditure imbalance have led to an increase in obesity prevalence all over the world. Although this trend is driven by the “obesogenic” environment, evidence demonstrated that it is facilitated by genetic susceptibility, being the heritability in familial and twin studies of around 40-70%, [1,2].
Although many genetic variants have consistently been associated with obesity, the individual impact on body weight seems to be small. Using genome-wide association studies (GWAS), several genes have been associated with obesity, especially the fat mass and obesity associated gene (FTO). Initially found in a GWAS of type 2 diabetes, FTO has been consistently associated with obesity and BMI in many studies [3-6]. Moreover, a published meta-analysis confirmed the association in 32387 individuals of European ancestry from 15 cohorts [7]. According to this study, the top hit within the intron 1 of FTO gene was the rs1421085 although all the SNPs in high linkage disequilibrium (LD) with this variant were also strongly associated with BMI, including the rs9939609 the most replicated that increase 31% the risk of obesity [4].
Other candidate genes have also been linked with obesity with varying degrees of association. The melanocortin 4 receptor (MC4R) gene, linked with monogenic obesity [8], also has a polygenic effect with the minor allele of the c.307G > A (rs2229616, p.Val103Ile) being protective against obesity [9,10]. Other loci, including the INSIG2, TMEM18, KCTD15, SH2B1, MTCH2, GNPDA2, BDNF, or CHST8 genes, have also been associated with obesity. Furthermore, two other loci, one close to the NEGR1 gene and another near STK33 were also associated to obesity in some studies and the NEGR1 gene has been independently associated with obesity in a pediatric cohort [11].
A genetic risk score constructed with the FTO, MC4R and six of the newly discovered loci performed poorly as a predictor of obesity and the explained variance of BMI was less than 1% [7]. Due to the heterogeneity of the trait, however, the predictive value may vary with the characteristics of the population studied. The objective of the present study was to assess the association between individual genetic variants and haplotypes of several obesity-related genes (FTO, MC4R, MTCH2, NEGR1, SEC16B, INSIG2, TMEM18, KCTD15, SH2B1, FAIM2, ATXN2L, BDNF, BDNFOS, GNPDA2, ADRB2, PRL, PTER, ADIPOQ, ETV5, MAF, NPC1, CTNNBL1, and HTR2C) and BMI in two Spanish populations. We analyzed interactions between the associated genetic variants in order to identify functional relationships between genes and metabolic pathways for BMI regulation. The potential predictive value of a weighted genetic risk score to predict obesity was also evaluated.
Methods
Two adult general population samples were used in the present study, one from Pizarra, a village of 6600 inhabitants which is located in the south of Spain, and the other from an urban area of Valladolid located in the center of Spain. All the participants provided written informed consent and the local ethical committees approved the studies.
The Pizarra study is a population-based survey of cardiovascular risk factors. The characteristics of this population have been previously published [12,13]. Briefly, 2090 subjects aged 18-65 years were randomly selected from the municipal register. For 1119 out of 2090 individuals demographic, anthropometric and DNA were available. Persons with severe clinical or psychological problems, pregnant women and those who were institutionalized were excluded. This population has a prevalence of obesity higher than that reported for other Spanish communities. From this population, we were only able to use 869 subjects for the final analysis (131 were excluded because of lack of complete information and 119 due to that they did not pass the quality thresholds).
The selection process of the Hortega study has also been previously published [14,15]. Briefly, subjects older than 18 years old were randomly selected from the public register of the western medical area of Valladolid (Spanish National Statistical Institute; http://ine.es). Subjects were invited to participate in the study by phone. Individuals with serious concomitant diseases or psychiatric disorders were excluded. A second list of subjects was selected to replace those who reclined to take part in the study. The percentage of replacement was 32%. The calculated minimal sample size required to be representative of the population was 1400, and 1504 individuals were finally recruited. From this population, we were only able to use 1425 subjects for the final analysis (2 were excluded because of technical reasons and 77 due to that they did not pass the quality thresholds).
The names of the institutional boards which approved the study were: Institutional board of the Clinical Hospital Río Hortega in Valladolid. Institutional board of the Carlos Haya Hospital in Malaga.
Anthropometric measurements
The anthropometric parameters were measured for all the individuals according to standard procedures in the two studies. Weight was assessed with a precise scale while the subjects were wearing light clothes and barefoot. Height was assessed in a similar way. Body mass index (BMI) was calculated as weight divided by height2 expressed in kg/m2. Obesity was defined as a BMI ≥ 30 kg/m2 and overweight as a BMI of 25.0–29.9 kg/m2[16]. Blood pressure was assessed with an automatic device following the recommendations of the European Society of Hypertension. Fasting blood samples for blood count and serum biochemistry were analyzed by an auto-analyzer.
Type 2 diabetes definition
In Hortega subjects were considered as diabetics if they were already diagnosed of type 2 diabetes by a physician or if the plasma glucose remained equal or higher than 126 mg/dl after the extraction of a second sample in fasting conditions in those subjects with glucose equal or higher than 140 mg/dl in non fasting conditions [17].
In Pizarra, the WHO 1998 criteria were used to classify the people with diabetes, IGT and IFG [12,18]. People were also considered to have diabetes if they were already receiving treatment with oral anti-diabetics. Those people being treated with diet only received an OGTT to verify the diagnosis.
Genotyping
Blood for the genotyping was taken into tubes with 15% of ethylenediaminetetraacetic acid (EDTA) and was kept at 4ºC to process in 5 days. Those samples which were not processed in five days after the extraction were frozen at -80ºC. DNA was isolated from peripheral blood cells using Realpure Genomic DNA extraction kit (Real Pure, Paterna, Spain) and the samples were diluted to a final concentration of 100 ng/μl. SNPs were selected based on a search in the PubMed database of previous reports about association with obesity in GWAS studies between the years 2007-2009. Selected SNPs were genotyped using an oligo-ligation assay (SNPlex; Applied Biosystems, Foster City, CA) following the manufacturer’s instructions. The characteristics of the selected SNPs and related genes are shown in Table 1. In both populations the minor allele frequencies (MAF) were quite similar to the MAF in the HapMap CEU samples. One SNP, rs7561317, close to the TMEM18 gene, was excluded because of being monomorphic.
Table 1.
Characteristics of the selected SNPs
| Locus | GENE name | HGN | SNP | TAG-SNP | CHR position | HGVS names | HORTEGA GEN% | PIZARRA GEN% | HORTEGA MAF | PIZARRA MAF | HWE p-value (All/controls) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1p31.1 |
NEURONAL GROWTH REGULATOR 1 |
NEGR1 |
rs3101336 |
|
72523773 |
NT_032977.8:g.42723104C > T |
96.80 |
100 |
0.237 |
0.252 |
0.472/0.663 |
|
rs2568958 |
|
72537704 |
NT_032977.8:g.42737035A > G |
98.86 |
100 |
0.115 |
0.095 |
0.858/0.303 |
|||
|
rs2815752 |
|
72585028 |
NT_032977.8:g.42784359A > G |
98.93 |
100 |
0.355 |
0.349 |
0.929/0.304 |
|||
| 1q25.2 |
SEC16 HOMOLOG B (S. CEREVISIAE) |
SEC16B |
rs10913469 |
|
176180142 |
NM_033127.2:c.1881 + 177A > G |
97.80 |
100 |
0.154 |
0.143 |
0.293/1 |
| 2q14.1 |
INSULIN INDUCED GENE 2 |
INSIG2 |
rs7566605 |
|
118552495 |
NT_022135.15:g.7543947C > G |
98.20 |
99.77 |
0.454 |
0.439 |
0.706/0.643 |
| 2p25.3 |
TRANSMEMBRANE PROTEIN 18 |
TMEM18 |
rs2867125 |
|
612827 |
NT_022327.14:g.612827T > C |
97.47 |
100 |
0.351 |
0.397 |
0.078/0.186 |
|
rs6548238 |
|
624905 |
NT_022327.14:g.624905T > C |
98.93 |
99.88 |
0.449 |
0.419 |
0.043/0.141 |
|||
|
rs4854344 |
|
628144 |
NT_022327.14:g.628144G > T |
98.73 |
99.88 |
0.380 |
0.387 |
0.0521/0.164 |
|||
| 3q27 |
ADIPONECTIN, C1Q AND COLLAGEN DOMAIN |
ADIPOQ |
rs17300539 |
|
188042154 |
NT_005612.15:g.93054610G > A |
98.53 |
99.54 |
0.433 |
0.403 |
0.032/0.342 |
|
rs3774261 |
1 |
188054253 |
NM_004797.2:c.215-414A > G |
98.20 |
99.54 |
0.496 |
0.484 |
0.967/0.726 |
|||
| 3q28 |
ETS VARIANT 5 |
ETV5 |
rs7647305 |
|
187316984 |
|
95 |
98.85 |
0.118 |
0.125 |
0.902/0.655 |
| 4p13 |
GLUCOSAMINE-6-PHOSPHATE DEAMINASE 2 |
GNPDA2 |
rs10938397 |
|
44877284 |
NT_006238.10:g.4884493A > G |
97.80 |
100 |
0.195 |
0.198 |
0.382/0.340 |
| 5q31-q32 |
ADRENERGIC, BETA-2-, RECEPTOR, SURFACE |
ADRB2 |
rs12654778 |
1 |
148185934 |
NT_029289.10:g.9368677G > A |
98.13 |
100 |
0.363 |
0.330 |
0.207/0.318 |
| 6p22.2-p21.3 |
PROLACTIN |
PRL |
rs4712652 |
|
22186594 |
NT_007592.14:g.12936866G > A |
97.07 |
97.58 |
0.346 |
0.375 |
0.804/0.516 |
| 10p12 |
PHOSPHOTRIESTERASE RELATED |
PTER |
rs10508503 |
|
16339957 |
NT_077569.2:g.10662847C > T |
96.27 |
99.19 |
0.346 |
0.375 |
0.006/0.015 |
| 11p13 |
BRAIN-DERIVED NEUROTROPHIC FACTOR |
BDNF |
rs4923461 |
|
27613486 |
NR_002832.1:c.244-4482G > A |
98.33 |
99.88 |
0.169 |
0.172 |
0.955/0.637 |
|
rs925946 |
|
27623778 |
NR_002832.1:c.404 + 5650G > T |
98.47 |
100 |
0.347 |
0.375 |
0.0001/0.003 |
|||
|
rs10501087 |
|
27626684 |
NR_002832.1:c.404 + 8556G > T |
98.00 |
99.65 |
0.424 |
0.384 |
0.779/0.634 |
|||
|
rs6265 |
|
27636492 |
NM_001709.3:c.196G > A (p.V66M) |
98.13 |
100 |
0.461 |
0.489 |
0.373/0.426 |
|||
| 11p14.1 |
BDNF OPPOSITE STRAND (NON-PROTEIN CODING) |
BDNFOS |
rs4074134 |
|
27603861 |
NR_002832.1:c.244-14107G > C |
98.86 |
99.54 |
0.185 |
0.160 |
0.956/0.841 |
| 11p11.2 |
MITOCHONDRIAL CARRIER HOMOLOG 2 |
MTCH2 |
rs4752856 |
1 |
47604618 |
NM_014342.2:c.681 + 590C > T |
98.40 |
99.77 |
0.240 |
0.261 |
0.928/0.699 |
|
rs10838738 |
|
47619625 |
NM_014342.2:c.87 + 882T > C |
97.87 |
99.65 |
0.489 |
0.455 |
1/0.545 |
|||
| 12q13 |
FAS APOPTOTIC INHIBITORY MOLECULE 2 |
FAIM2 |
rs7138803 |
|
48533735 |
NT_029419.11:g.12390774G > A |
98.66 |
99.42 |
0.354 |
0.344 |
0.391/0.111 |
| 16p11 |
ATAXIN 2-LIKE |
ATXN2L |
rs8049439 |
|
28745016 |
NM_007245.2:c.466-46T > C |
96.53 |
100 |
0.327 |
0.356 |
0.619/0.512 |
| 16q12.2 |
FAT MASS AND OBESITY ASSOCIATED |
FTO |
rs6499640 |
|
52327178 |
NM_001080432.1:c.45 + 31536A > G |
98.07 |
99.88 |
0.173 |
0.173 |
0.694/0.488 |
|
rs1421085 |
1 |
52358455 |
NM_001080432.1:c.46-43098T > C |
97.87 |
100 |
0.237 |
0.261 |
1/0.719 |
|||
|
rs1121980 |
1 |
52366748 |
NM_001080432.1:c.46-34805G > A |
98.93 |
100 |
0.073 |
0.092 |
0.617/0.389 |
|||
|
rs8050136 |
1 |
52373776 |
NM_001080432.1:c.46-27777C > A |
98.86 |
99.19 |
0.220 |
0.228 |
0.420/0.470 |
|||
|
rs3751812 |
|
52375961 |
NM_001080432.1:c.46-25592G > T |
98.80 |
100 |
0.381 |
0.376 |
0.471/0.471 |
|||
|
rs9939609 |
1 |
52378028 |
NM_001080432.1:c.46-23525T > A |
98.33 |
99.88 |
0.174 |
0.170 |
0.966/0.876 |
|||
|
rs7190492 |
|
52386253 |
NM_001080432.1:c.46-15300G > A |
98.73 |
99.65 |
0.351 |
0.351 |
0.782/0.438 |
|||
|
rs8044769 |
|
52396636 |
NM_001080432.1:c.46-4917C > T |
97.33 |
100 |
0.339 |
0.329 |
0.967/0.880 |
|||
| 16q22-q23 |
V-MAF MUSCULOAPONEUROTIC FIBROSARCOMA |
MAF |
rs1424233 |
|
78240252 |
|
99.13 |
99.77 |
0.321 |
0.321 |
0.805/0.52 |
| 16p11.2 |
SH2B ADAPTOR PROTEIN 1 |
SH2B1 |
rs4788102 |
|
28780899 |
NT_010393.15:g.20186477G > A |
98.27 |
99.77 |
0.224 |
0.204 |
0.890/0.466 |
| 18q22 |
MELANOCORTIN 4 RECEPTOR |
MC4R |
rs17782313 |
|
56002077 |
NT_025028.13:g.5641943T > C |
98.66 |
100 |
0.455 |
0.464 |
0.215/0.382 |
| 18q11-q12 |
NIEMANN-PICK DISEASE, TYPE C1 |
NPC1 |
rs1805081 |
|
19394430 |
NM_000271.3:c.644A > G (p.H215R) |
98.93 |
99.65 |
0.343 |
0.376 |
0.012/0.017 |
| 19q13.11 |
POTASSIUM CHANNEL TETRAMERISATION |
KCTD15 |
rs11084753 |
|
39013977 |
NT_011109.15:g.6590355A > G |
97.93 |
99.65 |
0.428 |
0.390 |
0.184/0.336 |
| 20q11.23-q12 |
CATENIN, BETA LIKE 1 |
CTNNBL1 |
rs6013029 |
|
35832994 |
NM_030877.3:c.750 + 3134G > T |
98.27 |
99.88 |
0.267 |
0.283 |
0.574/0.482 |
| Xq24 | 5-HYDROXYTRYPTAMINE RECEPTOR 2C | HTR2C | rs3813929 | 113724776 | NT_028405.11:g.250852C > T | 98.47 | 99.88 | 0.434 | 0.399 | 0.168/0.292 |
HGN: Human genome nomenclature; SNP: single nucleotide polymorphism; CHR position: chromosomal position; HGVS: human genome variation society; Gen%: genotyping call rate; MAF: minor allele frequency.
Statistical analysis
Quantitative variables are expressed as the mean ± standard deviation (SD). Qualitative variables are expressed as entire counts or percentage. We used the free software Quanto v1.2.4 to calculate the statistical power with the continuous and qualitative traits under an additive genetic model taking into account the minor allele frequency of the selected SNPs, the sample size, the mean and standard deviation of BMI in our sample and the magnitude of the association for that variants in the literature with a type I error of 0.05.
To assess the association between genotypes/haplotypes and BMI, we used linear regression models adjusted by age and gender under the additive inheritance genetic model. Because of the relationship of BMI with type 2 diabetes, we also performed the analysis including type 2 diabetes as co-variable. The interaction between genetic variants and BMI was made by adding a multiplicative term within the linear regression model. The Bonferroni correction adjusted for 26 independent tests (see Additional file 1: Figure S1 about the LD patterns) was used to correct for multiple comparisons, being the corrected p-value for significance of 0.00192.
The r2 was used to measure the linkage disequilibrium (LD). Haplotype frequencies were estimated by the Expectation Maximization Algorithm (EM). Tag-SNPs, LD and haploblocks were calculated using Haploview version 3.32. The individual SNP and haplotype analysis was performed with the program PLINK v.1.06 developed by Purcell (http://pngu.mgh.harvard.edu/purcell/plink/). The average genotyping call rate to filter individuals and SNPs was 95%. We also filter those SNPs with a MAF lower than 1% and with a HWE p-value lower than 0.001. A weighted genetic risk score (wGRS) was constructed using the risk alleles of the Tag-SNPs which showed a positive Beta parameter in Hortega, Pizarra and the pooled analysis. For each individual, the number of risk alleles (0,1,2) per SNP was weighted for their effect sizes and re-scaled by dividing by the average of the all the effect sizes. The weighted risk alleles for the selected SNPs were summed for each individual, and the overall individual sum was rounded to the nearest integer to represent the individual’s risk allele score. The comparison of BMI among the quintiles of the score adjusted for age and sex and the 95% confidence interval for the means were calculated from the linear regression estimates. We used the area under the curve (AUC) from the receiver operating characteristic (ROC) curves to assess the capability of the score to predict obesity in the two populations. These procedures were performed with StataIC 11 (StataCorp4905 Lakeway drive, College Station, Texas, 77845, USA).
Finally, the statistical heterogeneity of the results for the two populations was analysed using the p-value for Cochrane’s Q statistic and the I2 heterogeneity index. This meta-analysis was also performed with PLINK considering both fixed and random effect models. Two SNPs, rs4712652 and rs1424233, were excluded from the meta-analysis because allele mismatch.
Results
The general characteristics of the individuals from the two populations after removal of subjects with low genotyping call rate are shown in Table 2. Subjects from Pizarra population were significantly younger and had higher BMI and greater prevalence of obesity than those from the Hortega study. The gender distribution was balanced in the Hortega population whereas in the Pizarra study there were more women than men.
Table 2.
Characteristics of the individuals included in the study for Hortega and Pizarra populations after removing those individuals with low genotyping call rate
| Variables | HORTEGA study (N = 1425) | PIZARRA study (N = 869) |
|---|---|---|
| Age (years) |
54.4 ± 19.3 |
46.2 ± 13.8*** |
| Gender, M(%)/F(%) |
718 (50.4) / 707 (49.6) |
322 (37.2) / 543 (62.8)*** |
| Weight (kg) |
70.8 ± 12.9 |
74.0 ± 14.1*** |
| Height (m) |
1.63 ± 0.1 |
1.6 ± 0.1*** |
| Waist perimeter (cm) (M/F) |
95.70 ± 10.15 /83.4 ±12.8 |
100.6 ± 10.6/ 97.8 ±14.7*** |
| BMI (kg/m2) |
26.4 ± 4.2 |
28.6 ± 5.2*** |
| SBP (mmHg) |
130.8 ± 21.5 |
129.1 ± 21.8 |
| DBP (mmHg) |
79.3 ± 10.7 |
77.7 ± 12.4*** |
| Total Cholesterol (mg/dl) |
201.6 ± 38.2 |
202.6 ± 39.7 |
| LDL Cholesterol (mg/dl)¶ |
114.3 ± 34.5 |
124.2 ± 34.1*** |
| HDL Cholesterol (mg/dl) |
51.6 ± 4.2 |
58.5 ± 13.2*** |
| Triglycerides (mg/dl) |
178.1 ± 114.4 |
99.5 ± 67.0*** |
| Glucose (mg/dl) |
92.6 ± 20.7 |
106.7 ± 30.3*** |
| Obesity (N/%) |
253/18. 5 |
307/36.0 *** |
| Overweight (N/%) |
574/41.9 |
306/ 35.9* |
| Abdominal obesity (N/%)+ |
388/28.6 |
514/60.1 *** |
| DM2 (N/%) |
109/7.7 |
145/18.9 *** |
| HTN (N/%) | 602/42.3 | 365/42.8 |
*p-values denote differences between Hortega study and Pizarra study (p < 0.05); <0.01; ***p-values <0.001; M: Male, F: Female, BMI: Body mass index, SBP: Systolic blood pressure, DBP: Diastolic blood pressure, DM2: Type 2 Diabetes Mellitus, HTN: Hypertension; ¶Calculated with the Friedwald formula; +Abdominal obesity was defined as waist >102 cm in men and >88 cm in women.
Association analysis with BMI
Pizarra population: From 988 individuals with complete information, 119 subjects were excluded because of low genotyping rate. The genotyping call rate for the remaining individuals was 99.7 ± 0.9%. Except for the rs7561317, the rest of SNPs passed the thresholds for HWE, MAF or call rate. The SNP genotyping call rate for the remaining SNPs was 99.7 ± 0.45%. The results of the genetic association study in Pizarra are presented in Table 3.
Table 3.
Individual TagSNP association analysis with BMI adjusted by age and gender under an additive genetic model in Pizarra, Hortega and the pooled analysis
| |
|
|
|
|
|
|
BMI |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | A1 | MAF | Population | Prior statistical power | N* | Beta | Standard error | STAT** | P |
|
FTO |
rs9939609* |
A |
0.398 |
PIZARRA |
32.77 |
848 |
0.875 |
0.23 |
3.79 |
0.0001573 |
| |
|
|
0.435 |
HORTEGA |
66.65 |
1345 |
0.1348 |
0.15 |
0.90 |
0.368 |
| |
|
|
0.420 |
POOLED |
77.88 |
2215 |
0.2976 |
0.13 |
2.19 |
0.0287 |
| |
rs8044769 |
T |
0.463 |
PIZARRA |
30.96 |
848 |
−0.582 |
0.23 |
−2.54 |
0.0112 |
| |
|
|
0.455 |
HORTEGA |
62.61 |
1331 |
−0.1019 |
0.15 |
−0.68 |
0.495 |
| |
|
|
0.458 |
POOLED |
73.98 |
2198 |
−0.257 |
0.13 |
−1.90 |
0.0571 |
| |
rs7190492 |
A |
0.328 |
PIZARRA |
5.06 |
845 |
−0.559 |
0.23 |
−2.34 |
0.0192 |
| |
|
|
0.337 |
HORTEGA |
5.16 |
1346 |
0.2065 |
0.16 |
1.29 |
0.195 |
| |
|
|
0.333 |
POOLED |
5.21 |
2213 |
−0.07336 |
0.14 |
−0.51 |
0.6076 |
| |
rs6499640 |
G |
0.376 |
PIZARRA |
5.01 |
847 |
0.336 |
0.23 |
1.43 |
0.1511 |
| |
|
|
0.381 |
HORTEGA |
5.02 |
1347 |
0.09312 |
0.15 |
0.60 |
0.543 |
| |
|
|
0.379 |
POOLED |
5.02 |
2216 |
0.2335 |
0.13 |
1.69 |
0.0911 |
|
MC4R |
rs17782313* |
C |
0.199 |
PIZARRA |
9.69 |
848 |
0.575 |
0.28 |
1.98 |
0.0478 |
| |
|
|
0.191 |
HORTEGA |
16.33 |
1348 |
0.1167 |
0.19 |
0.60 |
0.544 |
| |
|
|
0.195 |
POOLED |
20.23 |
2218 |
0.3111 |
0.17 |
1.80 |
0.0716 |
|
MTCH2 |
rs10838738 |
G |
0.352 |
PIZARRA |
5.80 |
845 |
0.453 |
0.24 |
1.88 |
0.0603 |
| |
|
|
0.353 |
HORTEGA |
6.98 |
1348 |
0.0972 |
0.15 |
0.62 |
0.536 |
| |
|
|
0.353 |
POOLED |
7.61 |
2215 |
0.1698 |
0.14 |
1.19 |
0.2318 |
|
NEGR1 |
rs3101336 |
A |
0.375 |
PIZARRA |
7.89 |
848 |
−0.084 |
0.24 |
−0.34 |
0.7277 |
| |
|
|
0.343 |
HORTEGA |
11.89 |
1348 |
−0.403 |
0.15 |
−2.64 |
0.0083 |
| |
|
|
0.355 |
POOLED |
14.23 |
2210 |
−0.1586 |
0.14 |
−1.13 |
0.2561 |
|
ATXN2L |
rs8049439 |
C |
0.375 |
PIZARRA |
8.87 |
848 |
−0.139 |
0.23 |
−0.58 |
0.5576 |
| |
|
|
0.338 |
HORTEGA |
14.15 |
1344 |
0.3292 |
0.15 |
2.13 |
0.0331 |
| |
|
|
0.353 |
POOLED |
17.35 |
2209 |
0.1715 |
0.14 |
1.22 |
0.2198 |
|
HTR2C |
rs3813929 |
T |
0.157 |
PIZARRA |
10.16 |
847 |
−0.406 |
0.35 |
−1.16 |
0.2455 |
| |
|
|
0.182 |
HORTEGA |
19.46 |
1342 |
−0.4365 |
0.23 |
−1.89 |
0.0577 |
| |
|
|
0.172 |
POOLED |
23.29 |
2211 |
−0.5551 |
0.20 |
−2.68 |
0.0073 |
|
ADRB2 |
rs12654778 |
A |
0.388 |
PIZARRA |
3 5.35 |
848 |
0.271 |
0.23 |
1.16 |
0.2452 |
| |
|
|
0.380 |
HORTEGA |
69.22 |
1347 |
−0.1452 |
0.15 |
−0.95 |
0.339 |
| |
|
|
0.382 |
POOLED |
80.65 |
2217 |
0.01463 |
0.13 |
0.10 |
0.9152 |
|
SEC16B |
rs10913469 |
C |
0.142 |
PIZARRA |
9.33 |
848 |
0.392 |
0.32 |
1.21 |
0.2266 |
| |
|
|
0.152 |
HORTEGA |
16.43 |
1347 |
−0.1609 |
0.20 |
−0.78 |
0.432 |
| |
|
|
0.148 |
POOLED |
19.78 |
2216 |
0.00939 |
0.18 |
0.05 |
0.96 |
|
TMEM18 |
rs4854344 |
G |
0.172 |
PIZARRA |
15.26 |
847 |
−0.198 |
0.30 |
−0.64 |
0.5173 |
| |
|
|
0.169 |
HORTEGA |
30.08 |
1348 |
−0.3012 |
0.20 |
−1.48 |
0.138 |
| |
|
|
0.170 |
POOLED |
37.86 |
2217 |
−0.243 |
0.18 |
−1.33 |
0.1815 |
|
INSIG2 |
rs7566605 |
C |
0.327 |
PIZARRA |
13.68 |
846 |
0.101 |
0.24 |
0.41 |
0.676 |
| |
|
|
0.321 |
HORTEGA |
26.42 |
1348 |
−0.0301 |
0.16 |
−0.18 |
0.8514 |
| |
|
|
0.322 |
POOLED |
33.12 |
2216 |
0.1152 |
0.14 |
0.79 |
0.4265 |
|
ADIPOQ |
rs17300539 |
A |
0.126 |
PIZARRA |
7.59 |
844 |
−0.072 |
0.33 |
−0.21 |
0.8298 |
| |
|
|
0.118 |
HORTEGA |
11.08 |
1348 |
0.1146 |
0.22 |
0.51 |
0.6082 |
| |
|
|
0.120 |
POOLED |
13.15 |
2214 |
0.0818 |
0.20 |
0.40 |
0.6838 |
| |
rs3774261 |
A |
0.495 |
PIZARRA |
10.96 |
844 |
−0.036 |
0.22 |
−0.16 |
0.8727 |
| |
|
|
0.461 |
HORTEGA |
19.84 |
1346 |
−0.0549 |
0.15 |
−0.36 |
0.7138 |
| |
|
|
0.473 |
POOLED |
26.42 |
2210 |
0.0609 |
0.13 |
0.45 |
0.6503 |
|
ETV5 |
rs7647305 |
T |
0.205 |
PIZARRA |
10.76 |
838 |
0.022 |
0.28 |
0.079 |
0.9364 |
| |
|
|
0.215 |
HORTEGA |
19.82 |
1325 |
0.2402 |
0.18 |
1.30 |
0.1927 |
| |
|
|
0.213 |
POOLED |
24.39 |
2175 |
0.2106 |
0.16 |
1.26 |
0.2077 |
|
GNPDA2 |
rs10938397 |
G |
0.440 |
PIZARRA |
11.60 |
848 |
−0.338 |
0.22 |
−1.52 |
0.1278 |
| |
|
|
0.455 |
HORTEGA |
21.56 |
1348 |
−0.048 |
0.15 |
−0.32 |
0.7492 |
| |
|
|
0.449 |
POOLED |
26.80 |
2218 |
−0.1687 |
0.13 |
−1.26 |
0.2078 |
|
PRL |
rs4712652 |
A |
0.453 |
PIZARRA |
5.28 |
828 |
0.203 |
0.23 |
0.87 |
0.3846 |
| |
|
G |
0.487 |
HORTEGA |
5.69 |
1326 |
0.0367 |
0.15 |
0.24 |
0.8056 |
| |
|
A |
0.489 |
POOLED |
5.91 |
2175 |
−0.115 |
0.13 |
−0.84 |
0.3976 |
|
PTER |
rs10508503 |
T |
0.093 |
PIZARRA |
5.10 |
841 |
−0.119 |
0.39 |
−0.30 |
0.7617 |
| |
|
|
0.112 |
HORTEGA |
5.27 |
1315 |
−0.279 |
0.23 |
−1.21 |
0.2238 |
| |
|
|
0.105 |
POOLED |
5.34 |
2174 |
−0.3695 |
0.21 |
−1.71 |
0.0866 |
|
BDNF |
rs925946 |
T |
0.282 |
PIZARRA |
10.40 |
848 |
0.128 |
0.24 |
0.51 |
0.6046 |
| |
|
|
0.266 |
HORTEGA |
17.97 |
1348 |
−0.071 |
0.16 |
−0.44 |
0.6578 |
| |
|
|
0.272 |
POOLED |
22.37 |
2215 |
0.0795 |
0.14 |
0.54 |
0.5855 |
| |
rs10501087 |
C |
0.253 |
PIZARRA |
10.01 |
845 |
0.033 |
0.26 |
0.12 |
0.8988 |
| |
|
|
0.231 |
HORTEGA |
16.77 |
1348 |
0.0875 |
0.17 |
0.49 |
0.6231 |
| |
|
|
0.240 |
POOLED |
20.96 |
2214 |
0.1472 |
0.15 |
0.93 |
0.3493 |
|
FAIM2 |
rs7138803 |
A |
0.352 |
PIZARRA |
7.38 |
844 |
0.155 |
0.24 |
0.65 |
0.5159 |
| |
|
|
0.352 |
HORTEGA |
10.92 |
1348 |
−0.0643 |
0.15 |
−0.42 |
0.675 |
| |
|
|
0.351 |
POOLED |
12.81 |
2214 |
0.0513 |
0.14 |
0.36 |
0.7129 |
|
MAF |
rs1424233 |
A |
0.483 |
PIZARRA |
5.29 |
846 |
−0.192 |
0.23 |
−0.83 |
0.4074 |
| |
|
G |
0.499 |
HORTEGA |
5.70 |
1348 |
0.2336 |
0.14 |
1.57 |
0.1158 |
| |
|
A |
0.495 |
POOLED |
5.92 |
2216 |
−0.2422 |
0.13 |
−1.79 |
0.0725 |
|
NPC1 |
rs1805081 |
G |
0.329 |
PIZARRA |
5.25 |
845 |
−0.014 |
0.24 |
−0.05 |
0.9553 |
| |
|
|
0.352 |
HORTEGA |
5.64 |
1345 |
0.0046 |
0.15 |
0.03 |
0.9758 |
| |
|
|
0.344 |
POOLED |
5.83 |
2212 |
−0.0463 |
0.14 |
−0.32 |
0.7432 |
|
KCTD15 |
rs11084753 |
A |
0.395 |
PIZARRA |
5.62 |
846 |
0.093 |
0.23 |
0.39 |
0.6897 |
| |
|
|
0.352 |
HORTEGA |
6.44 |
1343 |
−0.1562 |
0.15 |
−1.01 |
0.31 |
| |
|
|
0.367 |
POOLED |
6.94 |
2210 |
0.0573 |
0.13 |
0.41 |
0.6781 |
|
CTNNBL1 |
rs6013029 |
T |
0.091 |
PIZARRA |
5.79 |
847 |
0.014 |
0.39 |
0.036 |
0.9708 |
| |
|
|
0.066 |
HORTEGA |
6.44 |
1348 |
−0.1522 |
0.30 |
−0.50 |
0.6133 |
| 0.076 | POOLED | 7.16 | 2216 | 0.1139 | 0.25 | 0.44 | 0.6547 | |||
A1: minor allele, *Number of non-missing individuals included in the analysis (top) and frequency of the haplotype in the sample (bottom); **t statistic coefficient; p-values are not corrected for multiple testing; Bold type indicates significant association after Bonferroni correction. **The SNPs, rs4788102 in the SH2B1 and rs4074134 of the BDNFOS genes are not included because are in high LD with the SNPs, rs8049439 of the ATXN2L gene (r2 0.94) and rs10501087of the BDNF gene (r2 0.95), respectively.
All SNPs of the FTO gene within a block of high LD in intron 1 (r2 > 0.8) were significantly associated with BMI (beta 0.87, p-value <0.001, for the most associated SNP, rs9939609). Their impact on BMI, based on the confident intervals for the beta parameter ranged from 0.31 to 1.34 (average 0.83) as the lowest and highest limits for the confident intervals (average 1.46). None of the other variants analysed were significantly associated with BMI after Bonferroni correction. The only SNPs, other than FTO which had a p-value lower than 0.05 for BMI, were the rs17782313 close to the MC4R gene and the rs10838738 within the MTCH2 gene. When DM2 was included as co-variable the results did not change.
Hortega population
This study included 1502 subjects but 77 were excluded because of low genotyping rate (<95%). The average genotyping call rate after removing them was 99.7 ± 0.7%. As it was in Pizarra population only the rs7561317 polymorphism had to be excluded. The average SNP call rate was 99.7 ± 0.5%.
In this population none of the SNPs or haplotypes reached the significance after Bonferroni correction (Table 3). The most strongly associated SNPs were on chromosome 1 near the neuronal growth regulator 1 precursor (NEGR1) gene and are in high LD (beta -0.40, p-value 0.008 for the association of rs3101336 with BMI). Other SNPs around nominal p-values for significance were rs8049439 located in the ATXN2L gene on chromosome 16 [beta 0.33 (0.02-0.63), p-value 0.03] and rs3813929 within the HTR2C gene on chromosome X [beta -0.43 (-0.88-0.01), p-value 0.05]. The addition of DM2 did not affect the results of the individual SNPs analysis.
Pooled analyses
The pooled sample size included 2490 individuals but 196 were excluded because of low genotyping call rate (<95%). All the SNPs in high LD within the FTO gene (Additional file 1: Figure S1) were nominally associated with BMI, Table 3. The allele A of the rs9939609 produced an increase of 0.29 in BMI [beta 0.29 (0.03-0.56), p-value 0.02]. In this case, the most associated SNP was close to de HTR2C gene on chromosome X. The allele T of this SNP produced a decrease of about 0.5 in BMI [beta -0.55 (-0.96 - -0.15), p-value 0.007].
Although a formal test for interaction did not reveal significant interaction between the SNPs, rs6499640 and rs1421085 of FTO, however the double homozygotes for the minor allele had significantly higher BMI values than double homozygotes for the wild allele (Additional file 2: Figure S2).
The information about the association of the non-Tag SNPs with BMI can be consulted in the Additional file 3: Table S1. Information about the LD and haploblocks can be consulted in the Additional file 1: Figure S1. The haplotype association analysis can be consulted in the Additional file 4: Table S2.
Meta-analysis and statistical heterogeneity
For those SNPs in the FTO gene in high LD, the level of statistical heterogeneity based on the p-value for Cochrane’s Q statistic and the I2 heterogeneity index, was very high.
For the rest of the SNPs in other loci, the level of heterogeneity was low or moderate.
None of the SNPs reached the statistical significance after Bonferroni correction.
Only three SNPs were nominally associated with BMI in the meta-analysis, rs9939609 of FTO (in the fixed effect model), rs3101336 of the NEGR1 gene (both fixed and random effects models) and rs3813929 of the HTR2C gene (both fixed and random effects models).
The results for the meta-analysis and the statistical heterogeneity are shown in Table 4.
Table 4.
Meta-analysis for the association of BMI with individual SNPs, for Pizarra and Hortega studies, and level of statistical heterogeneity for each SNP
| |
|
|
|
|
|
BMI |
|||
|---|---|---|---|---|---|---|---|---|---|
| CHR | Gene | SNP | A1 | P | P(R) | Beta | Beta(R) | Q | I |
| 16 |
FTO |
rs9939609 |
A |
0.004768 |
0.1902 |
0.3544 |
0.4843 |
0.0071 |
86.22 |
| |
|
rs8044769 |
T |
0.05001 |
0.1921 |
−0.245 |
−0.3108 |
0.0788 |
67.64 |
| |
|
rs7190492 |
A |
0.8216 |
0.6838 |
−0.0299 |
−0.1556 |
0.0076 |
85.97 |
| |
|
rs6499640 |
G |
0.1948 |
0.1948 |
0.1659 |
0.1659 |
0.3853 |
0 |
| 18 |
MC4R |
rs17782313 |
C |
0.1092 |
0.1872 |
0.2563 |
0.2941 |
0.1879 |
42.34 |
| 11 |
MTCH2 |
rs10838738 |
G |
0.1222 |
0.1837 |
0.2031 |
0.2281 |
0.2157 |
34.75 |
| 1 |
NEGR1 |
rs3101336 |
A |
0.0154 |
0.04555 |
−0.3124 |
−0.2989 |
0.2647 |
19.6 |
| 16 |
ATXN2L |
rs8049439 |
C |
0.1431 |
0.5759 |
0.1893 |
0.1295 |
0.0975 |
63.58 |
| 23 |
HTR2C |
rs3813929 |
T |
0.02603 |
0.02603 |
−0.4272 |
−0.4272 |
0.9414 |
0 |
| 5 |
ADRB2 |
rs12654778 |
A |
0.8699 |
0.9013 |
−0.0208 |
0.0254 |
0.1347 |
55.32 |
| 1 |
SEC16B |
rs10913469 |
C |
0.9855 |
0.8286 |
−0.0031 |
0.0586 |
0.149 |
51.97 |
| 2 |
TMEM18 |
rs4854344 |
G |
0.1106 |
0.1106 |
−0.2701 |
−0.2701 |
0.781 |
0 |
| |
INSIG2 |
rs7566605 |
C |
0.9423 |
0.9423 |
0.0097 |
0.0097 |
0.652 |
0 |
| 3 |
ADIPOQ |
rs17300539 |
A |
0.758 |
0.758 |
0.0573 |
0.0573 |
0.6433 |
0 |
| |
|
rs3774261 |
A |
0.6931 |
0.6931 |
−0.0494 |
−0.0494 |
0.9466 |
0 |
| |
ETV5 |
rs7647305 |
T |
0.2549 |
0.2549 |
0.1763 |
0.1763 |
0.5226 |
0 |
| 4 |
GNPDA2 |
rs10938397 |
G |
0.2632 |
0.2853 |
−0.1393 |
−0.1474 |
0.2784 |
14.87 |
| 10 |
PTER |
rs10508503 |
T |
0.2284 |
0.2284 |
−0.2387 |
−0.2387 |
0.7263 |
0 |
| 11 |
BDNF |
rs925946 |
T |
0.9293 |
0.9293 |
−0.012 |
−0.012 |
0.4992 |
0 |
| |
|
rs10501087 |
C |
0.633 |
0.633 |
0.0701 |
0.0701 |
0.8626 |
0 |
| 12 |
FAIM2 |
rs7138803 |
A |
0.9989 |
0.9989 |
−0.0002 |
−0.0002 |
0.4393 |
0 |
| 18 |
NPC1 |
rs1805081 |
G |
0.9971 |
0.9971 |
−0.0005 |
−0.0005 |
0.9492 |
0 |
| 19 |
KCTD15 |
rs11084753 |
A |
0.5295 |
0.5295 |
−0.0808 |
−0.0808 |
0.3724 |
0 |
| 20 | CTNNBL1 | rs6013029 | T | 0.7034 | 0.7034 | −0.0914 | −0.0914 | 0.738 | 0 |
CHR: Chromosome code; SNP: SNP identifier; A1: Minor allele code; P: Fixed-effects meta-analysis p-value; P(R): Random-effects meta-analysis p-value; BETA: Fixed-effects BETA estimate; BETA(R): Random-effects BETA estimate; Q: p-value for Cochrane’s Q statistic; I: I2heterogeneity index (0-100).
Weighted genetic risk score
A genetic risk score was constructed with the risk alleles of the six tagSNPs with a positive Beta in Hortega, Pizarra and the Pooled analysis, that is: rs9939609 and rs6499640 of the FTO gene; rs17782313 of the MC4R gene; rs10838738 of the MTCH2 gene; rs7647305 of the ETV5 gene; and rs10501087 of the BDNF gene.
The score constructed with the sum of the weighed risk alleles was positively correlated with BMI in Pizarra and in the pooled sample (r = 0.15 in Pizarra, p-value < 0.001; r = 0.082 in the pooled sample, p-value < 0.001). From the first to the fifth quintile of the score, the BMI increases 2 Kg/m2 in Pizarra, 0.45 Kg/m2 in Hortega and 0.93 Kg/m2 in the pooled analysis. The BMI values for each quintile of the score as well as the regression line between BMI and the risk score is shown in Figure 1. However, the variance of BMI associated to individual SNPs or to the score was very low (less than 1%). The predictive value of the score for obesity was poor [area under the curve (AUC) 0.515 and 0.594 in Hortega and Pizarra respectively] (Additional file 5: Figure S3).
Figure 1.
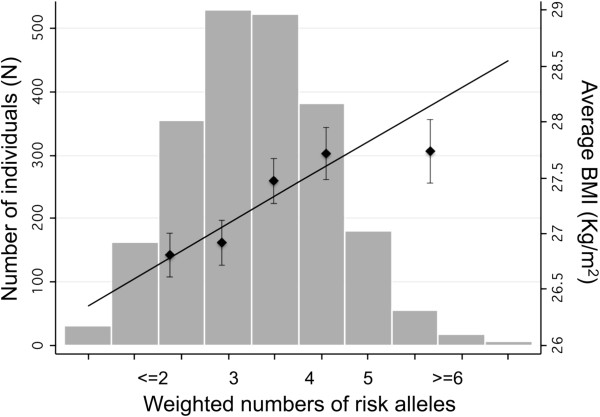
Combined impact of risk alleles on average BMI in the pooled analysis. For each individual, the number of risk alleles (0,1,2) per SNP was weighted for their effect sizes and re-scaled by dividing by the average of the all the effect sizes. The weighted risk alleles for the selected SNPs were summed for each individual, and the overall individual sum was rounded to the nearest integer to represent the individual’s risk allele score. Along the x axis, individuals in each risk allele category are shown (grouped ≤2 and ≥6 at the extremes), and the mean BMI (± s.e.m.) is plotted (y axis on right), with the line representing the regression of the mean BMI values across the risk allele scores. The histogram (y axis on left) represents the number of individuals for each risk-score category.
Discussion
The present study, carried out in two Spanish populations with different characteristics, confirms the association of BMI with some of the genes previously described and provides further evidence of the influence of the population characteristics on the association level. Variants in the FTO gene were significantly associated with BMI in one of the populations, Pizarra, whereas none of the selected variants were significantly associated in Hortega population or in the pooled analysis.
A large battery of previously described genes associated with BMI and obesity were selected for the present study. They included not only the best characterized FTO gene but also others recently described for which the association was not as strong or was less consistently reproduced. Among them, FTO, MC4R, MTCH2 and HTR2C are the main associated loci with BMI and obesity. Among the new discovered loci, ATXN2L, NEGR1 and SH2B1 have also shown a relationship with obesity.
Virtually all the SNPs in LD in the intron 1 of FTO were associated with BMI in Pizarra population. Furthermore, another SNP of FTO, rs6499640, not in LD with the others was also associated with BMI when it was included in the haplotypes. The estimated effect of the minor allele of these SNPs was very low in concordance with published studies in which carriers of the minor allele of rs9939609 weighed about three kg more than carriers of the other allele [4]. In the Pizarra population, the strongest association was with rs9939609 although the majority of polymorphisms in high LD with it reached the significance even after adjusting by confounding factors and by multiple comparisons. In the meta-analysis performed by Willer et al [7], the estimated effect for these variants ranged from 0.06 to 0.33 kg/m2, what means a change in weight between 173-954 g per allele for adults who are 1.60-1.80 m height [7]. The estimated maximum effect of FTO gene alleles was higher in Pizarra population as compare to the results of the meta-analysis although in the pooled analysis the overall effect was similar.
In contrast with the strong association of FTO in the Pizarra population, there was a weak association in Hortega population. This difference could be attributed to the characteristics of the two populations. The Pizarra population was younger and had significantly higher mean BMI and prevalence of obesity than Hortega population. In fact, it has been suggested that the contribution of FTO may be more evident in very obese and younger populations [19]. There is also a trend toward less association with increase in age in some studies [4,20].
As a consequence of the low estimated effect of the variants, none of the other loci out of FTO reached the statistical significance after multiple comparisons adjustments. However, some of them which were close to the nominal p-value merit some comments. After FTO, the most associated SNP with BMI in Pizarra population was the rs17782313 of MC4R which it is in agreement with previous meta-analysis [7,21]. Located 188 kb downstream of the MC4R, it was first identified using GWAS in 16876 individuals of European descent and replicated in 60352 adults. MC4R is a strong candidate gene for obesity because functional mutations of this gene are associated with monogenic forms of obesity [22]. The reported effect associated with this variant was lower than the effect reported for the FTO variants. The power to detect a true association decreases when the associated effect of that variant decrease.
The integration of the information of relevant SNPs into a genetic risk score might be a way to select those subjects at high risk to develop obesity in the future. By using the information provided in the present study, a weighted genetic risk score (wGRS) was constructed by using those Tag-SNPs with a positive Beta in Hortega, Pizarra and the pooled analysis. This wGRS include the following SNPs: rs9939609 and rs6499640 of the FTO gene; rs17782313 of the MC4R gene; rs10838738 of the MTCH2 gene; rs7647305 of the ETV5 gene; and rs10501087 of the BDNF gene.
From the first to the fifth quintile of the score, the BMI increased 0.45 kg/m2 in Hortega, 2.00 kg/m2 in Pizarra and 0.94 kg/m2 in the pooled analysis. These data are in agreement to those obtained by the Genetic Investigation of Anthropometric Traits (GIANT) project in which subjects with the highest score weighed on average 1.46 kg/m2 more than those with the lowest score [7].
Despite the significant differences observed in BMI and in the prevalence of obesity according to the score categories, the explained variance for BMI was less than 1%, similar to the one predicted in a previous study for Willer and colleagues [7]. Several potential explanations can be offered for the low predictive value of the wGRS but are mainly related with the marginal effect sizes of the tested variants and the skewed distribution of the effect sizes [23]. Other potential explanations for the low predictive value of the wGRS could be related with the gene-gene or especially with the gene-environment interactions which were not considered in the present study [23]. Because the majority of these genes are expressed in the central nervous system, acting in appetite regulation, behaviour and basal energy expenditure [7,24-26], the importance of environmental factors, mainly high energy intake and low physical activity, should be considered. Estimation of energy intake and physical activity, however, are unreliable because of under- or over-reported [27]. Since the effect of these genetic variants might be due to an increase of energy intake [28], inclusion of energy intake could lead to a marked improvement in prediction. This could have clinical impact if we are able to identify those individuals in which energy restriction below some threshold should be strongly recommended.
The main limitation of the present study is that our sample size was too small to detect association for the majority of the tested variants with low estimated effect, although we pooled two different populations to overcome this problem. This may have influenced the strength of the association but not the size of the effect, which was similar to that reported in one meta-analysis [7]. The appropriateness of combining these two populations in one is somehow controversial and merits some comments. Both samples share similar genetic backgrounds due to the low immigration rate and are supposed to be homogeneous ethnic populations. Because of this, we did not expect population stratification in our samples what can favour the pooled strategy. However, they belong to different geographical regions of Spain and they have completely different clinical characteristics. This clinical heterogeneity probably has had a great influence in the pooled analysis results. For this reason we also decided to perform a meta-analysis to assess the statistical heterogeneity. For some of the SNPs such as those of the FTO gene, the level of statistical heterogeneity based on the p-value for Cochrane’s Q statistic and the I2 heterogeneity index, was very high. For other SNPs, such as the rs12654778 at ADRB2 with a priori adequate statistical power to detect significant association, we did not find any. The clinical heterogeneity and the level of statistical heterogeneity, at least moderate, can justify the lack of association. Besides we cannot be sure for certain that the prior statistical power for that SNP is that high due to the previously commented considerations.
The different lipid and carbohydrate metabolic profiles observed between populations may be related with the different characteristics of the target populations with markedly different lifestyles. Several studies have shown that the effects of FTO alleles are attenuated by exercise [29,30]. Individuals from Pizarra population belonged to a rural area and were significantly younger than those from the Hortega study. The individuals from the latter study were recruited in the area covered by a tertiary hospital and the majority of them lived in urban areas. This population was also even regarding to gender distribution as compare with Pizarra population, which included mostly females. Because of the potential influence of the population to which each individual belongs, we also adjusted the analysis by this factor, and the results remained unchanged.
Conclusions
In conclusion, baseline characteristics of the populations, mainly age and grade of obesity, have a strong influence in the genetic association results. FTO was the only locus that was clearly associated with BMI in this study. None of the other loci including the MC4R, MTCH2 or the newly discovered ones, such as ATXN2L, NEGR1 and SH2B1 were associated with BMI in this study. The risk associated with these polymorphisms is low and the overall effect in BMI is minimal. Considering the high heritability of obesity, new variants remain to be discovered. As commented previously, the majority of the analysed loci are related to central nervous system mechanisms of obesity but many other mechanisms can influence body weight and their contribution have not been elucidated yet. New strategies, like the study of lean individuals [31-34], translational information from animal models [35-38], nutrigenomics [39,40], as well as the interaction with energy intake and physical activity may lead to a better understanding of the genetic component in the physiopathology of obesity.
Competing interests
We declare that we have not received reimbursements, fees, funding, or salary from organizations that may in any way gain or lose financially from the publication of this manuscript, either now or in the future.
We declare that we do not hold any stocks or shares in organizations that may in any way gain or lose financially from the publication of this manuscript, either now or in the future.
We declare that we do not hold and that we are not applying for any patents relating to the content of the manuscript.
We declare that we have not received reimbursements, fees, funding, or salary from an organization that holds or has applied for patents relating to the content of the manuscript.
We declare that we do not have any other financial or non-financial competing interests.
Authors’ contributions
FM: Develop the main analysis, wrote and edited the manuscript. MLM, DdM and FJC: Designed the study, made the genotyping and edited the manuscript. JR: Designed the study and reviewed the manuscript. GR; SM; and FS are the main investigators of the Pizarra study. Edited and reviewed the article. MP and JCE, are the main investigator of the Hortega study. Edited and reviewed the article. RSC: Reviewed and edited the article. All authors read and approved the final manuscript.
Supplementary Material
Information about the LD and haploblocks for the following genes: NEGR1, TMEM18, ADIPOQ, BDNF, MTCH2 and FTO.
Mean of BMI according to genotypes of rs1421085 and rs6499640 of FTO adjusted by age and gender in the pooled sample.
Individual association for the NonTag-SNPs with BMI adjusted by age and gender under an additive genetic model in Pizarra, Hortega and the pooled analysis.
Haplotype association analysis with BMI adjusted by age and gender in Pizarra, Hortega and in the pooled analysis.
Areas under the curve for the obesity prediction for the weighted genetic risk score in Pizarra and Hortega populations.
Contributor Information
Fernando Martínez-García, Email: fermar23@uv.es.
María L Mansego, Email: mlmansego@unav.es.
Gemma Rojo-Martínez, Email: gemma.rojo.m@gmail.com.
Griselda De Marco-Solar, Email: gridemar@alumni.uv.es.
Sonsoles Morcillo, Email: sonsoles.morcillo.exts@juntadeandalucia.es.
Federico Soriguer, Email: federico.soriguer.sspa@juntadeandalucia.es.
Josep Redón, Email: josep.redon@uv.es.
Monica Pineda Alonso, Email: monicpi16@gmail.com.
Juan C Martín-Escudero, Email: juancarlos.martinescudero@gmail.com.
Richard S Cooper, Email: rcooper@lumc.edu.
Felipe J Chaves, Email: felipe.chaves@uv.es.
Acknowledgement
Funding
This work was supported by CIBER of Diabetes and metabolic diseases (CIBERDEM) and CIBER obesity and nutrition (CIBEROB). CIBERDEM and CIBEROBN are initiatives of the “Fondo de Investigaciones Sanitarias”; INGENFRED (Ref. CIBER-02-08-2009) grant from CIBERDEM; PI070497 from the “Fondo de Investigaciones Sanitarias”; ACOMP/2009/201 and PROMETEO/2009/029 from the Valencian Government; SAF2005-02883 from Ministerio de Educación y Ciencia; P06-CTS-01684 from Andalusian Governement; Gema Rojo-Martínez [CP06/00133], and F.Javier Chaves [FIS01/3047] are contracted by the Health Institute Carlos III.
References
- Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27(4):325–351. doi: 10.1023/A:1025635913927. [DOI] [PubMed] [Google Scholar]
- Atwood LD, Heard-Costa NL, Cupples LA, Jaquish CE, Wilson PW, D’Agostino RB. Genomewide linkage analysis of body mass index across 28 years of the Framingham Heart Study. Am J Hum Genet. 2002;71(5):1044–1050. doi: 10.1086/343822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G. et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C. et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(6):724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- Renstrom F, Payne F, Nordstrom A, Brito EC, Rolandsson O, Hallmans G, Barroso I, Nordstrom P, Franks PW. Replication and extension of genome-wide association study results for obesity in 4923 adults from northern Sweden. Hum Mol Genet. 2009;18(8):1489–1496. doi: 10.1093/hmg/ddp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C. et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisse C, Clement K, Guy-Grand B, Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet. 1998;20(2):113–114. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- Young EH, Wareham NJ, Farooqi S, Hinney A, Hebebrand J, Scherag A, O’Rahilly S, Barroso I, Sandhu MS. The V103I polymorphism of the MC4R gene and obesity: population based studies and meta-analysis of 29 563 individuals. Int J Obes (Lond) 2007;31(9):1437–1441. doi: 10.1038/sj.ijo.0803609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutzmann F, Cauchi S, Durand E, Calvacanti-Proenca C, Pigeyre M, Hartikainen AL, Sovio U, Tichet J, Marre M, Weill J. et al. Common genetic variation near MC4R is associated with eating behaviour patterns in European populations. Int J Obes (Lond) 2009;33(3):373–378. doi: 10.1038/ijo.2008.279. [DOI] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I. et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Soriguer-Escofet F, Esteva I, Rojo-Martinez G, Ruiz de Adana S, Catala M, Merelo MJ, Aguilar M, Tinahones F, Garcia-Almeida JM, Gomez-Zumaquero JM. et al. Prevalence of latent autoimmune diabetes of adults (LADA) in Southern Spain. Diabetes Res Clin Pract. 2002;56(3):213–220. doi: 10.1016/S0168-8227(02)00002-5. [DOI] [PubMed] [Google Scholar]
- Soriguer F, Rojo-Martinez G, Almaraz MC, Esteva I, Ruiz de Adana MS, Morcillo S, Valdes S, Garcia-Fuentes E, Garcia-Escobar E, Cardona I. et al. Incidence of type 2 diabetes in southern Spain (Pizarra Study) Eur J Clin Invest. 2008;38(2):126–133. doi: 10.1111/j.1365-2362.2007.01910.x. [DOI] [PubMed] [Google Scholar]
- Mena-Martin FJ, Martin-Escudero JC, Simal-Blanco F, Carretero-Ares JL, Arzua-Mouronte D, Herreros-Fernandez V. Health-related quality of life of subjects with known and unknown hypertension: results from the population-based Hortega study. J Hypertens. 2003;21(7):1283–1289. doi: 10.1097/00004872-200307000-00015. [DOI] [PubMed] [Google Scholar]
- Martin Escudero JC, Simal Blanco F, Carretero Ares JL. et al. Prevalencia, grado de deteccion, tratamiento y control de la hipertension arterial en la poblacion general. Estudio Hortega. Hypertension. 2003;20:148–154. [Google Scholar]
- Clinical guidelines on the identification, evaluation, and treatment of overweight and obesity in adults: executive summary. Expert Panel on the Identification, Evaluation, and Treatment of Overweight in Adults. Am J Clin Nutr. 1998;68(4):899–917. doi: 10.1093/ajcn/68.4.899. [DOI] [PubMed] [Google Scholar]
- Mena Martin FJ, Martin Escudero JC, Simal Blanco F, Carretero Ares JL, Herreros Fernandez V. Cardiovascular risk factors in diabetic patients. Cross-sectional study in general population: Hortega study. An Med Interna. 2003;20(6):292–296. [PubMed] [Google Scholar]
- Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15(7):539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Jacobsson JA, Riserus U, Axelsson T, Lannfelt L, Schioth HB, Fredriksson R. The common FTO variant rs9939609 is not associated with BMI in a longitudinal study on a cohort of Swedish men born 1920-1924. BMC Med Genet. 2009;10:131. doi: 10.1186/1471-2350-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Kang K, Zhang C, van Dam RM, Kraft P, Hunter D, Lee CH, Hu FB. Fat mass-and obesity-associated (FTO) gene variant is associated with obesity: longitudinal analyses in two cohort studies and functional test. Diabetes. 2008;57(11):3145–3151. doi: 10.2337/db08-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS. et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini F, Maffei M, Pelosini C, Salvetti G, Scartabelli G, Pinchera A. Melanocortin-4 receptor mutations in obesity. Adv Clin Chem. 2009;48:95–109. [PubMed] [Google Scholar]
- Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187(2):367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MW, Porte D Jr. Diabetes, obesity, and the brain. Science. 2005;307(5708):375–379. doi: 10.1126/science.1104344. [DOI] [PubMed] [Google Scholar]
- Berthoud HR, Sutton GM, Townsend RL, Patterson LM, Zheng H. Brainstem mechanisms integrating gut-derived satiety signals and descending forebrain information in the control of meal size. Physiol Behav. 2006;89(4):517–524. doi: 10.1016/j.physbeh.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Berthoud HR, Morrison C. The brain, appetite, and obesity. Annu Rev Psychol. 2008;59:55–92. doi: 10.1146/annurev.psych.59.103006.093551. [DOI] [PubMed] [Google Scholar]
- Huang TT, Howarth NC, Lin BH, Roberts SB, McCrory MA. Energy intake and meal portions: associations with BMI percentile in U.S. children. Obes Res. 2004;12(11):1875–1885. doi: 10.1038/oby.2004.233. [DOI] [PubMed] [Google Scholar]
- Martinez-Gomez D, Welk GJ, Calle ME, Marcos A, Veiga OL. Preliminary evidence of physical activity levels measured by accelerometer in Spanish adolescents: the AFINOS Study. Nutr Hosp. 2009;24(2):226–232. [PubMed] [Google Scholar]
- Rampersaud E, Mitchell BD, Pollin TI, Fu M, Shen H, O’Connell JR, Ducharme JL, Hines S, Sack P, Naglieri R. et al. Physical activity and the association of common FTO gene variants with body mass index and obesity. Arch Intern Med. 2008;168(16):1791–1797. doi: 10.1001/archinte.168.16.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimaleswaran KS, Li S, Zhao JH, Luan J, Bingham SA, Khaw KT, Ekelund U, Wareham NJ, Loos RJ. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene. Am J Clin Nutr. 2009;90(2):425–428. doi: 10.3945/ajcn.2009.27652. [DOI] [PubMed] [Google Scholar]
- Lee YS. The role of genes in the current obesity epidemic. Ann Acad Med Singapore. 2009;38(1):45–53. [PubMed] [Google Scholar]
- Farooqi S, O’Rahilly S. Genetics of obesity in humans. Endocr Rev. 2006;27(7):710–718. doi: 10.1210/er.2006-0040. [DOI] [PubMed] [Google Scholar]
- Laskarzewski PM, Khoury P, Morrison JA, Kelly K, Mellies MJ, Glueck CJ. Familial obesity and leanness. Int J Obes. 1983;7(6):505–527. [PubMed] [Google Scholar]
- Forbes GB, Sauer EP, Weitkamp LR. Lean body mass in twins. Metabolism. 1995;44(11):1442–1446. doi: 10.1016/0026-0495(95)90144-2. [DOI] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA. et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372(6505):425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- Pomp D, Nehrenberg D, Estrada-Smith D. Complex genetics of obesity in mouse models. Annu Rev Nutr. 2008;28:331–345. doi: 10.1146/annurev.nutr.27.061406.093552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomp D, Mohlke KL. Obesity genes: so close and yet so far. J Biol. 2008;7(9):36. doi: 10.1186/jbiol93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afman L, Muller M. Nutrigenomics: from molecular nutrition to prevention of disease. J Am Diet Assoc. 2006;106(4):569–576. doi: 10.1016/j.jada.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Elliott RM, Johnson IT. Nutrigenomic approaches for obesity research. Obes Rev. 2007;8(Suppl 1):77–81. doi: 10.1111/j.1467-789X.2007.00323.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information about the LD and haploblocks for the following genes: NEGR1, TMEM18, ADIPOQ, BDNF, MTCH2 and FTO.
Mean of BMI according to genotypes of rs1421085 and rs6499640 of FTO adjusted by age and gender in the pooled sample.
Individual association for the NonTag-SNPs with BMI adjusted by age and gender under an additive genetic model in Pizarra, Hortega and the pooled analysis.
Haplotype association analysis with BMI adjusted by age and gender in Pizarra, Hortega and in the pooled analysis.
Areas under the curve for the obesity prediction for the weighted genetic risk score in Pizarra and Hortega populations.


