Abstract
Identification of vulnerability in the HIV-1 envelope (Env) will aid in Env-based vaccine design. We recently found an HIV-1 clade C Env clone (4-2.J45) amplified from a recently infected Indian patient showing exceptional neutralization sensitivity to autologous plasma in contrast to other autologous Envs obtained at the same time point. By constructing chimeric Envs and fine mapping between sensitive and resistant Env clones, we found that substitution of highly conserved isoleucine (I) with methionine (M) (ATA to ATG) at position 424 in the C4 domain conferred enhanced neutralization sensitivity of Env-pseudotyped viruses to autologous and heterologous plasma antibodies. When tested against monoclonal antibodies targeting different sites in gp120 and gp41, Envs expressing M424 showed significant sensitivity only to anti-V3 monoclonal antibodies and modestly to sCD4 and b12. Substitution of I424M in unrelated Envs also showed similar neutralization phenotype, indicating that M424 in C4 region induces exposure of neutralizing epitopes particularly in CD4 binding sites and V3 loop.
Keywords: HIV-1, neutralizing antibody, V3 loop, CD4bs, clade C, envelope, C4 domain, recent infection
Introduction
The remarkable variations in the genetic diversity, glycosylation patterns and conformational flexibility of the Human Immunodeficiency Virus Type 1 (HIV-1) envelope (Env), provide means for evading antibodies that neutralize and abrogate virus infectivity (Doria-Rose et al., 2010; Korber et al., 2001; Kwong et al., 2002; Seaman et al., 2010; Zhang et al., 2010). Humoral immunity in particular exerts substantial pressure on the gp120 HIV-1 envelope surface protein, leading to escape from the autologous neutralizing antibody (NAb) response by introducing specific mutations and/or compensatory substitutions in the env gene (Blish, Nguyen, and Overbaugh, 2008; Duenas-Decamp and Clapham, 2010; Duenas-Decamp et al., 2008; Duenas-Decamp et al., 2009; Gray et al., 2008; Gray et al., 2007b; O’Rourke et al., 2010; O’Rourke et al., 2009; Rong et al., 2007a; Rong et al., 2007b; Rong et al., 2009; Shen et al., 2010). While an AIDS vaccine is urgently needed, lack of understanding of how to precisely elicit potent and cross-reactive neutralizing antibodies (NAbs) poses a major hurdle to vaccine development (Barouch, 2008; Fauci et al., 2008; Walker, 2010). HIV-1 has evolved mechanisms to overcome neutralization by autologous antibodies during the natural course of infection wherein Env goes through substantial genetic drift due to antibody pressure giving rise to neutralization escape variants (Blay et al., 2006; Korber et al., 2001; Korber and Gnanakaran, 2009; Lynch et al., 2009; Zhang et al., 2010). Thus, elucidating Env modifications that modulate virus neutralization could illuminate how certain changes such as glycan positioning (Duenas-Decamp and Clapham, 2010; Duenas-Decamp et al., 2008) and specific substitutions expose neutralizing epitopes on Env for better presentation towards comprehensive antibody binding and neutralization (Li, Rey-Cuille, and Hu, 2001; Stamatatos et al., 2009; Wei et al., 2003; Zolla-Pazner, 2004).
It was found that only a limited number of monoclonal antibodies (MAbs) obtained from HIV-1-infected individuals have been found to exhibit potent neutralization against a wide range of primary isolates (Mascola, 2010; Walker, 2010; Zolla-Pazner and Cardozo, 2010). Very recently, it was proposed that quaternary structure-specific antibodies likely target antigenic variants specific for similar epitopes, with neutralization breadth determined by the prevalence of recognized variants among circulating isolates (Wu et al., 2011). In addition, there are many well studied broadly neutralizing human MAbs that recognize the CD4 binding site on Env such as b12 and VRC01 (Burton, 1994; Wu et al., 2010; Zhou et al., 2010), N-linked glycans on the gp120 outer domain such as 2G12 (Calarese, 2003; Trkola et al., 1996), and the recently described PG9 and PG16 MAbs targeting V1V2 regions in gp120 (Walker et al., 2009), In addition, three broadly neutralizing antibodies (bNAbs) viz., 2F5, 4E10 and Z13, that target the membrane-proximal external region (MPER) of gp41 showed extensive neutralization breadth and implicate the MPER as a possible vaccine target (Cardoso et al., 2005; Kunert et al., 2004; Muster, 1993; Ofek et al., 2004). MAbs and polyclonal serum antibodies directed to the V3 loop, which are commonly found during the course of natural infection, also are able to neutralize diverse HIV-1 strains (Bell et al., 2008; Hioe et al., 2010; Zolla-Pazner and Cardozo, 2010). Very recently, Xiang et al (Xiang et al., 2010) demonstrated cross talk between the V3 loop and adjacent beta strands, contributing to stabilizing the unliganded trimer. There are a number of changes to the HIV-1 Env that affect V3 exposure. These can result from CD4 binding as well as mutations (Kolchinsky et al., 2001; Laakso et al., 2007; Moore and Sodroski, 1996; Moore et al., 1993; Sullivan et al., 1998; Walker et al., 2009; Wyatt et al., 1992; Xiang et al., 2010).
In HIV-1 clade C infections, NAbs develop during early infection to high titer and have little or no cross neutralizing activity (Gray et al., 2007a; Li et al., 2006). Experiments showed that antibodies elicited by Env early in clade C infection recognize predominantly the variable loops (Li et al., 2006; Moore et al., 2008). However, in clade C infection, antibodies targeting the V3 loop were found to play a minimal role in neutralization of primary viruses (Binley and Burton., 2004; Li et al., 2005), possibly due to the occlusion of the V3 loop (Hartley et al., 2005; Krachmarov et al., 2006; Pinter et al., 2004). Recent studies on subtype C infection showed that the V1 through V4 domains were found to modulate neutralization sensitivity of Env through occlusion of conserved epitopes such as the coreceptor binding site (Gray et al., 2007a; Krachmarov et al., 2005; Krachmarov et al., 2006; Kwong et al., 1998; Moore et al., 2008; Pinter et al., 2004; Sagar et al., 2006; Wyatt et al., 1995) and by shielding of neutralization determinants (Derdeyn et al., 2004; Rademeyer et al., 2007). Unlike V1V2, the role of V4 and V5 in neutralization resistance is not clear (Kinsey et al., 1996; Rong et al., 2009; Wei et al., 2003).
We have recently characterized HIV-1 clade C Envs amplified from Indian patients with recent infection, wherein we found considerable variations in their sensitivities to autologous plasma antibodies (Ringe, Thakar, and Bhattacharya, 2010). On further investigation, we found one Env clone (4-2.J45) obtained from a patient (NARI-IVC4) which showed exceptional neutralization sensitivity compared to other Envs obtained at the same time point from the same patient. This sensitivity was noted with both contemporaneous plasma and with autologous plasma at baseline and from follow up visits in a longitudinal study. This prompted us to investigate the determinants conferring such shifts in neutralization sensitivity by making chimeric Env constructs and specific point substitutions. We found that substitution of methionine (M) in place of a highly conserved isoleucine (I) at position 424 in the C4 domain of gp120, proximal to the Phe43 cavity (Kwong et al., 1998; Madani et al., 2008; Repik and Clapham, 2008), conferred increased neutralization to autologous and heterologous plasma antibodies obtained from clade C infected Indian and South African donors. Further studies indicated that I424M substitution significantly increased sensitivity of Envs to anti-V3 monoclonal antibodies by greater exposure of neutralizing epitopes in V3 loop and also showed modest sensitivity to reagents targeting CD4 binding sites (CD4bs).
Results
A clade C Env clone showed exceptional neutralization sensitivity to autologous plasma antibodies compared to other contemporaneous Envs from the same patient
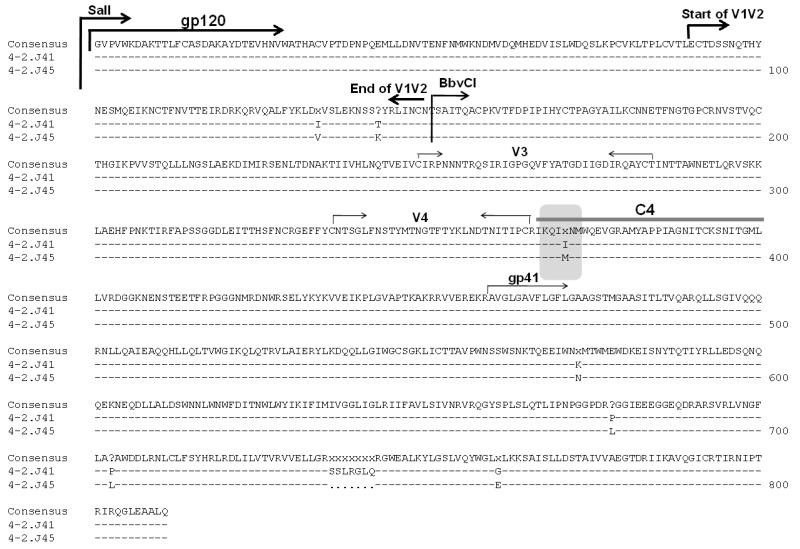
We previously described one clade C Env clone, 4-2.J45 which, in comparison to other contemporaneous Env clones (4-2.J41, 4-2.J42b, 4-2.J45b, 4-2.J46b and 4-2.J47b) (Ringe, Thakar, and Bhattacharya, 2010) with distinct sequences obtained from a recently infected Indian patient, showed greater than an 80-fold increase in neutralization sensitivity when tested against its autologous plasma (Ringe, Thakar, and Bhattacharya, 2010). These Env clones were amplified from plasma of the patient within one year of infection (Ringe, Thakar, and Bhattacharya, 2010). We first sought to investigate if V1V2 played any role in this increased autologous neutralization sensitivity, as we found two changes in the amino acid residues in the V2 region between the sensitive Env clone 4-2.J45 and a resistant Env clone 4-2.J41 that has >95% similarity in protein sequence (Figure 1). Thus, we exchanged an Env fragment between SalI and BbvCI sites from the resistant (4-2.J41) to the sensitive Env clone 4-2.J45 (Figure 1) to give rise to a chimeric Env with the 4-2.J45 backbone but with V1V2 from the resistant Env 4-2.J41. In essence, this chimeric Env [4-2.J45/J41V1V2)] was identical to 4-2.J45 where two residues (I186V and T195K) were substituted (found to be present in the resistant Env 4-2.J41). However, when this chimeric 4-2.J45 (J41/V1V2) Env was tested for its sensitivity to contemporaneous autologous plasma, it showed sensitivity similar to the 4-2.J45 wild type (WT) (data not shown here) indicating that the V1V2 loop did not play any role in enhanced sensitivity of 4-2.J45 Env to autologous antibodies.
Figure 1. Amino acid alignment between HIV-1 clade C sensitive Env (4-2.J45) and resistant Env (4-2.J41).
These two Env clones were obtained from the plasma of the same patient obtained at the same time point and found to differ significantly in their sensitivities to autologous plasma antibodies.
Presence of a methionine at the 424 position (M424) in the C4 domain of gp120 conferred enhanced neutralization sensitivity to autologous and heterologous plasma antibodies
Since we found that V1V2 did not confer increased neutralization sensitivity to autologous plasma antibodies, we next sought to map the determinants in other regions of the Env. Using the domain exchange strategy as described in the Materials and Methods, we exchanged gp41 between the sensitive Env (4-2.J45) and the resistant Envs; however when tested for the neutralization sensitivity, none of the chimeric viruses became sensitive to autologous plasma antibodies, suggesting that differences in residues between 4-2.J45 and other Envs was not basis for greater sensitivity of 4-2.J45 to autologous neutralization. Further fine mapping revealed one amino acid change (isoleucine to methionine) in the C4 region of gp120 at position 424 between 4-2.J45 and rest of the contemporaneous Envs (Figure 1). Interestingly, we found that I424 was present in all the resistant Envs, and isoleucine is highly conserved at the 424 position in the HIV Los Alamos database (www.hiv.lanl.gov) across all clades in group M HIV-1 and in SIV strains.
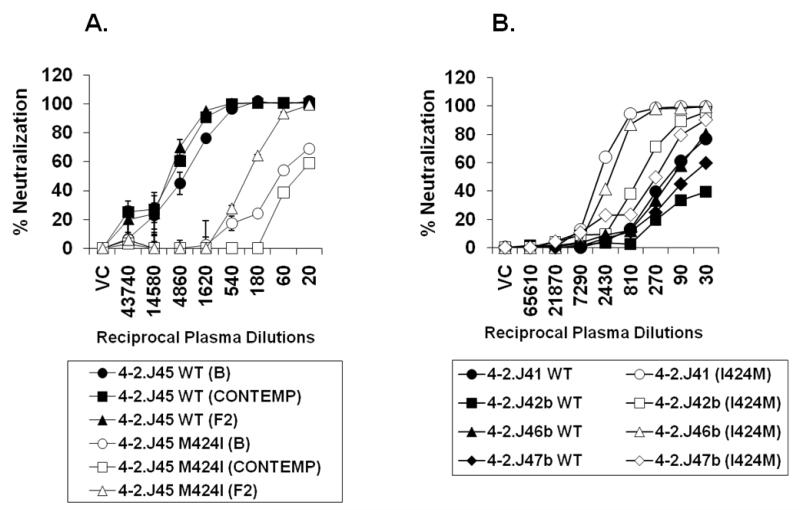
We next investigated whether substitution of methionine in place of highly conserved isoleucine at position 424 would alter the overall neutralization sensitivity to autologous plasma antibodies. By site-directed mutagenesis, we substituted M424I in the sensitive 4-2.J45 Env and tested its sensitivity along with the 4-2.J45 WT containing M424 to autologous plasma antibodies. As shown in Figure 2A, with M424I, 4-2.J45 Env became resistant to autologous plasma antibodies collected at three different times by approximately 80-fold. Our data suggested that indeed the presence of M424 resulted in increased sensitivity of 4-2.J45 Env to autologous antibodies. We further tested if I424M substitutions in four resistant Envs (obtained simultaneously from this patient at the same time point) altered sensitivities to autologous plasma. As shown in Figure 2B, Env-pseudotyped viruses containing M424 became more sensitive to autologous plasma antibodies compared to their counterparts containing I424.
Figure 2. Effect of I424M substitution on Env sensitivity to autologous neutralization.
A. Effect of I424M substitution towards increased sensitivity of 4-2.J45 Envs to autologous plasma obtained at baseline (B), contemporaneous (Contemp) and follow up (F2) B. Effect of I424M substitution in autologous Envs obtained from the same patient at the same time point towards their sensitivities to contemporaneous autologous plasma antibodies. Note that M424 conferred enhanced neutralization to autologous plasma antibodies. VC refers to virus control.
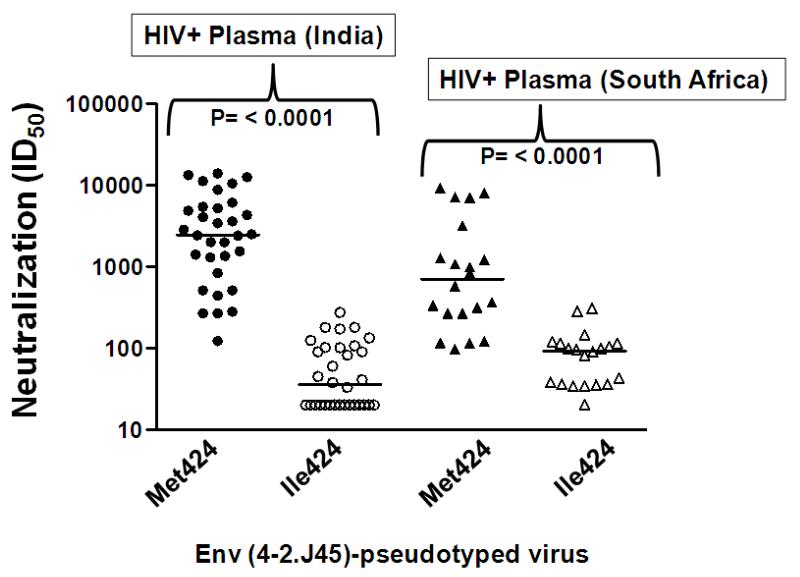
To further investigate whether M424 globally affected Env sensitivity to heterologous plasma antibodies in addition to autologous antibodies, we tested both 4-2.J45 WT (containing M424) and 4-2.J45 (containing I424) against several HIV+ plasma collected from asymptomatic patients both from India and South Africa where clade C infection is also prevalent. As shown in Figure 3, we observed significant differences (P<0.0001) in the sensitivities between 4-2.J45 (M424) and 4-2.J45 (I424) to these heterologous plasma and serum antibodies obtained from HIV+ Indian and South African donors, respectively. Comparable results were obtained with unrelated HIV-1 clade C Env 11-3.J3 (Ringe, Thakar, and Bhattacharya, 2010) (P = 0.0006) and clade B JRFL (P = 0.008) when tested against the same panel of HIV+ plasma specimens (data not shown). Thus, our data showed Envs expressing M424 conferred increased sensitivity to heterologous plasma and serum antibodies as opposed to Envs expressing I424.
Figure 3. Effect of Met424 and Ile424 on neutralization of Env-pseudotyped viruses by heterologous serum and plasma antibodies obtained from asymptomatic HIV+ Indian and South African donors.
Median inhibitory dilution (ID50) of HIV+ plasma and sera against Env-pseudotyped viruses containing Met424 and Ile424 were plotted using GraphPad Prism software. Note that a significant increase (P<0.00001) in sensitivity of Env-pseudotyped viruses occurred in presence of Met424 in C4 domain to HIV+ plasma antibodies obtained from both Indian and South African individuals.
M424 significantly enhanced Env sensitivity to anti-V3 MAbs without altering infectivity
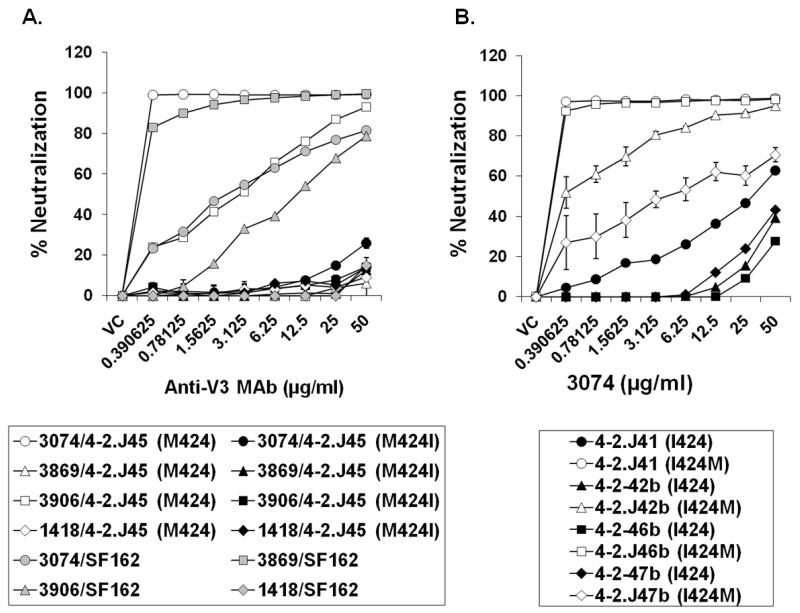
Since presence of M424 conferred enhanced sensitivity to an array of heterologous plasma and sera collected from positive donors from distinct geographical location, we hypothesized that this particular substitution possibly acted upon V3 loop. Our hypothesis was based on previous observation by different groups that majority of HIV+ plasmas predominantly target V3 loop (Carrow et al., 1991; Smith et al., 1994; Vogel, Kurth, and Norley, 1994; Walker et al., 2010). In addition, we previously found that the presence of the M424 residue in the 4-2.J45 Env did not significantly alter its sensitivity to MAbs targeting the MPER and CD4i epitopes (Ringe, Thakar, and Bhattacharya, 2010), compared to its contemporary neutralization-resistant Env clones containing I424. We therefore sought to examine whether I424M had affected the V3 loop which is part of the outer domain of the Env. To test this, the sensitivities of the Env-pseudotyped viruses containing either M424 or I424 to anti-V3 MAbs 3074, 3869 and 3906 were measured using TZM-bl target cells. As shown in Figure 4A, the 4-2.J45 Env containing M424 was found to be significantly more sensitive to anti-V3 MAbs 3074 and 3906 in contrast to the 4-2.J45 Env containing I424. Similarly, when we substituted methionine with isoleucine at position 424 (I424M) in related contemporaneous Envs 4-2.J41, 4-2.J42b, 4-2.J46b and 4-2.J47b obtained from the same patient, a significant increase in their sensitivity to anti-V3 MAb 3074 (that showed best neutralization above) was shown compared to the same Envs expressing I424 (Figure 4B). Additionally, we found that I424M substitution did not confer 4-2.J45 Env with CXCR4 usage. These data coupled with the observation that M424 did not alter the infectivity of Envs tested here (Figure S1) clearly indicate that Met424 modulated the Env conformation of the V3 loop and conferred enhanced neutralization sensitivity to plasma antibodies without compromising infectivity and without switching to CXCR4 usage.
Figure 4. Effect of presence of Met424 on sensitivity of Env-pseudotyped viruses to anti-V3 MAbs.
A. Met424 in 4-2.J45 Env was substituted with Ile424 and its effect on virus neutralization by three anti-MAbs was tested in TZM-bl cells. Note that anti-V3 MAB 3074 showed maximum neutralization. B. I424M substitution in the resistant Envs, pseudoviruses carrying Met424 conferred enhanced sensitivity to anti-V3 MAb 3074. VC refers to virus control.
M424 conferred increased neutralization by plasma antibodies and anti-V3 MAbs of heterologous clade C and clade B Envs
To further investigate whether Met424 could also enhance neutralization sensitivity of viruses carrying heterologous HIV-1 Envs, we first made the I424M substitution in an unrelated clade C Env (11-3.J3) obtained from the plasma of a different Indian patient with recent infection, reported previously (Ringe, Thakar, and Bhattacharya, 2010) as well as three clade B Envs (JRFL, YU2 and RHPA4259.7). Env-pseudotyped viruses containing both I424 and M424 were tested for their sensitivities to anti-V3 MAbs as well as plasma antibodies. Since the reactivity of 447-52D MAb was shown to be restricted by a polar interaction with the side chain of the R residue in the GPGR motif found mainly in clade B Envs but infrequently in other subtypes (Hioe et al., 2010), 447-52D was used to test clade B Envs. As shown in Figure 5, a significant increase in Env to anti-V3 MAbs of Envs expressing M424 was found. Thus, we found that I424M significantly enhanced 11-3.J3 Env (unrelated clade C) (Ringe, Thakar, and Bhattacharya, 2010) sensitivity greater than 250-fold to all three anti-V3 MAbs (3074, 3869 and 3906) tested here (Table 1 and Figure 5A). We also tested the effect of I424M substitution in three clade B Envs (RHPA4259.7, JRFL and YU2) and found 2-45-fold increase in their sensitivities to anti-V3 MAbs (3074, 3869, 3906 and 447-52D) (Table 1 and Figure 5B and 5C). Presence of M424 also showed enhanced sensitivity of heterologous Envs to plasma antibodies, highest being 11-3.J3 (with M424) (Table 2). Collectively, our data suggest that I424 M substitution alters the conformation of the V3 loop possibly through spatial and/or torsional displacement of V3 base (data not shown) towards unmasking of discontinuous epitopes to anti-V3 MAb in unliganded state (Xiang et al., 2010).
Figure 5. Effect of I424M substitution on neutralization sensitivities of unrelated clade C (A) and clade B (B and C) Envs.
Env pseudoviruses expressing M424 showed significant enhancement of Env sensitivity to anti-V3 MAbs. 11-3.J3 (M424) showed greater than 200-fold neutralization by anti-V3 MAbs 3074, 3869 and 3906 than that of I424, while clade B Envs showed 20-30% greater neutralization in presence of M424 with 3869 and 442-52D anti-V3 MAbs. VC refers to virus control.
Table 1.
Neutralization sensitivities of Envs to different anti-V3 MAbs
| Neutralization sensitivity to anti-V3 MAbs | |||
|---|---|---|---|
| IC50 (μg/ml) | |||
| Envelope | Clade | Ile424 | Met424 |
| 4.2J45 | C | >50 (3074, 3869, 3906) | <0.390625 (3074); >50 (3869); 3.125 (3906) |
| 11-3.J3 | C | 50 (3074, 3869, 3906) | <0.390625 (3074; 3869; 3906) |
| RHPA4259.7 | B | >50 (3074, 3869, 3906, 447-52D) | >50 (3074; 3906); 20 (3869); 26 (447-52D) |
| YU2 | B | >50 (3074, 3869, 3906, 447-52D) | >50 (3074; 3906); 8 (3869); 1.1 (447-52D) |
| JRFL | B | >50 (3074, 3869, 3906, 447-52D) | >50 (3074; 3906); 15 (3869); 1.56 (447-52D) |
IC50 values indicate the concentration of anti-V3 MAb resulting in 50% reduction in infectivity of Env-pseudotyped viruses in the TZM-bl cells. The anti-V3 MAbs used in this study are given in parenthesis next to their respective IC50 values.
Table 2.
Effect of I424M substitution on Env sensitivity to PG9, PG16, VRC01 and pooled plasma. Values represent inhibitory concentration (IC50) of MAbs tested. For plasma specimens, values represent reciprocal dilution at which 50% neutralization was observed.
| Envelope | PG9 | PG16 | VRC01 | b12 | Pooled plasma |
|---|---|---|---|---|---|
| 4-2.J45 (I424) | 0.018 | 0.16 | 0.42 | >10 | 67 |
| 4-2.J45 (M424) | 0.125 | 6 | 0.6 | >10 | 7046 |
| 11-3.J3 (I424) | 0.44 | 0.47 | 0.8 | >10 | 443 |
| 11-3.J3 (M424) | 0.49 | 0.92 | 0.51 | >10 | 2938 |
| JRFL (I424) | >10 | >10 | 0.1 | 0.1 | 97 |
| JRFL (M424) | >10 | >10 | 0.05 | 0.05 | 132 |
| YU2 (I424) | >10 | 0.15 | 0.06 | 3.55 | 134 |
| YU2 (M424) | 1.26 | 0.05 | 0.05 | 0.11 | 324 |
| RHPA4259.7 (I424) | >10 | 0.12 | <0.03 | 0.23 | 108 |
| RHPA4259.7 (M424) | >10 | 0.21 | <0.03 | 0.06 | 280 |
Presence of M424 in the C4 domain affects gp120 interaction with CD4 but not with CCR5
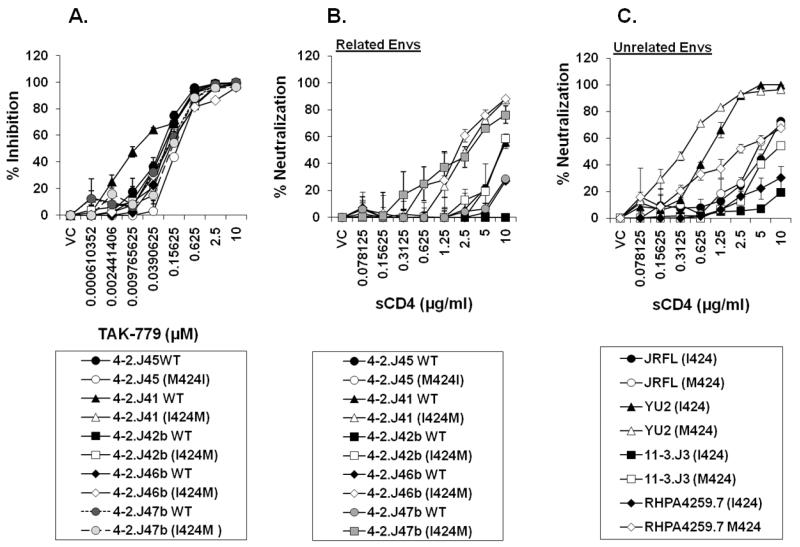
Since Met424 is very close to the 17b antibody binding site and influences the conformation of V3 as evidenced by imparting increased sensitivity to anti-V3 antibodies, we investigated if this residue also influences coreceptor binding. To test this, we incubated TZM-bl cells with different molar concentrations of CCR5 ligand TAK-779 for 1 hour following which Env-pseudotyped viruses expressing M424 and I424 were added and incubated further for two days. As shown in Figure 6A, no significant differences in inhibition of infectivity by TAK-779 were observed between viruses expressing either Met424 or Ile424, although, only 4-2.J41 Env containing Ile424 showed relatively greater sensitivity to TAK-779 than 4-2.J41 with Met424 (~14-folds). These data demonstrate that although the M424I substitution affected V3 conformation, it generally showed <4-fold impact on Env interaction with CCR5.
Figure 6. Effect of presence of Met424 on Env interaction with CD4 and CCR5.
A. Effect of Env-pseudotyped viruses carrying I424 and M424 on their degree of inhibition TAK-779. No significant difference in TAK-779 mediated inhibition was found due to presence of M424. However, presence of M424 showed modest increase in Env sensitivity to sCD4 both in related Envs (obtained from same patient at the same time point) (B) and unrelated clade B and C Envs (C) indicating that M424 imparted conformational shifts towards better binding with CD4. VC refers to virus control.
The 424 position in Env also resides in the vicinity of the structurally conserved Phe43 cavity on gp120 (Kwong et al., 1998; Madani et al., 2008; Repik and Clapham, 2008). To test whether I424M substitution could also alter gp120-CD4 interaction, a clone of HeLa cells expressing low cell surface CD4 and high CCR5 (RC49 clone) (Platt et al., 1998) was infected with Env-pseudotyped viruses expressing M424 and I424, and infectivity was measured by immunostaining of intracellular p24 as described earlier (Ringe, Thakar, and Bhattacharya, 2010). Env-pseudotyped viruses expressing M424 showed increases in infection of HeLa RC49 cells (Figure S2). In addition, M424 also showed a modest increase in Env sensitivity to sCD4 both in contemporaneous (related) Envs and in unrelated clade B and C Envs (Figure 6B and 6C). Our data provided evidence that alteration at position 424 in gp120 affects its interaction with CD4 but not with CCR5.
I424M substitution did not impact on quaternary epitopes in gp120 but showed modest shift in sensitivity to CD4bs antibodies
We next wanted to test if I424M substitution altered β20 sheet towards influencing the conformation of the outer domain of gp120 besides V3 loop. Thus, we examined whether I424M can alter the neutralization sensitivity of Envs to the MAbs that recognizes the CD4bs and to the conformational quaternary neutralizing epitopes (QNE). As shown in Table 2, we did not find any significant shifts in the neutralization sensitivity of Envs to PG9 and PG16 with I424M substitution, except YU2, where we found >8 and 3-fold increase in sensitivity (IC50) of Env expressing M424 to PG9 and PG16 respectively. Interestingly, 4-2.J45 Env expressing M424 showed relative resistance to PG9 and PG16 over 4-2.J45 expressing I424 (Table 2), wherein we found comparable sensitivities of other Envs to PG9 and PG16 except YU2, which showed approximately 8 and 3 fold increase in neutralization sensitivity to PG9 and PG16 MAbs respectively (Table 2). The indistinctness in PG9/PG16 sensitivities of 4-2.J45 and YU2 Envs expressing M424 was possibly due to some compensatory and conformational changes elsewhere within Env. Overall, we did not find any association between I424M substitution and neutralization sensitivity.
We next assessed if presence of M424 confers Env with increased sensitivity to MAbs targeting CDbs. Since we previously found that the clade C Envs were resistant to b12 (Ringe, Thakar, and Bhattacharya, 2010), we tested effect of I424M substitution of Env susceptibility to MAbs to CD4bs i.e., b12 and VRC01. As shown in Table 2, I424M substitution in JRFL, RHPA4259.7 and YU2 Envs conferred increase in b12 sensitivity by 2, 3.8 and 32.27 folds respectively. Thus, although overall the shift in b12 sensitivity due to I424M was modest (P=0.14), the 2 and approximately 4-fold increase in b12 sensitivity by JRFL (Tier 3) and RHPA4259.7 (Tier 2) expressing M424 was noted. However, when tested against recently discovered CD4bs MAb, VRC01 MAb, both Envs expressing M424 and I424 showed comparable sensitivity to VRC01; thus no significant difference (P = 0.2) in the VRC01 sensitivity (Table 2). Our data suggest that I424M while appeared to modulate CD4bs epitopes modestly, it does not have definitive effect on quaternary epitopes targeted by PG9 and PG16 MAbs. Additionally, I424M was not found to have any significant difference in the sensitivity of Envs to 4E10 MAb targeting MPER in gp41 (Ringe, Thakar, and Bhattacharya, 2010) even at higher concentrations up to 10 μg/ml (data not shown), negating any significant exposure of neutralizing epitopes in MPER due to I424M substitution.
Discussion
Characterization of the NAb targets on the HIV-1 Env early in infection will allow a better understanding of the epitopes vulnerable for virus neutralization and will have implications for rational Env-based vaccine design. In the present study, we sought to understand the basis of the exceptional sensitivity of a clade C Env to autologous plasma compared to other Envs obtained at the same time point from a recently infected Indian patient. Using Env chimeras constructed with sensitive and resistant Envs with closely related protein sequences followed by fine mapping we found a single substitution of methionine in place of the relatively conserved isoleucine at position 424 in the C4 region of Env conferred increased virus neutralization sensitivity to HIV+ plasma antibodies. Interestingly, when we examined the HIV database (www.hiv.lanl.gov), we found that methionine is completely absent in viruses at the 424 position in the HIV-1 M, N and O groups. It is possible that the 4-2.J45 Env variant was selected in vivo to acquire this rare residue early in the infection although this change was not found in other Env clones obtained either from the same time point or from the baseline (collected six month prior) as well as follow up plasma specimens (collected six months after) of the same individual (Ringe, Thakar, and Bhattacharya, 2010). Although we do not rule out the possibility of introduction of this unusual I424M substitution due to PCR error (albeit we used high fidelity proof reading polymerase during Env amplification and except 4-2.J45 Env, other Env clones amplified from the same patient at the same time contained the conserved I424), nonetheless, M424 was found to confer comparable infectivity to those containing I424, suggesting substitution of methionine at this position was not associated with defective production of infectious virions.
Interestingly, with introduction of M424 replacing the exceptionally conserved I424 (www.hiv.lanl.gov), all the neutralization resistant pseudotyped viruses containing contemporaneous Envs from the same patient became significantly sensitive to autologous plasma antibodies, suggesting that M424 indeed induced conformational shifts in Env towards greater exposure of neutralizing epitopes that are otherwise obscured. Notably, the fact that Env variants substituted with I424M in the C4 region of gp120 showed increased sensitivity to plasma antibodies from heterologous sources indicated that despite other differences between Env sequences, I424M substitution alone is capable of modulating Env conformation resulting greater exposure of neutralizing epitopes. Presence of such circulating envelope in plasma is perplexing and it is expected that such envelopes would perish quickly. We did not find such Env variants (M424) obtained at later time points in this particular patient support this view and hence we assume that 4-2.J45 might have certain advantage for example resistance to cell-mediate immune response or higher infectivity to persist for a small period of time (Bunnik et al., 2008; Rong et al., 2009). Since M424 conferred enhanced neutralization sensitivity to autologous plasma antibodies collected at different time points from this particular (IVC4) patient, we further sought to understand if epitopes that are targeted by autologous plasma antibodies in this patient constitute major targets of neutralizing antibodies during early infection. Thus, when we compared the vulnerabilities between Env containing M424 and I424 towards effective neutralization by several heterologous HIV+ plasma and serum antibodies essentially collected at early infection from Indian and South African donors, we found that Env-pseudotyped viruses containing Met424 in distinct genetic backgrounds (both in clade B and clade C Envs) became increasingly sensitive to heterologous plasma antibodies. The modest increase in neutralization sensitivity of unrelated Envs compared to 4-2.J45 due to I424M substitution to pooled plasma as shown in Table 2 was probably due to presence of residues elsewhere in 4-2.J45 Env that provided 4-2.J45 with conformation for enhanced susceptibility to plasma antibodies. Nonetheless, increase in the sensitivity of JRFL (subtype B) which is a Tier 2 virus (Mascola et al., 2005) after I424M substitution to plasma and serum antibodies obtained from clade C individuals was remarkable. Overall, our study indicated that substitution of highly conserved isoleucine with methionine at 424 position in C4 domain in gp120 resulted in exposure of neutralizing epitopes.
Did Met424 induce conformational shifts in Env towards enhanced sensitivity of Env-pseudotyped viruses to plasma antibodies by acting on particular sites on Env? Nakamura et al (Nakamura et al., 1993) previously reported antibodies that targets neutralizing epitopes in C4 domain and prevented CD4 binding with gp120; they specifically showed that mutation of residues at position 423 and 429 in C4 region in gp120 governs inhibition by a population of CD4 blocking antibodies. Based on our observation that I424M substitution showed enhanced susceptibility of Envs tested in the current study to plasma antibodies, it is possible presence of M424 affects population of antibodies targeting C4 region in gp120. Also, we found that M424 is located in close proximity to the Phe43 cavity, a site shown to be vulnerable for neutralizing antibodies and agents that interferes with gp120-CD4 interaction and has been implicated as a potentially attractive target for blocking virus entry (Kwong et al., 2002; Kwong et al., 1998; Madani et al., 2008; Repik and Clapham, 2008). Indeed, we found that I424M substitution impacted on exposure of discontinuous CD4bs epitopes that was indicated by increased sensitivity of Envs to sCD4 and moderate enhancement of clade B Envs to b12 MAb. However, VRC01, another CD4bs antibody showed comparable Env sensitivity and did not show any significant shifts in Env sensitivity due to presence of M424. This was possibly due to the fact that I424M do not impact on conformational masking of VRC01 epitope. Interestingly, the modest increase in b12 sensitivity also correlated with the increased sensitivity of Envs to sCD4 and modest increase in HeLa RC49 cells (expressing low surface CD4) due to I424M substitution which suggest structural alteration at CD4bs with I424M substitution resulting in exposure of neutralizing epitopes important for gp120-CD4 interaction. The position 424 in C4 region in gp120 is also in close proximity to the epitope recognized by 17b, a MAb that targets CD4i region (data not shown here). However, we recently showed that the 4-2.J45 Env was completely resistant to 17b (Ringe, Thakar, and Bhattacharya, 2010) thus eliminating any effect on alteration of CD4i region due to presence of M424. In addition, we did not find significant inhibition of Env-pseudoviruses to CCR5 antagonist TAK-779 due to I424M substitution. Recently, Xiang et al (Xiang et al., 2010) demonstrated that I424S change in YU2 Env has no effect on CCR5 binding. This indicates I424M substitution did not affect the structural integrity of coreceptor binding site. Nonetheless, 4-2.J45 Env expressing M424 did show increased sensitivity to sCD4 compared to contemporaneous Env clones at a concentration greater than 6.66μg/ml (Ringe, Thakar, and Bhattacharya, 2010). In the present study, we further assessed whether Met424 indeed confers greater sensitivity of other Envs substituted with M424 in place of I424 to sCD4. As expected, with I424M substitution, pseudotyped viruses containing contemporaneous Envs and Envs obtained from unrelated patients showed enhanced sensitivity to sCD4. In particular 4-2.J41, 4-2.J46b and 4-2.J47b Envs showed remarkable shifts towards enhanced sensitivity to sCD4. In addition, these Env-pseudotyped viruses also showed modest utilization of HeLa cells expressing low but high CCR5 on their surface for efficient entry. Collectively, these observations suggest that M424 modulated conformation of gp120 improving gp120-CD4 interaction. To further narrow down the region in the Env outer domain that conferred significant sensitivity to plasma antibodies, we tested Env-pseudoviruses containing M424 and I424 to three well characterized anti-V3 MAbs that were previously shown to be cross-reactive (Jiang et al., 2010). To our surprise, we found that Env-pseudotyped viruses Envs containing M424 showed >100-fold increased sensitivity to the anti-V3 MAbs (notably with MAb 3074) in contrast to Env-pseudotyped viruses containing I424 and was closely comparable to that we observed with plasma antibodies. This was probably due to increased accessibility of critical neutralizing epitopes in V3, resulting in better antibody binding, resulting in virus neutralization. The I424M substitution although was found to modulate the V3 loop conformation it does not appear to influence quaternary epitopes in gp120 as we found no association of I424M substitution with shift in the Env sensitivity to PG9 and PG16 MAbs that targets the quaternary epitopes. Also, the I424M substitution was not found to have significant difference in the sensitivity to MAbs targeting MPER such as 4E10 (Ringe, Thakar, and Bhattacharya, 2010) even at higher concentrations (data not shown). Thus, we conclude that I424M substitutions predominantly modulated V3 loop conformation in addition to exposition of discontinuous neutralizing epitopes on CD4bs on Env which predominantly accounted for greater neutralization by autologous and heterologous plasma antibodies.. The anti-V3 antibody titer produced in vivo may not be potent enough (possibly due to inefficient exposure of neutralizing epitopes in primary virus) to bind and neutralize efficiently to the virus post CD4 engagement which causes exposure of V3 loop for coreceptor binding (Huang et al., 2005; Xiang et al., 2010) – a small window of opportunity for antibody to bind to V3 loop. It is possible that increased exposure of neutralizing epitopes due to presence of M424 would be able to efficiently present these epitopes (which otherwise remains occluded in primary viruses) to B cells and elicit potent antibodies, which might interfere virus entry in the small window period post CD4 bound state.
We sought to understand how alteration in gp120 conformation due to specific mutation such as I424M in C4 domain exposes the neutralizing epitopes for effective antibody recognition? It was reported earlier (Bhattacharya, Peters, and Clapham, 2003; Clapham et al., 1989; Hart et al., 1991; Moore et al., 1992; Moore et al., 1990; Sattentau, 1993) that pre-treatment of HIV-1, HIV-2 and SIV with sCD4 confers increased exposure of V2 and V3 loops, significant reduction in infection of target cells, and increased neutralization by anti-V3 antibodies. In addition, laboratory adapted viruses such as HIV-1 IIIb, LAV-2ROD and SIVmac251 were demonstrated to have exposed V3 loop prior to sCD4 binding (Sattentau, 1993). These data suggest that Envs that are less CD4-dependent for productive entry tends to shift V3 loop away from the Env core thereby exposing sites vulnerable for neutralization. In our study, we found that Envs expressing M424 showed modest increase in their sensitivity to sCD4, supporting earlier findings that suggest an association between increased Env sensitivity to sCD4 and enhanced neutralization by anti-V3 antibodies. Although I424M showed modulation in V3 loop, however it did not appear to modulate quaternary epitopes in V1/V2 and V3 regions that are targeted by PG9 and PG16 MAbs. Interestingly, 4-2.J45 Env in presence of M424 showed relative resistance to PG9 and PG16, possibly due to presence in compensatory residues elsewhere in this particular Env. It has been shown that many primary envelopes with closely similar V3 sequence were found to be resistant to anti-V3 antibodies (Hioe et al., 2010) suggesting that V3 loop is well poised to evade from such antibodies in the trimeric Env structure while maintaining the primary function of coreceptor binding. This suggests that apart from changes within V3, other proximal elements of gp120 that interacts with V3 loop for example stem of V1V2 and bridging sheet modulate the binding of anti V3 antibodies to V3 loop structure. Such alteration likely resulted in unmasking of neutralizing discontinuous epitopes by shifting V3 loop away from Env core that were universally targeted by plasma antibodies tested here. In summary, our present study indicated that changes in β20 strand in C4 domain of gp120 due to I424M substitution resulted in alteration in Env conformation towards exposure of discontinuous neutralizing epitopes in V3 and CD4bs that form the basis for enhanced virus neutralization by plasma antibodies.
Materials and Methods
Patient materials, antibodies and inhibitors
Plasma and serum samples from individuals infected with HIV-1 were obtained from National AIDS Research Institute (NARI) clinics, Pune, India, following ethical approval by the institutional review board (IRB). Serum and plasma that were tested for neutralization assays were obtained from blood samples donated by the HIV+ individuals during their routine testing at heir first visit for acquisition of HIV-1 at the NARI clinics and were later confirmed for p24 antibody positivity by serological testing. Plasma samples of patients with recent infection (estimated through detuned ELISA) from which HIV-1 gp160 clones were obtained have been described recently (Ringe, Thakar, and Bhattacharya, 2010). 20 HIV+ plasma samples from the South African donors infected with HIV clade C were generously provided by Prof Lynn Morris, National Institute of Communicable Diseases, Johannesburg, South Africa. Plasma and serum samples were heat inactivated at 55° C for 1 hour before use in neutralization assays. The MAbs specific for the V3 loop, 3074, 3869 and 3906, and for parvovirus (MAb 1418) used in this study were described previously (Gorny et al., 2009; Hioe et al., 2010). VRC01 MAb was kindly provided by Dr John Mascola, VRC, NIH, USA. PG9 and PG16 MAbs obtained from International AIDS Vaccine Initiative (IAVI) and were described before (Walker et al., 2009). CCR5 antagonist, TAK-779, TZM-bl cells, 447-52D (MAb to V3) and anti-p24 hybridoma 183-H12-5C (Chesebro et al., 1992) were obtained from the NIH Research Reagents and Reference Program. Soluble CD4 (sCD4) was purchased from Progenics, Inc. and was provided by Dr David Montefiori, Duke University, Durham, NC, USA.
Construction of Env chimera
To find specific regions in Env modulating neutralization sensitivity, fragments were swapped between resistant (4-2.J41) and sensitive (4-2.J45) Env clones. Primers were designed to specifically amplify the region in Env to be inserted and also the primers that amplified entire backbone of the plasmid except the insert. These two PCR products generated amplicons having at least 15 base pair homologous sequences at 5′ and 3′ ends of the DNA fragments. The insert and backbone fragments mixed in a 1: 2 molar ratio with a cocktail containing pox DNA polymerase provided in Dry Down PCR cloning kit (Clontech Inc.) according to manufacture’s instructions. This In-Fusion enzyme produces single stranded overhangs by its exonuclease activity which is then annealed due to 15 base pair homology.
Site-directed mutagenesis
Specific primers were designed to introduce desired residue both in insert and in backbone DNA by PCR. Amplicons were purified and further digested with DpnI to remove any residual plasmid DNA. Ligations were carried out using dry-down PCR kit (Clontech Inc.) as described above.
Neutralization assays
Neutralization assays were carried out in TZM-bl cells using Env-pseudotyped viruses as described previously (Ringe, Thakar, and Bhattacharya, 2010). Briefly, 200 TCID50 of Env-pseudotyped viruses were incubated with various dilutions and concentrations of heat-inactivated patient’s plasma/sera and MAbs in a total volume of 150 μl (for plasma samples) or 100 μl (for MAbs) for 1 hour at 37°C in 96-well tissue culture plates (Corning, Inc.). Freshly trypsinized TZM-bl cells (1 × 104 per well) were subsequently added to each well with 25 μg/ml DEAE Dextran and further incubated for additional 2 days at 37°C in a CO2 incubator. One set of control wells received TZM-bl cells plus Env-pseudotyped virus (virus control) and another set received cells only (cell control). The reduction in infectivity was calculated by measuring the relative light units (RLU) of the infected TZM-bl cells in a luminometer (Perkin Elmer, Inc).
Infectivity assays in HeLa cells
Single round infectivity assays in HeLa cells were done as described before (Ringe, Thakar, and Bhattacharya, 2010). Briefly, Env-pseudotyped viruses were normalized against their TZM-bl infectivity titers and subsequently 200TCID50 (relative to TZM-bl cells) were mixed with 1 × 104/ well of HeLa (RC49) cells in a 96-well tissue culture plate without DEAE-Dextran. The entire mixture was then incubated further for 2 days in a CO2 incubator at 37°C. Virus infectivity was measured by immunostaining intracellular p24 as described previously (Peters et al., 2004).
Supplementary Material
Figure S1. Effect of I424M on infectivity of genetically Env-pseudotyped viruses. Pseudotyped viruses carrying Envs from different genetic backgrounds expressing either I424 or M424 were tested for their degree of infectivity in TZM-bl cells. The infectivity was assessed by measuring relative luminescence unit (RLU) relative to reverse transcriptase (RT) activity. Note that no significant association was found between I424M substitution and degree of infectivity in TZM-bl cells.
Figure S2. Effect of I424M on CD4 dependence. Infectivity of Env-pseudotyped viruses were tested in HeLa cells expressing low CD4 but CCR5 on surface (RC49) (Platt et al., 1998). RC49 cells were infected with equal TCID50 of Env-pseudotyped viruses and incubated for 72 hours at 37°C in a CO2 incubator. Infectivity was measured by immunostaining of intracellular p24 as described previously (Ringe, Thakar, and Bhattacharya, 2010).
Acknowledgements
This work was primarily supported by extramural grants from the Department of Biotechnology (BT/PR7829/MED/14/1133/2006 and BT/PR12853/MED/29/141/2009), Government of India to JB and partly by the Collaboration of AIDS Vaccine Discovery (CAVD) and Vaccine Immune Monitoring Consortium (VIMC) (Grant # 38619). SZP was supported by grants from the USA National Institutes of Health (AI 36085, HL59725, and AI27747), from the Bill and Melinda Gates Foundation and from research funds from the USA Department of Veterans Affairs. We thank Prof Lynn Morris, National Institute of Communicable Diseases (NICD), Johannesburg, South Africa for the generous gift of the HIV+ plasma samples obtained from South African donors, Dr Paul Clapham, UMASS Medical School, Worcester, Massachusetts for the pSVIIIenv-SF162, pSVIIIenv-YU2, pSVIIIenv-JRFL plasmids, Dr David Montefiori, Duke University for RHPA4259.7 Env plasmid and Dr David Kabat, University of Portland, Oregon for HeLa (RC49) cell line. We thank Dr Wayne Koff, IAVI for providing us with PG9 and PG16 MAbs, Dr Dennis Burton, Scripps Research Institute, La Jolla, CA, USA for b12 MAb and Dr John Mascola, Vaccine Research Center, National Institute of Health, USA for VRC01 MAb. We are appreciative of the NIH AIDS Research Reagent Reference Program for providing many reagents including TZM-bl cells, anti-p24 MAb (18-H12-5C) and TAK-779 and 447-42D MAb used in this study. RR and DS are supported by the Senior Research Fellowships from the Council of Scientific and Industrial Research (CSIR) and University grants Commission (UGC), Government of India respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barouch DH. Challenges in the development of an HIV-1 vaccine. Nature. 2008;455(7213):613–9. doi: 10.1038/nature07352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CH, Pantophlet R, Schiefner A, Cavacini LA, Stanfield RL, Burton DR, Wilson IA. Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization. J Mol Biol. 2008;375(4):969–78. doi: 10.1016/j.jmb.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya J, Peters PJ, Clapham PR. CD4-independent infection of HIV and SIV: implications for envelope conformation and cell tropism in vivo. Aids. 2003;17(Suppl 4):S35–43. doi: 10.1097/00002030-200317004-00004. [DOI] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blay WM, Gnanakaran S, Foley B, Doria-Rose NA, Korber BT, Haigwood NL. Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques. J Virol. 2006;80(2):999–1014. doi: 10.1128/JVI.80.2.999-1014.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blish CA, Nguyen MA, Overbaugh J. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 2008;5(1):e9. doi: 10.1371/journal.pmed.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J Virol. 2008;82(16):7932–41. doi: 10.1128/JVI.00757-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, Wilson IA. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22(2):163–73. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Carrow EW, Vujcic LK, Glass WL, Seamon KB, Rastogi SC, Hendry RM, Boulos R, Nzila N, Quinnan GV., Jr. High prevalence of antibodies to the gp120 V3 region principal neutralizing determinant of HIV-1MN in sera from Africa and the Americas. AIDS Res Hum Retroviruses. 1991;7(10):831–8. doi: 10.1089/aid.1991.7.831. [DOI] [PubMed] [Google Scholar]
- Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66(11):6547–54. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham PR, Weber JN, Whitby D, McIntosh K, Dalgleish AG, Maddon PJ, Deen KC, Sweet RW, Weiss RA. Soluble CD4 blocks the infectivity of diverse strains of HIV and SIV for T cells and monocytes but not for brain and muscle cells. Nature. 1989;337(6205):368–70. doi: 10.1038/337368a0. [DOI] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303(5666):2019–22. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- Doria-Rose NA, Klein RM, Daniels MG, O’Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, Mascola JR, Connors M. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol. 2010;84(3):1631–6. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duenas-Decamp MJ, Clapham PR. HIV-1 gp120 determinants proximal to the CD4 binding site shift protective glycans that are targeted by monoclonal antibody 2G12. J Virol. 2010;84(18):9608–12. doi: 10.1128/JVI.00185-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duenas-Decamp MJ, Peters P, Burton D, Clapham PR. Natural resistance of human immunodeficiency virus type 1 to the CD4bs antibody b12 conferred by a glycan and an arginine residue close to the CD4 binding loop. J Virol. 2008;82(12):5807–14. doi: 10.1128/JVI.02585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duenas-Decamp MJ, Peters PJ, Burton D, Clapham PR. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J Virol. 2009;83(6):2575–83. doi: 10.1128/JVI.02133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci AS, Johnston MI, Dieffenbach CW, Burton DR, Hammer SM, Hoxie JA, Martin M, Overbaugh J, Watkins DI, Mahmoud A, Greene WC. HIV vaccine research: the way forward. Science. 2008;321(5888):530–2. doi: 10.1126/science.1161000. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Wang XH, Williams C, Volsky B, Revesz K, Witover B, Burda S, Urbanski M, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S, Nadas A. Preferential use of the VH5-51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol Immunol. 2009;46(5):917–26. doi: 10.1016/j.molimm.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Moore PL, Bibollet-Ruche F, Li H, Decker JM, Meyers T, Shaw GM, Morris L. 4E10-resistant variants in a human immunodeficiency virus type 1 subtype C-infected individual with an anti-membrane-proximal external region-neutralizing antibody response. J Virol. 2008;82(5):2367–75. doi: 10.1128/JVI.02161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Moore PL, Choge IA, Decker JM, Bibollet-Ruche F, Li H, Leseka N, Treurnicht F, Mlisana K, Shaw GM, Karim SS, Williamson C, Morris L. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol. 2007a;81(12):6187–96. doi: 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Moore PL, Pantophlet RA, Morris L. N-linked glycan modifications in gp120 of human immunodeficiency virus type 1 subtype C render partial sensitivity to 2G12 antibody neutralization. J Virol. 2007b;81(19):10769–76. doi: 10.1128/JVI.01106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart TK, Kirsh R, Ellens H, Sweet RW, Lambert DM, Petteway SR, Jr., Leary J, Bugelski PJ. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc Natl Acad Sci U S A. 1991;88(6):2189–93. doi: 10.1073/pnas.88.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV’s switch-hitter. AIDS Res Hum Retroviruses. 2005;21(2):171–89. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- Hioe CE, Wrin T, Seaman MS, Yu X, Wood B, Self S, Williams C, Gorny MK, Zolla-Pazner S. Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS One. 2010;5(4):e10254. doi: 10.1371/journal.pone.0010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–8. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, Zolla-Pazner S, Kong XP. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat Struct Mol Biol. 2010;17(8):955–61. doi: 10.1038/nsmb.1861. [DOI] [PubMed] [Google Scholar]
- Kinsey NE, Anderson MG, Unangst TJ, Joag SV, Narayan O, Zink MC, Clements JE. Antigenic variation of SIV: mutations in V4 alter the neutralization profile. Virology. 1996;221(1):14–21. doi: 10.1006/viro.1996.0348. [DOI] [PubMed] [Google Scholar]
- Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J Virol. 2001;75(7):3435–43. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- Korber B, Gnanakaran S. The implications of patterns in HIV diversity for neutralizing antibody induction and susceptibility. Curr Opin HIV AIDS. 2009;4(5):408–17. doi: 10.1097/COH.0b013e32832f129e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov C, Pinter A, Honnen WJ, Gorny MK, Nyambi PN, Zolla-Pazner S, Kayman SC. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade a and clade B v3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J Virol. 2005;79(2):780–90. doi: 10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov CP, Honnen WJ, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J Virol. 2006;80(14):7127–35. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunert R, Wolbank S, Stiegler G, Weik R, Katinger H. Characterization of molecular features, antigen-binding, and in vitro properties of IgG and IgM variants of 4E10, an anti-HIV type 1 neutralizing monoclonal antibody. AIDS Res Hum Retroviruses. 2004;20(7):755–62. doi: 10.1089/0889222041524571. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420(6916):678–82. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakso MM, Lee FH, Haggarty B, Agrawal C, Nolan KM, Biscone M, Romano J, Jordan AP, Leslie GJ, Meissner EG, Su L, Hoxie JA, Doms RW. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 2007;3(8):e117. doi: 10.1371/journal.ppat.0030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Decker JM, Johnson RW, Bibollet-Ruche F, Wei X, Mulenga J, Allen S, Hunter E, Hahn BH, Shaw GM, Blackwell JL, Derdeyn CA. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J Virol. 2006;80(11):5211–8. doi: 10.1128/JVI.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79(16):10108–25. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rey-Cuille MA, Hu SL. N-linked glycosylation in the V3 region of HIV type 1 surface antigen modulates coreceptor usage in viral infection. AIDS Res Hum Retroviruses. 2001;17(16):1473–9. doi: 10.1089/08892220152644179. [DOI] [PubMed] [Google Scholar]
- Lynch RM, Shen T, Gnanakaran S, Derdeyn CA. Appreciating HIV type 1 diversity: subtype differences in Env. AIDS Res Hum Retroviruses. 2009;25(3):237–48. doi: 10.1089/aid.2008.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani N, Schon A, Princiotto AM, Lalonde JM, Courter JR, Soeta T, Ng D, Wang L, Brower ET, Xiang SH, Kwon YD, Huang CC, Wyatt R, Kwong PD, Freire E, Smith AB, 3rd, Sodroski J. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure. 2008;16(11):1689–701. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, D’Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, Petropoulos CJ, Polonis VR, Sarzotti M, Montefiori DC. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol. 2005;79(16):10103–7. doi: 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol. 2010;28:413–44. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- Moore JP, McKeating JA, Huang YX, Ashkenazi A, Ho DD. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J Virol. 1992;66(1):235–43. doi: 10.1128/jvi.66.1.235-243.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, McKeating JA, Weiss RA, Sattentau QJ. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250(4984):1139–42. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70(3):1863–72. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Thali M, Jameson BA, Vignaux F, Lewis GK, Poon SW, Charles M, Fung MS, Sun B, Durda PJ, et al. Immunochemical analysis of the gp120 surface glycoprotein of human immunodeficiency virus type 1: probing the structure of the C4 and V4 domains and the interaction of the C4 domain with the V3 loop. J Virol. 1993;67(8):4785–96. doi: 10.1128/jvi.67.8.4785-4796.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Gray ES, Choge IA, Ranchobe N, Mlisana K, Abdool Karim SS, Williamson C, Morris L. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J Virol. 2008;82(4):1860–9. doi: 10.1128/JVI.02187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura GR, Byrn R, Wilkes DM, Fox JA, Hobbs MR, Hastings R, Wessling HC, Norcross MA, Fendly BM, Berman PW. Strain specificity and binding affinity requirements of neutralizing monoclonal antibodies to the C4 domain of gp120 from human immunodeficiency virus type 1. J Virol. 1993;67(10):6179–91. doi: 10.1128/jvi.67.10.6179-6191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke SM, Schweighardt B, Phung P, Fonseca DP, Terry K, Wrin T, Sinangil F, Berman PW. Mutation at a single position in the V2 domain of the HIV-1 envelope protein confers neutralization sensitivity to a highly neutralization-resistant virus. J Virol. 2010;84(21):11200–9. doi: 10.1128/JVI.00790-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke SM, Schweighardt B, Scott WG, Wrin T, Fonseca DP, Sinangil F, Berman PW. Novel ring structure in the gp41 trimer of human immunodeficiency virus type 1 that modulates sensitivity and resistance to broadly neutralizing antibodies. J Virol. 2009;83(15):7728–38. doi: 10.1128/JVI.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78(19):10724–37. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, Simmonds P, Clapham PR. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J Virol. 2004;78(13):6915–26. doi: 10.1128/JVI.78.13.6915-6926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol. 2004;78(10):5205–15. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72(4):2855–64. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademeyer C, Moore PL, Taylor N, Martin DP, Choge IA, Gray ES, Sheppard HW, Gray C, Morris L, Williamson C. Genetic characteristics of HIV-1 subtype C envelopes inducing cross-neutralizing antibodies. Virology. 2007;368(1):172–81. doi: 10.1016/j.virol.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Repik A, Clapham PR. Plugging gp120s cavity. Structure. 2008;16(11):1603–4. doi: 10.1016/j.str.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Ringe R, Thakar M, Bhattacharya J. Variations in autologous neutralization and CD4 dependence of b12 resistant HIV-1 clade C env clones obtained at different time points from antiretroviral naive Indian patients with recent infection. Retrovirology. 2010;7(1):76. doi: 10.1186/1742-4690-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong R, Bibollet-Ruche F, Mulenga J, Allen S, Blackwell JL, Derdeyn CA. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J Virol. 2007a;81(3):1350–9. doi: 10.1128/JVI.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong R, Gnanakaran S, Decker JM, Bibollet-Ruche F, Taylor J, Sfakianos JN, Mokili JL, Muldoon M, Mulenga J, Allen S, Hahn BH, Shaw GM, Blackwell JL, Korber BT, Hunter E, Derdeyn CA. Unique mutational patterns in the envelope alpha 2 amphipathic helix and acquisition of length in gp120 hypervariable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J Virol. 2007b;81(11):5658–68. doi: 10.1128/JVI.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong R, Li B, Lynch RM, Haaland RE, Murphy MK, Mulenga J, Allen SA, Pinter A, Shaw GM, Hunter E, Robinson JE, Gnanakaran S, Derdeyn CA. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 2009;5(9):e1000594. doi: 10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Wu X, Lee S, Overbaugh J. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J Virol. 2006;80(19):9586–98. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau QJ, Moore JP, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84(3):1439–52. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Dennison SM, Liu P, Gao F, Jaeger F, Montefiori DC, Verkoczy L, Haynes BF, Alam SM, Tomaras GD. Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc Natl Acad Sci U S A. 2010;107:5972–5977. doi: 10.1073/pnas.0912381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Bruce CB, Featherstone AS, Downing RG, Biryahawaho B, Clegg JC, Carswell JW, Oram JD. Reactions of Ugandan antisera with peptides encoded by V3 loop epitopes of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1994;10(5):577–83. doi: 10.1089/aid.1994.10.577. [DOI] [PubMed] [Google Scholar]
- Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15(8):866–70. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- Sullivan N, Sun Y, Binley J, Lee J, Barbas CF, 3rd, Parren PW, Burton DR, Sodroski J. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J Virol. 1998;72(8):6332–8. doi: 10.1128/jvi.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70(2):1100–8. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T, Kurth R, Norley S. The majority of neutralizing Abs in HIV-1-infected patients recognize linear V3 loop sequences. Studies using HIV-1MN multiple antigenic peptides. J Immunol. 1994;153(4):1895–904. [PubMed] [Google Scholar]
- Walker LM, Burton DR. Rational antibody-based HIV-1 vaccine design: current approaches and future directions. Curr Opin Immunol. 2010;22:358–366. doi: 10.1016/j.coi.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–9. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6(8):e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wu X, Changela A, O’Dell S, Schmidt SD, Pancera M, Yang Y, Zhang B, Gorny MK, Phogat S, Robinson JE, Stamatatos L, Zolla-Pazner S, Kwong PD, Mascola JR. Immunotypes of a Quaternary Site of HIV-1 Vulnerability and Their Recognition by Antibodies. J Virol. 2011;85(9):4578–4585. doi: 10.1128/JVI.02585-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–61. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69(9):5723–33. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Thali M, Tilley S, Pinter A, Posner M, Ho D, Robinson J, Sodroski J. Relationship of the human immunodeficiency virus type 1 gp120 third variable loop to a component of the CD4 binding site in the fourth conserved region. J Virol. 1992;66(12):6997–7004. doi: 10.1128/jvi.66.12.6997-7004.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang SH, Finzi A, Pacheco B, Alexander K, Yuan W, Rizzuto C, Huang CC, Kwong PD, Sodroski J. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J Virol. 2010;84(7):3147–61. doi: 10.1128/JVI.02587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Foley B, Schultz AK, Macke JP, Bulla I, Stanke M, Morgenstern B, Korber B, Leitner T. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology. 2010;7:25. doi: 10.1186/1742-4690-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329(5993):811–7. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol. 2004;4(3):199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Cardozo T. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat Rev Immunol. 2010;10(7):527–35. doi: 10.1038/nri2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Effect of I424M on infectivity of genetically Env-pseudotyped viruses. Pseudotyped viruses carrying Envs from different genetic backgrounds expressing either I424 or M424 were tested for their degree of infectivity in TZM-bl cells. The infectivity was assessed by measuring relative luminescence unit (RLU) relative to reverse transcriptase (RT) activity. Note that no significant association was found between I424M substitution and degree of infectivity in TZM-bl cells.
Figure S2. Effect of I424M on CD4 dependence. Infectivity of Env-pseudotyped viruses were tested in HeLa cells expressing low CD4 but CCR5 on surface (RC49) (Platt et al., 1998). RC49 cells were infected with equal TCID50 of Env-pseudotyped viruses and incubated for 72 hours at 37°C in a CO2 incubator. Infectivity was measured by immunostaining of intracellular p24 as described previously (Ringe, Thakar, and Bhattacharya, 2010).